Abstract
Rate of recombination is a powerful variable affecting several aspects of molecular variation and evolution. A nonrecombining portion of the genome of most Drosophila species, the “dot” chromosome or F element, exhibits very low levels of variation and unusual codon usage. One lineage of Drosophila, the willistoni/saltans groups, has the F element fused to a normally recombining E element. Here, we present polymorphism data for genes on the F element in two Drosophila willistoni and one D. insularis populations, genes previously studied in D. melanogaster. The D. willistoni populations were known to be very low in inversion polymorphism, thus minimizing the recombination suppression effect of inversions. We first confirmed, by in situ hybridization, that D. insularis has the same E + F fusion as D. willistoni, implying this was a monophyletic event. A clear gradient in codon usage exists along the willistoni F element, from the centromere distally to the fusion with E; estimates of recombination rates parallel this gradient and also indicate D. insularis has greater recombination than D. willistoni. In contrast to D. melanogaster, genes on the F element exhibit moderate levels of nucleotide polymorphism not distinguishable from two genes elsewhere in the genome. Although some linkage disequilibrium (LD) was detected between polymorphic sites within genes (generally <500 bp apart), no long-range LD between F element loci exists in the two willistoni group species. In general, the distribution of allele frequencies of F element genes display the typical pattern of expectations of neutral variation at equilibrium. These results are consistent with the hypothesis that recombination allows the accumulation of nucleotide variation as well as allows selection to act on synonymous codon usage. It is estimated that the fusion occurred ∼20 Mya and while the F element in the willistoni lineage has evolved “normal” levels and patterns of nucleotide variation, equilibrium may not have been reached for codon usage.
Keywords: recombination, molecular variation, Drosophila willistoni group, F element, codon usage
Introduction
Since the seminal work of Kreitman (1983), numerous studies of DNA variation in Drosophila have revealed high levels of polymorphism (Moriyama and Powell 1996; Powell 1997; Begun et al. 2007). Generally, genes have nucleotide heterozygosity between 0.1% and 2% (proportion of nucleotide differences between two randomly drawn alleles) depending on function of the gene, exon, intron, flanking regions, etc. It was, therefore, notable when Berry et al. (1991) reported virtually no variation in Drosophila melanogaster and D. simulans in a gene (cubitus interuptus, ci) located on the fourth or “dot” chromosome, also known as the F element (Muller 1940). The lack of variation in ci was confirmed in two other species of the melanogaster subgroup (Hilton et al. 1994). This was followed by more detailed study of genes along the fourth chromosome of D. melanogaster (Wang et al. 2002). These authors found a 200 kb polymorphic section of this element with three major alleles, blocks of DNA in complete linkage disequilibrium (LD), throughout the species. Heterozygosity at the nucleotide level is high due to differences between just three ∼200 kb alleles each of which occurs in relatively high frequency. Further work on the melanogaster group species confirmed this low level of variation in F element genes and strong LD (Sheldahl et al. 2003; Wang et al. 2004; Arguello et al. 2010).
An exceptional pattern of codon usage has also been documented for genes on the F element compared with the rest of the Drosophila genome (Powell and Moriyama 1997; Vicario et al. 2007; Arguello et al. 2010). Codon usage in most of the genome exhibits strong bias indicative of selection for synonymous codon use, whereas genes on the F element appear to be under very little selection and codon usage reflects mutation bias.
These observations are consistent with there being virtually no recombination in the F element when it is present as a very short dot. In classical mapping experiments, no recombination has been detected in this chromosome in D. melanogaster (Hochman 1976). However, results from DNA variation studies (Wang et al. 2002, 2004; Arguello et al. 2010) imply rare recombination estimated to be about 1/100 that of the rest of the Drosophila nuclear genome. Selective sweeps or background selection (Charlesworth et al. 1993) of nonrecombining DNA induces monomorphism and low recombination promotes LD. This makes selection at specific sites, such as among synonymous codons, virtually impossible (the Hill–Robertson effect, 1966), especially if selection is weak as appears to be the case for synonymous mutations (Hartl et al. 1994; Akashi 1995). Theoretical analyses of data from the dot chromosome of Drosophila indicate that the Hill–Robertson effect is very strong in influencing patterns and levels of nucleotide polymorphisms (Kaiser and Charlesworth 2008; Betancourt et al. 2009; Charlesworth et al. 2009).
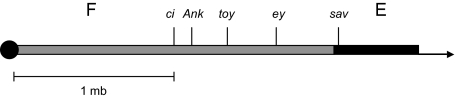
Most species of Drosophila have a small, independently segregating, dot chromosome (F element); exceptions are the D. willistoni and D. saltans groups (Clayton and Wheeler 1975). In D. willistoni, the F element has been incorporated into another arm, element E, adjacent to the centromere (fig. 1). This has been confirmed by in situ gene hybridization to polytene chromosomes (Papaceit and Juan 1998). We confirm in this paper that this same fusion occurs in another member of the willistoni group (D. insularis) indicating the fusion very likely predates the radiation of the willistoni group and is a monophyletic event.
FIG. 1.
Schematic map of E + F fusion in the Drosophila willistoni group for the genes studied in this report. Left solid dot represents the centromere. Data are from Papaceit and Juan (1998), this report, and the complete genome sequence (Drosophila 12 Genomes Consortium 2007).
