Abstract
Mycobacterium tuberculosis uses a proteasome system that is analogous to the eukaryotic ubiquitin-proteasome pathway and is required for pathogenesis. However, the bacterial analogue of ubiquitin, prokaryotic ubiquitin-like protein (Pup), is an intrinsically disordered protein bearing little sequence or structural resemblance to the highly structured ubiquitin. Thus it was unknown how pupylated proteins were recruited to the proteasome. Here, we show that the Mycobacterium proteasomal ATPase (Mpa) has three pairs of tentacle-like coiled-coils that recognize Pup. Mpa binds unstructured Pup via hydrophobic interactions and a network of hydrogen bonds, leading to the formation of an α-helix in Pup. Our work revealed a binding-induced folding recognition mechanism in the Pup-proteasome system that differs mechanistically from substrate recognition in the ubiquitin-proteasome system. This critical difference between the prokaryotic and eukaryotic systems could be exploited for the development of a small molecule-based treatment of tuberculosis.
INTRODUCTION
Proteasomes are ubiquitous in archaea and eukaryotes and found in some bacteria of the order Actinomycetales1,2. As in eukaryotes, the Mycobacterium tuberculosis (Mtb) proteasome system consists of a 20S proteolytic core particle and the Mycobacterium proteasomal ATPase Mpa, the structures of which appear to be conserved with their eukaryotic counterparts 3-5. Importantly, the proteasome is essential for Mtb to cause lethal infections in a mammalian host 6. Distinctions between the bacterial and eukaryotic systems also exist1,5,7, thus efforts are focused on developing Mycobacterium-specific proteasome inhibitors as anti-tuberculosis agents 8.
Mpa contains an N-terminal coiled-coil domain with a predicted α-helix, an intermediate domain with a double oligosaccharide/oligonucleotide binding (OB) fold, and an ATPase associated with various activities (AAA) domain, and forms hexamers 5,9. The architecture of Mpa conforms to other proteasomal ATPases, including the archaeal proteasome-activating nucleotidease (PAN) and the Rhodococcus ATPase forming ring-shaped complexes (ARC) 10,11. Mpa and ARC contain two OB folds in tandem, but in neither case had the coiled-coil domain structure been determined. The native coiled-coil structure in the Archaeoglobus fulgidus PAN was not determined; GCN4 leucine zippers substituted for the coiled-coils in PAN to produce a hybrid structure for crystallization11. A partial coiled-coil in the archaeal Methanocaldococcus jannaschii PAN containing 16 residues with two heptad repeats was also reported 10.
Proteasome substrates in Mycobacteria are covalently tagged with a 64 amino acid degradation signal called Pup 12,13. Pup covalently links to substrate lysines via an isopeptide bond with a carboxy (C)-terminal glutamate12,14. Production of a linear fusion between Pup and a non-proteasomal substrate confirmed that the C-terminal half of Pup is required to interact with Mpa, and the amino (N)-terminal half is required to facilitate substrate unfolding and degradation15,16. Thus, pupylated substrates are likely recruited to the proteasome via the specific recognition of Pup by Mpa 14,16, the precise molecular mechanism of which was unknown. In this study, we used biochemical, structural and genetic approaches to show that Pup forms a helical structure upon binding to Mpa in order to deliver proteins into the mycobacterial proteasome for destruction.
RESULTS
Crystal structure of Mpa1-234 revealed tentacle-like coiled-coils
To begin to understand how Pup targets proteins for degradation by the mycobacterial proteasome, we determined the extent of the full-length Mpa coiled-coil by solving the structure of the Mpa1-234 hexamer, which includes the entire coiled-coil and double OB domains (Fig. 1a). The crystals were large (0.7 mm), but diffracted poorly (~ 8 Å in the synchrotron beam line, NSLS X29) due to the high solvent content (85%) of the long coiled-coils, an amount that was nearly twice as much as seen in most protein crystals 17. We improved the diffraction quality by dehydrating the crystals and solved the structure at a resolution of 3.9 Å (Fig. 1a, Table 1). The crystals belong to space group P21 with two hexamers per asymmetric unit. The structure was solved by the molecular replacement method using the Mpa double OB fold structure 5.
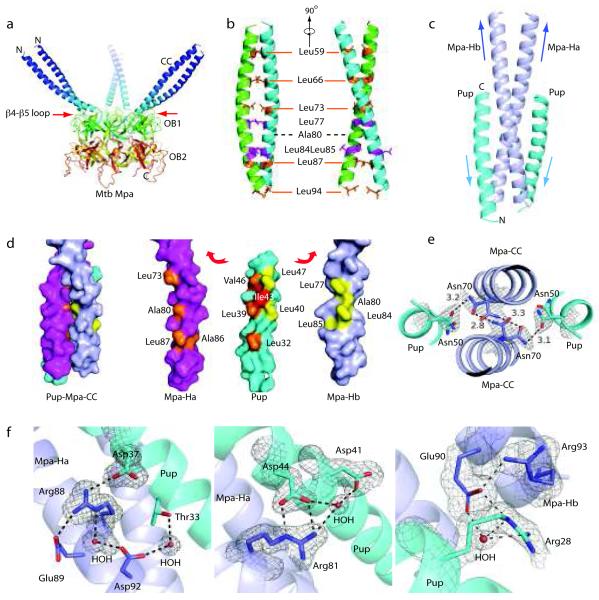
Figure 1.
