Abstract
Common fragile sites are regions that show elevated susceptibility to DNA damage, leading to alterations that can contribute to cancer development. FRA3B, located at chromosome region 3p14.2, is the most frequently expressed human common fragile site, and allelic losses at FRA3B have been observed in many types of cancer. The FHIT gene, encompassing the FRA3B region, is a tumor-suppressor gene. To identify the features of FHIT/FRA3B that might contribute to fragility, sequences of the human FHIT and the flanking PTPRG gene were compared with those of murine Fhit and Ptprg. Human and mouse orthologous genes, FHIT and Fhit, are more highly conserved through evolution than PTPRG/Ptprg and yet contain more sequence elements that are exquisitely sensitive to genomic rearrangements, such as high-flexibility regions and long interspersed nuclear element 1s, suggesting that common fragile sites serve a function. The conserved AT-rich high-flexibility regions are the most characteristic of common fragile sites.
Chromosome fragile sites are specific regions that show gaps, breaks, or rearrangements in metaphase chromosomes (1). These breaks are induced under specific culture conditions, generally by inhibiting or delaying DNA replication. Fragile sites are classified into two categories by their frequency in the population and by the chemistry of induction. Rare (or heritable) fragile sites are observed in <5% of individuals and most are induced by folic acid deficiency, whereas common (or constitutive) fragile sites are found in all individuals and most are induced by aphidicolin, an inhibitor of DNA polymerases (2). For rare fragile sites, the breakage is caused by unstable repeat expansion; for example, FRA11B is associated with CCG triplet repeats, and FRA10B and FRA16B with AT-rich minisatellite repeats (3–5). Chromosome locations of several common fragile sites coincide with locations of cancer breakpoints and/or cancer-associated genes, and the hypothesis that they play an important role in chromosomal instability in cancer development was proposed (6). This hypothesis preceded a wave of studies of recombinogenicity involving common fragile sites, reporting observations of sister chromatid exchanges, translocations, deletions, viral integrations, and intrachromosomal gene amplifications, at or near fragile sites in cancer (7–10). The reasons why common fragile regions are fragile, that is, so highly susceptible to breaks, is the subject of active investigation.
FRA3B at chromosomal band 3p14.2 is the most sensitive common fragile site in the human genome, and chromosomal abnormalities in this region are observed in a wide variety of human malignant diseases (reviewed in refs. 11 and 12). Breaks at common fragile sites are now known to occur spontaneously in ATR-deficient cells, and it has been proposed that they represent single-strand breaks that escape detection by the ATR-controlled S-phase/G2 checkpoint pathway (13). FRA3B exhibits hallmarks of fragile-site recombinogenicity, such as the translocation, t(3;8)(p14.2;q24), breakpoint at 3p14.2 in familial clear-cell renal carcinoma (14), plasmid integration sites (15), and a papilloma virus integration site (16). The fragile histidine triad (FHIT) gene was found to span FRA3B (17). The FHIT locus spans >1.5 megabase pairs (Mbp) of human genome, including FRA3B, but the mRNA is only 1.1 kb in size and consists of 10 small exons; exons 5–9 encode the Fhit protein (17, 18). FHIT is frequently deleted in numerous cancers and cancer cell lines, such as lung, digestive tract, kidney, breast, liver, and pancreatic cancers; and Fhit protein is absent or reduced in most cancers (reviewed in refs. 11, 12, and 19). The mouse Fhit ortholog also encompasses a common fragile site, Fra14A2, on murine chromosome 14 (20) and sustains homozygous deletions in murine cancer cell lines (21). Exogenous FHIT can suppress tumor growth (22), and exogenous Fhit protein expression induces apoptosis, directly or indirectly, in cancer cells (23, 24). Furthermore, Fhit knockout mice are more susceptible to tumor formation (25, 26), and delivery of FHIT in viral vectors prevents and reverses cancer development (27–29). Thus, Fhit functions as a tumor suppressor, consistently with the idea that fragile sites may harbor genes that, when altered, contribute to cancer development.
The human receptor protein tyrosine phosphatase γ (PTPRG) gene is located about 1.5 Mbp centromeric to FHIT on chromosome 3; and the mouse ortholog Ptprg is located 1.7 Mbp centromeric to Fhit on chromosome 14. We have reported the sequence and features of FRA3B; most cancer-associated rearrangements thus far defined occur within introns 3, 4, and 5 of the FHIT gene, and the orthologous fragile sites, human FRA3B and mouse Fra14A2, are highly conserved (30–32). The region from exon 3 through exon 6 has been considered the epicenter of fragility for FRA3B, and, thus, this region is referred to as FRA3B and Fra14A2. In a continuation of our analysis of the role of the DNA sequences at fragile regions in their susceptibility to rearrangement, we compared the complete genomic sequences of two genes in a human and murine orthologous chromosome region, FHIT vs. Fhit, containing fragile sites, and PTPRG vs. Ptprg, flanking the “epicenter” of fragility.
