Abstract
In Saccharomyces cerevisiae, association between the Est1 telomerase subunit and the telomere-binding protein Cdc13 is essential for telomerase to be recruited to its site of action. A current model proposes that Tel1 binding to telomeres marks them for elongation, as the result of phosphorylation of a proposed S/TQ cluster in the telomerase recruitment domain of Cdc13. However, three observations presented here argue against one key aspect of this model. First, the pattern of Cdc13 phosphatase-sensitive isoforms is not altered by loss of Tel1 function or by mutations introduced into two conserved serines (S249 and S255) in the Cdc13 recruitment domain. Second, an interaction between Cdc13 and Est1, as monitored by a two-hybrid assay, is dependent on S255 but Tel1-independent. Finally, a derivative of Cdc13, cdc13–(S/TQ)11→(S/TA)11, in which every potential consensus phosphorylation site for Tel1 has been eliminated, confers nearly wild-type telomere length. These results are inconsistent with a model in which the Cdc13–Est1 interaction is regulated by Tel1-mediated phosphorylation of the Cdc13 telomerase recruitment domain. We propose an alternative model for the role of Tel1 in telomere homeostasis, which is based on the assumption that Tel1 performs the same molecular task at double-strand breaks (DSBs) and chromosome termini.
TELOMERE length homeostasis is a highly regulated process that must balance telomere loss (as the result of incomplete replication and/or nucleolytic degradation) with telomeric repeat addition (through the action of telomerase and/or recombination). In the budding yeast Saccharomyces cerevisiae, a key regulatory event is recruitment of telomerase to chromosome ends by the telomere end-binding protein Cdc13 (Nugent et al. 1996; Evans and Lundblad 1999; Pennock et al. 2001; Bianchi et al. 2004; Chan et al. 2008). Recruitment relies on a direct interaction between Cdc13 and the Est1 subunit of telomerase (Pennock et al. 2001), which brings the catalytic core of the enzyme to its site of action. Disruption of this interaction, due to mutations in either CDC13 (cdc13-2) or EST1 (est1-60), results in an Est (ever-shorter-telomere) phenotype, as manifested by progressive telomere shortening and an eventual senescence phenotype. The recruitment activity of Cdc13, which resides in a 15-kDa N-terminal domain (Pennock et al. 2001), is sufficient to direct telomerase even to nontelomeric sites (Bianchi et al. 2004). As predicted by the recruitment model, association of telomerase with telomeres is greatly reduced in strains expressing the recruitment-defective cdc13-2 allele (Chan et al. 2008).
Telomerase action at individual telomeres is highly regulated. Using an assay that monitors telomere addition at single nucleotide resolution (single telomere extension, STEX), Lingner and colleagues showed that only ∼7% of telomeres with wild-type (i.e., 300 bp) length are elongated by telomerase during a single cell cycle (Teixeira et al. 2004). However, as telomere length declines, the extension frequency increases: ∼20% of telomeres 200 bp in length and >40% of 100-bp-long telomeres are elongated (Teixeira et al. 2004; Arneric and Lingner 2007). The mechanism by which telomerase distinguishes short from long telomeres has been the subject of intense investigation. Work from a number of laboratories has led to the proposal that Tel1-dependent phosphorylation of Cdc13 at underelongated telomeres mediates the interaction between Cdc13 and the telomerase-associated Est1 protein, thus ensuring that telomerase is directed to the shortest telomeres in a population. In support of this model, the Est1 and Est2 telomerase subunits exhibit enhanced association with telomeres that have been artificially shortened, whereas Cdc13 displays length-independent association with telomeres (Bianchi and Shore 2007; Sabourin et al. 2007). This suggests that the preferential elongation of shorter telomeres is controlled at the level of recruitment of the telomerase holoenzyme by Cdc13. Furthermore, efficient association of Est1 and Est2 with chromosome ends requires Tel1 and Mre11 (which acts in the same pathway as Tel1 for telomere length regulation; Nugent et al. 1998; Ritchie and Petes 2000) but not Mec1 (Takata et al. 2005; Goudsouzian et al. 2006). Tel1 itself is also telomere bound (Takata et al. 2004), with enhanced binding to shorter telomeres (Bianchi and Shore 2007; Hector et al. 2007; Sabourin et al. 2007; Abdallah et al. 2009), although there is considerable controversy over the degree and timing of Tel1 association with chromosome termini during the cell cycle. As expected for a key regulator of the interaction between Cdc13 and a telomerase subunit, a tel1-Δ strain has short telomeres (Lustig and Petes 1986), although telomere length is not impaired enough to confer the Est phenotype displayed by cdc13-2 and est1-60 strains.
Implicit in the above proposal is that Cdc13 must be a direct substrate of Tel1. In support of this, Teng and colleagues reported several years ago that the recruitment domain of Cdc13 has a cluster of potential Tel1 (and/or Mec1) phosphorylation sites (Tseng et al. 2006). Substrates of the DNA damage kinases often contain several closely spaced phosphorylation sites, termed S/TQ cluster domains (SCDs), which are considered a structural hallmark of many DNA damage-response proteins (Traven and Heierhorst 2005). On the basis of in vitro kinase assays with GST fusions to 75- to 90-amino-acid portions of the Cdc13 recruitment domain, Tseng et al. 2006 concluded that four SQ sites in the recruitment domain of Cdc13 are overlapping substrates for both Tel1 and Mec1, leading to the proposal that telomerase recruitment in S. cerevisiae is regulated by Tel1-dependent phosphorylation of Cdc13.
The above model makes a key prediction: in a tel1-Δ strain, telomerase should no longer exhibit a length-dependent pattern of elongation. However, preferential elongation of short telomeres still occurs at native chromosome ends in the absence of Tel1 (Arneric and Lingner 2007). In addition, Petes and colleagues have argued, on the basis of epistasis data, that Tel1 performs an indirect role in the telomerase pathway, rather than directly targeting a telomerase regulator (Ritchie et al. 1999; Ritchie and Petes 2000). These observations are not easily explained, if preferential recognition of short telomeres by telomerase is mediated by Tel1-dependent phosphorylation of Cdc13. In this current study, we have re-examined the evidence for phosphorylation of Cdc13 as a regulatory mechanism for telomere length homeostasis. We report on a series of observations that indicate that Tel1 contributes to telomere length control through a mechanism other than phosphorylation of the Cdc13 S/TQ cluster.
