Abstract
BAK1 is a co-receptor of brassinosteroid (BR) receptor BRI1, and plays a well-characterized role in BR signalling. BAK1 also physically interacts with the flagellin receptor FLS2 and regulates pathogen resistance. The role of BAK1 in mediating Nicotiana attenuata's resistance responses to its specialist herbivore, Manduca sexta, was examined here. A virus-induced gene-silencing system was used to generate empty vector (EV) and NaBAK1-silenced plants. The wounding- and herbivory-induced responses were examined on EV and NaBAK1-silenced plants by wounding plants or simulating herbivory by treating wounds with larval oral secretions (OS). After wounding or OS elicitation, NaBAK1-silenced plants showed attenuated jasmonic acid (JA) and JA-isoleucine bursts, phytohormone responses important in mediating plant defences against herbivores. However, these decreased JA and JA-Ile levels did not result from compromised MAPK activity or elevated SA levels. After simulated herbivory, NaBAK1-silenced plants had EV levels of defensive secondary metabolites, namely, trypsin proteinase inhibitors (TPIs), and similar levels of resistance to Manduca sexta larvae. Additional experiments demonstrated that decreased JA levels in NaBAK1-VIGS plants, rather than the enzymatic activity of JAR proteins or Ile levels, were responsible for the reduced JA-Ile levels observed in these plants. Methyl jasmonate application elicited higher levels of TPI activity in NaBAK1-silenced plants than in EV plants, suggesting that silencing NaBAK1 enhances the accumulation of TPIs induced by a given level of JA. Thus NaBAK1 is involved in modulating herbivory-induced JA accumulation and how JA levels are transduced into TPI levels in N. attenuata.
Keywords: BAK1, defence, herbivory, jasmonic acid (JA), jasmonic acid-isoleucine (JA-Ile), Nicotiana attenuata, SERK, trypsin proteinase inhibitor
Introduction
Plants have evolved sophisticated regulatory networks to modulate their growth and to cope with environmental stresses. Within these networks, gibberellins, auxins, cytokinins, jasmonates, ethylene, brassinosteroids (BRs), salicylic acid (SA), abscisic acid (ABA), and strigolactone play roles as regulators of almost every aspect of a plant's life (Chow and McCourt, 2006; Santner et al., 2009). These hormone molecules exist in very low concentrations but alter plant physiology dramatically. Accordingly, plants possess elaborate systems to tightly control both the accumulation and perception of these hormones. In recent years, new genetic and biochemical tools have greatly augmented our understanding of the biosynthesis, transport, degradation, signal transduction, and physiological functions of these phytohormones in various plant species. Now it has become clear that, although an individual hormone has specific roles in modulating growth and stress responses, complex antagonistic and synergistic interactions among them enable plants to fine-tune their cellular processes (Chow and McCourt, 2006; Grant and Jones, 2009).
Among these phytohormones, jasmonic acid (JA) is one of the best studied. JA is involved in plant development and responses to various stresses, such as attack from herbivores and necrotrophic fungi (Wasternack, 2007; Howe and Jander, 2008; Browse, 2009; Wu and Baldwin, 2009). JA is synthesized by the oxylipin pathway in which several enzymes, including 13-lipoxygenase (13-LOX), allene-oxide synthase (AOS), allene-oxide cyclase (AOC), OPDA reductase (OPR), and acyl-coenzyme A oxidase (ACX) play pivotal roles (Wasternack, 2007; Liechti and Farmer, 2002). Importantly, instead of JA, JA-Ile [the Ile conjugate of JA that is catalysed by jasmonate-resistant proteins (JARs)] is responsible for eliciting most of the JA-induced responses (Staswick et al., 2002; Wang et al., 2008). Recent work has identified COI1, an F-box protein, as the JA-Ile receptor; JA-Ile interacts with COI1 and promotes the binding of COI1 to the jasmonate-ZIM domain (JAZ) proteins, which repress JA signalling. JAZs are subsequently ubiquitinated by SCFCOI1 E3 ubiquitin ligase and are degraded by the 26S proteasome. The degradation of the JAZ repressors releases MYC2 transcription factors, which are responsible for activating JA response in Arabidopsis (Xie et al., 1998; Chini et al., 2007; Thines et al., 2007; Yan et al., 2009).
The ecological significance of JA biosynthesis and signalling in plant resistance to herbivores has been intensively studied in Nicotiana attenuata, a species of wild tobacco (Halitschke and Baldwin, 2003; Paschold et al., 2007). N. attenuata perceives fatty acid–amino acid conjugates (FACs) in the oral secretions (OS) (mixtures of caterpillar saliva and regurgitants) of its specialist herbivore, Manduca sexta, and rapidly activates the salicylic acid-induced protein kinase (SIPK) and the wound-induced protein kinase (WIPK), two mitogen-activated protein kinases (MAPKs); subsequently, JA, JA-Ile, and ethylene biosynthesis are induced (Halitschke et al., 2001; Wu et al., 2007). In N. attenuata, silencing the expression of LOX3, JAR4, JAR6, and COI1 greatly impairs the plants' direct and indirect defences and increases the plants' susceptibility to M. sexta, demonstrating the central role of JA/JA-Ile biosynthesis and signalling in plant–herbivore interactions (Halitschke and Baldwin, 2003; Paschold et al., 2007; Wang et al., 2007, 2008). Herbivory-induced JA accumulation and signalling finally activates the biosynthesis of anti-herbivore compounds, such as nicotine, trypsin proteinase inhibitors (TPIs), and diterpene glycosides (DTGs) (Halitschke and Baldwin, 2003; Paschold et al., 2007). The important ecological roles of these defensive compounds have been demonstrated in plants that are impaired in their biosynthesis. Silencing putrescine N-methyl transferase (PMT) in nicotine biosynthesis, TPI gene, and geranylgeranyl diphosphate synthase (GGPPS) in DTG biosynthesis diminish herbivory-induced nicotine, TPI, and DTG accumulation and thus compromise N. attenuata's resistance to herbivores (Steppuhn et al., 2004; Zavala et al., 2004; Jassbi et al., 2008; Heiling et al., 2010).
BRs are a class of phytohormones that regulate seed germination, stem and root elongation, vascular differentiation, leaf expansion, and apical dominance (Halliday, 2004). Genetic analysis revealed that BR insensitive 1 (BRI1), a leucine-rich repeat receptor-like kinase (LRR-RLK), is the BR receptor (Li and Chory, 1997; Kinoshita et al., 2005). Another LRR-RLK, the Somatic Embryogenesis Receptor Kinase 3 (SERK3), also named BRI1-associated Kinase 1 (BAK1), physically interacts with BRI1 and plays an essential role in BR signalling (Li et al., 2002; Nam and Li, 2002; Belkhadir and Chory, 2006; Karlova and de Vries, 2006). Apart from its function in BR signalling, SERK3/BAK1 has been shown to play an important role in plants' defence against pathogens: SERK3/BAK1 binds to FLS2, the flagellin receptor, and positively regulates FLS2-mediated innate immunity in Arabidopsis (Chinchilla et al., 2007). In N. benthamiana, SERK3/BAK1 is required for multiple pathogen-associated molecular patterns (PAMPs)-elicited responses and for resistance to bacteria and oomycetes (Heese et al., 2007). Therefore SERK3/BAK1 has been proposed to have multiple functions as a common co-receptor and is thought to co-operate with other receptors (Gendron and Wang, 2007). Whether SERK3/BAK1 is involved in plants' defence responses to herbivory has not been examined.
