Summary
Natural small compounds comprise most cellular molecules and bind proteins as substrates, products, cofactors and ligands. However, a large scale investigation of in vivo protein-small metabolite interactions has not been performed. We developed a mass spectrometry assay for the large scale identification of in vivo protein-hydrophobic small metabolite interactions in yeast and analyzed compounds that bind ergosterol biosynthetic proteins and protein kinases. Many of these proteins bind small metabolites; a few interactions were previously known, but the vast majority are novel. Importantly, many key regulatory proteins such as protein kinases bind metabolites. Ergosterol was found to bind many proteins and may function as a general regulator. It is required for the activity of Ypk1, a mammalian AKT/SGK1 kinase homolog. Our study defines potential key regulatory steps in lipid biosynthetic pathways and suggests small metabolites may play a more general role as regulators of protein activity and function than previously appreciated.
Introduction
Over the past decade considerable effort has been devoted to analyzing biological networks, particularly protein-protein, expression, transcription factor binding and even protein phosphorylation networks (reviewed in Snyder and Gallagher, 2009). These studies have provided a wealth of information for understanding protein function, which components work together and the basic principles of regulatory network organization.
In total numbers, small metabolites comprise the vast majority of cellular components, and like proteins, they are present in a broad range of cellular concentrations and participate in a wide variety of biochemical and regulatory functions. They serve as metabolic components, cofactors for enzymes, forms of energy for biochemical reactions and regulators of protein function (Forster et al., 2003). As regulators of protein function, metabolites can act globally to control many proteins or specifically target a limited number of proteins. Examples of the wide variety of small metabolite-protein associations include the binding of galactose to a sensor protein (Yano and Fukasawa, 1997), steroid hormones to transcription factors (Evans, 1988) and second messengers including phospholipids, cyclic nucleotides and arachidonic acids to specific cellular targets. In spite of their importance in mediating protein function and regulation, systematic approaches for analyzing in vivo interactions have not been performed. Such information is expected to be valuable not only for elucidating the biochemical activities and regulation of individual proteins, but also for assembling and understanding regulatory networks and connections between biological pathways. Furthermore, since metabolite levels can be adjusted by dietary intake of nutrients, understanding the regulation of cellular processes by metabolites has potential therapeutic value in correcting defects in biochemical pathways.
Saccharomyces cerevisiae has served as an important model organism for many large-scale studies including analysis of protein-protein interactions, phenotypes, genetic interactions, protein localization, gene expression, and transcription factor binding (reviewed in Horak and Snyder, 2002; Snyder and Gallagher, 2009). To date, over 682 metabolic compounds have been identified in yeast (Forster et al., 2003) and many are known to be hydrophobic; 52% have a logP greater than methanol (Fig. S1A). Many models of yeast metabolism have been generated and capable of predicting key regulatory metabolic steps (Cascante and Marin, 2008; Herrgard et al., 2008).
Several previous studies have analyzed small molecules and small molecule-protein interactions in yeast. Metabolites have been profiled from yeast extracts using mass spectrometry (Allen et al., 2003), and limited studies to identify metabolites that bind proteins have been performed (Lee et al., 2007). Protein and small molecule microarrays have been used to discover several in vitro interactions (Beloqui et al., 2009; Kuruvilla et al., 2002; Morozov et al., 2003; Zhu et al., 2001). Assays to examine small molecule-protein interactions have been developed (Maynard et al., 2009; Tagore et al., 2008); however, a systematic effort to identify the small metabolites that bind to large numbers of proteins in vivo has not been performed. Thus, the number and types of proteins that bind small molecules in the cell are not known. Such information is expected to both help inform potential regulatory interactions and elucidate the function and regulation of proteins and pathways.