Mpa1-234 hexamer has three 75 Å long coiled-coils needed for Pup recognition. (a) Crystal structure of Mpa1-234 revealed three long coiled-coils formed by six helices that sit atop the hexameric double OB-fold domain. Mpa1-46 was disordered in the crystal structure, and is thus not shown. (b) Two orthogonal views of the Mpa coiled-coil in cartoon representation. The coiled-coil was stabilized via a leucine zipper-like mechanism. Ala80 replaces the expected leucine at the 4th heptad repeat position. Presence of two adjacent leucines at positions 84 and 85 forced Leu85 to face the solvent. (c) Cartoon view of the co-crystal structure of the Pup21-64 in complex with the Mpa46-96 coiled-coil. Pup folds into an α-helix upon binding to the C-terminal half of the coiled-coil formed by Mpa-Ha and Mpa-Hb (see text for details). Arrows indicate helix direction. (d) Surface view of Pup21-64 on the Mpa46-96 coiled-coil (left). “Pulled apart” view of Pup21-64 and the Mpa46-96 coils (right). Hydrophobic residues in Pup that interact with Mpa Ha are colored in orange, and with Mpa Hb in yellow. The conserved Pup Ile43 (red) was centrally located and interacted with residues in both the Ha and Hb helices of Mpa. (e) The electron density map revealed that the conserved Asn50 of Pup and Asn70 of Mpa assume dual conformations. The conformations of Mpa Asn70 and Pup Asn50 enable H-bond formation within the Mpa coiled-coil and between Pup and Mpa. (f) Several hydrogen bonds further stabilize the chiefly hydrophobic interaction between Mpa (blue) and Pup (cyan). The σA-weighted 2Fo-Fc density map is contoured at 1σlevelaroundtheconservedresidues.
Table 1.
Data collection and refinement statistics
| Mpa1-234 | Mpa46-96 | Mpa46-96 Pup21-64 |
Mpa1-234 Pup1-64 |
|
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | F222 | P21 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 176.78, 176.65, 176.63 |
85.75,114.16, 114.49 |
44.75, 28.09, 96.23 |
176.78, 176.96, 176.61 |
| α, β, γ (°) | 90, 90.04, 90 | 90, 90, 90 | 90, 103.43, 90 | 90, 89.94, 90 |
| Resolution (Å) | 25-3.9 (4.10- 3.94) |
25-2.0 (2.03-2.0) | 25-1.9 (1.94- 1.90) |
25-4.5 (4.56- 4.48) |
| R merge | 6.6 (65.3) | 7.4 (58.6) | 7.6 (57.6) | 8.4 (60.9) |
| I/σI | 26.5 (2.5) | 54.2 (5.7) | 29.1 (2.0) | 16.7 (1.7) |
| Completeness (%) | 99.8 (100) | 99.4 (100) | 88.9 (75.6) | 99.0 (99.) |
| Redundancy | 4.6 (4.7) | 14.5 (14.5) | 6.9(5.8) | 3.8 (3.7) |
| Refinement | ||||
| Resolution (Å) | 25-3.9 | 25-2.0 | 25-1.9 | 25-4.5 |
| No. reflections | 93,255 | 17,050 | 18,548 | 65,041 |
| Rwork / Rfree | 29.3/32.4 | 20.9/27.4 | 20.3/23.7 | 27.3/30.4 |
| No. atoms | ||||
| Protein | 17,208 | 2,088 | 1,210 | 18,672 |
| Ligand/ion | - | - | - | - |
| Water | - | 60 | 77 | - |
| B-factors | ||||
| Protein | 204.1 | 38.7 | 35.9 | 187.3 |
| Ligand/ion | - | - | - | - |
| Water | - | 66.3 | 55.9 | - |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.023 | 0.023 | 0.027 | 0.017 |
| Bond angles (°) | 2.150 | 1.842 | 1.992 | 1.857 |
The N-terminal 51 residues were unstructured, but residues 52-96 formed a contiguous ~75 Å long α-helix in the Mpa crystal. Like ARC and PAN, the six α-helices of the Mpa hexamer formed three pairs of coiled-coils that sat atop three alternating OB domains, thus reducing the six-fold symmetry to three-fold. Strikingly, the coiled-coils protruded like tentacles from the main body of the Mpa hexamer (Fig. 1a). The Mpa coiled-coils were in a similar orientation to those in ARC and PAN 10,11, although the coiled-coils are much shorter in the latter two structures due to truncation or replacement of the coiled-coils in order to facilitate crystallization (Supplementary Fig. 1a,b).