Materials and Methods
DNA-Sequencing Templates for Mouse Fhit. Bacterial artificial chromosome (BAC) clones RPCI-23-275G17, RPCI-23-334H8, RPCI-23-331I7, RPCI-23-89D3, RPCI-23-387P7, RPCI-23-252M2, RPCI-23-278C20, and RPCI-23-26D3 were obtained from Research Genetics (Huntsville, AL). These BACs overlapped the previously sequenced region (GenBank accession nos. AF332859–AF332862 sequenced from CITB-228L5, RPCI-23-255P3, RPCI-23-258D17, CITB-225H3, and RPCI-23-409H24; ref. 32). Overlaps of the BACs were identified by using the BAC end database in the Institute for Genomic Research (Rockville, MD; www.tigr.org/tdb/bac_ends/mouse/bac_end_intro.html). BAC DNAs were prepared by Big BAC DNA Kit (Princeton Separations, Adelphia, NJ) according to the manufacturer's protocol. Shotgun libraries of BACs were constructed, sequenced, and assembled as described (30).
GenBank FHIT DNA Sequences. The FHIT locus is composed of genomic contigs, AC097357, AC098480, AC138071, AC096917, AF152363, AC104164, AC093556, AC099536, AF152365, AF152364, AC099780, and AC093418 in Homo sapiens chromosome 3 genomic contig NT_005999. To determine the sequence of the human FHIT locus, the contigs were assembled with U76262, AC21400, U66722, AC093413, AC093418, and AC016944 as described (30).
GenBank Human PTPRG and Mouse Ptprg DNA Sequences. Published sequences AC103587, AC103921, AC096919, AC098482, AC004695, AC104849, AC105939, and AC092502 were aligned for determination of PTPRG sequence. In the same way, CAAA0104192, CAAA01041295, CAAA01041298, CAAA01041301, CAAA01041304, CAAA01041307, CAAA01121361, CAAA01041310, CAAA01198442, CAAA01041313, CAAA01041316, CAAA01041319, CAAA01191495, CAAA01041322, CAAA01041325, CAAA01187830, CAAA01041328, CAAA01139854, CAAA01041331, CAAA01210983, CAAA01041334, CAAA01041338, CAAA01041341, CAAA01168673, CAAA01223606, CAAA01210009, CAAA01041344, CAAA01041347, CAAA01041351, CAAA01190846, CAAA01041354, CAAA01041357, CAAA01202745, CAAA01207798, CAAA01041360, AC102427, AC102619, AC102543, AC102542, AC102575, AC132881, AC113486, and AC102459 were assembled for Ptprg.
Computer Analysis of Sequences. The final assembled sequences were analyzed by using genefinder (33), genescan (34), blastn, and tblastn (35) programs to seek other genes or homologous sequences. Repetitive elements were masked by the repeatmasker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) under the slow (most-sensitive) setting that screens DNA sequences against a library of repetitive elements. The flexstab program (http://bioinfo.md.huji.ac.il/marg/Flexstab) was used for calculating helix flexibility, expressed as fluctuations in the twist angle of DNA sequences (10). The window size was 100 bp, and dinucleotide values were summed along the window and averaged by the window size. The matrix-association regions (MARs) within DNA sequences were predicted by MAR-WIZ (www.futuresoft.org/MAR-Wiz; ref. 36) under the default setting of parameters. The pipmaker program (http://bio.cse.psu.edu/pipmaker), which computes alignments of similar regions in two DNA sequences (37), was applied for the large-scale comparison between human and mouse sequences. After masking the repetitive sequences, DNA sequences were input and analyzed with the parameters, advanced pipmaker, search both strands, single coverage, and high-sensitivity and low-time limits.
Results
Sequences of Mouse Fhit and Human FHIT Regions. Shotgun sequencing was performed for the entire genomic sequence of mouse Fhit, including 5′ to 3′ UTRs. Eight BAC clones, 275G17, 334H8, 331I7, 89D3, 387P7, 252M2, 278C20, and 26D3, were sequenced and aligned, along with published region, from 228L5 through 409H24 (32); the Fhit genomic locus was covered by 13 BACs (Fig. 5, which is published as supporting information on the PNAS web site). For complete FHIT sequence, from 5′- to 3′-UTRs, 18 published human DNA contigs were assembled (Fig. 5). The sequences reported in this article were submitted to the GenBank database (accession nos. AY363102 and AY363103). Total lengths of the sequences were 1,605,735 bp in FHIT and 1,702,543 bp in Fhit (Table 1). By using database analyses and prediction programs, no other putative genes were found in either sequenced locus. The organizations of both loci are also diagrammed in Fig. 5. The genomic structure, locations of exons, and sizes of introns are quite similar for the two orthologs; both have large introns 4 and 5, 285 and 522 kbp in human and 308 and 556 kbp in mouse, but the location of mouse exon 3 is different from human. In both genes exons 5–9 encode the protein (17), whereas mouse exon 3 exhibits an additional Met codon (21); we have observed usage only of the exon 5 Met codon as start site.
