The authors show enhanced digestibility of cinnamoyl CoA-reductase and caffeic acid O-methyltransferase-deficient perennial ryegrass plants grown under glasshouse and field conditions. This indicates that both of these lignin biosynthetic genes are promising targets for transgenic approaches aiming to enhance forage quality and improve feedstock plants for biofuel production.
Abstract
Cinnamoyl CoA-reductase (CCR) and caffeic acid O-methyltransferase (COMT) catalyze key steps in the biosynthesis of monolignols, which serve as building blocks in the formation of plant lignin. We identified candidate genes encoding these two enzymes in perennial ryegrass (Lolium perenne) and show that the spatio-temporal expression patterns of these genes in planta correlate well with the developmental profile of lignin deposition. Downregulation of CCR1 and caffeic acid O-methyltransferase 1 (OMT1) using an RNA interference–mediated silencing strategy caused dramatic changes in lignin level and composition in transgenic perennial ryegrass plants grown under both glasshouse and field conditions. In CCR1-deficient perennial ryegrass plants, metabolic profiling indicates the redirection of intermediates both within and beyond the core phenylpropanoid pathway. The combined results strongly support a key role for the OMT1 gene product in the biosynthesis of both syringyl- and guaiacyl-lignin subunits in perennial ryegrass. Both field-grown OMT1-deficient and CCR1-deficient perennial ryegrass plants showed enhanced digestibility without obvious detrimental effects on either plant fitness or biomass production. This highlights the potential of metabolic engineering not only to enhance the forage quality of grasses but also to produce optimal feedstock plants for biofuel production.
INTRODUCTION
Lignin is a polymeric macromolecule that is most commonly found in cell walls of plants. The constituent subunits of lignins, the monolignols, are produced by a branch of the phenylpropanoid pathway (Whetten and Sederoff, 1995). Lignin plays multiple roles in plants by contributing to the hydrophobicity of plant vasculature (Kubitzki, 1987), by providing mechanical support for plant tissues (Zhong et al., 1997; Boerjan et al., 2003) and by protecting the plant from pathogen invasion (Hammond-Kosack and Jones, 1996). In most angiosperms, lignin is composed mainly of guaiacyl (G) and syringyl (S) monolignol subunits, which are derived from the coniferyl and sinapyl alcohol core monolignols, respectively. A third type, the p-hydroxyphenyl (H) subunit, is derived from the p-coumaryl core monolignol and is relatively abundant in monocotyledonous plants but rare in dicotyledonous plants (Vanholme et al., 2008). Lignin composition in monocots is often characterized by an increase in S subunit content and a decrease in G subunit content during the transition from vegetative to reproductive stages of development. This change is associated with a decrease in the digestibility and forage quality of plant tissues (Anterola and Lewis, 2002; Chen et al., 2002, 2004; Guillaumie et al., 2008).
The hydroxylation and methoxylation states of the C3 and C5 positions of lignin subunits determine the physical and chemical properties of polymeric lignin and consequentially the digestibility and forage quality of plant tissues (reviewed in Boerjan et al., 2003; Raes et al., 2003). The initial hydroxylation and O-methylation at the C3 position leads to the biosynthesis of G subunits (see Figure 9). Caffeoyl CoA 3-O-methyltransferase (EC 2.1.1.104) is generally regarded as the key catalyst of the methylation step. Downregulation of caffeoyl CoA 3-O-methyltransferase in angiosperms results in reduced lignin content and an increased S/G ratio, due mainly to reduced levels of G subunits (Meyermans et al., 2000; Zhong et al., 2000; Guo et al., 2001; Pinçon et al., 2001).
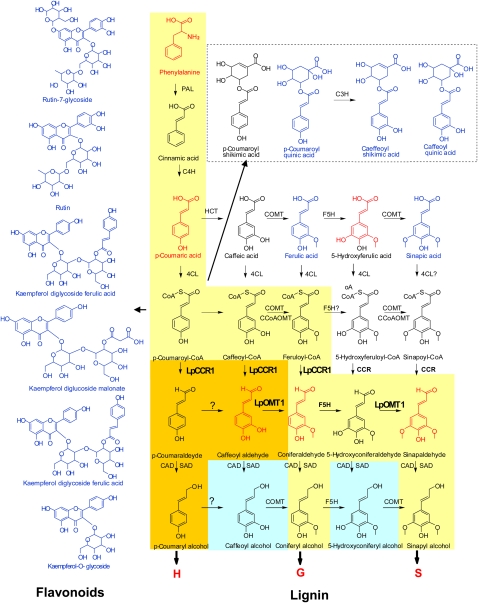
Figure 9.
Phenylpropanoid and Monolignol Biosynthetic Pathways.
The yellow route toward the production of monolignols is conserved in angiosperms. The orange route is found in some species, including perennial ryegrass (this article). The blue route is found in some species. CAD, cinnamyl alcohol dehydrogenase; 4CL, 4-coumarate:CoA ligase; C3H, p-coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CCoAOMT, caffeoyl-CoA O-methyltransferase; HCT, p-hydroxycinnamoyl-CoA:quinate shikimate p-hydroxycinnamoyltransferase; F5H, ferulate 5-hydroxylase; PAL, phenylalanine ammonia-lyase; SAD, sinapyl alcohol dehydrogenase. Compounds marked in red were downregulated in hpCCR1-1 lines. Those marked in blue were upregulated in hpCCR1-1 lines.
Caffeic acid O-methyltransferase (COMT; EC 2.1.1.68) catalyzes O-methylation of the C5 hydroxyl moiety of suitably hydroxylated phenolic rings of monolignols, leading to the preferential formation of S subunits. In general, most COMTs possess broad substrate permissiveness and therefore can potentially participate in multiple branches of the monolignol biosynthetic grid (Parvathi et al., 2001). COMTs from angiosperms, such as alfalfa (Medicago sativa), wheat (Triticum aestivum), and aspen (Populus tremula x Populus alba), exhibit a stronger preference for 5-hydroxyconiferaldehyde than for caffeoylaldehyde. COMTs generally exhibit a greater selectivity and higher methylation rate for 5-hydroxy, relative to 3-hydroxy, groups (Li et al., 2000; Parvathi et al., 2001; Zubieta et al., 2002; Ma and Xu, 2008).
In tandem with the downstream enzyme COMT, cinnamoyl CoA-reductase (CCR) catalyzes the committed step in the monolignol-specific branch of the phenylpropanoid pathway (Lacombe et al., 1997; Boerjan et al., 2003). Phylogenetic analysis shows that CCRs from monocots and dicots cluster into different clades. CCRs are represented by two functionally distinct genes in most of these species, termed CCR1 and CCR2.
In monocots, small multigene families of CCR genes have been identified, including gene families in maize (Zea mays), rice (Oryza sativa), sugarcane (Saccharum officinarum), perennial ryegrass (Lolium perenne), and switchgrass (Panicum virgatum) (Pichon et al., 1998; McInnes et al., 2002; Larsen, 2004; Ma and Tian, 2005; Escamilla-Treviño et al., 2010). The nucleotide sequences of the CCR1 and CCR2 transcripts from maize exhibit 73% sequence similarity in their coding regions (Pichon et al., 1998). Maize CCR2 is expressed at very low levels in many organs and has been recently shown to be strongly induced in the root elongation zone of maize seedlings during water-deficit conditions, suggesting that CCR2 may facilitate root acclimation to water stress (Fan et al., 2006). Maize CCR1 is also expressed in many different organs, with high transcript levels in adventitious roots, seminal roots, and leaves. More specifically, a high level of CCR1 expression in maize stems suggests that the CCR1 is likely involved in constitutive lignification. In addition to a key role in the formation of monolignol precursors, rice CCR was recently reported to act as an important regulator in a defense response through a GTP-dependent interaction with a Rac family GTPase (Kawasaki et al., 2006). Interestingly, the interaction between the rice CCR1 and Rac1 proteins was found both to stimulate CCR activity in vitro and to increase lignin accumulation in rice cell cultures.
Forage grasses currently provide 75% of feed requirements for livestock (Wilkins and Humphreys, 2003). The structural and chemical properties of monolignol subunits, including their capacity to form these cross-links and their hydrophobicity, are the primary determinants of the digestibility of forage species (Buxton and Russell, 1988; Jung, 1989; Vogel and Jung, 2001). Because the digestibility of grasses is negatively affected by increases in overall lignification and by high S/G subunit ratios associated with the vegetative–floral transition, there is significant commercial interest in altering the chemical structure of the heterogeneous lignin polymer by modifying subunit composition or by incorporating novel monolignol subunits (Anterola and Lewis, 2002; Boudet et al., 2003).
The comprehensive study reported here involved functional characterization of perennial ryegrass OMT1 and CCR1 in vivo and examined the consequences of modifying the expression of these genes on forage quality in transgenic perennial ryegrass plants grown under glasshouse and field conditions. Downregulation of CCR expression is reported in the forage grasses. Quantitative analyses and qualitative observations of changes in soluble phenolic content showed that phenylpropanoid-associated biosynthetic intermediates, made available by reduced CCR1 expression, were redirected to biosynthetic pathways outside the core general phenylpropanoid pathway. The combined findings provide strong evidence that OMT1 plays a role in G and S subunit biosynthesis in perennial ryegrass.
RESULTS
Lignin Deposition Patterns in Perennial Ryegrass Plants
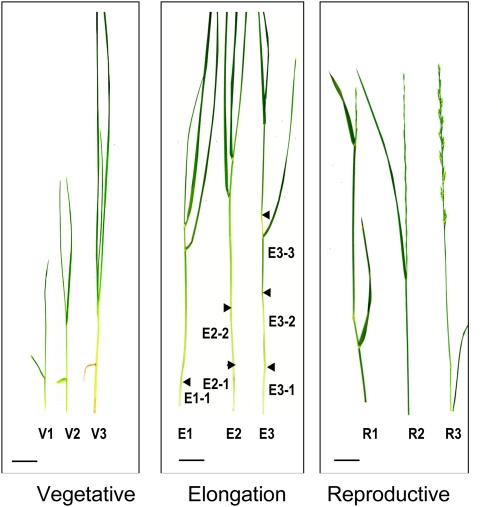
Three stages of development were chosen for analysis of lignin deposition in perennial ryegrass plants: (1) vegetative (V), comprising the early stages of leaf development prior to stem formation; (2) elongation (E), during which stems were present, the culm was elongated, and the inflorescence was enclosed in the uppermost leaf; and (3) reproductive (R), when the inflorescence began to emerge (Moore et al., 1991). Each of the three stages was further divided into three substages: V1, V2, and V3, reflecting the number of mature leaves; E1, E2, and E3, reflecting the developmental stages with one, two, and three palpable internodes, respectively; and R1, R2, and R3, reflecting inflorescence emergence, complete emergence of spikelets, and anthesis, respectively (Moore et al., 1991) (Figure 1).
Figure 1.
Developmental Stages of Perennial Ryegrass Plants.
Three substages within each of the vegetative (V), elongation (E), and reproductive (R) growth stages are shown: V1, first leaf collared; V2, leaf collared; V3, third leaf collared; E1, first node visible; E2, two nodes visible; E3, three nodes visible; R1, inflorescence emerging; R2, spikelets fully emerged; R3, full anthesis. Positions of nodes are shown by arrowheads. Internodes are shown (following the dash) for stages E1-E3. Bars = 2 cm (vegetative), 3 cm (elongation), and 2 cm (reproduction).
