Abstract
Phosphorylation-dependent ubiquitination combined with proteasomal degradation of transcriptional regulators is a recently appreciated mechanism for control of a number of inflammatory genes. Far less is known about the counterregulatory mechanisms that repress transcriptional activity in these pathways during resolution. Here, we investigated the transient nature of hypoxia-induced tumor necrosis factor (TNF)α in T84 cells, a process we have previously shown to involve phosphorylation-dependent degradation of the cAMP-response element-binding protein (CREB). Initial studies indicate hypoxia-induced TNFα to be a transient event, the resolution of which is associated with the appearance of a higher molecular weight modified form of CREB. Gene array analysis of mRNA derived from hypoxic cells identified a time-dependent induction of small ubiquitin-related modifier (SUMO)-1 mRNA. In prolonged hypoxia, CREB is posttranslationally modified by SUMO-1. Furthermore, SUMO-1 overexpression stabilizes CREB in hypoxia and enhances CREB-dependent reporter gene activity. Site-directed mutagenesis of lysine residues K285 and K304 identifies them as SUMO acceptors in vivo and in vitro. Mutation of K304 also results in loss of CREB nuclear localization, implying a role for SUMO-1 modification at this site in the subcellular localization of CREB. Thus, in prolonged hypoxia, CREB is modified by association with SUMO-1. Furthermore, we hypothesize that such an event stabilizes and promotes nuclear localization of CREB and thus complements an endogenous resolution phase for hypoxia-induced inflammatory processes.
Diminished tissue oxygen delivery (hypoxia) is common in a number of diseases (1). Initial transcriptional responses to hypoxia facilitate adaptation consistent with cell, tissue, and whole-animal survival (2). We have previously described a temporally downstream hypoxic phenotype, in intestinal epithelia, which actively contributes to inflammatory processes through transcriptional up-regulation of proinflammatory genes, including tumor necrosis factor (TNF)α (3). This phenotypic switch depends pivotally on the degradation of cAMP-response element-binding protein (CREB) (4). Hypoxia elicits CREB degradation in intestinal epithelial cells through phosphorylation-dependent targeting to the ubiquitin/proteasome pathway of proteolysis (5, 6). Although the induction of proinflammatory genes in hypoxia is transient, little is known about the pathways that repress ongoing transcriptional activity and whether endogenous “braking” mechanisms exist to counterregulate the transcriptional machinery in resolution. It has recently been appreciated that posttranslational modification of proteins through association with small ubiquitin-related modifier (SUMO)-1 mediates protein trafficking and protein–protein interactions and may represent a decoy mechanism for ubiquitination and subsequent proteasomal degradation (7). Such a pathway has been studied with regard to control of inflammatory gene transcriptional regulators. For example, SUMO-1 modification of IκΒ has been demonstrated to inhibit ubiquitination and degradation and subsequently results in the inhibition of nuclear factor κB (NFκB) activation (8, 9).
Stimuli demonstrated to induce or repress SUMO-1 modification of various proteins include DNA damage (10, 11), temperature (12), arsenic trioxide (13), and c-Jun activation (14). SUMO-1 modification is a three-step process involving SUMO-1 activation by the E1 enzyme Aos1/Uba2, protein-SUMO-1 conjugation by the E2 enzyme Ubc9, and SUMO ligation via E3-like ligases such as the nucleoporin RanBP2/Nup 358 and members of the PIAS family of proteins (15–20). RanGAP1 was the first protein identified as a target for SUMO-1 modification (21–23). However, a number of functionally diverse proteins have also been identified as SUMO-1 targets. As well as IκB, other endogenous proteins include PML, HIPK2, c-Jun, p53, p73α, topoisomerase II, PCNA, axin, Daxx, the androgen receptor, HSF-1, TEL, and c-Myb (10, 11, 13, 14, 19, 24–34). Furthermore, utilization of the SUMO-1 conjugation pathway to stabilize proteins from invading pathogens has been hypothesized for the viral proteins E1, E3L, IE2-p86, and E1B-55kDa (35–37) and the bacterial protein YopJ/P from Yersinia (38–40). Although modification of proteins by SUMO-1 is a covalent process, it is reversible through the activity of a number of specific isopeptidase enzymes (41).
Although the specific targeting mechanism for SUMO-1 modification remains unknown, interaction with target proteins depends on a ψKxE consensus motif in the target protein, where ψ represents a hydrophobic amino acid, and x represents any amino acid (42). Furthermore, a number of proteins, including PCNA, Daxx, and axin, have been recently demonstrated to be SUMO-modified at nonconsensus motifs (11, 33, 34). We identified potential SUMO-1 modification motifs in CREB between amino acids 303 and 306 and between 284 and 287, which are similar but do not fully conform to the consensus motif. Here, we demonstrate that prolonged hypoxia leads to a resolution of inflammatory gene expression, an event that we hypothesize to be mediated through SUMO-1 modification of CREB at one or both of these sites.
Materials and Methods
Cell Culture and Hypoxia.
T84, bovine aortic endothelial (BAE), and HeLa cells were maintained and exposed to hypoxia (pO2, 20 torr; pCO2, 35 torr; balance N2 and water vapor; 1 torr = 133 Pa) as described (3, 4, 6). Normoxic controls were exposed to the same protocols under conditions of atmospheric O2 (pO2, 147 torr; pCO2, 35 torr).