Table 1. Comparison of components in human and mouse orthologous regions.
| FHIT/FRA3B | Fhit/Fra14A2 | PTPRG | Ptprg | |
|---|---|---|---|---|
| Total length, bp | 1,605,735 | 1,702,543 | 733,390 | 696,902 |
| GC content, % | 38.9 | 40.1 | 40.8 | 43.4 |
| Element type | Elements, n (% of sequence) | |||
| SINEs | 702 (9.3) | 580 (5.1) | 506 (15.6) | 414 (8.9) |
| LINEs | 511 (19.3) | 352 (18.9) | 186 (8.0) | 73 (5.3) |
| LINE1 | 241 (15.1) | 309 (18.5) | 83 (5.3) | 63 (5.1) |
| LTR | 300 (7.8) | 372 (8.3) | 73 (3.3) | 108 (4.7) |
| MER | 278 (4.3) | 90 (1.0) | 150 (4.4) | 46 (1.2) |
| Total repeats | (40.8) | (33.4) | (31.5) | (20.2) |
| Regions, n (frequency per 100 kb) | ||||
| MARs | 44 (2.74) | 51 (3.00) | 24 (3.27) | 34 (4.88) |
| HFRs | 46 (2.86) | 46 (2.70) | 4 (0.55) | 8 (1.15) |
SINE, short interspersed nuclear element; LINE, long interspersed nuclear element; LTR, long terminal repeat; MER, medium reiteration frequency element.
Comparison of FHIT and Fhit Sequences. The overall GC content of human and mouse FHIT loci was 38.9% and 40.1%, respectively, over 1.5 Mbp of sequence (Table 1). Previously, we reported that the GC content in ≈600 kbp of FRA3B and Fra14A2 was 38.9% and 35.1%, respectively. The distributions of GC content in the two loci were not biased throughout sequences (Fig. 1). In Fhit, the percentages of nucleotides A, T, C, and G were 28.5%, 31.4%, 19.7%, and 20.3%, respectively, and in FHIT the percentages were 29.6%, 31.4%, 19.0%, and 20.0%, respectively. Those fractions were similar in any portion of the two loci. FHIT and Fhit contain higher percentages of A and T nucleotides than C and G, as do most common fragile sites thus far studied, such as FRA3B (30–32), FRA7H (10), FRA7G (38), FRA16D (39), and Fra14A (20, 32). Thus, the AT-rich sequence is associated with structural instability and might contribute to fragility.
Fig. 1.
The distribution of repetitive elements. The percentage of the repetitive elements and GC content in every 50-kbp sequence are diagrammed in the bar charts. The numbering of the horizontal axes indicates nucleotide positions of sequences in human FHIT and mouse Fhit. The positions of exons are shown by arrowheads with numbers.
The entire length, >1.6 Mbp, of FHIT and Fhit gene sequences were compared by using the advanced pipmaker program. The linear pattern in the dotplot chart means that homologous regions are conserved in conserved locations (Fig. 2A), and 81.8% of FHIT sequence, excluding repetitive elements, was homologous to Fhit sequence in conserved positions (green regions in the bottom row). Exons 3 and 10 in FHIT, both noncoding exons, did not have homologous regions in Fhit sequence, whereas other exons in human sequences had homologous exons in mouse sequences. The strongly aligned regions (at least 100 bp without a gap and with at least 70% nucleotide identity) were considered highly conserved regions (HCRs) and are represented in red in the bottom row. There were 577 HCRs spread over the FHIT sequence. The weight percent identity (WPI) was 69.3%, calculated by ΣCiFi/N, where Ci is the length in base pairs of each aligned sequence, Fi is the percent identity of that alignment, and N is the sum of Ci for all alignments (40). WPI indicates the percent identity for all aligned sequences. The WPI for FRA3B and Fra14A2, 72.6% (32), is slightly higher than for FHIT, but FHIT and Fhit are also highly conserved across species. To measure the conservation of potential regulatory motifs that might be relevant to splicing, the 500-bp regions in introns adjacent to 5′ and 3′ sides of human exons were compared with similar regions at mouse exons. In practice, this comparison was done by choosing the 500 bp flanking the human exons and searching the 1 kbp on either side of each mouse exon to find the conserved flanking segments. For example, the 5′ flanking region of the human exon 5 was conserved at base pairs 34–226 and 568–871 5′ of mouse exon 5. Intron regions flanking both sides of the nonconserved exons 3 and 10 and the 3′ flanking region of exon 9 were not at all conserved. Other regions, such as the 1.5 kbp 5′ of exon 1, including the promoter region, were well conserved. The average WPI in these conserved regions was 72.1%, slightly higher than the WPI of the entire sequences of FHIT/Fhit, 69.3%. This difference is quite small, suggesting that more refined methods would be required to identify bona fide regulatory sequences.
