Abstract
Human p14 (SF3b14), a component of the spliceosomal U2 snRNP, interacts directly with the pre-mRNA branch adenosine within the context of the bulged duplex formed between the pre-mRNA branch region and U2 snRNA. This association occurs early in spliceosome assembly and persists within the fully assembled spliceosome. Analysis of the crystal structure of a complex containing p14 and a peptide derived from p14-associated SF3b155 combined with the results of cross-linking studies has suggested that the branch nucleotide interacts with a pocket on a non-canonical RNA binding surface formed by the complex. Here we report a structural model of the p14•bulged duplex interaction based on a combination of X-ray crystallography of an adenine p14/SF3b155 peptide complex, biochemical comparison of a panel of disulfide cross-linked protein–RNA complexes, and small-angle X-ray scattering (SAXS). These studies reveal specific recognition of the branch adenosine within the p14 pocket and establish the orientation of the bulged duplex RNA bound on the protein surface. The intimate association of one surface of the bulged duplex with the p14/SF3b155 peptide complex described by this model buries the branch nucleotide at the interface and suggests that p14•duplex interaction must be disrupted before the first step of splicing.
Keywords: protein–RNA interaction, p14, branch duplex, splicing
INTRODUCTION
Pre-mRNA splicing by the human spliceosome requires the sequential recognition by RNA and protein factors of the 5′ and 3′ splice sites, the polypyrimidine tract, and the branch region (Burge et al. 1999). The dynamic nature of spliceosome assembly with respect to these recognition events has been established by a wealth of studies describing a complex series of RNA•RNA and RNA•protein associations during the course of the assembly pathway (Staley and Guthrie 1998). This sequence is characterized by the stepwise association of the U1, U2, and U4/U5/U6 snRNP particles as well as non-snRNP factors with the pre-mRNA substrate through the formation of E, A, B complexes into the mature spliceosomal C complex (Brow 2002).
One of the key steps in spliceosome assembly is recognition of the branch region. In contrast to the yeast branch sequence, 5′-UACUAAC-3′ which is very highly conserved, the human branch region sequence is variable (Burge et al. 1999; Gao et al. 2008); branch sequence plasticity in humans is important in splicing regulation, and mutations are associated with a variety of disease states resulting from aberrant splicing (for review, see Hartmann and Valcárcel 2009). In humans, initial recognition of the single-stranded branch sequence within the pre-mRNA is mediated by the branch-binding protein SF1, which is part of a network of factors that define the E or commitment complex (Reed 1990). The stable association of U2 snRNP with the pre-mRNA in A complex includes the formation of a bulged duplex structure between the pre-mRNA branch sequence and U2 snRNA that specifies the bulged nucleotide as the nucleophile for the first transesterification step of splicing (Query et al. 1994).
Sequential association of proteins with the pre-mRNA branch region has been demonstrated by site-specific modification of the branch adenosine with the photo-reagent benzophenone followed by cross-linking and analysis by SDS-PAGE (MacMillan et al. 1994). This study detected the early association of SF1 with the branch region but also revealed the specific, stepwise association of several other factors with the branch nucleotide from very early in spliceosome assembly into the mature spliceosome. Of particular interest was a strong cross-link to p14 that appeared in A complex and persisted within the fully assembled spliceosome. The kinetics of the appearance of this cross-link suggest that p14 is associated with the bulged branch region•U2 snRNA duplex (Fig. 1A; Query et al. 1994). The benzophenone reagent probed pre-mRNA–protein interactions within a radius of 15 Å; subsequent experiments showed that p14 directly contacted the branch nucleotide in A through C complexes indicating an intimate association between protein and RNA at the heart of the mammalian spliceosome (Query et al. 1996).
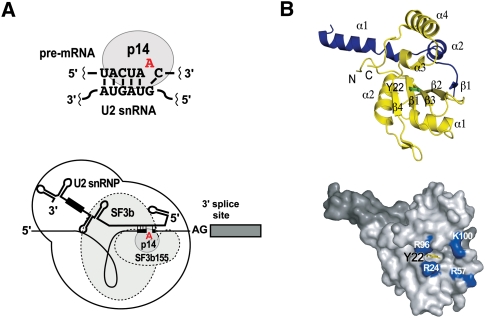
FIGURE 1.
Protein–protein and protein–RNA interactions of p14 in the human spliceosome. (A, upper) p14 directly contacts the bulged duplex formed between the pre-mRNA branch sequence and U2 snRNA; (lower) p14 as a constituent of the U2 snRNP-associated SF3b particle. (B, upper) Ribbon diagram showing the p14•SF3b155 peptide complex based on the X-ray structure (Schellenberg et al. 2006). (Yellow) p14; (blue) the SF3b155 peptide; (green) Y22. (Lower) Surface representation of the p14•SF3b155 peptide complex showing Y22 exposed within the surface pocket surrounded by conserved basic residues; (light gray) p14; (dark gray) SF3b155 peptide; (yellow) Y22; and (blue) R24, R57, R96, and K100.
p14 was initially isolated from purified mammalian spliceosomes and subsequently identified as a constituent of both the U2 and U12 snRNPs of the major and minor spliceosomes, respectively (Will et al. 2001). More specifically, p14 is part of SF3b, a salt-dissociable multi-protein complex conserved between the U2 and U12 snRNPs, and a strong interaction between p14 and the SF3b protein SF3b155 has been demonstrated (Fig. 1A). p14 is an evolutionarily highly conserved protein with orthologs across diverse species; the human protein is a 125-amino-acid polypeptide containing a central region with strong homology with the well-characterized RRM domain (Will et al. 2001). In particular, p14 contains consensus RNP1 and RNP2 motifs that mediate RRM•RNA interactions as evidenced by high-resolution structural analyses (Oubridge et al. 1994; Deo et al. 1999; Handa et al. 1999; Allain et al. 2000; Wang and Hall 2001; Clery et al. 2008).
As an initial step toward understanding the molecular basis of p14•branch duplex interaction, we solved the X-ray structure of p14 in a complex with a short peptide derived from SF3b155 (Fig. 1B; Schellenberg et al. 2006). As predicted, p14 contains a central RRM domain (residues 20 to 91). As well, the C terminus of the protein features two additional α-helices of four and 12 amino acids. The bound SF3b155 peptide forms a long N-terminal α-helix (amino acids 380–396) and a second shorter helix (amino acids 401–407). The C terminus of the SF3b155 fragment contains a short β-strand interacting with β-2 of the p14 RRM β-sheet; this is connected to the shorter α-helix by a loop that makes extensive contacts with both the shorter C-terminal helix and RRM of p14 (Schellenberg et al. 2006).
