Abstract
Rapid advances in DNA sequencing and genotyping technologies are beginning to reveal the scope and pattern of human genomic variation. Although single nucleotide polymorphisms (SNPs) have been intensively studied, the extent and form of variation at other types of molecular variants remain poorly understood. Polymorphism at the most variable loci in the human genome, microsatellites, has rarely been examined on a genomic scale without the ascertainment biases that attend typical genotyping studies. We conducted a genomic survey of variation at microsatellites with at least three perfect repeats by comparing two complete genome sequences, the Human Genome Reference sequence and the sequence of J. Craig Venter. The genomic proportion of polymorphic loci was 2.7%, much higher than the rate of SNP variation, with marked heterogeneity among classes of loci. The proportion of variable loci increased substantially with repeat number. Repeat lengths differed in levels of variation, with longer repeat lengths generally showing higher polymorphism at the same repeat number. Microsatellite variation was weakly correlated with regional SNP number, indicating modest effects of shared genealogical history. Reductions in variation were detected at microsatellites located in introns, in untranslated regions, in coding exons, and just upstream of transcription start sites, suggesting the presence of selective constraints. Our results provide new insights into microsatellite mutational processes and yield a preview of patterns of variation that will be obtained in genomic surveys of larger numbers of individuals.
Keywords: microsatellites, tandem repeats, population genomics, mutation, human genome
Introduction
Advances in genotyping and sequencing technologies have made feasible the measurement of human genetic variation on the genomic scale. Genomic variation is now routinely surveyed in large numbers of humans to reveal the relatedness among individuals and populations, historical migration routes, population expansions and declines, genetic determinants of phenotypic variation, and targets of natural selection. These inferences have been primarily based on single nucleotide polymorphisms (SNPs), the most common type of genetic variant in the human genome. Although large-scale copy number variants have seen increased attention in recent years (Conrad et al. 2006; Fiegler et al. 2006; Hinds et al. 2006; Locke et al. 2006; Redon et al. 2006; Perry 2008; Perry et al. 2008), understanding of the magnitude and form of alternative modes of DNA variation continues to lag far behind knowledge of SNP variation.
Microsatellites, or short tandem repeats, are among the most variable loci in the human genome. In human population genetics, microsatellites have been especially useful for characterizing genetic variation, providing detailed portraits of population structure and demographic history (Bowcock et al. 1994; Jorde et al. 1995, 1997; Kimmel et al. 1998; Rosenberg et al. 2002, 2005, 2006; Manica et al. 2005; Wang et al. 2007; Friedlaender et al. 2008; Tishkoff et al. 2009). Patterns of microsatellite polymorphism are intimately tied to the mutational process. The high levels of variation are attributable to rapid mutation, occurring at rates (10−3–10−5 per generation) (Weber and Wong 1993; Ellegren 2000) that are orders of magnitude higher than those at single nucleotides (10−8–10−9) (Nachman and Crowell 2000). Microsatellite mutation occurs primarily by replication slippage (Levinson and Gutman 1987; Ellegren 2000) and can return allele sizes to states already present in the population, complicating historical interpretations (Estoup et al. 2002). The most popular framework for interpreting microsatellite variation is the stepwise mutation model in which each new mutation causes the addition or subtraction of one repeat with equal probability (Ohta and Kimura 1973; Kimmel and Chakraborty 1996). Although aspects of human microsatellite variation appear to be adequately described by this model (Shriver et al. 1993; Valdes et al. 1993; Weber and Wong 1993; Kayser et al. 2000), additional complexities to the mutational process have been revealed, including multi-step mutations (Di Rienzo et al. 1994, 1998; Ellegren 2000; Huang et al. 2002), higher mutation rates at longer microsatellites (Weber and Wong 1993; Brinkmann et al. 1998; Webster et al. 2002; Whittaker et al. 2003; Ellegren 2004; Sainudiin et al. 2004; Legendre et al. 2007; Kelkar et al. 2008), biases toward expansion and contraction (Amos et al. 1996; Xu et al. 2000; Ellegren 2004), and effects of repeat sequence and length on mutation (Weber and Wong 1993; Chakraborty et al. 1997; Kelkar et al. 2008).
Conclusions about the microsatellite mutation process in humans have been primarily based on three approaches: analysis of pedigrees, comparison with chimpanzee genome sequences, and survey of polymorphism in populations. Each strategy has advantages and disadvantages. Studies of human pedigrees allow direct observation of mutations but usually tabulate a small number of mutational events due to limitations in the number of generations surveyed. Although comparisons between human and chimpanzee sequences yield a genomic view of factors affecting microsatellite evolution, they are complicated by the accumulation of multiple mutations per locus (homoplasy) due to the combination of long divergence time and high mutation rate. Polymorphism surveys, which provide useful descriptions of variation within and among human populations, have been focused thus far on relatively small numbers of loci that were previously ascertained to be highly variable (but see Brandström et al. 2008). Recently, Molla et al. (2009) circumvented this ascertainment bias, comparing trinucleotide microsatellites found in genic regions (exons and introns) of the human reference genome sequence to those in the Venter sequence, the Watson sequence, and the chimpanzee reference sequence. Molla et al. (2009) found a positive relationship between variation (polymorphism within humans and divergence with chimpanzee) and repeat number, suggesting that longer loci mutate faster. Additionally, a comparison between polymorphism and divergence revealed no strong evidence for natural selection affecting trinucleotide variation in exons. Importantly, patterns of variation at the majority of microsatellites in the human genome (which fall outside of genes) have yet to be analyzed.
Here, we take advantage of two high-quality genome sequences to examine human microsatellite variation at loci throughout the genome. Our results provide a new portrait of human microsatellite polymorphism and encourage the inclusion of these loci in population genetic research on the genomic scale.
Materials and Methods
We focused on two well-annotated genome sequences that were completed using Sanger sequencing. Although additional human genome sequences have been reported (Bentley et al. 2008; Ley et al. 2008; Wang et al. 2008; Wheeler et al. 2008; Ahn et al. 2009), they were derived from next generation technologies for which determination of microsatellite sequences will often be unreliable. The human genome reference sequence (v. 36.1) was downloaded from the UCSC genome browser (http://genome.ucsc.edu). The genome sequence of J. Craig Venter was downloaded from NCBI (http://www.ncbi.nlm.nih.gov/sites/entrez?db=Nucleotide&cmd=Search&term=CM000462:CM000485[PACC]). The reference sequence is a well-annotated consensus of sequences from multiple individuals. The Venter sequence is a diploid consensus of a man of English descent (Levy et al. 2007). We treated these two sequences as random draws from a population.