Here, we present data on the DNA polymorphism in some of the same F element genes studied in D. melanogaster in two species of the willistoni group, D. willistoni and D. insularis. We also examined one E element gene just distal to the fusion. In essence, we ask the question of how the molecular evolution of ancestrally nonrecombining (or rarely recombining) genes has changed when moved to a recombining chromosome. We examine codon usage, estimates of recombination, and levels and patterns of nucleotide polymorphism.
Materials and Methods
Material
D. willistoni and D. insularis were collected in St. Lucia in December 2006. D. willistoni was also collected in Pichilingue, Ecuador, in May 2008. Species identifications were confirmed by species-specific DNA sequences in the Alcohol dehydrogenase (Adh) locus (Griffith and Powell 1997). DNA extractions were made from flies directly from the field or a single F1 of isofemale lines. Thus, the alleles sampled were directly from the natural populations.
In Situ Hybridization
Polytene chromosome preparations for in situ hybridization were performed according to Montgomery et al. (1987). Prehybridization, hybridization, and detection were as described in Segarra and Aguadé (1992). The location of the hybridization signals in both D. willistoni and D. insularis was determined using the Schaeffer et al. (2008) photographic map of D. willistoni, given the high chromosomal similarity between species. Probes from the eyeless (ey), cubitus interruptus (ci), and Ankyrin (Ank) genes were obtained through biotin-16-dUTP labeling by nick translation of either gel-purified polymerase chain reaction (PCR) amplicons of D. willistoni (ey and ci) or cDNA (Ank) of D. melanogaster (Papaceit and Juan 1998).
DNA Isolation and Sequencing
In addition to Adh sequences to confirm identification of the sibling species of the willistoni group (Griffith and Powell 1997), five genes on the E + F chromosome, cubitus interuptus (ci), Ankyrin (Ank), eyeless (ey), toy, and salvador (sav), were amplified and sequenced for the D. willistoni and D. insularis samples. Sav, on element E, lies just distal to element F in D. willistoni and D. insularis (fig. 1). Primers used to amplify the above genes are detailed in supplementary table S1, Supplementary Material online, and were designed using the Comparative Assembly Freeze 1 of the D. willistoni genome (FlyBase) and software program Primer 3 (Rozen and Skaletsky 2000). PCR amplification and sequencing details are in supplementary materials, Supplementary Material online.
Data Analyses
To analyze codon usage along the F element in D. melanogaster and D. willistoni, we used genome annotation from the FlyBase repository (www.flybase.org) version 5.22 and 1.3, respectively. We calculated the FOP statistics (the frequency of optimal codon) from the coding sequence of the longest transcript of each gene, using the optimal codon for each species identified in Vicario et al. (2007). Extraction of annotations from the gff files and construction of the codon usage table and FOP calculations were performed with Python script written by Saverio Vicario and using the parsing routine of BioPython (Cock et al. 2009). Trends of FOP statistics along the F element were analyzed with a linear model approach within R statistical environment (R Development Core Team 2009).
Phase of segregating sites in nuclear allele sequences obtained from diploid PCR products was estimated using PHASE v2.1.1 (Stephens et al. 2001; Stephens and Donnelly 2003), with files formatted in SeqPHASE (Flot 2010). The “MR” model was used because it accommodates the possibility of intragenic recombination. For triallelic single nucleotide polymorphisms, the parent-independent mutation model was used. Search settings were: 500 iterations as burn-in, 1,000 main iterations, and thinning interval = 1. PHASE has very low false–positive rates even when haplotype pairs have confidence probabilities as low as 0.60 and/or some coalescent assumptions are violated and systematic bias in population parameter estimates seems to occur only when unresolved (low confidence) genotypes are omitted from the data set (Garrick et al. 2010). Initially, PHASE was run to identify multisite heterozygotes with confidence scores <0.95, and PCR products from those individuals were cloned using the pGEM-T Easy Vector System (Promega) and resulting colonies were screened for the target insert using primers SP6 and T7 (Promega). For each individual, two positive colonies were sequenced. Additionally, indels within the ey locus prevented a number of D. willistoni and D. insularis individuals from being directly sequenced. The above cloning procedure was therefore employed to rectify this issue. Here, however, five to ten positive colonies per individual were sequenced, thus ensuring the accurate identification of alleles despite inherent PCR error. In subsequent runs of PHASE, cloned alleles were treated as “known” haplotypes. Each data set was run three times with a different starting seed, and consistency across runs was checked by eye. The haplotype pair with the highest confidence probability score from the replicate run with the best average goodness-of-fit value was accepted and used for calculation of summary statistics and assessment of LD.
DNA sequence diversity summary statistics were calculated for exons and introns separately for each locus, species, and population. Fu’s (1997) F and Tajima’s (1989) D neutrality tests were performed on the same data partitions. Significance of F and D was assessed by comparing the observed value with a null distribution simulated via the coalescent process under constant population size (1,000 replicates). Following Fu (1997), F was considered significant at the 5% level only if P < 0.02. Neutrality of exons was also assessed by calculating dN/dS (ω), the ratio of nonsynonymous to synonymous polymorphisms. Nonrandom associations between polymorphisms at segregating sites within a gene were examined via tests of LD including both exons and introns for each locus; significance of LD was assessed using Fisher’s exact test (two tailed) and Bonferroni correction for multiple comparisons. All calculations described in this paragraph were performed using DnaSP V5.10 (Librado and Rozas 2009).