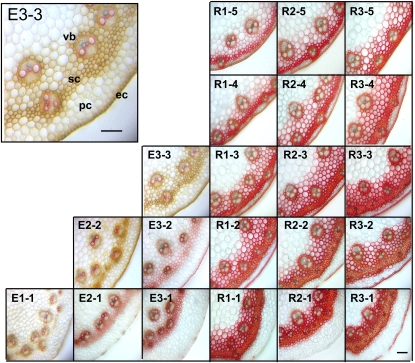
Transverse sections of internodes from stems collected at the E1-E3 and R1-R3 stages, with the basal internodes defined as the 1st internode, were stained with Maüle reagent, which stains guaiacyl (G) monolignol subunits brown and syringyl (S) monolignol subunits red. Lignin accumulation was observed in xylem, in sclerenchyma, and in parenchyma cells between vascular bundles as well as in epidermal cells (Figure 2). Reprogramming of lignin metabolism during the transition from the elongation to reproductive stages of development was associated with a dramatic increase in the number of heavily S-lignified cells within the sclerenchyma ring, vascular bundles, and epidermal cells and this was most pronounced in basal parts of tillers (Figure 2). Lignin accumulation gradually increased between the E1 and R3 stages with an increase in the S/G lignin ratio and an acropetal decrease in lignin content within each stem from basal to upper internodes (see Supplemental Figure 1A online). Near infrared reflectance spectroscopy (NIRS) was used to estimate the quality trait such as in vivo dry matter digestibility (IVVDMD) of stems. IVVDMD as a metabolized (digestible) energy was estimated using established calibration equations generated by measuring NIRS and correlating this to analytical measures of IVVDMD for a subset of samples (Flinn, 2003). Stem tissue from R1-1 internodes was almost 50% more digestible than stem tissue from R2-1 internodes (see Supplemental Figure 2A online). An in vitro digestibility experiment demonstrated that almost all cells in 2-mm sections of stem collected at the R1-1 stage were digested after a 24-h incubation in bovine rumen fluid (see Supplemental Figure 2B online). The heavily lignified ring of schlerenchyma cells showed virtually no digestion over this time course, remaining intact even after 48 h.
Figure 2.
Mäule Staining of Lignin in Transverse Sections of Developing Perennial Ryegrass Stems.
Transverse sections were prepared from different internodes of perennial ryegrass plants at the elongation (E) and reproductive (R) stages of development. Mäule staining was used to detect lignin rich in guaiacyl (G) and syringyl (S) monolignol subunits, which stain brown and red, respectively. Developmental stages are as for Figure 1. Micrographs are representative of at least six separate stems from different plants. vb, vascular bundle; sc, sclerenchyma; pc, parenchyma; ec, epidermal cells. Bars = 90 μm for all sections.
Characterization of COMT and CCR Genes from Perennial Ryegrass
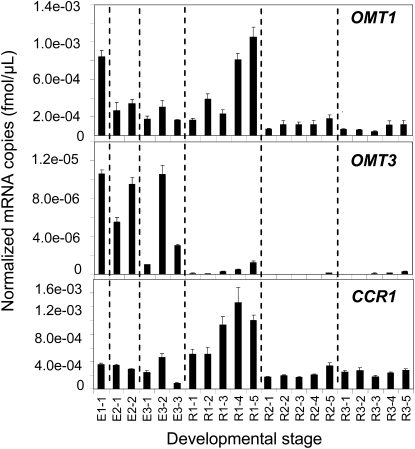
Two COMT-like genes from perennial ryegrass, namely, OMT1 and OMT3, analyzed in this work, have been previously described (Heath et al., 1998). Comparisons of deduced amino acid sequences with COMT sequences from other plants are shown in Supplemental Figures 3 and 4 and Supplemental Table 1 online. Expression of both OMT1 and OMT3 genes was elevated during the early elongation (E1) stage (Figure 3). However, these genes displayed different expression profiles during later stages of development. OMT1 showed low expression at E2 and E3 stages and was strongly upregulated at the R1 stage, with the highest level of expression in the uppermost (5th) internode. OMT3 showed almost complementary expression profiles at the E2, E3, and R1 stages with low expression at R1 stage. Despite marked differences in the expression levels of OMT1 and OMT3 at the E2, E3, and R1 stages, both genes showed elevated levels of expression at the E3-2 and R1-5 stages. Both genes showed a low level of expression at R2 and R3 developmental stages.
Figure 3.
Normalized Expression Levels of Perennial Ryegrass OMT1, OMT3, and CCR1 Genes in Different Internodes during Stem Development.
Data are normalized using expression values of three internal control genes encoding glyceraldehyde 3-phosphate dehydrogenase (GAPDH), histone (His), and tubulin (Tub) (see Supplemental Table 3 online). Average values and standard errors from three technical replicates derived from a single pooled sample of 10 tillers from the same stage of development are shown.
Perennial ryegrass CCR1, encoding a putative cinnamoyl CoA reductase, and showing strong expression in lignified tissues of shoots, stems, and roots, has been previously characterized (McInnes et al., 2002). Comparisons of the deduced CCR1 amino acid sequence with homologs from other plants are shown in Supplemental Figure 5 and Supplemental Table 2 online. Expression of CCR1 peaked during the R1 stage and was higher in the upper parts of the plants compared with the basal internodes (Figure 3). Similar to OMT1 and OMT3, CCR1 showed a reduced level of expression at R2 and R3 developmental stages.
Downregulation of OMT1 Expression in Transgenic Perennial Ryegrass Plants
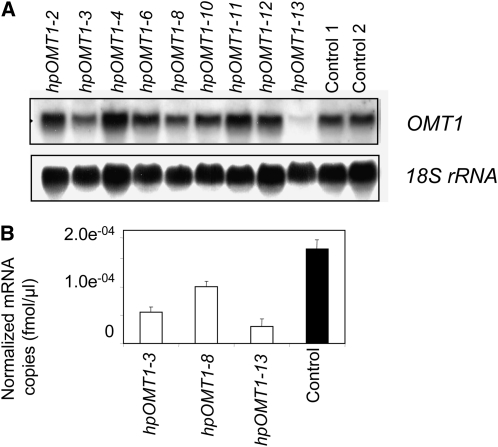
The in planta function of OMT1 was analyzed in transgenic perennial ryegrass plants (L. perenne cv Grasslands Impact 566) in which OMT1 expression was downregulated using a double-stranded RNA interference (dsRNAi)-mediated posttranscriptional gene silencing strategy. Nine putative transgenic lines with hygromycin resistance were obtained, and DNA gel blot hybridization analysis confirmed that these lines contained the Ubi:hpOMT1:nos cassette, referred to as the hpOMT1 transgene (see Supplemental Figure 6 online). RNA gel blot hybridization and real-time RT-PCR analysis was used to characterize the expression of OMT1 and OMT3 genes (Figure 4). In comparison to controls (isogenic lines of the same ‘Grasslands Impact 566’ genotype) the hpOMT1-3, hpOMT1-8, and hpOMT1-13 lines possessed decreased levels of endogenous Lp OMT1 expression by 44, 16, and 80%, respectively, and this was well correlated with RNA gel blot hybridization data (Figures 4A and 4B).
Figure 4.
Analysis of OMT1 Expression in Transgenic Perennial Ryegrass Lines Containing Hairpin Construct for Silencing of the OMT1 Gene.
(A) RNA gel blot analysis of Lp OMT1 (top panel) and 18S rRNA (bottom panel) gene expression in nine transgenic and two wild-type control (no construct) lines at the R1 stage.
(B) Quantitative RT-PCR analysis of Lp OMT1 expression in transgenic and wild-type plants at the R1 stage. Data are normalized using expression values of three control genes encoding glyceraldehyde 3-phosphate dehydrogenase (GAPDH), histone (His), and tubulin (Tub) (see Supplemental Table 3 online). Average values and standard errors from three technical replicates derived from a single pooled sample of 10 tillers from the same stage of development are shown.
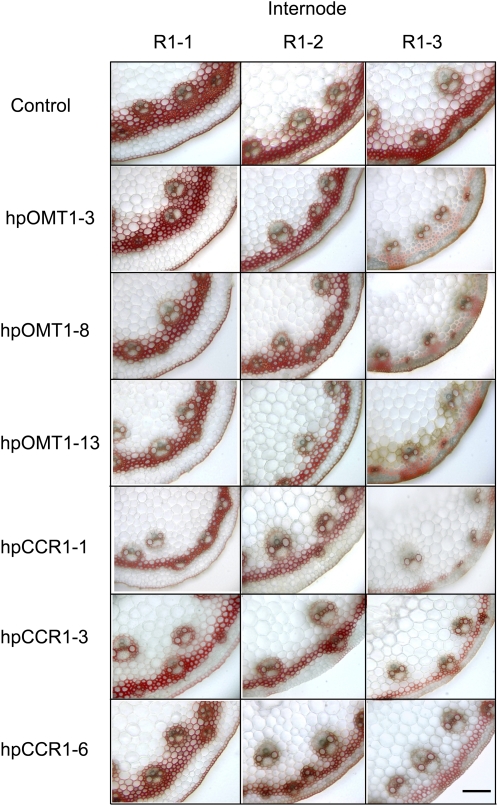
Histochemical staining of the three basal internodes at R1 stage showed a significant decrease in the number of lignified sclerenchyma and epidermal cells in the R1-3 (upper) internodes in transgenics compared with controls (Figure 5). Lignin level and composition (relative proportions of S-, G-, and H-type monomers released by thioacidolysis) of hpOMT1 transgenic and control plants are presented in Table 1. The levels of acetyl bromide–solubilized lignin at the R1 stage were reduced in the hpOMT1-3, hpOMT1-8, and hpOMT1-13 transgenic lines by 22, 17, and 10%, respectively, relative to the control lines (Table 1). All three hpOMT1 transgenic lines (hpOMT1-3, hpOMT1-8, and hpOMT1-13) had reduced levels of released S subunits (by 35, 29, and 22%, respectively) and G subunits (by 20, 13, and 11%, respectively) in stems (without leaves) relative to control plants at the R1 stage (Table 1). The more dramatically reduced S subunit levels compared with G subunits resulted in lowered S/G ratios in the transgenic lines (from 0.77 for control plants to 0.63, 0.62, and 0.72 for hpOMT1-3, hpOMT1-8, and hpOMT1-13, respectively; Table 1). The levels of H subunits, on the other hand, were found to be almost unchanged in the transgenic lines in comparison to control plants (down by 2.5 to 3%; Table 1).
Figure 5.
Mäule Staining of Lignin in Transverse Sections of Stems from hpOMT1 and hpCCR1 and Control Plants.
The three internodes (R1-1, R1-2, and R1-3) from stems of plants at the R1 stage were stained with Mäule reagent to detect lignins rich in guaiacyl (G) and syringyl (S) monolignol subunits, which stain brown and red, respectively. Bars = 90 μm for all sections.
Table 1.