To investigate the function of SERK3/BAK1 in plant–herbivore interactions, BAK1 was cloned in N. attenuata (NaBAK1) and its transcripts levels were silenced using a virus-induced gene-silencing (VIGS) approach. Traits important for herbivore defence in these plants were examined. Silencing NaBAK1 did not result in compromised wounding- or herbivory-induced MAPK activation, but impaired the accumulation of JA and JA-Ile after these stresses. However, NaBAK1-silenced plants still accumulated normal levels of herbivory-induced defensive compounds, TPIs. TPI activity analysis in methyl jasmonate (MeJA)-treated control and NaBAK1-silenced plants confirmed that NaBAK1 negatively modulates JA-induced TPI levels.
Materials and methods
Plant growth and sample treatments
Nicotiana attenuata (Solanaceae) seeds were from a line maintained in our laboratory that was originally collected in Utah (USA) and inbred for 30 generations in the greenhouse. Seed germination and plant cultivation followed Krügel et al. (2002). After being individually transferred into 1.0 l pots, plants were grown at 20–22 °C under 16 h of light. A virus-induced gene silencing (VIGS) system was used to silence the accumulation of NaBAK1 transcripts following a VIGS procedure optimized for N. attenuata (Ratcliff et al., 2001; Saedler and Baldwin, 2004). For wounding and water treatments (W+W), leaves were wounded with a pattern wheel, and 20 μl of water were immediately rubbed onto the wounded leaf; for wounding and OS treatments (W+OS), 20 μl of M. sexta OS (one-fifth diluted) were rubbed onto the wounded leaf; for wounding and FAC treatments (W+FAC), 20 μl of N-linolenoyl-L-Gln (FAC A) solution (27.6 ng μl−1) was rubbed onto the wounded leaf (Halitschke and Baldwin, 2003; Halitschke et al., 2001). For methyl jasmonate (MeJA) treatments, MeJA was dissolved in heat-liquefied lanolin at a concentration of 7.5 mg ml−1; 20 μl of the paste were applied to the base of a leaf, and 20 μl of pure lanolin were applied as a control. For the assays of JAR activity and substrate availability, the procedure described in Paschold et al. (2008) was followed. Samples were harvested in liquid nitrogen and stored at –80 °C in a freezer until analyses.
Cloning of NaBAK1 and NaSERK1, VIGS construct preparation, and sequence analysis
NaBAK1 full-length open reading frame (accession number HM639279) and NaSERK1 (accession number HM639280) partial sequence were amplified by PCR from a cDNA sample prepared from N. attenuata leaf tissue, using primers NaBAK1-1 and NaBAK1-2, and NaSERK1-1 and NaSERK1-2, respectively (see Supplementary Table S1 at JXB online). PCR products were cloned and sequenced.
Partial NaBAK1 sequence was cloned into pTV00 to form pTV-NaBAK1 using BamH1 and SalI enzymes (see Supplementary Table S1 at JXB online). The identity of pTV-NaBAK1 was confirmed by sequencing. pTV-NaBAK1 was transformed to an Agrobacterium strain for VIGS (Ratcliff et al., 2001).
Nucleotide and protein sequences were aligned in MegAlign (DNASTAR, Lasergene 8) using Clustal W algorithm. For phylogeny analysis, the unrooted Neighbor–Joining tree and bootstrap values were obtained using MEGA 4 software using default parameters and 1000 replications (Tamura et al., 2007) (www.megasoftware.net).
RNA extraction and quantitative real-time PCR (qPCR)
Total RNA was extracted from ground leaf samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer's instructions. For qPCR analysis, five replicated biological samples were used. 0.5 μg of total RNA sample was reverse-transcribed with oligo(dT) and Superscript II reverse transcriptase (Invitrogen). qPCR was performed on an ABI PRISM 7700 sequence detection system (Applied Biosystems, Foster City, CA, USA) using qPCR Core kits (Eurogentec, Seraing, Belgium). An N. attenuata actin2 gene was employed as the internal standard for normalizing cDNA concentration variations. The primer sequences for qPCR analysis are provided in Supplementary Table S2 at JXB online.
Protein extraction, kinase activity assay, and SDS-PAGE
Each protein sample was extracted from pooled leaves of five replicated plants. Protein extraction and MAPK in-gel kinase activity assay were done following Zhang and Klessig (1997). In-gel kinase images were obtained on an FLA-3000 phosphor imager system (Fujifilm, Tokyo, Japan). SDS-PAGE was done on a Bio-Rad Mini-PROTEAN 3 Cell using 10% gels. Gel staining was done using GelCode Blue Safe Stain reagent (Thermo Scientific).
Analysis of JA, JA-Ile, and SA concentrations
JA, JA-Ile, and SA concentrations were measured in five biological replicates following Wu et al. (2007). One milliliter of ethyl acetate spiked with 200 ng of D2-JA ,40 ng of 13C6-JA-Ile, and 40 ng of D4-SA, the internal standards for JA, JA-Ile, and SA, respectively, was added to each briefly ground leaf sample (∼150 mg). Samples were then ground on a FastPrep homogenizer (Thermo Electron, Waltham, MA, USA). After being centrifuged at maximum speed for 10 min at 4 °C, supernatants were transferred to fresh Eppendorf tubes and evaporated to dryness on a vacuum concentrator (Eppendorf, Hamburg, Germany). Each residue was resuspended in 0.5 ml of 70% methanol (v/v) and centrifuged to clarify phases. The supernatants were analysed on a HPLC-MS/MS (Varian, Palo Alto, CA, USA).
Analysis of secondary metabolites
The concentrations of nicotine and DTGs were analysed by HPLC as described in Keinanen et al. (2001). TPI activity was measured from five biological replicates using a radial diffusion assay described in Van Dam et al. (2001).
Measurement of ethylene emission
Five leaves treated with W+OS were immediately sealed in a three-neck 250 ml round bottom flask and kept in the plant growth chamber for 5 h. The headspace was flushed into a photoacoustic laser spectrometer with hydrocarbon-free clean air (INVIVO, Bonn, Germany), and the ethylene concentration was quantified by comparing ethylene peak areas with peak areas generated by a standard ethylene gas (von Dahl et al., 2007). Data were obtained from five replicated measurements.
Herbivore growth assay
The specialist herbivore M. sexta was obtained from a colony maintained in our laboratory. Each plant was infested with one neonate larva. Larval mass was recorded on days 6, 8, and 11.
Statistical analysis
Data were analysed by analysis of variance (ANOVA) or unpaired t test using StatView, version 5.0 (SAS, Cary, NC, USA).
