Abstract
We report herein that trefoil factor 3 (TFF3) is oncogenic and mediates anti-estrogen resistance in human mammary carcinoma. Forced expression of TFF3 in mammary carcinoma cells increased cell proliferation and survival, enhanced anchorage-independent growth, and promoted migration and invasion. Moreover, forced expression of TFF3 increased tumor size in xenograft models. Conversely, depletion of endogenous TFF3 with small interfering RNA (siRNA) decreased the oncogenicity and invasiveness of mammary carcinoma cells. Neutralization of secreted TFF3 by antibody promoted apoptosis, decreased cell growth in vitro, and arrested mammary carcinoma xenograft growth. TFF3 expression was significantly correlated to decreased survival of estrogen receptor (ER)-positive breast cancer patients treated with tamoxifen. Forced expression of TFF3 in mammary carcinoma cells increased ER transcriptional activity, promoted estrogen-independent growth, and produced resistance to tamoxifen and fulvestrant in vitro and to tamoxifen in xenograft models. siRNA-mediated depletion or antibody inhibition of TFF3 significantly enhanced the efficacy of antiestrogens. Increased TFF3 expression was observed in tamoxifen-resistant (TAMR) cells and antibody inhibition of TFF3 in TAMR cells improved tamoxifen sensitivity. Functional antagonism of TFF3 therefore warrants consideration as a novel therapeutic strategy for mammary carcinoma.
Introduction
Trefoil factor 3 (TFF3), previously designated as intestinal trefoil factor, belongs to the trefoil factor (TFF) family, which includes two other members, namely TFF1 and TFF2 [1]. All three TFF proteins are expressed in the epithelial cells that line mucous membranes, usually from mucin-secreting goblet cells. TFF1 is predominantly expressed in the stomach and colon, TFF2 expression is mainly localized in the stomach, whereas TFF3 expression is observed predominantly in the intestine [2,3]. TFF3 and other TFF members are classically involved in the protection of gastrointestinal tract against mucosal injury and subsequent repair [4]. TFF peptides act as motogen to facilitate cell migration and also inhibit apoptosis and prevent anoikis in the process of cell migration [4,5].
In addition to the protective and restorative effects of TFF3 in the gastrointestinal tract, compelling evidence has emerged from experimental and clinical studies to indicate a pivotal role of TFF3 in neoplastic diseases. TFF3 is overexpressed in a variety of human malignancies including mammary [6], gastric [7,8], prostate [9], hepatocellular [10], and endometrial [11] carcinomas, and it has demonstrated prosurvival, proinvasive, and proangiogenic activities in cells derived from several common human solid tumors [12–17].
TFF3 messenger RNA (mRNA) is focally expressed in duct luminal cells of normal mammary gland and exhibits increased expression in both in situ and invasive carcinomas [6]. Although the role of TFF3 in mammary carcinoma remains undefined, the prognostic or predictive value of TFF3 expression in mammary carcinoma has been indicated by several clinical studies. TFF3 mRNA expression has been demonstrated to predict micrometastatic breast cancer [18] and is strongly correlated with breast cancer metastatic to bone [19]. Serial analysis of gene expression has included TFF3 in a signature of genes that are expressed in mammary carcinoma but absent from blood and bone marrow [20]. TFF3 has been identified as one of a panel of four genes that accurately detected minimal residual disease in blood and predict survival in breast cancer patients with metastatic disease [21]. In addition, TFF3 along with TFF1 has been used as a marker for the detection of disseminated breast cancer cells [22]. Notably, in malignancies of the human mammary gland, TFF3 and TFF1 are observed to coexpress [6,23] and coregulate each other in a positive feedback loop [24]. Moreover, both TFF1 and TFF3 are estrogen-regulated genes [23]. TFF1 has recently been demonstrated to be oncogenic in human mammary carcinoma cells [25]. We therefore speculated that TFF3 may possess similar functions as TFF1 and contribute to the malignant behavior of mammary carcinoma cells. This hypothesis is also supported by our previous study, which demonstrated that TFF3 partially mediated oncogenic transformation stimulated by autocrine human growth hormone [26].
The purpose of this study was to systematically delineate the functional role of TFF3 in mammary carcinoma and investigate the effects of TFF3 on the cellular response to estrogen and antiestrogenic agents. We report herein that TFF3 is oncogenic, predicts outcome of estrogen receptor (ER)-positive breast cancer patients treated with tamoxifen, and mediates anti-estrogen resistance in mammary carcinoma.
Materials and Methods
Plasmid Constructs
The coding sequence for human TFF3 (GenBank accession number NM_003226) was cloned into the mammalian expression vector pIRESneo3 (Invitrogen, Carlsbad, CA), designated as pIRESneo3-TFF3. The human TFF3 complementary DNA (cDNA) coding for the mature peptide was cloned into the pGEX-4T1 vector (Amersham Biosciences, Piscataway, NJ) in frame with the N-terminal glutathione-S-transferase to generate pGEX-4T1-TFF3 plasmid for the expression of glutathione-S-transferase-TFF3 fusion proteins in Escherichia coli.
To generate small interfering RNA (siRNA) oligonucleotides targeting TFF3, the DNA sequence AAACAACGGTGCATAAATGAG was selected to construct an siRNA expression plasmid using the pSilencer 2.1-U6 hygro vector (Ambion, Austin, TX) according to the manufacturer's protocol. The resultant vector was designated as pSilencer-TFF3. The control siRNA plasmid (pSilencer-control) encodes an siRNA that has no significant sequence similarity to human gene sequences.
Cell Culture
Human mammary carcinoma cell lines MCF-7 and T47D were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in conditions as recommended. MCF-7 cells were stably transfected with pIRESneo3-TFF3 or pIRESneo3 plasmids using FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN). After 4 weeks of selection in the medium containing 800 µg/ml G418 (Invitrogen), pooled stable transfectants were designated as MCF7-TFF3 and MCF7-Vec, respectively. Similarly, MCF-7 cells were stably transfected with pSilencer-TFF3 or pSilencer-control plasmids and then selected in the medium containing 100 µg/ml hygromycin B (RocheDiagnostics) to obtain MCF7-siTFF3 and MCF7-siCK cells, respectively.
Reagents
β-Estradiol (E2), tamoxifen, and fulvestrant were purchased from Sigma-Aldrich (St Louis, MO). BCL-2 inhibitor YC137 was purchased from Calbiochem (San Diego, CA).
Semiquantitative Reverse Transcription-Polymerase Chain Reaction and Quantitative Polymerase Chain Reaction
Semiquantitative reverse transcription-polymerase chain reaction (RT-PCR) and quantitative PCR (qPCR) were performed as described previously [27]. The primer sequences used for semiquantitative RT-PCR and qPCR are listed in Tables W1 and W2, respectively.
Microarray Data Sets and Survival Analysis of ER-Positive Breast Cancer Patients Treated with Tamoxifen
TFF3 gene expression characteristics in primary human breast tumors were examined by microarray analysis in a cohort of 68 ER positive cases treated by surgery and/or radiation followed by adjuvant tamoxifen monotherapy. This cohort represents a subset of the previously published Uppsala cohort [28] for which the microarray data set is accessible at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) through accession number GSE3494. Briefly, the microarray data were processed using the MAS5.0 algorithm (Affymetrix AGCC software), scaled to a mean target signal intensity of 500, and log2-transformed. Using the probe set corresponding to the TFF3 transcript (204623_at), breast cancer cases were divided by TFF3 transcript levels into “low” (below mean) and “high” (above mean) gene expression groups or into three expression groups based on expression tertiles (lowest 33% n = 23, middle 33% n = 22, highest 33% n = 23), then group survival characteristics were compared by Kaplan-Meier analysis (Sigma Plot 11.0; Systat Software Inc., San Jose, CA). Three survival end points were considered: 1) distant metastasis-free survival (DMFS), with an event defined as breast cancer metastasis to a distant organ site; 2) disease-specific survival, with an event defined as death owing to breast cancer; and 3) disease-free survival, with an event defined as a local, regional, or distant recurrence.
Meta-analysis of Breast Cancer Microarray Data Sets
The method for meta-analysis of breast cancer microarray data sets to determine TFF3 expression and ER status in breast cancer patients is described in the Supplementary Data.
Luciferase Reporter Assay
MCF-7 cells were plated in six-well plates and cultured in phenol red-free RPMI medium supplemented with 10% charcoal-stripped fetal bovine serum (CS-FBS) for 72 hours before transfection. Total DNA 1 µg per well containing 0.5 µg of pGL2-ERE-luciferase plasmids and 0.5 µg of pcDNA3-β-galactosidase reporter plasmids were transfected using FuGENE 6 reagent. After 24 hours, cells were then treated with either DMSO or 10 nM E2 in 1% CS-FBS for another 24 hours. The luciferase reporter assay was performed using Luciferase Assay System (Promega, Madison, WI) following the manufacturer's protocols. The luciferase values were normalized for β-galactosidase activity, which was determined using β-galactosidase Enzyme Assay System (Promega).