An intriguing feature of the p14•peptide complex, as revealed by the X-ray structure, is that the canonical β-sheet of the RRM is occluded by one of the C-terminal helices of p14, the central helix of the SF3b155 peptide, and the loop connecting the peptide to the C-terminal β-strand, resulting in a large buried surface. The interface between the two proteins is extensive including a hydrophobic core surrounded by a set of hydrogen bonds and salt bridges.
The blocking of one face of p14 is significant because the β-sheet of the RRM represents the canonical RNA binding surface of the domain including the highly conserved RNP1 and RNP2 motifs (Oubridge et al. 1994; Deo et al. 1999; Handa et al. 1999; Allain et al. 2000; Wang and Hall 2001; Clery et al. 2008). In the p14/SF3b155 peptide complex, residues of RNP1 and RNP2 are largely buried. Intriguingly, a portion of RNP2 is exposed within a pocket on the otherwise occluded surface: A highly conserved aromatic residue within RNP2, Y22 forms the base of this pocket on the p14•peptide surface (Fig. 1B). The surface surrounding this pocket includes four basic residues—R24, R57, R96, and K100. The side chains of two of these (R24, R57) project from the surface of the p14 β-sheet, the third (R96) is found at the end of α3, and the fourth (K100) in the loop between α3 and α4. The identities of R96 and K100 are highly conserved among p14 orthologs but not between p14 and other RRMs (Schellenberg et al. 2006).
In biochemical studies, we were able to characterize a cross-link between the bulged adenosine of a model branch region duplex and the mutant Y22M p14/SF3b155 peptide complex where the introduced methionine forms the floor of the pocket on the p14 surface. Mapping the RNA cross-link to M22 in the mutant allowed us to infer that the bulged nucleotide is bound within the p14 pocket and that the surrounding surface represents the RNA-binding face of p14 (Schellenberg et al. 2006).
In order to further characterize the interaction of p14 with the pre-mRNA branch duplex, we have used a disulfide tethering approach to compare the stabilities of different RNA•protein complexes. The results of these studies, combined with X-ray crystallographic and solution-scattering studies, allow us to propose a model of the branch duplex•p14 interaction with significant implications with respect to the role of p14 in the assembled spliceosome.
RESULTS
Interrogating the p14•RNA complex using a disulfide tethering strategy
The affinity of the isolated p14/SF3b155 peptide complex for short model RNA duplexes is not strong (>100 μM); neither is there a marked specificity for a specific bulged duplex over double-stranded or single-stranded RNA (Spadaccini et al. 2006; MJ Schellenberg and AM MacMillan, unpubl.). These observations are not surprising and may reflect several different features of branch duplex recognition within the spliceosome. These include the cooperative nature of RNA–protein interactions—SF3b155 has been shown to directly interact with nucleotides at the −6 position, just 5′ to the branch region U2 duplex, and at the +5 position, 3′ to the branch (Gozani et al. 1998)—as well as the fact that p14 likely interacts with multiple bulged duplex structures due to the variable nature of the branch sequence in humans (Gao et al. 2008).
To address problems of affinity/specificity and to facilitate biochemical and structural characterization of p14•RNA interaction, we chose to use a disulfide tethering approach to stabilize a complex between thiol-derivatized RNA and p14 containing a single exposed Cys residue (Fig. 2). This approach, first developed by Verdine and coworkers, has been very successfully used in the characterization of protein•DNA complexes (Huang et al. 1998; Huang et al. 2000; He and Verdine 2002; Fromme et al. 2004; Banerjee and Verdine 2006; Johnson et al. 2006; Corn and Berger 2007; Komazin-Meredith et al. 2008; Lee et al. 2008; Zhao et al. 2008) and has been adopted by others to accelerate screening of drug–target interactions (Erlanson et al. 2000, 2003a,b; Cancilla et al. 2008). Sequence-specific and non-specific complexes of the Escherichia coli Ada protein with DNA have been trapped via intramolecular disulfides (He and Verdine 2002). X-ray structures of the DNA repair protein MutY bound to a DNA lesion (Fromme et al. 2004) and a complex of HIV reverse transcriptase with a DNA template primer were also based on a disulfide trapping strategy (Huang et al. 1998).
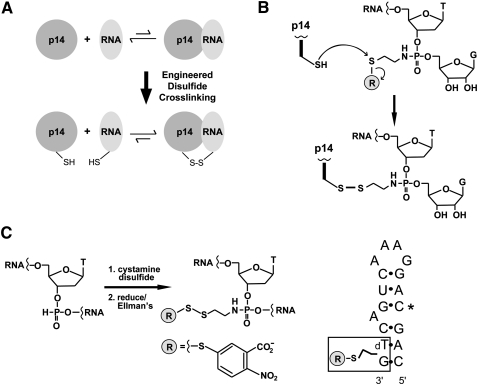
FIGURE 2.
Disulfide cross-linking strategy for interrogating the p14/SF3b155 peptide•RNA complex. (A) Cross-linking of engineered Cys containing p14/SF3b155 peptide to thiol-derivatized RNA generates a tethered RNA–protein complex in which cognate association is predicted to stabilize the disulfide bond to reduction. (B) p14•RNA cross-linking reaction. Chemistry of disulfide bond formation between single Cys containing p14/SF3b155 peptide and an N-thioalkyl modified RNA, where attachment to the backbone is mediated by single phosphoramidate subtitution for a backbone phosphate. (C) Synthesis of thiol-modified RNA. (Left) Oxidation of an H-phosphonate monomer with cystamine disulfide is followed by reduction and trapping with Ellman's reagent during the course of automated synthesis. (Right) Sequence of thiol-modifed RNA synthesized as a mimic of the pre-mRNA•U2 snRNA bulged duplex containing C in place of pseudouridine at position 4 (*).