The two-stranded parallel coiled-coils in Mpa were formed by one α-helix (trans) that extended to, and dimerized with, its neighboring α-helix (cis) (Fig. 1b). The cis α-helix connected to the OB domain via a cis peptide bond between the highly conserved Pro97 and Pro98. The length of the Mpa coiled-coil could accommodate six heptad repeats, but only five were in the structure, leaving a gap between heptads 3 (Leu73) and 4 (Leu87). At the predicted heptad position (residue 80), an alanine took the place of the expected leucine. However, there were three leucines nearby (77, 84, and 85); and Leu85 faced away from the hydrophobic core of the coiled-coil, making it solvent exposed (Fig. 1b).
The coiled-coil region of Mpa is needed for Pup recognition
The crystal structure of Mpa1-234 revealed that Mpa1-96 could be further divided into unstructured (Mpa1-45) and coiled-coil (Mpa46-96) domains, thus we wanted to determine if either part were responsible for Pup recognition. We produced Mpa1-46, Mpa46-96, and Mpa1-96 and investigated their binding to various hexahistidine (His6)-tagged Pup constructs in vitro. We found that both Mpa1-96 and Mpa 46-96 interacted with Pup whereas Mpa1-46 did not (Supplementary Fig. 2a). Taken together, these data indicated that the Mpa coiled-coil was sufficient for the recognition of Pup.
Previous studies have shown that the N-terminal 20 residues of Pup (Pup1-20) are required for in vivo and in vitro proteolysis by the mycobacterial proteasome 15,16. We thus asked if Pup1-20 was required for binding to the Mpa coiled-coil. We produced N-terminal truncated Pup21-64 and full-length Pup, and examined binding with various N-terminal fragments of Mpa in vitro. We found that Pup21-64 was sufficient to bind to the Mpa coiled-coil (Supplementary Fig. 2b), thus Pup1-20 is not required for binding to the Mpa coiled-coil, although it is absolutely required for proteasome-mediated proteolysis.
Binding-induced folding of Pup with the Mpa coiled-coil
In order to elucidate how Mpa recognizes Pup, we crystallized Mpa46-96 alone and in complex with Pup21-64, and solved the structures by molecular replacement, with a single α-helix extracted from the Mpa coiled-coil structure (Supplementary Fig. 3, Fig. 1c; Table 1). In the complex, Mpa46-96 formed a native two-stranded parallel coiled-coil that is essentially the same as observed in Mpa1-234 (Fig. 1b). The two helices are designated as Ha and Hb (Fig. 1c). Upon binding to the Mpa46-96 coiled-coil, Pup21-64 formed an α-helix (Fig. 1c). Pup interacted with the Mpa46-96 coiled-coil with a 1:1 stoichiometry, because both sides of the Mpa46-96 coiled-coil were equivalent and available for Pup binding. In the context of the Mpa hexamer, however, the inner and the outer surfaces of the coiled-coil were not equivalent: the lower part of the coiled-coil at the outside surface was blocked by a crossing loop between β4 and β5 of the first OB domain (Fig. 1a, red arrows), thus we predicted that Pup should not be able to bind the outside surface of the coiled-coil in an Mpa hexamer.
The Pup region extending from Ser21 to Ala51 folded into an α-helix, apparently using the Mpa coiled-coil as a template. The C-terminal 13 residues of Pup were disordered in the crystal. The Pup helix interacted in an anti-parallel fashion with the lower half of the Mpa coiled-coil (Fig. 1c). The Pup surface interacting with the Mpa coiled-coil was mainly hydrophobic and involved two patches of leucine zipper-like interactions: a smaller patch between Leu32 of Pup and Leu87 and Ala86 of Mpa-Ha; and an extensive patch formed by Leu39, Leu40, Ile43, Val46, and Leu47 of Pup, Ala80, Leu73 of Mpa Ha, and Leu85, Leu84, Ala80, and Leu77 of Mpa Hb (Fig. 1d).
Polar residues such as asparagine and aspartate have been shown to drive helix-helix associations through the formation of hydrogen bonds19,20. A key feature of the Pup and Mpa-coiled-coil interaction is the conserved Asn70 in the Mpa coiled-coil and the Asn50 in Pup: both residues were at the same height in the Mpa-Pup coiled-coil and assumed dual conformations that enabled simultaneous hydrogen bonding within the Mpa coiled-coil, and between Mpa and Pup (Fig. 1e). Furthermore, the Pup interaction with the two Mpa helices was asymmetric. Pup contacted one Mpa helix (Ha) extensively via a series of H-bonds among conserved residues, including: Mpa Arg88 with Pup Asp37 (Fig. 1f, left); Mpa Asp92 with Pup Thr33 via a water (Fig. 1f, left); and Mpa Arg81 with Pup Asp44 and with Asp41 via a water (Fig. 1f, center). The Pup interaction with the other Mpa helix (Hb) was weaker: Mpa Arg93 oriented the Glu90 via a H-bond to form two water-mediated H-bonds with Arg28 in Pup (Fig. 1f, right).