Fig. 2.
Comparison of human and murine orthologous sequences. The dotplot views of the alignments were produced by the advanced pipmaker program. (A) The dotplot comparison of FHIT vs. Fhit. (B) The dotplot comparison of PTPRG vs. Ptprg. In A and B, the horizontal and vertical axes represent the nucleotide number of human and mouse sequences, respectively, and the bottom rows show aligned regions in green and strongly aligned regions (at least 100 bp without a gap and with at least 70% nucleotide identity) in red. The locations of exons in human and mouse Fhit are also shown by arrowheads in A.
Repetitive Elements in FHIT and Fhit Loci. The repeat content of FHIT and Fhit sequences are summarized in Table 1, and the distribution of elements is diagrammed in Fig. 1. Every type of element was spread over the entire FHIT and Fhit sequences. FHIT contains more repetitive element sequence (40.8% of total sequence) than Fhit (33.4%), whereas both are rich in LINE1 and poor in SINEs, like FRA3B and Fra14A2 (30–32). The percent of interspersed repeat and LINE1 sequence is higher in the first half of the FHIT sequence, containing the fragile epicenter, than in the second half; however, this is not true for Fhit.
Features of FHIT and Fhit Sequences. Complete FHIT and Fhit loci were analyzed for components associated with instability by using computer programs. The FLEXSTAB program calculated the helical-twist angle, a potential local variation (10). Forty-six high-flexibility regions (HFRs) were spread over the entire FHIT and Fhit sequences (Table 1 and Fig. 3). The average GC contents of HFRs in human FHIT and mouse Fhit were 23.0% and 27.2%, respectively, much lower than the total GC contents (38.9% and 40.1%). Thus HFRs are rich in A and T. The MAR-WIZ program predicted 44 and 51 MARs scattered in FHIT and Fhit sequences, respectively (Table 1). MARs are regions where chromosomes are attached to the nuclear matrix, creating looped domains between attachment regions, and are considered to be related to replication origins. As an example, the MAR potential score around exons 6 and 7 of FHIT is shown in Fig. 6A, which is published as supporting information on the PNAS web site.
Fig. 3.
The transition of flexibility in sequences of FHIT and Fhit. Whole sequences of human FHIT and mouse Fhit were analyzed by flexstab. The flexstab program calculates helix flexibility. The peaks scoring >14 were considered HFRs. The horizontal axes indicate the nucleotide positions of sequences in human FHIT and mouse Fhit. The positions and the numbers of exons are shown by arrowheads.
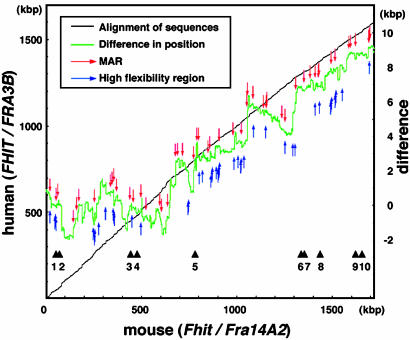
HFRs, MARs, and Positional Difference Between FHIT and Fhit Loci. The pipmaker program also provided a percent identity plot (PIP) chart to visualize the small regions in more detail (Fig. 6B). The PIP demonstrated that exons, except exons 3 and 10, were highly conserved and that regions in introns were also conserved; nevertheless, an ≈100-kbp difference occurred in total length of FHIT and Fhit sequences. This difference has been produced in the history of rearrangements, such as deletion and insertion, over evolutionary time. Therefore, to investigate the influence of HFRs and MARs on rearrangements, the positional differences between the conserved regions in FHIT and Fhit loci were calculated (Fig. 4). Significant changes of position (>10 kbp) were observed throughout the locus, and HFRs and MARs were located not only around the changed positions but everywhere in the locus. The result suggested that HFRs and MARs may play roles in fragility.
Fig. 4.
The difference in position of conserved sequences and the distribution of elements. x and left y axes indicate the nucleotide number of mouse Fhit and human FHIT sequences, respectively, and the gray line represents the alignment of these two orthologs. The differences in positions of conserved regions, the green line, were calculated on the assumption that the difference is 0 at the first nucleotide of exon 1 in human and mouse Fhit sequences. The difference in position is scaled on the right y axis. The elements that are prone to be affected by rearrangement, MARs and HFRs, are highlighted by red and blue arrows, respectively. The positions of exons in mouse Fhit are provided.