Measurement of Lignin in Stems of Transgenic Perennial Ryegrass hpOMT1 and hpCCR1 Lines at the R1 Stage Using the Acetyl Bromide Method and GC-MS
| Glasshouse | Field Trial | |||||||||||||
| Line | AcBr Lignina | G Ligninb | S Ligninb | H Ligninb | S/G | S/H | IVVDMD | AcBr Lignina | G Ligninb | S Ligninb | H Ligninb | S/G | S/H | IVVDMD |
| Control | 16.94 ± 0.7 | 24.6 ± 1.8 | 19.0 ± 1.5 | 0.81 ± 0.04 | 0.77 | 23.4 | 61.0 | 13.3 ± 2.8 | 12.1 ± 3.5 | 10.8 ± 0.1 | 0.49 ± 0.01 | 0.89 | 22.0 | 75.65 |
| hpOMT1-2 | 16.04 ± 1.2 | 24.5 ± 1.6 | 21.5 ± 1.8 | 0.74 ± 0.04 | 0.88 | 29.4 | ||||||||
| hpOMT1-3 | 14.24 ± 1.3 | 19.6 ± 2.5 | 12.5 ± 1.8 | 0.79 ± 0.04 | 0.63 | 15.8 | 67.1 | 12.0 ± 0.8 | 8.6 ± 0.8 | 7.9 ± 0.1 | 0.43 ± 0.01 | 0.91 | 18.3 | 82.60 |
| hpOMT1-6 | 14.51 ± 0.6 | 25.5 ± 1.5 | 20.5 ± 1.9 | 0.67 ± 0.02 | 0.80 | 30.5 | ||||||||
| hpOMT1-8 | 15.72 ± 0.8 | 21.4 ± 1.5 | 13.4 ± 2.5 | 0.78 ± 0.04 | 0.62 | 17.1 | 67.6 | |||||||
| hpOMT1-10 | 15.84 ± 0.4 | 26.7 ± 1.8 | 21.5 ± 1.2 | 0.69 ± 0.05 | 0.80 | 31.0 | ||||||||
| hpOMT1-11 | 16.21 ± 0.5 | 27.9 ± 1.0 | 21.4 ± 1.4 | 0.83 ± 0.04 | 0.76 | 25.7 | ||||||||
| hpOMT1-12 | 16.09 ± 0.4 | 24.3 ± 1.2 | 20.5 ± 1.8 | 0.80 ± 0.01 | 0.84 | 25.6 | ||||||||
| hpOMT1-13 | 15.64 ± 0.2 | 21.9 ± 1.4 | 15.9 ± 1.8 | 0.79 ± 0.04 | 0.72 | 20.1 | 63.5 | |||||||
| hpCCR1-1 | 11.64 ± 0.27 | 14.3 ± 1.6 | 10.3 ± 1.4 | 0.42 ± 0.09 | 0.70 | 24.5 | 68.5 | 8.5 ± 1.6 | 3.0 ± 0.9 | 2.80 ± 0.4 | 0.17 ± 0.01 | 0.96 | 16.4 | 87.43 |
| hpCCR1-2 | 16.60 ± 0.17 | 24.4 ± 2.9 | 23.8 ± 2.1 | 0.79 ± 0.12 | 0.97 | 29.8 | ||||||||
| hpCCR1-3 | 12.64 ± 0.20 | 17.5 ± 2.6 | 13.9 ± 1.6 | 0.57 ± 0.04 | 0.79 | 24.3 | 68.3 | |||||||
| hpCCR1-4 | 14.64 ± 0.17 | 18.1 ± 1.4 | 15.1 ± 1.7 | 0.71 ± 0.11 | 0.83 | 21.3 | ||||||||
| hpCCR1-5 | 16.04 ± 0.21 | 23.1 ± 2.4 | 20.8 ± 2.4 | 0.73 ± 0.15 | 0.9 | 28.4 | ||||||||
| hpCCR1-6 | 13.04 ± 0.29 | 17.9 ± 1.7 | 14.0 ± 1.3 | 0.55 ± 0.09 | 0.78 | 24.4 | 67.9 | |||||||
Lines shown in bold had the greatest reduction in the level of CCR1 or OMT1 gene expression.
Soluble lignin, percentage of dry cell walls.
mg/g dry cell walls.
NIRS was used to estimate IVVDMD of R1-stage stems from transgenic and control plants. The hpOMT1-3, hpOMT1-8, and hpOMT1-13 lines showed 9.8, 11.8, and 5.0% higher IVVDMD levels, respectively, than the average IVVDMD of control plants (Table 1).
In the 3rd internode at the R1 stage, the hpOMT1-3, hpOMT1-8, and hpOMT1-13 lines also had reduced levels of acetyl bromide lignin (by 39, 17, and 14%, respectively), released S subunits (by 51, 32, and 34%, respectively), and released G subunits (by 32, 25, and 22%, respectively) compared with controls. The decrease in S/G ratio was therefore more pronounced for lignin from the R1-3 stage (0.68 for wild-type plants compared with 0.44, 0.57, and 0.52 for hpOMT1-3, hpOMT1-8, and hpOMT1-13, respectively; Table 2) than from whole stems. Again the levels of thioacidolytically released H subunits were found to be almost unchanged in the hpOMT1-3, hpOMT1-8, and hpOMT1-13 lines in comparison to control plants (Table 1).
Table 2.
Measurement of Lignin in the 3rd Internode of Perennial Ryegrass hpOMT1 and hpCCR1 Lines at the R1 Stage Using the Acetyl Bromide Method and GC-MS
| Linea | AcBrb Lignin | G Ligninb | S Ligninc | H Ligninc | S/G | S/H |
| Control | 11.29 ± 1.3 | 17.8 ± 2.8 | 11.2 ± 1.5 | 0.32 ± 0.1 | 0.68 | 35.0 |
| hpOMT1-2 | 11.49 ± 1.2 | 18.8 ± 2.6 | 13.1 ± 1.8 | 0.31 ± 0.1 | 0.63 | 42.2 |
| hpOMT1-3 | 6.90 ± 0.3 | 12.1 ± 0.8 | 5.4 ± 0.8 | 0.36 ± 0.1 | 0.44 | 15.0 |
| hpOMT1-6 | 12.05 ± 0.8 | 20.0 ± 1.9 | 12.9 ± 1.9 | 0.30 ± 0.1 | 0.71 | 43.0 |
| hpOMT1-8 | 9.33 ± 0.7 | 13.3 ± 2.5 | 7.7 ± 1.5 | 0.29 ± 0.1 | 0.57 | 26.5 |
| hpOMT1-10 | 10.56 ± 0.8 | 20.6 ± 2.8 | 13.4 ± 1.2 | 0.35 ± 0.1 | 0.65 | 38.2 |
| hpOMT1-11 | 11.60 ± 0.8 | 20.1 ± 2.1 | 12.6 ± 1.6 | 0.25 ± 0.1 | 0.62 | 50.1 |
| hpOMT1-12 | 10.73 ± 0.9 | 19.5 ± 2.2 | 12.4 ± 1.7 | 0.27 ± 0.1 | 0.63 | 45.9 |
| hpOMT1-13 | 9.68 ± 0.7 | 13.8 ± 1.6 | 7.3 ± 1.6 | 0.30 ± 0.1 | 0.52 | 24.3 |
| hpCCR1-1 | 4.58 ± 0.7 | 9.5 ± 1.6 | 5.4 ± 1.9 | 0.20 ± 0.1 | 0.56 | 27.1 |
| hpCCR1-2 | 10.08 ± 1.7 | 18.9 ± 2.6 | 12.2 ± 1.6 | 0.30 ± 0.1 | 0.64 | 40.0 |
| hpCCR1-3 | 7.01 ± 1.8 | 13.6 ± 1.6 | 7.4 ± 2.6 | 0.23 ± 0.1 | 0.54 | 32.1 |
| hpCCR1-4 | 9.78 ± 0.6 | 16.9 ± 3.0 | 11.2 ± 3.6 | 0.29 ± 0.1 | 0.66 | 38.6 |
| hpCCR1-5 | 11.60 ± 2.7 | 17.1 ± 3.2 | 10.2 ± 2.9 | 0.26 ± 0.1 | 0.59 | 39.3 |
| hpCCR1-6 | 7.98 ± 0.3 | 14.8 ± 1.9 | 9.1 ± 2.6 | 0.25 ± 0.1 | 0.61 | 36.4 |
Lines shown in bold had the greatest reduction in the level of CCR1 or OMT1 gene expression.
Plants were grown under glasshouse conditions.
Soluble lignin, percentage of dry cell walls.
mg/g dry cell walls.
Downregulation of CCR1 Expression in Transgenic Perennial Ryegrass Plants
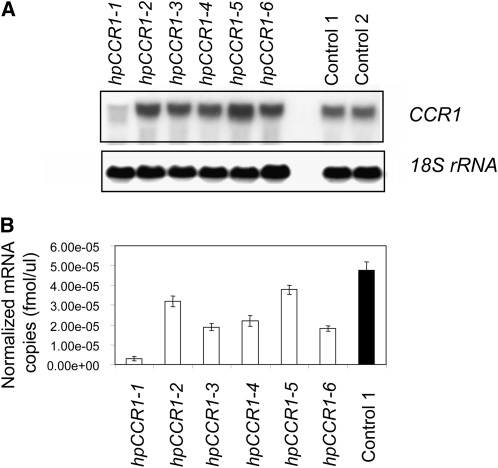
The functional analyses of CCR1 used transgenic perennial ryegrass plants in which CCR1 expression was downregulated using a dsRNAi-mediated gene silencing strategy. DNA gel blot hybridization analysis identified six transgenic lines possessing a transgene that encodes an hph selectable marker for hygromycin resistance and hpCCR1 (see Supplemental Figure 7 online). RNA gel blot analysis and RT-PCR revealed that endogenous CCR1-specific expression decreased by 94% in the hpCCR1-1 transgenic line in comparison to wild-type control plants (Figure 6). CCR1 expression fell by ~50% in the hpCCR1-3, hpCCR1-4, and hpCCR1-6 lines but was only slightly lower in the hpCCR1-2 and hpCCR1-5 lines compared to wild-type control plants.
Figure 6.
Analysis of CCR1 Gene Expression in Transgenic Perennial Ryegrass Lines Containing hpCCR1.
(A) RNA gel blot analysis of CCR1 (top panel) and 18S rRNA (bottom panel) gene expression in nine transgenic and wild-type control lines at the R1 stage.
(B) Quantitative RT-PCR analysis of Lp CCR1 expression in transgenic and wild-type plants at the R1 stage. Data are normalized using expression values of three control genes encoding glyceraldehyde 3-phosphate dehydrogenase (GAPDH), histone (His), and tubulin (Tub) (see Supplemental Table 3 online). Average values and standard errors from three technical replicates derived from a single pooled sample of 10 tillers from the same stage of development are shown.
Phenotypic analysis of the hpCCR1 lines showed no significant physical differences in comparison to control plants. Detailed microscopy analysis of stem cross sections of hpCCR1-1, hpCCR1-3, and hpCCR1-6 plants stained with Mäule reagent also showed no obvious changes in the size, shape, and compactness of the vasculature cells (Figure 5). Mäule staining of the first three internodes at the R1 stage showed a decrease in the number of stained sclerenchyma and epidermal cells in all three internodes (Figure 5).
The acetyl bromide–soluble lignin content of stems at the R1 stage was significantly reduced in hpCCR1-1, hpCCR1-3, and hpCCR1-6 lines by 32, 29, and 26%, respectively, with respect to isogenic controls (Table 1). Thioacidolysis analyses demonstrated that the lignin composition of R1-stage stems from the hpCCR1 lines was characterized by reduced levels of all three types (S, G, and H) of subunits (Table 1). In the hpCCR1-1, hpCCR1-3, and hpCCR1-6 lines, the level of G subunits was decreased by 43, 30, and 29%, respectively; the level of S subunits was decreased by 44, 28, and 27%, respectively; and the level of H units also fell by 50, 30, and 33%, respectively. As a result of reductions in the levels of both S and G subunits, the S/G ratio was not significantly altered in the hpCCR1-1, hpCCR1-3, and hpCCR1-6 lines (0.70, 0.79, and 0.78, respectively) in comparison to wild-type plants (0.77; Table 1).
NIRS analysis of transgenic and control plants at the R1 stage showed 12, 11, and 10% increase in digestibility of hpCCR1-1, hpCCR1-3, and hpCCR1-6 lines, respectively, relative to control plants (Table 1).