Production of Recombinant Human TFF3 and Generation of TFF3 Polyclonal Antibody
The recombinant mature human TFF3 (rhTFF3) protein was expressed from pGEX-4T1-TFF3 plasmid in E. coli and purified using glutathione-S-transferase tag under nondenaturing conditions and was subsequently used to generate a rabbit TFF3 polyclonal antibody (TFF3-pAb) as described previously [25].
Western Blot Analysis
Cells were grown to 70% to 80% confluence, then serum starved overnight before lysed in the fresh lysis buffer (2% SDS, 20% glycerol, 60 mM Tris-HCl pH 6.8) supplemented with complete protease inhibitor tablets (Roche Diagnostics). The whole cell lysate was sonicated followed by centrifugation at 10,000g for 10 minutes. The supernatant was collected, and protein concentration was determined by Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, CA). Protein samples were boiled for 5 minutes in the presence of 0.1 mM dithiothreitol. Sodium dodecylsulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) Western blot analysis was performed as described previously [29] using the following antibodies: mouse BCL-2 monoclonal antibody (BD Pharmingen, San Diego, CA), rabbit BAX polyclonal antibody (Calbiochem, San Diego, CA), mouse β-ACTIN monoclonal antibody (Sigma-Aldrich). Proteins were visualized using horseradish peroxidase-conjugated secondary antibody with enhanced chemiluminescence. The image was scanned by GS-800 calibrated densitometer and analyzed by software Quantity One (Bio-Rad Laboratories, Hercules, CA).
Cell Function Assays
Total cell number, apoptosis assay using Hoechst 33258 staining, bromodeoxyuridine (BrdU) incorporation, 96-well colony formation in soft agar assay, three-dimensional Matrigel assay, transwell migration, and invasion assays were performed as previously described [30]. Apoptosis was also assessed using fluorescein isothiocyanate-conjugated annexin V and propidium iodide (PI) double staining. After serum deprivation or treatment with 200 µg/ml TFF3-pAb or rabbit IgG (Sigma) in 0.5% FBS for 24 hours, cells were washed with PBS and labeled using Annexin V-FLUOS staining kit (Roche Diagnostics) according to the manufacturer's instructions and analyzed by fluorescence microscopy. For 3-(4,5-dimethylathiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) cell viability assay, 3 x 103 cells per well were plated in 175 µl of culture medium containing 10% FBS or 0.5% FBS in a 96-well plate, and 4 mg/ml TFF3-pAb or rabbit IgG at the volume of 25 µl was added into each well. After incubation for 3 days, cell viability was measured by MTT assay as described previously [31].
Cell Cycle Analysis
MCF7-Vec and MCF7-TFF3 cells were cultured in serum-free medium for 24 hours, trypsinized, fixed in ice-cold 80% ethanol, and stored in -20°C. Fixed cells were incubated with 1% Nonidet-P40/PBS solution containing 4′,6-diamidino-2-phenylindole (DAPI, 1 µg/ml) for 15 minutes at room temperature before being assessed by BD FACSAriaII flow cytometer (BD Biosciences, Rockville, MD). Data were analyzed using BD FACSDIVA software.
Tumor Xenografts in Nude Mice
TFF3-pAb treatment in tumor xenografts was performed essentially as described previously [25]. For the study of tamoxifen treatment in tumor xenografts, MCF7-Vec and MCF7-TFF3 xenografts were established in 3- to 4-week-old BALBc athymic nude mice (Shanghai Slaccas Co, Shanghai, China) supplemented with 0.72 mg of E2 pellets (Innovative Research, Sarasota, FL) as previously described [30]. On day 10 after inoculation (tumor reached the size of 80 mm3 approximately), mice were randomized to receive either continued estrogen supplementation or estrogen plus 5 mg of tamoxifen base pellet (Innovative Research; n = 6 per group). Tumor volume measurement, evaluation of tumor proliferation by BrdU incorporation and apoptosis by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay, and histologic staining with hematoxylin and eosin were performed as described [30].
Statistics
All experiments were repeated at least three times. All numerical data are expressed as mean ± SEM from a representative experiment performed in triplicate. Data were analyzed using an unpaired two-tailed t test or analysis of variance. P < .05 was considered significant. Kaplan-Meier analyses were performed using Sigma Plot 11.0 software, and the significance of the hazard ratios was ascertained by the likelihood ratio test of P value.
Results
Forced Expression of TFF3 in MCF-7 Cells Increases Proliferation and Survival
Using a previously published microarray data set (Gene Expression Omnibus accession number GSE3494) [28], we analyzed TFF3 mRNA expression in breast cancer of 251 patients to determine the clinical relevance of TFF3 expression in breast cancer. Kaplan-Meier analysis demonstrated a highly significant correlation of increased TFF3 mRNA expression in breast cancer to decreased DMFS (P = .005; Figure W1 and Supplementary Data).
A cell model was therefore established to determine the functional role of TFF3 in human mammary carcinoma cells. MCF-7 cells were stably transfected with a TFF3 expression vector (designated as MCF7-TFF3) or with the empty vector (designated as MCF7-Vec). Increased expression of TFF3 protein in MCF7-TFF3 cells compared with MCF7-Vec cells was verified by Western blot analysis (Figure 1A).
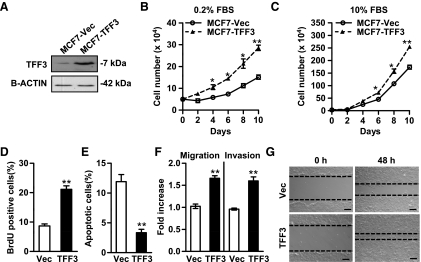
Figure 1.
Forced expression of TFF3 in MCF-7 cells increases cell proliferation and survival, and enhances cell migration and invasion. MCF-7 cells were stably transfected with the expression plasmid containing TFF3 cDNA (designated as MCF7-TFF3) or the empty vector (designated as MCF7-Vec) as control. (A) Western blot analysis of TFF3 protein level in cell lysates of MCF7-Vec and MCF7-TFF3 cells. β-ACTIN was used as a loading control for cell lysates. (B and C) Total cell number assays: MCF7-Vec and MCF7-TFF3 cells were cultured in the medium containing 0.2% FBS (B) or 10% FBS (C). Cell numbers were counted at the indicated time points. (D) BrdU incorporation assay: The percentages of BrdU-positive cell nuclei relative to the total number of cells in MCF7-Vec and MCF7-TFF3 cells were determined. (E) Apoptosis assay: MCF7-Vec and MCF7-TFF3 cells were cultured in serum-free medium for 24 hours, and cell apoptosis was measured by fluorescent microscopic analysis of nuclear staining patterns with Hoechst 33258 dye. (F) Migration and invasion assays: MCF7-Vec and MCF7-TFF3 cell migration after 24 hours and invasion after 48 hours in the transwell chambers. (G) Wound healing assay. Bar, 100 µm in G. *P < .05. **P < .01.
Forced expression of TFF3 in MCF-7 cells increased the total cell number in monolayer in both serum-depleted (Figure 1B) and serum-replete (Figure 1C) conditions. Increased cell number is achieved by increased cell proliferation and/or decreased cell death. Forced expression of TFF3 increased S-phase entry as assessed by BrdU incorporation (Figure 1D). Concordantly, cell cycle profiles determined by flow cytometric analysis revealed a higher percentage of MCF7-TFF3 cells in S-phase (25%) than MCF7-Vec cells (15%) in serum-free medium (Figure W2A and Supplementary Data). The effect of forced expression of TFF3 on apoptosis after serum deprivation for 24 hours was determined by two methods. Fluorescent microscopic analysis of cell nuclear staining patterns with Hoechst 33258 dye demonstrated that forced expression of TFF3 reduced apoptosis compared with vector cells (Figure 1E). Annexin V and PI double staining revealed that both early (annexin V-positive and PI-negative) and late apoptosis (annexin V-positive and PI-positive) were decreased in MCF7-TFF3 cells (3% and 2%, respectively) compared with MCF7-Vec cells (5% and 3%, respectively; Figure W2B and Supplementary Data). We further examined the effect of forced expression of TFF3 on cell survival after treatment with doxorubicin, an inhibitor of topoisomerase II. Cell viability after treatment with doxorubicin at various concentrations for 3 days was determined by MTT assay. A significant enhancement of cell viability was observed in MCF7-TFF3 cells compared with MCF7-Vec cells in the presence of doxorubicin (Figure W2C and Supplementary Data).
qPCR analysis (Table W3 and Supplementary Data) demonstrated that forced expression of TFF3 in MCF-7 cells dramatically increased the expression levels of cell cycle-related genes including CCNA1 (Cyclin A1), which promotes G1- to S-phase transition [32], and CDK1 (Cyclin-dependent kinase 1), which plays an essential role in S-phase entry and mitosis [33]. Moreover, a significant decrease in the mRNA level of CDKN1A (p21), a negative regulator of cell growth [34], was observed in MCF7-TFF3 cells.