TNFα Release Assay.
Basolateral TNFα release from T84 monolayers grown on permeable support inserts was measured as described (3, 4). Confluent monolayers were exposed to hypoxia for 24 h, and media were removed and replaced with fresh preequilibrated hypoxic media. This procedure was repeated at 48 and 72 h. TNFα release in each period (0–24, 24–48, and 48–72 h) was assessed independently by ELISA (BioSource International, Camarillo, CA).
On-Bead Biotinylation.
T84 monolayers were exposed to normoxia or 48-h hypoxia, lysed in low-stringency lysis buffer, and immunoprecipitated overnight with polyclonal CREB antibody. Beads were washed three times in low-stringency lysis buffer, biotinylated (1 mM biotin, 25 min, 4°C), washed again, and resuspended in 150 mM NH4Cl (in lysis buffer) for 30 min at 4°C. Washed beads were resuspended in sample buffer, boiled (5 min), and separated on 10% SDS/PAGE. Proteins were transferred to nitrocellulose membrane and probed for biotinylated proteins by using avidin–peroxidase.
Gene Array Analysis.
Gene array analysis was carried out as described (6). T84 cells were exposed to hypoxia (0, 6, 18 h), total RNA was extracted, and mRNA was isolated and DNase treated (6). The mRNA profile was assessed by using quantitative gene chip expression arrays (Affymetrix, Santa Clara, CA; ref. 43).
Analysis of mRNA Levels by PCR.
RT-PCR analysis of mRNA levels was performed by using DNase-treated total RNA as described (4) with primers specific for SUMO-1 (5′-CGTCATCATGTCTGACCAGGA-3′ and 5′-CACTGAAAGTCACAGTCCAGG-3′) or for human β-actin (5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′ and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′).
Northern Blot Analysis.
RNA was isolated and treated as described above, denatured (58°C for 15 min), loaded on a 1% agarose-1 M 4-morpholinepropanesulfonic (Mops) gel in 1 M Mops running buffer, and run at 80 V for 90 min. RNA was transferred to a positively charged nylon membrane (Roche Diagnostics) by capillary action for 180 min. Crosslinking was achieved by using UV irradiation. A probe was generated by random primer biotinylation by using a commercially available kit (Kirkegaard & Perry Laboratories). The membrane was prehybridized in 10 ml of formamide buffer/100 μg/ml herring sperm DNA hybridization solution for 1 h at 42°C. The denatured probe was then added for 16 h in hybridization solution. Detection was achieved by using a commercially available kit and exposing membranes to photographic film.
Immunoprecipitation/Western Blotting.
To examine CREB/IκΒ ubiquitination and SUMO-1 modification, whole-cell lysates were prepared as described (4). CREB or IκΒ was immunoprecipitated from these lysates by using antibodies from New England Biolabs and Upstate Biotechnology (Lake Placid, NY), respectively. Immunoprecipitates were separated by 10% SDS/PAGE, transferred to nitrocellulose, and probed with anti-ubiquitin (StressGen Biotechnologies, Victoria, Canada) or anti-SUMO-1 (Zymed). After washing, a species-matched peroxidase-conjugated secondary antibody was added (Cappell). Labeled bands were detected by enhanced chemiluminesence (Amersham Pharmacia).
Peptide Treatment.
HIV-tat peptide-facilitated loading of cells with bioactive peptides was carried out as described (6). Synthetic peptides were generated by SynPep (Dublin, CA). The HIV-tat-conjugated SUMO-target sequence was YGRKKRR-QRRRGECRRKKKEYVKC. The peptide was made to a stock concentration of 50 mM in H2O. Peptides were added to the indicated final concentrations 2 h before incubation in hypoxia.
Transfections.
The SUMO-1 expression plasmid which encoded for a His (6)-tagged SUMO-1 protein was a kind gift from Ron Hay (University of St. Andrews, Fife, Scotland). One microgram of plasmid was transfected into BAE cells by using Effectene transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions.
A luciferase gene reporter assay was used to investigate the impact of hypoxia on transcriptional events under the control of CREB as described (6). Briefly, BAE cells were grown to ≈60% confluence and transfected with a luciferase reporter plasmid under the control of a basic promoter element (TATA) plus a defined inducible cis-enhancer element containing CRE response element motifs (Stratagene). Where appropriate, cells were cotransfected with positive control plasmids.
Site-Directed Mutagenesis.
The plasmid used for expression and mutagenesis was pCREB–enhanced GFP (EGFP) (CLONTECH). All site-directed mutagenesis was carried out by using the QuikChange XL Site-Directed Mutagenesis kit according to the manufacturer's instructions (Stratagene). Mutant plasmids were confirmed by DNA sequencing. Mutations made were K155T, K155R, K285I, K285R, K304T, and K304R. Intracellular localization of EGFP-CREB was visualized by fluorescence microscopy.
In Vivo and in Vitro CREB SUMO Modification Assays.