Microsatellites were identified by applying Tandem Repeats Finder (Benson 1999) using the following parameters: match = 2, mismatch = 7, indel = 7, minimum alignment score = 12, and maximum period size = 500. The program was applied separately to each autosome in each sequence. We focused on autosomal loci to avoid complications of comparing loci with different inheritances and effective population sizes. Only perfect matches were retained. Microsatellites in the two sequences were first aligned according to sequence position. To confirm homology, 250 bp of flanking sequence on each side of each microsatellite was compared using JAligner (http://jaligner.sourceforge.net). Only microsatellites with at least 95% flanking sequence identity on one side were retained. Identical microsatellite sequences situated within 200 bp on the same chromosome were removed to reduce the possibility of including tandemly duplicated loci. Genomic locations of microsatellites were classified as intergenic, intronic, UTR, or coding exonic by comparing the positions with the human reference gene list downloaded from the UCSC Genome Browser. The number of SNP differences between the reference sequence and the Venter sequence were counted in 5, 10, 20, and 50 kb windows using the list provided (Levy et al. 2007).
All statistical analyses were conducted in R (R Core Development Team 2009). Each microsatellite could only take on a heterozygosity value of 0 or 1 (with n = 2 sequences); as a result, we used the proportion of polymorphic loci within a particular category as the primary measure of variation. Ninety-five percent confidence intervals for proportions of polymorphic loci were estimated by bootstrapping across loci with 1,000 replicates. We considered the absolute difference in repeat number among polymorphic loci as a second measure of variation. Multiple logistic regressions with the proportion of polymorphic loci as the dependent variable were fit using the R glm function, specifying a binomial error structure with the family = binomial option. Multiple linear regressions with the absolute difference in copy number as the dependent variable were fit using the R lm function.
To examine whether the set of genes that contained variable microsatellites were enriched for particular biological functions, we conducted a gene ontology analysis. We used the Functional Annotation Tool in the DAVID Bioinformatics package (http://david.abcc.ncifcrf.gov/summary.jsp), treating the list of genes containing variable microsatellites as the “sample gene list” and the list of genes containing invariant microsatellites as the “background gene list.” Separate analyses were performed for microsatellites in coding regions and microsatellites within 1 kb upstream of transcription start sites. We used P = 0.01 as the significance cutoff.
Results
We identified 2,862,022 perfect microsatellites with at least three repeats that could be reliably compared between the NCBI Reference sequence and the Venter sequence. A total of 78,429 (2.7%) of these microsatellites were different in the two sequences. We compared subsets of loci to identify factors underlying this polymorphism.
The Effects of Repeat Number on Microsatellite Variation
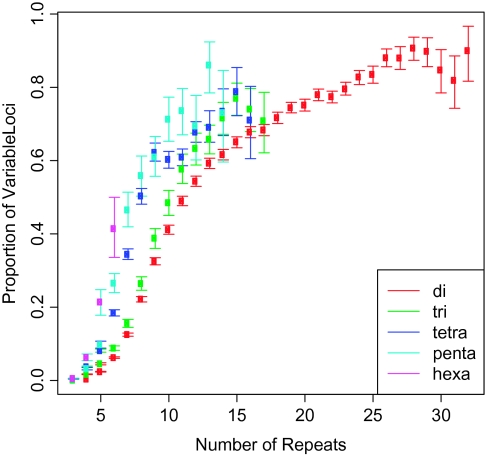
The proportion of variable loci exhibited a striking positive dependence on repeat number (the Venter sequence was used as the standard in this and subsequent comparisons with repeat number; fig. 1). Short microsatellites, which were the most common, showed low levels of polymorphism. For example, more than half of all loci surveyed had three repeats, and less than 0.2% of these loci were variable (a fact that severely reduced the overall genomic proportion of polymorphic loci [2.7%]). Long microsatellites were less common and highly variable. For example, 877 out of 1,050 (83.5%) loci with 25 repeats were polymorphic. Visual inspection of figure 1 suggested that most of the change in polymorphism with repeat number was concentrated in the range between 5 (where polymorphism began to increase rapidly) and 12 (where the rate of increase began to decline) repeats. Similar dependences on repeat number were observed when microsatellites with different repeat lengths (dinucleotides through hexanucleotides) were analyzed separately (supplementary tables 1–5, Supplementary Material online).
FIG. 1.
Genomic proportions of variable loci by repeat number and repeat length. Error bars represent 95% confidence intervals estimated by bootstrapping. The repeat number in the Venter sequence is used as the reference. Only repeat numbers with at least 50 loci (repeat range 3–32) are shown. Complete details and sample sizes appear in supplementary tables 1–6, Supplementary Material online.
The Effects of Repeat Length and Sequence on Microsatellite Variation
We compared the proportion of variable loci at microsatellites with different repeat lengths (dinucleotides through hexanucleotides). The most variable motifs were tetranucleotides (4.5% variable) and the least variable motifs were trinucleotides (0.8%). Dinucleotides and pentanucleotides exhibited very similar levels of polymorphism (both 3.7%); hexanucleotides showed a slight reduction (2.5%). The large numbers of loci in each category led to tight bootstrap confidence intervals on proportions of variable microsatellites, which only overlapped for dinucleotides and pentanucleotides.