Results
In Situ Mapping
Because only D. willistoni had been studied for position of F element genes (Papaceit and Juan 1998), we performed in situ hybridization of these genes in D. insularis. Supplementary figure S1, Supplementary Material online, has photographs of results for three F element genes, ci, ey, and Ank. The results confirm that the F element is in the same position in D. insularis, attached to element E adjacent to the centromere. In addition, the order of the F element genes are the same as in D. willistoni (fig. 1 and supplementary fig. S1, Supplementary Material online here and fig. 2 in Papaceit and Juan 1998), indicating that the attachment of the F and E elements was a single event in the ancestral lineage of the present day willistoni group.
Codon Usage
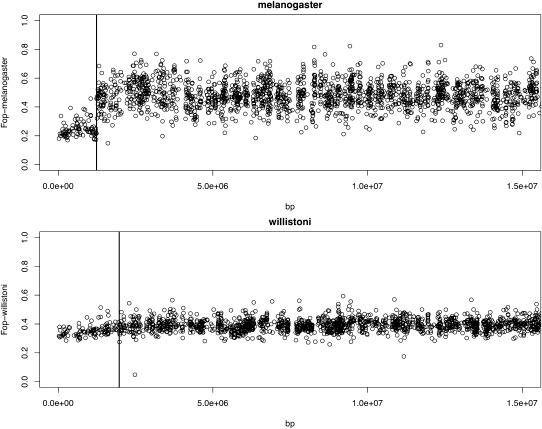
Figure 2 shows the pattern of codon usage along the lengths of elements E and F in D. melanogaster and D. willistoni based on the genome sequences (Drosophila 12 Genomes Consortium 2007). The frequency of the “optimal” codon (FOP) for each species (defined in Vicario et al. 2007) is along the length of the chromosomal elements measured in base pairs. As we have shown previously (Powell and Moriyama 1997; Vicario et al. 2007) in species that have this element as a small dot such as D. melanogaster, genes on the nonrecombining F element show reduced usage of optimal codons. In D. willistoni, the F element genes on the E + F chromosome exhibit FOP more like the E element genes in this species, although with a clear gradient from the centromere to the fusion point (fig. 2).
FIG. 2.
Frequency of optimal codons defined in Vicario et al. (2007) for protein-coding genes along the length of the E and F elements. In Drosophila melanogaster, these two elements exist as separate chromosomes (separated by vertical line) and are fused in D. willistoni with the junction noted by the vertical line. The telomere is to the left for both the E and F elements of D. melanogaster and E + F of D. willistoni. Only about half the E element is shown as assembly does not allow unambiguous continuation beyond this point for D. willistoni.
It is well known that the degree of usage of optimal codons for a species is directly proportional to recombination rate (Kliman and Hey 1993; Comeron et al. 1999; Comeron and Kreitman 2002; Hey and Kliman 2002). Furthermore, in Drosophila, it is well established that recombination rate falls off along arms of chromosomes toward both ends, the centromere and telomere. Table 1 presents the regressions of FOP and position along the chromosomes for approximately 2 Mb adjacent to the centromere and 2 Mb at the telomere (2 Mb is the approximate size of the F element euchromatin). As expected, there is a significant positive correlation going from the centromere out 2 Mb and a negative correlation in the last 2 Mb toward the telomere for D. melanogaster. For D. willistoni, there is a highly significant correlation between position of the F element genes and FOP, but unlike in D. melanogaster, the centromeric end of the E element in D. willistoni exhibits no significant gradient in FOP. There remains a significant negative gradient at the telomeric end of element E in D. willistoni comparable in magnitude with that in D. melanogaster.
Table 1.
Regression Coefficients (R) between Frequency of Optimal Codon (FOP) and Position in Base Pairs along Elements Starting from the Centromere. 2 Mb Is Used for Comparison as That Is the Approximate Length of the F Element.
| Species | F Element | First 2 Mb Element E | Last 2 Mb Element E |
| willistoni | +0.362*** | +0.105 NS | −0.164* |
| melanogaster | +0.022 NS | +0.155* | −0.187** |
NOTE.—NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
These results confirm that codon usage in F element genes in D. willistoni has begun to evolve to higher usage of optimal codons and that there still exists a steep gradient from the centromere toward the fusion point, most likely due to a gradient in recombination. Because the centromeric end of the E element shifted with the fusion event, the formerly centromeric end of the E element has lost its gradient in codon usage in D. willistoni.
Recombination Estimations
We used PHASE to estimate recombination between the four genes studied in D. willistoni and D. insularis. Table 2 presents the numerical results based on detailed data in supplementary table S2, Supplementary Material online. These estimates are for c which is 4Ner, so it is conceivable that the difference between species is due to a difference in Ne. Given the very restricted distribution of D. insularis (only known from two Caribbean islands) and the wide distribution of D. willistoni (throughout the Neotropics), it is highly unlikely the difference between species in the estimates of c are due to D. insularis having a greater Ne than D. willistoni, at least for contemporary population sizes. Second, although the confidence intervals on these estimates are large and overlapping, it is notable that in both species, the pattern is in the expected direction of a gradient of recombination from the centromere outward. This is consistent with the gradient in codon usage documented above.
Table 2.
