Abstract
CD4+ T cells are required for immunity against many viral infections, including HIV-1 where a positive correlation has been observed between strong recall responses and low HIV-1 viral loads. Some HIV-1-specific CD4+ T cells are preferentially infected with HIV-1, whereas others escape infection by unknown mechanisms. One possibility is that some CD4+ T cells are protected from infection by the secretion of soluble HIV-suppressive factors, although it is not known whether these factors are produced during primary antigen-specific responses. Here, we show that soluble suppressive factors are produced against CXCR4 and CCR5 isolates of HIV-1 during the primary immune response of human CD4+ T cells. This activity requires antigenic stimulation of naïve CD4+ T cells. One anti-CXCR4 factor is macrophage-derived chemokine (chemokine ligand 22, CCL22), and anti-CCR5 factors include macrophage inflammatory protein-1α (CCL3), macrophage inflammatory protein-1β (CCL4), and RANTES (regulated upon activation of normal T cells expressed and secreted) (CCL5). Intracellular staining confirms that CD3+CD4+ T cells are the source of the prototype HIV-1-inhibiting chemokines CCL22 and CCL4. These results show that CD4+ T cells secrete an evolving HIV-1-suppressive activity during the primary immune response and that this activity is comprised primarily of CC chemokines. The data also suggest that production of such factors should be considered in the design of vaccines against HIV-1 and as a mechanism whereby the host can control infections with this virus.
CD4+ T cells are required for immunity against many viral infections including HIV-1 (reviewed in ref. 1). In this regard, a positive correlation has been observed between strong recall responses of CD4+ T cells and low HIV-1 viral loads (2–4). Some HIV-1-specific CD4+ T cells are preferentially infected with this virus (5), whereas others escape HIV infection by unknown mechanisms. One possibility is that some CD4+ T cells are protected from infection by virtue of their ability to secrete soluble HIV-suppressive factors. The production of HIV-1-suppressive factors is well established for CD8+ T cells (6) and has been correlated with decreased viral loads (7), resistance of hemophiliacs repeatedly exposed to HIV-1 to infection (8), and favorable prognosis in HIV-infected subjects (9, 10). This information raises the question of whether similar factors are produced by CD4+ T cells during antigen-specific responses. Although there are several reports that CD4+ T cell lines and clones can secrete anti-HIV-1-suppressor factors (11–14), it is not known whether CD4+ T cells produce anti-HIV-1-suppressor factors in an immunologically clear context such as the primary immune response.
To address this question, we developed an in vitro system that mimics the conditions of primary antigen-specific CD4+ T cell responses. Primary T cell responses are initiated in secondary lymphoid tissues by antigen-loaded dendritic cells, which are the only cells capable of priming naïve T cells (15–17). We used negative selection to obtain highly purified naïve CD4+ T cells from the peripheral blood of healthy donors and also monocytes to generate immature monocyte-derived dendritic cells (MDDC) to initiate a primary T cell response in vitro. This system faithfully mimics the early stages of the antigen-specific response of naïve human CD4+ T cells, including clonal activation, expansion, contraction, and maintenance (unpublished work and ref. 18). We used this system to study the kinetics of production of HIV-1-suppressive factors against both CXCR4 (X4) and CCR5 (R5) viruses. CD4+ T cells, starting as naïve cells and differentiating in response to different antigens, secreted factors that inhibited both HIV-1IIIB (X4) and HIV-1BaL (R5). The anti-X4 factor is predominantly the CC chemokine CCL22 (macrophage-derived chemokine), whereas the anti-R5 factors are CCL3 (macrophage inflammatory protein-1α), CCL4 (macrophage inflammatory protein-1β), and CCL5 [regulated upon activation of normal T cells expressed and secreted (RANTES)]. Intracellular staining indicates that CD3+CD4+ cells are the source of the prototype HIV-1 inhibiting chemokines CCL22 and CCL4. Taken together, these studies demonstrate the production of suppressive factors against X4 and R5 HIV-1 isolates during the primary immune response of CD4+ T cells. The significance of these studies for HIV-1 pathogenesis and vaccine development is discussed.