Sequences of Human PTPRG and Mouse Ptprg Regions. For determination of complete sequences of human PTPRG and mouse Ptprg, including the 5′ to 3′ UTRs, eight human and 35 murine published DNA contigs were aligned, respectively. The 15 gaps in Ptprg sequence (maximum length, 590 bp, and total length, 3 kbp) were ignored for analysis. The total length of sequence was 733,390 bp in PTPRG and 696,902 bp in Ptprg (Table 1).
Comparison of PTPRG and Ptprg Sequences. The GC contents of human and mouse Ptprg loci were 40.8% and 43.4%, respectively (Table 1). The percentages of G and C nucleotides were slightly higher than in FHIT and Fhit. The percentages of nucleotides A, T, C, and G were 31.3%, 28.0%, 20.8%, and 19.9% in PTPRG and 26.4%, 30.2%, 21.2%, and 22.3% in Ptprg. The percentages were similar throughout the loci. In a comparison of sequences, 81.2% of PTPRG sequence, excluding repetitive elements, was homologous to Ptprg sequence at conserved positions (green regions) and the WPI was 69.7%. These percentages were almost the same as in FHIT and Fhit. Therefore, the sequences of both genes were well conserved (Fig. 2B). The human PTPRG gene contains more repetitive elements than mouse, similarly to FHIT, and the fractions were respectively lower, 31.5% in PTPRG and 20.2% in Ptprg. Unlike the FHIT/Fhit loci, sequences of human PTPRG and mouse Ptprg are rich in SINEs and poor in LINE1s (Table 1). Regarding features proposed to contribute to rearrangements, human PTPRG contains 24 MARs and 4 HFRs, and mouse Ptprg contains 34 MARs and 8 HFRs (Table 1).
Comparison of FHIT and PTPRG Loci. FHIT loci in human and mouse contain higher percentages of the nucleosides adenosine and thymidine, and repetitive elements, especially LINE1, than PTPRG loci, and lower proportions of SINEs. MARs are spread in similar numbers in FHIT and PTPRG loci. However, the frequency of HFRs in human and mouse Fhit are 2.86 and 2.70 per 100 kbp, respectively; these fractions are far higher than the 0.55 and 1.15 observed in PTPRG and Ptprg. Both genes, FHIT/Fhit and PTPRG/Ptprg were conserved well across the two species. Because FHIT/Fhit exhibits more HCRs (red regions in Fig. 2), it is more highly conserved than PTPRG/Ptprg.
Discussion
We have compared complete sequences of two human and mouse orthologous genes, FHIT and PTPRG, and the results illustrate features that differentiate the fragile FHIT gene from the flanking PTPRG gene: (i) both large genes are well conserved and both consist of 20–40% repetitive sequences, but the fragile genes exhibit ≈19% LINE sequences compared with 8% and 5% for human and mouse PTPRG, respectively; (ii) the PTPRG/Ptprg genes exhibit three to five MARs per 100 kbp compared with ≈3 for the FHIT/Fhit genes, whereas the FHIT/Fhit genes have 5- and 2.35-fold more HFRs per 100 kbp than the human and mouse PTPRG genes, respectively. Thus, the outstanding features of the fragile genes are a high frequency of LINEs, LTRs, and HFRs. Recently, it was reported that the FRA3B region extends 4 Mbp, spreading ≈2.5 Mbp centromeric to FHIT (41). This extended region also contains the PTPRG locus. Because a clear difference exists in frequency of AT-rich HFRs in the sequence of the FHIT/Fhit loci compared with PTPRG/Ptprg loci, we favor the interpretation that FHIT/Fhit loci encompass most of the FRA3B and that PTPRG/Ptprg loci are not fragile, although it is possible that absence of one PTPRG locus, through frequent loss-of-heterozygosity events (42, 43), contributes to tumor development.