MATERIALS AND METHODS
Strains and plasmids:
The yeast strains and plasmids used in this study are shown in Tables 1 and 2, respectively. All plasmids containing missense mutations were generated by QuickChange mutagenesis, and a sequenced restriction fragment was subcloned back into an unmutagenized backbone, to eliminate the possibility of unlinked mutations. The diploid strains used in Figure 7 were constructed by introducing the relevant mutation into a haploid strain, using the URA3 integrating constructs listed in Table 2, and selecting for “pop-outs” on 5-FOA–containing media. The genotype of the CDC13 or TEL2 locus in the resulting 5-FOA–resistant strains was determined by molecular diagnostics as well as by telomere length, and mutant haploids were subsequently mated to an isogenic tlc1-Δ haploid. The resulting heterozygous diploid strains were completely isogenic, with the exception of the relevant loci (TLC1, CDC13, TEL1, or TEL2).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| YVL2540 | MATacdc13-Δ∷LYS2 ura3-52lys2-801trp1-Δ1 his3-Δ200 leu2-Δ1 ade2-101/pVL438 |
| YVL3006 | MATαcdc13-Δ∷LYS2 ura3-52lys2-801trp1-Δ1 his3-Δ200 leu2-Δ1/pVL438 |
| YVL2985a | MATa/αtlc1-Δ∷HIS3/TLC1 tel2-1/TEL2 |
| YVL2993a | MATa/αtlc1-Δ∷HIS3/TLC1 cdc13-2/CDC13 |
| YVL3007a | MATa/αtlc1-Δ∷HIS3/TLC1 cdc13-S249A,S255A/CDC13 |
| YVL3009a | MATa/αtlc1-Δ∷HIS3/TLC1 cdc13-S255A/CDC13 |
| YVL3015a | MATa/αtlc1-Δ∷HIS3/TLC1 tel1-Δ∷KANMX6/TEL1 |
| YAB208 | mat-Δ ade2-Δ LEU2∷GAL-HO lys2-Δ mnt2-Δ∷LYS2 adh4-Δ∷IM-3 rad52-Δ∷SpHIS5 (ADE2-4XUASg-HO site-URA3) |
| PJ69-4A | MATatrp1-901 leu2-3,112 ura3-52his3-200 gal4-Δ gal80-Δ GAL2-ADE2 LYS2∷GAL1-HIS3 met2∷GAL7-lacZ |
| YVL2929 | MATatrp1-901 leu2-3,112 ura3-52his3-200 gal4-Δ gal80-Δ GAL2-ADE2 LYS2∷GAL1-HIS3 met2∷GAL7-lacZ tel1-Δ∷KANMX6 |
| YVL3052 | MATaleu2 trp1 ura3-52 prb− prc− pep4-3 sml1-Δ∷NAT |
| YVL3053 | MATaleu2 trp1 ura3-52 prb− prc− pep4-3 sml1-Δ∷NAT mec1-Δ∷TRP1 |
| YVL3054 | MATaleu2 trp1 ura3-52 prb− prc− pep4-3 sml1-Δ∷NAT tel1-Δ∷KANMX6 |
Additional genotype: ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pVL438 | CEN URA3 CDC13 | Evans and Lundblad (1999) |
| pVL648 | CEN LEU2 CDC13 | Evans and Lundblad (1999) |
| pVL1084 | CEN LEU2 CDC13 | Pennock et al. (2001) |
| pVL690 | CEN LEU2 cdc13–E252K (cdc13-2) | Pennock et al. (2001) |
| pVL1519 | CEN LEU2 cdc13–S225A | This work |
| pVL3512 | CEN LEU2 cdc13–P235A | This work |
| pVL3516 | CEN LEU2 cdc13–F237A | This work |
| pVL1521 | CEN LEU2 cdc13–S249A | This work |
| pVL2570 | CEN LEU2 cdc13–S249A | This work |
| pVL3518 | CEN LEU2 cdc13–Q250A | This work |
| pVL3513 | CEN LEU2 cdc13–N251A | This work |
| pVL1727 | CEN LEU2 cdc13–E252A | This work |
| pVL3515 | CEN LEU2 cdc13–F253A | This work |
| pVL3517 | CEN LEU2 cdc13–N254A | This work |
| pVL2571 | CEN LEU2 cdc13–S255A | This work |
| pVL4351 | CEN LEU2 cdc13–Q256A | This work |
| pVL3335 | CEN LEU2 cdc13–S306A | This work |
| pVL4140 | CEN LEU2 cdc13–4Q→ 4Aa | This work |
| pVL5005 | CEN LEU2 cdc13–11Q→ 11Ab | This work |
| pVL2691 | URA3 cdc13-2 (integrating vector) | This work |
| pVL2619 | URA3 cdc13–S255A (integrating vector) | This work |
| pVL2693 | URA3 cdc13–S249A, S255A (integrating vector) | This work |
| pVL2859 | URA3 tel2-1 (integrating vector) | This work |
| pVL3277 | 2μ LEU2 CDC13-(myc)18 | This work |
| pVL2640 | 2μ LEU2 cdc13–S249A, S255A-(myc)18 | This work |
| pAB301 | CEN TRP1 ADH–GBD–CDC13–RDaa211-331 | Bianchi et al. (2004) |
| pAB313 | CEN TRP1 ADH–GBD–CDC13–RDaa211-331–E252K | Bianchi et al. (2004) |
| pVL3068 | CEN TRP1 ADH–GBD–CDC13–RDaa211-331–S255A | This work |
| pVL3070 | CEN TRP1 ADH–GBD–CDC13–RDaa211-331–S249A | This work |
| pVL693 | 2μ LEU2 ADH–GAL4–AD–EST1 | This work |
| pVL859 | 2μ LEU2 ADH–GAL4–AD–STN1aa5-494 | Chandra et al. (2001) |
| pVL3125 | 2μ TRP1 ADH–GAL4–DBD–CDC13R635A | Gao et al. (2007) |
| pVL3346 | 2μ TRP1 ADH–GAL4–DBD–CDC13R635A E252K | This work |
| pVL3345 | 2μ TRP1 ADH–GAL4–DBD–CDC13R635A S255A | This work |
| pVL3484 | 2μ TRP1 ADH–GAL4–DBD–CDC13R635A S249A | This work |
| pVL3527 | 2μ TRP1 ADH–GAL4–DBD–CDC13R635A P235S | This work |
| pVL3524 | 2μ TRP1 ADH–GAL4–DBD–CDC13R635A F237V | This work |
| pVL3525 | 2μ TRP1 ADH–GAL4–DBD–CDC13R635A F253A | This work |
| pVL3526 | 2μ TRP1 ADH–GAL4–DBD–CDC13R635A N254A | This work |
4Q→4A: cdc13–Q226A, Q250A, Q256A, Q307A.
11Q→11A: cdc13–Q66A, Q226A, Q250A, Q256A, Q307A, Q391A, Q476A, Q612A, Q644A, Q653A, Q828A.
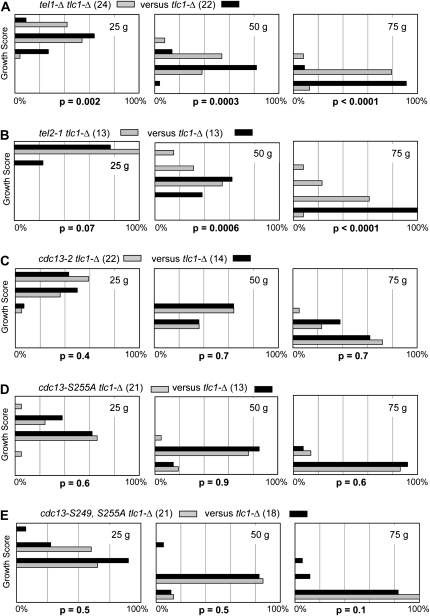
Figure 7.—
(A–E) Epistasis analysis of tlc1-Δ strains, with or without mutations in the Tel1/Tel2 pathway or the Cdc13 recruitment domain. For each epistasis test, a heterozygous diploid strain was freshly dissected and tlc1-Δ isolates were streaked for three successive streak-outs (corresponding to ∼25, ∼50, and ∼75 generations of growth); numbers in parentheses represent the number of isolates of each genotype analyzed. Streak-outs were incubated for 48 hr at 30°, followed by a genotype-blind assessment of the growth phenotype. Each histogram displays the percentage of isolates (x-axis) that fell into five phenotypic categories (y-axis), ranging from a scale of 1 (severe senescence) to 5 (comparable to wild-type growth); see Figure S6 for examples of each of these five categories, as well as an evaluation of the validity of the analysis used to evaluate the statistical significance. The cdc13–S249A, S255A epistasis test was only performed once; all other epistasis tests were performed at least twice, with similar results.
Genetic methods:
Telomere length and senescence phenotypes were analyzed following dissection of strains bearing mutations integrated into the genome or as plasmid-borne mutations transformed into a cdc13-Δ/p URA3CDC13 shuffle strain. In the latter case, transformants were streaked for single colonies on 5-FOA–containing media to identify isolates that had lost the covering CDC13 plasmid and subsequently monitored for the appearance of a senescence phenotype on successive streak-outs, as described previously (Lundblad and Szostak 1989; Lendvay et al. 1996). For the epistasis analysis shown in Figure 7, senescence was assayed through a genotype-blind assessment of the growth characteristics of multiple isolates of a tlc1-Δ strain, compared to multiple isolates of a tlc1-Δ strain bearing a second mutation (Rizki and Lundblad 2001). Following dissection of the relevant diploid, haploid spores containing the tlc1-Δ mutation were identified and subsequently streaked for single colonies three successive times. After 48 hr incubation at 30°, growth was scored on a scale of 1 (severe senescence) to 5 (comparable to wild-type growth), and the CDC13, TEL1, or TEL2 genotype of each haploid strain was determined at the conclusion of the experiment. Data for each genotype at each of the three time points were displayed on a histogram, and a Student's t-test was used to assess the statistical significance between genotypes at each time point.