In vivo SUMO modification of CREB was evaluated in BAE cells transfected with wild-type or mutated EGFP-CREB. After EGFP immunoprecipitation with the Full Length Living Colors antibody (CLONTECH), samples were resolved by SDS/PAGE, transferred to nitrocellulose, and Western blotted with a CREB or SUMO antibody. For in vitro SUMO modification assays, wild-type and mutant EGFP-CREB was expressed in HeLa cells, immunoprecipitated (as above), and incubated for 2 h at 37°C in the presence or absence of SUMO and the enzymes necessary to catalyze the SUMO modification (25). Samples were then resuspended in reducing sample buffer, and SUMO-modification of CREB was assayed as described above.
Data Presentation.
Cytokine ELISA and luciferase reporter assay data were compared by Student's t test with P < 0.05 considered statistically significant. All values are given as mean ± SEM for n experiments.
Results
Resolution of Hypoxia-Elicited TNFα Expression.
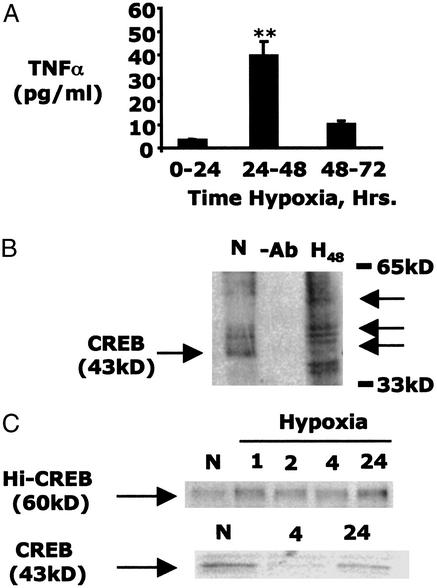
Previously, we demonstrated that hypoxia-induced TNFα expression depended on the ubiquitin-mediated degradation of CREB (3, 4, 6). Here, we examined the resolution of this response in prolonged periods of exposure. Subjection of T84 cells to hypoxia resulted in the temporally segmental release of TNFα, which was maximal between 24 and 48 h (42.4 ± 5.7 pg per monolayer; Fig. 1A) of exposure and resolved between 48 and 72 h (8.4 ± 1.7 pg/monolayer; Fig. 1A). This resolution phase was not associated with cell death as assessed by transepithelial electrical resistance (data not shown).
Figure 1.
Hypoxia-induced TNFα is transient. (A) ELISA was used to measure temporally segmental, basolateral release of TNFα from T84 cells. Hypoxia-elicited TNFα release was transient and resolved between 48- and 72-h hypoxia (n = 3; P < 0.05). (B) On-bead biotinylation was used to investigate the impact of 48-h hypoxia on CREB expression and modification. Higher molecular weight modified forms of CREB, which evolve in hypoxia, are indicated (black arrows). (C) T84 cells exposed to increasing periods of hypoxia demonstrate decreased native 43-kDa CREB expression as previously described (Lower; ref. 6). During the same period, a higher molecular weight band (60 kDa) appears, which is recognized by the CREB antibody, implicating a modified form of CREB.
Because previous studies implicated the ubiquitin-dependent degradation of CREB in TNFα induction (6), we searched for the existence of alternatively modified stable forms of CREB in prolonged periods of hypoxia. To do this, CREB immunoprecipitates were biotinylated, resolved, and probed with peroxidase-conjugated avidin. As shown in Fig. 1B, and consistent with previous work (4, 6), prolonged hypoxia was associated with the near-complete loss of native CREB (43 kDa). However, the present studies also revealed the appearance of higher molecular weight stable forms of CREB in cells subjected to hypoxia (Fig. 1B). Furthermore, CREB immunoprecipitation/Western blot analysis also revealed the evolution of a higher molecular weight modified CREB (≈60 kDa) associated with prolonged hypoxia, suggesting the existence of a hypoxia-specific modification of CREB (Fig. 1C). Native (43 kDa) CREB was diminished under these conditions, as described (Fig. 1C).
Hypoxia Increases SUMO-1 Expression.
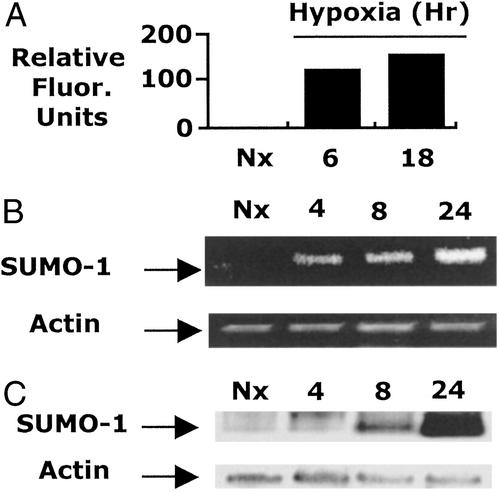
A broad search of hypoxia-regulated genes using microarray technologies revealed the vast majority of genes remained unaltered in hypoxia (>96%), including many in the protein modification family (e.g., ubiquitin, ubiquitin conjugating enzymes, ubiquitin activating enzymes, ubiquitin carrier proteins, etc.). However, this analysis revealed the time-dependent induction of SUMO-1 mRNA (Fig. 2A). Because SUMO-1 conjugation is potentially inhibitory to ubiquitination and subsequent proteasomal degradation (8, 9, 11) and results in an ≈17-kDa modification in targeted proteins (consistent with our findings in Fig. 1), we considered the possibility that the late hypoxia-elicited induction of SUMO-1 may represent an “offswitch” to the early ubiquitin-dependent hypoxia-elicited inflammatory phenotype. We confirmed the transcriptional induction of SUMO-1 mRNA in response to hypoxia by using RT-PCR and Northern blot analysis and determined that hypoxia induces SUMO-1 mRNA expression within 4–8 h of exposure (Fig. 2 B and C). These findings suggest the possibility that transcriptional induction of SUMO-1 and subsequent SUMO-1 modification of CREB could provide a repression mechanism for CREB degradation-dependent transcriptional responses (e.g., TNFα).
