Abstract
Motivation: MicroRNAs (miRNAs) are small non-coding RNAs that cause mRNA degradation and translational inhibition. They are important regulators of development and cellular homeostasis through their control of diverse processes. Recently, great efforts have been made to elucidate their regulatory mechanism, but the functions of most miRNAs and their precise regulatory mechanisms remain elusive. With more and more matched expression profiles of miRNAs and mRNAs having been made available, it is of great interest to utilize both expression profiles to discover the functional regulatory networks of miRNAs and their target mRNAs for potential biological processes that they may participate in.
Results: We present a probabilistic graphical model to discover functional miRNA regulatory modules at potential biological levels by integrating heterogeneous datasets, including expression profiles of miRNAs and mRNAs, with or without the prior target binding information. We applied this model to a mouse mammary dataset. It effectively captured several biological process specific modules involving miRNAs and their target mRNAs. Furthermore, without using prior target binding information, the identified miRNAs and mRNAs in each module show a large proportion of overlap with predicted miRNA target relationships, suggesting that expression profiles are crucial for both target identification and discovery of regulatory modules.
Contact: bing.liu@unisa.edu.au; jiuyong.li@unisa.edu.au
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
MicroRNAs (miRNAs) are non-protein-coding RNAs that are expressed from longer transcripts encoded in animals, plants, viruses and single-celled eukaryotes (Zhao and Srivastava, 2007). They cause mRNA degradation, translational inhibition or a combination of the two by completely or partially complementary base binding to their target mRNAs (He and Hannon, 2004).
miRNAs are pivotal regulators of development and cellular homeostasis through their control of diverse processes, including cell differentiation, proliferation, growth, mobility, and apoptosis (Du and Zamore, 2007). Consequently, dysregulation of miRNA functions may lead to diseases. Recent studies have reported differentially expressed miRNAs in diverse cancer types such as breast cancer (Iorio et al., 2005), lung cancer (Yanaihara, 2006), prostate cancer (Porkka et al., 2007), colon cancer (Akao et al., 2007) and ovarian cancer (Yang et al., 2008). Therefore, the understanding of miRNA is critical in understanding basic biological processes, elucidating the development and inhibition of pathogenesis of many diseases, and facilitating biotechnology projects.
Many computational approaches have been proposed to elucidate miRNA functions in recent years. We classify these works into three categories: (i) miRNA target prediction (Bentwich et al., 2005; Griffiths-Jones et al., 2008; Hatzigeorgiou., 2007; Krek et al., 2005), that is, to identify which mRNAs are targeted by which miRNAs; (ii) discovering miRNA regulatory modules (MRMs), that is, to identify a group of co-expressed miRNAs and mRNAs, either at sequence level (Yoon and De Micheli, 2005) or by integrating sequence and expression profiles of miRNAs and mRNAs (Huang et al., 2007; Joung et al., 2007; Peng et al., 2009; Tran et al., 2008); (iii) prediction of functional miRNA regulatory modules (FMRMs), which are regulatory networks of miRNAs and their target miRNAs for specific biological processes (Joung and Fei, 2009; Liu et al., 2009a, b).
The identification of FMRMs is critical in understanding the biological pathways and the development and inhibition of pathogenesis of many diseases. It also has a great potential for the development of gene therapeutic treatments and miRNA-based drugs (Croce, 2009).
For FMRM discovery, Liu et al. (2009a) proposed a method based on association rule mining. It associates the reverse expression patterns of miRNAs and mRNAs with biological conditions. A novel method is further proposed (Liu et al., 2009b) using Bayesian Network structure learning. This method was designed to explore all the possible miRNA regulatory patterns for biological conditions under the comparative experiment designs. These two methods are supervised methods where biological conditions are directly applied to the search for the FMRMs. An unsupervised method was proposed by Joung and Fei (2009) for FMRM discovery. It is an innovative application of the author-topic model (Steyvers et al., 2004) in bioinformatics that makes use of the expression profiles of mRNAs and the putative miRNA target information, without considering the expression profiles of miRNAs. Therefore, the regulatory relationships of miRNAs and mRNAs are determined largely based on the miRNA target information which is predicted at the sequence level. Thus, it encounters difficulties in answering the question of how miRNAs regulate their target mRNAs in the identified modules.