De novo telomere assays were performed as described previously (Bianchi et al. 2004), with several slight modifications: strains expressing the GBD–Cdc13–RD protein fusions were grown in liquid selective media containing 2% raffinose, galactose was added to a final concentration of 3% for 3.5 hr, and serial dilutions were plated and allowed to grow for 2 days to form visible colonies. Colonies were replica plated onto plates containing 5-FOA or α-aminoadipate to detect loss of the distal URA3 or LYS2 genes, respectively.
Biochemical analysis:
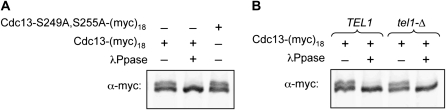
Protease-deficient yeast strains (either TEL1 or tel1-Δ) expressing Ccd13-(myc)18 were grown in selective media to OD600 = 0.8. Washed pellets were resuspended in cold TMG200 (10 mm Tris–HCl pH 8.0, 1 mm MgCl2, 10% glycerol, 200 mm NaCl), lysed using a mortar and pestle in the presence of liquid N2, and extracts were clarified by spinning two times at 14,000 rpm for 20 min. For phosphatase-treated samples, 80 μg of protein lysate was incubated at 30° for 1 hr with 80 units of λ-phosphatase (NEB) in 50 mm HEPES pH 7.5, 100 mm NaCl, 2 mm DTT, 0.01% Brij 35, 1 mm MgCl2 and clarified by spinning at 14,000 rpm for 10 min. Samples were resolved on 6% PAGE (30:0.39 acrylamide:bis) and subjected to Western analysis with 9E10 anti-myc antibody.
RESULTS
Analysis of mutations in two conserved serines in the recruitment domain of Cdc13:
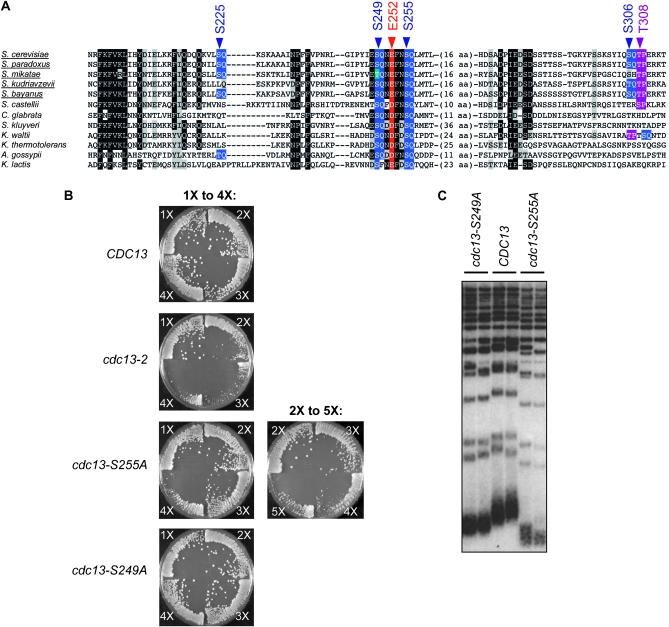
The S. cerevisiae Cdc13 protein contains 11 potential consensus phosphorylation sites for the Tel1 and/or Mec1 kinases; four of these motifs (S225Q, S249Q, S255Q, and S306Q) reside in the telomerase recruitment domain of Cdc13 and have been proposed to form an S/TQ cluster domain (Tseng et al. 2006). To evaluate the potential role of phosphorylation in recruitment of telomerase by Cdc13, we constructed an alignment of full-length Cdc13 proteins from a dozen fungal species belonging to the Saccharomyces clade. Examination of this alignment revealed that only 2 of the 11 S/TQ motifs (S249Q and S255Q) in the S. cerevisiae Cdc13 protein were highly conserved (Figure 1A and data not shown). The S255Q motif was invariant among the entire collection of 12 species, and the S249Q motif was similarly conserved (although not invariant). In contrast, the remaining 9 SQ or TQ motifs were not conserved, including the 2 other members of the proposed S/TQ cluster domain (S225Q and S306Q), even among species within the Saccharomyces sensu stricto group (Figure 1A).
Figure 1.—
Phenotypic analysis of cdc13–S249A and cdc13–S255A strains. (A) Alignment of the recruitment domain of Cdc13, with the sensu stricto species underlined; amino acids 199 to 314 are shown for the S. cerevisiae protein. Potential SQ phosphorylation sites are indicted in blue, a Cdk1-dependent phosphorylation site (Li et al. 2009) is highlighted in purple, and the residue mutated in the recruitment-defective cdc13-2 allele is indicated in red. (B) Isogenic diploid cdc13-2/CDC13, cdc13–S255A/CDC13 and cdc13–S249A/CDC13 strains were dissected in parallel, and senescence of the indicated haploid strains was assessed as described in materials and methods; 10–12 isolates were examined for each genotype and representative examples are shown (see Figure S1 for additional examples). Plates were photographed after 3 days incubation at 30°; each quadrant corresponds to ∼25 generations of growth, such that 1X = ∼25 generations and 4X = ∼100 generations. (C) Telomere length of the haploid mutant strains generated in B was examined after ∼50 generations of growth. The phenotypic analysis of the severity of the cdc13–S249A and cdc13–S255A defects shown in B and C differs in several aspects from a previous report (Tseng et al. 2006). In particular, this group did not observe a senescence phenotype associated with the cdc13–S255A strain, and they also reported that the cdc13–S249A and cdc13–S255A mutant strains exhibited similar telomere lengths. In the results shown here, strains in which the mutations had been integrated into the genome were analyzed, thereby alleviating possible problems with fluctuations in plasmid copy number. We also examined growth and telomere length phenotypes for up to 100 generations (see also Figure S1). These protocol differences presumably explain the difference in the results obtained in this study vs. those reported previously.
The two potential phosphorylation sites that were highly conserved (S249Q and S255Q) bracket residue E252 (Figure 1A), which has been previously shown to be essential for the interaction between Cdc13 and Est1 to mediate recruitment of telomerase to chromosome ends (Evans and Lundblad 1999; Pennock et al. 2001; Bianchi et al. 2004). We therefore compared alleles of CDC13, which replaced either S255 or S249 with alanine for their effects on telomere replication with the previously well-characterized cdc13-2 mutation (bearing a charge swap mutation at residue E252). The cdc13–S255A strain displayed a pronounced telomere replication defect, as evidenced by a senescence phenotype (although the appearance of senescence was delayed, relative to the cdc13-2 strain) and extremely short telomeres (Figure 1, B and C and supporting information, Figure S1). Furthermore, continued propagation of the cdc13–S255A strain gave rise to telomeric rearrangements (Figure S1), which are characteristic of a recombination-based pathway for telomere maintenance that can be employed in the absence of telomerase. In contrast, the cdc13–S249A strain displayed only a modest decline in telomere length, with no accompanying senescence (Figure 1, B and C), indicating that residue S249 makes a more minimal contribution to telomere length regulation.