Estimates from PHASE for Recombination between Genes. Figures are Log10 4Ner, where r Is the Actual Estimated Recombination. Regions Are Listed from Centromere Toward the Distal Fusion Point.
| Species | ci-Ank | Ank-toy | toy-Sav | |
| inuslaris | Upper 95% | −1.585 | −1.404 | −1.268 |
| Mode | −2.063 | −1.844 | −1.637 | |
| Lower 95% | −2.923 | −2.285 | −2.002 | |
| willistoni | Upper 95% | −4.207 | −1.919 | −1.878 |
| Mode | −6.018 | −3.698 | −2.611 | |
| Lower 95% | −7.995 | −5.052 | −3.846 |
Levels of Molecular Variation
Table 3 summarizes the levels of nucleotide variation found in D. willistoni and D. insularis (detailed data in supplementary material, Supplementary Material online). In both species, all genes studied on the E + F chromosome (three or four in the F element and one in the E element adjacent to the fusion) exhibit a moderate amount of nucleotide polymorphism (with the exception of ci in the St. Lucia D. willistoni sample). These π and θ estimates are about an order of magnitude greater than those found for F element genes in melanogaster species (Sheldahl et al. 2003; Wang et al. 2004; Arguello et al. 2010). Adh on element B in D. willistoni and D. insularis is comparable with these element F genes in level of nucleotide variation with the exception of Adh in the Ecuador population of D. willistoni. This locus in this population sample is a clear outlier exhibiting much higher exon variation than any other locus, 23 haplotypes in a sample of only 44 alleles (supplementary table S2, Supplementary Material online). dN/dS ratios do not indicate this excess in exon variation is due to nonsynonymous variation (supplementary table S3, Supplementary Material online). There is no obvious explanation for this exceptional sample.
Table 3.
Summary of Variation in Six Genes Studied in this Paper.
| Exon(s) | Intron(s) | ||||||||||||||
| Locus | Species | Poplulation | N | bps | Haps | S | Hd | θ | π | bps | Haps | S | Hd | θ | π |
| ci | Drosophila insularis | STL | 166 | 781 | 7 | 7 | 0.750 | 1.6 | 2.1 | 55 | 3 | 2 | 0.247 | 6.4 | 5.4 |
| D. willistoni | STL + ECU | 148 | 781 | 3 | 2 | 0.440 | 0.5 | 0.8 | 55 | 3 | 2 | 0.027 | 6.5 | 0.5 | |
| STL | 104 | 781 | 1 | 0 | — | — | — | 55 | 2 | 1 | 0.019 | 3.5 | 0.4 | ||
| ECU | 44 | 781 | 3 | 2 | 0.553 | 0.6 | 0.8 | 55 | 2 | 1 | 0.045 | 4.2 | 0.8 | ||
| Ank | D. insularis | STL | 96 | 371 | 10 | 7 | 0.694 | 3.7 | 3.2 | 106 | 4 | 4 | 0.656 | 7.3 | 14.3 |
| D. willistoni | STL + ECU | 134 | 371 | 5 | 4 | 0.059 | 2.0 | 0.2 | 106 | 2 | 1 | 0.382 | 2.3 | 4.8 | |
| STL | 90 | 371 | 3 | 2 | 0.044 | 1.1 | 0.1 | 106 | 2 | 1 | 0.470 | 2.5 | 6.0 | ||
| ECU | 44 | 371 | 4 | 3 | 0.133 | 1.9 | 0.5 | 106 | 2 | 1 | 0.045 | 2.9 | 0.6 | ||
| toy | D. insularis | STL | 104 | 590 | 8 | 8 | 0.647 | 2.6 | 2.2 | 258 | 11 | 8 | 0.728 | 6.4 | 6.1 |
| D. willistoni | STL + ECU | 140 | 590 | 8 | 9 | 0.455 | 2.8 | 1.9 | 261 | 6 | 6 | 0.297 | 4.8 | 1.5 | |
| STL | 96 | 590 | 7 | 8 | 0.506 | 2.6 | 2.0 | 261 | 3 | 2 | 0.120 | 1.7 | 0.5 | ||
| ECU | 44 | 590 | 3 | 5 | 0.321 | 2.0 | 1.4 | 261 | 5 | 5 | 0.559 | 5.1 | 3.2 | ||
| ey | D. willistoni | STL + ECU | 92 | 748 | 12 | 10 | 0.767 | 2.6 | 3.6 | 569 | 8 | 9 | 0.728 | 3.1 | 3.8 |
| STL | 48 | 748 | 9 | 10 | 0.715 | 3.0 | 3.6 | 569 | 6 | 7 | 0.616 | 2.8 | 3.2 | ||
| ECU | 44 | 748 | 7 | 6 | 0.707 | 1.8 | 2.5 | 569 | 7 | 8 | 0.789 | 3.2 | 4.3 | ||
| sav | D. insularis | STL | 92 | 983 | 13 | 9 | 0.779 | 1.8 | 1.8 | 80 | 3 | 3 | 0.382 | 7.5 | 8.1 |
| D. willistoni | STL + ECU | 78 | 983 | 9 | 10 | 0.728 | 2.1 | 1.3 | 187 | 4 | 3 | 0.124 | 3.3 | 0.7 | |
| STL | 38 | 983 | 7 | 8 | 0.694 | 2.0 | 1.4 | 187 | 2 | 1 | 0.053 | 1.3 | 0.3 | ||
| ECU | 40 | 983 | 5 | 4 | 0.731 | 1.0 | 1.1 | 187 | 3 | 2 | 0.188 | 2.5 | 1.0 | ||
| Adh | D. insularis | STL | 132 | 612 | 10 | 9 | 0.694 | 2.7 | 1.7 | — | — | — | — | — | — |
| D. willistoni | STL + ECU | 144 | 612 | 33 | 28 | 0.797 | 8.3 | 3.9 | — | — | — | — | — | — | |
| STL | 100 | 612 | 13 | 11 | 0.609 | 3.5 | 2.2 | — | — | — | — | — | — | ||
| ECU | 44 | 612 | 23 | 24 | 0.874 | 9.0 | 4.9 | — | — | — | — | — | — | ||
NOTE.—STL, St. Lucia, ECU, Ecuador; N, number of alleles sampled; bps, number of aligned base pairs; Haps, number of haplotypes; S, number of segregating sites; Hd, haplotype (gene) diversity; θ, Watterson’s estimate × 103; π = obs. nucleotide heterozygosity × 103.