Figure 2.
Expression of SUMO-1 mRNA in hypoxia. (A) mRNA microarray analysis was used to investigate global gene expression in T84 cells exposed to increasing periods of hypoxia. Although 96% of genes studied remained stable, the expression of SUMO-1 was increased in a time-dependent manner. (B) Microarray data were confirmed by RT-PCR analysis, which demonstrates a time-dependent increase in SUMO-1 mRNA in hypoxia. (C) Northern blot analysis was used directly to assess SUMO-1 expression in RNA derived from cells exposed to indicated periods of hypoxia.
Hypoxia Induces SUMO Modification of CREB.
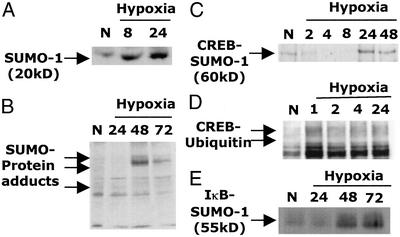
We next investigated whether SUMO-1 protein modification was associated with hypoxia. First, we confirmed induction at the protein level by Western blot analysis. SUMO-1 protein was minimally expressed in control cells, induced by 8 h of hypoxia, and remained elevated by 24 h (Fig. 3A). We next used Western blotting to probe whole-cell lysates for changes in overall protein SUMO-modification patterns. T84 cells displayed significant increases in protein SUMO modification, apparent within 24 h and particularly at 48 h (Fig. 3B). In data not shown, increased SUMO modification was evident in diverse cell types subjected to hypoxia, including microvascular endothelia and renal tubular epithelia, and was also demonstrable in vivo (colonic mucosal scrapings from hypoxic mice). Anti-SUMO-1 immunoblotting of SUMO-1 immunoprecipitates confirmed increased protein SUMO-1 modification by 48 h of hypoxia (data not shown).
Figure 3.
Hypoxia enhances protein SUMO-1 modification. (A) Western blot analysis was used to confirm hypoxia-elicited up-regulation of SUMO-1 protein expression. (B) Western blot analysis of whole-cell lysates was used to investigate SUMO-1 modification of proteins in 0- to 72-h hypoxia. Arrows indicate SUMO-1-modified proteins. (C) SUMO-1 modification of CREB in hypoxia was investigated by CREB immunoprecipitation and Western blot with an anti-SUMO-1 antibody. A band at ≈60 kDa increased with prolonged hypoxia, consistent with SUMO-1-modified CREB. (D) CREB ubiquitination in hypoxia was investigated by immunoprecipitation of CREB and Western blot with an anti-ubiquitin antibody. Bands consistent with ubiquitin-modified CREB were rapidly detected by 1 h of hypoxic exposure. (E) SUMO-1 modification of IκB in hypoxia was investigated by IκB immunoprecipitation and Western blot with an anti-SUMO-1 antibody. A band at ≈55 kDa increased with prolonged hypoxia, consistent with SUMO-1-modified IκB.
Both IκΒα and CREB are negatively associated with the regulation of inflammatory gene transcription in hypoxia. We have proposed that the phosphorylation-dependent targeting of IκΒ and CREB to ubiquitination and degradation is a potential activation pathway for an inflammatory phenotype-induced in hypoxia (4, 6). IκΒα has previously been identified as a substrate for SUMO-1 modification (9), an event dependent on the presence of a SUMO-binding motif (LKKE). SUMO-1 modification of IκΒ inhibits ubiquitination and thus may be an important endogenous anti-inflammatory signal. We hypothesized that SUMO-1 modification of CREB may represent an anti-inflammatory signal in prolonged hypoxia. Thus, we addressed whether CREB is directly SUMO modified in hypoxia.