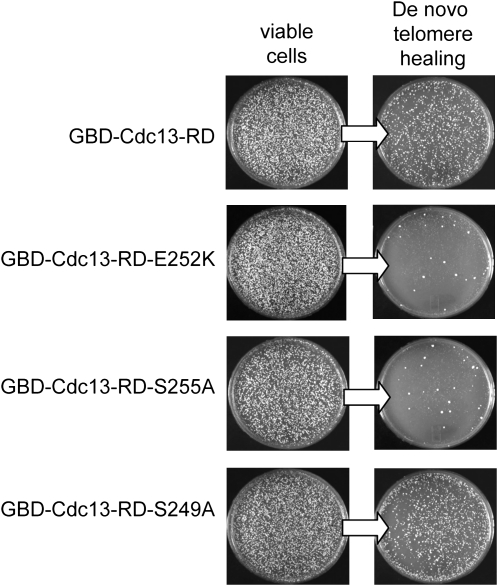
The effects of mutations in S249 and S255 were also examined in a de novo telomere healing assay in which telomerase is ectopically recruited to a nontelomeric DNA end (Bianchi et al. 2004). In this assay, the recruitment domain of Cdc13 (Cdc13–RD) is placed adjacent to a newly created double-strand break (DSB), as a GBD–Cdc13–RD fusion protein, which is capable of binding to a Gal4 DNA binding site located next to an HO-induced DSB. Telomere healing was detected by monitoring the loss of either of two genes, URA3 or LYS2, which are distal to the HO-induced break. As shown in Figure 2, the GBD–Cdc13–RD–S255A fusion was incapable of mediating telomere healing following induction of a DSB. In cells expressing this fusion, very few colonies were able to grow following HO induction on media that detected loss of the distal URA3 or LYS2 genes (Figure 2 and data not shown), compared to the efficient healing observed with the wild-type GBD–Cdc13–RD fusion. As expected, de novo telomere formation was also defective in the presence of the GBD–Cdc13–RD–E252K fusion (referred to as GBD–Cdc13–RDest in Bianchi et al. 2004). In contrast, telomere healing occurred at roughly wild-type levels with a GBD–Cdc13–RD–S249A fusion (Figure 2), which parallels the minimal effect that this allele had on telomere function at native chromosome termini.
Figure 2.—
Residue S255, but not S249, is required in a de novo telomere addition assay. Strains expressing the indicated GBD–Cdc13–RD protein fusions were grown in selective media containing 2% raffinose, exposed to 3% galactose for 3.5 hr to induce cleavage by the HO endonuclease to generate a double-strand break adjacent to four copies of the Gal4 DNA binding site, and aliquots were plated for single colonies. Plates were incubated for 3 days at 30° to allow colony growth and subsequently replica plated onto media that only allows growth of viable Ura− cells, which requires healing of the HO-induced break by conversion into a functional telomere; the Ura− phenotype is due to loss of the URA3-containing distal portion of the chromosome.
A two-hybrid interaction between Cdc13 and Est1 is dependent on the recruitment domain of Cdc13, but does not require Tel1:
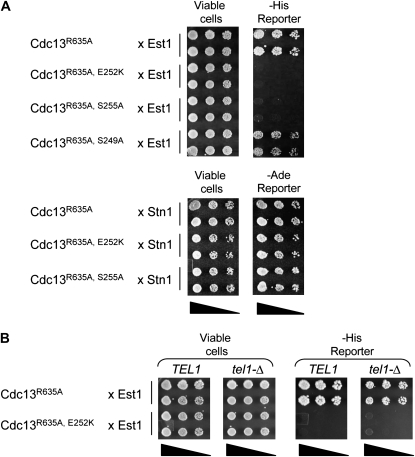
The above analysis demonstrates that residue S255, like E252, is crucial for telomere replication, and the proximity of these two residues suggests that S255 also mediates telomerase recruitment by contributing to the Cdc13–Est1 interaction. Since the in vivo experiments shown in Figures 1 and 2 do not directly test this interaction, we turned to an alternative means of examining whether the association between Est1 and the mutant Cdc13–S255A protein was impaired. In previous efforts by our lab and others, it has not been possible to detect an interaction between Cdc13 and Est1 using either coimmunoprecipitation from yeast extracts under endogenous expression conditions or with purified proteins in vitro. This precluded a direct biochemical examination of the Cdc13–S255A mutant protein. Therefore, we developed a two-hybrid assay to probe the Cdc13–Est1 interaction, using the full-length Cdc13 and Est1 proteins fused to Gal4DBD or Gal4AD, respectively (see Figure S2 for a comparison of these reagents with a previously published set of Cdc13 and Est1 two-hybrid constructs used by Qi and Zakian 2000). In this assay, a weak interaction between Cdc13 and Est1 could be observed, as exhibited by the ability to activate the His reporter but not the more stringent Ade reporter (Figure 3 and data not shown). As predicted, the Cdc13–Est1 association was disrupted by the cdc13-2 (E252K) and est1-60 mutations, arguing that this assay was probing a biologically valid interaction (Figure 3A and data not shown). The Gal4DBD–Cdc13–S255A construct was also defective for interaction with Est1, indicating that the telomere replication defect of the cdc13–S255A strain was due to a reduced interaction with telomerase. In contrast, the cdc13–S249A mutation, which only made a minor contribution to telomere length (Figure 1), had little effect in this two-hybrid test. Neither the cdc13-2 nor the cdc13–S255A mutations altered the interaction with Stn1 in this assay (Figure 3A), indicating that the inability to interact with Est1 was not due to nonspecific destabilization of the Gal4DBD–Cdc13–E252K or Gal4DBD–Cdc13–S255A constructs. The correlation between in vivo telomere length maintenance and a two-hybrid interaction between Cdc13 and Est1 was further supported by the behavior of additional mutations in the recruitment domain of Cdc13, as discussed in a later section.
Figure 3.—
A two-hybrid interaction between Cdc13 and Est1 is dependent on S255 but not on Tel1. (A) Two-hybrid analysis of the interaction between Cdc13–R635A (wild-type or mutant versions of pVL3125) and either Est1 (top) or Stn1 (bottom); dilutions of the S. cerevisiae strain pJ69-4A, coexpressing Gal4DBD–Cdc13 and either Gal4AD–Est1 or Gal4AD–Stn1, were plated on appropriate selective plates to monitor viability and the ability to activate either the GAL1–HIS3 or GAL2–ADE2 reporter genes, respectively. The Gal4AD–Est1 fusion protein complemented an est1-Δ strain (data not shown). The R635A mutation in the DNA binding domain of Cdc13, which eliminates DNA binding (Anderson et al. 2003), was necessary to alleviate the lethality due to overexpression of Cdc13. (B) Two-hybrid analysis of the interaction between Gal4DBD–Cdc13 or Gal4DBD–Cdc13-2 (E252K) and Gal4AD–Est1 in pJ69-4A vs. an isogenic tel1-Δ derivative of pJ69-4A, performed as in A.
This two-hybrid assay also provided a means of evaluating the potential role of Tel1 in mediating the interaction between Cdc13 and Est1, by comparing the behavior of TEL1 and tel1-Δ strains in this test. As shown in Figure 3B, an interaction between Cdc13 and Est1 could still be observed in a tel1-Δ strain. Furthermore, this Cdc13–Est1 interaction was still mediated by the Cdc13 recruitment domain, as it was abolished by the cdc13-2 mutation, similar to the behavior in the TEL1 strain. These results indicate that the Cdc13–Est1 two-hybrid interaction was dependent on the serine residue at position 255, but not dependent on Tel1 function.
We also attempted to ask whether this two-hybrid interaction could be maintained in the absence of both Tel1 and Mec1. Unexpectedly, the two-hybrid strain was substantially more sensitive than other strain backgrounds to the simultaneous loss of both Tel1 and Mec1 function, as a mec1-Δ tel1-Δ sml1-Δ derivative of this strain was essentially inviable after propagation for ∼25 generations (Figure S2), which precluded a test of the potential two-hybrid interaction between Cdc13 and Est1 in the absence of both kinases. Therefore, we cannot rule out the possibility that phosphorylation by Mec1 in the absence of Tel1 contributes to the Cdc13–Est1 interaction. However, it is worth pointing out that strains expressing kinase-dead or null alleles of MEC1 do not display a defect in telomere elongation (Mallory and Petes 2000; Frank et al. 2006), indicating that Mec1 does not contribute to telomerase recruitment in vivo.