The members of the Bcl-2 family were also differentially regulated by TFF3. BCL-2 mRNA level was increased by forced expression of TFF3 in MCF-7 cells, whereas BAX, a cell death mediator inducing mitochondrial damage [35], was significantly downregulated. In addition, decreased mRNA levels of CASP7 (Caspase7) and HTATIP2 (also known as TIP30, CC3, a metastasis suppressor that inhibits metastasis and promotes apoptosis) were observed in MCF-7 cells consequent to forced expression of TFF3. Therefore, forced expression of TFF3 promoted proliferation and reduced apoptosis of human mammary carcinoma cells, which was associated with the differential regulation of cell cycle and apoptosis-related genes.
Forced Expression of TFF3 in MCF-7 Cells Promotes Migration and Invasion
The effect of TFF3 on cell motility was examined by the transwell migration and wound healing assays. Forced expression of TFF3 in MCF-7 cells resulted in a 1.7-fold increased cell migration compared with vector cells (Figure 1F). Similarly, in the wound healing assay, MCF7-TFF3 cells closed monolayer wounds significantly faster than MCF7-Vec cells (Figure 1G). Furthermore, in the transwell chambers coated with Matrigel, the invasive capacity of MCF7-TFF3 cells was enhanced 1.6-fold compared with the respective control cells (Figure 1F).
qPCR analysis (Table W3 and Supplementary Data) concordantly demonstrated that MCF7-TFF3 cells expressed lower levels of CDH1 and CTNNG, which encode E-CADHERIN and γ-CATENIN, respectively. γ-CATENIN mediates E-CADHERIN-dependent intercellular adhesions. Decreased expression of E-CADHERIN and γ-CATENIN is thought to contribute to metastasis of human mammary carcinoma [36,37]. In contrast, increased expression of FN1, which encodes the extracellular matrix protein FIBRONECTIN, and VIM, which encodes the intermediate filament VIMENTIN, was observed in MCF7-TFF3 cells relative to MCF7-Vec cells. Decreased expression of epithelial markers E-CADHERIN and γ-CATENIN and induction of mesenchymal markers FIBRONECTIN and VIMENTIN are associated with epithelial-mesenchymal transition [38].
Forced Expression of TFF3 Enhances Oncogenicity of MCF-7 Cells
Anchorage-independent growth is a hallmark of oncogenic transformation. We have previously demonstrated that transient transfection of TFF3 conferred the ability of capacity for immortalized but otherwise normal human mammary epithelial cells to form colonies in soft agar and also resulted in an increase in soft agar colony formation in mammary carcinoma cells [26]. Stable forced expression of TFF3 in MCF-7 cells also stimulated anchorage-independent growth as indicated by increased colony formation (Figure 2A), foci formation (Figure 2B), and total cell number in suspension culture (Figure 2C) compared with MCF7-Vec cells.
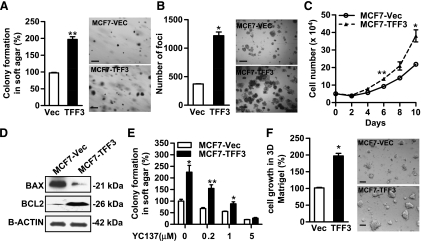
Figure 2.
Forced expression of TFF3 enhances oncogenicity of MCF-7 cells. Anchorage-independent growth was evaluated by colony formation in soft agar (A), foci formation (B), and growth in suspension culture (C) in the medium supplemented with 10% FBS. (D) Western blot analysis of BAX and BCL-2 protein levels in cell lysates of MCF7-Vec and MCF7-TFF3 cells. β-ACTIN was used as a loading control for cell lysates. (E) Colony formation in soft agar: MCF7-Vec and MCF7-TFF3 cells were seeded in 0.35% agar in a 96-well plate and treated with BCL-2 inhibitor YC137 at the indicated concentrations for 10 days. (F) Three-dimensional Matrigel assay: MCF7-Vec and MCF7-TFF3 cells were cultured in 4% Matrigel in 10% FBS medium for 7 days. Bar, 100 µm in A, B, and F. *P < .05. **P < .01.
We subsequently determined the involvement of BCL-2 in TFF3-stimulated anchorage-independent growth. We observed that, concordant with the qPCR results described above, forced expression of TFF3 markedly increased the protein level of BCL-2 and decreased the expression of BAX (Figure 2D). BCL-2 specific inhibitor YC137 reduced soft agar colony formation by MCF7-Vec cells in a dose-dependent manner. YC137 also abrogated TFF3-enhanced colony formation in soft agar, suggesting that TFF3-stimulated anchorage-independent growth is BCL-2-dependent (Figure 2E).
We further evaluated the effect of forced expression of TFF3 on cell growth in the three-dimensional culture of laminin-rich extracellular matrix gels (Matrigel; BD Biosciences, Franklin Lakes, NJ). The colonies formed by MCF7-TFF3 cells were significantly larger and appeared more invasive compared with those formed by MCF7-Vec cells (Figure 2F). Consistent with the in vitro analyses, forced expression of TFF3 also promoted MCF-7 cell-derived xenograft growth (see below).
Depletion of TFF3 by siRNA Reduces Oncogenicity of Mammary Carcinoma Cells
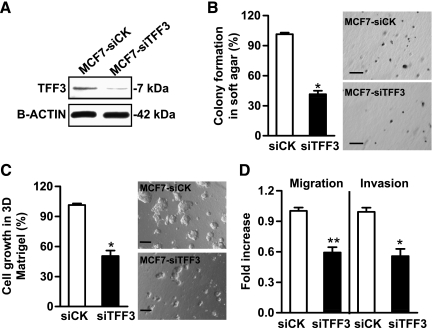
An siRNA construct targeting TFF3 or a scrambled siRNA as control was stably transfected into MCF-7 cells (designated as MCF7-siTFF3 and MCF7-siCK, respectively). Figure 3A demonstrated that the TFF3 protein level was efficiently decreased in MCF7-siTFF3 cells compared with MCF7-siCK cells. siRNA-mediated depletion of TFF3 produced a 59% decrease in anchorage-independent growth (Figure 3B) and a 50% decrease in cell growth in three-dimensional Matrigel (Figure 3C). Furthermore, depletion of TFF3 reduced MCF-7 cell migration and invasion by 41% and 44%, respectively (Figure 3D).
Figure 3.
Depletion of TFF3 by siRNA inhibits anchorage-independent growth and invasiveness of mammary carcinoma cells. (A) Western blot analysis of the efficiency of siRNA-mediated depletion of TFF3 protein in MCF-7 cells. β-ACTIN was used as a loading control for cell lysates. (B) Colony formation in soft agar: MCF7-siCK and MCF7-siTFF3 cells were seeded in 0.35% agar and cultured for 10 days. (C) Three-dimensional Matrigel assay: MCF7-Vec and MCF7-TFF3 cells were cultured in 4% Matrigel in 10% FBS medium for 7 days. (D) Migration and invasion assays: MCF7-siCK and MCF7-siTFF3 cell migration after 24 hours and invasion after 48 hours in the transwell chambers. Bar, 100 µm in B and C. *P < .05. **P < .01.
Functional Antagonism of TFF3 by TFF3 Polyclonal Antibody Reduces Cell Viability In Vitro and Inhibits Xenograft Growth
The rhTFF3 protein was produced under nondenaturing conditions. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) determined the presence of dimeric rhTFF3 under non-reducing conditions (∼14 kDa) and monomeric rhTFF3 under reducing conditions (∼7 kDa; Figure W3A and Supplementary Data). A polyclonal antibody against rhTFF3 was generated in rabbits and was affinity-purified against antigen. ELISA confirmed that TFF3-pAb bound to rhTFF3 with high affinity (Kd = 53.3 pM) and at the detection limit of 10 pg (Figure W3, B and C, and Supplementary Data). Western blot analysis confirmed that TFF3-pAb did not exhibit cross-reactivity with rhTFF1 protein (Figure W3D and Supplementary Data).