Epithelial cells were exposed to increasing periods of hypoxia, and IκΒ or CREB was immunoprecipitated, resolved by SDS/PAGE, and immunoprobed with anti-SUMO-1. We found that CREB immunoprecipitates demonstrated an evolving SUMO-1-modified band in hypoxia (at a molecular weight of ≈60 kDa, consistent with a CREB–SUMO conjugate), indicating a CREB-SUMO-1 association in hypoxia (Fig. 3C). IκBα was similarly SUMO-1 modified by these conditions of hypoxia (Fig. 3E), and as such, served as a control protein for SUMO-1 modification. Importantly, maximal SUMO-1 modification of both IκΒ and CREB occurred at time points between 24 h and maximally at 48 h. In separate experiments, CREB immunoprecipitates were probed with anti-ubiquitin. These studies demonstrated, as we described previously (6), that CREB ubiquitination occurred transiently at earlier time points, with detectable ubiquitination by 1–2 h of hypoxia (Fig. 3D). As described earlier in Fig. 1C, native CREB degradation is detectable by 4 h of hypoxia, whereas a SUMO-modified form evolves at later time points. Combined, these data support our hypothesis that CREB is a SUMO-1 substrate and that prolonged hypoxia promotes posttranslational modification of CREB by SUMO-1.
SUMO Overexpression Stabilizes CREB.
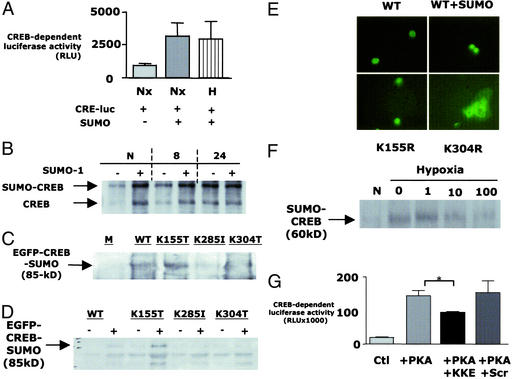
To investigate whether SUMO-1 functionally stabilizes CREB, we overexpressed SUMO-1 by transient transfection and assessed CREB activity by using a CREB-dependent luciferase reporter assay. As shown in Fig. 4A, overexpression of SUMO-1 in BAE cells resulted in a functionally enhanced basal CREB activity. Furthermore, the SUMO-1-modified form of CREB remained stable in hypoxia, whereas the native form was degraded (Fig. 4B). Together, these data demonstrate that SUMO-1 modification of CREB results in stabilization of the active protein in both normoxia and hypoxia.
Figure 4.
SUMO-1 stabilizes CREB through modification at a specific binding motif. (A) A CREB-dependent luciferase reporter assay was used to determine the impact of SUMO-1 overexpression on CREB activity in BAE cells. SUMO-1 overexpression significantly enhanced CREB-dependent activity under normoxic (N) and 48-h hypoxic (48) conditions. (B) Western blot analysis was used to investigate the impact of SUMO-1 overexpression on CREB levels in the nuclear lysates derived from normoxic and hypoxic cells. SUMO-1 overexpression increased the expression of SUMO-1-modified and nonmodified CREB in normoxia. SUMO-1 overexpression resulted in the stabilization of SUMO-1-modified but not nonmodified CREB in hypoxia. (C) Site-directed mutagenesis of lysines 285 and 304 but not 155 resulted in decreased EGFP-CREB SUMO modification in vivo. (D) Site-directed mutagenesis of lysines 285 and 304 but not 155 resulted in decreased EGFP-CREB SUMO modification in vitro. (E) Wild-type CREB is localized in the nuclear compartment and is enhanced by cotransfection with SUMO. Mutation of the SUMO acceptor residue (K304R) resulted in a loss of nuclear localization of EGFP-CREB, whereas mutation of the nonaccepting lysine residue (K155R) did not alter CREB nuclear localization. (F) Western blot analysis of CREB immunoprecipitates was used to investigate the impact of the KKKE-containing peptide on SUMO-1 modification of CREB in hypoxia. The bioactive peptide diminished SUMO-1 modification of CREB in a concentration-dependent manner. (G) A CREB-dependent luciferase reporter assay was used to investigate the impact of inhibition of CREB SUMO-1 modification on CREB activity. Cotransfection of CRE-luciferase and protein kinase A resulted in a significant increase in CREB-dependent activity, which was unaltered by pretreatment of cells with a scrambled control peptide but was significantly diminished by pretreatment with the KKKE-containing peptide.
The consensus SUMO modification motif is ψKxE, where ψ represents a large hydrophobic amino acid and x represents any amino acid (42). Although no exact consensus motif exists in CREB, three similar sequences exist between amino acids 303 and 306 (KKKE), 284 and 287 (RKRE), and 154 and 157 (EKSE), which we hypothesized to be potential acceptor sites for SUMO on CREB. We used site-directed mutagenesis of CREB to mutate lysine residues on each of these sites. Mutation of lys (K285) and lys (K304) resulted in decreased SUMO-modification of CREB both in vivo and in vitro. (Fig. 4 C and D, respectively). Interestingly, we found that K304 lies within the putative nuclear localization sequence of CREB. Mutation of K304 to an amino acid with similar properties (R; to maintain nuclear localization sequence activity but lose SUMO modification) resulted in a loss in the nuclear localization of CREB, implicating a role for SUMO modification at this motif in the nuclear localization of CREB (Fig. 4E).