Pup recognition by the Mpa1-234 hexamer
We next asked if the above-described interaction between the Pup fragment and the Mpa coiled-coil fragment could occur between Pup and Mpa hexamer. We co-crystallized full-length Pup with Mpa1-234. Although the crystals were large, they diffracted X-rays only to a resolution of 4.5 Å in the beam line (X29 of the National Synchrotron Light Source). This was most likely due to high solvent content (81%). In order to prevent the potential phase bias at the coiled-coil region, we solved the Pup:Mpa1-234 complex structure by the molecular replacement method using the Mpa double OB fold structure, which does not contain the coiled-coil region.
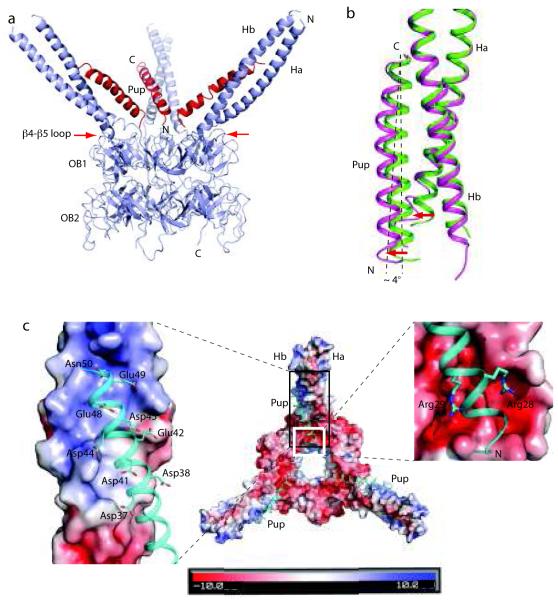
In the Pup:Mpa1-234 structure, Pup residues 21-51 formed an α-helix that bound to the lower half of the Mpa coiled-coil in an anti-parallel manner (Fig. 2a, Table 1), similar to what was observed in the Pup21-64:Mpa46-96 complex (Fig. 1c). Although full-length Pup was used for crystallization, the N-terminal 20 residues were disordered, consistent with our finding that Pup1-20 was not required for binding to the Mpa coiled-coil (Supplementary Fig. 2). The C-terminal 52-64 residues of Pup were also disordered in the crystal, similar to what we observed in the Pup21-64:Mpa46-96 crystal structure; this unstructured region might serve as a flexible linker between the induced α-helix and the C-terminus that forms an isopeptide bond with a substrate. Pup was not involved in crystalline packing, and binding of Pup did not markedly change the position of the coiled-coils in Mpa. Consistent with our earlier prediction, Pup bound only to the interior side of the coiled-coil in the Mpa hexamer, most likely because the outside surface was partially blocked by cross-barrel loops in Mpa (Fig. 2a, red arrows).
Figure 2.
Full-length Pup in the context of hexameric Mpa1-234. (a) Overall structure of the Pup:Mpa1-234 complex showing three Pups (red) apparently bound to all three Mpa coiled-coils. The red arrows point to the cross loops of the OB folds, which partially block the outside surfaces of the Mpa coiled-coils and prevent Pup binding. Pup formed an α-helix upon binding to the hexameric Mpa1-234, and interacted with the Mpa coiled-coil in an anti-parallel fashion, as observed in the Pup21-64:Mpa46-96 structure. Mpa1-51, Pup1-20, and Pup52-64 were disordered in the Pup:Mpa1-234 complex structure. (b) Alignment at the Mpa coiled-coil region of Pup:Mpa1-234 (magenta) with that of Pup21-64:Mpa46-96 (green) revealed that the presence of Pup1-20 in the hexameric structure resulted in the N-terminus of the Pup helix to point away from the Mpa coiled-coil (red arrows). This movement can be approximated by the Pup helix tilt at about 4° around its C-terminal end (dashed lines). (c) High-resolution view of Pup with the Mpa1-234 coiled-coil. Pup (cyan) is shown in cartoon view and hexameric Mpa is shown in surface charge view. The positively charged middle region of the Mpa coiled-coil was neutralized by the negatively charged C-terminal half of the Pup helix (the black boxed region of the middle panel and in the left panel). Conversely, two arginines (28 and 29) in Pup neutralized the two negatively charged pockets at the root of the Mpa coiled-coil (white boxed region in the middle panel, enlarged on the right).
The electron density of the Pup α-helix was clear but weak, and disappeared at the 3σ threshold, corresponding to approximately a third of the electron density of the Mpa coiled-coil (Supplementary Fig. 4). While this could be interpreted as disorder in Pup, it is more likely that the weak density was due to Pup binding to only one of the three coiled-coils in any given Mpa hexamer. This binding mode agreed with the one Pup per Mpa hexamer stoichiometry as reported previously 18,21.