In recent years, more and more sample matched expression data having been profiled for multiple classes of conditions or tissues with both miRNAs and mRNAs, providing the opportunity to investigate potential FMRMs systematically for various biological processes by integrating the available data. In addition, some researchers have suggested that algorithms that do not consider known targets may avoid biases (Bartel, 2009; Lewis et al., 2003, 2005). Hence, it is of great interest to discover FMRMs with expression profiles of miRNAs and mRNAs without using target binding information. Therefore, in this article, we propose a FMRM discovery method that integrates heterogeneous datasets, including expression profiles of both miRNAs and mRNAs, with or without using the prior target binding information.
Our method is inspired by the Correspondence Latent Dirichlet Allocation (Corr-LDA) (Blei and Jordan 2003), a probabilistic graphical model that has been successfully applied to automatic image annotation with caption words. Given observations of image and caption words, Corr-LDA captures the correspondence between them by modeling topics described by both images and caption words with latent variables. In our question, FMRMs are dependent groups of miRNAs and mRNAs linked to latent functions. Our aim is to capture the correspondence between miRNAs and mRNAs, assuming that they participate in the same latent functions. Therefore, we apply the idea of annotating images with caption words to FMRM discovery by mapping topics to functional modules, images to miRNAs and words to mRNAs, respectively.
In this work, we firstly modify the Corr-LDA and derive the solution to discover FMRMs. Then, we apply our method to mouse model expression datasets for human breast cancer research. The result shows that our model is able to capture several biologically meaningful modules. Furthermore, without using the prior target binding information, the identified miRNAs and mRNAs in each FMRMs show a large proportion of overlap with predicted miRNA target relationships, suggesting that targets and FMRMs can be predicted from expression profiles alone, and providing an independent verification of the underlying strategy.
2 METHODS
We begin with the assumption that functional modules governing miRNA and mRNA expression, which are associated with a variety of biological functions, are reflected by the data from microarray experiments. We model functional modules with latent random variables which act as a bridge between miRNAs and mRNAs. By inferring the latent variables, we can identify FMRMs.
2.1 Modeling FMRM discovery
More specifically, we model FMRMs with a probabilistic generative process. Given the K latent functions presented in the samples, our method considers miRNAs and mRNAs as observations generated from a probabilistic process over these K functions. Thus, each sample is a random mixture of miRNAs and mRNAs associated with K functional modules. By inferring the probability distributions of the latent variables, we are able to obtain the probabilities of how samples, miRNAs and mRNAs are related to functional modules.
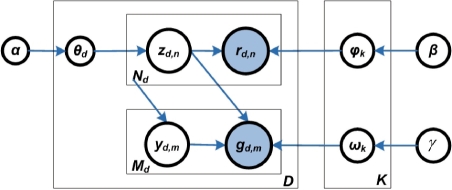
We depict the model in Figure 1 with a plate notation. In this notation, nodes stand for random variables (observed variables are shaded and latent ones are unshaded); edges denote conditional dependency between random variables; and plates denote replications of a substructure with the number of repetitions given in the bottom corner (either right or left side).
Fig. 1.
Generative model of FMRM discovery. Given expression data of miRNAs and mRNAs of D samples, each sample d is a mixture of random miRNAs and mRNAs. Each miRNA rd,n and mRNA gd,m are generated from one of the K latent functional modules, selected by zd,n.
In Figure 1, the D samples were profiled with a set of miRNAs V and a set of mRNAs T. Random variable rd,n and gd,m denote the indexes of a miRNA and mRNA expressed in the d-th sample, respectively, with d ∈ {1,···, D}, n ∈ {1,···, Nd}, and m ∈ {1,···, Md}. Nd and Md are the total numbers of times the miRNAs and mRNAs which are expressed in the d-th sample. Random variable zd,n stands for the latent functional module associating with the n-th miRNA in the d-th sample. We assume that zd,n, rd,n and gd,m all have multinomial distributions with parameters θd, φk and ωk, respectively. Each parameter has a Dirichlet prior with hyperparameters α, β and γ, correspondingly.
Without considering the putative target constraints, the generative procedure for each sample d can be illustrated by the following hierarchical sampling process: to generate the d-th sample, (i) a latent module zd,n is drawn from its multinomial distribution θd; (ii) then, a miRNA rd,n is drawn from its multinomial distribution φk, given the selected module zd,n; (iii) for each mRNA gd,m, one of the miRNAs, indexed by yd,m, is selected from Rd = {rd,n} and a corresponding mRNA gd,m is drawn from its multinomial distribution ωk, conditional upon the same module that generates the selected miRNA rd,n.