Results
Cloning of NaBAK1 in N. attenuata
A 696 bp fragment of the NaBAK1 gene in N. attenuata was cloned from cDNA according to the N. benthamiana NbSERK3/BAK1 sequence (EST accession number: CK291393) (Heese et al., 2007). This partial NaBAK1 sequence showed 95% homology to NbSERK3/BAK1. Using this cloned NaBAK1 fragment as the bait, the NaBAK1 full-length open reading frame was identified from an N. attenuata transcriptome database prepared by 454 sequencing; this full-length sequence was further confirmed by cloning and sequencing (accession number: HM639279). Arabidopsis has five SERK gene family members, which are involved in different signalling pathways (Albrecht et al., 2008). Cloning and searching an N. attenuata transcriptome database prepared by 454 sequencing resulted in identification of another SERK gene in N. attenuata (accession number: HM639280), whose deduced protein sequence showed high similarity to SERK1 proteins (92%, 90%, and 87% similarity to Solanum tuberosum StSERK1, Medicago truncatula MtSERK1, and Arabidopsis AtSERK1 protein, respectively) (see Supplementary Fig. S1 at JXB online). Therefore, this protein is designated NaSERK1.
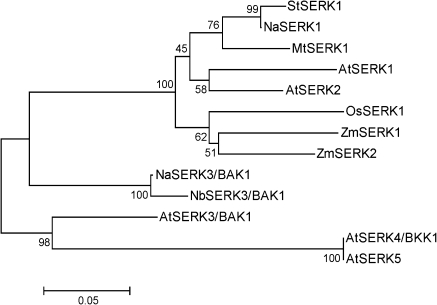
To examine these SERK protein sequences further, an unrooted phylogenetic tree was constructed using the deduced peptide and protein sequences (Fig. 1). Consistent with the sequence similarity data, NaBAK1 and NaSERK1 closely clustered with NbSERK3/BAK1 and StSERK1, respectively.
Fig. 1.
Phylogenetic tree of SERK proteins. Phylogenetic tree (unrooted) was constructed using protein sequences of several SERK genes from different plant species. NCBI (GenBank) accession numbers of SERK genes and SERK proteins included in this analysis are: N. attenuata: NaBAK1 (HM639279), NaSERK1 (HM639280); N. benthamiana: NbSERK3/BAK1 (CK291393); Arabidopsis thaliana: AtSERK1 (NP_177328), AtSERK2 (NP_174683), AtSERK3 (NP_567920), AtSERK4 (NP_178999), AtSERK5 (NP_179000); Solanum tuberosum: StSERK1 (ABO14172); Medicago truncatula: MtSERK1 (AAN64294); Oryza sativa: OsSERK1 (BAD86793); Zea mays: ZmSERK1 (CAC37638); ZmSERK2 (CAC37639).
Herbivory specifically induces transcript accumulation of NaBAK1 in N. attenuata
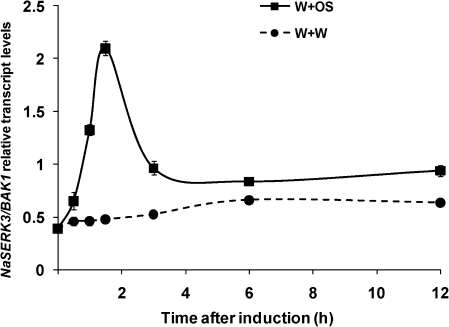
To examine whether herbivory induces the accumulation of NaBAK1 transcripts, N. attenuata rosette leaves were wounded with a pattern wheel and 20 μl of M. sexta OS was immediately applied to each leaf (W+OS), since this treatment effectively elicits herbivory-induced responses in N. attenuata (Halitschke et al., 2001). For a comparison, 20 μl of water was applied to wounds (W+W). Using gene-specific primers, quantitative real-time PCR (qPCR) was used to measure the transcript levels of NaBAK1 at different times (Fig. 2). NaBAK1 transcript levels quickly increased about 5 fold 1.5 h after W+OS treatment and gradually decreased to near basal levels after 6 h. By contrast, after W+W treatment, NaBAK1 transcript levels increased only marginally (Fig. 2). These data suggest that NaBAK1 is probably involved in N. attenuata's defence against herbivores.
Fig. 2.
Herbivory specifically induces the accumulation of NaBAK1 transcripts in N. attenuata. N. attenuata leaves were wounded with a pattern wheel and 20 μl of water (W+W) or M. sexta oral secretions (W+OS) were immediately applied to each leaf. Samples were harvested at the indicated times after treatments. The transcript levels (±SE) of NaBAK1 were analyzed by quantitative real-time PCR.
Silencing of NaBAK1 using VIGS
To study the function of NaBAK1 in N. attenuata's responses to herbivory, the transcript levels of the NaBAK1 gene were knocked down using a VIGS approach (Saedler and Baldwin, 2004). A 283 bp fragment of NaBAK1 cDNA was cloned into the pTV00 vector to form pTV-NaBAK1. Plants were inoculated with Agrobacterium carrying pTV-NaBAK1 to generate NaBAK1-silenced (hereafter, NaBAK1-VIGS) plants; plants inoculated with Agrobacterium carrying pTV00 empty vector were used for comparison (hereafter, EV plants).
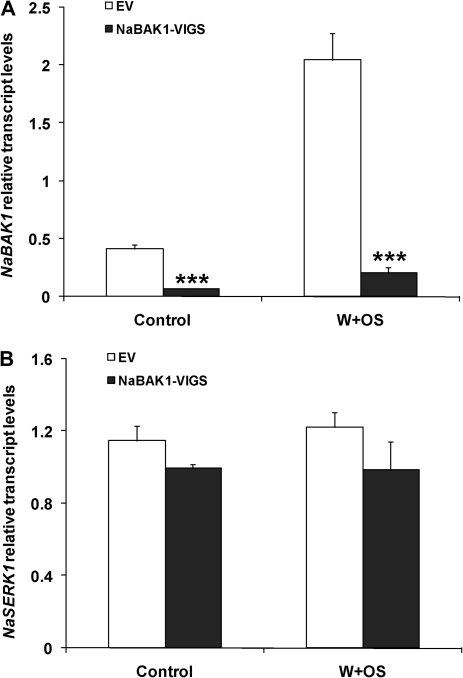
To confirm the efficiency and specificity of gene silencing, NaBAK1 and NaSERK1 transcript levels were quantified in EV and NaBAK1-VIGS plants using qPCR. Transcript levels of NaBAK1 in NaBAK1-VIGS plants were only 15% and 10% as high as those in EV plants before and 1 h after W+OS treatment, respectively (Fig. 3A). By contrast, no significant difference in the transcript levels of NaSERK1 was found between EV and NaBAK1-VIGS plants before and 1 h after W+OS treatment (unpaired t test; P=0.1299 for control and P=0.1808 for W+OS treatment) (Fig. 3B). These data suggest that the pTV-NaBAK1 construct efficiently and specifically silenced transcripts of NaBAK1 without co-silencing NaSERK1.
Fig. 3.