Excluding D. willistoni Adh in Ecuador, it appears that the genes on the F element in the willistoni lineage have equivalent levels of nucleotide variation as the two non-F element genes. Mean θ (× 103) for exons of F element genes is 1.9 (SD = 1.07), whereas Adh (excluding Ecuador) and Sav have mean θ of 2.2 (SD = 0.95). This level of variation in F element genes in D. willistoni and D. insularis is much higher than the exceptionally low variation found in these genes in species of the melanogaster subgroup (Berry et al. 1991; Hilton et al. 1994; Wang et al. 2002). (Note that we do not include data on the ey locus for D. insularis because it became clear that this gene is in multiple copies in this species and thus unambiguously determining orthology and paralogy is difficult. We simply exclude these data.)
It is also notable that for all loci studied, both on the F element and sav, D. insularis introns display considerably greater nucleotide diversity than that found in D. willistoni introns. This is consistent with the higher rate of recombination estimated for D. insularis compared with D. willistoni (table 2).
Pattern of Molecular Variation
The pattern of nucleotide variation is also very different in the fused E + F in D. willistoni and D. insularis compared with species where these elements are separate chromosomes. Wang et al. (2002) documented very strong LD along the length of the F element in D. melanogaster. In tests of LD, we found several cases of significant (after Bonferroni correction for multiple tests) associations within genes, sites that averaged 50–400 bps apart (supplementary table S4, Supplementary Material online). However, we found no significant LD between loci.
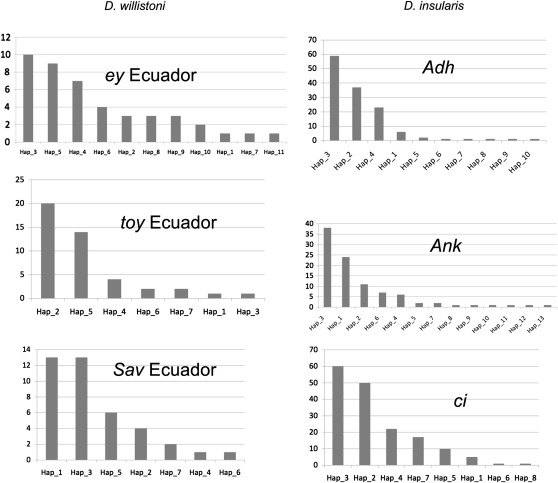
Examples of the distribution of allele frequencies are in figure 3 with similar figures for all loci in all populations in supplementary figure S2, Supplementary Material online. All loci exhibit the expected pattern of a most frequent allele followed by a few alleles in moderate frequency trailing off to singletons. The exception is ci in D. willistoni that exhibits very little variation. The patterns of distributions in figure 3 are the expected at equilibrium for neutral alleles. Tests of the conformance to these expectations are Tajima (1989) and Fu (1997), results of which are in supplementary table S5, Supplementary Material online. The Ank locus exons (but not introns) give some indication of nonconformance to neutral expectations as do toy introns. The exceptional variation in Adh in the Ecuador sample also yields tests at odds with neutrality. However, as to whether these tests indicate recent bottleneck or some other demographic perturbation is not at all clear; for example, the negative Ank F’s and D for exons are twice accompanied by positive values for introns in the same genes. Generally, the F element genes in the willistoni group species display levels of accumulated nucleotide variation and its distribution (pattern) along haplotypes is not distinct from the two genes on other elements in these species (fig. 3).
FIG. 3.
Examples of distribution of allele frequencies determined by concatinating exons and introns. One example is given for each of the six genes studied, three for each species. The counts are shown on the ordinate and haplotypes arbitrarily numbered along the abscissa. Details in supplementary material, Supplementary Material online including similar plots for all loci in all samples and haplotype sequences.
Discussion
Timing of the Fusion
The willistoni group is part of the willistoni–saltans lineage in subgenus Sophophora. All members of this lineage lack the dot chromosome (Clayton and Wheeler 1975). Here, we show that two members of the willistoni group have the fusion of elements E and F and the order of genes is identical. Because D. insularis and D. willistoni have the deepest branch within the sibling species of the willistoni group (Gleason et al. 1998; Robe et al. 2010), it is most parsimonious to assume the fusion was a monophyletic event in the ancestral lineage of this group. Given the absence of the dot in the saltans group, it is likely the fusion predated the split between the willistoni and saltans groups. These groups are estimated to have last shared a common ancestor ∼20 Mya (Powell et al. 2003; O’Grady P, personal communication).
Codon Usage
As first documented by Kliman and Hey (1993) and since confirmed by a number of studies, codon usage bias is directly proportional to recombination rate, that is, genes in regions of higher recombination have higher codon usage bias than genes in regions of low recombination. This is likely due to the ineffectiveness of selection at single sites due to genetic hitchhiking (Hill and Robertson 1966). Our results are consistent with this. Because the F element became attached to the E element, codon usage has begun to evolve to that optimal for D. willistoni, although evidently the ∼20 My since this fusion has not been long enough to bring the F element genes into equilibrium. The frequencies of optimal codons in the D. willistoni F element genes have not yet reached the pattern typical of ∼2Mb adjacent to centromeres, for example, compare with element E in D. melanogaster (fig. 2). The slope of F element genes is much steeper in D. willistoni compared with either the E element telomeric region or centromeric regions in D. melanogaster.