Analysis of the R1-3 internode of hpCCR1-1, hpCCR1-3, and hpCCR1-6 lines showed that the total acetyl bromide lignin level was reduced by 60, 33, and 30%, respectively, with respect to control plants (Table 2). In comparison to R1-3 lignin of control plants, the hpCCR1-1, hpCCR1-3, and hpCCR1-6 lines showed reductions in G subunit levels by 47, 25, and 28%, respectively; reductions in S subunit levels by 52, 34, and 21%, respectively; and reductions in H subunit levels by 36, 27, and 19%, respectively. The S/G ratios were slightly lower for the R1-3 lignin of the hpCCR1 transgenic lines in comparison to control plants, as a result of more significant reduction in the levels of S subunits.
Field Evaluation of hpOMT1 and hpCCR1 Perennial Ryegrass Plants
The impact of the modified lignin composition of hpOMT1 and hpCCR1 lines on plant fitness, lignin composition, and digestibility was assessed under natural field conditions. Transgenic hpOMT1-3 and hpCCR1-1 lines with high IVVDMD scores and control isogenic lines of the same ‘Grasslands Impact 566’ genotype were transplanted to a field site in Hamilton, Victoria, Australia during June 2009. All plants in the field site were monitored weekly over the July 2009 to January 2010 period and no supplemental watering was provided. No significant phenotypic differences in growth vigor were observed among the hpOMT1-3, hpCCR1-1, and control plants during the cold August (daily average 14°C) and mild September (daily average 18°C) conditions (see Supplemental Figure 8 online). Under warmer November (daily average 22°C) and December conditions (daily average 25°C), hpCCR1-1 plants showed slightly reduced vigor score compare with both control and hpOMT1 plants. The vigor scores of all transgenic lines declined compared with control plants during January, when temperatures of >40°C were recorded on several days. The hpOMT1-3 lines showed more susceptibility to infection by rust fungi (Puccinia species) (scored 3.2), in comparison to control plants and hpCCR1-1 plants (both scored 1.3).
The acetyl bromide method of analysis of whole tillers at the R1 stage showed a 6% reduction in lignin content in the hpOMT1-3 line, in comparison to control plants. Thioacidolysis of these samples revealed 34, 28, and 14% reductions in G, S, and H unit-derived products, respectively, relative to control plants, with no significant change in the S/G ratio (Table 1). The hpCCR1-1 plants grown under field conditions showed up to a 37% reduction in acetyl bromide soluble lignin, in comparison to control plants. Thioacidolysis of samples from hpCCR1-3 lines revealed reductions of 70, 71, and 71% in the release of H, G, and S unit-derived products, respectively, relative to control plants. The S/G ratio in samples from field-grown hpCCR1-1 and hpOMT1-3 transgenic plants was not significantly different from that of control plants because of similar reductions in the levels of the G and S monolignol unit-derived thioacidolysis products (Table 1). The lower levels of total lignin measured in plants grown under field conditions in comparison to glasshouse conditions could be explained by differences in sampling methods. Only stems were sampled from glasshouse-grown plants, while whole tillers, including leaf blades, were sampled from field-grown plants.
The tillers of plants at the early reproductive (R1) stage were collected and assessed for digestibility using NIRS. The IVVDMD values of the hpCCR1-1 and hpOMT1-3 plants were found to be 14 and 9% higher relative to control plants (Table 1).
Biochemical Differences between hpCCR1 and Control Perennial Ryegrass Plants Grown under Field Conditions.
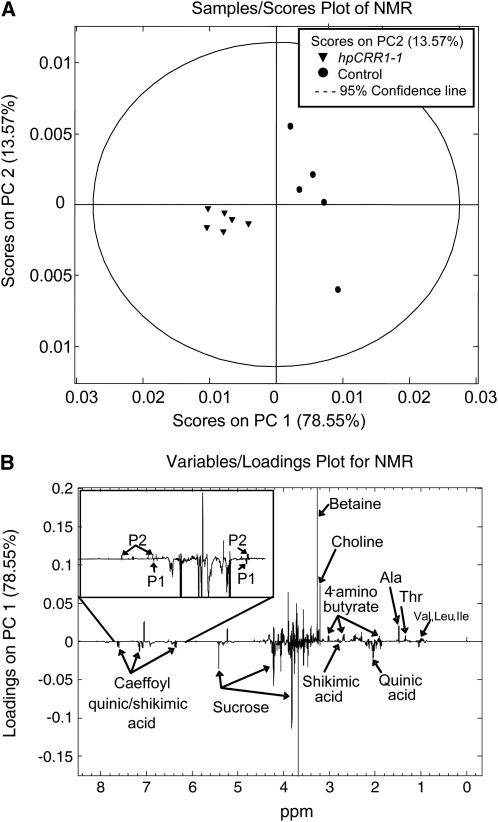
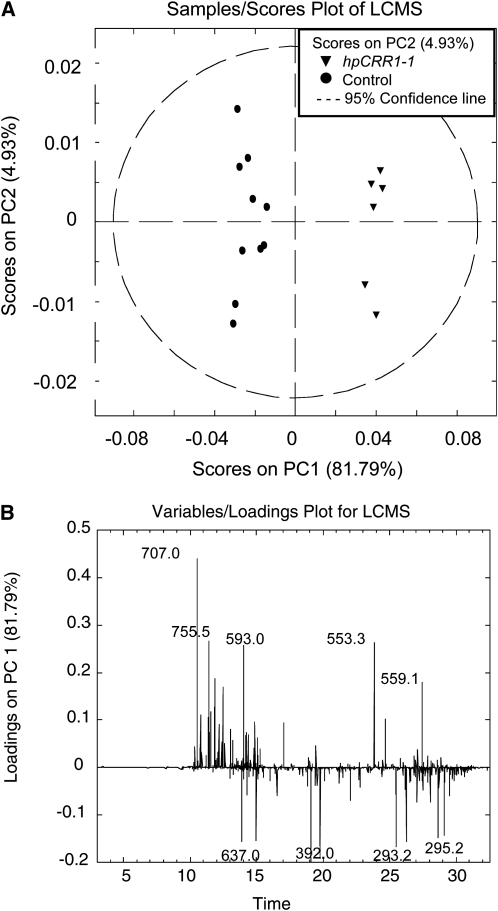
An unbiased statistical analysis of both NMR and liquid chromatography–mass spectrometry (LC-MS) data for identification of differentially accumulated metabolites within and outside the lignin biosynthetic grid was undertaken for the hpCCR1-1 plants and control plants grown under field conditions. One-dimensional NMR data were acquired for methanolic water extracts of the hpCCR1-1 and control plants and the data analyzed statistically. Principle component analysis (PCA) afforded clear separation of the wild-type and hpCCR1-1 spectra (Figure 7A). Examination of the loadings plot that describes the variables causing separation in the PCA plot led to the identification of key metabolites contributing to the differences in the two spectral data sets. While most carbohydrates remained unchanged, sucrose was significantly higher in the hpCCR1-1 plants. Pro, Ala, Val, Thr, Leu, amino butyrate, and choline fell in the hpCCR1-1 plants compared with wild-type cultivars (Table 3). The most significantly reduced metabolite was the osmolyte betaine. Inspection of PC1 loadings (Figure 7B) revealed that O-caffeoyl quinic acid (confirmed by two-dimensional NMR analysis) was by far the most significant differentiator in the phenolic region of the NMR spectra. This metabolite was 13-fold higher in the hpCCR1-1 plants. In addition, several additional currently unidentified phenolic metabolites increased in hpCCR1-1 plants (P1; Figure 7B; see Supplemental Figure 9 online), and this increase appeared to occur as levels of a separate pool of phenolic metabolites fell (P2; Figure 7B; see Supplemental Figure 9 online). Quantitative analyses of each spectrum provided fold changes for these metabolites (Table 3).
Figure 7.
NMR Analysis of Aqueous Soluble Metabolites from hpCCR1-1 and Control Plants.
(A) PCA analysis of 1H NMR data (spectra acquired in D2O for maximum organic acid and carbohydrate solubility). Wild-type (circles) and CCR1-1 plants (triangles) separate on the PC1 axis.
(B) PC1 loadings plot describing the most significantly different metabolites between control and hpCCR1-1 plants. Negative loadings are indicative of metabolites upregulated in CCR1-1 plants, while positive loadings indicate metabolites are more abundant in wild-type plants.
Table 3.
Differentially Accumulating Metabolites in hpCCR1-1 Plants, as Identified by NMR
| No. | Metabolite | hpCCR1-1 versus Control (Fold Change) | t Test (P Value) |
| 1 | 4-Aminobutyrate | −2.5 | 0.007 |
| 2 | Ala | −3.6 | 0.008 |
| 3 | Choline | −2.0 | 0.001 |
| 4 | Fructose | −1.1 | 0.568 |
| 5 | Fumarate | −2.1 | 0.121 |
| 6 | Glucose | −1.4 | 0.108 |
| 7 | Isobutyrate | 1.8 | 0.018 |
| 8 | Ile | −1.2 | 0.207 |
| 9 | Leu | −2.9 | 0.002 |
| 10 | Pro | −2.9 | 0.015 |
| 11 | Sucrose | 1.7 | 0.050 |
| 12 | Thr | −3.0 | 0.0002 |
| 13 | Val | −2.2 | 0.016 |
| 14 | Betaine | −16.7 | 0.000004 |
P value ≤ 0.05 is considered significant.
To further examine the phenolic pool, LC-MS analyses of the extracts were performed. Two approaches were used: an unbiased and more comprehensive analysis of the LC-MS data and a more targeted approach in which intermediates of the lignin biosynthetic grid were targeted for quantification. A representative chromatogram is shown in Supplemental Figure 10 online. Data alignment and feature selection generated a list of nearly 3000 traits, and subsequent statistical analysis of this data set again showed clear separation of the two plant types. The PCA plot of the negative LC-MS data, shown in Figure 8A, leads to clear separation on PC1, indicating that there are significant biochemical differences between the plants. Subsequent analysis of the loadings plots in Figure 8B indicates that this segregation is due to higher and lower levels of several key metabolites relative to control plants. In positive mode MS analysis, one metabolite (mass-to-charge ratio [m/z] 333) dominates the loadings (significant reductions in hpCCR1-1), whereas in negative mode MS analysis, a multitude of metabolites appear to be differentially regulated (Figure 8B, Table 4).
Figure 8.
Statistical Analysis of Electrospray Ionization Negative LC-MS Data.
(A) PCA plot demonstrates excellent separation between the plants. Wild-type (circles) and CCR1-1 plants (triangles) separate on the PC1 axis, which describes 82% of the LC-MS detected metabolite variation between the plants.
(B) Loadings plot for PC1 depicting the m/z values and retention times for significantly up- and downregulated metabolites.
Table 4.