Here we present a systematic large scale investigation of the endogenous protein-metabolite interactome in yeast. We focused on the interaction of hydrophobic metabolites with components of the ergosterol biosynthesis pathway and protein kinases. Ergosterol biosynthetic enzymes were studied because we expected that these might bind hydrophobic metabolites, and protein kinases were chosen because of their importance in global regulation of protein function. We found that a large number of proteins bound to hydrophobic metabolites and described many novel interactions. Further analysis has revealed that the yeast sterol, ergosterol, binds to many protein kinases, often with 1:1 stoichiometry, and is important for the activity of a highly conserved kinase, Ypk1, a member of the Akt/SGK1 family, and for the protein levels of Ssk22, a MAPKKK involved in osmotic responses. Ergosterol is the major sterol in yeast and, analogous to cholesterol in mammals, it is an abundant component of plasma membranes. Overall, our results demonstrate that a variety of small metabolite-protein interactions occur in eukaryotes and suggesting an extensive role in the global regulation of protein activities.
Results
A large scale assay to identify hydrophobic compounds associated with proteins: application to ergosterol biosynthetic enzymes
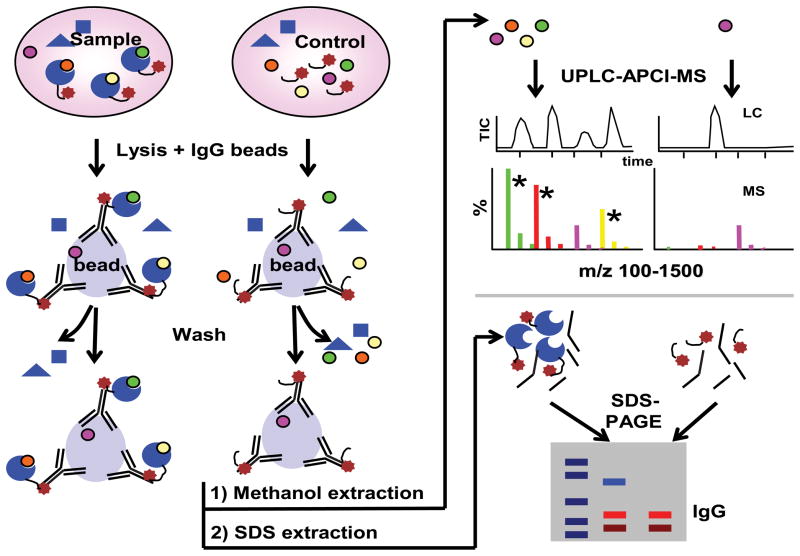
We developed a sensitive and scalable method to systematically identify small metabolites bound to proteins using affinity protein purification and mass spectrometry (Fig. 1). Briefly, proteins tagged with an IgG-binding protein domain (Gelperin et al., 2005) were isolated from lysates using magnetic beads. After washing, the small metabolites were then extracted in methanol and analyzed using a reverse phase C18 column-equipped Ultra Performance Liquid Chromatography (UPLC) column coupled to a Quadrupole-Time-of-flight (Q-TOF) mass spectrometer. As a negative control, parallel experiments were performed using a yeast strain lacking the fusion protein (Y258). The metabolites significantly enriched in the presence of the fusion relative to the control strains and methanol solvent were identified. We focused on hydrophobic molecules because they are less likely to be removed from proteins during washes and can readily be detected by atmospheric pressure chemical ionization (APCI). The resultant mass spectra are comprised mostly of the protonated precursor ions ([M+H]+) which readily allow small metabolite identification. The mass spectrometry assay was first developed and standardized using 12 diverse compounds, including lipid-soluble vitamins from A, D, E and K families, and two sterols (ergosterol and lanosterol) which allowed us to optimize the sensitive detection (~200 femtomole in a mixture in profile scan mode) and separation of each of these compounds (Fig. S1B). We also found that this method detected more than 340 features in a methanol extract of yeast cells (An LC profile and list of peaks enriched in the extract relative to the solvent are in Figure S1C and Table S1), indicating its scope is sufficiently broad to cover at least hundreds of metabolites in a single experiment.
Figure 1. Flow chart for the identification small metabolites bound to proteins.
Molecules bound to a strain expressing a protein of interest relative to a control stain are identified using the scheme presented. See also Figure S1. and Table S1.