The phosphatase-sensitive mobility of Cdc13 is not affected by mutations in the recruitment domain or by loss of Tel1 or Mec1 function:
To monitor phosphorylation, the mobility of Cdc13 bearing a C-terminal (myc)18 epitope was examined on 6% SDS–PAGE gels using an altered acrylamide-bis ratio to enhance separation of potential phospho-isoforms. Under these conditions, the Cdc13-(myc)18 protein exhibited an altered mobility, which was sensitive to λ-phosphatase (Figure 4A). This provided a reagent for testing whether Mec1- and/or Tel1-mediated phosphorylation occurs at either S249Q or S255Q, as previously proposed, by examining the mobility of the Cdc13-(myc)18 protein bearing mutations in both S249 and S255. As shown in Figure 4A, no alteration in the pattern of the phosphatase-sensitive isoform(s) of the Cdc13-(myc)18-S249A, S255A protein was observed. Furthermore, the altered mobility exhibited by the Cdc13-(myc)18 protein was unchanged in a tel1-Δ strain (Figure 4B) or in a tel1-Δ mec1-Δ strain (Figure S3). Although these observations are subject to the caveat that not all phosphorylation events result in a mobility shift, our assay conditions nevertheless were sensitive enough to detect a change in mobility when a previously described Cdk phosphorylation site in Cdc13 at T308P (Li et al. 2009) was eliminated (Figure S3).
Figure 4.—
Cdc13 phospho-isoform(s) are unaffected by mutations in the recruitment domain or loss of Tel1 function. (A) Extracts were prepared from a protease-deficient yeast strain expressing either Cdc13-(myc)18 or Cdc13–S249A, S255A-(myc)18 treated/ mock-treated with λ-phosphatase, resolved on 6% PAGE and examined for the presence of the Cdc13-(myc)18 protein by anti-myc Western. (B) Extracts prepared from isogenic TEL1 and tel1-Δ strains expressing Cdc13-(myc)18 were analyzed as in A.
As a more direct test of the phosphorylation status of S255, we raised phospho-specific antibodies that specifically recognized the proposed phospho-isoform of S255 in vitro, as assessed using an affinity-purified domain of Cdc13 containing S255 (see Figure S4 for more details). This antibody preparation failed to recognize S255-specific phosphorylation of Cdc13-(myc)18 protein expressed in yeast (data not shown), even though immunoprecipitated Cdc13 protein was examined to maximize detection of S255 phosphorylation; nevertheless, we cannot exclude the possibility that this negative result was a consequence of the detection limits of this particular antibody. Collectively, these attempts to obtain direct evidence that S249 or S255 were phosphorylated in vivo were unsuccessful.
A variant of Cdc13 lacking all 11 SQ or TQ phosphorylation consensus sites confers wild-type telomere length:
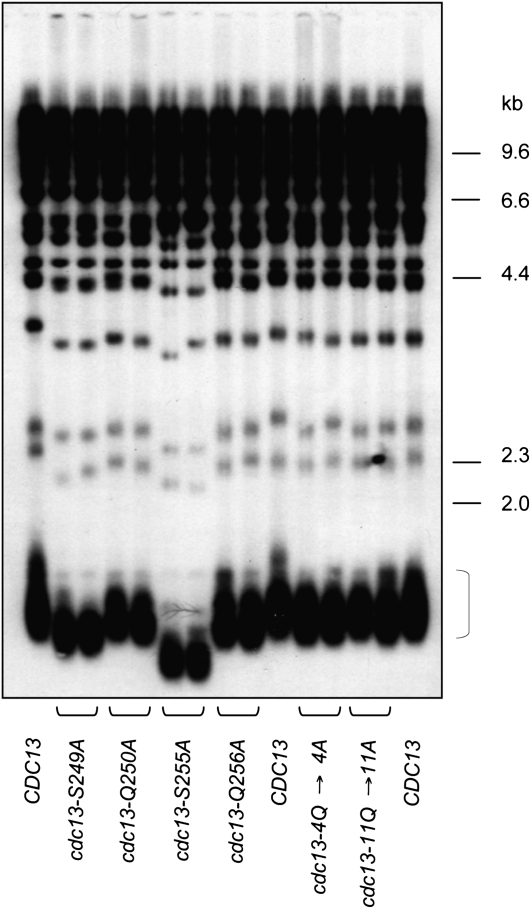
The above results raised the possibility that S255 and S249 might perform a structural role in telomerase recruitment, rather than as targets for a kinase. To differentiate between these two possibilities, we asked whether the adjacent glutamine residues in the S249Q and S255Q motifs might be dispensable for in vivo function, by mutating Q250 and Q256 to alanine residues. Several studies have demonstrated that a glutamine amino acid in the +1 position relative to the targeted serine is critical for substrate recognition by the DNA damage kinases (Kim et al. 1999; Smolka et al. 2007; Chen et al. 2010). Thus, if S249Q and S255Q are in fact targets for phosphorylation by Tel1 (or Mec1), this predicts that the cdc13–Q250A and cdc13–Q256A mutant strains should exhibit defects in telomere length regulation similar to that of the cdc13–S249A and cdc13–S255A strains. However, the cdc13–Q250A and cdc13–Q256A mutations had little to no effect on telomere length (Figure 5), in contrast to the more substantial telomere shortening observed in the cdc13–S249A and cdc13–S255A strains.
Figure 5.—
Telomere length of a cdc13–11Q→11A strain, which lacks all consensus Tel1/Mec1 phosphorylation sites. Telomere length analysis of the indicated mutations in CDC13 transformed into the cdc13-Δ/pCDC13 URA3 shuffle strain (YVL3006); strains were propagated for ∼50 generations following eviction of the covering CDC13 plasmid, prior to preparing genomic DNA for analysis of telomere length. The bracket indicates the 1.2 to 1.5 kb Y′-containing telomeric restriction fragment, which corresponds to approximately two-thirds of the telomeres in this strain background.
A potential caveat to this result is that binding of Tel1 (or Mec1) to an adjacent SQ motif in the proposed S/TQ cluster might be sufficient to phosphorylate either S255 or S249 in the cdc13–Q250A and cdc13–Q256A strains. Therefore, two additional mutant alleles of CDC13 were constructed, in which the glutamine residue was mutated to alanine at additional candidate phosphorylation motifs. As shown in Figure 5, a strain bearing mutations in four glutamine residues encompassing all four SQ motifs in the recruitment domain of Cdc13 (cdc13–Q226A, Q250A, Q256A, Q307A, referred to as cdc13–4Q→4A) exhibited a telomere length just slightly shorter than that of wild type. Finally, to eliminate the possibility that Tel1 or Mec1 were accessing the telomerase recruitment domain by binding a consensus SQ or TQ site anywhere in the Cdc13 protein, an allele of CDC13 was constructed in which all 11 SQ and TQ consensus sites were mutated to SA or TA, respectively. The telomere length of the resulting cdc13–11Q→11A strain again was only slightly reduced, relative to wild type (Figure 5).
Thus, telomere homeostasis is essentially unaffected in a strain expressing a variant of Cdc13 that does not contain even a single consensus phosphorylation site for the Tel1/Mec1 kinases. The validity of this experiment depends on the specificity of Tel1 and/or Mec1 kinases for substrates that contain glutamine in the +1 position. This has been addressed by a quantitative phosphoproteomics study, which identified 30 phosphorylation sites, in 28 different proteins, which were direct targets of Mec1 and/or Tel1 (see Table 2; Smolka et al. 2007). The majority of these sites (25 events) occurred at an S/TQ motif, with an additional 3 sites with a conservative replacement for glutamine (glutamic acid or asparagine) at the +1 position. The two remaining phosphorylation events occurred at sites with either serine or isoleucine at +1. This unbiased study demonstrates that Mec1 and Tel1 have a very high specificity for S/TQ motifs. Furthermore, on the basis of a survey of the published literature, we have not identified a single example of Mec1- or Tel1-dependent phosphorylation of serine or threonine residues with alanine in the +1 position. Taken together, the results shown in Figures 3, 4, and 5 argue against the proposal that in vivo telomere length is regulated by Tel1-dependent (or Mec1-dependent) phosphorylation of the proposed S/TQ cluster in the Cdc13 recruitment domain.