When the constraint of the putative target relationship between miRNAs and mRNAs is preferred, for each mRNA, one of the miRNAs from the set of hosting miRNAs of that mRNAs is selected, and a corresponding mRNA is drawn from the multinomial distribution of mRNAs, conditional upon the same module that generates the selected miRNA.
From the above generative process, the parameter Θ = {θd} associates samples with modules, Φ = {φk} assigns the probability of miRNAs expressed in module Z = {zd,n}, and Ω = {ωk} indicates the probability of mRNAs expressed in Z corresponding to the miRNAs. Therefore, by estimating Θ, Φ and Ω, we can identify FMRMs (details in Sections 2.3–2.5).
Under this model, miRNAs can associate with any modules, but mRNAs may only associate with the modules that produce the miRNAs. In effect, this model captures the hierarchical notion that miRNAs are generated under specific FMRMs, and mRNAs are regulated by the miRNAs.
2.2 Data conversion
In order to apply the above model to the expression profiles of miRNAs and mRNAs, we convert the expression values to the counts of miRNAs and mRNAs present in the samples.
Given a microarray experiment profiled D′ samples, similar to Joung and Fei (2009), we considered that miRNAs and mRNAs have events of their expression in every sample that are likely to be associated with functional modules. Therefore, each miRNA or mRNA can be represented as a vector of variables, {s1+, s2−,···, sD−1+, sD−}. It corresponds to the expression events of a miRNA or mRNA in all samples, where duplex {s2d−1+, s2d−} indicates an over- and underexpressed miRNA or mRNA of sample d, d ∈ {1,···, D′}, thus, D = 2D′. To get the integer counts (σ2d−1,i, σ2d,i) for the duplex expression status, we convert the expression value of a miRNA or mRNA of sample d with,
| (1) |
where ed,i is the expression value of a miRNA or mRNA in the d-th sample, ε is a scaling constant and medd denotes the median of all miRNAs or mRNAs in the d-th sample.
Then, the counts of miRNAs and mRNAs are replaced by the indexes from the set of miRNAs, Vand the set of mRNAs, T. The indexes, therefore, are the random variables rd,n and gd,m used in the model (Fig. 1).
2.3 Estimating model parameters
Because the exact inference for the parameters of our model is intractable, we used the collapsed Gibbs sampling method (Liu, 1994) to estimate parameters.
This method iteratively generates samples that converge to draws from a target distribution of random variables Z through integrating out the parameters Θ, Φ and Ω for each sampling. For the d-th sample and the n-th miRNA, the sampling is expressed as a conditional probability:
| (2) |
where zd,n is the current module assignment of the n-th miRNA of the d-th sample. Z−(d,n) is the current module assignment of all miRNAs in all samples excluding that of the n-th miRNA of d-th sample. nd,−(d,n)k is the number of times that the k-th FMRM has been observed with miRNAs across samples excluding that of the n-th miRNA of the d-th sample. nk,−(d,n)v is the number of times that miRNA v is assigned to the k-th FMRM excluding that of the n-th miRNA of d-th sample. mk,−(d,n)t is the number of times that mRNA t is assigned to the k-th FMRM excluding the current assignment.
After sufficient sampling, the distribution of zd,n converges to the target distribution of Z and then we estimate the parameters Θ, Φ and Ω based on the values of the module assignments produced from the sampling:
| (3) |
Unlike the preceding sampling procedure, here nd(k), nk(v) and mk(t) are calculated from the assignment results for all data without excluding the current module. Using Equations (2) and (3), the Gibbs sampling procedure can be designed. The algorithm includes three stages: initialization, sampling and reading out of parameters. It is provided in Supplementary Algorithm 1.
2.4 Assigning biological conditions to modules
The parameters inferred from this model provide insights into the datasets at several levels. Θ clusters samples into modules that should relate to the biological conditions of the experiments.
We conceive a statistic model to identify the connection between biological conditions and modules. Let C be the number of biological conditions of the D samples in the dataset, and ci be the number of samples belonging to condition i, where ∑i=1Cci = D. For each module, assume there are x samples among the n highest probability samples that belong to same condition i. The random variable x follows a hypergeometric distribution with parameters D, ci and n, denoted as
| (4) |
We assign the biological condition i to module k when x is at a statistically significant level, for example, P < 0.05.
2.5 Identifying miRNAs and mRNAs for modules
The parameters Φ and Ω indicate the probabilities of each miRNA and mRNA participating in a FMRM. For a K-FMRMs, Φ is a K × P probability matrix where the element φk,v indicates the likelihood that miRNA v belongs to the k-th FMRM. Similarly, Ω is a K × Q probability matrix where the element ωk,t indicates the belief of mRNA t participating in the k-th FMRM, and Q is the number of mRNAs under investigation.