We investigated several potential sources of the reduced diversity at trinucleotides. First, this reduction was not caused simply by the relatively higher incidence of these loci in coding exons (0.8% of trinucleotides in intergenic regions were variable). Second, we asked whether misannotation of coding trinucleotides as intergenic was responsible for reduced diversity by analyzing the coding potential of all intergenic trinucleotides using HMMgene (v. 1.1; http://www.cbs.dtu.dk/services/HMMgene/). Although coding potential was detected for 1.3% (7,967 out of 586,129) of intergenic trinucleotides, there was no difference in diversity between loci with and without coding potential (P = 0.40; Fisher's exact test [FET]), suggesting that unannotated coding regions were not responsible for the reduced variation at trinucleotides. Finally, because repeat number was an important determinant of polymorphism levels, we asked how this variable affected differences in variation among loci with different repeat lengths. Trinucleotides were more variable than dinucleotides and less variable than longer repeats at most repeat numbers (fig. 1). At the repeat number of three, however, trinucleotides were less variable than dinucleotides, and loci with repeat number of three comprised a much greater fraction of the total trinucleotides (89%) than the total dinucleotides (36%). Collectively, these results indicate higher polymorphism rates for longer repeat lengths at the same repeat number.
We also investigated the effects of repeat sequence characteristics on microsatellite variation. Sequence motifs exhibited a range of polymorphism levels (supplementary table 6, Supplementary Material online). Restricting consideration to those sequence classes with at least 25 instances in the genome, the most variable motifs for each repeat length were AC (8.5% polymorphic), AAC (3.2%), TCTA (23.3%), CTATT (26.9%), and TATCTA (32.1%). The least variable sequences for dinucleotides and trinucleotides were CT (1.1%) and CCA (0.1%), whereas many tetranucleotide, pentanucleotide, and hexanucleotide sequences showed no variation (partly because they were less common in the genome). Variable loci tended to have repeats with lower GC content (mean GC = 0.312) than did invariant loci (mean GC = 0.363; Mann–Whitney U; P < 10−15). This relationship was particularly strong for trinucleotides (mean GC at variable loci = 0.207; mean GC at invariant loci = 0.397), although a different relationship was observed for the subset of trinucleotides found in coding exons (mean GC at variable loci = 0.734; mean GC at invariant loci = 0.583; Mann–Whitney U; P < 10−6).
The Effects of Genomic Location on Microsatellite Variation
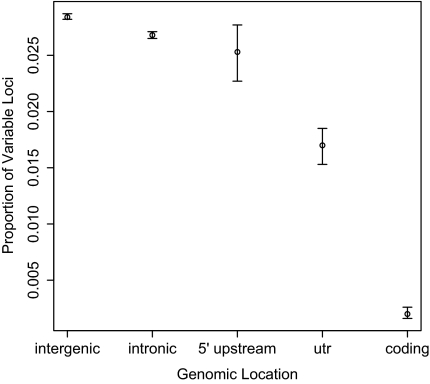
To understand selective constraints that might shape microsatellite variation, we estimated proportions of polymorphic loci separately for intergenic regions, introns, regions just upstream of the transcription start site, 5′ and 3′ UTRs, and coding exons (fig. 2 and table 1). Microsatellites in introns were slightly but significantly less variable than those in intergenic regions (FET; P < 10−15). Microsatellites in UTRs showed 40% less variation than those in intergenic regions (FET; P < 10−15). Variation at microsatellites in coding exons was substantially reduced relative to intergenic microsatellites (FET; P < 10−15), with 93% less polymorphism. Microsatellites located within 5 kb upstream of transcription start sites showed similar levels of variation to other intergenic microsatellites (FET; P = 0.31), but microsatellites located within 1 kb showed a significant reduction in variation (FET; P = 0.01). Among the group of microsatellites located within 1 kb upstream of transcription start sites, variable microsatellites were significantly farther away from transcription start sites than were invariant microsatellites (Mann–Whitney U; P = 0.005; medianvariable = 522 bp; medianinvariant = 469 bp).
FIG. 2.
Genomic proportions of variable loci by location. Error bars represent 95% confidence intervals estimated by bootstrapping. “5′ upstream” = loci within 1 kb upstream of transcription start sites.
Table 1.
Genomic Proportions of Variable Loci by Location.
| Location | Number of Loci | Proportion Variable | Proportion Variable Bootstrap Lower 95% Confidence Limit | Proportion Variable Bootstrap Upper 95% Confidence Limit |
| Intergenic | 1,673,380 | 0.0284 | 0.0282 | 0.0287 |
| Intronic | 1,054,152 | 0.0268 | 0.0265 | 0.0271 |
| Within 1 kb upstream of transcription start site | 17,118 | 0.0253 | 0.0227 | 0.0277 |
| UTR | 27,286 | 0.0170 | 0.0153 | 0.0185 |
| Coding exonic | 31,764 | 0.0020 | 0.0016 | 0.0026 |
To determine whether genes containing variable microsatellites were differentially associated with particular functional classes, we conducted a gene ontology analysis. In coding regions, variable microsatellites were biased toward genes involved in transcription and development (table 2), matching the results from recent genomic examinations of coding trinucleotides (Molla et al. 2009; Kozlowski et al. 2010). Alternatively, genes with variable microsatellites located within 1 kb upstream of transcription start sites were enriched for functions related to signal transduction, with an emphasis on G-protein coupled receptors (table 3).
Table 2.
Gene Ontology Analysis Comparing Variable and Invariant Microsatellites in Coding Exons.
| Gene Ontology Categorya | Number of Genes with Variable Microsatellites in Category | Number of Genes with Variable Microsatellites in at Least One Other Category | Number of Genes with Invariant Microsatellites in Category | Number of Genes with Invariant Microsatellites in at Least One Other Category | Fold Enrichment among Variable Microsatellites | P Value |
| Triplet repeat expansion | 3 | 50 | 16 | 9,890 | 37.09 | 0.0028 |
| DNA-directed RNA polymerase complex (GO:0000428) | 3 | 44 | 17 | 9,118 | 36.57 | 0.0028 |
| DNA-directed RNA polymerase activity (GO:0003899) | 3 | 49 | 24 | 9,323 | 23.78 | 0.0067 |
| Nucleotidyltransferase activity (GO:0016779) | 4 | 49 | 75 | 9,323 | 10.15 | 0.0067 |
| Compositionally biased region: Poly-Gly | 6 | 42 | 155 | 7,461 | 6.88 | 0.0015 |
| Compositionally biased region: Poly-Ser | 7 | 42 | 230 | 7,461 | 5.41 | 0.0015 |
| Compositionally biased region: Poly-Ala | 6 | 42 | 209 | 7,461 | 5.10 | 0.0054 |
| Compositionally biased region: Poly-Glu | 6 | 42 | 220 | 7,461 | 4.84 | 0.0067 |
| Cellular developmental process (GO:0048869) | 14 | 45 | 1256 | 8,567 | 2.12 | 0.0085 |
| Transcription (GO:0006350) | 18 | 45 | 1713 | 8,567 | 2.00 | 0.0033 |
| Developmental process (GO:0032502) | 20 | 45 | 2207 | 8,567 | 1.73 | 0.0088 |
| Gene expression (GO:0010467) | 20 | 45 | 2212 | 8,567 | 1.72 | 0.0090 |
| Binding (GO:0005488) | 47 | 49 | 7656 | 9,323 | 1.17 | 0.0050 |
Only one representative of redundant functional categories that included exactly the same genes is listed. Only categories with P < 0.01 are shown. Total number of queried genes with annotation was 61; total number of background genes with annotation was 11,500.