Identification of the Major Soluble Phenolic Compounds Differentially Expressed in hpCCR1-1 Compared to Controls by LC-MS Analysis
| No. | hpCCR1 versus Controls (Fold Change) | RTa | m/z [M] | Accurate MS | MS2 | Identityb |
| 15 | Down (3.2) | 4.4 | 166 [M+H]+ | 166.0805 | 149, 120 | Phe |
| 16 | Up (13.3) | 10.4 | 353 [M-H]− 707 [2M-H− | 353.0852 | 353, 191 | Caffeoylquinic acid |
| 17 | Up (5.1) | 10.7 | 771 [M-H]− | 771.1908 | 609, 462, 301 | Rutin-7-glycoside |
| 18 | Up (3.9) | 11.4 | 755 [M-H]− | 755.1962 | 593, 285 | Kaempferol-7,3-glycoside ferulic acid |
| 19 | Up (3.6) | 11.8 | 695 [M-H]− (697 [M+H]+) | 695.1393 | 651, 530, 488, 447, 287 (534, 448, 287) | Kaempferol diglucoside malonate |
| 20 | Up (5.9) | 12.1 | 335 [M-H]− | 335.0852 | NA | Caffeoyl shikimic acid |
| 21 | Down (384) | 12.5 | 434 [M+H]+ | 434.2014 | 288 | U1 |
| 22 | Up (14.1) | 12.7 | 337 [M-H]− | 337.0908 | 191 | Coumaroyl quinic acid |
| 23 | Down (9.5) | 13.1 | 163 [M-H]− | 163.0399 | NA | Coumaric acid |
| 24 | Down (220) | 13.3 | 333 [M+H]+ | 333.1244 | 318, 289 | U2 |
| 25 | Up (3.1) | 13.4 | 609 [M-H]− | 609.1394 | 447, 301 | Rutin |
| 26 | Down (4.8) | 13.5 | 165 [M+H]+ | 165.0196 | NA | Caffeoyl aldehyde |
| 27 | Up (4.3) | 13.9 | 593 [M-H]− | 593.1446 | 285 | Kaempferol-O-rutinoside |
| 28 | Up (3.6) | 13.9 | 193 [M-H]− | 193.0497 | NA | Ferulic aicd |
| 29 | Up (15.0) | 14.0 | 223 [M-H]− | 223.0597 | NA | Sinapic acid |
| 30 | Up (2.9) | 14.7 | 447 [M-H]− | 447.0906 | 285 | Kaempferol-O- glycoside |
| 31 | Up (2.9) | 14.9 | 535 [M+H]+ 533 [M-H]− | 535.0891 533.0879 | 287 447, 285 | U3 |
| 32 | Down (74.4) | 15.0 | 637 [M-H]− | 637.1335 | 331 | U4 |
| 33 | Down (3.9) | 15.2 | 209 [M-H]− | 209.0785 | 165 | 5-Hydroxyferulic acid |
| 34 | Downc | 15.4 | 177 [M-H]− | 177.0549 | NA | Coniferaldehyde |
| 35 | Down (3.4) | 15.5 | 207 [M-H]− | 207.0653 | NA | Sinapaldehyde |
| 36 | Down (2.1) | 18.9 | 327 [M-H]− | 327.2130 | 309, 291, 229, 211, 201, 171, 155 | U5 |
| 37 | Down (3.1) | 19.7 | 329 [M-H]− | 329.2287 | 314 | U6 |
| 38 | Up (48.2) | 24.7 | 721 [M-H]− | 721.3574 | 675, 415, 397 | U7 |
| 39 | Up (2.9) | 27.4 | 559 [M-H]− | 559.3062 | 513, 277, 253 | U8 |
| 40 | Down (2.1) | 27.5 | 293 [M-H]− | 293.2079 | 275, 265, 249, 231, 224, 205, 195, 179 | U9 |
Retention time (RT) is expressed in minutes and denotes the peak of the eluting metabolite by MS ion extraction.
U denotes an unidentified metabolite
There was no detectable peak in the hpCCR1-1 chromatograms (average response for control plants was 59,899).
Direct correlation of these LC-MS data with the previously described NMR data is apparent, and again, O-caffeoyl quinic acid stands out as a major metabolic differentiator between hpCCR1-1 plants and wild-type controls (Table 4). Of the lignin biosynthetic intermediates detected, both ferulic acid and sinapic acid showed significant increases (up 3.6- and 15.0-fold, respectively). Phe, coumaric acid, 5-hydroxyferulic acid, sinapldehyde, and caffeoyl aldehyde showed significant decreases (3.2-, 9.5-, 3.9-, 3.4-, and 4.8-fold, respectively). Coniferaldehyde was not detectable in the hpCCR1-1 chromatograms but readily observed in control plants with an average peak area of 59,899 and with excellent signal-to-noise ratio of 39:1.
To identity the metabolites of significance, accurate mass and MSn fragmentation data were acquired. Several of these notable compounds were flavonols, including coumaroyl quinic acid (up 14.1-fold), caffeoyl shikimic acid (up 5.9-fold), caffeoyl quinic acid (14.1-fold), rutin-7-glycoside (up 5.1-fold), kaempferol-O-rutinoside (up 4.3-fold), kaempferol-7,3-glycoside ferulic acid (up 3.9-fold), kaempferol diglucoside malonate (up 3.6-fold), rutin (up 3.1-fold), and kaempferol-O-glycoside (up 2.9-fold) (Table 4). The metabolite kaempferol diglucoside malonate is described here. The proposed structure was determined as described previously (Rochfort et al., 2006) and is supported by the MSn and accurate mass data, although the position of the malonate ester cannot be determined unambiguously and may in fact be attached to C2 or C3 of the second hexose. Nine unidentified (U) compounds showed different levels of increases or decreases in hpCCR1-1 compared with controls. Some of them, U1 (m/z 434), U2 (m/z 220), and U4 (m/z 637), showed significant levels of downregulation in transgenic plants (384-, 220-, and 74-fold, respectively).
DISCUSSION
Functional Analysis of the OMT1 Gene in Perennial Ryegrass Plants
Changes in the spatio-temporal expression profile of OMT1 from the E1 to R3 stages of development showed that OMT1 expression correlates well with increasing lignification and the production of S subunits within stems of developing perennial ryegrass plants. The COMT1b gene from tall fescue (Festuca arundinaceae) is also expressed at a high level between the E3 and R1 stages of development (Chen et al., 2004). However, the expression level of Fa COMT1b remains high during later reproductive stages (R2-R3), whereas the Lp OMT1 expression level appears to decline during the R2 and R3 stages. It is possible that the RNA gel blot hybridization analysis used in the tall fescue study detected transcripts of additional members of the COMT gene family that were differentially expressed. The lower expression levels of both Lp OMT1 and Fa COMT1a in basal internodes compared with upper internodes of stems, where lignin biosynthesis is most active, may reflect a negative feedback loop caused by the accumulation of monolignols.
Transgenic approaches have been widely used for modification of lignin content and composition in different species often by downregulating enzymes catalyzing steps early and late in the monolignol biosynthetic grid (Chabbert et al., 1994; Atanassova et al., 1995; Ralph et al., 1997, 2001; Zhong et al., 1998; Jouanin et al., 2000; Guo et al., 2001). The most striking effects on lignin composition in plants deficient in COMT are reduced levels of S subunits without significant changes in G subunit levels that coincide with the incorporation of 5-hydroxyconiferyl alcohol into the lignin polymer.
Transgenic maize lines in which COMT is downregulated show lignin levels and composition profiles that are comparable with bm3 (brown midrib mutant from Z. mays) lines (Barnes et al., 1971; Muller et al., 1971; Grand et al., 1985; Vignols et al., 1995; Piquemal et al., 2002). A similar phenotype, characterized by a reduced level of S lignin and a lower S/G ratio, is seen in transgenic tall fescue plants in which the COMT1a gene has been downregulated (Chen et al., 2004). Transgenic maize and tall fescue lines show enhanced cell wall digestibility.
Correlations between spatio-temporal patterns of Lp OMT1 expression and lignin deposition, as well as lignin composition in OMT1-deficient transgenic plants, in this study provide strong evidence that OMT1 is involved in S-lignin biosynthesis in perennial ryegrass (Figure 9).
Although the key role of COMT in methylation of the 5-OH group in 5-OH-coniferyl aldehyde is widely accepted, it has been suggested that COMT could alternatively or additionally catalyze the 3-O-methylation of caffeoyl aldehyde and caffeoyl alcohol (Maury et al., 1999; Dixon et al., 2001; Parvathi et al., 2001; Do et al., 2007; Ma and Xu, 2007; Guillaumie et al., 2008; Ma and Xu, 2008). Consistent with this, alfalfa COMT has a strong kinetic preference for both of these caffeoyl substrates (Parvathi et al., 2001), and radiochemical labeling studies demonstrated the incorporation of caffeoyl alcohol and caffeoyl aldehyde into both G and S subunits (Matsui et al., 2000). Recombinant COMT protein from wheat showed the highest level of catalytic activity toward both caffeoyl aldehyde and 5-hydroxyconiferaldehyde (Ma and Xu, 2008). The Vmax and Km values for recombinant Fa COMT1 were also high for both caffeoyl aldehyde and 5-hydroxyconiferaldehyde (Chen et al., 2004). Moreover, a reduced level of G subunits was observed in transgenic tall fescue plants in which COMT1a had been downregulated (Chen et al., 2004; this study). Downregulation of OMT1 in transgenic perennial ryegrass plants results in a reduction of ~30% in the level of G subunits in whole stem and the upper internode at the R1 stage. When these transgenic plants were grown under field conditions, equally strong reductions were seen in the levels of S and G subunits. Therefore, coniferyl aldehyde, a precursor of both G and S subunits, could be the product of either COMT-mediated 3-O-methylation of caffeoyl aldehyde or CCR-mediated reduction of feruloyl CoA molecules (Figure 9).
Functional Analysis of the CCR1 Gene in Perennial Ryegrass Plants
Similar to CCR genes from wheat, maize, Arabidopsis, and eucalyptus (Eucalyptus gunnii), Lp CCR1 is expressed at the highest levels in shoots, stems, and roots (Lacombe et al., 1997; Pichon et al., 1998; Lauvergeat et al., 2001). Transcripts of all of these genes were detected in tissues undergoing active lignification. In spite of the close phylogenetic relationship between the perennial ryegrass and switchgrass CCR1 proteins, the corresponding genes exhibit differences in expression profiles within developing stems. A high level of Lp CCR1 expression was detected in actively growing perennial ryegrass tissues in upper internodes of stems at later stages of development (E2, E3, and R1). The highest level of switchgrass CCR1 expression was detected in the basal internode of plants at the E4 stage (Escamilla-Treviño et al., 2010). It is interesting that the highest levels of maize CCR1 transcripts occur in both upper and basal internodes of R stage plants (Pichon et al., 1998). Lower levels of Lp CCR1 expression at the R2 and R3 stages suggest that this gene is functionally complemented by other members of the CCR gene family in L. perenne.
Downregulation of CCR expression commonly results in a significant reduction in lignin content, disappearance of monolignol coupling products, accumulation of lignin pathway intermediates and derivatives, and the alteration of cell wall structure (Piquemal et al., 1998; Jones et al., 2001; O’Connell et al., 2002; Goujon et al., 2003; van der Rest et al., 2006; Wadenbäck et al., 2008). Some plants in which CCR genes have been profoundly downregulated also display phenotypic abnormalities, including delayed senescence, retarded seed development, and reduced size (Piquemal et al., 1998; Jones et al., 2001; O’Connell et al., 2002; Goujon et al., 2003; van der Rest et al., 2006; Wadenbäck et al., 2008). Dwarf phenotypes were observed in tobacco (Nicotiana tabacum) and tomato (Solanum lycopersicum) plants in which CCR expression was downregulated using antisense and RNAi approaches, respectively (Piquemal et al., 1998; Chabannes et al., 2001; van der Rest et al., 2006; Dauwe et al., 2007). Two Arabidopsis mutations affecting the CCR1 gene associate with a dwarf phenotype, delayed senescence and reduced lignin content (Mir Derikvand et al., 2008). Downregulation of a CCR gene in transgenic poplar plants (P. tremula x P. alba) is associated with up to a 50% reduction in lignin content and an orange-brown, often patchy, coloration of the outer xylem (Leplé et al., 2007).
Downregulation of CCR1 in transgenic perennial ryegrass plants grown under glasshouse and field conditions led to reduced total lignin levels and lower levels of both S and G subunits, and no significant changes in the S/G ratio. Reduced lignin content and improved digestibility in these plants was not associated with significant penalties in terms of biomass production, health, or vigor under glasshouse or field conditions. Very little is known about the control of H unit incorporation into lignin in angiosperms. In transgenic maize, tall fescue and perennial ryegrass plants in which COMT genes were downregulated the level of H units fell slightly. However, the level of H units in hpCCR1 perennial ryegrass plants fell by up to 50%, suggesting involvement of Lp CCR1 in p-coumaryl alcohol biosynthesis (Figure 9).