The positioning of the branch adenosine base within the pocket on p14 allows us to crudely model the interaction of the protein complex with a cognate RNA based on X-ray and NMR structures of bulged duplex RNAs (Berglund et al. 2001; Newby and Greenbaum 2002; Lin and Kielkopf 2008). Given the positioning of adenine within the pocket and minimizing steric clashes with the p14/SF3b155 peptide surface, two possible orientations of the RNA, related by a 180° rotation, may be modeled (1 and 2 in Fig. 3A; see Supplemental Material). We created a Cys-less version of p14 (C83S; C74V) based on phylogenetic comparisons (Will et al. 2001) and determined the structure of the mutant p14•SF3b155 peptide complex to show that it is not perturbed by these changes (data not shown). From Cys-less p14, we then created a panel of single-Cys-containing p14 mutants where the introduced residues were designed, based on our X-ray structure and crude model, to sample the phosphodiester backbone with respect to both possible RNA orientations on the proposed RNA binding surface (Fig. 3A). One mutant, D37C, is located on the opposite, “rear,” face of the p14/SF3b155 peptide complex to serve as a negative control. This combination of modified proteins, especially the D37C mutant, is important with respect to addressing any concern that the tethering strategy might force a nonbiological interaction; screening and comparison of a panel of disulfide linked complexes serve as a control for the validity of the approach (see below).
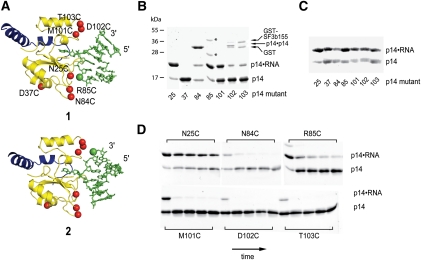
FIGURE 3.
Formation and reduction of p14/SF3b155 peptide•RNA disulfides. (A) Two models of p14/SF3b155 peptide•RNA interaction (1 and 2), related by a 180° rotation about the branch adenosine glycosidic bond, based on cross-linking of a bulged nucleotide to the pocket on the p14 surface (Schellenberg et al. 2006). (Yellow) p14; (blue) the SF3b155 peptide; and (green) RNA. Shown are sites of single-Cys mutations introduced into Cys-less p14 (red balls) and site of RNA backbone modification with the C2 thiol functionality (green ball). Modeling based on the p14/SF3b155 peptide X-ray structure (PDB: 2F9D) and a bulged RNA X-ray structure (PDB: 1I9X) (Berglund et al. 2001). (B) Coomassie-stained SDS-PAGE gel showing formation of p14•RNA disulfide cross-links under mild equilibrating conditions with panel of single Cys mutants (p14•p14 disulfide is formed upon dialysis of p14 samples into nonreducing buffer before RNA addition); (*) uncharacterized contaminant. (C) Coomassie-stained SDS-PAGE gel showing initial formation of p14•RNA disulfide cross-links. (D) Coomassie-stained SDS-PAGE gel showing time course of p14•RNA reduction (0, 10, 30, 60, 120 min).
We chemically synthesized model bulged RNA hairpins, containing a single, commercially available 2′-deoxy-H-phosphonate precursor (Froehler 1986), to mimic the pre-mRNA•U2 snRNA duplex (Fig. 2C). These were derivitized with a thiol by oxidation of the H-phosphonate with cystamine disulfide during the synthesis, which we then reduced and protected with Ellman's reagent. The position of RNA modification was based on an initial model of RNA–protein interaction where placement of the bulged nucleotide within the p14 pocket sets the register of duplex association with that face of the protein. The length of RNA was similarly based on the initial crude model. A highly conserved pseudouridine in U2 snRNA base-pairs with the nucleotide 5′ to the branch position and has been proposed to stabilize an extrahelical disposition of the branch adenosine (Newby and Greenbaum 2002). Therefore, two RNAs were initially synthesized, one containing a single pseudouridine to match the position of this modified base within the U2 snRNA, and another containing cytidine at the same position (Fig. 2C). Disulfide bond formations between these RNAs and p14 cysteine mutants were essentially the same, suggesting that the replacement of pseudouridine with cytidine is a valid model of the bulged duplex (see Supplemental Material).
Allowing Cys mutant protein complexes to equilibrate with the mixed Ellman's•RNA disulfides under mild reducing conditions (60 μM β-mercaptoethanol) overnight produced the desired RNA–protein disulfide in variable yield as assayed by SDS-PAGE (Fig. 3B) and confirmed by RNase treatment (see Supplemental Material). The highest yields of complex were those involving formation of RNA disulfides with the N25C and R85C proteins, suggesting that these are the most thermodynamically stable linkages. A roughly 50% yield of the M101C and N84C disulfides was obtained; the yields of all the D102C and T103C complexes were much lower. Significantly, almost no D37C disulfide was observed. In several cases, intermolecular p14 disulfide formation, upon dialysis into non-reducing buffer and before RNA addition, was observed; this was significant in the case of N84C, likely reflecting the location of the introduced cysteine on a flexible loop of p14. Initial formation of disulfide linkages, as assayed by quenching an aliquot of the above reactions after 20 min (Fig. 3C), produced a roughly equivalent yield of almost all of the RNA–protein disulfides, showing that no single Cys mutant formed the initial disulfide with significantly greater or less (e.g., due to steric constraints) efficiency. We were also able to show that the N25C protein–RNA disulfide is kinetically stable to reduction with respect to the other disulfides by treatment with a high concentration of β-mercaptoethanol followed by SDS-PAGE analysis (Fig. 3D).
The results of the tethering and reduction experiments are very informative, immediately suggesting that the bulged duplex interacts with one face of the p14/SF3b155 peptide complex, and furthermore indicating the preferred binding orientation of the bulged duplex on this face. The lack of D37C disulfide under reducing, equilibrating conditions is a control for non-specific protein–RNA disulfide formation on the rear face of the complex and the varying yields of the other disulfides support one of the two proposed orientations on the front face (1 in Fig. 3A). Finally, the kinetic stability of the N25C disulfide with respect to all of the others but especially the R85C complex (Fig. 3D) suggests that the N25C•RNA disulfide most represents a cognate RNA–protein association. The relatively high amount of M101C disulfide formed under mildly equilibrating conditions (Fig. 3B) shows that an ensemble of p14•RNA complexes (specific and non-specific) is likely present under the conditions of the experiment; however, this disulfide and those to D102C and T103C are rapidly reduced, suggesting that they are unstable/strained and do not represent cognate complexes.
Adenine recognition by the p14/SF3b155 peptide complex
In parallel with the tethering experiments described above, we attempted co-crystallization and soaks of p14/SF3b155 peptide crystals with a variety of nucleobases, nucleosides, and mononucleotides. Following a soak of Cys-less p14/SF3b155 peptide crystals with adenine alone, we solved the 2.4 Å resolution structure of an adenine•p14/SF3b155 peptide complex (Fig. 4A; Table 1). The structure reveals adenine bound within the pocket on the p14 surface as predicted on the basis of cross-linking to the Y22M mutant (Schellenberg et al. 2006).