Additional residues in the Cdc13 recruitment domain are required for interaction with Est1:
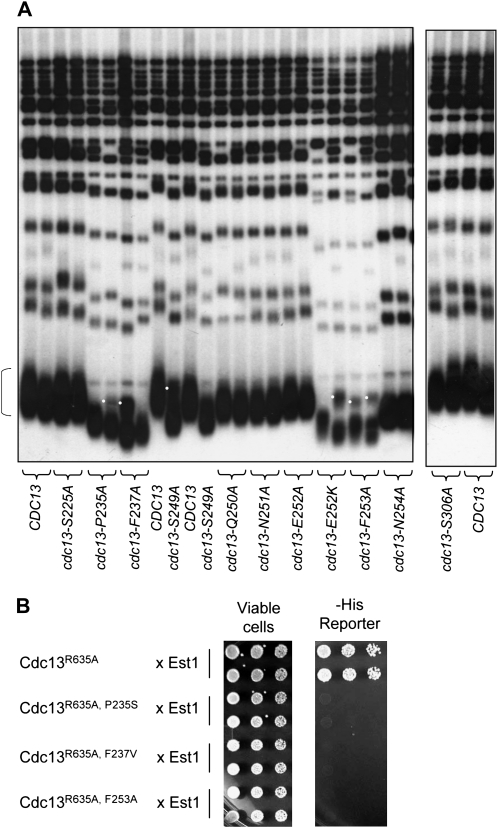
As a further step in our analysis of the Cdc13 recruitment domain, we introduced alanine mutations into conserved residues in the region spanning amino acids 235 to 254, on the basis of the alignment shown in Figure 1A. Strains expressing alanine mutations in three conserved residues—P235, F237, and F253—resulted in a substantial reduction in telomere length, whereas mutations in other residues in this domain had little (N254) or no (S225, N251, and S306) effect (Figure 6A). Mutations in residues P235, F237, and F253 also exhibited impaired ability to interact with Est1 as assessed by a two-hybrid test (Figure 6B), indicating that the telomere length defect exhibited by mutations in these three residues is due to a reduced interaction with the Est1 telomerase subunit. In contrast, a Cdc13–N254A two-hybrid fusion protein retained interaction with Est1 in this assay (data not shown), consistent with the slight in vivo defect displayed by the cdc13–N254A strain.
Figure 6.—
Additional residues in the Cdc13 recruitment domain mediate the interaction with Est1. (A) Telomere length analysis of the indicated mutations in CDC13, analyzed as described in Figure 5; white dots indicate a restriction fragment polymorphism that frequently occurs in these strains in the 1.2–1.5 kb Y′-containing telomeric restriction fragment (indicated by a bracket). (B) Two-hybrid analysis of the interaction between Cdc13–R635A (wild-type or mutant versions of pVL3125) and Est1 (pVL693), as monitored by the ability to activate the GAL1–HIS3 reporter gene; Figure S2 shows that these same Gal4DBD–Cdc13 proteins are not impaired, as assessed by their ability to interact with Stn1 in a two-hybrid assay.
We also examined the effects of several double mutant combinations in this region. The cdc13–S249A, S255A mutant strain exhibited a more severe telomere replication defect than the cdc13–S255A single mutant strain, displaying a senescence phenotype that was comparable to that of the cdc13-2 strain (Figure S1 and Figure S5). This additive effect was not specific to these two SQ motifs, however, as a second double mutant strain (cdc13–F253A, S255A) also exhibited a more severe telomere replication defect than the cdc13–S255A single mutant strain, which was again comparable to that of the cdc13-2 strain. Mutations in the two other proposed SQ motifs in this domain (cdc13–S225A and cdc13–S306A) did not enhance the severity of the phenotype displayed by a cdc13–S255A mutation (Figure S5), consistent with the wild-type telomere length of strains bearing either of these two mutations (Figure 6A). We conclude that the S225Q and S306Q sites are completely dispensable for telomere length regulation at native chromosome termini.
Epistasis analysis between the TEL1 pathway and the telomerase recruitment pathway:
Epistasis analysis has previously demonstrated that a tel1-Δ mutation partially attenuates the senescence phenotype of a tlc1-Δ strain (Ritchie et al. 1999; Abdallah et al. 2009). As pointed out by Petes and colleagues (Ritchie et al. 1999), this result is not easily explained by a model in which the primary role of Tel1 at telomeres is to target a regulatory component of the telomerase pathway. We revisited this epistasis result, by comparing how the senescence progression of a tlc1-Δ strain was influenced by the presence of tel1-Δ or tel2-1 mutations vs. mutations in the recruitment domain of CDC13. Senescence was assayed through a genotype-blind assessment of the growth characteristics of multiple isolates of a tlc1-Δ strain, compared to multiple isolates of a tlc1-Δ strain bearing an additional mutation. This protocol relies on a large number of samples to deal with the inherent variability of the senescence phenotype of telomerase-defective strains, with genotypes determined only at the conclusion of the experiment (see materials and methods and Figure S6 for more details). We have previously validated this approach by complementary techniques, establishing it as a semiquantitative approach (Rizki and Lundblad 2001).
As expected, on the basis of prior observations (Ritchie et al. 1999; Abdallah et al. 2009), the senescence progression of the tel1-Δ tlc1-Δ strain was slightly attenuated relative to that of a TEL1 tlc1-Δ strain (Figure 7A), with a difference between the two genotypes that was statistically significant. Similarly, the tel2-1 mutation, which functions in the same pathway as Tel1 for DNA damage signaling (Anderson et al. 2008), also partially alleviated the senescence phenotype resulting from loss of telomerase (Figure 7B). In contrast, there was no difference between the senescence phenotypes of tlc1-Δ and tlc1-Δ cdc13-2 strains, consistent with previous observations (Nugent et al. 1996). Similarly, neither cdc13–S255A nor the cdc13–S249A, S255A double mutation altered the senescence progression of a tlc1-Δ strain (Figure 7, C, D, and E). These observations confirm that the recruitment function of Cdc13, as defined by mutations in E252 and S255, acts in the telomerase pathway. In contrast, even though tel1-Δ and tel2-1 mutations confer short telomeres (Lustig and Petes 1986), defects in these two genes can partially rescue the growth defect of a telomerase null strain, indicating that the Tel1/Tel2-mediated pathway performs an activity that is distinct from regulating telomerase function in vivo.
DISCUSSION
Association between Est1 and the recruitment domain of Cdc13 is essential for telomerase to be recruited to its site of action (Evans and Lundblad 1999; Pennock et al. 2001; Bianchi et al. 2004; Chan et al. 2008). This recruitment step has been proposed to be regulated by phosphorylation of a proposed S/TQ cluster in the recruitment domain of Cdc13 (Tseng et al. 2006). However, three observations presented in this study argue against this conclusion. First, the pattern of Cdc13 phosphatase-sensitive isoforms is not altered by loss of Tel1 or Mec1 function or by mutations introduced into two conserved serines (S249 and S255) in this domain. Second, an interaction between Cdc13 and Est1, as monitored by a two-hybrid assay, is dependent on S255 but Tel1 independent. Finally, a derivative of Cdc13, cdc13–(S/TQ)11→(S/TA)11, in which every potential consensus phosphorylation site for Tel1 or Mec1 has been eliminated, retains nearly wild-type telomere length. Collectively, these results are not consistent with Tel1-mediated phosphorylation of the Cdc13 S/TQ cluster regulating its interaction with Est1.