Effect of CCR1 Downregulation on Biosynthesis of Phenylpropanoid Intermediates
Downregulation of the CCR genes in a variety of species has been shown to redirect metabolic flux away from developmentally related lignification in CCR1-deficient plants. Silencing of the CCR gene in tobacco, tomato, and poplar results in decreased flux from feruloyl-CoA to G and S units, respectively, leading to the observed reduction in the level of lignin-specific phenolic molecules (van der Rest et al., 2006; Dauwe et al., 2007; Leplé et al., 2007). By contrast, the levels and composition of the some of the stress-related phenylpropanoid intermediates and derivatives were strongly enhanced in these transgenic lines. Most of these accumulating metabolites were modified through quination and glucosylation by p-hydroxycinnamoyl-CoA:D quinate and a family of glycosyl transferases, respectively (van der Rest et al., 2006; Dauwe et al., 2007; Leplé et al., 2007). Some molecules, such as coumaroyl-CoA, can be also conjugated to shikimate producing p-coumaroyl-shikimate by p-hydroxycinnamoyl-CoA:shikimate p-hydroxycinnamoyltransferase (Figure 9; Schoch et al., 2001; Boerjan et al., 2003; Hoffmann et al., 2004; Dauwe et al., 2007).
Downregulation of Lp CCR1 results in a decreased level of the downstream products in the lignin biosynthesis pathway, such as H, G, and S units, caffeoyl aldehyde, and coniferaldehyde (these compounds are shown in red in Figure 9). This suggests that coumaroyl-CoA and caffeoyl-CoA, along with feruloyl-CoA, may serve as the primary substrates for CCR1 in perennial ryegrass. This also indicates that coniferaldehyde, a common intermediate of both G- and S-lignin biosynthesis, may be synthesized from feruloyl-CoA or from caffeyl aldehyde. The latter scenario correlates with predicted roles of Lp OMT1 in methylation of 3-OH in caffeoyl aldehyde, suggesting an additional, caffeoyl aldehyde–mediated branch in both G- and S-related monolignol biosynthesis (Figure 9).
Reduced flux from coumaroyl-CoA, caffeoyl-CoA, and feruloyl-CoA to H, G, and S units, respectively, would therefore lead to enhanced accumulation of coumaric acid, cinnamic acid, and ferulic acid. As a result, an elevation in the levels of p-courmaroyl quinic acid, caeffoyl quinic acid, and caeffoyl shikimic acid would occur in hpCCR1-1 plants. Caffeoyl shikimate and caffeoyl quinate can be stored or, alternatively, converted back into caffeoyl-CoA and feruloyl-CoA (Ulbrich and Zenk, 1980; Schoch et al., 2001). Low levels of CCR1 also diverts coumaroyl CoA esters into the flavonoid biosynthetic pathway, resulting in the accumulation of flavonol conjugates, as seen in poplar, tobacco, and tomato plants with genetic alterations in the expression of lignin biosynthetic genes (van der Rest et al., 2006; Besseau et al., 2007; Dauwe et al., 2007; Leplé et al., 2007). Enhanced levels of flavonol glycosides and acylated anthocyanins were previously seen in Arabidopsis plants in which hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase was silenced, presumably resulting in the redirection of metabolic flux from monolignol to flavonoid production due to elevated levels of coumaroyl CoA esters (Besseau et al., 2007). The soluble phenolic fraction of CCR-deficient tomato plants contained flavonoids that were not detected in wild-type plants, including rutin (quercetin-3-rutinoside) and kaempferol rutinoside (van der Rest et al., 2006). In the hpCCR1-1 plants described here, the enhanced amounts of quinylated and glucosylated derivatives of quercetin and kaempferol, rutin-7-glycoside, quercetin-3-rutinoside, kaempferol-7,3-glycoside ferrulic acid, kaempferol-rutinoside, kaempferol diglucoside malonate, and kaempferol-O-glycoside may trigger a concomitant induction of the detoxification and storage mechanisms suggested by Dauwe et al. (2007).
Modification of Lignin Biosynthesis for Improvement of Forage Quality and Biofuel Production
The influence of lignin composition on cell wall digestibility has been studied in a number of forage grasses, including bluestem grass (Andropogon gerardii), timothy grass (Phleum pratense), orchard grass (Dactylis glomerata), smooth bromegrass (Bromus inermis), perennial ryegrass, switchgrass, Italian ryegrass (Lolium multiflorum), and tall fescue (Smith and Flinn, 1985, 1991; Chesson et al., 1986; Jung and Vogel, 1986; Buxton and Russell, 1988; Buxton and Martin, 1989; Jung, 1989; Grabber et al., 1992). The bm3 mutant of maize, in which COMT activity is reduced by 70 to 90%, shows lower levels of cell wall constituents, acid detergent fiber, acid detergent lignin, as well as increased digestibility (Grand et al., 1985; Vignols et al., 1995; Guillaumie et al., 2008). In transgenic maize plants in which COMT expression and S-lignin subunit biosynthesis are downregulated, the digestibility of leaves and stems is 2 and 7% higher, respectively, in comparison to wild-type plants (Piquemal et al., 1998). Reduced levels of S-lignin subunits in transgenic tall fescue plants correlate with an up to 10.8% increase in digestibility (Chen et al., 2004). Field-grown hpCCR1-1 and hpOMT1-3 plants showed up to 13.6 and 8.6% increase in digestibility, respectively, at reproductive stages. This is noteworthy since a 5 to 6% increase in the digestibility in perennial ryegrass and the concomitant availability of nutrients is estimated to increase summer milk production in southern Australia by up to 27% (Smith et al., 1998).
Perennial forage grasses with low annual energy inputs and high soluble sugar content are also ideal crops for bioethanol and biogas production (up to 4500 liters of ethanol and 375 m3 of methane per ha, respectively) (Murphy and Power, 2009). Furthermore, biofuel from grass need not displace food production. Because of these combined properties, forage grasses offer greater potential as a feedstock for bioethanol and biomethane production than many other energy crops, such as oilseed rape (Brassica napus) for biodiesel, wheat and maize grains for starch, and sugar beet (Beta vulgaris) for bioethanol and other alcohols.
In summary, our results provide evidence that in planta OMT1 can methylate both syringyl- and guaiacyl-lignin subunits at various stages of perennial ryegrass development. Downregulation of CCR1 gene expression in perennial ryegrass is directly associated with a dramatic decrease in the levels of total lignin and S, G, and H subunits, redirection of metabolic flux away from lignifications, and enhanced detoxification of the phenylpropanoid intermediates through glycosylation and quinylation. The enhanced digestibility of OMT1- and CCR1-deficient perennial ryegrass plants grown under field conditions in comparison to nontransgenic plants, without consequences for fitness and biomass production, indicate that this gene is a promising target for the improvement of forage quality in grasses using a transgenic approach. Finally, our results showed a plasticity of the plant’s metabolism in response to metabolic modifications, revealing that the intermediates of lignin biosynthesis are part of a complex regulatory network that extends beyond phenylpropanoid metabolism. This metabolic plasticity presumably enables plants to respond and adapt to different environmental conditions.
METHODS
Plant Material
Lolium perenne cv Grasslands Impact plants were propagated under glasshouse conditions. Natural daylight was supplemented with sodium vapor lamps to provide a 16-h photoperiod, and a 24°C day/20°C night temperature regime was used. Plants were vernalized for 12 weeks at 4°C with an 8-h photoperiod, and flowering was induced by transferring the plants to glasshouse conditions as above. Whole stems (without leaves) or internodes at V, E, and R stages from six representative plants were frozen in liquid nitrogen and stored at −80°C for expression analysis and for gas chromatography–mass spectrometry (GC-MS) and NIRS analysis.
Histochemical Staining of Lignin
Mäule reagent specifically stains syringyl (S) lignin subunits red and guaiacyl (G) subunits brown (Lin and Dence, 1992; Chen et al., 2002). Images were captured using a Leica MZ FLIII light microscope fitted with a digital camera. Micrographs shown in Figures 2 and 7 are representative of at least six different stems from different plants.
Construction of Transformation Vectors
To prepare vectors for dsRNAi-mediated posttranscriptional gene silencing, the PCR products containing the 265 bp of the Lp CCR1 and 1080 bp of the Lp OMT1 genes were amplified from the cDNA clones using the primers shown in Supplemental Table 3 online. PCR fragments were introduced into the pDONR221 entry vector (Invitrogen) using the BP recombinase reaction. Fully sequenced entry clones were transferred from pDONR221 to a destination vector with recombination sites oriented such that the GATEWAY-enabled transgene would form an inverted CCR1 and OMT1 repeats separated by the Lp CCR1 gene intron under the control of the maize (Zea mays) ubiquitin (Ubi) promoter and the nopaline synthase (nos) terminator.
Perennial Ryegrass Transformation
Transgenic perennial ryegrass plants (L. perenne cv Grasslands Impact 566) expressing the plasmid pAcH1, containing the hph (for hygromycin phosphotransferase) selectable marker gene for hygromycin resistance (Bilang et al., 1991) and Ubi:hpLp OMT1:nos (hpOMT1) and Ubi:hpLp CCR1:nos (hpCCR1) were generated according to Spangenberg et al. (1995). Transgenic hpOMT1 and hpCCR1 and a nontransgenic control of the same, L. perenne cv Grasslands Impact, genetic background were propagated in six copies under glasshouse conditions.
Expression Analysis
RNA was extracted from perennial ryegrass stems using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA gel blot hybridization was performed according to Sambrook et al. (1989). The probes for detection of the OMT1 and CCR1 genes are shown in Supplemental Table 3 online. The transcript abundance of OMT1, OMT3, and CCR1 genes was measured by quantitative RT-PCR using specific oligonucleotide primer pairs and SYBR Green chemistry with absolute quantification. Cycling conditions (Mx3005P; Stratagene) were as follows: 95°C for 10 min, 40 cycles of 95°C for 30 s, and 60°C for 1 min. A dissociation curve was obtained after amplification by heating products from 60 to 95°C. A 10-fold dilution series of standard templates was prepared for absolute quantification, with DNA concentrations ranging between 1 ng/μL and 1 fg/μL. Results were analyzed using MxPro software (Stratagene). Data are normalized using expression values of three internal control genes from perennial ryegrass encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH), histone (His), and tubulin (Tub) (see Supplemental Table 3 online). Standard error statistics are calculated from three technical replicates as per the geNorm manual (http://medgen.ugent.be/genorm/).
Field Trials
Transgenic hpOMT1-3 and hpCCR1-1 populations and a nontransgenic control of the same ‘Grasslands Impact 566’ genetic background were propagated, and six copies of each plant were acclimatized in a glasshouse from June 2008 to June 2009. In June 2009, upon evaluation by the Office of the Gene Technology Regulator, DIR 082/2007 plants were transplanted to the field. A field trial was established in Hamilton Victoria, Australia, on June 2009. The field isolation zone was sown with triticale on May 2009. Plants were planted at 0.5-m intervals. All plants in the field site were monitored weekly over the July 2009 to January 2010 period with no supplemental watering used for the field-grown plants. The tiller samples from six representative plants were frozen in liquid nitrogen and stored at −80°C for GC-MS, NIRS, LC-MS, and NMR analysis. Between September 2009 and January 2010, transgenic founder plants hpOMT1-3 and hpCCR1-1 and control plants were phenotypically scored under field conditions for rust resistance (scores from 0 [lowest] to 5 [highest]) and growth vigor (scores from 0 [lowest] to 5 [highest]) and sampled for forage quality assessment using NIRS and lignin level and composition analysis.