For each FMRM, we consider the top ranked miRNAs and mRNAs with the highest probabilities to be the participants of the FMRM.
2.6 Reconstructing miRNA–mRNA target relationships
We query a miRNA target database to reconstruct the target relationship of the miRNAs and mRNAs in each module. Hypothesis tests are conducted on the identified miRNAs and mRNAs to evaluate whether they are likely to have been identified by chance or not.
2.7 Function and pathway analysis of FMRMs
Function and pathway analysis of the identified FMRMs is conducted by reviewing literature and querying the Ingenuity Pathway Analysis (IPA, www.ingenuity.com) database of functional biological pathways to identify the significantly enriched functions and pathways.
3 RESULTS
In this section, we present the results and analysis of applying our model to a mouse mammary dataset (Zhu et al., 2010).
The dataset were profiled with 46 samples derived from nine classes of mouse models, representing one normal type and two breast cancer subtypes: basal and luminal. The expression data were screened with 1336 probes of miRNAs (corresponding to 334 unique miRNAs) and 22 626 probes of mRNAs. For each type of the conditions, 3–7 samples were profiled with miRNAs and mRNAs (details in Supplementary Material).
In order to compare with the target prediction, the expression datasets of miRNAs and mRNAs were further filtered with MicroCosm Targets V5.0 (Griffiths-Jones et al., 2008), and only those in MicroCosm were maintained for analysis. Consequently, 1112 probes of miRNAs and 19 223 probes of mRNAs were used in our experiment.
3.1 Implementation
Given the above expression data of miRNAs and mRNAs, the input data for our model include a 1112 × 46 matrix of miRNA expression values and a 19 223 × 46 matrix of mRNA expression values. In the following discussion, we do not consider the putative target information to avoid the bias probably incurred by the prior prediction (Bartel, 2009).
In the experiment, the constant ε for converting the expression values was 30. After the data conversion, the number of samples D is 92. We set the number of FMRMs, K, to 20. This value is determined by the number of sample types. Our datasets were profiled with nine classes of mouse models. miRNAs and mRNAs could be over- or underexpressed in the samples so the number of sample types is 18. In addition, two extra types were added to allow the redundancy as our model could discover subtypes of classes. We set the hyperparameters α, β and γ to 10. The number of iterations of Gibbs sampling is 2500. These value settings are based on empirical experiments.
3.2 Associating FMRMs with biological conditions
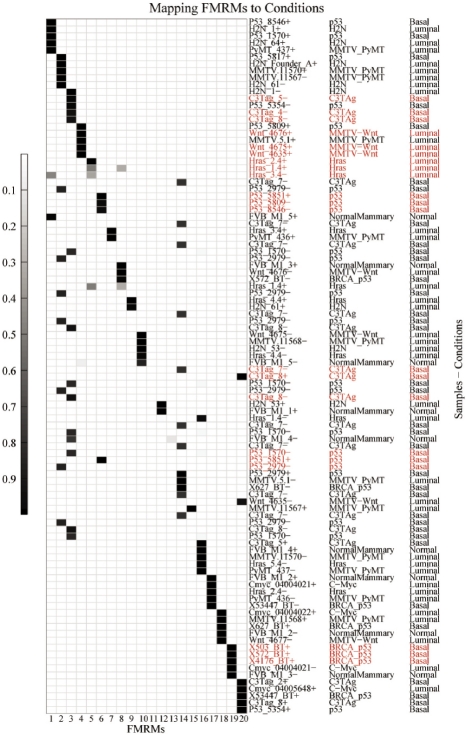
The parameter Θ obtained with our method is a 92 × 20 probability matrix. Referring to Section 2.4, the element θd,k of Θ is the belief of sample d belonging to module k. We extracted the top 5% (5) ranked samples with highest probabilities in each module, and assigned biological conditions to each module according to these samples as discussed in Section 2.4. Figure 2 illustrates this mapping procedure. The five highest probability samples extracted from each module are arranged on the y-axis. Their associations with other modules are also shown in the map.
Fig. 2.