Materials and Methods
Generation of MDDC. Heparinized peripheral blood was obtained from healthy volunteers under informed consent approved by the Institutional Review Board of the University of Maryland. Mononuclear cells were isolated by centrifugation over Ficoll-Hypaque (Sigma). MDDC were generated as described (19) with minor modifications (20, 21). Briefly, monocytes were negatively selected from peripheral blood mononuclear cells, adhered to plastic for 2 h, washed, and then cultured for 7 days in MDDC culture medium [StemSpan serum-free medium, StemCell Technologies, Vancouver, supplemented with 100 μg/ml streptomycin, 100 units/ml of penicillin G (Life Technologies, Rockville, MD), 50 ng/ml granulocyte–macrophage colony-stimulating factor, and 1,000 units/ml of IL-4 (R & D Systems)]. The MDDC displayed clear dendritic morphology and had the expected phenotype as determined by flow cytometry (data not shown).
Flow Cytometry. Cells of the types indicated in the text were washed in wash buffer [PBS with 2% heat-inactivated human AB serum (Sigma) and 0.1% sodium azide] and incubated for 30 min at 4°C with fluorochrome-labeled mAbs. The following mAbs were purchased from Becton Dickinson–Pharmingen: FITC anti-CD1a, peridinin chlorophyll protein (PerCP) anti-CD3, PerCP anti-CD4, allophycocyanin (APC) anti-CD4, APC anti-CD14, PerCP anti-CD19, APC anti-CD45RO, APC anti-CD56, phycoerythrin (PE) anti-CD62L, FITC anti-CD83, PE anti-CD86, APC anti-R5, and PE anti-HLA-DR. Additional mAbs were purchased from Beckman Coulter, including biotin-conjugated anti-T cell receptor (TCR) Vβ2, and PE anti-TCR Vβ17. The samples were analyzed by four-color flow cytometry using a FACScalibur flow cytometer (Becton Dickinson). Data analysis was performed with flowjo software (Tree Star, San Carlos, CA).
Isolation of Naïve CD4+ T Cells. Naïve CD4+ T cells were isolated from peripheral blood mononuclear cells by negative selection with a mixture of mAbs from StemCell Technologies according to the manufacturer's instructions. The mixture included mAbs to CD8, CD14, CD16, CD19, CD45RO, CD56, and glycophorin A. An anti-human HLA-DR tetrameric mAb (StemCell Technologies) was also included to increase the purity of the naïve CD4+ T cells. The isolated cells were completely depleted of monocytes, NK, B, and CD8+ T cells and were >95% CD3+ CD4+. Generally, 92–99% of the isolated CD4+ T cells were naïve as judged by surface expression of CD62L (CD62L+), low expression of CD11a, and the lack of expression of CD25, CD45R0, and HLA-DR (data not shown).
Antigen-Specific Stimulation of Naïve CD4+ T Cells in Vitro. Carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes) labeling of naïve CD4+ T cells was carried out as described (22). Naïve CD4+ T cell proliferation medium consists of α-MEM without ribonucleosides and deoxyribonucleosides supplemented with 10% heat-inactivated human AB serum, 4 mM l-glutamine, 20 mM Hepes buffer, 100 μg/ml streptomycin, 100 units/ml penicillin G, 1 mM sodium pyruvate, and 50 μM 2-mercaptoethanol, hereafter referred to as complete α-MEM. Lipopolysaccharide contamination of the antigens was monitored routinely by the limulus assay (BioWhittaker) and was always <100 pg/ml. Immature MDDC were harvested on day 7 after monocyte culture in MDDC culture medium and washed three times in complete α-MEM. The washed MDDC (2 × 103 per well for antigen-specific responses and 1 × 104 per well for mixed lymphocyte reaction) were used for priming naïve CD4+ T cells (2 × 105 per well), which were labeled or not with CFSE, in 200 μl of total culture volume in 96-well U-bottom plates with or without antigen. Unless indicated otherwise, the following antigen concentrations were used: 100 ng/ml Staphylococcus enterotoxin B (SEB; Sigma) or 100 ng/ml toxic shock syndrome toxin 1 (TSST-1; Toxin Technologies, Sarasota, FL). The cultures were incubated in a humidified incubator in the presence of 5% CO2 at 37°C for the times indicated in the text. The day the antigen-specific response was initiated is designated as day zero. Proliferation was estimated by CFSE dilution using flow cytometry. Background proliferation was determined by omitting antigen from the cultures. For flow cytometry, cultured cells were harvested, washed in wash buffer, and incubated for 30 min at 4°C with mAbs, washed, and fixed for flow cytometric analysis. The instrument settings used for acquisition of samples for different days in the same experiment were identical. A total of 50,000–400,000 events were acquired for data analysis. In some cases, dead cells were excluded from the analysis by using ethidium monoazide staining (Molecular Probes).