We first established the profiling assay using a group of 21 enzymes involved in ergosterol biosynthesis (Parks and Casey, 1995) whose known substrates and products, most of which are non-polar hydrophobic molecules, are readily detectable by LC-APCI-Q-TOF (Fig. 2A). The assays for the Erg proteins were performed using 2–3 separate protein preparations (biological replicates) each containing 0.5–5 picomoles of protein; for each sample preparation 4–6 technical replicates were analyzed in parallel. The mass spectra data were analyzed in MarkerLynx or XCMS to identify molecules based on retention times and accurate molecular mass (see Methods and Fig. S1D for details). Since the background for the peak regions is very low, the correspondence between both technical and biological replicates was extremely high (mean of r.s.d. = 7.5± 4% for 3 technical replicates; for biological replicates see next section). We therefore used a stringent threshold for calling positive signals. Finally, protein purity was examined using SDS-gel electrophoresis after metabolite extraction. In general a single or major band of the expected size is present in the strain expressing the fusions relative to the negative control, although 14% of preparations contained more than one band indicative of either associated proteins or degradation products (Fig S2A, Fig. S3A).
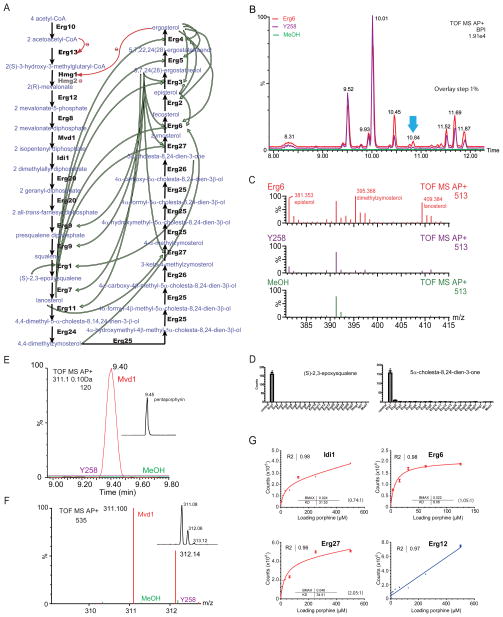
Figure 2. Identification of small metabolites associated with ergosterol biosynthetic proteins.
(A) An overview of ergosterol biosynthesis pathway. Substrates and products of the yeast ergosterol biosynthetic pathway (retrieved from MetaCyc with modification) are in blue whereas protein enzymes are in black (included in this study) or gray (not included in this study). Known interactions are linked by red curve labelled with Θ for inhibitory effects. Interactions discovered in this study are indicated by green arrows from a metabolite to a binding protein.
(B) LC plots of the small metabolites extracted from a protein (Erg6, red), the negative control (Y258, purple), and the methanol solvent (green), respectively. Base peak intensity (BPI, %) is plotted with retention time (in minute) of corresponding mass spectra (shifted by 1% for clarity). Note BPI peaks are composite, not a good indicator of the intensity of single molecular masses. The 100% BPI in counts is indicated on the graph. All traces were smoothed by the Savitzky-Golay method using 2 passes of window size of 3 scans.
(C) Combined average mass spectra of the 10.80–11.20 min region in B (indicated by a blue block arrow). The masses of 3 Erg5-bound small metabolites are indicated along with their chemical identities. The X-axis is the peak mass (amu); the Y-axis is the peak intensity (%).
(D) Summary of the average peak intensity of two small metabolites listed in Table 1 extracted from each of the 21 ergosterol biosynthetic proteins (n=5). * indicates statistical significance (two tail t-test, P value<0.01) in comparison with the negative control Y258. Error bar = SEM.
(E) An LC plot showing detection of pentaporphyrin I (311.100 amu at 9.40 min) from Mvd1 (red) but not from Y258 (purple) or methanol samples (green). Indent shows the LC of pure pentaporphyrin I.
(F) Combined mass spectra of the LC peak region in E. Color labels are as in E. X-axis is shifted by 0.05 amu for clarity. The indent profile shows the mass spectrum of pure pentaporphyrin.