To confirm whether nonconsensus sites such as 303–306 (KKKE) could function as SUMO-1 modification motifs, we generated a synthetic peptide sequence spanning the putative target amino acid sequence containing lysine 304. This peptide was made cell permeant by synthesis of an N-terminal HIV-tat sequence, which we have used successfully in the past (6). Initially, we investigated the impact of preloading cells with this peptide on protein SUMO-1 modification in hypoxia. As shown in Fig. 4F, preloading cells with increasing concentrations of the KKKE-containing (but not the scrambled control) peptide and subsequent assessment of CREB SUMO modification by immunoprecipitation/Western blot revealed a loss of SUMO-1 modification with increasing KKKE peptide concentration, suggesting that the decoy peptide functionally diminishes SUMO modification of CREB.
Additional studies were performed to determine the impact of the decoy peptide on CREB activity. To do this, protein kinase A-activated CREB luciferase reporter assays were used, and as can be seen in Fig. 4G, preloading of cells with HIV-tat-KKKE peptide (10 μM final concentration) significantly decreased CREB-dependent luciferase activity compared with control (P < 0.05; n = 3), Taken together, these results indicate that the KKKE sequence represents a functional SUMO-1 targeting domain on CREB.
Discussion
Hypoxia is commonly associated with inflammation in a range of pathologies, and hypoxia-elicited transcriptional events may contribute significantly to the extent of inflammatory disease (1). Although the vast majority of inflammatory processes are self-limiting and resolve with little or no consequence, it is now appreciated that this latter phase of resolution may be more an active response than a passive process. In the present studies, we addressed the late-phase repression of a modeled inflammatory event, typified by the transcriptional induction of TNFα (3, 4), to identify potential targets for resolution. This approach identified SUMO-1 modification as a potentially important posttranslation modification for resolution of an inflammatory phenotype.
Initial studies directed at addressing potential posttranslational modification of CREB by hypoxia revealed the appearance of an ≈17-kDa modification associated with prolonged hypoxia. An unbiased search for protein modifiers by microarray analysis revealed that, whereas the majority of genes, including other protein modifiers, remained unaltered in hypoxia, SUMO-1 was transcriptionally induced. Of interest in this regard, a number of bona fide SUMO-1 substrates (7) have been associated with either transcriptional or posttranslational regulation by hypoxia, including p53, Mdm2, c-jun, Glut-1, and Glut-4 (44–46). In addition, NFκB transcriptional activity is enhanced by hypoxia, and the expression and function of IκBα have been tightly linked to SUMO-1 conjugation (8). Thus, significant evidence places SUMO-1 as a central player in the generalized hypoxic response.
A number of functions have been attributed to SUMO-1 conjugation of proteins, most notably the stabilization of protein structure, the regulation of protein–protein interactions, and the organization of subcellular compartmentalization (47). Most recent studies indicate that the enzymes implicated in SUMO-1 conjugation are expressed predominantly in the nucleus. For example, transfection studies using epitope-tagged SUMO-1, SUMO-1-activating enzymes, and Ubc9 indicated dominant nuclear/nuclear membrane localization within dot-like structures (48). Our present findings that CREB is a SUMO-1 substrate are thus consistent within this localization, because CREB is expressed predominantly within the nuclear domain (49) and has been previously observed to be associated with Ubc9 (50). Interestingly, we found that, whereas ubiquitination of CREB is a relatively early event (≈2 h), SUMO-1 conjugation of CREB occurs comparatively late in the hypoxic response (24 and 48 h after onset). This may be related to the necessity for transcriptional up-regulation of SUMO-1 in hypoxia, because little or no SUMO-1 expression (either RNA or protein) was evident in normoxic cells. Such observations suggest a self-regulating mechanism for ubiquitination and SUMO-1 modification of transcriptionally active CREB. At present, it is not known whether this is unique to CREB or a more general mechanism for other SUMO-1 conjugates. It is likely that this self-regulating path is a more general mechanism, because the expression of other SUMO-1 conjugates (e.g., IκBα) is also tightly regulated through proteolysis. More work will be necessary to define the relationship between SUMO-1 expression levels and relative conjugation activity.
Although the exact targeting mechanism(s) for proteins to conjugation by SUMO-1 is not precisely detailed, it is clear that acceptor proteins express a minimal consensus site for SUMO-1 modification. Recent mutagenesis-based mapping studies have more clearly defined this consensus site to include ψKxE, where ψ is a hydrophobic amino acid and x is any amino acid (42). Recently, the exact SUMO-targeting sequence for a number of proteins has been defined as containing the consensus motif (7). Importantly, a number of SUMO-1-modified proteins including PCNA, Daxx, and Axin, also contain active nonconsensus-binding sites (11, 33, 34). Analysis of CREB identified no consensus modification site; however, putative SUMO-1 modification sites were located at amino acid positions 304, 285, and 155, which exist within sequences similar to the consensus motif although lacking the hydrophobicity of the ψ residue. Site-directed mutagenesis of CREB using a transfection-based approach revealed that mutation of lysines 285 and 304 resulted in decreased association between CREB and SUMO both in vivo and in vitro. Multiple sumoylation sites have been described elsewhere for other proteins (51, 52). Mutation of lysine 155 actually resulted in increased CREB modification by SUMO-1. Further experiments will determine whether this site may represent the ubiquitin acceptor residue. The CREB K304R mutation, which should maintain nuclear localization sequence (NLS) activity but lose SUMO-1 modification, loses its nuclear localization, implicating a role for SUMO modification of the NLS in the subcellular localization of CREB.