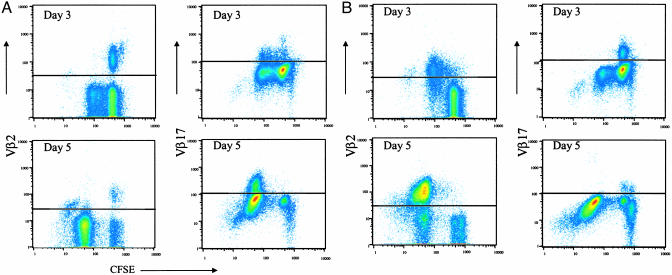
TCR Vβ Usage. Naïve CFSE-labeled CD4+ T cells (2 × 105 cells per well) were stimulated with 1 μg/ml SEB or TSST-1 in the presence of autologous MDDC (2 × 103 cells per well). At days 3 and 5, the cells were harvested and analyzed for Vβ2 and Vβ17 expression by flow cytometry.
Viral Inhibition Assay. Cells were cultured with or without antigens as indicated in the text, and the tissue culture supernatants were harvested at the indicated time points. An acute viral inhibition assay was used to test the HIV-1-suppressive activity of these supernatants as described (9). Briefly, medium (complete α-MEM), test supernatants (50% vol/vol), or positive controls were added to HIV-1-infected peripheral blood mononuclear cells (9). On the third day of culture, half of the medium was replaced with the corresponding medium or supernatant. On day 6, p24 was quantified by ELISA (NEN) as a measure of HIV-1 replication. The p24 concentration in the presence of complete α-MEM was considered as 100% viral growth, and the percentage of inhibition of HIV-1 was calculated by using the p24 concentration obtained in the presence of the supernatants harvested from the test cultures. For the reversal of HIV-1-suppressor activity with antibodies, supernatants harvested from activated CD4+ T cells were incubated with a polyclonal anti-CCL22 antibody (PeproTech, Rocky Hill, NJ), a mixture of polyclonal antibodies against CCL3, CCL4, and CCL5, or isotype controls (R & D Systems) for 45 min at 37°C before addition to the infected cultures. The final concentration of each antibody was 50 μg/ml. The isotype control antibodies did not affect the p24 value of the medium by >5%.
Chemokine ELISA. The levels of CCL1 (I309), CCL3, CCL4, CCL5, and CCL22 in the supernatants of activated and nonactivated cells were determined by ELISA according to the manufacturer's instructions (R & D Systems).
Analysis of Intracellular Chemokines Using Flow Cytometry. Four days after priming of CFSE-labeled naïve CD4+ T cells with SEB (100 ng/ml), they were collected, washed, and restimulated (or not) with phorbol myristate acetate (PMA; 25 ng/ml) and ionomycin (1 μg/ml) (both from Sigma) for a total of 5 h. Brefeldin A (Sigma) was added at 10 μg/ml immediately after the addition of PMA/ionomycin. The cells were washed, permeablized with FACS permeablizing solution (Becton Dickinson–Pharmingen), washed again, and incubated with a combination of PE-labeled CCL22-specific (R & D Systems), or PE-labeled CCL4-specific mAbs in addition to APC anti-CD4- and peridinin chlorophyll protein anti-CD3-specific mAbs (all from Becton Dickinson–Pharmingen). The cells were washed and fixed for flow cytometric analysis.
Results and Discussion
To address whether CD4+ T cells produce HIV-1-suppressor factors during primary responses, we developed an in vitro model system that mimics the conditions of primary antigen-specific CD4+ T cell responses. We used negative selection to obtain highly purified MDDC and naïve CD4+ T cells from the peripheral blood of healthy donors to initiate a primary T cell response in vitro. In this system, there are no qualitative differences in the responses elicited by soluble proteins, superantigens, and alloantigens, although there are quantitative differences with respect to the magnitude and kinetics of the responses that are determined by differences in antigen-specific precursor frequency (unpublished work and ref. 18). For example, naïve CD4+ T cells show the same evolution pattern of forward and orthogonal light scatter populations regardless of the antigenic stimulus. This is also true for the evolution of CD4+ T cells subsets defined by the conventional activation/memory makers CD45R0 and CD62L (23–25). Indeed, the data strongly suggest that once an immune response is initiated in our system, it evolves in a deterministic fashion irrespective of whether the immunogen was keyhole limpet hemocyanin, superantigen, or alloantigen. Thus, the superantigen response in our system is a reasonable model for a generic primary immune response of human CD4+ T cells.