Silencing NaBAK1, but not NaSERK1 by virus-induced gene silencing. N. attenuata plants were infiltrated with Agrobacterium carrying pTV00 or pTV-NaBAK1 to generate EV and NaBAK1-VIGS plants, respectively. Mean (±SE) levels of NaBAK1 (A) and NaSERK1 (B) transcripts in EV and NaBAK1-VIGS plants were measured with qPCR in untreated (control) and 1 h W+OS-treated samples. Asterisks represent significantly different transcript levels between EV and NaBAK1-VIGS plants (unpaired t test; *P <0.05; **P <0.01; ***P <0.001; n=5).
Compared with those of EV plants, the leaves of NaBAK1-VIGS plants were somewhat darker green and moderately curly; in addition, by the late elongation stage, the NaBAK1-VIGS plants showed a semi-dwarf stature similar to that of Arabidopsis bak1 mutants (Li et al., 2002; Nam and Li, 2002) (see Supplementary Fig. S2 at JXB online). All experiments were performed on the leaves of plants at the early elongation stage, when EV and NaBAK1-VIGS plants showed no substantial morphological difference (see Supplementary Fig. S2 at JXB online).
Silencing NaBAK1 impairs the wounding- and herbivory-induced accumulation of JA and JA-Ile, but not the accumulation of SA and ethylene
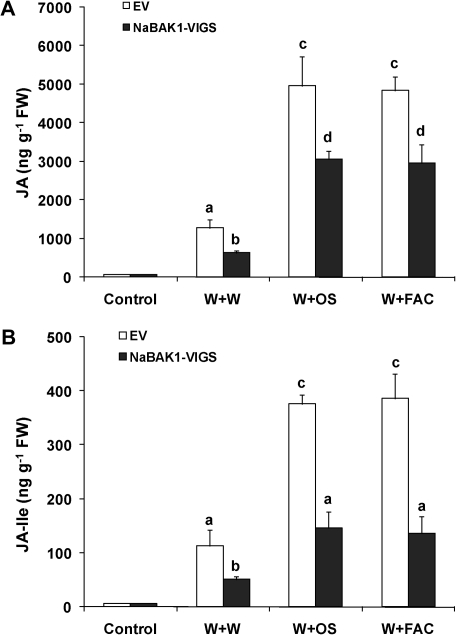
To examine whether NaBAK1 regulates the wounding- and herbivory-induced accumulation of JA and JA-Ile, EV and NaBAK1-VIGS plants were elicited with W+W and W+OS, and their JA and JA-Ile contents were measured after 1.5 h. W+W and W+OS elicited highly increased levels of JA in EV plants, and silencing NaBAK1 greatly diminished the accumulation of JA after these treatments (Fig. 4A). Similarly, 1.5 h after either treatment, the concentrations of JA-Ile in NaBAK1-VIGS plants were also highly reduced (Fig. 4B). Examining the JA and JA-Ile bursts in EV and NaBAK1-VIGS plants throughout the duration of the burst revealed similar results (see Supplementary Fig. S3 at JXB online). Notably, the differences of JA and JA-Ile levels in EV and NaBAK1-VIGS plants 1 h after W+W treatment were alleviated in the plants treated with W+OS (see Supplementary Fig. S3 at JXB online 3), suggesting that NaBAK1 has somewhat different roles in modulating wounding- and herbivory-induced JA accumulation.
Fig. 4.
NaBAK1 positively regulates wounding- and herbivory-induced accumulation of JA and JA-Ile. Leaves of EV and NaBAK1-VIGS plants were wounded with a pattern wheel and 20 μl of water (W+W), M. sexta oral secretions (W+OS), or FAC A solution (W+FAC) were immediately applied to each leaf. Individual leaves from five replicate plants were harvested after 1.5 h. Mean JA (A) and JA-Ile (B) concentrations (±SE) in EV and NaBAK1-VIGS plants were analysed with HPLC-MS/MS. Different letters indicate significant differences (two-way ANOVA, Fisher's PLSD test; P <0.05; n=5).
NaBAK1-silenced plants may have compromised perception of pathogens or pathogen-derived elicitors in M. sexta OS, and this could result in the decreased JA accumulation if these pathogens or pathogen-derived elicitors contribute to the elicitation of JA. To rule out this possibility, plants were wounded with a pattern wheel and a synthetic FAC, N-linolenoyl-L-Gln (18:3-Gln, one of the most abundant FACs in M. sexta OS) was applied (W+FAC), and the JA and JA-Ile levels were quantified 1.5 h after W+FAC treatment. Similar to W+OS treatment, W+FAC induced the same JA and JA-Ile levels in EV plants, confirming that FACs are required and sufficient to elicit herbivory-induced JA and JA-Ile accumulation; nevertheless, W+FAC treatment also elicited significantly reduced JA and JA-Ile levels in NaBAK1-VIGS plants compared with those in EV plants (Fig. 4A, B). These data argue against the possibility that the decreased herbivory-induced JA and JA-Ile levels resulted from compromised pathogen- (or pathogen-derived elicitor)-induced responses in NaBAK1-VIGS plants.
It is well known that SA antagonizes JA accumulation (Penacortes et al., 1993; Doares et al., 1995). Therefore, the SA content was measured in EV and NaBAK1-VIGS plants. No significant differences were found in the SA levels of EV and NaBAK1-VIGS plants after W+OS or W+FAC treatments, suggesting that the reduced herbivory-induced JA levels were not due to the antagonistic effect of SA levels in NaBAK1-VIGS plants (see Supplementary Fig. S4A at JXB online). Herbivory strongly increases ethylene biosynthesis and emissions, which is correlated with elevated defence levels in N. attenuata plants (von Dahl et al., 2007). The levels of W+OS-induced ethylene were determined in both EV and NaBAK1-VIGS plants (see Supplementary Fig. S4B at JXB online), which, unlike the levels of JA and JA-Ile, were not affected by NaBAK1 silencing. Therefore, NaBAK1 is not involved in regulating herbivory-induced ethylene production in N. attenuata.
To explore further whether the attenuated wounding- and herbivory-induced JA accumulation in NaBAK1-VIGS plants was associated with altered transcript levels of genes in the JA biosynthetic pathway, the transcript abundance of NaLOX3, NaAOS, NaAOC, NaOPR3, and NaACX1, was measured by qPCR. Comparing EV with NaBAK1-VIGS plants within 3 h, no large differences in transcript levels for any of these genes were found, except that minor differences were detected in NaLOX3 (3 h after W+OS), NaAOS (1 h after W+W), NaOPR3 (1 h after W+W), and NaACX1 (1 h after W+OS and 3 h after W+W) (see Supplementary Fig. S5 at JXB online). Therefore, the large differences of wounding- and herbivory-induced JA levels between EV and NaBAK1-VIGS plants probably resulted from post-transcriptional modifications of certain genes/enzymes involved in JA biosynthesis, for example, protein abundance or enzyme activity, rather than from changes in transcript abundance. Furthermore, these data suggest that NaBAK1 modulates the transcript levels of JA biosynthetic genes in a treatment- and gene-specific manner.