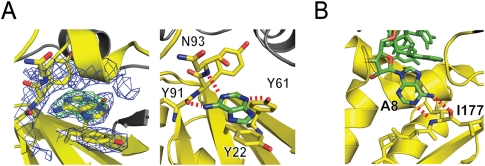
FIGURE 4.
Recognition of the branch nucleotide base during spliceosome assembly. (A) X-ray structure of adenine bound to p14/SF3b155 peptide. (Left) 2Fo − Fc map at 2.4 Å resolution using phases calculated from the final, refined model and contoured at 1σ (blue) and Fo − Fc map using phases calculated from a model lacking adenine contoured at +5σ (green). (Right) Interactions between adenine and the SF3b14 pocket. The purine stacks on Y22 of RNP2, features Watson-Crick-like hydrogen-bonding interactions between the N-6 exocylic amine and the main-chain carbonyl of Y91 and between N-1 and the main-chain N-H of N93, as well as hydrogen-bonding between N-3 and the hydroxyl of otherwise buried Y61 of RNP1. (B) Recognition of human branch region by the KH domain of SF1. Detail from the NMR structure showing specific recognition of the branch nucleotide base and SF1 mediated by Watson-Crick like interactions with the main chain of I177 (Liu et al. 2001).
TABLE 1.
Data collection, phasing, and refinement statistics
The resolution of the adenine•p14/SF3b155 peptide structure allows us to unambiguously model the specific disposition of adenine within the pocket (Fig. 4A). The base stacks on the conserved Y22 of RNP2 consistent with the cross-linking results. Specific recognition of the base within the pocket is mediated by a Watson-Crick-like interaction with main-chain functionalities (the N-6 exocyclic amine with the carbonyl of Y91 and N-1 with the amide N-H of N93), as well as hydrogen-bonding of N-3 to the phenolic hydroxyl of the otherwise buried Y61 of RNP1 (Fig. 4A).
Overall, upon adenine binding, the structure of the p14•SF3b155 peptide complex is unperturbed with the exception of the side chain of R96. In the absence of adenine, this lies along the side of the pocket, forming a hydrogen bond with the main chain carbonyl of Y91. As noted above, adenine binding replaces this with an N6 interaction; the R96 side chain is swung out from the surface in the complex, a conformation that would be suitable to interact with the phosphate backbone of a bound RNA duplex.
The placement of adenine within the structure corroborates the RNA•protein orientation proposed on the basis of the tethering experiments (1 in Fig. 3A) since, with adenine fixed, the alternative disposition would not only require the energetically unfavorable syn orientation of the bulged base but also be subject to significant steric clashes of the duplex•protein interface.
SAXS analysis of tethered p14/SF3b155 peptide•RNA complex
The results of the tethering experiments supported by the X-ray structure suggest that the N25C complex represents a cognate model of a bulged RNA duplex•p14/SF3b155 interaction. We therefore decided to characterize this specific complex further using small-angle X-ray scattering (SAXS). This method has several advantages over others in the structural characterization of macromolecular structure. As a solution technique, it does not require crystallization; because of small sample volumes, relatively small absolute amounts of sample are required; and finally, a significant amount of structural information is contained within the scattering data.
We performed SAXS on samples of both the N25C p14/SF3b155 peptide complex and purified N25C p14/SF3b155 peptide•RNA disulfide. Upon scale-up, in the final purification of the N25C p14/SF3b155•RNA complex by anion exchange chromatography, we were able to separate two species present in a roughly 20:1 ratio. The minor species showed a scattering curve that was relatively featureless and did not flatten at low angles, consistent with a disordered or unfolded structure (data not shown). We interpret this minor cross-linked species, which we did not characterize further, to represent a strained disulfide formed with one of the two possible diastereomers at phosphorous that result from the oxidation of the H-phosphonate precursor during the chemical synthesis. We examined both the p14/SF3b155 peptide and p14/SF3b155 peptide•RNA samples by dynamic light scattering and detected no evidence of higher-order multimers or aggregates (data not shown).
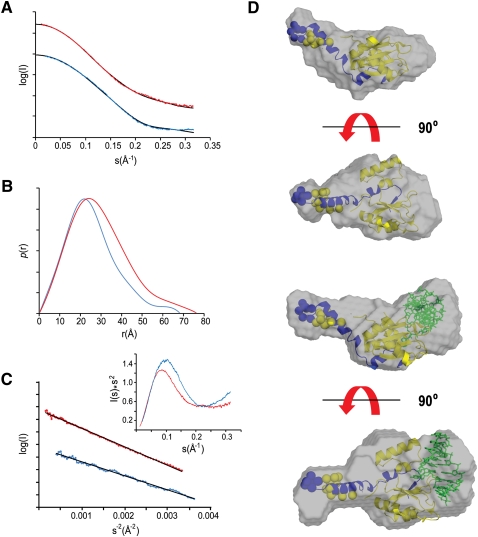
With respect to the N25C p14/SF3b155 peptide complex and the major purified N25C p14/SF3b155 peptide•RNA disulfide, the SAXS scattering plot suggests the presence of ordered, folded structures (Fig. 5A). The flattening of the raw scattering plots at low angles shows that there is minimal aggregation in these samples. The bell-shaped distribution function, P(r), for both is representative of a folded, essentially globular structure (Fig. 5B); the asymmetry represented by a shoulder at ∼50 Å is consistent with the protrusion of the long helix, α1, of the SF3b155 peptide observed in the X-ray structure (Fig. 1B). Linear Guinier plots (Fig. 5C) indicate the absence of aggregation in both samples. The bell-shaped structure of the Kratky plot (Fig. 5C, inset) is also consistent with an essentially globular structure; the plateau of this plot at higher s-values again may be representative of scattering from the helical protrusion.
FIGURE 5.
Small-angle X-ray scattering (SAXS) analysis of p14/SF3b155 peptide and tethered p14/SF3b155 peptide•RNA complexes. (A) Experimental SAXS curves for p14/SF3b155 peptide (blue) and p14/SF3b155 peptide•RNA (red); also shown in black are scattering curves calculated from the CHADD (p14/SF3b155 peptide) or SASREF models (p14/SF3b155 peptide•RNA). (B) Distance distribution functions, P(r), for p14/SF3b155 peptide (blue) and p14/SF3b155 peptide•RNA (red) complexes calculated from the experimental scattering data using GNOM. (C) Guinier plots of p14/SF3b155 peptide (blue) and p14/SF3b155 peptide•RNA data (red); (inset) Kratky plots. (D, upper) SAXS envelope for the p14/SF3b155 peptide complex derived from the DAMMIN refined ab initio model superimposed on the model from CHADD. (Lower) SAXS envelope for the p14/SF3b155 peptide•RNA complex derived from the DAMMIN refined ab initio model superimposed on the model from SASREF. Dummy residues added for (yellow) p14 and (blue) SF3b155 using CHADD.