An example of system performance is shown in Fig. 1 by demonstrating specific clonal expansion in response to two superantigens. Two TCR Vβ markers were used to trace the clonal expansion of naïve CD4+ T cells responding selectively to the superantigens SEB and TSST-1. Vβ2 is the only Vβ known to be used by TSST-1, whereas Vβ17 is one of six Vβs used by SEB (reviewed in refs. 26 and 27). CFSE-labeled naïve CD4+ T cells were stimulated with SEB or TSST-1 and Vβ expression plotted versus CFSE intensity as an indicator of cell division for each of the stimuli. Fig. 1 A Right shows that ≈8% (day 3) and ≈16% (day 5) of the naïve CD4+ cells responding to SEB were Vβ17+. As expected (26, 27), Vβ2+ cells did not respond to SEB (Fig. 1 A Left). By contrast, essentially all Vβ2+ cells responded to TSST-1 by days 3 and 5 (Fig. 1B Left). Again, as expected (26, 27), Vβ17+ cells did not respond to TSST-1 (Fig. 1B Right). Approximately 17% of the naïve CD4+ T cells responding to TSST-1 were Vβ2-, suggesting additional TCR Vβ(s) are used in response to this superantigen. These results demonstrate efficient clonal activation and expansion of naïve CD4+ T cells in this system without activation of bystander clones. Additional aspects of this system such as clonal contraction and maintenance are described in detail elsewhere (unpublished work and ref. 18).
Fig. 1.
TCR Vβ usage suggests that the responses are strictly antigen-specific. Naïve CD4+ T cells labeled with CFSE were stimulated with 1 μg/ml SEB (A) or TSST-1 (B) in the presence of autologous MDDC as described in Materials and Methods. At indicated time points, the cells were harvested and stained with Vβ2 and Vβ17 mAbs for flow cytometric analysis. Dead cells were excluded from this analysis by ethidium monoazide staining. The horizontal lines represent the cutoff of specific Vβ staining as determined by unstimulated cells and isotype controls. Data are representative of independent experiments carried out with cells derived from three normal donors.
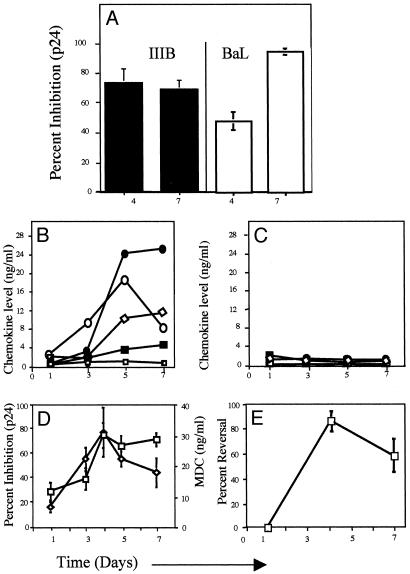
The production of soluble anti-HIV suppressive factors was determined by using supernatants collected after 1–7 days of stimulation of naïve CD4+ T cells with autologous MDDC and superantigens (SEB or TSST-1) or with allogeneic MDDC. Supernatants were tested for suppression of X4 (HIV-1IIIB) or R5 (HIV-1BaL) infection in a phytohemagglutinin-activated peripheral blood mononuclear cells-based infectivity assay as described in Materials and Methods. As shown in Fig. 2A, anti-X4 and anti-R5 activities were detected. In the data shown in Fig. 2 A, the anti-X4 activity was 75% inhibition and 71% inhibition at days 4 and 7 poststimulation, respectively (data derived from eight independent donors). Anti-R5 activities increased from ≈40% inhibition on day 4 to ≈95% inhibition by day 7 (data derived from three different donors). These activities were present at equivalent levels regardless of whether the cells were activated with the superantigens SEB, TSST-1, or allogeneic MDDC (data not shown).
Fig. 2.