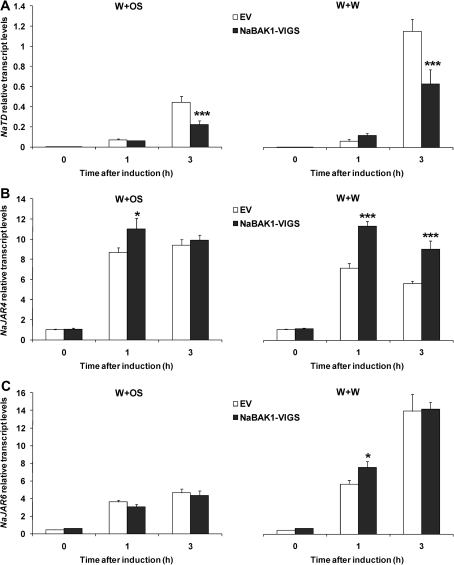
In N. attenuata, threonine deaminase (NaTD) plays an important role in converting Thr to Ile to supply the Ile used by NaJAR4 and NaJAR6 to conjugate Ile with JA to form JA-Ile (Kang et al., 2006; Wang et al., 2007, 2008). The transcript abundance of these JA-Ile biosynthetic genes was quantified and it was found that transcript levels of NaTD were significantly reduced in NaBAK1-VIGS plants compared with EV plants only 3 h after treatments with both W+W and W+OS (Fig. 5A). Conversely, NaJAR4 and NaJAR6 transcript levels were somewhat higher in NaBAK1-VIGS plants than in EV plants (Fig. 5B, C). This suggests that the reduced levels of JA-Ile in NaBAK1-VIGS plants are not likely due to decreased NaJAR4 and NaJAR6 activity.
Fig. 5.
Transcript accumulation of NaTD, NaJAR4, and NaJAR6 in wounding- and herbivory-induced EV and NaBAK1-VIGS plants. Leaves of EV and NaBAK1-VIGS plants were wounded with a pattern wheel and 20 μl of water (W+W) or M. sexta oral secretions (W+OS) were immediately applied to each leaf. Leaves from replicate plants were harvested at the indicated times. Mean transcript levels (±SE) of N. attenuata NaTD (A), NaJAR4 (B), and NaJAR6 (C) were measured with quantitative real-time PCR. Asterisks represent significantly different levels between EV and NaBAK1-VIGS plants at the indicated times (unpaired t test; *P <0.05; **P <0.01; ***P <0.001; n=5).
Together, these results indicate that NaBAK1 is required for the wounding- and herbivory-induced accumulation of JA and JA-Ile, and this regulation does not occur on a transcriptional level.
JA levels, rather than JAR activity, limit the accumulation of JA-Ile in NaBAK1-VIGS plants
In order to investigate whether impaired JAR activity and/or the limited availability of JA-Ile biosynthesis substrates (Ile and/or JA) account for the reduced JA-Ile levels in NaBAK1-VIGS plants, the substrates for JA-Ile biosynthesis were supplied to the wound site of EV and NaBAK1-VIGS plants during W+W and W+OS treatments and the resulting JA-Ile levels were quantified (Kang et al., 2006; Paschold et al., 2008).
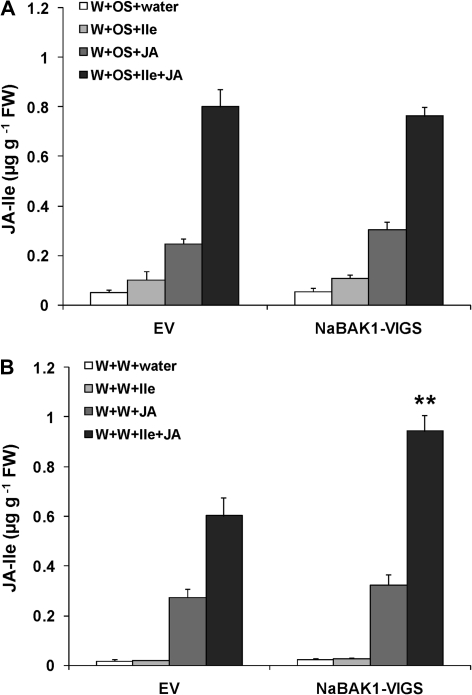
The water and OS used for W+W and W+OS treatments were supplemented with either 0.625 μmol of Ile (W+W+Ile and W+OS+Ile), JA (W+W+JA and W+OS+JA), or both (W+W+Ile+JA and W+OS+Ile+JA). Compared with W+OS, treating EV plants with W+OS+Ile only slightly affected the levels of JA-Ile, whereas treating EV with W+OS+JA increased JA-Ile levels 3.8-fold (Fig. 6A). This suggests that JA but not Ile is the limiting factor for JA-Ile biosynthesis. Dramatically increased JA-Ile levels (15-fold) were detected when both Ile and JA were supplied (W+OS+Ile+JA); therefore only when an excess amount of JA is applied (higher than the endogenous levels of JA), Ile becomes the limiting substrate (Fig. 6A). Similar results were obtained from EV plants that were treated with W+W and supplied with these substrates (Fig. 6B). Importantly, NaBAK1-VIGS plants had similar or somewhat elevated JA-Ile levels compared with EV plants after being supplied with Ile and JA (Fig. 6A, B). Apparently, when substrates are sufficiently available, NaBAK1-VIGS and EV plants have similar JAR activity to synthesize W+W- and W+OS-induced JA-Ile.
Fig. 6.
Ile and JA, rather than JAR activity, limit the accumulation of JA-Ile in EV and NaBAK1-VIGS plants. Leaves of EV and NaBAK1-VIGS plants were wounded with a pattern wheel and treated with 20 μl of M. sexta oral secretions (OS) or water (W+OS and W+W, respectively) or an excess (0.625 nmol) of Ile or JA (W+OS+Ile, W+OS+JA, W+W+Ile, and W+W+JA) or both (W+OS+Ile+JA and W+W+Ile+JA). Mean JA-Ile levels (±SE) were analysed 2 h after plants were OS elicited and supplied with an excess of JA-Ile biosynthesis substrates (A) and wounded and supplied with an excess of JA-Ile biosynthesis substrates (B). Asterisks represent significantly different levels between EV and NaBAK1-VIGS plants after indicated treatments (unpaired t test; *P <0.05; **P <0.01; ***P <0.001; n=5)
It is concluded that substrate availability, most likely the decreased JA levels in NaBAK1-VIGS plants, rather than the enzymatic activity of JAR proteins, are responsible for the reduced JA-Ile levels observed in these plants.
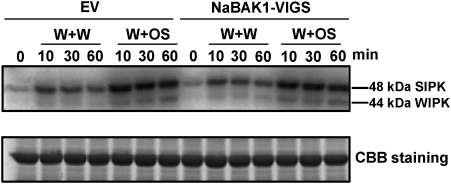
Silencing NaBAK1 does not impair W+W- and W+OS-induced MAPK activity
Compared with wild-type plants, treating Arabidopsis bak1 mutants with the pathogen elicitor, flg22, leads to partially compromised MAPK activation (Chinchilla et al., 2007; Heese et al., 2007). To test whether decreased JA levels after W+OS treatment were due to attenuated MAPK activity, MAPK activity was determined in NaBAK1-VIGS and EV plants with an in-gel kinase assay. Both W+W and W+OS treatment induced similar levels of SIPK and WIPK activity in EV and NaBAK1-VIGS plants (Fig. 7). Therefore, it is concluded that NaBAK1 mediates wounding- and herbivory-elicited JA and JA-Ile accumulation downstream of SIPK and WIPK or through a SIPK- and WIPK-independent pathway in N. attenuata.