Table 3.
Gene Ontology Analysis Comparing Variable and Invariant Microsatellites in Regions within 1 kb Upstream of Transcription Start Sites.
| Gene Ontology Categorya | Number of Genes with Variable Microsatellites in Category | Number of Genes with Variable Microsatellites in at Least One Other Category | Number of Genes with Invariant Microsatellites in Category | Number of Genes with Invariant Microsatellites in at Least One Other Category | Fold Enrichment Among Variable Microsatellites | P Value |
| IPR000276: Rhodopsin-like GPCR superfamily | 22 | 222 | 391 | 7,601 | 1.93 | 0.0047 |
| g-protein-coupled receptor | 25 | 232 | 443 | 7,810 | 1.90 | 0.0029 |
| GO:0001584 – rhodopsin-like receptor activity | 23 | 220 | 431 | 7,344 | 1.78 | 0.0092 |
| GO:0004888 – transmembrane receptor activity | 36 | 220 | 705 | 7,344 | 1.70 | 0.0017 |
| Receptor | 40 | 232 | 839 | 7,810 | 1.60 | 0.0027 |
| GO:0004872 – receptor activity | 48 | 220 | 1,013 | 7,344 | 1.58 | 0.0011 |
| GO:0004871 – signal transducer activity | 58 | 220 | 1,246 | 7,344 | 1.55 | 0.0004 |
Only one representative of redundant functional categories that included exactly the same genes is listed. Only categories with P < 0.01 are shown. Total number of queried genes with annotation was 407; the number of background genes with annotation was 9,186.
We also compared repeat number differences among genomic locations. Variable microsatellites in intergenic regions and introns differed by similar numbers of repeats (Mann–Whitney U; P > 0.05). Alternatively, repeat number differences at variable microsatellites in UTRs and coding exons were significantly smaller than in intergenic regions (Mann–Whitney U; P = 0.019 and P = 0.0005, respectively). These patterns suggest that several types of microsatellites in genes experience selective constraints.
We asked whether flanking base composition was associated with microsatellite variation. GC content was slightly but significantly lower near variable loci. The four scales we tested showed a weakening relationship with increasing window size: 5 kb (Mann–Whitney U; P < 10−15), 10 kb (P < 10−14), 20 kb (P < 10−7), and 50 kb (P < 10−4). In all cases, the magnitude of the difference was very small (e.g., 5 kb windows: median GC near variable loci = 0.397 and median GC near invariant loci = 0.399). The difference in repeat number among variable loci was also higher in regions with lower GC content (Spearman's correlation; P < 10−3 on all scales), although the effect was again modest (r = −0.015 on all scales).
The Genomic Distribution of Repeat Number Differences
The distribution of repeat number differences at variable loci is shown in figure 3. The absolute difference in repeat number had a median of 1.0, a mean of 2.5, and a standard deviation of 2.8. The majority (53%) of variable loci differed by one repeat, 19% differed by two copies, and 9% differed by three copies, with larger differences occurring progressively less frequently.
FIG. 3.
Differences in repeat number among variable loci. Repeat number differences are shown as absolute values. Although small numbers of loci showed larger repeat number differences, the upper limit on the x axis is fixed at 20 to improve the clarity of presentation.
Separate analyses by repeat length revealed stronger biases toward repeat number differences of one for trinucleotides (69%), tetranucleotides (66%), pentanucleotides (63%), and hexanucleotides (78%). Dinucleotides showed the lowest percentage of loci differing by one repeat (47%). The relatively large number of dinucleotides reduced the genomic fraction of loci differing by one repeat. In addition to varying more frequently, longer loci showed larger differences in repeat number (Spearman's ρ = 0.44; P < 10−15).
The Relationship between Microsatellite Variation and SNP Variation
We compared the absolute difference in microsatellite repeat number with the number of SNPs between the two consensus sequences (Levy et al. 2007) in windows of 5, 10, 20, and 50 kb centered on each microsatellite. Microsatellite variation was positively correlated with the number of SNPs, with window size having little effect on correlation strength (5 kb: Spearman's ρ = 0.06; 10 kb: ρ = 0.05; 20 kb: ρ = 0.05; 50 kb: ρ = 0.04; P < 10−15 in all tests). Separate analyses by repeat length yielded correlations with similar magnitudes for dinucleotides through hexanucleotides. Additionally, more SNPs were found near variable microsatellites (mean = 3.8 in 5 kb window) than near monomorphic microsatellites (mean = 2.5 in 5 kb window; P < 10−15; Mann–Whitney U). This effect was again observed across the four window sizes.
We compared correlations between squared repeat number difference and SNP number with analytical predictions for the case of two sequences (Payseur and Cutter 2006). Assuming an effective population size of 104, no recombination, a per-generation SNP mutation rate of 10−8, and a per-generation microsatellite mutation rate of 10−4 (yielding a microsatellite population mutation rate of 4), the predicted Pearson's correlation for a 5 kb region was 0.019. The observed correlation was 0.037. This similarity between predictions and observations was seen across a range of parameter values (data not shown) and confirmed the theoretical expectation of weak correlations for loci with contrasting mutation rates and mechanisms (Payseur and Cutter 2006).