Several studies reported the GC content of common fragile sites (10, 30, 31, 38, 39), and common fragile sites have been considered AT-rich regions. Referring to the standard genomic sequences, GC content is 40–45% on average in the human and mouse genome (44, 45). LINE elements are abundant retrotransposons that constitute ≈17% of the draft human sequence, and another ≈15% is made up of Alu elements (44). Therefore, AT content in fragile sites and the proportion of LINE sequences in FRA3B are slightly higher than average. We reported that many cancer cell deletion end points were located near or in LINE1 elements and proposed that homologous LINE1s participate in repair of fragile breaks (30, 31). Khodarev et al. (46) suggested that LINE1 retrotransposable elements might be involved in the formation of loop structures, providing the framework for periodic DNA interactions with MARs. And two studies reported the association between LINE1 elements and genome instability, with LINE1 retrotransposition associated with large genomic deletions and inversions (47, 48). The percentage of total LINE1 length in human and mouse Fhit was over twice as high as in human and mouse Ptprg, although similar GC percents were observed in the FHIT/Fhit and PTPRG/Ptprg genes (Table 1). The average length of individual LINE1 elements in FHIT and Fhit was ≈1 kbp, as opposed to ≈0.5 kbp in PTPRG and Ptprg; 64 of 241 and 69 of 309 LINE1s were long elements (>1 kbp) in FHIT and Fhit, respectively. The results suggest that FHIT/Fhit contains more fragments of younger LINE1 elements than PTPRG/Ptprg and that FHIT/Fhit have been influenced by LINE1 quite recently. In addition, the frequency of HFRs in FHIT and PTPRG were 2.86 and 0.55 per 100 kbp (Table 1), respectively, indicating that FHIT includes many more unstable regions within its sequence. Thus, the FHIT gene contains many features that are sensitive to genomic aberrations, and the sequence itself indicates the history of rearrangements, through LINE1 insertions, for example. The pipmaker program found 577 HCRs between human and mouse Fhit loci. The average GC content of HCRs in human FHIT was 37.9%, a little lower than the total GC content in FHIT (38.9%). We searched for HCR homologous sequences in GenBank, but homologs were not found in other common fragile sites, FRA7H, FRA7G, and FRA16D, indicating that common fragile sites may not have consensus sequences other than high AT content and the attendant HFRs.
Our study revealed the apparently contradictory character of the FHIT genes straddling fragile regions, evolutionary conservation accompanied by susceptibility to recombination events. Since large amounts of sequence data have become available, many common fragile-region sequences have been combed for clues to the cause of fragility. Thus far, two common themes have emerged, that common fragile regions (i) are late-replicating or can become late- or later-replicating in the presence of the DNA polymerase α inhibitor, aphidicolin, the fragile-site inducer and (ii) exhibit a high density of AT-rich islands of high flexibility relative to nonfragile regions. The distribution of HFRs throughout the mouse and human FHIT genes, relative to the paucity of HFRs in the PTPRG/Ptprg loci, was the chief distinction between the fragile and nonfragile loci, we hypothesize that HFRs are the distinguishing sequence feature associated with fragility. Recently, Casper et al. (13) discovered that fragility at common fragile regions occurs spontaneously at a very high rate in cells deficient in the replication checkpoint kinase, Atr. Thus, stalling of DNA replication forks, not sensed by the Atr complex, could lead to persistence of single-stranded regions through G2 and mitosis, appearing as the characteristic fragile gaps in mitotic chromosomes. If fragility is based mainly on specific sequences, then the specific hallmark sequences of fragile regions, the HFRs, might be related to the late/delayed replication phenomenon and the occurrence of single-strandedness that escapes detection by the ATR DNA-damage checkpoint. A mechanistic relation between multiple HFRs and the response to aphidicolin inhibition could be investigated.
For evolutionary conservation of a genomic region, presumably the region must be conserved through the germ line, suggesting that fragile gaps/breaks must occur infrequently in germ-line meiotic divisions. Does the relative stability of the orthologous fragile sites over evolutionary time vs. the demonstrable mitotic fragility suggest that DNA-damage checkpoints during meiosis use a stricter Atr pathway so that the germ line is better protected from DNA alterations? Maybe, yet it is clear that the fragile regions must be susceptible to integrations of new LINE and LTR sequences in the germ line. The paradox of evolutionary conservation of these orthologous fragile regions despite their recombinogenicity also suggests that fragile regions provide some useful function at the species level.
Supplementary Material
Acknowledgments
We thank Dr. Webb Miller (Department of Computer Science and Engineering, Pennsylvania State University, University Park) for careful reading of our manuscript and useful comments. Work for this research was supported in part by National Cancer Institute Grant P01 CA77738 and Cancer Center Support Grant P30 CA56036 (to C.M.C. and K.H.).
Abbreviations: HCR, highly conserved region; MAR, matrix-association region; HFR, high-flexibility region; LTR, long terminal repeat; SINE, short interspersed nuclear element; LINE, long interspersed nuclear element; BAC, bacterial artificial chromosome; WPI, weight percent identity; Mbp, megabase pairs.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY363102 and AY363103).