(G) In vitro binding curves of pentaporphyrin and Erg proteins. Each binding curve was subject to fitting comparison (P value< 0.01) to a saturable binding curve (specific) or a straight line (non-specific). Binding constants Kd, Bmax and curve fitting R2 are indicated (in μM) on each graph. Stoichiometry (metabolite:protein) is also indicated.
See also Fig. S2.
Analysis of the LC-MS results revealed that 16 of the 21 purified Erg proteins associated with small metabolites (Table 1). One example shown in Figure 2 contains several compounds associated with Erg6 that eluted from the LC column at retention time 10.84 min (Fig. 2B); this peak contained 3 mass peaks which were significantly lower (t test P value<0.01, >10 fold) in the Y258 yeast control or methanol solvent (Fig. 2C). These 3 peaks were identified as episterol (381.353 atomic mass unit (amu)), dimethylzymosterol (395.368 amu) and lanosterol (409.384 amu), respectively, by elemental composition analysis and hydrophobicity matching in retention time with known chemicals (See Fig. S1B). Nine other proteins also bound small metabolites related to ergosterol biosynthesis. A large number of Erg and other proteins analyzed in this study that were just as abundant in the protein preparations as the metabolite binding protein did not bind any metabolites (Fig. S3D). For each of the ten proteins that bound ergosterol related metabolites a specific set of associating molecules were observed, and similarly each metabolite had a distinct profile. For example, (S)-2,3-epoxysqualene and 5α-cholesta-8,24-dien-3-one specifically associated only with Erg1 (Fig. 2D), whereas lanosterol, ergosterol and episterol consistently co-purified with 5, 5 and 3 Erg proteins, respectively, above the control (Table 1). One Erg proteins was found to bind known substrates (e.g. Erg3 bound episterol) and three other Erg proteins bound known products (e.g. (S)-2,3-epoxysqualene for Erg1) suggesting that these substrates and products are tightly associated with their metabolizing/biosynthetic enzymes.
Table 1.
Summary of identified small metabolites associated with ergosterol biosynthetic proteins.
| Ergosterol | 5,7,24(28)- ergostatrienol | lanosterol | 4,4-dimethyl- zymosterol | 5a-cholesta-8,24-dien-3-one | episterol | (S)-2,3- epoxylsqualene | Pentaporphyrin | |
|---|---|---|---|---|---|---|---|---|
| RT (min) | 10.84 | 13.47 | 11.06 | 11.03 | 11.89 | 10.99 | 9.95 | 9.40 |
| Mass (amu) | 379.337 | 379.337 | 409.384 | 395.368 | 383.328 | 381.353 | 425.379 | 311.100 |
| Element composition | C28H43* | C28H43* | C30H49* | C29H47* | C27H43O | C28H45* | C30H49O | C20H15N4 |
| Erg1 | P | |||||||
| Erg2 | P | |||||||
| Erg3 | P | S | ||||||
| Erg4 | P | |||||||
| Erg5 | S | |||||||
| Erg6 | ||||||||
| Erg7 | P | S | ||||||
| Erg9 | ||||||||
| Erg11 | S | |||||||
| Erg24 | P | |||||||
| Erg25 | S | |||||||
| Erg26 | P | |||||||
| Erg27 | ||||||||
| Hmg1 | ||||||||
| Idi1 | ||||||||
| Mvd1 | S |
Erg enzymes not bound to any known intermediates or pentaporphyrin: Erg 8 Erg10 Erg12 Erg13 Erg20
Gray shade denotes a small metabolite-protein association identified in this study. “P” and “S” indicates known product and substrate, respectively. Bound proteins for ergosterol and pentaporphyrin had an enrichment greater than 3-fold and 1000-fold, respectively, and the remainder were enriched greater than 5-fold. All bound metabolites had a t-test (two tailed unequal variance in relative to the negative control) p-value below 0.05; for ergosterol and pentaporphyrin p-values of less than 0.01 and 1E-10 was used.