Multivariate Analyses of Microsatellite Variation
We asked whether the factors considered above were still associated with differences in microsatellite variation after adjusting for the effects of other variables in two sets of analyses. First, we used the fraction of polymorphic loci as the measure of variation in a multiple logistic regression. Second, we used the logarithm of the absolute difference in repeat number at polymorphic loci in a multiple linear regression.
Multiple logistic regression demonstrated significant effects of all variables tested—repeat number (P < 10−263), repeat length (P < 10−56 for all lengths), repeat GC content (P < 10−263), flanking GC content (P < 10−28), SNP number (P < 10−263), and genomic location (P < 10−10 for all locations)—on the proportion of polymorphic loci. All effects were in the directions observed in bivariate analyses, except flanking GC content, which increased with the proportion of variable loci in multivariate analyses.
Multiple linear regression revealed significant effects of repeat number (P < 10−263), repeat length (P < 10−3 for all lengths), repeat GC content (P < 10−263), flanking GC content (P < 10−4), and SNP number (P < 10−190), but not genomic location (P > 0.10 for all locations) on the absolute difference in repeat number. Again, effects were in the directions observed in bivariate analyses, with the exception of flanking GC content, which increased with the difference in repeat number in multivariate analyses. The adjusted R2 was 0.212, leaving unexplained the majority of variation in repeat number difference.
Discussion
Genomic Patterns of Microsatellite Variation and Implications for Mutational Mechanisms
The microsatellites commonly used in human population genetics represent a useful but small subset of the total genomic complement of loci. As a result, levels of variation at randomly chosen microsatellites—as well as differences among classes of these loci—have remained poorly understood. Our study begins to fill this void. Overall, the patterns we observed both bolster previous conclusions and add new information about microsatellite mutational mechanisms.
About 2.7% of the perfect microsatellites we surveyed were polymorphic, a value 45 times as large as the amount of SNP variation (0.06%) in this data set (Levy et al. 2007). Nevertheless, this level of polymorphism is much lower than heterozygosity at microsatellites typically analyzed in human population genetics, which often exceeds 70% (Jorde et al. 1997; Payseur et al. 2002; Rosenberg et al. 2002; Tishkoff et al. 2009). Although panels of widely used microsatellites are routinely chosen for their high heterozygosities (Ghebranious et al. 2003), we included all perfect repeats with repeat numbers of at least three (without regard to variation) in our analyses. Additionally, our genome-wide survey only used two sequences in contrast to the larger population samples usually employed in microsatellite polymorphism studies. Both these factors contribute to the discrepancy in levels of variation. The finding that the human genome harbors a large class of shorter microsatellites that shows no polymorphism in a comparison between two randomly chosen sequences is an important contribution of our study.
Statements about average microsatellite polymorphism mask remarkable heterogeneity in the levels of variation among loci. We uncovered differences between categories of microsatellites that agree with previous findings from population genetic, molecular evolution, and pedigree studies in humans. The strongest effect we observed—increasing polymorphism with higher repeat number—suggests that rates of mutation are elevated by the addition of repeats. Higher mutation rates at longer microsatellites have been observed in human pedigrees (Weber and Wong 1993; Amos et al. 1996; Brinkmann et al. 1998; Whittaker et al. 2003; Ellegren 2004) and inferred from comparisons between humans and chimpanzees (Amos and Rubinstzein 1996; Webster et al. 2002; Sainudiin et al. 2004; Legendre et al. 2007; Kelkar et al. 2008) as well as human population genetic studies (Molla et al. 2009; Pemberton et al. 2009). We estimate that polymorphism begins to increase rapidly at approximately five repeats, close to the threshold value predicted from genomic distributions of microsatellite lengths (Lai and Sun 2003). The overall relationship between microsatellite variation and repeat number in humans bears a striking resemblance (in both shape and actual frequencies) to that observed in a similar study in which genomic shotgun sequences from three chickens were compared (Brandström and Ellegren 2008). This similarity suggests that a common mutational mechanism underlies length-dependent mutation in two distantly related vertebrates. The most likely explanation for this set of observations is that microsatellites with higher repeat numbers undergo increased rates of replication slippage (Levinson and Gutman 1987; Ellegren 2004; Pearson et al. 2005). Our results support the claim that different categories of microsatellites can be used for different purposes in human population genetics (Jorde et al. 1997; Brinkmann et al. 1998). An abundance of new mutations at longer microsatellites might provide access to evolutionary events in the near past (e.g., bottlenecks associated with recent colonizations), whereas allele frequency differences at shorter loci could be more reliable guides to earlier human population history (e.g., separation of major population groups). These results also motivate further research connecting microsatellite length distributions to their physical properties (Bacolla et al. 2008).
Our results reveal that in addition to repeat number, microsatellite repeat length affects polymorphism levels. The ranking of overall proportions of polymorphic loci by repeat length in humans was similar to the pattern in chickens (Brandström and Ellegren 2008), with trinucleotides exhibiting the strongest decrease in relative levels of variation. The overall reduction in variation at trinucleotides appears to have been caused by a disproportionately large number of trinucleotides in the smallest repeat number category (three), a group which was also less variable among trinucleotides. The overall positive relationship between repeat length and diversity we observed suggests that loci with longer repeats mutate more rapidly. Using parent-offspring comparisons at 28 microsatellites in CEPH families, Weber and Wong (1993) estimated higher mutation rates for tetranucleotides than dinucleotides. Di Rienzo et al. (1998) also found that tetranucleotides were more mutable than dinucleotides and trinucleotides in patients with sporadic colon cancers. However, population genetic and comparative genomic studies in humans have described an inverse relationship between diversity or divergence and repeat length (Chakraborty et al. 1997; Kelkar et al. 2008; Pemberton et al. 2009). It is difficult to reconcile this pattern with our results, but several factors may contribute to the discrepancy. Because Chakraborty et al. (1997) and Pemberton et al. (2009) focused on microsatellites that were intentionally chosen for high heterozygosity, uneven ascertainment biases among loci with different repeat lengths could have generated the observed patterns. For example, the dinucleotides, trinucleotides, and tetranucleotides analyzed by Pemberton et al. (2009) had different repeat number distributions; loci with shorter repeat lengths had higher average repeat numbers. Hence, the inverse relationship between diversity and repeat length observed by Pemberton et al. (2009) might be explained by the strong positive association between polymorphism and repeat number rather than reflecting inherent differences between repeat lengths. In contrast, we selected loci without regard to variation, minimizing the contribution of these ascertainment biases. Previous studies also included loci with imperfect repeats, whereas we focused exclusively on perfect repeats. Differences in mutation dynamics between imperfect and perfect repeats could generate disparities between studies.