Activated antigen-specific CD4+ T cells inhibit X4 and R5 HIV-1 isolates, and this activity is mediated by the CC chemokines. (A) Individual supernatants harvested on days 4 and 7 from SEB-, TSST-1-, or allogeneic MDDC-activated cells were tested for inhibition of X4 (HIV-1IIIB) and R5 (HIV-1BaL) viruses in the viral inhibition assay as described in Materials and Methods. The data are shown as the average percentage of inhibition by the individual supernatants. Anti-HIV-1 chemokine concentrations (□, CCL1; •, CCL3; ⋄, CCL4; ▪, CCL5, and ○, CCL22), determined by ELISA in the supernatants, are shown for naïve CD4+ T cells cultured with autologous MDDC in the presence of TSST-1 (B) and for naïve CD4+ T cells cultured with MDDC alone (C) (see Materials and Methods for details). Data are representative of 10 experiments. (D) Temporal correlation between CCL22 concentration (⋄) and HIV-1IIIB inhibition (□) for eight different individuals in supernatants from naïve CD4+ T cells cultured with autologous MDDC and superantigens (SEB or TSST-1) or with allogeneic MDDC. (E) CCL22-neutralizing antibodies reverse the antiviral activity of activated CD4+ T cell supernatants against HIV-1IIIB. The reversal of HIV-1IIIB suppressor activity by CCL22-neutralizing antibodies is shown as the average percentage of reversal in three independent experiments (using independent donors) in which naïve CD4+ T cells were cultured with autologous MDDC and SEB. Error bars represent the SEM.
Because CD4+ T cell lines and clones are known to secrete antiviral chemokines (11–14), it was important to determine whether these factors are secreted during the primary immune response. To examine this possibility, we used ELISA to quantify the two X4-suppressive CC chemokines, CCL1 (28, 29) and CCL22 (30), and the three R5-suppressive β-chemokines, CCL3, CCL4, and CCL5. Chemokines were quantified in supernatants taken on days 1–7 from mixtures of naïve CD4+ T cells and MDDC cultured with (Fig. 2B) or without (Fig. 2C) antigenic stimulation. As shown in Fig. 2B, the concentrations of CCL3, CCL4, CCL5, and CCL22 increased with time of culture, reaching a plateau by ≈day 5. By contrast, CCL1 concentrations did not increase. On the other hand, these chemokines were found at very low levels in the supernatants of unstimulated cells (Fig. 2C). Thus, we speculated that CCL22 comprises the anti-X4 activity and that CCL3, CCL4, and CCL5 comprise the anti-R5 activity. This idea is supported for CCL22 and anti-X4 activity by the close temporal relationship between HIV-1IIIB inhibition and CCL22 levels in the supernatants of the activated cells (Fig. 2D) and by the reversal of anti-X4 activity (see Materials and Methods for details) of the supernatants by anti-CCL22 polyclonal antibodies (Fig. 2E). Although, these data suggest that CCL22 is the predominant anti-X4 factor in the supernatants at times of peak antiviral activity, the inability to completely reverse the antiviral effect by anti-CCL22 (days 1 and 7, Fig. 2E) leaves open the possibility that additional X4 HIV-suppressor factors are elaborated in our system. Although such putative factors remain unidentified they do not appear to be α-defensins (ref. 31 and data not shown).
There was also a temporal correlation between the levels of CCL3, CCL4, and CCL5 in the supernatants and their ability to suppress R5 viruses (data not shown). In addition, treatment of day 4 supernatants with a mixture of anti-CCL3, CCL4, and CCL5 polyclonal antibodies completely reversed HIV-1BaL suppression (Supporting Text and Fig. 4, which are published as supporting information on the PNAS web site) confirming that R5 ligands are responsible for the anti-R5 activity. Notably, CCL22 did not significantly inhibit R5 HIV-1 in the context of our system because such activity would have been apparent upon neutralization of the R5 ligands. Such findings contrast with the properties of recombinant or purified native forms of CCL22, which are R5 suppressive (30, 32) and emphasize a complex interplay among HIV-suppressive factors released during immune responses that is not reflected in the direct analyses of isolated chemokines. On the other hand, a recent study showed that the addition of naïve CD4+ T cells suppressed replication of HIV-1BaL (R5 isolate) in infected CD62L-/CD45RA- memory CD4+ T cells after anti-CD3/CD28 or anti-CD3/CD80 stimulation (33). This activity was also seen in TCR-activated total (naïve and memory), HIV-1-infected CD4+ T cells (33). However, the suppressor activity was not attributed to the presence of CCL3, CCL4, and CCL5 in the supernatants of the activated cells and seems to require cell-to-cell contact (33). Despite differences in the techniques, methods of activation and identity of the suppressor factor, the data presented in ref. 33 and herein strongly suggest that naïve cells can suppress R5 HIV-1 infection upon activation in vitro.