We next used the ab initio modeling program GASBOR (Svergun et al. 2001) in real space mode to generate models refined against the P(r) function. Ten rounds of model-building from different and random starting positions for dummy atoms were averaged using the program DAMAVER (Volkova and Svergun 2003). The unfiltered model from DAMAVER was further refined by using it as a starting model for the program DAMMIN (Svergun 1999) to yield the final calculated envelope. The ab inito envelope for the p14/SF3b155 peptide complex is bipartite featuring a globular density and long extension within which we fit the p14/SF3b155 peptide crystal structure using the program SUPCOMB (Fig. 5D, upper; Kozin and Svergun 2001). The globular portion of the crystallographic X-ray density fits well and was positioned by virtue of the close correspondence of the helical extension of SF3b155 α1 with the protrusion from the calculated envelope, which leaves room for the five amino acids N-terminal to α1 that are disordered in the crystal structure.
In comparing the ab initio model to the p14/SF3b peptide X-ray structure, we first used the program CHADD (Petoukhov et al. 2002) to add dummy residues to that structure representing the poorly ordered/disordered N-terminal residues (11 and seven residues of p14 and the SF3b155 peptide, respectively). The radius of gyration, Rg, derived from this model, 19.3 Å, is close to that derived from the experimental scattering data (19.7 Å); the theoretical scattering curve calculated from this model using CRYSOL (Svergun et al. 1995) fits the experimental with a χ2 of 2.7 (Fig. 5A; see Supplemental Material).
We next generated an ab initio envelope for the tethered N25C p14/SF3b155•RNA complex using the protocol described above for the p14/SF3b155 peptide complex alone. The resulting envelope is similar to that derived for the protein complex alone, featuring, again, a globular region and a narrow protrusion that we assign to α1 of the SF3b155 peptide. Fixing the p14/SF3b155 peptide crystal structure within the envelope using this constraint reveals significant additional density in the ab initio envelope corresponding to the proposed RNA-binding face of the protein complex (Fig. 5B, right).
We used the program SASREF (Petoukhov and Svergun 2005) to generate a model of the p14/SF3b155 peptide•RNA complex. The protein model was derived from the crystal structure with the missing residues added using CHADD, and the RNA model was a 6-bp bulged duplex based on the larger X-ray structure of Schultz and colleagues capped with a canonical GAAA tetraloop. Ten rounds of SASREF docking using restraints corresponding to the N25C disulfide and bulged-adenosine binding pocket were performed. The models were quite similar, and the one with the best fit to the experimental data (χ2 of 2.0) (see Supplemental Material) is shown in Figure 5D. Using SUPCOMB, we were able to fit this model within the ab initio envelope, once again positioning the helical extension of the SF3b155 peptide within the protrusion from the globular main body of the calculated mass.
DISCUSSION
A complete understanding of p14 function in branch region recognition will include a high-resolution structure of p14 in a complex with a bulged RNA duplex. Nevertheless, the studies described here provide a satisfactory low-resolution model of p14•bulged duplex interaction with implications for spliceosome assembly and activation. Specifically, they argue that the p14•branch duplex association precedes incorporation of that RNA structure into the fully activated spliceosome.
Disulfide tethering as a probe of p14•RNA interaction
The disulfide tethering approach described here confirms a model of RNA interaction with one face of p14 and allows us to distinguish between two proposed orientations of RNA on the p14 surface. Enhanced stability of disulfides to reduction has been clearly shown to reflect the specificity of protein•ligand interaction in tethered complexes. Relative disulfide stability is a measure of relative binding affinity (Gilbert 1995; Stanojevic and Verdine 1995; O'Shea et al. 1989) and reflects the fact the disulfide bond formation/reformation in specific complexes is essentially an intramolecular process that is entropically favored. The high efficiency of disulfide cross-linking of N25C and R85C under mild reducing conditions suggests the specific orientation of RNA on the p14 surface (1 in Fig. 3A). Furthermore, the kinetic stability of the N25C complex to reduction suggests that tethering at this position was optimal in terms of representing a cognate complex. The different efficiencies of disulfide formation strongly support this model and serve as a control for the approach.
The methodology described here should be of general utility in the characterization of RNA–protein interactions, especially where there is uncertainty with respect to the nature of the cognate interaction. As an example, Corn and Berger have successfully described a high-throughput protocol using biotinylated thiol-derivatized DNA to screen for specific protein–DNA complexes from a library of Cys-containing mutants of bacterial DNA primase (Dnag) (Corn and Berger 2007). Because, as in the case with p14, RNA–protein interactions are often cooperative, and isolated complexes may only have a weak affinity, this approach may be particularly useful in screening RNA–protein complexes.
Recognition of the branch adenosine
Specific recognition of adenine, revealed by the X-ray structure reported here, supports our previous suggestion that the unpaired adenosine of the pre-mRNA•U2 snRNA bulged duplex is bound within a pocket on the p14 surface. This interaction is specific and stabilized by three hydrogen-bonding interactions. This includes the interaction of the adenine N6 exocyclic amine with the main chain of Y91 that is dependent on the displacement of the R96 side chain. The hydrogen bond between Y61 and N3 of adenine may only be representative of a subset of branch nucleotide p14 interactions since phenylalanine frequently occupies this position in p14 orthologs. The NMR structure of a single-stranded RNA, representing the human branch sequence, bound to the SF1 KH domain has also been described (Fig. 4B; Liu et al. 2001). Interestingly, recognition of the nucleotide corresponding to the branch adenosine is also specified by a Watson-Crick-like interaction, in this case with the backbone amide and carbonyl of I177.
Together, the SF1•RNA and p14•adenine structures reveal specific recognition of the branch adenosine at multiple stages of spliceosome assembly from the E complex (SF1) through the A, B, and C complexes (p14). The observed specific interactions with adenine provide a molecular explanation for the pronounced preference for that residue at the branch position and may function along with a spliceosomal proofreading mechanism (Xu and Query 2007) to help ensure fidelity of branch selection.