We found that sequences of variable microsatellites were lower in GC content than sequences of invariant microsatellites. This result conflicts with Pemberton et al. (2009), who discovered a positive correlation between GC content and heterozygosity among tetranucleotides (restricting our comparisons to tetranucleotides does not change our result). However, variable tetranucleotides show lower GC content in chickens (Brandström and Ellegren 2008), whereas human–chimp repeat size differences at tetranucleotides are uncorrelated with GC content (Kelkar et al. 2008). Differences in study design likely contributed to these discordant patterns (Pemberton et al. 2009). Pemberton et al. (2009) sampled more than 1,000 individuals at several hundred loci, whereas Brandström and Ellegren (2008), Kelkar et al. (2008), and our study surveyed only a few individuals at many more loci.
Several caveats accompany our conclusions about microsatellite mutational processes and patterns of polymorphism. First, we have focused on microsatellites with perfect repeats. Consideration of imperfect repeats would introduce additional heterogeneity in patterns of mutation and polymorphism. Second, our results do not address some aspects of the mutational process, including the existence of biases toward expansions or contractions (Cooper et al. 1999; Xu et al. 2000; Ellegren 2004). Third, our approach assumes that the two sequences we have compared are accurate. Although errors accompany all genome sequence comparisons, highly repetitive regions pose special challenges in sequencing and assembly. Some loci that show very large differences in repeat number between the two sequences (fig. 3) might reflect errors in one or both genome sequences. The median difference in repeat number was not statistically different from 0 (P > 0.05; Wilcoxon signed rank test), suggesting that error differences between the two sequences did not impart strong directional biases on the results. Additionally, our analyses focused on the consensus sequences available for the Venter and reference genomes. We underestimated microsatellite diversity by ignoring within-individual variation.
Finally, in contrast to pedigree studies in which mutations can be observed as they arise, population studies of unrelated individuals cannot easily separate the contributions of mutational and genealogical processes to levels of polymorphism. Nevertheless, our results suggest large interlocus heterogeneity in the effects of mutation that should be considered when interpreting genomic patterns of microsatellite variation.
Selective Constraints on Microsatellite Variation
Microsatellites in introns, UTRs, and coding exons showed reduced variation relative to those in intergenic regions. If we assume that microsatellites are randomly distributed with regard to genomic location and intergenic loci provide a neutral benchmark, we can roughly estimate levels of selective constraint by comparing the proportion of polymorphic loci in each class to that for intergenic loci. These comparisons reveal constraint levels (relative reductions in polymorphism) of 5.6% in introns, 34.5% in UTRs, and 90.8% in coding exons. However, these estimates ignore the possibility that microsatellites with different characteristics are nonrandomly located in these regions. To inspect selective constraints on a more homogeneous class of loci, we focused on trinucleotides with at least 10 repeats. Among this subset, 65.3% (1,376 out of 2,107) of intergenic loci, 60.9% (852 out of 1,400) of intronic loci, 45.8% (11 out of 24) of UTR loci, and 40.7% (11 out of 27) of loci in coding exons were variable. In this case, constraints are estimated to be 6.7% in introns, 29.9% in UTRs, and 37.7% in coding exons.
Our estimates of selective constraint are rough but they serve to illustrate two points. First, selection against microsatellite mutations in human populations may be strong, especially in genic regions. Second, inferences about selection on microsatellites are likely to be sensitive to assumptions about the mutation process. Fortunately, genomic data can now be used to compare microsatellites with similar mutational properties by selecting loci with similar sequences and modal lengths.
In contrast to our results, Molla et al. (2009) reported little evidence for selection on trinucleotides in genic regions. Two differences between our studies likely contribute to this disparity. Whereas Molla et al. (2009) used intronic loci as neutral benchmarks, we used intergenic microsatellites. We noticed a reduction in variation in introns relative to intergenic regions, suggesting that intergenic microsatellites might provide better neutral standards. We focused solely on polymorphism, whereas Molla et al. (2009) compared polymorphism with divergence. Incorporating divergence into the analysis should help to account for variation in the neutral mutation rate (our inference of selective constraints assumes similar mutation rates across genomic regions), but the properties of the modified McDonald–Kreitman test used by Molla et al. (2009) have yet to be explored in the context of microsatellites.
Molla et al. (2009) noted a similar fraction of variable loci in exons and introns for longer microsatellites, but a reduction in the fraction of variable loci in exons for shorter microsatellites. This interesting pattern suggests that diversity at coding microsatellites reflects a balance between mutation and selection. Selection removes diversity at both short and long microsatellites, but this reduction only persists at short loci, which experience low mutation rates. Conversely, rapid mutation at longer microsatellites erases the signature of selection. We used our data to ask whether a similar interplay between mutation and selection was evident for microsatellites in UTRs, another category of loci that seems to be selectively constrained. The ratio of polymorphism for microsatellites with fewer than ten repeats located in UTRs versus intergenic regions was 0.62 (0.008/0.013). The comparable ratio for microsatellites with ten or more repeats was 0.95 (0.61/0.64). This simple test agrees with the results of Molla et al. (2009) for coding exons and suggests that high rates of mutation at longer microsatellites can obscure the signal of selective constraints. These considerations emphasize the need for new models and tests of neutrality for microsatellites that account for mutational processes to accompany the existing battery of methods available for SNPs (Nielsen 2005).