Alignment of the Mpa coiled-coil in the Pup:Mpa1-234 structure with that in the Pup21-64:Mpa46-96 structure showed that the N-terminal portion of the Pup helix was 3-5 Å farther away from the Mpa coiled-coil in context with the Mpa hexamer (Fig. 2a-b). This was caused by a ~ 4° tilt of the Pup helix around its C-terminus (Fig. 2b), which shifted the Pup N-terminus towards the central channel of the Mpa hexamer.
Finally, we observed a large positively charged patch at the middle section of the coiled-coil (Fig. 2c, left panel). Positively charged side chains in Mpa were counter-balanced by eight negatively charged residues in Pup (Asp37, Asp38, Asp41, Asp44, Asp45, Glu42, Glu48, and Glu49). At the root of the Mpa coiled-coil, two negatively charged pockets were occupied by Arg28 and Arg29 in Pup (Fig. 2c, right panel). Thus, electrostatic interactions also appeared to play a role in the recognition of Pup by Mpa.
Pup-Mpa interacting residues are critical for proteolysis
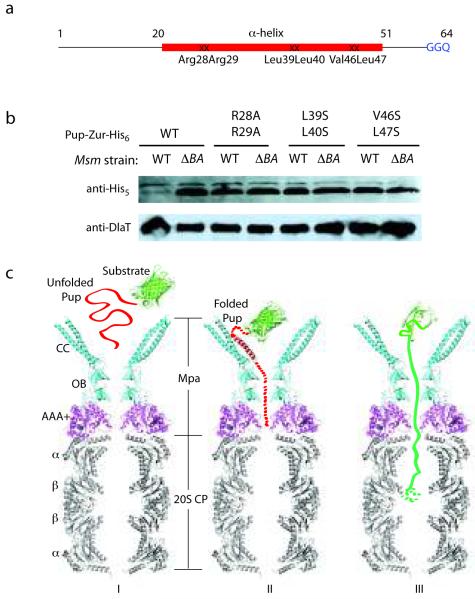
The presence of negatively charged pockets in the Mpa coiled-coil that seemed to make robust interactions with Pup Arg28 and Arg29 (Fig. 2c) prompted us to test the importance of the positively charged residues in protein degradation. Additionally, based on the crystal structure of Pup21-64:Mpa46-96 (Fig. 1d), several conserved hydrophobic residues within the α-helical region of Pup appeared to be important for interacting with Mpa. We introduced three pairs of double mutations into a Pup reporter fusion construct (Pup-Zur-His6) 16: one pair mutated arginines 28 and 29 to alanine, and two pairs disrupted hydrophobic regions of Pup (L39S L40S and V46S L47S) (Fig. 1d and Fig. 3a). Strikingly, all three double mutations in Pup nearly or completely abolished degradation of the reporter in M. smegmatis (Msm) (Fig. 3b). This result demonstrated the essentiality of the Pup helical region for the recognition of Pup by Mpa for the proteolysis of a substrate by the mycobacterial proteasome.
Figure 3.
Essentiality of the Pup helical region for proteasomal degradation supports a binding-induced folding recognition mechanism by Mpa. (a) Site-directed mutations in Pup resulted in abrogated degradation of a Pup-linear fusion by the Msm proteasome. The binding-induced helical region is in red. Mutated residues are indicated. (b) Equivalent cell numbers were analyzed from stationary phase cultures of wild type (WT) or proteasome-deleted (ΔprcBA, ΔBA) M. smegmatis synthesizing WT or mutated Pup-Zur-His6. Detection of the linear fusion proteins was done with anti-His5. DlaT (dihydrolipoamide acyltransferase) was the loading control. (c) Model for the targeting of pupylated proteins for degradation by Mpa and the mycobacterial proteasome. The Pup:Mpa1-234 complex structure (red and cyan) was placed over the homologous PAN AAA+ domain structure (PDB ID 3H4M, magenta), which was further overlaid on the Mtb proteasome core structure (PDB ID 2FHH, gray). Only a vertical central slice of the complex structure is shown for clarity. Pup is in red, and a model substrate (GFP) in green. See main text for details.
DISCUSSION
In this study, we solved the crystal structure of the Mpa1-234 hexamer alone and two co-crystal structures of Pup:Mpa. The co-crystal structures include the interacting fragments at a high resolution (1.9 Å), and full-length Pup in the Mpa hexamer at a lower resolution (4.5 Å). These structures revealed a binding-induced recognition mechanism for Pup-Mpa-mediated proteasomal degradation.