Fig. 7.
Silencing NaBAK1 does not impair wounding- and herbivory-induced MAPK activity in N. attenuata leaves. Leaves of EV and NaBAK1-VIGS plants were treated with W+W or W+OS and harvested at the indicated times. MAPK activity was analysed by an in-gel kinase assay using myelin basic protein as the substrate (upper panel). Same amount of each protein sample was run on another PAGE gel and stained with Coomassie Brilliant Blue (CBB) to confirm equal loading (bottom panel).
NaBAK1-silenced plants have increased levels of basal and MeJA-induced TPI activity
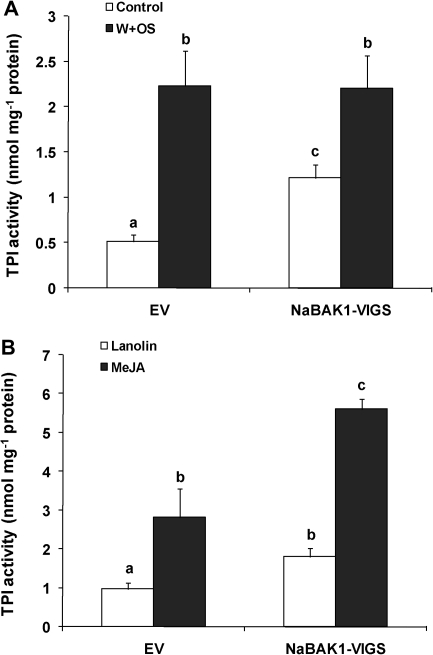
Next it was examined whether, after herbivory, the impaired JA and JA-Ile accumulation in NaBAK1-VIGS plants led to reduced levels of defensive compounds against herbivores. Under greenhouse conditions, herbivore attack or W+OS treatment increases the contents of TPIs and DTGs in N. attenuata (Zavala et al., 2004; Paschold et al., 2007). However, under the growth condition optimized for VIGS, neither EV nor NaBAK1-VIGS plants showed enhanced levels of DTGs after W+OS treatment; only TPIs were induced after this treatment (Fig. 8A; see Supplementary Fig. S6 at JXB online). We speculate that this was due to the relatively low temperature and light intensity which was needed for the VIGS. Therefore, our analyses was focused on TPIs.
Fig. 8.
NaBAK1-silenced plants have increased levels of basal and MeJA-induced TPI activity. Leaves from EV and NaBAK1-VIGS plants were left untreated (control) or wounded with a pattern wheel and 20 μl of M. sexta oral secretions (OS) were immediately applied to each leaf (W+OS). Samples were harvested 3 d after treatment. TPI activity (±SE) (A) was determined. Each EV and NaBAK1-VIGS plants was treated with 20 μl of lanolin paste or lanolin paste containing MeJA (7.5 μg μl−1). After 3 d, samples were harvested, and TPI activity (±SE) (B) was analysed. Error bars represent standard errors; different letters indicate significant differences (two-way ANOVA, Fisher's PLSD test; P <0.05; n=5).
When untreated, EV and NaBAK1-VIGS had similar levels of JA and JA-Ile (Fig. 4); however, compared with the basal TPI activity in EV plants, NaBAK1-VIGS plants exhibited around 100% higher levels of TPI activity; moreover, after W+OS treatment, although NaBAK1-VIGS plants had lower JA and JA-Ile than did EV plants, similar levels of TPI activity were detected in EV and NaBAK1-VIGS plants (Fig. 8A). These data suggest that silencing NaBAK1 enhances JA-induced TPI activity. To test this hypothesis, 150 μg of MeJA dissolved in 20 μl of lanolin paste was applied to one leaf of each EV and NaBAK1-VIGS plant (the same amount of lanolin was used for controls). In agreement with our hypothesis, after 3 d, MeJA induced higher levels of TPIs activity in NaBAK1-VIGS plants than in EV plants (Fig. 8B). Moreover, similar MeJA-induced DTG levels were seen in EV and NaBAK1-VIGS plants (see Supplementary Fig. S6 at JXB online).
Bioassays were performed to determine whether silencing NaBAK1 altered the performance of the specialist insect herbivore M. sexta on N. attenuata plants. Consistent with the equal levels of W+OS-induced defensive secondary metabolites in EV and NaBAK1-VIGS plants, M. sexta larvae gained similar masses on these plants (see Supplementary Fig. S7 at JXB online).
Discussion
In both N. benthamiana and Arabidopsis, SERK3/BAK1 is required for maintaining plant innate immunity to pathogens (Chinchilla et al., 2007; Heese et al., 2007). A reverse genetic approach was used here to examine the functions of SERK3/BAK1 in plants' resistance to herbivores. Our data demonstrate that SERK3/BAK1 is involved in regulating N. attenuata's defence responses, such as the accumulation of JA/JA-Ile and the induction of TPI activity.
SERK3/BAK1 in SERK family
In Arabidopsis, AtSERK3/BAK1 belongs to a small gene family consisting of five members. Particular combinations of SERKs, such as AtSERK1 and AtSERK2, AtSERK1 and AtSERK3/BAK1, and AtSERK3/BAK1 and AtSERK4/BKK1, play important roles in BR-dependent and BR-independent signalling pathways, suggesting that closely related SERK members provide functional specificity (Albrecht et al., 2008). AtSERK3/BAK1 has multiple functions in BR signalling (Li et al., 2002; Nam and Li, 2002), innate immunity (Chinchilla et al., 2007; Heese et al., 2007), and cell death control (He et al., 2007; Kemmerling et al., 2007).
Two SERK genes in N. attenuata were cloned. Using a VIGS system, NaBAK1 (NaSERK3) was specifically silenced without co-silencing its homologue, NaSERK1. Our cloning and searching of a transcriptome database prepared by 454 sequencing did not result in finding other SERK genes. We suspect that either there are no other SERK genes in N. attenuata or they have extremely low transcript abundance. Whether other SERK genes exist in N. attenuata, and whether the pTV-NaBAK1 construct attenuates transcripts of these SERKs remains unclear.