The inference that microsatellite mutations can affect fitness is supported by comparative genomic and functional studies in humans and other species (Li et al. 2004). Microsatellites in open reading frames are biased toward trinucleotides and hexanucleotides (a pattern also observed in our results), presumably because mutations at other types of loci will often induce frameshifts (Metzgar et al. 2000; Borstnik and Pumpernik 2002). Coding microsatellites can also cause slippage of the transcriptional machinery (Fabre et al. 2002), leading to dysfunctional messenger RNAs that are degraded by the nonsense-mediated RNA decay pathway (Conti and Izaurralde 2005). Coding regions harbor fewer highly variable mononucleotide runs than expected under neutrality, indicating the action of purifying selection (Loire et al. 2009). Finally, the expansion of amino acid runs that can accompany microsatellite mutation can cause disease, including neurodegenerative disorders (Everett and Wood 2004). Selective constraints on microsatellites outside exons have been more difficult to gauge, but several lines of evidence suggest that they exist. Microsatellites in 5' and 3' UTRs have been shown to regulate gene expression (Kashi et al. 1997; Li et al. 2004) and contribute to disease (Kenneson et al. 2001). Promoter elements that contribute to variation in gene expression in humans are enriched for microsatellites (Rockman and Wray 2002). Our finding of reduced variation at microsatellites within 1 kb of transcription start sites, relative to other intergenic loci, also supports the existence of selective constraints on regulatory sequences.
Integrating Genomic Variation at Loci with Different Mutational Mechanisms
The rapid accumulation of resequencing and genotyping data is beginning to allow a meaningful comparison between the two largest pools of variation in the human genome, SNPs and microsatellites. We discovered that variable microsatellites tend to be located in regions with more SNPs (Brandström et al. 2008), confirming that shared genealogical history confers correlated levels of variation (Payseur and Cutter 2006). However, the correlations between microsatellite and SNP diversity were weak, mirroring patterns seen for population structure measured in larger samples of humans (Payseur and Jing 2009). Collectively, these results indicate that differences in the mutational process between microsatellites and SNPs effectively uncouple their diversity patterns, even when they share identical genealogical histories.
According to our survey, a typical 100 kb stretch in the genome contains a few microsatellites that differ between two randomly chosen human sequences, and this calculation ignores the large number of imperfect repeats that exist as well as microsatellites we were unable to confidently align. The genomic density of polymorphic microsatellites will be even higher in larger population samples. Although human genomes contain many more SNPs than microsatellites, the increased rate of polymorphism makes microsatellites a powerful resource for population genetics. Because microsatellites often harbor a large number of alleles, these loci should be especially useful in large-scale efforts to catalog rare variants in the human genome (including the 1000 Genomes Project; http://www.1000genomes.org).
Microsatellite variation also can be compared and combined with SNP variation to reconstruct human evolutionary history on different timescales (deKnijff 2000; Mountain et al. 2002; Payseur and Cutter 2006). The development of analytical tools for integrating patterns of polymorphism at loci with contrasting mutation rates and mechanisms will be required for building a synthetic view of human genomic variation in its many forms.
Supplementary Material
Supplementary tables 1–6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank Samuel Levy for providing information about the Venter sequence. We thank Aida M. Andrés for discussions during the course of this project, and Asher Cutter, Noah Rosenberg, Anne Stone, and several anonymous reviewers for thoughtful comments on the manuscript. This research was supported by National Institutes of Health (grant HG004498) and a Medical Education and Research Committee New Investigator grant from the University of Wisconsin School of Medicine and Public Health.
References
- Ahn SM, Kim TH, Lee S, et al. (21 co-authors) The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos W, Rubinstzein DC. Microsatellites are subject to directional evolution. Nat Genet. 1996;12:13–14. doi: 10.1038/ng0196-13. [DOI] [PubMed] [Google Scholar]
- Amos W, Sawcer SJ, Feakes RW, Rubinsztein DC. Microsatellites show mutational bias and heterozygote instability. Nat Genet. 1996;13:390–391. doi: 10.1038/ng0896-390. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Larson JE, Collins JR, Li J, Milosavljevic A, Stenson PD, Cooper DN, Wells RD. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008;18:1545–1553. doi: 10.1101/gr.078303.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D, Balasubramanian RS, Swerdlow HP, et al. (193 co-authors) Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borstnik B, Pumpernik D. Tandem repeats in protein coding regions of primate genes. Genome Res. 2002;12:909–915. doi: 10.1101/gr.138802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Brandström M, Bagshaw AT, Gemmell NJ, Ellegren H. The relationship between microsatellite polymorphism and recombination hot spots in the human genome. Mol Biol Evol. 2008;25:2579–2587. doi: 10.1093/molbev/msn201. [DOI] [PubMed] [Google Scholar]
- Brandström M, Ellegren H. Genome-wide analysis of microsatellite polymorphism in chicken circumventing the ascertainment bias. Genome Res. 2008;18:881–887. doi: 10.1101/gr.075242.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann B, Klintschar M, Neuhuber F, Huhne J, Rolf B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Kimmel M, Stivers DN, Davison LJ, Deka R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc Natl Acad Sci U S A. 1997;94:1041–1046. doi: 10.1073/pnas.94.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Cooper G, Burroughs NJ, Rand DA, Rubinsztein DC, Amos W. Markov chain Monte Carlo analysis of human Y-chromosome microsatellites provides evidence of biased mutation. Proc Natl Acad Sci U S A. 1999;96:11916–11921. doi: 10.1073/pnas.96.21.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deKnijff P. Messages through bottlenecks: on the combined use of slow and fast evolving polymorphic markers on the human Y chromosome. Am J Hum Genet. 2000;67:1055–1061. doi: 10.1016/s0002-9297(07)62935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo A, Donnelly P, Toomajian C, Sisk B, Hill A, Petzl-Erler ML, Haines GK, Barch DH. Heterogeneity of microsatellite mutations within and between loci, and implications for human demographic histories. Genetics. 1998;148:1269–1284. doi: 10.1093/genetics/148.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB. Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci U S A. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nat Genet. 2000;24:400–402. doi: 10.1038/74249. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Estoup A, Jarne P, Cornuet JM. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol. 2002;11:1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- Everett CM, Wood NW. Trinucleotide repeats and neurodegenerative disease. Brain. 2004;127:2385–2405. doi: 10.1093/brain/awh278. [DOI] [PubMed] [Google Scholar]