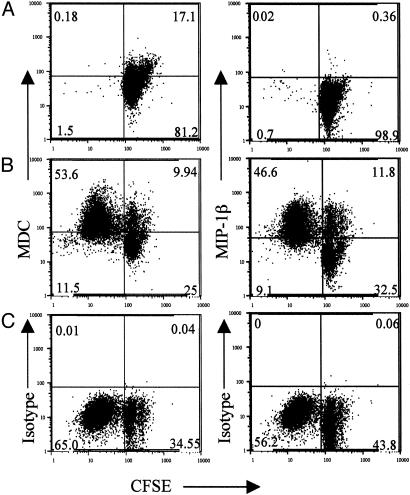
Although it is clear that secretion of these antiviral factors in our system requires antigen activation, their cellular source could be CD4+ T cells, MDDC, or both (11–14, 34). Because we postulate that the secretion of these antiviral chemokines is an important effector mechanism whereby CD4+ T cells temper HIV-1 infection, we used intracellular staining to verify that CD4+ T cells are in fact the primary sources of chemokines in our system. The data shown in Fig. 3 demonstrate directly that CCL22 (Left) and CCL4 (Right) are synthesized by the responding CD4+ T cells after 4 days of culture with SEB and autologous MDDC. It should be noted that the cells did produce both CCL22 and CCL4 with and without undergoing cell division upon antigenic stimulation, suggesting that they can produce these CC chemokines early during primary responses. We have not been able to recover sufficient MDDC in this system to determine whether they also synthesize these chemokines. Nevertheless, our data are most consistent with the CD4+ T cells being the primary source of antiviral chemokines in that MDDC are present at frequencies <1% of those of CD4+ T cells in this system. Taken together, these studies demonstrate and characterize the production of suppressive factors against X4 and R5 HIV-1 isolates during the primary antigen-specific CD4+ T cell response. Our data show that the activity produced during the primary CD4+ T cell response is comprised principally, if not exclusively, of antiviral β chemokines as it has been described for memory CD8+ T cells (6, 30, 35–37).
Fig. 3.
CD4+ T cells synthesize anti-X4 and anti-R5 chemokines in response to antigen. Naïve CD4+ T cells were labeled with CFSE and stimulated or not with 100 ng/ml SEB in the presence of autologous MDDC. On day 4 of culture, the cells were restimulated with phorbol myristate acetate/ionomycin, and the synthesis of anti-X4 (CCL22, Left) and anti-R5 (CCL4, Right) chemokines by unstimulated (A) and SEB-stimulated (B) cells was determined by intracellular staining as described in Materials and Methods. (C) Isotype control staining is shown only for SEB-stimulated cells. Shown is cell division, as determined by decreased CFSE intensities on the abscissa and CCL22, CCL4 (A and B), or isotype control staining (C) on the ordinate. These parameters were determined on populations gated for both resting and blasting lymphocytes by using forward light scatter versus orthogonal light scatter followed by gating on CD3+/CD4+ cells. Data are representative of four independent donors and experiments.
Summary and Conclusions
Our results demonstrate that CD4+ T cells produce HIV-1-suppressive factors against the X4 (HIV-1IIIB) and R5 (HIV-1BaL) viruses during primary responses in vitro. These activities were present at equivalent levels regardless of whether the cells were activated with SEB, TSST-1, or allogeneic MDDC. The anti-X4 activity was caused mainly by CCL22, and the anti-R5 activity was caused by the R5 ligands (CCL3, CCL4, and CCL5). This finding was confirmed by using neutralizing antibodies against CC chemokines to effectively reverse the anti-HIV-1-suppressor activity obtained from antigen-stimulated cells. We also showed that activated CD4+ T cells are the major source of these CC chemokines as determined by flow cytometry.
The HIV-1-suppressor activity of activated CD4+ T cells, during primary responses, paints a more complicated picture than expected for the means by which CD4+ T cells regulate HIV-1 infection. Our data suggest that if CD4+ T cells encounter HIV-1 in the right context, some of them will be protected against R5 and X4 viruses. This finding suggests a complicated outcome of acute infection in which some HIV-1-specific CD4+ T cells are infected and die, whereas others are resistant to HIV-1 infection by virtue of their ability to produce anti-X4 and anti-R5 factors. A major future effort will be to determine whenever the release of these factors, known and unknown, allows some degree of “autoprotection” in the CD4+ T cell population. It is also possible that the production of these antiviral factors drive the generation of the CD4+ T cells that function as the major lymphocytic reservoir of HIV-1 (reviewed in ref. 38). Most notably, our system makes it possible to address these issues in the context of the primary antigen-specific response of human CD4+ T cells. These studies identify targets for vaccine development to prevent either infection with HIV-1 or its ability to cause rapidly progressive disease.