The effects of TFF3-pAb on mammary carcinoma cell growth were evaluated in MCF-7 and T47D cells. T47D cells have previously been reported to express TFF3 [23], and we have confirmed this observation by Western blot analysis (data not shown). A significant reduction of cell viability was observed on TFF3-pAb treatment in serum-depleted conditions (0.5% FBS) with 90% and 43% inhibition compared to treatment with nonspecific rabbit IgG in MCF-7 and T47D cells, respectively (Figure 4A). Under serum-replete conditions (10% FBS), TFF3-pAb reduced MCF-7 cell viability by 31% and T47D cell viability by 25% (Figure 4A) compared with control rabbit IgG. Notably, treatment with TFF3-pAb resulted in a significant increase in apoptotic cell death in both MCF-7 and T47D cells as measured by Hoechst 33258 dye (Figure 4B) and annexin V and PI double staining (Figure W3E and Supplementary Data). In addition, TFF3-pAb also reduced cell growth in three-dimensional Matrigel by 53% in MCF-7 cells and 42% in T47D cells compared with control rabbit IgG (Figure 4C).
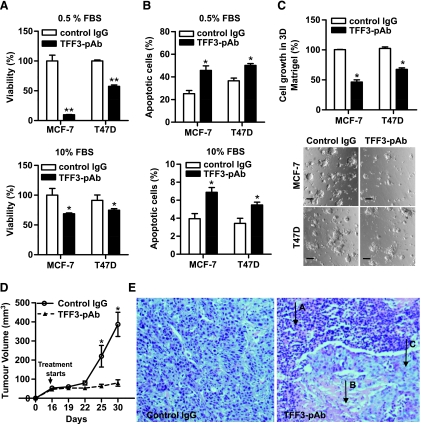
Figure 4.
Neutralization of TFF3 by TFF3 polyclonal antibody reduces viability of mammary carcinoma cells and inhibits xenograft growth. (A) MCF-7 and T47D cells were treated with 500 µg/ml control rabbit IgG or TFF3-pAb in either 0.5% FBS or 10% FBS medium for 3 days. Cell viability was measured by MTT assay as described in Materials and Methods. (B) Apoptotic cell death was assessed by fluorescent microscopic analysis of cell nuclear staining patterns with Hoechst 33258 dye after treatment for 24 hours. (C) Three-dimensional Matrigel: MCF-7 cells and T47D cells were cultured in 4% Matrigel in 10% FBS medium containing 500 µg/ml nonspecific rabbit IgG or TFF3-pAb for 7 days. MCF-7 xenografts in immunodeficient mice were treated with either control rabbit IgG or TFF3-pAb for 14 days. (D) Tumor growth of TFF3-pAb-treated mice and control rabbit IgG-treated mice. (E) Histologic staining of the tumors with hematoxylin and eosin. Arrow A indicates inflammatory granuloma; arrow B, necrotic tissue within the tumor; and arrow C, residual tumor. Bar, 100 µm in C. *P < .05. **P < .01.
We further examined the effect of TFF3-pAb treatment on the growth of human mammary carcinoma xenografts in nude mice. TFF3-pAb, dosed 16 days after MCF-7 cell implantation and there-after daily for 2 weeks, produced tumor growth arrest (Figure 4D). Histologic analyses showed large necrotic fields surrounded by inflammatory granuloma. The residual tumor cells were isolated from normal mouse tissue by a vascular fibrous capsule (Figure 4E). To investigate potential adverse effects of TFF3-pAb treatment, we examined the tissue integrity in several organs including liver, lung, intestine, kidney, and stomach of both TFF3-pAb and nonspecific rabbit IgG-treated mice. No histologic alterations were identified between the two groups (Figure W4 and Supplementary Data). Therefore, TFF3-pAb, as a single agent, effectively produced growth arrest of mammary carcinoma xenografts.
TFF3 Expression Correlates to Poor Outcome in ER-Positive Breast Cancer Patients Treated with Tamoxifen
TFF3 is an estrogen-regulated gene [23]. We therefore performed a meta-analysis to determine the association of TFF3 expression with ER status in breast cancer patients. We analyzed 15 breast cancer microarray data sets available in the cancer microarray database Oncomine (www.oncomine.org) and identified a strong correlation of TFF3 mRNA expression with ER-positive status in mammary carcinoma (P < .0001, t value of -1.45 with 95% confidence interval of -1.78 to -1.11; Figure W5 and Supplementary Data).
We further examined whether TFF3 expression would predict the outcome of ER-positive breast cancer patients treated with antiestrogens. Kaplan-Meier analysis of 68 ER-positive breast cancer patients treated with tamoxifen demonstrated a highly significant correlation of increased TFF3 mRNA expression to decreased DMFS (P = .00002; Figure W6A and Supplementary Data) and disease-specific survival (P = .001; Figure W6B and Supplementary Data). TFF3 expression was also strongly associated with disease-free survival (P = .00003; Figure W6C and Supplementary Data). To highlight the significant association of TFF3 expression with poor DMFS outcome, we further split the cohort into three groups: lowest 33% (n = 23), intermediate 33% (n = 22), and highest 33% (n = 23) of the expression level of TFF3 (Figure W6D and Supplementary Data). The groups with the intermediate and highest TFF3 expression have a particularly poor outcome in comparison to those with the lowest TFF3 expression (P = .008).
TFF3 Enhances ER Transcriptional Activity and Function
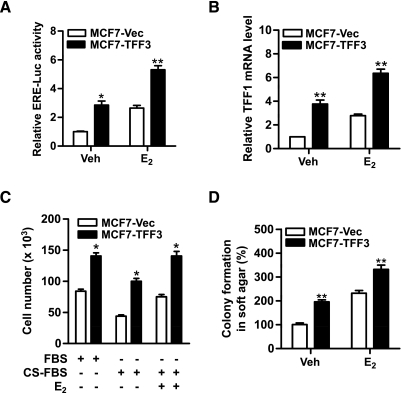
Because of the observed correlation between TFF3 expression and ER status, we subsequently investigated a potential functional interaction between TFF3 and ER signaling pathways. We first measured ER transcriptional activity with a luciferase reporter containing the ER element (ERE). MCF7-TFF3 cells exhibited a 2.8-fold higher basal ER transcriptional activity compared with MCF7-Vec cells. E2 increased ERE luciferase activity and forced expression of TFF3 additively increased ER transcriptional activity with E2 (Figure 5A). TFF1 is a classic estrogen-responsive gene [39]. qPCR analysis demonstrated that forced expression of TFF3 increased TFF1 mRNA level compared with vector cells (Figure 5B). We further examined the effect of forced expression of TFF3 on cell proliferation in monolayer and anchorage-independent growth under estrogen-deprived or estrogen-replete conditions. In the absence of estrogen, forced expression of TFF3 resulted in increased cell number in monolayer (Figure 5C) and colony formation in soft agar (Figure 5D) compared with vector cells. E2 promoted cell growth both in monolayer culture and in soft agar, and forced expression of TFF3 enhanced E2-stimulated cell proliferation under both culture conditions (Figure 5, C and D).
Figure 5.
Forced expression of TFF3 increases ER transcriptional activity in mammary carcinoma cells. (A) ERE-luciferase assay: MCF7-Vec and MCF7-TFF3 cells were transiently cotransfected with of pGL2-ERE-luciferase plasmid and pcDNA3 β-galactosidase plasmids in six-well plates in phenol red-free RPMI medium supplemented with 1% CS-FBS. Twenty-four hours later, the cells were treated with either DMSO (vehicle) or 10 nM E2 for another 24 hours. The cell lysates were collected and assessed for luciferase activity. (B) MCF7-Vec and MCF7-TFF3 cells were treated with either DMSO (vehicle) or 10 nM E2 in phenol red-free RPMI medium supplemented with 1% CS-FBS for 24 hours before RNA isolation. TFF1 mRNA levels were measured by qPCR. (C) Total cell number: MCF7-Vec and MCF7-TFF3 cells were cultured in 10% FBS, 10% CS-FBS, or 10% CS-FBS containing 10 nM E2 for 48 hours. (D) Colony formation in soft agar: MCF7-Vec and MCF7-TFF3 cells were seeded in 0.35% agar in a 96-well plate and treated with vehicle (Veh) or 10 nM E2 in 10% CS-FBS medium for 10 days. *P < .05, **P < .01 compared with vector control group for the same treatment.