Function of NaBAK1 in herbivory-induced early responses in plants
In Arabidopsis, the binding of bacterial flagellin to the FLS2 receptor activates MAPK signalling (Asai et al., 2002). SERK3/BAK1 as a component of plants' innate immunity has recently been identified: SERK3/BAK1 binds to FLS2 and is implicated in the activation of two MAPKs, MPK6 and MPK3, homologues of SIPK and WIPK in Nicotiana, respectively (Chinchilla et al., 2007; Heese et al., 2007). Plants may possess receptors that perceive herbivory by binding with herbivore-derived cues, such as FACs (Truitt et al., 2004); this hypothesis is further supported by the finding that FACs in herbivore OS rapidly activate MAPKs (Wu et al., 2007). Unlike Arabidopsis bak1 mutants, which have decreased flagellin-induced MAPK activity, NaBAK1-silenced plants have similar SIPK and WIPK activity as EV plants after W+OS treatment (Fig. 7). This suggests that NaBAK1 does not interact with the yet-to-be-identified FAC receptor. Furthermore, silencing NaBAK1 also decreases wounding-induced JA, supporting the notion that NaBAK1 does not function at the level of perception of FACs.
Herbivory rapidly elevates MAPK activity, which is positively associated with JA biosynthesis in N. attenuata (Wu et al., 2007). Although EV and NaBAK1-VIGS plants have similar levels of MAPK activity, NaBAK1-silenced plants have low levels of elicited JA, indicating that NaBAK1 modulates wounding- and herbivory-induced JA levels either downstream of MAPK signalling or in a MAPK-independent pathway. Our transcriptional analyses demonstrate that NaBAK1 is not associated with the transcriptional regulation of JA biosynthetic genes; therefore, it is likely that NaBAK1 is involved in positively regulating the activity of certain JA-biosynthetic enzyme(s). In Arabidopsis, wounding induces the production of AtPep1 (a 23-amino acid peptide) from its precursor protein, which is perceived by a LRR-RLK, PEPR1, and thereafter activates expression of PDF1.2 in a jasmonate-dependent manner (Huffaker et al., 2006; Yamaguchi et al., 2006). Importantly, application of AtPep1 to Arabidopsis cell suspension culture enhances the phosphorylation levels of BAK1 and induces the formation of heterocomplexes that consist of BAK1 and very likely PEPR1 (Schulze et al., 2010). Whether Pep1 and its receptor, PEPR1, in N. attenuata are also involved in wounding- and herbivory-induced JA accumulation and whether the binding of NaBAK1 to PEPR1 is required for Pep1-induced JA production deserve further study.
BR and JA signalling
JA signalling plays crucial roles in plant–herbivore interactions (Halitschke and Baldwin, 2003; Paschold et al., 2007; Wang et al., 2008). In untreated plants, which have similar levels of JA, NaBAK1-VIGS plants exhibit higher TPI activity. Although NaBAK1-VIGS plants have highly decreased JA levels after herbivory, the accumulation of herbivory-induced TPIs is not affected.
Analysis of MeJA-induced TPI levels confirmed that NaBAK1 negatively modulates levels of jasmonate-induced TPIs in N. attenuata. Conversely, the contents of DTGs are similar when EV and NaBAK1-VIGS are treated with MeJA. This suggests that the different MeJA-induced TPI levels in EV and NaBAK1-silenced plants are not due to differences in transport or metabolism of the applied MeJA, namely that MeJA diffuse more quickly into the leaves of NaBAK1-VIGS plants and MeJA is more efficiently hydrolysed to JA (and subsequently converted to JA-Ile) in NaBAK1-VIGS plants, both of which would lead to elevated levels of MeJA-induced TPIs in NaBAK1-VIGS plants. Moreover, the distinct patterns of TPI and DTG accumulation also indicated the specificity of NaBAK1 in regulating different JA-induced defences.
Recently, Campos et al. (2009) showed that the tomato BR biosynthesis mutant dpy has augmented levels of the anti-herbivore metabolite zingiberene, as well as elevated transcripts of the serine proteinase inhibitor PI-I, suggesting that BR negatively interacts with JA in the formation of anti-herbivory traits in tomato. We hypothesized that BR signalling might also negatively regulate certain JA responses in N. attenuata, such as TPI accumulation, under herbivore attack. Consistent with this hypothesis, the psc1 mutants have a leaky mutation of the DWARF4 gene that encodes a key enzyme in BR biosynthesis, and these mutants partially suppressed coi1 mutants' insensitivity to JA inhibition of root growth in Arabidopsis seedlings (Ren et al., 2009).
However, the possibility cannot be excluded that NaBAK1 regulates JA-induced TPI accumulation in a BR signalling-independent manner. In Arabidopsis SERK3/BAK1 mediates pathogen-induced programmed cell death independently of BR signalling (He et al., 2007; Kemmerling et al., 2007). Thus, it would be interesting to explore the dependence of SERK3/BAK1 on BR signalling in the regulation of herbivory-induced TPI responses using plants silenced in BR biosynthetic genes or BRI1, the BR receptor gene. Furthermore, these plants can also be used to investigate whether BR and its signalling play any roles in wounding- and herbivory-induced JA accumulation.
In summary, it is shown that NaBAK1 is a member of the herbivory-induced response network and plays multiple roles in plant–herbivore interactions: NaBAK1 is required for herbivory-induced JA and JA-Ile accumulation and NaBAK1 also plays a negative role in controlling TPI activity induced by herbivory or jasmonates. Further study should address the molecular mechanism by which NaBAK1 regulates herbivory-induced JA and TPI accumulation and whether other SERKs are also involved in plant-herbivore interactions.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Alignment of SERK proteins.
Supplementary Fig. S2. Morphology of EV and NaBAK1-VIGS plants.
Supplementary Fig. S3. NaBAK1 positively regulates W+OS- and W+W-induced accumulation of jasmonic acid (JA) and JA-isoleucine (JA-Ile).
Supplementary Fig. S4. EV and NaBAK1-VIGS plants have the same levels of SA and ethylene production after W+OS induction.
Supplementary Fig. S5. Transcript accumulation of JA biosynthetic genes in EV and NaBAK1-VIGS plants.
Supplementary Fig. S6. EV and NaBAK1-VIGS plants have similar levels of DTGs in response to W+OS and MeJA treatment.
Supplementary Fig. S7. M. sexta has similar growth rates on EV and NaBAK1-VIGS plants.
Supplementary Table S1. Primers used for cloning of NaBAK1 and NaSERK1, and preparation of pTV-NaBAK1 construct.
Supplementary Table S2. Primer sequences used for quantitative real-time PCR (SYBR Green analysis).
Supplementary Material
Acknowledgments
We thank Emily Wheeler for editorial assistance, Stefan Meldau, Meredith Schuman, and Hendrik Wünsche for discussion, Daniel Lindsay and Yvonn Stampnik for technical assistance, and the Max Planck Society for funding.