Assigning biological conditions to FMRMs. The y-axis on the right side of the figure denotes sample names, mouse model types and breast cancer subtypes in three columns. Using the parameter Θ, the likelihood that a particular sample is associated with a specific module, the top 5% samples associated with each module are displayed using the grey scale. These samples are considered to map modules to biological conditions. Samples may occur more than once in the y-axis because some samples are significantly associated with more than one module. Some modules, such as module-11, have only rather low probability of association with samples, and thus have nearly white shading even for their top five samples. Significant mapping of FMRMs to conditions is highlighted.
In order to assign biological conditions to modules at the statistically significant level, we conceived a statistical model to map modules to biological conditions by using the mouse model classes instead of tumor types directly (Table 1). From Table 1, seven modules have been mapped to specific mouse model classes at a significant level (P < 0.05). These mouse models can be further mapped to two human breast tumor subtypes (Blenkiron et al., 2007; Desai et al., 2002; Herschkowitz et al., 2007), suggesting that the identified modules are associated with those biological conditions. Other modules are clustered by samples with mixed biological conditions, suggesting that they may participate in several cellular processes.
Table 1.
Assigning biological conditions to FMRMs
| FMRM# | ci | x | Mouse model class | Tumor subtype | P-value |
|---|---|---|---|---|---|
| 3 | 10 | 3 | C3TAg | Basal | 0.0081 |
| 4 | 8 | 3 | MMTV-Wnt | Luminal | 0.004 |
| 5 | 10 | 3 | Hras | Luminal | 0.0081 |
| 6 | 14 | 3 | p53 | Basal | 0.0222 |
| 11 | 10 | 3 | C3TAg | Basal | 0.0081 |
| 13 | 14 | 3 | p53 | Basal | 0.0222 |
| 19 | 10 | 3 | BRCA_p53 | Basal | 0.0081 |
According to Equation (4), biological conditions are assigned to FMRMs based on a hypergeometric distribution. The significant results are given in this table. The size of population is 92, the number of each draw is 5% of the population, i.e. 5. ci is the number of samples belonging to each condition, which include both over- and underexpressed status. x denotes the observed number of samples with the assigned biological condition in each draw. FMRM# is the module number corresponding to the number in Figure 2.
Furthermore, the top 5% (56) ranked probes of miRNA and the top 0.1% (192) ranked probes of mRNA with the highest probabilities in each module were also extracted from the inferred parameters Φ and Ω. They are assigned to the same biological conditions according to the modules they belong to, respectively. The detailed information of each FMRM is given in Supplementary Material, including the miRNAs, predicated target mRNAs and the associated biological condition for each FMRM.
3.3 Target reconstruction
To reconstruct the target relationships between miRNAs and mRNAs, we use MicroCosm to link miRNAs and mRNAs identified in each FMRM. The numbers of linked miRNAs and mRNAs are given in Table 2.
Table 2.
Numbers of miRNA-mRNA pairs identified in FMRMs
| FMRM# | Subtype | miRNA# | mRNA# | target pair# | P-value |
|---|---|---|---|---|---|
| 3 | Basal | 33 | 190 | 273 | 1.70E-07 |
| 4 | Luminal | 18 | 190 | 147 | 3.23E-08 |
| 5 | Luminal | 16 | 191 | 144 | 2.98E-07 |
| 6 | Basal | 16 | 189 | 146 | 1.48E-06 |
| 11 | Basal | 17 | 190 | 122 | 1.13E-11 |
| 13 | Basal | 18 | 186 | 136 | 1.29E-10 |
| 19 | Basal | 18 | 188 | 133 | 2.71E-12 |
The miRNAs and mRNAs identified in each module are linked by MicroCosm. Compared with the number of pairs linked by MicroCosm given the same number of randomly chosen miRNAs and mRNAs, the miRNAs and mRNAs identified in each module are not identified by chance.
To investigate whether the miRNAs and mRNAs in each module were identified by chance, we randomly selected a group of miRNAs and a group of mRNAs from MicroCosm with the same numbers as those in the identified modules, and queried how many pairs that can be linked by MicroCosm. The distribution of the number of matched pairs was estimated by a simulation which was executed 10 000 times. Illustrated with Supplementary Figure S1, the estimated distribution shows that the numbers of target relationships of the randomly chosen miRNAs and mRNAs are significantly different from those of the identified miRNAs and mRNAs in each module (P < 0.05). It indicates that the miRNAs and mRNAs in each module are not identified by chance. The linked miRNAs and mRNAs of each FMRM are given in Supplementary Material.
3.4 Functional validation of miRNAs
To further validate that the identified miRNAs are relevant to cancers, we investigated the implications of miRNAs for cancers through literature review. We built a benchmark based on the current knowledge (details in Supplementary Material), and compared it with the miRNAs identified in the modules.