Forced Expression of TFF3 Reduces Anti-Estrogen Sensitivity of MCF-7 Cells in a BCL-2-Dependent Manner
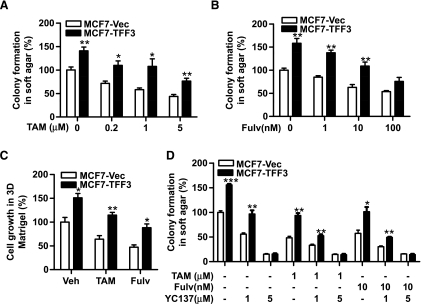
As our data indicated that TFF3 promoted estrogen-independent growth, we next examined the effect of TFF3 on the cellular response to antiestrogens in MCF-7 cells. Figure 6, A and B, show a dose-dependent decrease in colony formation in soft agar in MCF7-Vec cells after tamoxifen and fulvestrant treatment. Forced expression of TFF3 significantly abrogated the inhibitory effect of both antiestrogens on anchorage-independent growth. Similarly, MCF7-TFF3 cells displayed a reduced sensitivity to antiestrogen treatment in three-dimensional Matrigel with 51% and 41% greater growth relative to MCF7-Vec cells in the presence of tamoxifen and fulvestrant, respectively (Figure 6C).
Figure 6.
Forced expression of TFF3 reduces anti-estrogen sensitivity in mammary carcinoma cells. (A, B) Colony formation in soft agar: MCF7-Vec and MCF7-TFF3 cells were seeded in 0.35% agar in a 96-well plate and treated with tamoxifen (TAM) or fulvestrant (Fulv) in 10% FBS at the concentrations indicated for 10 days. (C) Three-dimensional Matrigel: MCF7-Vec and MCF7-TFF3 cells were seeded in 4% Matrigel in 10% FBS medium and treated with vehicle (Veh) or 1 µM tamoxifen or 10 nM fulvestrant for 7 days. (D) Colony formation in soft agar: MCF7-Vec and MCF7-TFF3 cells were seeded in 0.35% agar and treated with BCL-2 inhibitor YC137 at 1 or 5 µM alone or in combination with 1 µM tamoxifen or 10 nM fulvestrant for 10 days. *P < .05, **P < .01, ***P < .001 compared with vector control group for the same treatment.
We have shown that TFF3 increased BCL-2 expression to promote anchorage-independent growth. To determine whether BCL-2 is also involved in anti-estrogen resistance stimulated by TFF3, we examined the anchorage-independent growth of MCF7-Vec and MCF7-TFF3 cells in the presence of the BCL-2 inhibitor YC137 alone or in combination with tamoxifen or fulvestrant. As shown in Figure 6D, treatment of YC137 eliminated the survival advantage of MCF7-TFF3 cells in response to tamoxifen or fulvestrant, suggesting that decreased anti-estrogen sensitivity produced by forced expression of TFF3 is mediated by increased BCL-2 expression.
Forced Expression of TFF3 Reduces Tamoxifen Sensitivity of MCF-7 Cells In Vivo
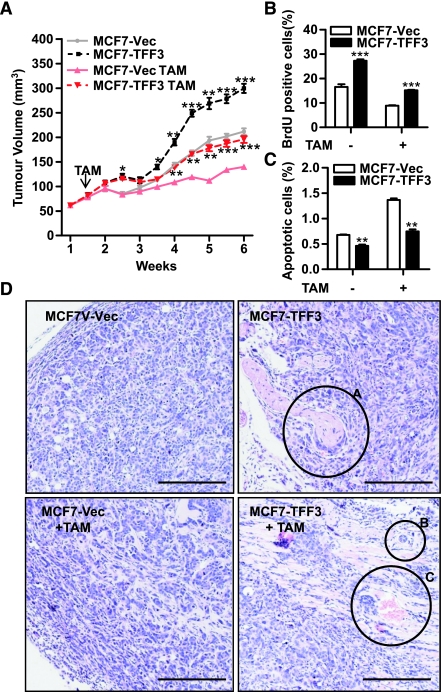
We next investigated the effect of forced expression of TFF3 on tamoxifen sensitivity in a mouse xenograft model. MCF-7 xenografts were grown for 10 days and subsequently treated with tamoxifen in the presence of estrogen supplement. In the untreated groups, tumors formed by MCF7-TFF3 cells were larger than those formed by MCF7-Vec cells, indicating that TFF3 enhanced tumorigenicity in vivo. Tamoxifen inhibited the growth of tumors derived from MCF7-Vec cells as expected. In contrast, forced expression of TFF3 attenuated the growth-inhibitory effect of tamoxifen, producing a tumor growth rate comparable with that observed in untreated MCF7-Vec tumors (Figure 7A).
Figure 7.
Forced expression of TFF3 reduces tamoxifen sensitivity of mammary carcinoma cells in vivo. MCF7-Vec and MCF7-TFF3 cells were implanted into the mammary fat pad of athymic nude mice. The tumor xenografts were treated with estrogen or estrogen plus tamoxifen on day 10 after inoculation. (A) The growth curves of MCF7-Vec and MCF7-TFF3 tumors in the presence and the absence of tamoxifen (TAM). (B and C) Evaluation of tumor proliferation by nuclear BrdU incorporation (B) and apoptosis by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay (C) in MCF7-Vec and MCF7-TFF3 xenografts. (D) Histologic staining of representative tumors with hematoxylin and eosin. Circle A indicates the region of perineural invasion; circles B and C, regions of vascular invasion. Bar, 100 µm in D. *P < .05, **P < .01, ***P < .001 compared with vector control group for the same treatment.
Tamoxifen suppressed BrdU incorporation and increased apoptosis in MCF7-Vec tumors. MCF7-TFF3 tumors exhibited 1.7-fold higher BrdU incorporation (Figure 7B) and 45% less apoptosis (Figure 7C) relative to MCF7-Vec cells after tamoxifen treatment, suggesting that forced expression of TFF3 reduced tamoxifen efficacy in xenografts by preventing cell cycle arrest and providing protection from apoptosis. Histologic examination revealed that MCF7-TFF3 tumors contained pleomorphic cells with irregular hyperchromatic nuclei and scanty cytoplasm compared with well-differentiated MCF7-Vec tumor cells (Figure 7D). Moreover, MCF7-TFF3 tumors were larger with ill-defined margins to the surrounding tissues and evident vascular and perineural invasion. Tamoxifen treatment resulted in a smaller tumor lesion in MCF7-Vec tumors, whereas MCF7-TFF3 tumors still exhibited invasive tendencies (Figure 7D).
Depletion of TFF3 by siRNA or Inhibition of TFF3 by Antibody Increases Anti-Estrogen Sensitivity in Mammary Carcinoma Cells
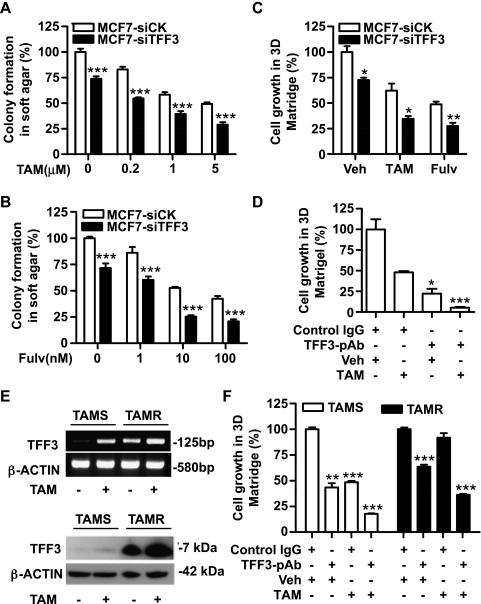
Tamoxifen and fulvestrant inhibited colony formation in soft agar of control siRNA transfected cells (Figure 8, A and B). Similar inhibitory effects of tamoxifen and fulvestrant were observed in three-dimensional Matrigel (Figure 8C). Depletion of TFF3 by siRNA increased the sensitivity of MCF-7 cells to both tamoxifen and fulvestrant, resulting in reduced growth in soft agar (Figure 8, A and B) and in three-dimensional Matrigel (Figure 8C). Consistent with the effect of depletion of TFF3 by siRNA, TFF3-pAb also enhanced the growth-inhibitory effect of tamoxifen. As shown in Figure 8D, TFF3-pAb reduced MCF-7 cell growth in three-dimensional Matrigel by 78.8%, whereas tamoxifen produced a 52% growth inhibition. The combination of TFF3-pAb with tamoxifen produced a total 95% inhibition of cell growth in three-dimensional Matrigel.
Figure 8.