SAXS model of p14•RNA interaction
The envelopes generated from the SAXS data for both the p14/SF3b155 peptide and major RNA•p14/SF3b155 peptide complexes correspond with the density expected on the basis of the p14/SF3b155 peptide X-ray structure, in the latter case with the positioning of bound RNA on one face of the protein complex. The p14/SF3b155 peptide•RNA model formed on the basis of these data is of low resolution but is fully consistent with our previous photo-cross-linking results (Schellenberg et al. 2006), the disulfide cross-linking reduction studies, and the pocket-bound adenine X-ray structure.
Chemical shift mapping has been used to examine the interaction of the p14•SF3b155 peptide complex with single-stranded and bulged duplex RNAs representing U2 snRNA and the pre-mRNA•U2 snRNA duplex, respectively (Spadaccini et al. 2006; Kuwasako et al. 2008). Overall, the NMR results are consistent with the essentials of the structural model proposed here, specifically with respect to the identification of the face of p14 involved in RNA binding and the occlusion of the canonical RNA-binding surface of the RRM. They suggest limited interaction with the RNP motifs (with the exception of Y22 and K24), consistent with their being largely buried and inaccessible; the principal chemical shift perturbations suggest RNA binding involving areas flanking the central β-sheet including the loop between α2 and β4 and the C-terminal portion of p14 proximal to α3. Although it is possible that association of p14 with single-stranded RNA occurs prior to the recruitment of U2 snRNP to the branch region, the p14•SF3b155•RNA interaction that occurs in the A and higher-order complexes must involve recognition of a bulged duplex structure (Query et al. 1994).
Implications for role of p14 and SF3b in spliceosome assembly and activation
The model of p14•duplex interaction described here suggests that an intimate complex is formed between p14 and the pre-mRNA•U2 snRNA duplex. One face of the RNA and, more specifically, the bulged branch nucleotide is bound on the protein surface, consistent with previous cross-linking results showing direct contact between the branch residue and p14 in the A through B and C complexes during spliceosome assembly.
In the model described here, the branch nucleotide, and more specifically the 2′ hydroxyl that is the nucleophile for the first step of splicing, is buried at the interface between the bulged duplex and p14 protein complex (Fig. 3A); this is true even in the mature spliceosome, as represented by the C complex, since photo-cross-linking shows a direct interaction between p14 and the bulged branch adenosine (Query et al. 1996). Thus, although the p14•RNA complex persists into the mature spliceosome, it must be disrupted to allow the first step of splicing to occur.
Luhrmann and coworkers have purified an active step 1 spliceosome and characterized its RNP core. Based on the under-representation of both the SF3a and SF3b complexes in this assembly, they have suggested that the association of both with the spliceosome must be disrupted at or during the first step of splicing (Bessonov et al. 2008). It has recently been suggested by Stevens and colleagues that a Prp2-dependent rearrangement displaces both SF3a and SF3b from the branch region prior to the first step of splicing in yeast. They further propose a model whereby SF3 association with the pre-mRNA prevents premature nucleophilic attack by the branch adenosine until the correct orientation of the proper substrate within the spliceosome (Lardelli et al. 2010). The structural model reported here and the resulting requirement for disruption of the bulged duplex p14 interaction are consistent with both of these observations. Interestingly, in Saccharomyces cerevisiae, which lacks a p14 homolog, the branch nucleophile can attack U2 snRNA in the presence of a weak 5′ splice site where presumably the positioning of the proper substrate has been decoupled from spliceosome activation (Smith et al. 2007). Finally, bulged RNA duplexes have also been shown to be susceptible to hydrolysis 3′ to the bulged residue by virtue of the backbone geometry (Portmann et al. 1996). Thus, the p14•branch duplex association may serve as a regulatory step preventing aberrant chemistry during the assembly of the correct spliceosome active site.
MATERIALS AND METHODS
Protein expression and purification
DNA fragments encoding mutant p14 were generated by overlapping PCR and cloned into pMALC2-X as described (Schellenberg et al. 2006). All mutants were verified by sequencing. Protein expression and purification of mutant p14•SF3b155 amino acids 373–415 were carried out as previously described (Schellenberg et al. 2006).
Protein crystallization and structure determination
Crystals of the Cys-less p14•SF3b155 peptide complex were grown using the hanging drop method with a reservoir solution of 12%–14% PEG 3350, 100 mM Tris (pH 7), and 200 mM NaCl. For adenine soaks, a 100 mM solution of adenine hemisulfate (Sigma) was heated to dissolve the adenine, quickly cooled to room temperature, and 0.2 μL was added to a 2-μL drop containing p14•SF3b155 peptide crystals. Crystals were cryoprotected in reservoir solution containing 15% glycerol and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at beamline 8.3.1 at the Advanced Light Source (Berkeley, CA). Data were processed using HKL2000 (Otwinowski and Minor 1997) and solved using REFMAC5 (Murshudov et al. 1997) for a rigid-body refinement with the coordinates from PDB entry 2F9D, followed by rounds of model building and refinement using XFIT (McRee 1999) and REFMAC5.
RNA synthesis
RNAs were synthesized on an ABI 394 synthesizer using 2′-ACE chemistry (Dharmacon) modified to incorporate the thiol tether during synthesis. After the first detritylation step, the RNA synthesis column was removed from the machine, and the following steps were performed manually with a syringe: (1) wash with 1 mL of 50:50 acetonitrile:pyridine; (2) coupling with 28 mg of 5′-trityl 3′-H phosphonate thymidine monomer (ChemGenes) dissolved in 1 mL of 50:50 acetonitrile:pyridine and 40 μL of pivaloyl chloride (Sigma) for 60 sec; (3) wash with 1 mL of 50:50 acetonitrile:pyridine; (4) wash with 5 mL of acetonitrile; (5) oxidation for 1 h with 50:25:25 carbon tetrachloride:pyridine:cystamine disulfide (freshly dried over MgSO4 in dichloromethane); (6) wash with 5 mL of pyridine; and finally, (7) wash with 5 mL of acetonitrile. The column was returned to the ABI 394 synthesizer and capped with acetic anhydride followed by detritylation, and the synthesis was continued using standard 2′-ACE chemistry. The oligonucleotides were deprotected according to the manufacturer's instructions with the added steps of addition of 0.1 M β-mercaptoethanol during base deprotection and 20 mM tris(2-carboxyethyl)phosphine (TCEP) during the 2′-ACE removal step. Crude oligonucleotides were then modified with 5,5′-dithiobis-(2-nitrobenzoic) acid (Ellman's regeant) prior to separation by denaturing PAGE (20%, 19:1 acrylamide:bis acrylamide, 8 M urea) in TBE running buffer. The band corresponding to the full-length oligonucleotide containing the thiol modification was identified, excised, and extracted from the gel slice.