In summary, the data presented above show that soluble HIV-suppressive factors are released during primary antigen-specific responses of CD4+ T cells and suggest that such responses should be considered in the design of vaccines against HIV-1 and as a mechanism whereby the host controls infections with this virus.
Supplementary Material
Acknowledgments
We thank Dr. Mario Roederer (Vaccine Research Center, National Institutes of Health, Bethesda), Dr. Donna Farber (University of Maryland, Baltimore), and Dr. Tarek M. Shata (New York Blood Bank, New York) for their helpful discussion during this work. This work is supported by National Institutes of Health Grants AI47490, AI43046, and AI3892 (to G.K.L.) and RO1 HL63647 (to A.D.) and the International AIDS Vaccine Initiative.
Abbreviations: APC, allophycocyanin; CFSE, carboxyfluorescein succinimidyl ester; MDDC, immature monocyte-derived dendritic cells; PE, phycoerythryin; R5, CCR5; SEB, Staphylococcus enterotoxin B; TCR, T cell receptor; TSST-1, toxic shock syndrome toxin 1; X4, CXCR4.
References
- 1.Altfeld, M. & Rosenberg, E. S. (2000) Curr. Opin. Immunol. 12, 375-380. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg, E. S., Billingsley, J. M., Caliendo, A. M., Boswell, S. L., Sax, P. E., Kalams, S. A. & Walker, B. D. (1997) Science 278, 1447-1450. [DOI] [PubMed] [Google Scholar]
- 3.Kalams, S. A. & Walker, B. D. (1998) J. Exp. Med. 188, 2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, D. P., Homann, D. & Oldstone, M. B. (2000) Virology 266, 257-263. [DOI] [PubMed] [Google Scholar]
- 5.Douek, D. C., Brenchley, J. M., Betts, M. R., Ambrozak, D. R., Hill, B. J., Okamoto, Y., Casazza, J. P., Kuruppu, J., Kunstman, K., Wolinsky, S., et al. (2002) Nature 417, 95-98. [DOI] [PubMed] [Google Scholar]
- 6.Walker, C. M., Moody, D. J., Stites, D. P. & Levy, J. A. (1986) Science 234, 1563-1566. [DOI] [PubMed] [Google Scholar]
- 7.Mackewicz, C. E., Yang, L. C., Lifson, J. D. & Levy, J. A. (1994) Lancet 344, 1671-1673. [DOI] [PubMed] [Google Scholar]
- 8.Zagury, D., Lachgar, A., Chams, V., Fall, L. S., Bernard, J., Zagury, J. F., Bizzini, B., Gringeri, A., Santagostino, E., Rappaport, J., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 3857-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchi, F., DeVico, A. L., Yarchoan, R., Redfield, R., Cleghorn, F., Blattner, W. A., Garzino-Demo, A., Colombini-Hatch, S., Margolis, D. & Gallo, R. C. (2000) Proc. Natl. Acad. Sci. USA 97, 13812-13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garzino-Demo, A., Moss, R. B., Margolick, J. B., Cleghorn, F., Sill, A., Blattner, W. A., Cocchi, F., Carlo, D. J., DeVico, A. L. & Gallo, R. C. (1999) Proc. Natl. Acad. Sci. USA 96, 11986-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dragic, T., Litwin, V., Allaway, G. P., Martin, S. R., Huang, Y., Nagashima, K. A., Cayanan, C., Maddon, P. J., Koup, R. A., Moore, J. P. & Paxton, W. A. (1996) Nature 381, 667-673. [DOI] [PubMed] [Google Scholar]
- 12.Furci, L., Scarlatti, G., Burastero, S., Tambussi, G., Colognesi, C., Quillent, C., Longhi, R., Loverro, P., Borgonovo, B., Gaffi, D., et al. (1997) J. Exp. Med. 186, 455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha, K., Volsky, D. J. & Matczak, E. (1999) J. Virol. 73, 7891-7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha, K., Bentsman, G., Chess, L. & Volsky, D. J. (1998) J. Virol. 72, 876-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell, D., Young, J. W. & Banchereau, J. (1999) Adv. Immunol. 72, 255-324. [DOI] [PubMed] [Google Scholar]
- 16.Hart, D. N. (1997) Blood 90, 3245-3287. [PubMed] [Google Scholar]
- 17.Banchereau, J. & Steinman, R. M. (1998) Nature 392, 245-252. [DOI] [PubMed] [Google Scholar]
- 18.Abdelwahab, S. F. (2002) Ph.D. thesis (University of Maryland, Baltimore).