References
- 1.Sutherland, G. R. (1979) Am. J. Hum. Genet. 31, 125-135. [PMC free article] [PubMed] [Google Scholar]
- 2.Glover, T. W., Berger, C., Coyle, J. & Echo, B. (1984) Hum. Genet. 67, 136-142. [DOI] [PubMed] [Google Scholar]
- 3.Jones, C., Penny, L., Mattina, T., Yu, S., Baker, E., Voullaire, L., Langdon, W. Y., Sutherland, G. R., Richards, R. I. & Tunnacliffe, A. (1995) Nature 376, 145-149. [DOI] [PubMed] [Google Scholar]
- 4.Hewett, D. R., Handt, O., Hobson, L., Mangelsdorf. M., Eyre, H. J., Baker, E., Sutherland, G. R., Schuffenhauer, S., Mao, J. I. & Richards, R. I. (1998) Mol. Cell 1, 773-781. [DOI] [PubMed] [Google Scholar]
- 5.Yu, S., Mangelsdorf, M., Hewett, D., Hobson, L., Baker, E., Eyre, H. J., Lapsys, N., Le Paslier, D., Doggett, N. A., Sutherland, G. R., et al. (1997) Cell 88, 367-374. [DOI] [PubMed] [Google Scholar]
- 6.Yunis, J. J. & Soreng, A. L. (1984) Science 226, 1199-1204. [DOI] [PubMed] [Google Scholar]
- 7.Glover, T. W. & Stein, C. K. (1988) Am. J. Hum. Genet. 43, 265-273. [PMC free article] [PubMed] [Google Scholar]
- 8.Popescu, N. C., Zimonjic, D. & DiPaolo, J. A. (1990) Hum. Genet. 84, 383-386. [DOI] [PubMed] [Google Scholar]
- 9.Coquelle, A., Pipiras, E., Toledo, F., Buttin, G. & Debatisse, M. (1997) Cell 89, 215-225. [DOI] [PubMed] [Google Scholar]
- 10.Mishmar, D., Rahat, A., Scherer, S. W., Nyakatura, G., Hinzmann, B., Kohwi, Y., Mandel-Gutfroind, Y., Lee, J. R., Drescher, B., Sas, D. E., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huebner, K., Garrison, P. N., Barnes, L. D. & Croce, C. M. (1998) Annu. Rev. Genet. 32, 7-31. [DOI] [PubMed] [Google Scholar]
- 12.Huebner, K. & Croce, C. M. (2001) Nat. Rev. Cancer 1, 214-221. [DOI] [PubMed] [Google Scholar]
- 13.Casper, A. M., Nghiem, P., Arlt, M. F. & Glover, T. W. (2002) Cell 111, 779-789. [DOI] [PubMed] [Google Scholar]
- 14.Cohen, A. J., Li, F. P., Berg, S., Marchetto, D. J., Tsai, S., Jacobs, S. C. & Brown, R. S. (1979) N. Engl. J. Med. 301, 592-595. [DOI] [PubMed] [Google Scholar]
- 15.Rassool, F. V., McKeithan, T. W., Neilly, M. E., van Melle, E., Espinosa, R., III, & Le Beau, M. M. (1991) Proc. Natl. Acad. Sci. USA 88, 6657-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilke, C. M., Hall, B. K., Hoge, A., Paradee, W., Smith, D. I. & Glover, T. W. (1996) Hum. Mol. Genet. 5, 187-195. [DOI] [PubMed] [Google Scholar]
- 17.Ohta, M., Inoue, H., Cotticelli, M. G., Kastury, K., Baffa, R., Palazzo, J., Siprashvili, Z., Mori, M., McCue, P., Druck, T., et al. (1996) Cell 84, 587-597. [DOI] [PubMed] [Google Scholar]
- 18.Zimonjic, D. B., Druck, T., Ohta, M., Kastury, K., Croce, C. M., Popescu, N. C. & Huebner, K. (1997) Cancer Res. 57, 1166-1170. [PubMed] [Google Scholar]
- 19.Richards, R. I. (2001) Trends Genet. 17, 339-345. [DOI] [PubMed] [Google Scholar]
- 20.Glover, T. W., Hoge, A. W., Miller, D. E., Ascara-Wilke, J. E., Adam, A. N., Dagenais, S. L., Wilke, C. M., Dierick, H. A. & Beer, D. G. (1998) Cancer Res. 58, 3409-3414. [PubMed] [Google Scholar]
- 21.Pekarsky, Y., Druck, T., Cotticelli, M. G., Ohta, M., Shou, J., Mendrola, J., Montgomery, J. C., Buchberg, A. M., Siracusa, L. D., Manenti, G., et al. (1998) Cancer Res. 58, 3401-3408. [PubMed] [Google Scholar]
- 22.Siprashvili, Z., Sozzi, G., Barnes, L. D., McCue, P., Robinson, A. K., Eryomin, V., Sard, L., Tagliabue, E., Greco, A., Fusetti, L., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 13771-13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sard, L., Accornero, P., Tornielli, S., Delia, D., Bunone, G., Campiglio, M., Colombo, M. P., Gramegna, M., Croce, C. M., Pierotti, M. A., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 8489-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji, L., Fang, B., Yen, N., Fong, K., Minna, J. D. & Roth, J. A. (1999) Cancer Res. 59, 3333-3339. [PubMed] [Google Scholar]