Depletion of TFF3 by siRNA or inhibition of TFF3 function by antibody increases anti-estrogen sensitivity in MCF-7 cells and improves tamoxifen sensitivity in tamoxifen-resistant cells. (A and B) Colony formation in soft agar: MCF7-siCK and MCF7-siTFF3 cells were seeded in 0.35% agar and treated with tamoxifen (TAM) and fulvestrant (Fulv) in 10% FBS at the concentrations indicated for 10 days. (C) Three-dimensional Matrigel: MCF7-siCK and MCF7-siTFF3 cells were seeded in 4% Matrigel and treated with vehicle (Veh) or 1 µM tamoxifen or 10 nM fulvestrant for 7 days. (D) Three-dimensional Matrigel: MCF-7 cells were seeded in 4% Matrigel and treated with 1 µM tamoxifen alone or in combination with 500 µg/ml TFF3-pAb or control rabbit IgG for 7 days. (E) TAMS and TAMR cells were cultured in phenol red-free RPMI medium containing 10% CS-FBS for 3 days and then treated with either vehicle or 1 µM tamoxifen for 24 hours. TFF3 expression was determined by semiquantitative RT-PCR and Western blot analysis. (F) TAMS and TAMR cells were seeded in 4% Matrigel in 10% CS-FBS medium and treated with 1 µM tamoxifen alone or in combination with 500 µg/ml TFF3-pAb or control rabbit IgG for 7 days. *P < .05, **P < .01, ***P < .001 compared with vector control group for the same treatment (A, B, and C) or compared with control IgG and vehicle-treated group (D and F).
Inhibition of TFF3 Improves Tamoxifen Sensitivity in Tamoxifen-Resistant Cells
We previously have generated a tamoxifen-resistant MCF-7 cell line (designated as TAMR) and a control tamoxifen-sensitive MCF-7 cell line (designated as TAMS) [40]. In an attempt to determine the role of TFF3 in acquired tamoxifen resistance, we examined TFF3 expression in TAMS and TAMR cells. In the absence of tamoxifen, both the mRNA and protein levels of TFF3 were higher in TAMR cells compared with TAMS cells (Figure 8E). Paradoxically, tamoxifen increased TFF3 expression at both mRNA and protein levels in TAMS cells, and TAMR cells exhibited further elevated TFF3 expression in response to tamoxifen (Figure 8E).
To investigate the function of TFF3 in TAMR cells, TAMR cells and TAMS cells were treated with tamoxifen, TFF3-pAb, or tamoxifen plus TFF3-pAb, and cell growth in three-dimensional Matrigel was measured. Tamoxifen did not significantly affect TAMR cell growth, whereas TFF3-pAb alone suppressed the growth of TAMR cells by 36% (Figure 8F). Combined treatment with TFF3-pAb and tamoxifen resulted in 64% growth inhibition, producing a percentage reduction similar to that observed in TAMS cells treated with tamoxifen.
Discussion
There is a growing body of evidence that supports a role for TFF3 in oncogenic transformation and neoplastic progression of mammary carcinoma. We herein reported a strong correlation of TFF3 mRNA expression with poor survival outcome not only in the cohort of breast cancer patients treated with tamoxifen but also in the wider population of patients with breast cancer. In support of the clinical relevance of elevated TFF3 expression in breast cancer, we further demonstrated that TFF3 stimulated oncogenicity and invasiveness of mammary carcinoma cells and was involved in anti-estrogen resistance both in vitro and in vivo.
TFF3 is a potent survival factor for mammary carcinoma cells. Forced expression of TFF3 significantly reduced serum deprivation and doxorubicin-induced apoptosis in MCF-7 cells. Concordantly, qPCR analyses revealed that TFF3 modulated the expression of some key apoptosis-related genes. We further demonstrated that TFF3-stimulated anchorage-independent growth is BCL-2-dependent. The antiapoptotic action of BCL-2 has been well established [41]. It is unlikely that up-regulation of BCL-2 is the sole mechanism accounting for the prosurvival effect of TFF3. For example, the expression of BAX, a proapoptotic member of the BCL-2 family, was dramatically reduced by TFF3. Moreover, other survival signaling pathways may also contribute to TFF3-stimulated oncogenicity in mammary carcinoma cells. TFF3 has been reported to activate mitogen-activated protein kinase [24], phosphatidylinositol-3-kinase-Akt [16,42], signal transducer and activator of transcription 3 [13], and nuclear factor kappa B (NF-κB) [43] survival pathways. Interestingly, BCL-2 seems to be a common final mediator as BCL-2 expression is regulated by all of the above-mentioned signaling pathways [44,45]. Moreover, we demonstrated herein functional cross talk between the TFF3 and ER pathways. Forced expression of TFF3 increased ER transcriptional activity and enhanced the estrogenic response. Given the prosurvival effect of ER signaling in mammary carcinoma cells, TFF3-mediated enhancement of ER function may be considered as one potential mechanism producing TFF3-stimulated cell survival.
The fundamental contribution of growth factor-driven signaling pathways to the development of either de novo or acquired endocrine resistance has been widely recognized. In this study, we identified TFF3 as a novel growth factor involved in anti-estrogen resistance. As an estrogen-inducible gene, TFF3 expression is stimulated by estrogen treatment [23]. Paradoxically, the selective ER modulator tamoxifen also increased TFF3 expression level in the ER-positive mammary carcinoma cells, which is consistent with previously published data [23]. The mechanisms of the agonist action of tamoxifen remain unclear, although gene expression profiling has identified partial agonistic activity of tamoxifen on 30% of E2-inducible genes [46]. Of particular note is the fact that in TAMR cells, TFF3 expression was substantially increased and further induced by tamoxifen. Functional antagonism of TFF3 by antibody effectively inhibited cell growth and improved tamoxifen sensitivity of TAMR cells. We therefore speculate that tamoxifen-induced TFF3 expression may provide a survival advantage in the presence of tamoxifen treatment and will contribute to both intrinsic and acquired tamoxifen resistance.
Although the mechanisms by which growth factor signaling pathways promote anti-estrogen resistance have not been fully elucidated, cumulative experimental data suggest that activated growth factor signaling pathways either cross talk with ER signaling pathway resulting in ligand-independent or tamoxifen-induced ER activation or function as alternative mitogenic and/or survival pathways, consequently promoting anti-estrogen resistant tumor cell growth [47–49]. Our data also suggested a potential functional interaction between TFF3 and ER signal transduction through which ER activation upregulated TFF3 expression and TFF3 stimulation increased ER transcriptional activity. Epidermal growth factor receptor (EGFR) and HER2 signaling pathways have long been implicated in anti-estrogen resistance. In support of TFF3-mediated anti-estrogen resistance, the potential involvement of EGFR activation in the TFF3 signaling pathway has been reported by several studies [16,42,50]. In addition, because induction of cell apoptosis mediates the growth-inhibitory effect of antiestrogens, activation of cell survival pathways and protection from apoptosis would attenuate anti-estrogen sensitivity [49,51]. We demonstrated that TFF3-stimulated resistance to antiestrogens is mediated by up-regulation of BCL-2 expression. The contribution of BCL-2 in anti-estrogen-induced apoptosis and resistance to antiestrogen therapy has been well studied in cell culture model [52–54], although conflicting data were reported from clinical studies, which revealed a correlation of decreased BCL-2 expression with reduced tamoxifen sensitivity [51]. This may partially reflect the estrogenic regulation of BCL-2 expression in estrogen-responsive tumors that are more likely to respond to antiestrogen therapy [55]. As previously discussed, BCL-2 is a common mediator of several survival signaling pathways. Therefore, increased BCL-2 is probably a consequence of activation of survival pathways, resulting from increased TFF3 expression.
Although TFF3 is an estrogen-responsive gene and its expression level is positively correlated to ER status in human mammary carcinoma, there is growing evidence that a subset of ER-negative mammary tumors also express high levels of TFF3 [56,57]. These mammary tumors have been characterized by expression of androgen receptor (AR) and HER2 and classified into molecular apocrine subtype [58]. Cross talk between AR and HER2 pathways have also been recently elucidated in mammary carcinoma cell lines with molecular apocrine features [57]. Whether the TFF3 signaling pathway contributes to AR and HER2 cross talk and affects molecular apocrine mammary carcinoma cell growth remains to be determined. Further investigation of TFF3 function in ER-negative mammary carcinoma cell lines is therefore required to provide a more comprehensive understanding of TFF3 actions in mammary carcinoma.
Herein we have identified TFF3 as a functionally and clinically validated molecule involved in progression of human mammary carcinoma. Forced expression of TFF3 enhanced the oncogenicity, and mediated anti-estrogen resistance, of mammary carcinoma cells. Conversely, inhibition of TFF3 by siRNA or antibody reduced the oncogenicity of mammary carcinoma cells and increased the efficacy of antiestrogens. Given low or undetectable TFF3 expression in normal mammary tissues and elevated expression of TFF3 in mammary carcinoma [6], targeting of TFF3 may therefore represent a novel therapeutic approach for breast cancer treatment with high specificity and efficacy.