Model for Pup-mediated proteolysis in Mycobacteria
Based on our crystal structures and the existing biochemical data, we propose the following mechanism of substrate recognition in the Pup-proteasome system (Fig. 3c): (I) Pup is initially in an unfolded state when covalently linked to a protein substrate by its C-terminus via an isopeptide bond 15,16,18,21; (II) unstructured Pup folds into an α-helix, using the Mpa coiled-coil as a template; and (III) the protein substrate, along with Pup, is pulled into the central channel of the proteasomal ATPase at the expense of ATP hydrolysis 5,22. At the interface between Mpa and the 20S core particle, Pup might either be removed by a “depupylase” 23, or be pushed by Mpa further into the 20S core and degraded along with the substrate 15.
Pup recognition by tentacle-like coiled-coils in Mpa hexamers
The crystal structure of the Mpa1-234 hexamer revealed three pairs of long coiled-coils on top of the proteasomal ATPase. Interestingly, the position of the coiled-coils is similar among the three prokaryotic proteasomal ATPase structures (Supplementary Fig. 1c), and the Mpa coiled-coils did not change their positions when bound to Pup. It is unclear whether these coiled-coils are relatively inflexible or become flexible during substrate recruitment and unfolding. The long coiled-coils of Mpa may have evolved because Mycobacteria use an extended degradation signal (Pup) distinct from eukaryotic Ub and archaeal SAMPs 24,25.
An Mpa hexamer binds to only one Pup, with KD ranging from 3.4 μM to 4.2 μM, based on fluorescence anisotropy and isothermal titration calorimetry 18,21. The reduced electron density of the Pup α-helix in the Pup:Mpa1-234 crystal structure is consistent with previous observations that one Pup binds to one Mpa hexamer, i.e. Pup binds to only one of the three Mpa coiled-coils. Why this stoichiometry? We propose that the narrow Mpa channel in the OB fold region is the limiting factor 5. The crystal structure predicts that the unstructured first 20 residues of Pup either converge in the space at the bottom of the three coiled-coils, or have already threaded into the OB channel (Fig. 3c, center). Neither the bottom region of coiled-coils nor the OB channel is large enough to accommodate two Pups. This one-substrate-at-a-time targeting mechanism could prevent multiple pupylated substrates from aggregating at the proteasomal ATPase.
We also showed that Pup binds to the Mpa coiled-coil in an anti-parallel manner, placing the unstructured Pup N-terminus at the bottom of the coiled-coils, and at the entrance of the central OB channel of the Mpa hexamer (Fig. 2). Although we showed that Pup binding does not require its N-terminal 20 amino acids, Pup1-20 is essential for degradation15,16. We presume that the disordered nature of these 20 residues facilitates the threading of Pup into the narrow ATPase channel (Fig. 3c).
Binding-induced folding of Pup is likely required for degradation
Site-directed mutagenesis of specific amino acids in Pup, chosen based on interactions discovered in our analysis, disrupted proteasomal degradation. This supported the hypothesis that residues important for the folding of Pup on the extended Mpa coiled-coil are critical for Pup recognition. That these mutations could virtually abolish degradation revealed important insight into how proteins are targeted for degradation by a post-translational modifier distinct from Ub.
Many intrinsically disordered eukaryotic proteins have been found to adopt folded structures upon binding to their biological targets26. The “fly-casting” mechanism was proposed to explain the widespread occurrence of unfolded domains in eukaryotic proteins27. This mechanism might also be applicable to the Pup-proteasome system. Full-length Pup alone is randomly coiled and extended under physiologically relevant conditions in vitro. The randomly coiled state of Pup may increase its target capture radius, allowing it to find the proteasomal ATPase more easily. Once the C-terminal part of Pup contacts the Mpa coiled-coil, the pupylated substrate is “reeled” into the proteasomal ATPase, and the Pup N-terminus is gradually forced towards the OB channel in Mpa. The pulling of Pup’s N-terminus into the OB channel might subsequently pull the Pup helix away from the Mpa coiled-coil, causing Pup to unfold and thread further into the channel to reach the Mpa AAA region. This notion is supported by our structure demonstrating the displacement of the N-terminal half of the Pup helix (residues 21-30) from the Mpa coiled-coil (Fig. 2a-b), and by the observation that this region of Pup interacts more weakly with Mpa than the C-terminal half 21.
The present work provides the first insights into the molecular mechanism of Pup recognition by the Mpa-proteasome system. Because of the high sequence conservation of Pup and proteasomal ATPases within the Actinomycetales, the binding-induced folding of Pup is likely a general recognition mechanism among bacteria with a proteasome system. Importantly, the Pup-Mpa interaction, in contrast to proteasome protease activity, is unique to bacteria. Thus, the Pup-Mpa interaction may provide a highly specific target for the development of anti-tuberculosis therapies.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Kristin Burns for critical review of this manuscript. We thank Carl Nathan for advice and encouragement. X-ray diffraction data for this study were collected at beam lines X25 and X29 of the National Synchrotron Light Source. Financial support was principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. This work was supported by NIH grant AI070285 and Brookhaven National Laboratory LDRD grant 10-016 awarded to H.L., and by NIH grant HL092774 awarded to K.H.D. K.H.D. was also supported by a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Diseases award.