We generated a cell-permeant peptide spanning the SUMO-1 consensus motif. This approach has been used widely to study protein structure–function in intact cells (53) and, because of its homology to nuclear localization sequences (54), is particularly effective at targeting nuclear substrates, such as CREB. Our studies revealed that loading of cells with this peptide efficiently inhibited CREB SUMO-1 modification in hypoxia, presumably through functioning as a decoy for SUMO-1 modification.
Because SUMO-1 has been previously associated with antagonism of ubiquitination (9, 42), we investigated its potential role in the resolution of hypoxia-elicited inflammatory gene expression. This hypothesis is consistent with a number of previous studies indicating that SUMO-1 may hold promise in modulating the inflammatory response. For example, given the importance of tightly regulated expression of the SUMO-1 conjugate c-Jun in stress and inflammation (55), it is quite probable that SUMO-1 is central to a number of key kinase activation pathways. Moreover, Desterro et al. (8) have directly implicated SUMO-1 in the inhibition of the proinflammatory transcription factor NFκB through stabilization of the inhibitory subunit IκBα. Of note in this regard, we and others have shown that hypoxia activates NFκB through as-yet-undefined mechanism(s). Our studies here revealed that IκBα is directly SUMO-modified in hypoxia (see Fig. 3G) and, given the importance of IκBα stabilization to NFκB control, such findings may contribute significantly to understanding transcriptional pathways in addition to those mediated by CREB. As such, we propose that SUMO-1 modification/stabilization and nuclear targeting of anti-inflammatory transcriptional regulators may represent a general antiinflammatory signal, and that hypoxia may be central to coordination of this response (see Fig. 5).
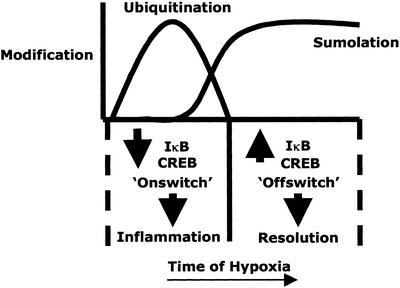
Figure 5.
Model of the temporal posttranslational modifications of transcriptional regulators in hypoxia. In early periods of hypoxia, IκB and CREB are ubiquitinated rapidly in a phosphorylation-dependent manner, resulting in targeting of these molecules to proteasomal degradation. Such removal of anti-inflammatory regulators results in the induction of a proinflammatory phenotype. More prolonged periods of hypoxia lead to the induction of SUMO-1 expression and SUMO-lation of IκB and CREB. This late event depends on the transcription and translation of SUMO and leads to the stabilization of these regulators, consequently mediating an inhibition of the hypoxia-elicited inflammatory phenotype.
Acknowledgments
We acknowledge technical assistance from Annemarie Griffin. The SUMO-1 plasmid was a kind gift of Dr. Ron Hay. This work was supported by grants from the National Institutes of Health to C.T.T. (DK02682), S.P.C. (DK50189, DE13499, and HL60569), and G.T.F. (DK02564), and by grants from The Wellcome Trust and the Science Foundation of Ireland (C.T.T.), the Health Research Board of Ireland (K.C.), and the Crohn's and Colitis Foundation (J.K.).
Abbreviations
- TNF
tumor necrosis factor
- EGFP
enhanced GFP
- SUMO
small ubiquitin-related modifier
- NFκB
nuclear factor κB
- CREB
cAMP-response element-binding protein
- BAE
bovine aortic endothelial
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Taylor C T, Colgan S P. Pharmacol Res. 1999;16:1498–1505. doi: 10.1023/a:1011936016833. [DOI] [PubMed] [Google Scholar]
- 2.Bunn H F, Poyton R O. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 3.Taylor C T, Dzus A L, Colgan S P. Gastroenterology. 1998;114:657–668. doi: 10.1016/s0016-5085(98)70579-7. [DOI] [PubMed] [Google Scholar]
- 4.Taylor C T, Fueki N, Agah A, Hershberg R M, Colgan S P. J Biol Chem. 1999;274:19447–19454. doi: 10.1074/jbc.274.27.19447. [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Ben-Neriah Y. Ann Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 6.Taylor C T, Furuta G T, Synnestvedt K, Colgan S P. Proc Natl Acad Sci USA. 2000;97:12091–12096. doi: 10.1073/pnas.220211797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melchoir F. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 8.Desterro J M, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 9.Hay R T, Vuillard L, Desterro J M, Rodriguez M S. Philos Trans R Soc London B. 1999;354:1601–1609. doi: 10.1098/rstb.1999.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao Y, Desai S D, Liu L F. J Biol Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 11.Hoege C, Pfander B, Moldovan G L, Pyrowolkis G, Jentsh S. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 12.Schwienhorst I, Johnson E S, Dohmen R J. Mol Gen Genet. 2000;263:771–786. doi: 10.1007/s004380000254. [DOI] [PubMed] [Google Scholar]