Unequivocal evidence that a protein is phosphorylated usually relies either on immunological detection with phosphoepitope-specific antibodies, or direct monitoring of protein phosphorylation status by phosphopeptide mapping, or mass spectrometry analysis. Initial indications that Cdc13 might be regulated by phosphorylation came from a quantitative phosphoproteomics study, which identified two candidate phosphorylation sites on Cdc13 at S306Q and T308P (Smolka et al. 2007). An in vivo role for phosphorylation of each of these two sites has been subsequently established by directed studies. The T308P motif of Cdc13 is targeted by the Cdk kinase, and defects in this phosphorylation step confer an ∼75-bp decline in telomere length (Li et al. 2009), whereas the S306Q site is phosphorylated by Mec1 in response to DNA damage, with subsequent consequences for access of Cdc13 to double-strand breaks (Zhang and Durocher 2010). In both cases, the development of phospho-specific antibodies directed at these two sites has provided direct evidence that Cdc13 is phosphorylated in vivo at T308P and S306Q. In contrast, a phospho-specific antibody directed at the phosphorylated version of S255Q was not capable of recognizing S255-specific phosphorylation of Cdc13 expressed in yeast (Figure S4 and data not shown). Therefore, the biochemical evidence that S249Q and S255Q are targets of Tel1 and/or Mec1 rests solely on in vitro kinase assays monitoring the phosphorylation status of GST fusions to 75- to 90-amino-acid portions of Cdc13 in Tel1 or Mec1 immunoprecipitates (Tseng et al. 2006); however, such in vitro approaches can be notoriously promiscuous with regard to target specificity and thus often do not reflect the status of in vivo phosphorylation.
It is worth emphasizing that we only examined the cdc13–(S/TQ)11→(S/TA)11 strain for telomere length, leading to the conclusion that phosphorylation of Cdc13 by Tel1 (or Mec1) is not required for telomere length regulation. However, this result does not exclude the possibility that S/TQ residues of Cdc13 (in addition to the S306Q site mentioned above) may be phosphorylated by Tel or Mec1 for other regulatory purposes, such as in response to DNA damage.
An alternative model for the role of Tel1 in telomere homeostasis:
Multiple studies have shown that association of telomerase with telomeres is highly regulated. Est1 and Est2 association peaks during late S phase, when telomerase elongation occurs (Marcand et al. 2000), and this association is partially Tel1 dependent (Sabourin et al. 2007). Est1 and Est2, as well as Tel1, also exhibit an enhanced interaction with a telomere which has been experimentally shortened (Bianchi and Shore 2007; Sabourin et al. 2007). In contrast, telomeric association of Cdc13 is independent of both Tel1 and telomere length, which suggests that the preferential recognition of short telomeres by telomerase occurs after Cdc13 binding. As a result of these observations, a prevailing model in the field proposes that Tel1 binding to telomeres marks them for elongation through phosphorylation of the recruitment domain of Cdc13. Enhanced association of Tel1 with shorter telomeres, with subsequent phosphorylation of Cdc13 at this population of telomeres, provides a satisfying explanation for the ability of telomerase to preferentially target shorter telomeres.
However, if Tel1 does not phosphorylate the telomerase recruitment domain of Cdc13, as we argue in this current study, this indicates that the molecular mechanism for Tel1-mediated length regulation has not yet been solved. We would like to repropose an alternative model for Tel1 at telomeres, which is one variant of a general model originally put forth by the Petes group (see Ritchie et al. 1999; Ritchie and Petes 2000 for a more detailed discussion). This model stems from the need to provide an explanation for why loss of Tel1 attenuates the senescence of a telomerase-defective strain. As pointed out by Ritchie et al. 1999, this result is inconsistent with the idea that the sole function of Tel1 at telomeres is to target a component of the telomerase pathway. When considering alternative models, we considered the numerous parallels between double-strand breaks and telomeres. In particular, there is a modest delay in the accumulation of single-stranded DNA resection products at experimentally induced double-strand breaks in a tel1-Δ strain (Mantiero et al. 2007). We propose that resection of chromosome ends is similarly slightly impaired in the absence of Tel1. Such a molecular defect at telomeres provides a straightforward explanation for the deferred senescence phenotype of a telomerase-defective strain that lacks Tel1, since reduced resection would decrease the telomere shortening rate (although telomeres still would ultimately become critically short, in the absence of telomerase).
In a telomerase-proficient strain that lacks Tel1, we suggest that the shorter G-rich overhang, which would be the consequence of reduced resection, provides a suboptimal substrate for elongation by telomerase. Thus, in this model, the telomere length defect observed in tel1-Δ cells is an indirect consequence of a resection defect, rather than a direct consequence of altered regulation of telomerase. Although there is no direct evidence in support of this second aspect of the model, it is nevertheless consistent with the reduction in the overall frequency of elongation events by telomerase in tel1-Δ cells (Arneric and Lingner 2007) as well as the reduced telomeric association of Est1 and Est2 in a tel1-Δ strain. Blackburn and colleagues have also previously proposed that the requirement for Tel1/Mec1 at telomeres could be bypassed by enhancing the extent of the single-stranded overhang to increase telomerase accessibility (Chan et al. 2001).
One observation that has to be reconciled with the above proposal is that the interaction of Cdc13 with chromosome termini during late S phase is not altered in the absence of Tel1. Virtually every model about Cdc13 assumes that Cdc13 interacts solely with the single-stranded G-rich overhang present at the extreme termini of chromosomes. However, biochemical studies have established that Cdc13 does not require a free 3′ DNA terminus for binding and can bind equally well to internal tracts of G-rich telomeric DNA (Nugent et al. 1996). A priori, Cdc13 could potentially interact with the G-rich single-stranded region that is exposed when the replication fork moves through the duplex region of telomeric repeats; if so, this would be consistent with the association of Cdc13 with chromosome termini that is observed during late S phase. In fact, the method used to monitor Cdc13 telomeric association (chromatin immunoprecipitation) cannot distinguish between binding to the terminal G-rich overhang vs. binding to single-stranded regions exposed by the replication fork. Therefore, the Tel1-independent association of Cdc13 with telomeres does not argue for or against a change in the length of the G-rich overhang in tel1-Δ strains. The recent demonstration that the Cdc13-associated Stn1 and Ten1 proteins exhibit notable similarities with subunits of the RPA complex (Gao et al. 2007; Gelinas et al. 2009; Sun et al. 2009; Paschini et al. 2010) also lends credence to the idea that a telomere-dedicated RPA complex could be a part of the replication machinery as it passes through the duplex telomeric tract.
This alternative model for Tel1 also extends to the Mre11 complex (MRX), which acts in the same pathway for telomere length as Tel1 (Nugent et al. 1998; Ritchie and Petes 2000). Like Tel1, association of Est1 and Est2 with telomeres is reduced in mrx-Δ strains (Takata et al. 2005; Goudsouzian et al. 2006), and mutations in the MRX complex partially alleviate the consequences of a tlc1-Δ mutation (T. B. Toro and V. Lundblad, data not shown). MRX-defective cells also exhibit altered resection at both double-strand breaks and native telomeres (Larrivee et al. 2004; Takata et al. 2005; Mantiero et al. 2007). Thus, our model argues that the primary defect at telomeres—reduced resection of the telomeric C strand—occurs when either Tel1 or the MRX complex is defective, analogous to the defect that is observed at double-strand breaks when either of these two activities is defective.
There is, however, one Tel1-related phenomenon that is not addressed by the above model, which is that the extent of telomerase-mediated elongation at extremely short telomeres (<125 bp) is attenuated in the absence of Tel1 (Chang et al. 2007). Specifically, the highly processive telomerase activity that is observed at these extremely short telomeres becomes nonprocessive in a tel1-Δ strain. Chang et al. (2007) have argued that this particular subpopulation cannot arise as the result of normal telomere erosion and instead may be the consequence of replication fork collapse. If so, such aberrant termini may be recognized and processed, at least initially, through regulatory events that are different than the events that occur at chromosome termini that have been fully replicated.
What regulates preferential association of telomerase with short telomeres?