Glossary
Abbreviations
- ABA
abscisic acid
- BR
brassinosteroid
- BRI1
brassinosteroid insensitive 1
- BAK1
BRI1-associated receptor kinase 1
- DTG
diterpene glycoside
- JA
jasmonic acid
- JA-Ile
jasmonic acid-isoleucine
- MAPK
mitogen-activated protein kinase
- OS
oral secretions
- SA
salicylic acid
- SERK
somatic embryogenesis receptor kinase
- TPI
trypsin proteinase inhibitor
References
- Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signalling pathways. Plant Physiology. 2008;148:611–619. doi: 10.1104/pp.108.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Chory J. Brassinosteroid signalling: a paradigm for steroid hormone signalling from the cell surface. Science. 2006;314:1410–1411. doi: 10.1126/science.1134040. [DOI] [PubMed] [Google Scholar]
- Browse J. Jasmonate passes muster: a receptor and targets for the defence hormone. Annual Review of Plant Biology. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- Campos ML, de Almeida M, Rossi ML, Martinelli AP, Litholdo Junior CG, Figueira A, Rampelotti-Ferreira FT, Vendramim JD, Benedito VA, Peres LE. Brassinosteroids interact negatively with jasmonates in the formation of anti-herbivory traits in tomato. Journal of Experimental Botany. 2009;60:4347–4361. doi: 10.1093/jxb/erp270. [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Chow B, McCourt P. Plant hormone receptors: perception is everything. Genes and Development. 2006;20:1998–2008. doi: 10.1101/gad.1432806. [DOI] [PubMed] [Google Scholar]
- Doares SH, Narvaez-Vasquez J, Conconi A, Ryan CA. Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiology. 1995;108:1741–1746. doi: 10.1104/pp.108.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Wang ZY. Multiple mechanisms modulate brassinosteroid signalling. Current Opinion in Plant Biology. 2007;10:436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Jones JD. Hormone (dis)harmony moulds plant health and disease. Science. 2009;324:750–752. doi: 10.1126/science.1173771. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT. Antisense LOX expression increases herbivore performance by decreasing defence responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. The Plant Journal. 2003;36:794–807. doi: 10.1046/j.1365-313x.2003.01921.x. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiology. 2001;125:711–717. doi: 10.1104/pp.125.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ. Plant hormones: the interplay of brassinosteroids and auxin. Current Biology. 2004;14:R1008–R1010. doi: 10.1016/j.cub.2004.11.025. [DOI] [PubMed] [Google Scholar]
- He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Current Biology. 2007;17:1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proceedings of the National Academy of Sciences, USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiling S, Schuman MC, Schoettner M, Mukerjee P, Berger B, Schneider B, Jassbi AR, Baldwin IT. Jasmonate and ppHsystemin regulate key malonylation steps in the biosynthesis of 17-hydroxygeranyllinalool diterpene glycosides, an abundant and effective direct defence against herbivores in Nicotiana attenuata. The Plant Cell. 2010;22:273–292. doi: 10.1105/tpc.109.071449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G. Plant immunity to insect herbivores. Annual Review of Plant Biology. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proceedings of the National Academy of Sciences, USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassbi AR, Gase K, Hettenhausen C, Schmidt A, Baldwin IT. Silencing geranylgeranyl diphosphate synthase in Nicotiana attenuata dramatically impairs resistance to tobacco hornworm. Plant Physiology. 2008;146:974–986. doi: 10.1104/pp.107.108811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT. Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defences against Manduca sexta. The Plant Cell. 2006;18:3303–3320. doi: 10.1105/tpc.106.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, de Vries SC. Advances in understanding brassinosteroid signalling. Science STKE. 2006;2006 doi: 10.1126/stke.3542006pe36. pe36. [DOI] [PubMed] [Google Scholar]
- Keinanen M, Oldham NJ, Baldwin IT. Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. Journal of Agricultural and Food Chemistry. 2001;49:3553–3558. doi: 10.1021/jf010200+. [DOI] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriguez P, et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Current Biology. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Cano-Delgado AC, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology. 2002;12:177–183. [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signalling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Li JM, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Liechti R, Farmer EE. The jasmonate pathway. Science. 2002;296:1649–1650. doi: 10.1126/science.1071547. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signalling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Paschold A, Bonaventure G, Kant MR, Baldwin IT. Jasmonate perception regulates jasmonate biosynthesis and JA-Ile metabolism: the case of COI1 in Nicotiana attenuata. Plant and Cell Physiology. 2008;49:1165–1175. doi: 10.1093/pcp/pcn091. [DOI] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT. Co(i)-ordinating defences: NaCOI1 mediates herbivore- induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defences. The Plant Journal. 2007;51:79–91. doi: 10.1111/j.1365-313X.2007.03119.x. [DOI] [PubMed] [Google Scholar]
- Penacortes H, Albrecht T, Prat S, Weiler EW, Willmitzer L. Aspirin prevents wound-induced gene-expression in tomato leaves by blocking jasmonic acid biosyntheis. Planta. 1993;191:123–128. [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC. Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. The Plant Journal. 2001;25:237–245. doi: 10.1046/j.0960-7412.2000.00942.x. [DOI] [PubMed] [Google Scholar]
- Ren CM, Han CY, Peng W, Huang Y, Peng ZH, Xiong XY, Zhu Q, Gao BD, Xie DX. A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis. Plant Physiology. 2009;151:1412–1420. doi: 10.1104/pp.109.140202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT. Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. Journal of Experimental Botany. 2004;55:151–157. doi: 10.1093/jxb/erh004. [DOI] [PubMed] [Google Scholar]
- Santner A, Calderon-Villalobos LI, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nature Chemical Biology. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. Journal of Biological Chemistry. 2010;285:9444–9451. doi: 10.1074/jbc.M109.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. The Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppuhn A, Gase K, Krock B, Halitschke R, Baldwin IT. Nicotine's defensive function in nature. PLoS Biology. 2004;2:E217. doi: 10.1371/journal.pbio.0020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Truitt CL, Wei HX, Pare PW. A plasma membrane protein from Zea mays binds with the herbivore elicitor volicitin. The Plant Cell. 2004;16:523–532. doi: 10.1105/tpc.017723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam NM, Horn M, Mares M, Baldwin IT. Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. Journal of Chemical Ecology. 2001;27:547–568. doi: 10.1023/a:1010341022761. [DOI] [PubMed] [Google Scholar]
- von Dahl CC, Winz RA, Halitschke R, Kuhnemann F, Gase K, Baldwin IT. Tuning the herbivore-induced ethylene burst: the role of transcript accumulation and ethylene perception in Nicotiana attenuata. The Plant Journal. 2007;51:293–307. doi: 10.1111/j.1365-313X.2007.03142.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Allmann S, Wu J, Baldwin IT. Nicotiana attenuataComparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiology. 2008;146:904–915. doi: 10.1104/pp.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Halitschke R, Kang JH, Berg A, Harnisch F, Baldwin IT. Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta. 2007;226:159–167. doi: 10.1007/s00425-007-0477-3. [DOI] [PubMed] [Google Scholar]
- Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Annals of Botany. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. Herbivory-induced signalling in plants: perception and action. Plant, Cell and Environment. 2009;32:1161–1174. doi: 10.1111/j.1365-3040.2009.01943.x. [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT. Herbivory rapidly activates MAPK signalling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. The Plant Cell. 2007;19:1096–1122. doi: 10.1105/tpc.106.049353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defence and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proceedings of the National Academy of Sciences, USA. 2006;103:10104–10109. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Hui D, Baldwin IT. Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defences. Plant Physiology. 2004;134:1181–1190. doi: 10.1104/pp.103.035634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF. Salicylic acid activates a 48 kDa MAP kinase in tobacco. The Plant Cell. 1997;9:809–824. doi: 10.1105/tpc.9.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.