- Fabre E, Dujon B, Richard GF. Transcription and nuclear transport of CAG/CTG trinucleotide repeats in yeast. Nucleic Acids Res. 2002;30:3540–3547. doi: 10.1093/nar/gkf483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H, Redon R, Andrews D, et al. (27 co-authors) Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res. 2006;16:1566–1574. doi: 10.1101/gr.5630906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlaender JS, Friedlaender FR, Reed FA, et al. (12 co-authors) The genetic structure of Pacific Islanders. PLoS Genet. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebranious N, Vaske D, Yu AD, Zhao CF, Marth G, Weber JL. STRP screening sets for the human genome at 5 cM density. BMC Genomics. 2003;4:6. doi: 10.1186/1471-2164-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds DA, Kloek AP, Jen M, Chen XY, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- Huang QY, Xu FH, Shen H, Deng HY, Liu YJ, Liu YZ, Li JL, Recker RR, Deng HW. Mutation patterns at dinucleotide microsatellite loci in humans. Am J Hum Genet. 2002;70:625–634. doi: 10.1086/338997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Bamshad MJ, Watkins WS, Zenger R, Fraley AE, Krakowiak PA, Carpenter KD, Soodyall H, Jenkins T, Rogers AR. Origins and affinities of modern humans: a comparison of mitochondrial and nuclear genetic data. Am J Hum Genet. 1995;57:523–538. doi: 10.1002/ajmg.1320570340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Rogers AR, Bamshad M, Watkins WS, Krakowiak P, Sung S, Kere J, Harpending HC. Microsatellite diversity and the demographic history of modern humans. Proc Natl Acad Sci U S A. 1997;94:3100–3103. doi: 10.1073/pnas.94.7.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashi Y, King D, Soller M. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. doi: 10.1016/s0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, et al. (14 co-authors) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet. 2000;66:1580–1588. doi: 10.1086/302905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–38. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- Kimmel M, Chakraborty R. Measures of variation at DNA repeat loci under a general stepwise mutation model. Theor Popul Biol. 1996;50:345–367. doi: 10.1006/tpbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- Kimmel M, Chakraborty R, King JP, Bamshad M, Watkins WS, Jorde LB. Signatures of population expansion in microsatellite repeat data. Genetics. 1998;148:1921–1930. doi: 10.1093/genetics/148.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski P, de Mezer M, Krzyzosiak WJ. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;1:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Sun F. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol Biol Evol. 2003;20:2123–2131. doi: 10.1093/molbev/msg228. [DOI] [PubMed] [Google Scholar]
- Legendre M, Pochet N, Pak T, Verstrepen KJ. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007;17:1787–1796. doi: 10.1101/gr.6554007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, et al. (31 co-authors) The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, et al. (48 co-authors) DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Locke DP, Sharp AJ, McCarroll SA, et al. (11 co-authors) Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loire E, Praz F, Higuet D, Netter P, Achaz G. Hypermutability of genes in Homo sapiens due to the hosting of long mono-SSR. Mol Biol Evol. 2009;26:111–121. doi: 10.1093/molbev/msn230. [DOI] [PubMed] [Google Scholar]
- Manica A, Prugnolle F, Balloux F. Geography is a better determinant of human genetic differentiation than ethnicity. Hum Genet. 2005;118:366–371. doi: 10.1007/s00439-005-0039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzgar D, Bytof J, Wills C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10:72–80. [PMC free article] [PubMed] [Google Scholar]
- Molla M, Delcher A, Sunyaev S, Cantor C, Kasif S. Triplet repeat length bias and variation in the human transcriptome. Proc Natl Acad Sci U S A. 2009;106:17095–17100. doi: 10.1073/pnas.0907112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain JL, Knight A, Jobin M, Gignoux C, Miller A, Lin AA, Underhill PA. SNPSTRs: empirically derived, rapidly typed, autosomal haplotypes for inference of population history and mutational processes. Genome Res. 2002;12:1766–1772. doi: 10.1101/gr.238602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Ohta T, Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet Res. 1973;22:201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Cutter AD. Integrating patterns of polymorphism at SNPs and STRs. Trends Genet. 2006;22:424–429. doi: 10.1016/j.tig.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Cutter AD, Nachman MW. Searching for evidence of positive selection in the human genome using patterns of microsatellite variability. Mol Biol Evol. 2002;19:1143–1153. doi: 10.1093/oxfordjournals.molbev.a004172. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Jing P. A genomewide comparison of population structure at STRPs and nearby SNPs in humans. Mol Biol Evol. 2009;26:1369–1377. doi: 10.1093/molbev/msp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Pemberton TJ, Sandefur CI, Jakobsson M, Rosenberg NA. Sequence determinants of human microsatellite variability. BMC Genomics. 2009;10:612. doi: 10.1186/1471-2164-10-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH. The evolutionary significance of copy number variation in the human genome. Cytogenet Genome Res. 2008;123:283–287. doi: 10.1159/000184719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Ben-Dor A, Tsalenko A, et al. (17 co-authors) The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: 2009. [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. (43 co-authors) Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Mol Biol Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Gonzalez-Quevedo C, et al. (13 co-authors) Low levels of genetic divergence across geographically and linguistically diverse populations from India. PLoS Genet. 2006;2:e215. doi: 10.1371/journal.pgen.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Sainudiin R, Durrett RT, Aquadro CF, Nielsen R. Microsatellite mutation models: insights from a comparison of humans and chimpanzees. Genetics. 2004;168:383–395. doi: 10.1534/genetics.103.022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Jin L, Chakraborty R, Boerwinkle E. VNTR allele frequency distributions under the stepwise mutation model: a computer simulation approach. Genetics. 1993;134:983–993. doi: 10.1093/genetics/134.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, et al. (25 co-authors) The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AM, Slatkin M, Freimer NB. Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, et al. (77 co-authors) The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lewis CM, Jakobsson M, et al. (27 co-authors) Genetic variation and population structure in native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Webster MT, Smith NG, Ellegren H. Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments. Proc Natl Acad Sci U S A. 2002;99:8748–8753. doi: 10.1073/pnas.122067599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, et al. (27 co-authors) The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Whittaker JC, Harbord RM, Boxall N, Mackay I, Dawson G, Sibly RM. Likelihood-based estimation of microsatellite mutation rates. Genetics. 2003;164:781–787. doi: 10.1093/genetics/164.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Peng M, Fang Z. The direction of microsatellite mutations is dependent upon allele length. Nat Genet. 2000;24:396–399. doi: 10.1038/74238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.