From the literature, 42 miRNAs have been validated to have implications for cancers. We identified a significant number of miRNAs covered by the benchmark shown in Table 3. The comparison shows that the miRNAs identified by our method are largely consistent with the current knowledge of miRNAs for cancers.
Table 3.
Validation of identified miRNAs in the FMRMs
| FMRM# | Supported miRNAs | Coverage | P-value |
|---|---|---|---|
| 3 | let-7a, let-7b, let-7c, | 8/33(22.24%) | 0.02641 |
| let-7d, let-7e, let-7f, | |||
| miR-221,miR-29a | |||
| 4 | let-7a, let-7b, let-7c,let-7d, | 10/18(55.56%) | 6.68E-06 |
| let-7e, let-7f, let-7g, let-7i, | |||
| miR-21,miR-221 | |||
| 5 | let-7b, let-7c, let-7d, | 9/17(52.94%) | 3.56E-05 |
| let-7i, miR-200b, miR-200c, | |||
| miR-29a, miR-29b, miR-30c | |||
| 6 | let-7a, let-7b, let-7c, | 8/16(50.00%) | 1.76E-04 |
| let-7d, let-7i, miR-103, | |||
| miR-21, miR-221 | |||
| 15 | let-7a, let-7c,let-7f, | 9/17(52.94%) | 3.56E-05 |
| let-7g, miR-141, miR-19b, | |||
| miR-21, miR-200a,miR-200b | |||
| 19 | let-7a, let-7b, let-7c, | 11/18(61.11%) | 5.45E-07 |
| let-7d, let-7e, let-7f, | |||
| miR-143, miR-145, | |||
| miR-21, miR-29a, miR-29b |
The comparison shows that significant numbers of miRNAs identified in the FMRMs are relevant to cancers. From the literature, 42 miRNAs have been validated as either oncogenes or tumor suppressors (details in Supplementary Material). Among the 334 miRNAs under investigation, a significant number of miRNAs in identified modules are supported by the current knowledge. The coverage is the percentage of the number of miRNAs in each module supported by literature. P-value is calculated by a hypergeometric probability density function at each of the numbers of miRNAs supported by the literature, using the corresponding size of the total miRNAs under investigation (334), numbers of miRNAs in each module and numbers of miRNAs identified from the literature (42). The modules with significant supports are given in this table.
It is worth noting that several miRNAs, such as the let-7 family and miR-21, are identified in multiple modules, suggesting they could be involved in multiple biological processes. The frequent occurrence of these particular miRNAs is consistent with their known strong association with multiple cancer types, including breast cancers. The identification of multiple modules containing different but overlapping sets of miRNAs is likely to be the consequence of activation of distinct subsets of common gene interaction networks in specific cancer subtypes. For example, Blenkiron et al. (2007) identified 31 miRNAs differentially expressed between basal and luminal tumors. Among them, let-7a, b and f are underexpressed in basal tumors but overexpressed in luminal tumors. These miRNAs were identified in module 3, 4, 5 and 6 using our method and show patterns that are consistent with their reported involvement in these tumors.
3.5 Functional validation of miRNA target genes
It is expected that the miRNA target genes are also relevant to the specific biological processes. To validate that the identified mRNAs are relevant to basal and luminal tumors, firstly we compared the identified mRNAs with a work conducted by Adelaide et al. (2007). Their results suggest the existence of potential oncogenes and tumor suppressor genes differentially associated with the basal and luminal subtype. As their results are largely consistent with many previous researches (Bergamaschi et al., 2006; Chin et al., 2006; Neve et al., 2006), we validate our analysis based on their results.
In our results, 18 genes have been identified by Adelaide et al. (2007) as in Supplementary Table S1. Among these genes, Ccdc77 identified in module-3 also is targeted by miR-29a and miR-221, Hspa14 identified in module-4 is targeted by miR-21 , and Cox4i1 identified in module-19 is targeted by let-7c and let-7e. It further confirms that let-7e, miR-21, miR-29a and miR-221 may have important regulatory functions towards basal and luminal tumors. In addition, Rbm4b identified in module-3 is targeted by miR-697 and miR-700, Rbx1 identified in module-5 is targeted by miR-709, Gspt1 identified in module-11 is targeted by miR-669c and miR-710 and Cox4i1 identified in module-19 is targeted by miR-709. It suggests that miR-669c, miR-697, miR-709 and miR-710 may also play important roles in regulating basal and luminal tumors. It is worth noting that many previously reported results were not recovered in the current study because the investigated data were profiled with mouse model while the results of Adelaide et al. (2007) were produced on breast cancer samples of humans.