Protein-RNA disulfide bond formation
Mutant p14•SF3b155 peptide complex was dialyzed overnight into buffer (10 mM Tris at pH 8.0, 60 mM KCl, 0.1 M EDTA) containing 1 mM β-mercaptoethanol, followed by dialysis in the same buffer containing 0.1 mM β-mercaptoethanol for 1 h. Protein complex was mixed with RNA (50 μM) in the same buffer such that the final concentration of β-mercaptoethanol was 60 μM, and disulfide bond formation was allowed to proceed overnight at 4°C. For analysis of the initial products of cross-linking, samples were removed after 20 min and treated as described below. All samples were analyzed by 16% (w/v) SDS-PAGE under non-reducing conditions after first quenching all free thiols with 2 mM iodoacetamide for 10 min and using a loading dye lacking β-mercaptoethanol. For SAXS analysis the p14•SF3b155 peptide RNA complex was purified on a Superdex 75 column (GE Healthcare Life Sciences) followed by ion exchange on a mono-Q HR5/5 column (GE Healthcare Life Sciences).
Protein-RNA disulfide reduction
p14•SF3b155 peptide RNA complex was added to reactions containing 5 mM β-mercaptoethanol and incubated at room temperature for between 10 and 120 min. At the end of each time point, free thiols were quenched with a twofold excess of iodoacetamide for 10 min followed by non-reducing 16% (w/v) SDS-PAGE using loading dye lacking β-mercaptoethanol.
Small-angle X-ray scattering
Small-angle X-ray scattering (SAXS) data were collected at beamline 12.3.1 of the Advanced Light Source, Lawrence Berkeley National Laboratory, at 12 keV on a MAR165 detector. Samples were maintained at 10°C during data collection using a thermostatically controlled cuvette. p14•SF3b155 peptide samples at 2.5 and 10 mg/mL and a p14•SF3b155 peptide–RNA sample at a concentration of 10 mg/mL were buffer-exchanged overnight at 4°C in 25-μL dialysis buttons, using 6–8 kDa nominal molecular weight cutoff regenerated cellulose dialysis tubing, against a 50-mL volume of the SAXS buffer (10 mM Tris at pH 8.0, 60 mM NaCl, 0.1 mM EDTA, and 5% glycerol). A series of exposures (6, 60, and 6 sec) was measured to assess for radiation sensitivity. Scattering of the dialysis medium alone from 6- or 60-sec exposures was subtracted from the scattering of the equivalently exposed samples to yield scattering curves for the macromolecules alone. Data were analyzed using the programs PRIMUS43 (Konarev et al. 2003), GNOM44 (Svergun 1992), and AUTORG45 (Konarev et al. 2003). No radiation damage was apparent as the first and last 6-sec exposures produced curves that were superimposable, no aggregation was visible to the program AUTORG45, and a linear dependence of log[I(s)] versus s2 in the range sRG < 1.3 was observed. The scattering curves for the 6-sec exposure (data from 0.012 sec to 0.163 sec) and the 60-sec exposure (data from 0.072 sec to 0.318 sec) of the 10 mg/mL p14•SF3b155 peptide RNA sample were merged. Scattering data from 0.012 sec to 0.163 sec for the 60-sec exposure of the 2.5 mg/mL p14•SF3b155 peptide and data from 0.072 sec to 0.318 sec for the 60-sec exposure of the 10 mg/mL sample were merged using PRIMUS43. The merged scattering curves were used for analysis with the program GNOM44 which generated a P(r) curve. Ten rounds of model building using the program GASBOR (Svergun et al. 2001) with 168 dummy atoms for the p14•SF3b155 peptide complex and 219 dummy atoms for the p14•SF3b155 peptide•RNA complex were averaged using DAMAVER (Volkova and Svergun 2003). The starting model from DAMAVER was used as an initial model for the program DAMMIN (Svergun 1999), which produced the final refined ab initio model.
The program CHADD (Petoukhov et al. 2002) was used to add dummy residues to the crystal structure of p14•SF3b155 peptide (PDB entry 2F9D, chains A and P) to model amino acids that were disordered in the crystal structure. The program SASREF (Petoukhov and Svergun, 2005) was used to model the complex between the p14•SF3b155 peptide model generated by CHADD and a model RNA duplex (PDB entry 1I9X: nucleotides 2–8 of chain A and nucleotides 7–12 of chain B) (Berglund et al. 2001), with a GAAA tetraloop at the end of the helix from PDB entry 1TLR (nucleotides 5–8) (Butcher et al. 1997), using restraints corresponding to the N25C-RNA disulfide tether and the bulged adenosine–Y22 interaction. Theoretical scattering from models was compared to the experimental scattering curves using the program CRYSOL27 (the solvent density was maintained at the default value of 0.334 e/Å3) (Svergun et al. 1995). The ab initio models were superimposed with the CHADD and SASREF models using the program SUPCOMB20 (Kozin and Svergun 2001).
Coordinates
Protein Data Bank: Coordinates for the p14/SF3b155 peptide•adenine complex have been deposited under accession code 3LQV.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Greg Hura and Dr. Michal Hammel (Advanced Light Source) and Dr. Ross Edwards (University of Alberta) for their help in designing the SAXS experiment and for advice on data processing. We also thank Emily Gesner for helpful comments on the manuscript. This work was supported by an Operating Grant from the Canadian Institutes of Health Research (CIHR).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2224411.