indicates a dehydrated form of the expected formula
Importantly the majority of metabolite-protein interactions detected in our assay were novel (e.g. dimethylzymosterol for Erg6) (Table 1). Of particular interest were lanosterol and ergosterol which each bound 5 Erg proteins. Ergosterol bound its natural synthesizing enzyme Erg4 and four other enzymes that control the last five steps in ergosterol biosynthesis starting from zymosterol, suggesting a multi-step feedback regulatory mechanism (Fig, S2B, and Table 1). Ergosterol was not detected with the known ergosterol-regulated enzyme Hmg1, probably due to a low level of protein in the protein preparation (Fig. S2A). Although low levels of protein may be an issue in several instances, it is unlikely to be a major problem overall as the distribution of protein levels of the 37 metabolite binding proteins identified in our entire study (Erg proteins and protein kinases) is similar to the distribution of the level of the majority of the 124 proteins analyzed (Fig. S3D). Lanosterol also bound five enzymes; these enzymes are located at different points in the biosynthetic pathway. Interestingly, unlike other erg mutant strains, yeast lacking Erg7, the enzyme that produces lanosterol, fails to grow in the absence of lanosterol (Karst and Lacroute, 1977), suggesting a major role for this lipid in yeast that might include modulation of protein function. Overall, these results raise the possibility that, in addition to the known inhibitory regulation of Hmg1 by ergosterol and of Erg13 by acetoacetyl-CoA (Parks and Casey, 1995), many steps in the ergosterol biosynthesis pathway may be regulated by biosynthetic products of the pathway (Fig. 2A).
Novel metabolites were discovered to bind yeast Erg proteins
In addition to well-characterized sterols and other lipids that bound Erg proteins was an unexpected metabolite, pentaporphyrin I. Pentaporphyrin is a heme-related intermediate that may bind noncovalently to proteins due to the lack of peripheral methyl groups. It was detected in 7 of 21 Erg enzymes and was identified using a known standard (Fig. 2E–F; Table 1). It is not clear whether this metabolite is “free” porphyrin or derived from a bound form after loss of the central coordinated metal ion during LC-MS detection. Nevertheless measurements of the binding affinity and stoichiometry revealed dissociation constants (Kd) of 8 to 34 μM and pentaporphyrin:protein stoichiometries of approximately 1:1 (for Idi1 and Erg6) to 2:1 (for Erg27) (Fig. 2G). The discovery that pentaporphyrin is associated with several ergosterol biosynthetic components may explain the observation that elimination of heme synthesis results in ergosterol auxotrophy (Parks and Casey, 1995) and suggests that pentaporphyrin-Erg protein interactions are important for protein function. Thus, our systematic analyses of small-protein interactions reveal novel interactions important for protein function and further help explain phenotypes for yeast strains lacking these different proteins.
Large scale analysis of hydrophobic small metabolites bound to yeast protein kinases
Protein kinases control cellular processes and regulate protein function at many levels. We next examined which of the yeast protein kinases bound hydrophobic metabolites. 103 protein kinases representing all functional branches in yeast (Hunter and Plowman, 1997) (Fig. 3A) were purified and analyzed as described above (Fig. S3A). Two biological replicates were performed for all 103 kinases, with at least 3 technical replicates per biological replicate. A total of 95 metabolite peaks (background and specific peaks) were identified in both experiments, and these peaks were highly correlated in both retention time and exact mass from the 2 batches (R2=0.78) (Fig. S3B). Using a stringent threshold, a total of 10 different peaks were found to be associated with 21 protein kinases, but not with negative (Y258) or methanol solvent controls (Table S2 and Fig. S3C,E) or with many other abundant yeast proteins (Fig. S3D). The specific peaks fell into two classes. One major class (11 of 14 analyzed) yielded reproducible peak intensity signals (R2 >0.9) whereas another set (3 of 14) exhibited less correlation of peak intensities (R2<0.75). It is likely that the compounds bound to the highly correlated peaks represent strong steady-state interactions, and those with differing levels of interacting metabolites interact transiently and/or weakly (Morozov et al., 2003).