Supplementary Data
Meta-analysis of Breast Cancer Microarray Data Sets
Data were collected from publicly available breast cancer microarray data sets in the cancer microarray database Oncomine (www.oncomine.org). TFF3 gene expression was log-transformed, median-centered per array, and SD normalized to one per array. A meta-analysis approach was used to determine whether higher expression levels of the gene TFF3 was associated with ER+ or ER-status in breast cancer patients. Using t values and sample sizes from each study, we calculated effect sizes by the standardized mean difference method. The within-study variance for the effect size was calculated as the inverse of the study sample size. The pooled effect size was estimated in an intercept-only model with the study identifiers as random effects; this model was weighted by the between-study variance and the estimated within-study variances that were held constant. The meta-analysis was carried out using the MIXED procedure in SAS version 9.2 (SAS Institute, Inc, Cary, NC).
Acknowledgments
The authors thank Carol Chelimo for the meta-analysis of breast cancer microarray data and Michael Steiner for the assistance in performing flow cytometry experiments.
Abbreviations
- BrdU
bromodeoxyuridine
- CS-FBS
charcoal-stripped fetal bovine serum
- DMFS
distant metastasis-free survival
- ER
estrogen receptor
- qPCR
quantitative polymerase chain reaction
- siRNA
small interfering RNA
- TFF3
trefoil factor 3
- TFF3-pAb
TFF3-polyclonal antibody
Footnotes
This work was funded by the Breast Cancer Research Trust (NZ), the Foundation for Research, Science and Technology of New Zealand, the Cancer Science Institute of Singapore, the Hundred-Talent Scheme of Chinese Academy of Sciences, National Natural Science Foundation of China (30571030), and National Basic Research Program of China (2007CB914503).
N.K., J.K., X.G.K., J.Z.T., J.K.P., M.K.M., L.D.M., E.T.L., H.C.M., P.M.G., and D.X.L. have no conflicts of interest to declare. P.E.L. and T.Z. consult for and P.E.L. has equity interest in Perseis Therapeutics Ltd. P.E.L. is also named on PCT application numbers WO 2006/69253 and WO 2008/042435 and US provisional application number 61/059558.
This article refers to supplementary materials, which are designated by Tables W1 to W3 and Figures W1 to W6 and are available online at www.neoplasia.com.
References
- 1.Schmitt H, Wundrack I, Beck S, Gött P, Welter C, Shizuya H, Simon MI, Blin N. A third P-domain peptide gene (TFF3), human intestinal trefoil factor, maps to 21q22.3. Cytogenet Cell Genet. 1996;72:299–302. doi: 10.1159/000134208. [DOI] [PubMed] [Google Scholar]
- 2.Regalo G, Wright NA, Machado JC. Trefoil factors: from ulceration to neoplasia. Cell Mol Life Sci. 2005;62:2910–2915. doi: 10.1007/s00018-005-5478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madsen J, Nielsen O, Tornøe I, Thim L, Holmskov U. Tissue localization of human trefoil factors 1, 2, and 3. J Histochem Cytochem. 2007;55:505–513. doi: 10.1369/jhc.6A7100.2007. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann W. TFF (trefoil factor family) peptide-triggered signals promoting mucosal restitution. Cell Mol Life Sci. 2005;62:2932–2938. doi: 10.1007/s00018-005-5481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taupin D, Podolsky DK. Trefoil factors: initiators of mucosal healing. Nat Rev Mol Cell Biol. 2003;4:721–732. doi: 10.1038/nrm1203. [DOI] [PubMed] [Google Scholar]
- 6.Poulsom R, Hanby AM, Lalani EN, Hauser F, Hoffmann W, Stamp GW. Intestinal trefoil factor (TFF 3) and pS2 (TFF 1), but not spasmolytic polypeptide (TFF 2) mRNAs are co-expressed in normal, hyperplastic, and neoplastic human breast epithelium. J Pathol. 1997;183:30–38. doi: 10.1002/(SICI)1096-9896(199709)183:1<30::AID-PATH1085>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Dhar DK, Wang TC, Tabara H, Tonomoto Y, Maruyama R, Tachibana M, Kubota H, Nagasue N. Expression of trefoil factor family members correlates with patient prognosis and neoangiogenesis. Clin Cancer Res. 2005;11:6472–6478. doi: 10.1158/1078-0432.CCR-05-0671. [DOI] [PubMed] [Google Scholar]
- 8.Yamachika T, Werther JL, Bodian C, Babyatsky M, Tatematsu M, Yamamura Y, Chen A, Itzkowitz S. Intestinal trefoil factor: a marker of poor prognosis in gastric carcinoma. Clin Cancer Res. 2002;8:1092–1099. [PubMed] [Google Scholar]
- 9.Faith DA, Isaacs WB, Morgan JD, Fedor HL, Hicks JL, Mangold LA, Walsh PC, Partin AW, Platz EA, Luo J, et al. Trefoil factor 3 overexpression in prostatic carcinoma: prognostic importance using tissue microarrays. Prostate. 2004;61:215–227. doi: 10.1002/pros.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okada H, Kimura MT, Tan D, Fujiwara K, Igarashi J, Makuuchi M, Hui AM, Tsurumaru M, Nagase H. Frequent trefoil factor 3 (TFF3) overexpression and promoter hypomethylation in mouse and human hepatocellular carcinomas. Int J Oncol. 2005;26:369–377. [PMC free article] [PubMed] [Google Scholar]
- 11.Bignotti E, Ravaggi A, Tassi RA, Calza S, Rossi E, Falchetti M, Romani C, Bandiera E, Odicino FE, Pecorelli S, et al. Trefoil factor 3: a novel serum marker identified by gene expression profiling in high-grade endometrial carcinomas. Br J Cancer. 2008;99:768–773. doi: 10.1038/sj.bjc.6604546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babyatsky M, Lin J, Yio X, Chen A, Zhang JY, Zheng Y, Twyman C, Bao X, Schwartz M, Thung S, et al. Trefoil factor-3 expression in human colon cancer liver metastasis. Clin Exp Metastasis. 2009;26:143–151. doi: 10.1007/s10585-008-9224-9. [DOI] [PubMed] [Google Scholar]
- 13.Christine R, Sylvie R, Erik B, Geneviève P, Amélie R, Gérard R, Marc B, Christian G, Samir A. Implication of STAT3 signaling in human colonic cancer cells during intestinal trefoil factor 3 (TFF3)- and vascular endothelial growth factor-mediated cellular invasion and tumor growth. Cancer Res. 2005;65:195–202. [PubMed] [Google Scholar]
- 14.Yio X, Zhang JY, Babyatsky M, Chen A, Lin J, Fan QX, Werther JL, Itzkowitz S. Trefoil factor family-3 is associated with aggressive behavior of colon cancer cells. Clin Exp Metastasis. 2005;22:157–165. doi: 10.1007/s10585-005-6615-z. [DOI] [PubMed] [Google Scholar]
- 15.Chan MW, Chan VY, Leung WK, Chan KK, To KF, Sung JJ, Chan FK. Anti-sense trefoil factor family-3 (intestinal trefoil factor) inhibits cell growth and induces chemosensitivity to adriamycin in human gastric cancer cells. Life Sci. 2005;76:2581–2592. doi: 10.1016/j.lfs.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Taupin DR, Kinoshita K, Podolsky DK. Intestinal trefoil factor confers colonic epithelial resistance to apoptosis. Proc Natl Acad Sci USA. 2000;97:799–804. doi: 10.1073/pnas.97.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry JK, Kannan N, Grandison PM, Mitchell MD, Lobie PE. Are trefoil factors oncogenic? Trends Endocrinol Metab. 2008;19:74–81. doi: 10.1016/j.tem.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Weigelt B, Verduijn P, Bosma AJ, Rutgers EJ, Peterse HL, Van't Veer LJ. Detection of metastases in sentinel lymph nodes of breast cancer patients by multiple mRNA markers. Br J Cancer. 2004;90:1531–1537. doi: 10.1038/sj.bjc.6601659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smid M, Wang Y, Klijn JGM, Sieuwerts AM, Zhang Y, Atkins D, Martens JWM, Foekens JA. Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006;24:2261–2267. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- 20.Bosma AJ, Weigelt B, Lambrechts AC, Verhagen OJHM, Pruntel R, Hart AAM, Rodenhuis S, Van't Veer LJ. Detection of circulating breast tumor cells by differential expression of marker genes. Clin Cancer Res. 2002;8:1871–1877. [PubMed] [Google Scholar]