REFERENCES
- Allain FHT, Bouvet P, Dieckmann T, Feigon J 2000. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J 19: 6870–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Verdine GL 2006. A nucleobase lesion remodels the interaction of its normal neighbor in a DNA glycosylase complex. Proc Natl Acad Sci 103: 15020–15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Rosbash M, Schultz SC 2001. Crystal structure of a model branchpoint–U2 snRNA duplex containing bulged adenosines. RNA 7: 682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R 2008. Isolation of an active site step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Brow DA 2002. Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl T, Sharp PA 1999. The RNA World, 2nd ed (ed. Gesteland RF et al. ), pp. 525–560 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Butcher SE, Dieckmann T, Feigon J 1997. Solution structure of a GAAA tetraloop receptor RNA. EMBO J 16: 7490–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancilla MT, He MM, Viswanathan N, Simmons RL, Taylor M, Fung AD, Cao K, Erlanson DA 2008. Discovery of an Aurora kinase inhibitor through site-specific dynamic combinatorial chemistry. Bioorg Med Chem Lett 18: 3978–3981 [DOI] [PubMed] [Google Scholar]
- Cléry A, Blatter M, Allain FH 2008. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18: 290–298 [DOI] [PubMed] [Google Scholar]
- Corn JE, Berger JM 2007. FASTDXL: A generalized screen to trap disulfide stabilized complexes for use in structural studies. Structure 15: 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98: 835–845 [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Braisted AC, Raphael DR, Randal M, Stroud RM, Gordon EM, Wells JA 2000. Site-directed ligand discovery. Proc Natl Acad Sci 97: 9367–9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanson DA, Lam JW, Wiesmann C, Luong TN, Simmons RL, DeLano WL, Choong IC, Burdett MT, Flanagan WM, Lee D, et al. 2003a. In situ assembly of enzyme inhibitors using extended tethering. Nat Biotechnol 21: 308–314 [DOI] [PubMed] [Google Scholar]
- Erlanson DA, McDowell RS, He MM, Randal M, Simmons RL, Kung J, Waight A, Hansen SK 2003b. Discovery of a new phosphotyrosine mimetic for PTP1B using breakaway tethering. J Am Chem Soc 125: 5602–5603 [DOI] [PubMed] [Google Scholar]
- Froehler BC 1986. Deoxynucleoside H-Phosphonate diester intermediates in the synthesis of internucleotide phosphate analogues. Tetrahedron Lett 27: 5575–5578 [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, Verdine GL 2004. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427: 652–656 [DOI] [PubMed] [Google Scholar]
- Gao K, Masuda A, Matsuura T, Ohno K 2008. Human branch point consensus sequence is yUnAy. Nucleic Acids Res 36: 2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert HF 1995. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol 251: 8–28 [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R 1998. A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S 1999. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398: 579–585 [DOI] [PubMed] [Google Scholar]
- Hartmann B, Valcárcel J 2009. Decrypting the genome's alternative messages. Curr Opin Cell Biol 21: 377–386 [DOI] [PubMed] [Google Scholar]
- He C, Verdine GL 2002. Trapping distinct structural states of a protein/DNA interaction through disulfide crosslinking. Chem Biol 9: 1297–1303 [DOI] [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, Harrison SC 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Huang H, Harrison SC, Verdine GL 2000. Trapping of a catalytic HIV reverse transcriptase template:primer complex through a disulfide bond. Chem Biol 7: 355–364 [DOI] [PubMed] [Google Scholar]
- Johnson AA, Santos W, Pais GC, Marchand C, Amin R, Burke TR, Verdine GL, Pommier Y 2006. Integration requires a specific interaction of the donor DNA terminal 5′-cytosine with glutamine 148 of the HIV-1 integrase flexible loop. J Biol Chem 281: 461–467 [DOI] [PubMed] [Google Scholar]
- Komazin-Meredith G, Santos WL, Filman DJ, Hogle JM, Verdine GL, Coen DM 2008. The positively charged surface of herpes simplex virus UL42 mediates DNA binding. J Biol Chem 283: 6154–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr 36: 1277–1282 [Google Scholar]
- Kozin MB, Svergun DI 2001. Automated matching of high- and low-resolution structural models. J Appl Crystallogr 34: 33–41 [Google Scholar]
- Kuwasako K, Dohmae N, Inoue M, Shirouzu M, Taguchi S, Güntert P, Séraphin B, Muto Y, Yokoyama S 2008. Complex assembly mechanism and an RNA-binding mode of the human p14-SF3b155 spliceosomal protein complex identified by NMR solution structure and functional analyses. Proteins 71: 1617–1636 [DOI] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR III, Stevens SW 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Radom CT, Verdine GL 2008. Trapping and structural elucidation of a very advanced intermediate in the lesion-extrusion pathway of hOGG1. J Am Chem Soc 130: 7784–7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Kielkopf CL 2008. X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines. Biochemistry 47: 5503–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M 2001. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098–1102 [DOI] [PubMed] [Google Scholar]
- MacMillan AM, Query CC, Allerson CA, Chen S, Verdine GL, Sharp PA 1994. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev 8: 3008–3020 [DOI] [PubMed] [Google Scholar]
- McRee DE 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL 2002. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol 9: 958–965 [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Rutkowski R, Stafford WF, Kim PS 1989. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science 245: 646–648 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K 1994. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372: 432–438 [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI 2005. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 89: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Eady NA, Brown KA, Svergun DI 2002. Addition of missing loops and domains to protein models by x-ray solution scattering. Biophys J 83: 3113–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portmann S, Grimm S, Workman C, Usman N, Egli M 1996. Crystal structures of an A-form duplex with single-adenosine bulges and a conformational basis for site-specific RNA self-cleavage. Chem Biol 3: 173–184 [DOI] [PubMed] [Google Scholar]
- Query CC, Moore MJ, Sharp PA 1994. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev 8: 587–597 [DOI] [PubMed] [Google Scholar]
- Query CC, Strobel SA, Sharp PA 1996. Three recognition events at the branch-site adenine. EMBO J 15: 1392–1402 [PMC free article] [PubMed] [Google Scholar]
- Reed R 1990. Protein composition of mammalian spliceosomes assembled in vitro. Proc Natl Acad Sci 87: 8031–8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Luhrmann R, Glover JN, MacMillan AM 2006. Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci 103: 1266–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2007. Trans-splicing to spliceosomal U2 snRNA suggests disruption of branch site-U2 pairing during pre-mRNA splicing. Mol Cell 26: 883–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaccini R, Reidt U, Dybkov O, Will C, Frank R, Stier G, Corsini L, Wahl MC, Lührmann R, Sattler M 2006. Biochemical and NMR analyses of an SF3b155–p14–U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA 12: 410–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Verdine GL 1995. Deconstruction of GCN4/GCRE into a monomeric peptide–DNA complex. Nat Struct Biol 2: 450–457 [DOI] [PubMed] [Google Scholar]
- Svergun DI 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- Svergun DI 1999. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76: 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Barberato C, Koch MHJ 1995. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28: 768–773 [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MHJ 2001. Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova VV, Svergun DI 2003. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hall TMT 2001. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol 8: 141–145 [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Lührmann R, Query CC 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J 20: 4536–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Query CC 2007. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell 28: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, McKee CJ, Kessl JJ, Santos WL, Daigle JE, Engelman A, Verdine GL, Kvaratskhelia M 2008. Subunit-specific protein footprinting reveals significant structural rearrangements and a role for N-terminal Lys-14 of HIV-1 integrase during viral DNA binding. J Biol Chem 283: 5632–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]