Appendix
METHODS
Plasmid construction and protein purification
Phusion® high-fidelity DNA polymerase (New England BioLabs, Inc.) was used for molecular cloning. The polymerase chain reactions (PCR) products were cloned into pET24b(+)/pET15b (Novagen) using enzymes from New England Biolabs. Plasmids were introduced into One Shot® TOP10 (Invitrogen) or chemically competent E. coli ER2566 (New England Biolabs)/BL21(DE3) (Invitrogen) for protein production (see Supplementary Table 1). All plasmids were sequenced.
We constructed a large number of Mpa/Pup truncations to search for those that synthesized soluble and crystallizable proteins. Bacteria producing various Mpa/Pup constructs were grown in Luria-Bertani medium. Gene expression was induced by IPTG at a final concentration of 0.2 mM. The supernatant of the bacterial lysates were loaded into 5 ml QIAGEN Ni2+ affinity columns and the target proteins were eluted with an imidazole step gradient. For His6-tag removal, the imidazole eluate was buffer exchanged to 15 PBS by dialysis and treatment with thrombin as per the manufacturer’s protocol (GE healthcare), then a second Ni2+ affinity chromatography step was performed and the flow-through was collected and further purified by ion exchange and gel filtration chromatography. For various truncated proteins, the purified proteins were dialyzed into suitable buffer (see Supplementary Table 2) and concentrated with Vivaspin® tubes (Sartorius). The purity of the protein was examined by SDS-PAGE using 10–20% linear-gradient Precast Gel (Bio-Rad), and stained by Coomassie blue. The protein concentration was determined by Coomassie Plus™ Protein Assay Reagent (Thermo Scientific).
In vitro binding experiments
For in vitro binding experiments, the His6-tags on all Mpa variants were cleaved by thrombin (GE healthcare). The N-terminal truncated version of Pup was Pup21-64-GGE with an N-terminal His6-tag; full length Pup was Pup1-64-GGQ with a C-terminal non-cleavable His6-tag. All protein samples were dialyzed in buffer A (5 mM HEPES, 500 mM NaCl, 2 mM MgCl2, pH 8.2). The final protein concentrations were adjusted to ~2 mg ml−1 for pull down experiments. Ni-NTA beads (Novagen) were washed four times in buffer B (5 mM HEPES, 500 mM NaCl, 2 mM MgCl2, pH 8.2, 20 mM imidazole) to completely remove the stock solution (20% v/v ethanol). 40 μl Pup protein solution was mixed with 40 μl Mpa variants and incubated on ice for 1 hour, then mixed with Ni-NTA beads, and incubated on ice with gentle shaking. The mixture was centrifuged at 18,000 g for 4 min, and the supernatant was carefully removed. The beads were washed in buffer B 3 times and the centrifugation step was repeated to remove non-specific binding. Finally, the beads were eluted by Buffer C (5 mM HEPES, 500 mM NaCl, 2 mM MgCl2, pH 8.2, 300 mM imidazole) to release the pull-down products. The control samples went through the same preparation steps, except that the loaded protein samples were Mpa variants without Pup. All samples were examined by SDS-PAGE using 10–20% w/v linear-gradient Precast Gel (Bio-Rad), and stained by Coomassie blue.
Site-directed mutagenesis
Site-directed mutations were introduced into pSYMP-pup-zur-his6 16 by sewing overlap extension (SOE) PCR 28 using hsp60f and pMNrev with SOE primers to introduce mutant sites. PCR products were cloned into NdeI/HindIII sites of the parental vector (see Supplementary Table 1 for primers). All plasmids were sequenced by GENEWIZ, Inc. Msm was transformed by electroporation using routine methods 29. Immunoblotting using antibodies to penta-histidine (anti-His5; Qiagen) was performed as described elsewhere 12,16.
Crystallization, data reduction, and structure determination
Hanging drop vapor diffusion method was used for crystallization of all Mpa/Pup variants and their complexes. Briefly, a 2 μl droplet of protein sample was mixed with 2μl of reservoir buffer, and the mixed droplet was incubation at 21 °C for diffusive equilibrium with 1 ml reservoir solution. For cryo-crystallography, the crystal solution was gradually replaced by artificial mother solution with suitable cryo-protectant and the crystals were picked up and flash frozen in liquid Nitrogen. Details for crystallization are listed in Supplementary Table 2. All crystal screening and data collection were carried out at X25 and X29 beam lines at National Synchrotron Light Source, Brookhaven National Laboratory.
Due to their high solvent contents, the diffraction ability of the Mpa1-234 and Pup:Mpa1-234 crystals were poor, reached only to ~8 Å at X29 beam line. We dehydrated the crystals in the air for about 10 min. This markedly improved the diffraction ability of the crystals. The crystals were fragile, and extreme care was required during manipulation. The Mpa46-96 and Pup21-64:Mpa46-96 crystals were also fragile and sensitive: Mpa46-96 crystals would crack with even slight vibration during microscope inspection; Pup21-64:Mpa46-96 crystals only lasted for a few days before they disappeared.