- 25.Fong, L. Y., Fidanza, V., Zanesi, N., Lock, L. F., Siracusa, L. D., Mancini, R., Siprashvili, Z., Ottey, M., Martin, S. E., Druck, T., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanesi, N., Fidanza, V., Fong, L. Y., Mancini, R., Druck, T., Valtieri, M., Rudiger, T., McCue, P. A., Croce, C. M. & Huebner, K. (2001) Proc. Natl. Acad. Sci. USA 98, 10250-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumon, K. R., Ishii, H., Fong, L. Y., Zanesi, N., Fidanza, V., Mancini, R., Vecchione, A., Baffa, R., Trapasso, F., During, M. J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishii, H., Dumon, K. R., Vecchione, A., Trapasso, F., Mimori, K., Alder, H., Mori, M., Sozzi, G., Baffa, R., Huebner, K., et al. (2001) Cancer Res. 61, 1578-1584. [PubMed] [Google Scholar]
- 29.Ishii, H., Zanesi, N., Vecchione, A., Trapasso, F., Yendamuri, S., Sarti, M., Baffa, R., During, M. J., Huebner, K., Fong, L. Y., et al. (2003) FASEB J. 17, 1768-1770. [DOI] [PubMed] [Google Scholar]
- 30.Inoue, H., Ishii, H., Alder, H., Snyder, E., Druck, T., Huebner, K. & Croce, C. M. (1997) Proc. Natl. Acad. Sci. USA 94, 14584-14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mimori, K., Druck, T., Inoue, H., Alder, H., Berk, L., Mori, M., Huebner, K. & Croce, C. M. (1999) Proc. Natl. Acad. Sci. USA 96, 7456-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiraishi, T., Druck, T., Mimori, K., Flomenberg, J., Berk, L., Alder, H., Miller, W., Huebner, K. & Croce, C. M. (2001) Proc. Natl. Acad. Sci. USA 98, 5722-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solovyev, V. V., Salamov, A. A. & Lawrence, C. B. (1994) Nucleic Acids Res. 22, 5156-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78-94. [DOI] [PubMed] [Google Scholar]
- 35.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 36.Singh, G. B., Kramer, J. A. & Krawetz, S. A. (1997) Nucleic Acids Res. 25, 1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz, S., Zhang, Z., Frazer, K. A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R. & Miller, W. (2000) Genome Res. 10, 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatarelli, C., Linnenbach, A., Mimori, K. & Croce, C. M. (2000) Genomics 68, 1-12. [DOI] [PubMed] [Google Scholar]
- 39.Ried, K., Finnis, M., Hobson, L., Mangelsdorf, M., Dayan, S., Nancarrow, J. K., Woollatt, E., Kremmidiotis, G., Gardner, A., Venter, D., et al. (2000) ) Hum. Mol. Genet. 9, 1651-1663. [DOI] [PubMed] [Google Scholar]
- 40.Oeltjen, J. C., Malley, T. M., Muzny, D. N., Miller, W., Gibbs, R. A. & Belmont J. W. (1997) Genome Res. 7, 315-329. [DOI] [PubMed] [Google Scholar]
- 41.Becker, N. A., Thorland, E. C., Denison, S. R., Phillips, L. A. & Smith, D. I. (2002) Oncogene 21, 8713-8722. [DOI] [PubMed] [Google Scholar]
- 42.Lubinski, J., Hadaczek, P., Podolski, J., Toloczko, A., Sikorski, A., McCue, P., Druck, T. & Huebner, K. (1994) Cancer Res. 54, 3710-3713. [PubMed] [Google Scholar]
- 43.LaForgia, S., Morse, B., Levy, J., Barnea, G., Cannizzaro, L. A., Li, F., Nowell, P. C., Boghosian-Sell, L., Glick, J., Weston, A., et al. (1991) Proc. Natl. Acad. Sci. USA 88, 5036-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 45.Smit, A. F. (1999) Curr. Opin. Genet. Dev. 9, 657-663. [DOI] [PubMed] [Google Scholar]
- 46.Khodarev, N. N., Bennett, T., Shearing, N., Sokolova, I., Koudelik, J., Walter, S., Villalobos, M. & Vaughan, A. T. (2000) J. Cell. Biochem. 79, 486-495. [PubMed] [Google Scholar]
- 47.Gilbert, N., Lutz-Prigge, S. & Moran, J. V. (2002) Cell 110, 315-325. [DOI] [PubMed] [Google Scholar]
- 48.Symer, D. E., Connelly, C., Szak, S. T., Caputo, E. M., Cost, G. J., Parmigiani, G. & Boeke, J. D. (2002) Cell 110, 327-338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.