Metabolite Profiling
Determination of Lignin Content and Composition
For lignin content and composition analysis, 300 mg of dry powder was extracted with water, ethanol, and acetone (subsequent extractions, 20 mL of each solvent), and the remaining cell wall material was used for both total lignin content and composition analysis. The lignin content of stems and internodes from six representative plants was quantified using the acetyl bromide–soluble lignin method (Iiyama and Wallis, 1990) (6 mg of the cell wall preparation was used). Trimethylsilyl (derivatives of monomers and dimers from lignin) were identified by GC-MS using the method of Lapierre and Rolando (1988), using 5 mg of the cell wall samples.
Near-Infrared Spectroscopy
The nutritive value of plants was determined by taking 70 g of vegetative herbage sample, cut 5 cm aboveground. Samples were frozen at −80°C before being freeze-dried using a VirTis Genesis 35 freeze dryer. Freeze-dried samples were ground using a Cyclotek mill with a 1-mm screen. A total of 0.3 g of ground herbage sample (<1-mm particle size) were scanned between 400 and 2500 nm, with data points every 2 nm, on a FOSS XDS near-infrared Rapid Content analyzer (Smith and Flinn, 1991). Spectroscopy data were analyzed using ISIscan version 3.60.0.11037, and nutritive value was estimated using existing calibration equations, where digestibility was assessed as the percentage difference in mass between feed intake and output of fecal matter and was expressed as IVVDMD (Flinn, 2003).
LC-MS Parameters
HPLC Analysis
The extracts were analyzed using a 150 × 2.1-mm Agilent Eclipse XDB 3.5 μm C18 HPLC column fitted to an Agilent series 1200 high-performance liquid chromatograph (Agilent). Metabolites were eluted from the column using a gradient mobile phase, A (0.1% formic acid in water; Riedel-de Haen) and B (0.1% formic acid in acetonitrile; Riedel-de Haen) at 0.2 mL/min. Initial conditions (95% A) were maintained for 5 min before initiating a linear gradient to 20% A over 25 min, and this was maintained for 5 min before returning to the initial gradient conditions. The compounds were detected with a Thermo Fisher LTQ Velos Orbitrap mass spectrometer operating in the electrospray ionization mode with a HESI probe for both positive and negative data acquisition.
MS Analysis
For LC-MS analysis, 50 mg of dry powder was extracted twice with 1 mL of methanol:water (80:20, v:v). The supernatants were combined and 10 μL injected. Each extract was analyzed using a LTQ Velos Orbitrap (Thermo Fischer Scientific) with a positive-negative switching protocol over a mass range of 80 to 2000 amu with data stored in profile mode.
For structure elucidation, negative mode accurate mass data and MSn information were obtained in six scan events over the 80 to 2000 amu range. Initial data processing was carried out using Expressionist Refiner MS (Genedata) to align MS data, carry out noise reduction, and for peak picking. The data were analyzed statistically in MatLab. Relative quantitation was performed in LCQuant 2.6 (Thermo Fischer Scientific).
Analysis of Standards
p-coumaric acid (Sigma-Aldrich), Phe (ICN Biomedicals), ferulic acid (Sigma-Aldrich), sinapic acid (Fluka, Sigma-Aldrich), sinapyl alcohol (Aldrich Chemical Co.), caffeic acid (Sigma-Aldrich), and coniferyl alcohol (Aldrich Chemical Co.) were analyzed under the conditions described above. Retention time data and m/z information were then used to develop the LCQuant method for data analysis.
NMR Analysis
Proton spectra were obtained on a Bruker 800 MHz instrument equipped with a cryoprobe. A standard Bruker zgesgp pulse sequence was used over a spectral range of −1.9 to 12.0 ppm with 256 scans collected after four dummy scans, using a 1-s pulse delay and total acquisition time of 1.5 s at a temperature of 300K. Processing and statistical analysis was performed as reported previously (Rochfort et al., 2009). For structure elucidation, the following two-dimensional NMR experiments were run: hsqcedetgpsisp2.2, hmbcgplpndqf, cosygpqf, and dipsi2esgpph. All spectra were analyzed in Topspin 2.1. (Bruker Biospin).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Lp OMT3, AAD10255; Lp OMT1, AAD10253; Lp COMT, AAC18623.1Sb; OMT, AAL57301.1; So COMT, CAA13175.1; Fa COMT, AAK68907.1; Hv OMT, BAC54275.1; Bo COMT, ABS18316.1; Eg OMT, CAA52814.1; Ms COMT, AAB46623.1; Os OMT, ABB90678.1; Zm OMT, AAB03364.1; Zv COMT, AAA86718.1; Ta COMT1, ABP63535.1; Nt COMT, AAL91506.1; At OMT, AAB96879.1; Ta COMT2, AAP23942.1; Pt COMT, AAB61731.1; Ta OMT-3, ABP93669.1; Lp CCR1 gene, AY061889; Sl CCR1, AAY41879.1; Sl CCR2, AAY41880.1; Eg CCR, CAA56103.1; Pt CCR, AAF43141.1; At CCR1, AAG46037.1; At CCR2, NP_178197.1; Hv CCR, AAN71760.1; Ta CCR2, AAX08107.1; Lp CCR, AAG09817.1; Lp CCR1, AAL47182.1; Pv CCR1, ACZ74580.1; So CCR, CAA13176.1; Zm CCR1, NP_001105488.1; Pv CCR2, ACZ74585.1; Ta CCR1, ABE01883.1; Zm CCR2, NP_001105715.1; Os CCR, CAD21520.1; Mt CCR1, TC 128336; and Mt CCR2, TC 132256.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantification of Lignin Content and Composition in Perennial Ryegrass Stems.
Supplemental Figure 2. Assessment of the Digestibility of Perennial Ryegrass Stems.
Supplemental Figure 3. Phylogenetic Tree Showing the Relationships between the Deduced Amino Acid Sequences of Plant Caffeic Acid O-Methyltransferases.
Supplemental Figure 4. Multiple Sequence Alignment of Caffeic Acid O-Methyltransferases from Perennial Ryegrass and Other Plant Species.
Supplemental Figure 5. Phylogenetic Tree Showing the Relationships between the Deduced Amino Acid Sequences of Plant Cinnamoyl CoA-Reductases.
Supplemental Figure 6. DNA Gel Blot Hybridization Analysis of Transgene Copy Number in Perennial Ryegrass Plants Containing an hpOMT1 Construct.
Supplemental Figure 7. DNA Gel Blot Hybridization Analysis of Transgene Copy Number in Perennial Ryegrass Plants Containing an hpCCR1 Construct.
Supplemental Figure 8. Phenotypic Analysis of the Plants Grown under Field Conditions.
Supplemental Figure 9. NMR Analysis of Aqueous Soluble Metabolites from hpCCR1-1 and Control Plants
Supplemental Figure 10. LC-MS Chromatograms of an hpCCR1-1 Mutant Compared with Control in Both Positive and Negative Acquisition Modes.
Supplemental Table 1. Homology between Members of COMT Proteins from Perennial Ryegrass and Tall Fescue.
Supplemental Table 2. Homology between Members of CCR Proteins from Monocots.
Supplemental Table 3. List of Primers.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 3.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 5.
Supplemental Methods. Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank Ulrik John, Matthew Hayes, and Stephen Panter for critical reading of the manuscript. We also acknowledge the assistance of Craige Trenerry for LC-MS data acquisition, David Allen for GC-MS data acquisition, and Vilnis Ezernieks for NMR and LC-MS data acquisition. We also thank Melinda Boyd, Larry Jewell, Carly Elliott, Junping Wang, Darren Pickett, Tania Wilkinson, Mei-Ching Liu, and Stacey Erb for their help preparing plants for field trial and for harvesting and NIRS scanning.
References
- Anterola A.M., Lewis N.G. (2002). Trends in lignin modification: A comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294 [DOI] [PubMed] [Google Scholar]
- Atanassova R., Favet N., Martz F., Chabbert B., Tollier M.-T., Monties B., Fritig B., Legrand M. (1995). Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 8: 465–477 [Google Scholar]
- Barnes R.F., Muller L.D., Bauman L.F., Colenbrander V.F. (1971). In vitro dry matter disappearance of brown-midrib mutants of maize (Zea mays L.). J. Anim. Sci. 33: 881–884 [Google Scholar]
- Besseau S., Hoffmann L., Geoffroy P., Lapierre C., Pollet B., Legrand M. (2007). Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilang R., Iida S., Peterhans A., Potrykus I., Paszkowski J. (1991). The 3′-terminal region of the hygromycin-B-resistance gene is important for its activity in Escherichia coli and Nicotiana tabacum. Gene 100: 247–250 [DOI] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Boudet A.M., Kajita S., Grima-Pettenati J., Goffner D. (2003). Lignins and lignocellulosics: A better control of synthesis for new and improved uses. Trends Plant Sci. 8: 576–581 [DOI] [PubMed] [Google Scholar]
- Buxton D.R., Martin G.C. (1989). Forage quality of plant parts of perennial grasses and relationship to phenology. Crop Sci. 29: 429–435 [Google Scholar]
- Buxton D.R., Russell J.R. (1988). Lignin constituents and cell-wall digestability of grass and legume stems. Crop Sci. 28: 553–558 [Google Scholar]
- Chabannes M., Barakate A., Lapierre C., Marita J.M., Ralph J., Pean M., Danoun S., Halpin C., Grima-Pettenati J., Boudet A.M. (2001). Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 28: 257–270 [DOI] [PubMed] [Google Scholar]
- Chabbert B., Monties B., Barriere Y., Argillier O. (1994). Biological variability in lignification of maize: expression of the brown midrib bm3 mutation. J. Sci. Food Agric. 64: 349–355 [Google Scholar]
- Chen L., Auh C., Chen F., Cheng X., Aljoe H., Dixon R.A., Wang Z. (2002). Lignin deposition and associated changes in anatomy, enzyme activity, gene expression, and ruminal degradability in stems of tall fescue at different developmental stages. J. Agric. Food Chem. 50: 5558–5565 [DOI] [PubMed] [Google Scholar]
- Chen L., Auh C., Dowling P., Bell J., Lehmann D., Wang Z. (2004). Transgenic down-regulation of caffeic acid O-methyltransferase (COMT) led to improved digestibility in tall fescue (Festuca arundinacea). Funct. Plant Biol. 31: 235–245 [DOI] [PubMed] [Google Scholar]
- Chesson A., Stewart C.S., Dalgarno K., King T.P. (1986). Degradation of isolated grass mesophyll, epidermis and fibre cell walls in the rumen and by cellulolytic rumen bacteria in axenic culture. J. Appl. Microbiol. 60: 327–336 [Google Scholar]
- Dauwe R., et al. (2007). Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 52: 263–285 [DOI] [PubMed] [Google Scholar]
- Dixon R.A., Chen F., Guo D., Parvathi K. (2001). The biosynthesis of monolignols: A “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57: 1069–1084 [DOI] [PubMed] [Google Scholar]
- Do C.-T., Pollet B., Thevenin J., Sibout R., Denoue D., Barriere Y., Lapierre C., Jouanin L. (2007). Both caffeoyl coenzyme A 3-O-methyltransferase1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoylmalate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Escamilla-Treviño L.L., Shen H., Uppalapati S.R., Ray T., Tang Y., Hernandez T., Yin Y., Xu Y., Dixon R.A. (2010). Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol. 185: 143–155 [DOI] [PubMed] [Google Scholar]
- Fan B.Y., Hu S.Y., Lu H., Jiang X.N. (2006). Soluble prokaryotic expression of 4-coumarate:coenzyme A ligase from Populus tomentosa and enzyme activity of recombinant 4CL1. J. Beijing Forest. Univ. 28: 1–8 [Google Scholar]
- Flinn P. (2003). Premium grains for livestock: An update on NIR calibration development and implementation. Grain Industries Centre for NIR, 8th Annual Meeting of Participants, Melbourne, pp. 18–25. http://ses.library.usyd.edu.au/handle/2123/2295