All datasets were indexed, integrated and scaled using HKL2000 30. The structures were determined with molecular replacement method in PHASER 31 and PHENIX suite 32. For low-resolution structure solution of Mpa1-234 and Pup:Mpa1-234, the hexameric Mpa OB domain (PDB: 3FP9) was used for molecular replacement, and the initial model was manually built in COOT 33 and refined by Refmac5 in CCP4 suite with strong NCS restraints 34. Initially, the Mpa1-234 and Pup:Mpa1-234: datasets were processed in the cubic P213 space group, but the R-factor is too high after refinement. We eventually reduced the space group to P21 and then the R factor could be refined to acceptable values. Only group B-factors (polypeptide chain) were refined for these lower resolution structures. For the higher resolution datasets of Mpa46-96 and Pup21-64:Mpa46-96, a single helix (residues 52-96) from the Mpa1-234 structure was used for molecular replacement, and the initial models were automatically rebuilt by PHENIX, further refinements were carried out in Refmac5 and examined in COOT. Data process and refinement statistics are listed in Supplementary Table 3. Mpa residues 1-51 were disordered in the crystal structure of Mpa1-234 alone; Residues Mpa1-51, Pup1-20, Pup52-64 were also disordered in the crystal structure of Pup:Mpa1-234 complex structure; Mpa residues 46-51, 95, and 96 were disordered in the crystal structure of Mpa46-96 alone; Mpa46-51, Pup52-64 residues are disordered in Mpa46-96:Pup21-64 complex structure. All figures were drawn in PyMOL (The PyMOL Molecular Graphics System, Version 1.2.x, Schrödinger, LLC).
Footnotes
Accession codes. Coordinates and structure factors have been deposited in Protein Data Bank with the following accession codes: 3M9B (Mpa1-234 hexamer); 3M9H (Mpa46-96 four helix bundle); 3M91 (Pup21-64:Mpa46-96 complex); and 3M9D (Pup:Mpa1-234 complex).
REFERENCES
- 1.Finley D. Recognition and Processing of Ubiquitin-Protein Conjugates by the Proteasome. Annual Review of Biochemistry. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupas A, et al. Eubacterial proteasomes. Mol Biol Rep. 1997;24:125–31. doi: 10.1023/a:1006803512761. [DOI] [PubMed] [Google Scholar]
- 3.Lin G, et al. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol Microbiol. 2006;59:1405–16. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 4.Hu G, et al. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 2006;59:1417–28. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, et al. Structural insights on the mycobacterium tuberculosis proteasomal ATPase Mpa. Structure. 2009;17:1377–1385. doi: 10.1016/j.str.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–20. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 8.Nathan C, et al. A philosophy of anti-infectives as a guide in the search for new drugs for tuberculosis. Tuberculosis (Edinb) 2008;88(Suppl 1):S25–33. doi: 10.1016/S1472-9792(08)70034-9. [DOI] [PubMed] [Google Scholar]
- 9.Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–71. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–84. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–90. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 12.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–7. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd. Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–75. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutter M, Damberger FF, Imkamp F, Allain FH, Weber-Ban E. Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J Am Chem Soc. 2010;132:5610–2. doi: 10.1021/ja910546x. [DOI] [PubMed] [Google Scholar]
- 15.Striebel F, Hunkeler M, Summer H, Weber-Ban E. The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup’s N-terminus. Embo J. 2010;29:1262–71. doi: 10.1038/emboj.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns KE, Pearce MJ, Darwin KH. Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J Bacteriol. 2010;192:2933–5. doi: 10.1128/JB.01639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–7. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 18.Sutter M, Striebel F, Damberger FF, Allain FH, Weber-Ban E. A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009;583:3151–7. doi: 10.1016/j.febslet.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 19.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci U S A. 2001;98:2250–5. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meindl-Beinker NM, Lundin C, Nilsson I, White SH, von Heijne G. Asn- and Asp-mediated interactions between transmembrane helices during translocon-mediated membrane protein assembly. EMBO Rep. 2006;7:1111–6. doi: 10.1038/sj.embor.7400818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, et al. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J Mol Biol. 2009;392:208–17. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008;15:1147–51. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns KE, et al. “Depupylation” of Prokaryotic Ubiquitin-like Protein from Mycobacterial Proteasome Substrates. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darwin KH, Hofmann K. SAMPyling proteins in archaea. Trends Biochem Sci. 2010;35:348–51. doi: 10.1016/j.tibs.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humbard MA, et al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 27.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A. 2000;97:8868–73. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–8. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 29.Hatfull GF, Jacobs JWR. Molecular Genetics of Mycobacteria. ASM Press; Washington, DC: 2000. [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods in Enzymology. Vol. 276. Academic Press; New York: 1997. Processing of X-ray Diffraction Data Collected in Oscillation Mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. The CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2002;58:1955–7. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.