- 19.Sallusto, F. & Lanzavecchia, A. (1994) J. Exp. Med. 179, 1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagley, K. C., Abdelwahab, S. F., Tuskan, R. G., Fouts, T. R. & Lewis, G. K. (2002) J. Leukocyte Biol. 72, 962-969. [PubMed] [Google Scholar]
- 21.Bagley, K. C., Abdelwahab, S. F., Tuskan, R. G., Fouts, T. R. & Lewis, G. K. (2002) Infect. Immun. 70, 5533-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyons, A. B. & Parish, C. R. (1994) J. Immunol. Methods 171, 131-137. [DOI] [PubMed] [Google Scholar]
- 23.Picker, L. J., Treer, J. R., Ferguson-Darnell, B., Collins, P. A., Buck, D. & Terstappen, L. W. (1993) J. Immunol. 150, 1105-1121. [PubMed] [Google Scholar]
- 24.Roederer, M., Raju, P. A., Mitra, D. K. & Herzenberg, L. A. (1997) J. Clin. Invest. 99, 1555-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roederer, M., Dubs, J. G., Anderson, M. T., Raju, P. A. & Herzenberg, L. A. (1995) J. Clin. Invest. 95, 2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marrack, P. & Kappler, J. (1990) Science 248, 705-711. [DOI] [PubMed] [Google Scholar]
- 27.Kotzin, B. L., Leung, D. Y., Kappler, J. & Marrack, P. (1993) Adv. Immunol. 54, 99-166. [DOI] [PubMed] [Google Scholar]
- 28.Horuk, R., Hesselgesser, J., Zhou, Y., Faulds, D., Halks-Miller, M., Harvey, S., Taub, D., Samson, M., Parmentier, M., Rucker, J., et al. (1998) J. Biol. Chem. 273, 386-391. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S., Tiffany, H. L., King, L., Murphy, P. M., Golding, H. & Zaitseva, M. B. (2000) J. Virol. 74, 6946-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal, R., Garzino-Demo, A., Markham, P. D., Burns, J., Brown, M., Gallo, R. C. & DeVico, A. L. (1997) Science 278, 695-698. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, L., Yu, W., He, T., Yu, J., Caffrey, R. E., Dalmasso, E. A., Fu, S., Pham, T., Mei, J., Ho, J. J., et al. (2002) Science 298, 995-1000. [DOI] [PubMed] [Google Scholar]
- 32.Agrawal, L., Vanhorn-Ali, Z. & Alkhatib, G. (2002) J. Leukocyte Biol. 72, 1063-1074. [PubMed] [Google Scholar]
- 33.Mengozzi, M., Malipatlolla, M., De Rosa, S. C., Herzenberg, L. A. & Roederer, M. (2001) Proc. Natl. Acad. Sci. USA 98, 11644-11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vulcano, M., Albanesi, C., Stoppacciaro, A., Bagnati, R., D'Amico, G., Struyf, S., Transidico, P., Bonecchi, R., Del Prete, A., Allavena, P., et al. (2001) Eur. J. Immunol. 31, 812-822. [DOI] [PubMed] [Google Scholar]
- 35.Cocchi, F., DeVico, A. L., Garzino-Demo, A., Arya, S. K., Gallo, R. C. & Lusso, P. (1995) Science 270, 1811-1815. [DOI] [PubMed] [Google Scholar]
- 36.Mackewicz, C. E., Barker, E. & Levy, J. A. (1996) Science 274, 1393-1395. [DOI] [PubMed] [Google Scholar]
- 37.Garzino-Demo, A., DeVico, A. L., Conant, K. E. & Gallo, R. C. (2000) Immunol. Rev. 177, 79-87. [DOI] [PubMed] [Google Scholar]
- 38.Blankson, J. N., Persaud, D. & Siliciano, R. F. (2002) Annu. Rev. Med. 53, 557-593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.