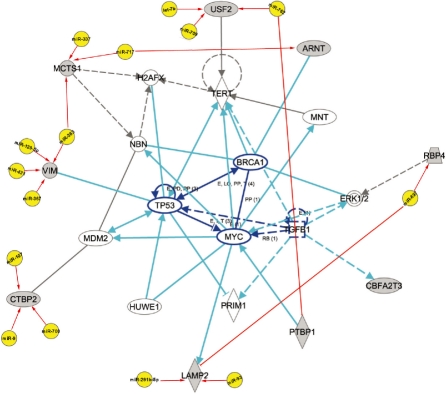
Furthermore, we have queried the mRNAs identified in each module against the IPA Database. We specifically focused on human species as we are interested in the networks of human cancers. The networks participated by the mRNAs identified in FMRMs are highly associated with cancers. Many genes are directly related to cancers and genetic disorders. They are co-targeted by a group of miRNAs identified from our method, suggesting the identified miRNAs and their target mRNAs have implications for cancers. For example, a network participated by the miRNAs and mRNAs identified by our method are associated with cancer, cellular compromise, DNA replication and repair (Fig. 3). The networks which are explicitly associated with cancers and within the top five networks of each module are given in Supplementary Table S2. The detailed networks are also given in Supplementary Figure S2–S6. The identified genes of FMRMs, which are relevant to cancers, are given in Table 4. The results indicate that our methods effectively identified many cancer-related genes. Those genes are targeted by a group of miRNAs, suggesting that those miRNAs also participate in the networks of cancers.
Fig. 3.
A network with the function of cancer, cellular compromise, DNA replication and repair. It is participated by a group of miRNAs and their target mRNAs identified by our method, suggesting that these miRNAs and their target mRNAs have the function of cancers.
Table 4.
Cancer-associated genes of FMRMs
| FMRM# | mRNAs | mRNA# | P-value (adj.) |
|---|---|---|---|
| 3 | CALR, RBP4, VIM, NDUFV2, | 12 | 4.89E-03 - |
| SDCBP, MCTS1, AP2S1, PRPF8, | 2.54E-02 | ||
| COL18A1, AK2, ARNT, RPS15 | |||
| 4 | DNMT1, NF2, RRM2 | 3 | 2.13E-03 - |
| 3.05E-02 | |||
| 5 | CEBPB, DDX39, HSP90AB1, | 8 | 2.01E-03 - |
| MT2A, NUP62, SQLE, TCP1, TRIO | 4.84E-02 | ||
| 11 | DICER1, ENO1, HSP90B1, | 5 | 5.26E-03 - |
| RXRB, SPRY2 | 4.84E-02 | ||
| 13 | IGF2R, LSM14B, NCOR2, SP110, | 14 | 6.88E-03 - |
| STX5, TOR2A, ACHE, HDAC3, | 4.56E-02 | ||
| PARP1, POSTN, SMAD4, UBE2I, | |||
| RNF6, BAK1 |
Many genes identified in FMRMs are relevant to cancers. Genes identified in FMRMs are directly assigned to diseases and disorders. The cancer-related genes of FMRMs within their top five biofunctions are listed.
4 CONCLUSION AND DISCUSSION
miRNAs have been regarded as one of the most important regulators. Identifying their functions and regulatory mechanisms is critical in understanding biological processes of organisms. Great efforts, in both biological experiments and computational methods, have been made to illustrate their functions. However, the precise regulatory functions of most miRNAs remain elusive due to the complexity of the regulatory mechanisms.
In this article, we have presented a model to discover functional miRNA regulatory modules (FMRMs), which are groups of miRNAs and mRNAs for specific biological conditions. This model is inspired by the Corr-LDA, which has been used to extract the correspondence patterns from heterogeneous data. We modified Corr-LDA and derived the solution for FMRM discovery.
Our method models FMRMs with a generative process. It makes use of the expression profiles of miRNAs and mRNAs, with or without using the target relationships between miRNAs and mRNAs based on the sequence binding information. It simultaneously identifies groups of interactive miRNAs and mRNAs, which are believed to participate in specific biological functions.