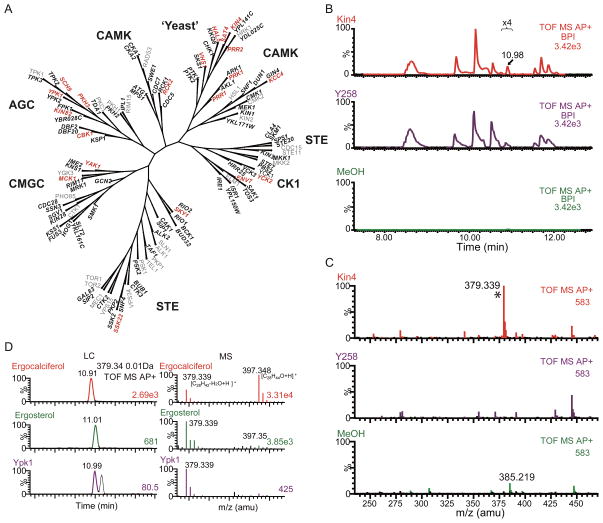
Figure 3. S. cerevisiae protein kinase-small metabolite interaction.
(A) A total of 103 of 129 kinases were analyzed in this study. Kinases not tested are indicated in gray. The 20 kinases that bound small metabolites are in red.
(B) An LC plot of the small metabolites extracted from a kinase (Kin4, red), the negative control (Y258, purple), and the methanol (green). The focused retention time region (in minute, zoomed in 4x) is indicated above the trace. Graph labels are as in Fig. 2B.
(C) Combined average mass spectra of the 10.9 to 11.1 min region in B. Graph label is as in Fig. 2C (n=3 for Kin4; n=9 for Y258 and methanol). The peak corresponding to ergosterol is marked by *.
(D) An example showing identification of a bound metabolite as ergosterol. The mass spectra of pure ergosterol, pure ergocalciferol, and one of the small metabolites extracted from protein kinase Ypk1 shown on right and their respective UPLC on left. The elemental composition is indicated respective mass peaks. Graph label is as in B,C.
An example of specific binding is shown in Fig. 3B–C for Kin4, a protein kinase regulating mitotic exit (Caydasi and Pereira, 2009). The methanol extract from Kin4 contained a singly charged ion of 379.337 amu at retention time 10.98 min. Analysis of standards revealed that this compound is ergosterol (dehydrated state) rather than ergocalciferol, a compound of identical molecular mass (Fig. 3D).
The 10 specific binding metabolites represent different lipids and sterols; however one kinase Ste20 was found to bind pentaporphyrin, albeit at reduced levels relative to the ergosterol enzymes, raising the possibility that Ste20 is associated with this molecule. The compound identified most often among different kinases was ergosterol which was associated with 15 different protein kinases. None of these proteins was previously known to bind ergosterol, and, except for Yck2, none were known to be membrane-associated. KEGG pathways analysis reveals an enrichment of ergosterol bound-kinases in metabolism of inositol phosphate, starch and sucrose, nicotinate and nicotinamide, and sphingoglycolipids (Table S3). The different ergosterol binding kinases belong to different families of protein kinases (Fig. 3A), suggesting a potential regulatory role of ergosterol in the regulation of many types of protein kinases and many different aspects of yeast biology.
Ergosterol-protein kinase interactions have binding affinities and stoichiometries in ranges expected for physiological relevance
To determine if the binding coefficients observed are likely to be physiologically relevant, the stoichiometry and affinity of the ergosterol-protein kinase association was determined for 3 kinases, Ypk1, Hal5, and Rck2, along with three non binding controls Atg1, Psk2 and denatured Ypk1, using an in vitro binding assay we developed (Fig. 4A). For a fixed amount of the ergosterol-bound protein kinases, ergosterol binding exhibited a saturable curve over an increasing concentration of free ergosterol, indicating specific binding. In contrast, the binding curve was close to linear for the same amount of denatured protein or control proteins (Fig. S4), indicating non-specific adsorption. The Kd for each ergosterol binding kinase was between 4.7 to 17.9 μM (Fig. 4A), figures significantly lower than the endogenous concentration of ~4.8 mM for ergosterol in yeast, assuming a uniform distribution throughout the cell (Ejsing et al., 2009). Although neither the kinase nor ergosterol is likely to be uniform in their cellular distribution, the 4.7–17.9 μM binding constant found for Ypk1 and the other kinases is well within a plausible range for biological relevance. The binding ratio of ergosterol to protein was found to be close to 1 (0.98–1.1, 95% confidence interval) for each protein kinase suggesting that one small metabolite binds to one protein (Fig. 4A). This ratio is consistent with an in vivo biological role for ergosterol in regulating kinase activity.