The current model in the field presumably answers this question: association of Tel1 with short telomeres, and subsequent phosphorylation of the telomerase recruitment domain of Cdc13, ensure that telomerase is directed to the shortest telomeres. However, telomerase can still distinguish short from long native telomeres in the absence of Tel1 (Arneric and Lingner 2007). Furthermore, the S-phase peak of association of Est1 and Est2 is diminished, but not eliminated, in the absence of Tel1 (Sabourin et al. 2007). Thus, although the overall frequency of telomerase elongation events is reduced in a tel1-Δ strain (Arneric and Lingner 2007), the basic regulatory mechanisms that direct telomerase to telomeres in S phase, and preferentially to short telomeres, remain intact. Combined with the results that we report here, the basis for preferential elongation of a particular subpopulation of telomeres remains an unresolved question.
The recruitment domain of Cdc13 has a single regulatory function:
In addition to residues immediately adjacent to the location of the cdc13-2 allele, this study also examined the effect of mutations across a conserved ∼25-amino-acid domain, including residue P235, which had previously been implicated in telomere length regulation. Analysis of the cdc13-4 mutation (P235S) led to the proposal that this mutation defined a separate function for Cdc13, distinct from the recruitment function impaired by the cdc13-2 mutation (Meier et al. 2001). If P235 is in fact required for a different biochemical activity, this predicts that the Cdc13–P235S mutant protein should still retain the ability to interact with Est1. However, as shown in Figure 5, the cdc13-4 mutation (P235S), as well as a mutation in an adjacent residue (F237), failed to interact with Est1 as assessed by two hybrid. Furthermore, a cdc13-2,4 strain exhibited a telomere replication defect that was indistinguishable from that of a cdc13-2 strain (Buckley 2005). We conclude that the region around P235 is required for telomerase recruitment, rather than for a separate function. More generally, our analysis of residues adjacent to the location of the original cdc13-2 mutation defines a minimal ∼25-amino-acid region that is required for interaction with the Est1 telomerase subunit.
Acknowledgments
We thank members of the Lundblad lab, Titia de Lange and Reuben Shaw for useful discussions during the course of this work, David Shore for strains, and Erin Ford and Leslie Ricks for technical assistance. This research was supported by a postdoctoral fellowship DAMD 17-02-1-0276 (to R.B.C), by grant GM55867 from the National Institutes of Health, and by funding from the F. M. Kirby Foundation and the G. Harold and Leila Y. Mathers Charitable Foundation.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.122044/DC1.
References
- Abdallah, P., P. Luciano, K. W. Runge, M. Lisby, V. Geli et al., 2009. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell. Biol. 11 988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, C. M., D. Korkin, D. L. Smith, S. Makovets, J. J. Seidel et al., 2008. Tel2 mediates activation and localization of ATM/Tel1 kinase to a double-strand break. Genes Dev. 22 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, E. M., W. A. Halsey and D. S. Wuttke, 2003. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry 42 3751–3758. [DOI] [PubMed] [Google Scholar]
- Arneric, M., and J. Lingner, 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 8 1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, A., and D. Shore, 2007. Increased association of telomerase with short telomeres in yeast. Genes Dev. 21 1726–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, A., S. Negrini and D. Shore, 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell 16 139–146. [DOI] [PubMed] [Google Scholar]
- Buckley, K., 2005. Investigating the recruitment of telomerase to the telomere in Saccharomyces cerevisiae. Ph.D. Thesis, Baylor College of Medicine, Houston, Texas.
- Chan, A., J. B. Boule and V. A. Zakian, 2008. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 4 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S. W., J. Chang, J. Prescott and E. H. Blackburn, 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11 1240–1250. [DOI] [PubMed] [Google Scholar]
- Chandra, A., T. R. Hughes, C. I. Nugent and V. Lundblad, 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, M., M. Arneric and J. Lingner, 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. H., C. P. Albuquerque, J. Liang, R. T. Suhandynata and H. Zhou, 2010. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 285 12803–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, S. K., and V. Lundblad, 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286 117–120. [DOI] [PubMed] [Google Scholar]
- Frank, C. J., M. Hyde and C. W. Greider, 2006. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell 24 423–432. [DOI] [PubMed] [Google Scholar]
- Gao, H., R. B. Cervantes, E. K. Mandell, J. H. Otero and V. Lundblad, 2007. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14 208–214. [DOI] [PubMed] [Google Scholar]
- Gelinas, A. D., M. Paschini, F. E. Reyes, A. Heroux, R. T. Batey et al., 2009. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl. Acad. Sci. USA 106 19298–19303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsouzian, L. K., C. T. Tuzon and V. A. Zakian, 2006. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24 603–610. [DOI] [PubMed] [Google Scholar]
- Hector, R. E., R. L. Shtofman, A. Ray, B. R. Chen, T. Nyun et al., 2007. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell 27 851–858. [DOI] [PubMed] [Google Scholar]
- Kim, S. T., D. S. Lim, C. E. Canman and M. B. Kastan, 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274 37538–37543. [DOI] [PubMed] [Google Scholar]
- Larrivee, M., C. LeBel and R. J. Wellinger, 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian and V. Lundblad, 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., S. Makovets, T. Matsuguchi, J. D. Blethrow, K. M. Shokat et al., 2009. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad, V., and J. W. Szostak, 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57 633–643. [DOI] [PubMed] [Google Scholar]
- Lustig, A. J., and T. D. Petes, 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83 1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, J. C., and T. D. Petes, 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins relates to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97 13749–13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero, D., M. Clerici, G. Lucchini and M. P. Longhese, 2007. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 8 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand, S., V. Brevet, C. Mann and E. Gilson, 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10 487–490. [DOI] [PubMed] [Google Scholar]
- Meier, B., L. Driller, S. Jaklin and H. M. Feldmann, 2001. New function of CDC13 in positive telomere length regulation. Mol. Cell. Biol. 21 4233–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent, C. I., T. R. Hughes, N. F. Lue and V. Lundblad, 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274 249–252. [DOI] [PubMed] [Google Scholar]
- Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger et al., 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8 657–660. [DOI] [PubMed] [Google Scholar]
- Paschini, M., E. K. Mandell and V. Lundblad, 2010. Structure prediction-driven genetics in Saccharomyces cerevisiae identifies an interface between the t-RPA proteins Stn1 and Ten1. Genetics 185 11–21. [DOI] [PMC free article] [PubMed]
- Pennock, E., K. Buckley and V. Lundblad, 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104 387–396. [DOI] [PubMed] [Google Scholar]
- Qi, H., and V. A. Zakian, 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev. 14 1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K. B., and T. D. Petes, 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K. B., J. C. Mallory and T. D. Petes, 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki, A., and V. Lundblad, 2001. Defects in mismatch repair promote telomerase-independent proliferation. Nature 411 713–716. [DOI] [PubMed] [Google Scholar]
- Sabourin, M., C. T. Tuzon and V. A. Zakian, 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell 27 550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka, M. B., C. P. Albuquerque, S. H. Chen and H. Zhou, 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA 104 10364–10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., E. Y. Yu, Y. Yang, L. A. Confer, S. H. Sun et al., 2009. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 23 2900–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata, H., Y. Kanoh, N. Gunge, K. Shirahige and A. Matsuura, 2004. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell 14 515–522. [DOI] [PubMed] [Google Scholar]
- Takata, H., Y. Tanaka and A. Matsuura, 2005. Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol. Cell 17 573–583. [DOI] [PubMed] [Google Scholar]
- Teixeira, M. T., M. Arneric, P. Sperisen and J. Lingner, 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117 323–335. [DOI] [PubMed] [Google Scholar]
- Traven, A., and J. Heierhorst, 2005. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays 27 397–407. [DOI] [PubMed] [Google Scholar]
- Tseng, S. F., J. J. Lin and S. C. Teng, 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 34 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., and D. Durocher, 2010. De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev. 24 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]