We have applied this method to a mouse model dataset for human breast cancer research. The method has effectively identified several modules related to breast cancer subtypes: basal and luminal. Since the datasets used were profiled from mouse tissues, many genes have been filtered out because we focus on human genes. Thus, previously reported results were not fully recovered in this work. However, a large proportion of miRNAs and mRNAs identified in the modules have been reported to have associations with basal and luminal subtypes. Many others have direct indications on cancers and genetic disorders. Furthermore, many novel associations among miRNAs, mRNAs and biological processes have been predicted by our model. Several miRNAs and mRNAs are highly related to cancers as reported by previous works, suggesting those modules may have roles in the corresponding development processes.
Our model allows discovering the FMRMs with or without using the target relationship between miRNAs and mRNAs. Some researchers have suggested that algorithms that do not consider known targets may avoid biases (Bartel, 2009; Lewis et al., 2003, 2005). Bonnet et al. (2010) also showed that expression profiles only can be used to infer miRNA regulatory networks. Our method provides the flexibility of inferring FMRMs with or without target relationships of miRNAs and mRNAs. We have demonstrated this model without using the prior target prediction. The results suggest that expression profiles of miRNAs and mRNAs are crucial for both target identification and regulatory module discovery.
Supplementary Material
ACKNOWLEDGEMENT
We thank Drs Chuxia Deng, Daniel Medina, Yi Li, Ming Yi and Robert Stephens for providing some tumor samples of the datasets.
Funding: Intramural Program, Center for Cancer Research, NIH, Bethesda, MD, USA (in part).
Conflict of Interest: none declared.
REFERENCES
- Adelaide J, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- Akao Y, et al. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bergamaschi A, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes, Chromosomes and Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- Blei DM, Jordan MI. SIGIR '03: Proceedings of the 26th annual international ACM SIGIR conference on Research and development in informaion retrieval. New York, NY, USA: ACM; 2003. Modeling annotated data; pp. 127–134. [Google Scholar]
- Blenkiron C, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, et al. Module network inference from a cancer gene expression dataset identifies microRNA regulated modules. PLoS ONE. 2010;5:e10162. doi: 10.1371/journal.pone.0010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai KV, et al. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc. Natl Acad. Sci. USA. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. Beginning to understand microRNA function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, et al. mirbase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Suppl. 1):D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzigeorgiou AG. Same computational analysis, different miRNA target predictions. Nature Methods. 2007;4:191. doi: 10.1038/nmeth0307-191a. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Herschkowitz J, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, et al. Using expression profiling data to identify human microRNA targets. Nat. Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Joung J-G, Fei Z. Identification of microRNA regulatory modules in arabidopsis via a probabilistic graphical model. Bioinformatics. 2009;25:387–393. doi: 10.1093/bioinformatics/btn626. [DOI] [PubMed] [Google Scholar]
- Joung J-G, et al. Discovery of microRNA-mRNA modules via population-based probabilistic learning. Bioinformatics. 2007;23:1141–1147. doi: 10.1093/bioinformatics/btm045. [DOI] [PubMed] [Google Scholar]
- Krek A, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lewis BP, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liu JS. The collapsed gibbs sampler in bayesian computations with applications to a gene regulation problem. J. Am. Stat. Assoc. 1994;89:930–958. [Google Scholar]
- Liu B, et al. Discovery of functional miRNA-mRNA regulatory modules with computational methods. J. Biomed. Infor. 2009a;42:685–691. doi: 10.1016/j.jbi.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Liu B, et al. Exploring complex miRNA-mRNA interactions with bayesian networks by splitting-averaging strategy. BMC Bioinformatics. 2009b;10:408. doi: 10.1186/1471-2105-10-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, et al. Computational identification of hepatitis c virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics. 2009;10:373. doi: 10.1186/1471-2164-10-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka KP, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Steyvers M, et al. KDD '04: Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining. New York, NY, USA: ACM; 2004. Probabilistic author-topic models for information discovery; pp. 306–315. [Google Scholar]
- Tran D, et al. Finding microRNA regulatory modules in human genome using rule induction. BMC Bioinformatics. 2008;9(Suppl. 12):S5. doi: 10.1186/1471-2105-9-S12-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yang H, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- Yoon S, De Micheli G. Prediction of regulatory modules comprising microRNAs and target genes. Bioinformatics. 2005;21(suppl. 2):ii93–ii100. doi: 10.1093/bioinformatics/bti1116. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem. Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Zhu M, et al. Comprehensive genomic profiling identifies miRNA signatures associated with mouse mammary tumor subtypes. In preparation. 2010 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.