That ∼20 My is not long enough to bring about complete conformance of F element genes to optimal codon usage confirms that very small selection coefficients are associated with selection among synonymous codons (e.g., Hartl et al. 1994; Akashi 1995; Maside et al. 2004). The fact that codon usage has shifted for some amino acids in D. willistoni (Anderson et al. 1993; Powell et al. 2003; Vicario et al. 2007) suggests that the shift in codon usage predates the E + F fusion, that is, the codon usage of the F element genes have not risen to the level of the shift in codon usage in the rest of the genome. The split of the saltans/willistoni lineage from the obscura/melanogaster lineage in subgenus Sophophora is estimated to have occurred 30–40 Mya (Russo et al. 1995; Powell et al. 2003; O’Grady P, personal communication). Thus, there were 10–20 My between the origin of the saltans/willistoni linage and the split between saltans and willistoni. Given that both these groups have the same pattern of codon usage shift (Rodriguez-Trelles et al. 2000; Powell et al. 2003), this shift must have begun during the 10–20 My when they shared a common ancestor. In other words, 30–40 My is long enough to evolve the unique codon usage pattern in genes in most of the genomes of the saltans/willistoni groups, but ∼20 My is long enough to only partially achieve this shift in the F element genes.
Nucleotide Diversity
Unlike codon usage, the amount and pattern of nucleotide diversity of F element genes in D. willistoni and D. insularis appear to have converged on a normal pattern for these species. Very few genes have been studied for polymorphism, but of those that have, there is little indication of differences between F element genes and the rest of the genome with regard to level of polymorphism. Similarly, except for linked sites within genes, there is no outstanding LD. There is no indication of LD between loci, unlike that found in D. melanogaster (Wang et al. 2002). For the most part, the distributions of allele frequencies (fig. 3 and supplementary material, Supplementary Material online) also indicate patterns consistent with expectations of accumulation of neutral variation at equilibrium.
Recombination and Population History
Our estimates of recombination (table 2) are consistent with the codon usage (fig. 2). There is a parallel gradient in estimated recombination rates with usage of optimal codons, although it is noted that these recombination estimates have large confidence intervals and this pattern is not statistically significant. Consistent with the higher estimated recombination rate in D. insularis compared with D. willistoni, the former species has higher levels of nucleotide diversity in introns compared with the latter species (table 3). What is not clear is why D. insularis has a higher recombination rate than D. willistoni, one to four orders of magnitude greater (table 2). This could be due to the high number of paracentric inversion known in D. willistoni. However, the populations of D. willistoni studied here, St. Lucia and Pichilingue, were chosen precisely because they were known to be low in inversion polymorphism (Dobzhansky 1957; da Cunha et al. 1959; Ayala et al. 1971) to minimize the effect of inversions suppressing recombination. Nevertheless, one inversion in the E + F element of D. willistoni (the III-A) is very close to the fusion point but appears not to include the F element (pictured in Dobzhansky et al. 1950; compared with fig. 10 in Schaeffer et al. 2008). This inversion is absent or rare in most populations of this species (including those studied in this report), although heterozygotes for the III “A or B” inversion does reach ∼50% in some Peruvian and Brazilian populations (da Cunha et al. 1959; these authors only report the frequencies of inversion heterozygotes rather than gene arrangement frequencies). Nothing is known of naturally occurring inversions in D. insularis.
Because PHASE estimates recombination as the parameter 4Ner, where r is the recombination value, it is possible that the difference between species for these estimates are due to differences in Ne. We argued above that it is very unlikely that D. insularis has a larger contemporary Ne than D. willistoni given the narrow distribution of the former and wide distribution of the latter (Ehrman and Powell 1982). However, it is conceivable that ancestral Nes are very different from those today. Comparisons of θ and π (table 3) and the allele frequency plots (fig. 3 and supplementary fig. S2, Supplementary Material online) are inconsistent among the loci in regard to whether they indicate a bottleneck (or expanding population size). The statistical comparison of θ and π, Tajima’s test, should be negative in a recently bottlenecked expanding population, yet as noted above, the loci are ambiguous in this regard with some loci exhibiting significantly negative measures in exons and positive measures in introns of the same genes. Previous findings of surprisingly high nonsynonymous polymorphism in the Adh locus (Griffith and Powell 1997) led these authors to speculate D. willistoni may have recently experienced a bottleneck; the present data do not confirm an excess of nonsynonymous Adh polymorphisms in this species (supplementary table S3, Supplementary Material online). One might expect to find F element genes to show patterns of bottleneck and expansion given that the fusion was monophyletic, which would have meant a bottleneck of one when the fusion occurred. Such a bottleneck is no longer evident in the data, not surprising if the fusion took place at least 20 Mya; with five generations per year, ∼108 generations have elapsed since the fusion.