- Goujon T., Ferret V., Mila I., Pollet B., Ruel K., Burlat V., Joseleau J.P., Barrière Y., Lapierre C., Jouanin L. (2003). Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: Effects on phenotype, lignins and cell wall degradability. Planta 217: 218–228 [DOI] [PubMed] [Google Scholar]
- Grabber J.H., Jung G.A., Abrams S.M., Howard D.B. (1992). Digestion kinetics of parenchyma and sclerenchyma cell walls isolated from orchardgrass and switchgrass. Crop Sci. 32: 806–810 [Google Scholar]
- Grand C., Parmentier P., Boudet A., Boudet A.M. (1985). Comparison of lignins and of enzymes involved in lignification in normal and brown midrib (bm3) mutant maize seedlings. Physiol. Veg. 23: 905–911 [Google Scholar]
- Guillaumie S., Goffner D., Barbier O., Martinant J.P., Pichon M., Barrière Y. (2008). Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT) down-regulated, and normal maize plants. BMC Plant Biol. 8: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Chen F., Inoue K., Blount J.W., Dixon R.A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack K.E., Jones J.D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8: 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R., Huxley H., Stone B., Spangenberg G. (1998). cDNA cloning and differential expression of three caffeic acid O-methyltransferase homologues from perennial ryegrass (Lolium perenne). J. Plant Physiol. 153: 649–657 [Google Scholar]
- Hoffmann L., Besseau S., Geoffroy P., Ritzenthaler C., Meyer D., Lapierre C., Pollet B., Legrand M. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iiyama K., Wallis A.F.A. (1990). Determination of lignin in herbaceous plants by an improved acetyl bromide procedure. J. Sci. Food Agric. 51: 145–161 [Google Scholar]
- Jones L., Ennos A.R., Turner S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Jouanin L., Goujon T., de Nadaï V., Martin M.T., Mila I., Vallet C., Pollet B., Yoshinaga A., Chabbert B., Petit-Conil M., Lapierre C. (2000). Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 123: 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H.G. (1989). Forage lignins and their effects on fiber digestibility. Agron. J. 81: 33–38 [Google Scholar]
- Jung H.G., Vogel , K.P. (1986). Influence of Lignin on Digestibility of Forage Cell Wall Material. J. Anim. Sci. 62: 1703–1712 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Koita H., Nakatsubo T., Hasegawa K., Wakabayashi K., Takahashi H., Umemura K., Umezawa T., Shimamoto K. (2006). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 103: 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitzki J. (1987). Phenylpropanoid metabolism in relation to land plant origin and diversification. J. Plant Physiol. 131: 17–24 [Google Scholar]
- Lacombe E., Hawkins S., Van Doorsselaere J., Piquemal J., Goffner D., Poeydomenge O., Boudet A.-M., Grima-Pettenati J. (1997). Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: Cloning, expression and phylogenetic relationships. Plant J. 11: 429–441 [DOI] [PubMed] [Google Scholar]
- Larsen K. (2004). Cloning and characterization of a ryegrass (Lolium perenne) gene encoding cinnamoyl-CoA reductase (CCR). Plant Sci. 166: 569–581 [DOI] [PubMed] [Google Scholar]
- Lauvergeat V., Lacomme C., Lacombe E., Lasserre E., Roby D., Grima-Pettenati J. (2001). Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Lapierre C., Rolando C. (1988). Thioacidolyses of pre-methylated lignin samples from pine compression and poplar woods. Holzforschung 42: 1–4 [Google Scholar]
- Leplé J.-C., et al. (2007). Downregulation of cinnamoyl-coenzyme A reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Popko J.L., Umezawa T., Chiang V.L. (2000). 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J. Biol. Chem. 275: 6537–6545 [DOI] [PubMed] [Google Scholar]
- Lin S.Y., Dence C.W. (1992). Methods in Lignin Chemistry. (Berlin: Springer-Verlag; ). [Google Scholar]
- Ma Q.H., Tian B. (2005). Biochemical characterization of a cinnamoyl-CoA reductase from wheat. Biol. Chem. 386: 553–560 [DOI] [PubMed] [Google Scholar]
- Ma Q.H., Xu Y. (2008). Characterization of a caffeic acid 3-O-methyltransferase from wheat and its function in lignin biosynthesis. Biochimie 90: 515–524 [DOI] [PubMed] [Google Scholar]
- Matsui N., Chen F., Yasuda S., Fukushima K. (2000). Conversion of guaiacyl to syringyl moieties on the cinnamyl alcohol pathway during the biosynthesis of lignin in angiosperms. Planta 210: 831–835 [DOI] [PubMed] [Google Scholar]
- Maury S., Geoffroy P., Legrand M. (1999). Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol. 121: 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes R., Lidgett A., Lynch D., Huxley H., Jones E., Mahoney N., Spangenberg G. (2002). Isolation and charasterization of a cinnamoyl-CoA reductase gene from perennial ryegrass (Lolium perenne). J. Plant. Physiol. 159: 415–422 [Google Scholar]
- Meyermans H., et al. (2000). Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275: 36899–36909 [DOI] [PubMed] [Google Scholar]
- Mir Derikvand M., Sierra J.B., Ruel K., Pollet B., Do C.-T., Thévenin J., Buffard D., Jouanin L., Lapierre C. (2008). Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956 [DOI] [PubMed] [Google Scholar]
- Moore K.J., Moser L.E., Vogel K.P., Waller S.S., Johnson B.E., Pedersen J.F.D. (1991). Describing and quantifying growth stages of perennial forage grasses. Agron. J. 83: 1073–1077 [Google Scholar]
- Muller L.D., Barnes R.F., Bauman L.F., Colenbrander V.F. (1971). Variations in lignin and other structural components of brown midrib mutants of maize (Zea mays L). Crop Sci. 11: 413–415 [Google Scholar]
- Murphy J., Power N. (2009). An argument for using biomethane generated from grass as a biofuel in Ireland. Biomass Bioenerg. 33: 504–512 [Google Scholar]
- O’Connell A., Holt K., Piquemal J., Grima-Pettenati J., Boudet A., Pollet B., Lapierre C., Petit-Conil M., Schuch W., Halpin C. (2002). Improved paper pulp from plants with suppressed cinnamoyl-CoA reductase or cinnamyl alcohol dehydrogenase. Transgenic Res. 11: 495–503 [DOI] [PubMed] [Google Scholar]
- Parvathi K., Chen F., Guo D.J., Blount J.W., Dixon R.A. (2001). Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Pichon M., Courbou I., Beckert M., Boudet A.M., Grima-Pettenati J. (1998). Cloning and characterization of two maize cDNAs encoding cinnamoyl-CoA reductase (CCR) and differential expression of the corresponding genes. Plant Mol. Biol. 38: 671–676 [DOI] [PubMed] [Google Scholar]
- Pinçon G., Maury S., Hoffmann L., Geoffroy P., Lapierre C., Pollet B., Legrand M. (2001). Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57: 1167–1176 [DOI] [PubMed] [Google Scholar]
- Piquemal J., et al. (2002). Down-regulation of caffeic acid o-methyltransferase in maize revisited using a transgenic approach. Plant Physiol. 130: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal J., Lapierre C., Myton K., O'Connell A., Schuch W., Grima-Pettenati J., Boudet A.M. (1998). Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 13: 71–83 [Google Scholar]
- Raes J., Rohde A., Christensen J.H., Van de Peer Y., Boerjan W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J., Lapierre C., Lu F., Marita J.M., Pilate G., Van Doorsselaere J., Boerjan W., Jouanin L. (2001). NMR evidence for benzodioxane structures resulting from incorporation of 5-hydroxyconiferyl alcohol into lignins of O-methyltransferase-deficient poplars. J. Agric. Food Chem. 49: 86–91 [DOI] [PubMed] [Google Scholar]
- Ralph J., MacKay J.J., Hatfield R.D., O’Malley D.M., Whetten R.W., Sederoff R.R. (1997). Abnormal lignin in a loblolly pine mutant. Science 277: 235–239 [DOI] [PubMed] [Google Scholar]
- Rochfort S.J., Ezernieks V., Yen A. (2009). NMR-based metabolomics using earthworms as potential indicators for soil health. Metabolomics 5: 95–107 [Google Scholar]
- Rochfort S.J., Imsic M., Jones R., Trenerry V.C., Tomkins B. (2006). Characterization of flavonol conjugates in immature leaves of pak choi [Brassica rapa L. Ssp. chinensis L. (Hanelt.)] by HPLC-DAD and LC-MS/MS. J. Agric. Food Chem. 54: 4855–4860 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schoch G., Goepfert S., Morant M., Hehn A., Meyer D., Ullmann P., Werck-Reichhart D. (2001). CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Smith K.F., Flinn P. (1991). Monitoring the performance of a broad-based calibrartion for measuring the nutritive value of two independent populations of pasture using near infrared reflectance (NIR) spectroscopy. Aust. J. Exp. Agric. 31: 205–210 [Google Scholar]
- Smith K.F., Kearney G.A., Culvenor R.A. (1998). The use of repeated measurements analysis for the evaluation of seasonal variation in the dry matter yield and nutritive value of perennial ryegrass (Lolium perenne L.) cultivars. Aust. J. Exp. Agric. 38: 145–154 [Google Scholar]
- Spangenberg G., Wang Z.Y., Wu X.L., Nagel J., Potrykus I. (1995). Transgenic perennial ryegrass (Lolium perenne) plants from microprojectile bombardment of embryogenic suspension cells. Plant Sci. 108: 209–217 [DOI] [PubMed] [Google Scholar]
- Ulbrich B., Zenk M.H. (1980). Partial purification and properties of para-hydro hydroxycinnamoyl-CoA-shikimate-para-hydroxycinnamoyl transferase from higher plants. Phytochemistry 19: 1625–1629 [Google Scholar]
- van der Rest B., Danoun S., Boudet A.-M., Rochange S.F. (2006). Down-regulation of cinnamoyl-CoA reductase in tomato (Solanum lycopersicum L.) induces dramatic changes in soluble phenolic pools. J. Exp. Bot. 57: 1399–1411 [DOI] [PubMed] [Google Scholar]
- Vanholme R., Morreel K., Ralph J., Boerjan W. (2008). Lignin engineering. Curr. Opin. Plant Biol. 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Vignols F., Rigau J., Torres M.A., Capellades M., Puigdomènech P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K.P., Jung H.J.G. (2001). Genetic modification of herbaceous plants for feed and fuel. Crit. Rev. Plant Sci. 20: 15–49 [Google Scholar]
- Wadenbäck J., von Arnold S., Egertsdotter U., Walter M.H., Grima-Pettenati J., Goffner D., Gellerstedt G., Gullion T., Clapham D. (2008). Lignin biosynthesis in transgenic Norway spruce plants harboring an antisense construct for cinnamoyl CoA reductase (CCR). Transgenic Res. 17: 379–392 [DOI] [PubMed] [Google Scholar]
- Whetten R., Sederoff R. (1995). Lignin biosynthesis. Plant Cell 7: 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins P.W., Humphreys M.O. (2003). Progress in breeding perennial forage grasses for temperate agriculture. J. Agric. Sci. Camb. 140: 129–150 [Google Scholar]
- Zhong R., Iii W.H., Negrel J., Ye Z.H. (1998). Dual methylation pathways in lignin biosynthesis. Plant Cell 10: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Morrison W.H., III, Himmelsbach D.S., Poole F.L., II, Ye Z.H. (2000). Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 124: 563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R.Q., Taylor J.J., Ye Z.H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C., Kota P., Ferrer J.L., Dixon R.A., Noel J.P. (2002). Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.