- 13.Muller S, Matunis M J, Dejean A. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller S, Berger M, Lehembre F, Seeler J S, Haupt Y, Dejean A. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 15.Desterro J M, Thomson J, Hay R T. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 16.Desterro J M, Rodriguez M S, Kemp G D, Hay R T. J Biol Chem. 1997;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 17.Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. Biochem Biophys Res Commun. 1999;254:693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- 18.Sampson D A, Wang M, Matunis M J. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 19.Pichler A, Gast A, Seeler J S, Dejean A, Melchior F. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt D, Muller S. Proc Natl Acad Sci USA. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 22.Mahajan R, Gerace L, Melchior F. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matunis M J, Wu J, Blobel G. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minty A, Dumont X, Kaghad M, Caput D. J Biol Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez M S, Desterro J M P, Lain S, Midgley C A, Lane D P, Hay R T. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwartz S E, Scheffner M, Del Sal G. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poukka H, Karvonen U, Janne O A, Palvimo J J. Proc Natl Acad Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y H, Choi C Y, Kim Y. Proc Natl Acad Sci USA. 1999;96:12350–12355. doi: 10.1073/pnas.96.22.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi P P, Freemont P, de The H. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong Y, Rogers R, Matunis M J, Mayhew C N, Goodson M, Park-Sarge O K, Sarge K D. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 31.Bies J, Markus J, Wolff L. J Biol Chem. 2002;277:8999–9009. doi: 10.1074/jbc.M110453200. [DOI] [PubMed] [Google Scholar]
- 32.Chakrabarti S R, Sood R, Nandi S, Nucifora G. Proc Natl Acad Sci USA. 2000;97:13281–13285. doi: 10.1073/pnas.240315897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rui H L, Fan E, Zhou H, Xu Z, Zhang Y, Lin S C. J Biol Chem. 2002;277:42981–42989. doi: 10.1074/jbc.M208099200. [DOI] [PubMed] [Google Scholar]
- 34.Jang M S, Ryu S W, Kim E. Biochem Biophys Res Commun. 2002;295:495–500. doi: 10.1016/s0006-291x(02)00699-x. [DOI] [PubMed] [Google Scholar]
- 35.Rangasamy D, Woytek K, Khan S A, Wilson V G. J Biol Chem. 2000;275:37999–38004. doi: 10.1074/jbc.M007777200. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann H, Floss S, Stamminger T. J Virol. 2000;74:2510–2524. doi: 10.1128/jvi.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogan S, Heaphy S. Virus Genes. 2000;21:193–195. doi: 10.1023/a:1008139514123. [DOI] [PubMed] [Google Scholar]
- 38.Orth K, Xu Z, Mudgett M B, Bao Z Q, Palmer L E, Bliska J B, Mangel W F, Staskawicz B, Dixon J E. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 39.Cornelis G R, Denecker G. Nat Med. 2001;7:21–23. doi: 10.1038/83298. [DOI] [PubMed] [Google Scholar]
- 40.Endter C, Kzhyshkowska J, Stauber R, Dobner T. Proc Natl Acad Sci USA. 2001;98:11312–11317. doi: 10.1073/pnas.191361798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishida T, Kaneko F, Kitagawa M, Yasuda H. J Biol Chem. 2001;276:39060–39066. doi: 10.1074/jbc.M103955200. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez M S, Dargemont C, Hay R T. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 43.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 44.Blagosklonny M V. Oncogene. 2001;20:395–398. doi: 10.1038/sj.onc.1204055. [DOI] [PubMed] [Google Scholar]
- 45.Salnikow K, Kluz T, Costa M, Piquemal D, Demidenko Z N, Xie K, Blagosklonny M V. Mol Cell Biol. 2002;22:1734–1741. doi: 10.1128/MCB.22.6.1734-1741.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behrooz A, Ismail-Beigi F. News Physiol Sci. 1999;14:105–110. doi: 10.1152/physiologyonline.1999.14.3.105. [DOI] [PubMed] [Google Scholar]
- 47.Muller S, Hoege C, Pyrowolakis G, Jentsh S. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 48.Sternsdorf T, Jensen K, Reich B, Will H. J Biol Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- 49.Mayr B, Montminy M. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 50.Firestein R, Feuerstein N. J Biol Chem. 1998;273:5892–5903. doi: 10.1074/jbc.273.10.5892. [DOI] [PubMed] [Google Scholar]
- 51.Tian S, Poukka H, Palvimo J J, Janne O A. Biochem J. 2002;367:907–911. doi: 10.1042/BJ20021085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Drean Y, Mincheneau N, Le Goff P, Michel D. Endochrinology. 2002;143:3482–3489. doi: 10.1210/en.2002-220135. [DOI] [PubMed] [Google Scholar]
- 53.Becker-Hapak M, McAllister S S, Dowdy S F. Methods. 2001;24:247–256. doi: 10.1006/meth.2001.1186. [DOI] [PubMed] [Google Scholar]
- 54.Efthymiadis A, Briggs L J, Pyrowolakis G, Jentsh S. J Biol Chem. 2001;273:1623–1628. doi: 10.1074/jbc.273.3.1623. [DOI] [PubMed] [Google Scholar]
- 55.Aggarwal B B. Ann Rheum Dis. 2000;59 Suppl 1:6–16. doi: 10.1136/ard.59.suppl_1.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]