- 21.Weigelt B, Bosma AJ, Hart AAM, Rodenhuis S, Van't Veer LJ. Marker genes for circulating tumour cells predict survival in metastasized breast cancer patients. Br J Cancer. 2003;88:1091–1094. doi: 10.1038/sj.bjc.6600868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer. 2006;13:1033–1067. doi: 10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 23.May FE, Westley BR. Expression of human intestinal trefoil factor in malignant cells and its regulation by oestrogen in breast cancer cells. J Pathol. 1997;182:404–413. doi: 10.1002/(SICI)1096-9896(199708)182:4<404::AID-PATH875>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Taupin D, Wu DC, Jeon WK, Devaney K, Wang TC, Podolsky DK. The trefoil gene family are coordinately expressed immediate-early genes: EGF receptor- and MAP kinase-dependent interregulation. J Clin Invest. 1999;103:R31–R38. doi: 10.1172/JCI3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amiry N, Kong X, Muniraj N, Kannan N, Grandison PM, Lin J, Yang Y, Vouyovitch CM, Borges S, Perry JK, et al. Trefoil factor-1 (TFF1) enhances oncogenicity of mammary carcinoma cells. Endocrinology. 2009;150:4473–4483. doi: 10.1210/en.2009-0066. [DOI] [PubMed] [Google Scholar]
- 26.Xu XQ, Emerald BS, Goh ELK, Kannan N, Miller LD, Gluckman PD, Liut ET, Lobie PE. Gene expression profiling to identify oncogenic determinants of autocrine human growth hormone in human mammary carcinoma. J Biol Chem. 2005;280:23987–24003. doi: 10.1074/jbc.M503869200. [DOI] [PubMed] [Google Scholar]
- 27.Pandey V, Perry JK, Mohankumar KM, Kong XJ, Liu SM, Wu ZS, Mitchell MD, Zhu T, Lobie PE. Autocrine human growth hormone stimulates oncogenicity of endometrial carcinoma cells. Endocrinology. 2008;149:3909–3919. doi: 10.1210/en.2008-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu DX, Lobie PE. Transcriptional activation of p53 by Pitx1. Cell Death Differ. 2007;14:1893–1907. doi: 10.1038/sj.cdd.4402209. [DOI] [PubMed] [Google Scholar]
- 30.Tang JZ, Zuo ZH, Kong XJ, Steiner M, Yin Z, Perry JK, Zhu T, Liu DX, Lobie PE. Signal transducer and activator of transcription (STAT)-5A and STAT5B differentially regulate human mammary carcinoma cell behavior. Endocrinology. 2010;151:43–55. doi: 10.1210/en.2009-0651. [DOI] [PubMed] [Google Scholar]
- 31.Van de Loosdrecht AA, Beelen RHJ, Ossenkoppele GJ, Broekhoven MG, Langenhuijsen MMAC. A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells. J Immunol Methods. 1994;174:311–320. doi: 10.1016/0022-1759(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 32.Ji P, Agrawal S, Diederichs S, Bäumer N, Becker A, Cauvet T, Kowski S, Beger C, Welte K, Berdel WE, et al. Cyclin A1, the alternative A-type cyclin, contributes to G1/S cell cycle progression in somatic cells. Oncogene. 2005;24:2739–2744. doi: 10.1038/sj.onc.1208356. [DOI] [PubMed] [Google Scholar]
- 33.Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 34.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 35.Czabotar PE, Colman PM, Huang DCS. Bax activation by Bim? Cell Death Differ. 2009;16:1187–1191. doi: 10.1038/cdd.2009.83. [DOI] [PubMed] [Google Scholar]
- 36.Cowin P, Rowlands TM, Hatsell SJ. Cadherins and catenins in breast cancer. Curr Opin Cell Biol. 2005;17:499–508. doi: 10.1016/j.ceb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 37.Erin N, Wang N, Xin P, Bui V, Weisz J, Barkan GA, Zhao W, Shearer D, Clawson GA. Altered gene expression in breast cancer liver metastases. Int J Cancer. 2009;124:1503–1516. doi: 10.1002/ijc.24131. [DOI] [PubMed] [Google Scholar]
- 38.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 39.Sun JM, Spencer VA, Li L, Yu Chen H, Yu J, Davie JR. Estrogen regulation of trefoil factor 1 expression by estrogen receptor a and Sp proteins. Exp Cell Res. 2005;302:96–107. doi: 10.1016/j.yexcr.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 40.Kang J, Qian PX, Pandey V, Perry JK, Miller LD, Liu ET, Zhu T, Liu DX, Lobie PE. Artemin is estrogen regulated and mediates antiestrogen resistance in mammary carcinoma. Oncogene. 2010;29:3228–3240. doi: 10.1038/onc.2010.71. [DOI] [PubMed] [Google Scholar]
- 41.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinoshita K, Taupin DR, Itoh H, Podolsky DK. Distinct pathways of cell migration and antiapoptotic response to epithelial injury: structure-function analysis of human intestinal trefoil factor. Mol Cell Biol. 2000;20:4680–4690. doi: 10.1128/mcb.20.13.4680-4690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen YH, Lu Y, De Plaen IG, Wang LY, Tan XD. Transcription factor NF-κB signals antianoikic function of trefoil factor 3 on intestinal epithelial cells. Biochem Biophys Res Commun. 2000;274:576–582. doi: 10.1006/bbrc.2000.3176. [DOI] [PubMed] [Google Scholar]
- 44.Brumatti G, Salmanidis M, Ekert PG. Crossing paths: interactions between the cell death machinery and growth factor survival signals. Cell Mol Life Sci. 2010;67:1619–1630. doi: 10.1007/s00018-010-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 46.Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–1533. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 47.Arpino G, Wiechmann L, Osborne CK, Schiff R. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29:217–233. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riggins RB, Schrecengost RS, Guerrero MS, Bouton AH. Pathways to tamoxifen resistance. Cancer Lett. 2007;256:1–24. doi: 10.1016/j.canlet.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 50.Liu D, El-Hariry I, Karayiannakis AJ, Wilding J, Chinery R, Kmiot W, McCrea PD, Gullick WJ, Pignatelli M. Phosphorylation of β-catenin and epidermal growth factor receptor by intestinal trefoil factor. Lab Invest. 1997;77:557–563. [PubMed] [Google Scholar]
- 51.Riggins RB, Bouton AH, Liu MC, Clarke R. Antiestrogens, aromatase inhibitors, and apoptosis in breast cancer. Vitam Horm. 2005;71:201–237. doi: 10.1016/S0083-6729(05)71007-4. [DOI] [PubMed] [Google Scholar]
- 52.Diel P, Smolnikar K, Michna H. The pure antiestrogen ICI 182780 is more effective in the induction of apoptosis and down regulation of BCL-2 than tamoxifen in MCF-7 cells. Breast Cancer Res Treat. 1999;58:87–97. doi: 10.1023/a:1006338123126. [DOI] [PubMed] [Google Scholar]
- 53.Zhang GJ, Kimijima I, Onda M, Kanno M, Sato H, Watanabe T, Tsuchiya A, Abe R, Takenoshita S. Tamoxifen-induced apoptosis in breast cancer cells relates to down-regulation of bcl-2, but not bax and bcl-X(L), without alteration of p53 protein levels. Clin Cancer Res. 1999;5:2971–2977. [PubMed] [Google Scholar]
- 54.Kim R, Tanabe K, Emi M, Uchida Y, Toge T. Modulation of tamoxifen sensitivity by antisense Bcl-2 and trastuzumab in breast carcinoma cells. Cancer. 2005;103:2199–2207. doi: 10.1002/cncr.21029. [DOI] [PubMed] [Google Scholar]
- 55.Nahta R, Esteva FJ. Bcl-2 antisense oligonucleotides: a potential novel strategy for the treatment of breast cancer. Semin Oncol. 2003;30:143–149. doi: 10.1053/j.seminoncol.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 56.Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, Gerald WL. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25:3994–4008. doi: 10.1038/sj.onc.1209415. [DOI] [PubMed] [Google Scholar]
- 57.Naderi A, Hughes-Davies L. A functionally significant cross-talk between androgen receptor and ErbB2 pathways in estrogen receptor negative breast cancer. Neoplasia. 2008;10:542–548. doi: 10.1593/neo.08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, MacGrogan G, Bergh J, Cameron D, Goldstein D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
References
- 1.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Klijn JGM, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-Van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 3.Van De Vijver MJ, He YD, Van't Veer LJ, Dai H, Hart AAM, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 4.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 8.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 9.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, Ferno M, Peterson C, Meltzer PS. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. [PubMed] [Google Scholar]
- 11.Ginestier C, Cervera N, Finetti P, Esteyries S, Esterni B, Adelailde J, Xerri L, Viens P, Jacquemier J, Charafe-Jauffret E, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12:4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 12.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Jr, Marks JR, Nevins JR. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci USA. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Van De Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 15.Zhao H, Langerød A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Kåresen R, Botstein D, Børresen-Dale AL, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.