Codon Usage Bias Intensity in willistoni and melanogaster
In figure 2, we presented the frequency of optimal codon usage along about half the length of the E element for D. melanogaster and D. willistoni. It is evident that the scatter of data points for D. melanogaster is greater than that for D. willistoni. In fact, what this shows is that the lower bound for both species is about the same, ∼0.35, but that the range above this lower bound is greater for D. melanogaster. That is, there are more genes with extreme bias in D. melanogaster than in D. willistoni. Because FOP is an average across all amino acids and there has been a change in optimal codon in D. willistoni for about a third of the amino acids (Powell et al. 2003; Vicario et al. 2007), the period of time that D. willistoni has been under selection for the shift in optimal codons for these amino acids may cause this parameter averaged across all amino acids not to reach extremes. Alternatively, it may indicate that the effective population size in the lineage leading to D. melanogaster was historically larger than the lineage leading to D. willistoni, meaning more effective selection for mutations with very small selection coefficients.
Conclusions and Caveats
It is clear that the level and pattern of variation in genes on the F element in the willistoni group is much different from that found in these genes in species where they are on a (nearly) nonrecombining chromosome, the dot. Because the genes studied are the same as those studied in D. melanogaster, it is unlikely the differences are due to gene-specific constraints. Moving these genes into a recombining part of the genome has allowed accumulation of variation with no LD over distances above about 500 bp as well as allowed these genes to evolve codon usage closer to the optimal for the species. Have willistoni group F element genes come to an equilibrium? There is no indication that F element genes are less variable than genes in other parts of the genome in the willistoni group (table 3) or that the pattern of distribution of allele frequencies is different (fig. 3). It is less clear if codon usage in F element genes has reached an equilibrium. Given that this element is adjacent to the centromere in the E + F chromosome, a strong gradient in codon usage exists (fig. 2) that likely reflects a gradient in recombination. The steepness of this gradient is larger for the E + F chromosome than that in comparable regions indicating equilibrium may not yet have been reached for codon usage.
Although we estimated that the fusion of F and E elements likely occurred ≥20 Mya, it is conceivable that this estimate is off. It is based on the assumptions of a molecular clock (Powell et al. 2003; O’Grady P, personal communication). In addition, we are assuming the most parsimonious interpretation, that this fusion was a monophyletic event predating separation of the willistoni and saltans groups, which may not be the case.
Supplementary Material
Supplementary tables S1–S5 and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
For assistance in collecting Drosophila, we thank Alessandro Powell, Sam Genecin, and Gisella Caccone for the St. Lucia sample, and Andrea Acurio and Violeta Raphael for the Ecuador sample. Patrick O’Grady shared his findings with regard to timing of the splits of Drosophila lineages. Richard Kliman made useful comments on a draft of this manuscript. This work was supported by the US National Institutes of Health (RO1 GM0077533).
References
- Akashi H. Inferring weak selection from patterns of polymorphism and divergences at “silent” sites in Drosophila DNA. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C, Carew EC, Powell JR. Evolution of the Adh locus in the Drosophila willistoni group: Loss of an intron and shift in codon usage. Mol Biol Evol. 1993;10:605–618. doi: 10.1093/oxfordjournals.molbev.a040027. [DOI] [PubMed] [Google Scholar]
- Arguello JR, Zhang Y, Kado T, Fan C, Zhao R, Innan H, Wang W, Long M. Recombination yet inefficient selection along the Drosophila melanogaster subgroup’s fourth chromosome. Mol Biol Evol. 2010;27:848–861. doi: 10.1093/molbev/msp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala FJ, Powell JR, Dobzhansky Th. Enzyme variability in the Drosophila willistoni group. II. Polymorphisms in continental and island populations of Drosophila willistoni. Proc Natl Acad Sci U S A. 1971;68:2480–2483. doi: 10.1073/pnas.68.10.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. (13 co-authors) Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry AJ, Ajioka JW, Kreitman M. Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics. 1991;129:1111–1117. doi: 10.1093/genetics/129.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ, Welch JJ, Charlesworth B. Reduced effectiveness of selection caused by lack of recombination. Curr Biol. 2009;19:655–660. doi: 10.1016/j.cub.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Betancourt AJ, Kaiser VB, Gordo I. Genetic recombination and molecular evolution. Cold Spring Harbor Symp Quant Biol. 2009;74:1–10. doi: 10.1101/sqb.2009.74.015. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton FE, Wheeler MR. A catalog of Drosophila metaphase chromosome configurations. In: King RC, editor. Handbook of genetics. Invertebrates of genetic interest. 1975. Vol. 3. New York: Plenum. p. 471–512. [Google Scholar]
- Cock PJA, Antao T, Chang JT, et al. (11 co-authors) Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M. Population, evolutionary and genomic consequences of interference selection. Genetics. 2002;161:389–410. doi: 10.1093/genetics/161.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M, Aguadé M (11 co-author) Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha AB, Dobzhansky Th, Pavlovsky O, Spassky B. Genetics of natural populations. XXVIII. Supplementary data on the chromosomal polymorphism in Drosophila willistoni in its relation to the environment. Evolution. 1959;13:389–404. [Google Scholar]
- Dobzhansky Th. Genetics of natural populations. XXVI. Chromosomal variability in island and continental populations of Drosophila willistoni from Central America and the West Indies. Evolution. 1957;11:280–293. [Google Scholar]
- Dobzhansky Th, Burla H, da Cunha AB. A comparative study of chromosomal polymorphism in sibling species of the willistoni group of Drosophila. Am Nat. 1950;84:229–246. [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes in the context of the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Ehrman L, Powell JR. The Drosophila willistoni species group. In: Ashburner M, Carson HL, Thompson J, editors. The genetics and biology of Drosophila. 1982. Vol. 3b. London: Academic Press. [Google Scholar]
- Flot J-F. SeqPHASE: a web tool for interconverting PHASE input/output files and FASTA sequence alignments. Mol Ecol Resour. 2010;10:162–166. doi: 10.1111/j.1755-0998.2009.02732.x. [DOI] [PubMed] [Google Scholar]
- Fu Y-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick RC, Sunnucks P, Dyer RJ. Nuclear gene phylogeography using PHASE: dealing with unresolved genotypes, lost alleles, and systematic bias in parameter estimation. BMC Evol Biol. 2010;10:118. doi: 10.1186/1471-2148-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JM, Griffith EC, Powell JR. A molecular phylogeny of the Drosophila willistoni group: conflicts between species concepts? Evolution. 1998;52:1093–1103. doi: 10.1111/j.1558-5646.1998.tb01836.x. [DOI] [PubMed] [Google Scholar]
- Griffith EC, Powell JR. Adh nucleotide variation in Drosophila willistoni: high replacement polymorphism in an electrophoretically monomorphic protein. J Mol Evol. 1997;45:232–237. doi: 10.1007/pl00006225. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Moriyama EN, Sawyer SA. Selection intensity for codon bias. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Kliman RM. Interactions between natural selection, recombination and gene density in the genes of Drosophila. Genetics. 2002;160:595–608. doi: 10.1093/genetics/160.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hilton H, Kliman RM, Hey J. Using hitchhiking genes to study adaptation and divergence during speciation within the Drososphila melanogaster species complex. Evolution. 1994;48:1900–1913. doi: 10.1111/j.1558-5646.1994.tb02222.x. [DOI] [PubMed] [Google Scholar]
- Hochman B. The fourth chromosome of Drosophila melanogaster. In: Ashburner M, Novitski E, editors. Genetics and biology of Drosophila. 1976. Vol. 1b. London: Academic Press. p. 903–928. [Google Scholar]
- Kaiser VB, Charlesworth B. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 2008;25:9–12. doi: 10.1016/j.tig.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Kliman RM, Hey J. Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol Biol Evol. 1993;10:1239–1258. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983;304:412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Maside X, Lee AW, Charlesworth B. Selection on codon usage in Drosophila americana. Curr Biol. 2004;14:150–154. doi: 10.1016/j.cub.2003.12.055. [DOI] [PubMed] [Google Scholar]
- Montgomery E, Charlesworth B, Langley CH. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 1987;49:31–41. doi: 10.1017/s0016672300026707. [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR. Intraspecific nuclear DNA variation in Drosophila. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Bearings of the ‘Drosophila’ work on systematics. In: Huxley J, editor. The new systematics. Oxford: Clarendon Press; 1940. pp. 185–268. [Google Scholar]
- Papaceit M, Juan E. Fate of dot chromosome genes in Drosophila willistoni and Scaptodrosophila lebononensis determined by in situ hybridization. Chrom Res. 1998;6:49–54. doi: 10.1023/a:1009218508672. [DOI] [PubMed] [Google Scholar]
- Powell JR. Progress and prospects in evolutionary biology: the Drosophila model. New York: Oxford University Press; 1997. [Google Scholar]
- Powell JR, Moriyama EN. Evolution of codon usage bias in Drosophila. Proc Natl Acad Sci U S A. 1997;94:7784–7790. doi: 10.1073/pnas.94.15.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Sezzi E, Moriyama EN, Gleason JM, Caccone A. Analysis of a shift in codon usage in Drosophila. J Mol Evol. 2003;57:s214–s225. doi: 10.1007/s00239-003-0030-3. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2009. ISBN 3-900051-07-0. Available from http://www.R-project.org. [Google Scholar]
- Robe LJ, Cordeiro J, Loreto ELS, Valente VLS. Taxonomic boundaries, phylogenetic relationships and biogeography of the Drosophila willistoni subgroup (Diptera: Drosophilidae) Genetica. 2010;138:601–617. doi: 10.1007/s10709-009-9432-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles FR, Tarrio F, Ayala FJ. Evidence for a high ancestral GC content in Drosophila. Mol Biol Evol. 2000;17:1710–1717. doi: 10.1093/oxfordjournals.molbev.a026269. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer 3 on the WWW for general users and for biologist programmers. 2000 doi: 10.1385/1-59259-192-2:365. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa (NJ): The Humana Press Inc. p. 365–386. [DOI] [PubMed] [Google Scholar]
- Russo CAM, Takezaki N, Nei M. Molecular phylogeny and divergence times of Drosophilid species. Mol Biol Evol. 1995;1:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW, Bhutkar A, McAllister BF, et al. (38 co-authors) Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra C, Aguadé M. Molecular organization of the X chromosome in different species of the obscura group of Drosophila. Genetics. 1992;130:513–521. doi: 10.1093/genetics/130.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldahl LA, Weinreich DM, Rand DM. Recombination, dominance and selection on amino acid polymorphism in the Drosophila genome: contrasting patterns on the X and fourth chromosomes. Genetics. 2003;165:1195–1208. doi: 10.1093/genetics/165.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila. BMC Evol Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Thornton WK, Berry A, Long M. Nucleotide variation along the Drosophila melanogaster fourth chromosome. Science. 2002;295:134–137. doi: 10.1126/science.1064521. [DOI] [PubMed] [Google Scholar]
- Wang W, Thornton K, Emerson JJ, Long M. Nucleotide variation and recombination along the fourth chromosome in Drosophila simulans. Genetics. 2004;166:1783–1794. doi: 10.1534/genetics.166.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.