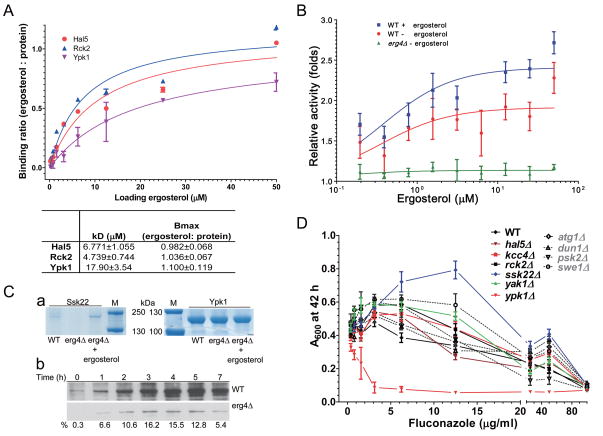
Figure 4. Detailed analysis of several kinase-small metabolite interactions.
(A) In vitro binding analysis of ergosterol and several protein kinases. The curve-fitting was done in GraphPad Prism 5. Error bars are SEM (n=3). Statistic comparison of curve fitting between a straight line for non-specific binding (null) vs. one-site specific binding was used to determine specific binding pattern (P value<0.05). The R2 (unweighted) was 0.918, 0.908, and 0.933 for Hal5, Rck2, and Ypk1 respectively. The ergosterol-binding characteristics of their protein kinases are listed below.
(B) Protein kinase activity of Ypk1 is stimulated by the addition of ergosterol. Ypk1 protein was purified from wild type (BY4741) cells grown in the presence or absence of 2 mM ergosterol during galactose induction of protein expression, or from ergosterol deficient yeast (erg4Δ). Equal amounts of purified protein were tested in each assay. The relative activity was determined using a Sgk1-specific kinase assay. Error bars=SEM, n=4.
(C) Levels of Ssk22 and Ypk1 in yeast. a) Ssk22 and Ypk1 were purified from equal amounts of wild type (BY4741) and ergosterol-lacking mutant (erg4Δ) cells with or without 0.4 mM ergosterol. In 3 independent experiments, Ssk22 cannot be detected in erg4Δ. b) Immunoblot of Ssk22 and Ypk1 from yeast cell lysates over 7-hour after galactose induction. Proteins were probed with rabbit IgG (1:10,000 dilution of 10 mg/ml stock). Equal amounts of protein were loaded; erg4Δ strains produce less protein as indicated by relative abundance listed below (% of wild type).
(D) Cell growth (absorption at 600 nm) is affected by the ergosterol-repressing drug fluconazole in mutants of ergosterol binding protein kinases. Error bars are SEM, n=8. Dotted lines and gray legends are mutants of protein kinases that did not bind ergosterol in this study.
See also Fig. S4.
Ergosterol regulates the activity of a highly conserved kinase, Ypk1, and influences the Ssk22 levels
To determine if ergosterol binding is important for kinase function, we tested the effect of ergosterol on Ypk1 activity using in vitro kinase assays (Fig. 4B). Ypk1 is a yeast homolog of the mammalian SGK/AKT protein kinases which are involved in many important cellular processes and human disease (Brazil and Hemmings, 2001); yeast Ypk1 has been implicated in receptor-mediated endocytosis and sphingolipid-mediated signalling (Jacquier and Schneiter, 2010). Ypk1 was purified from wild type cells grown in the absence and presence of 0.4 mM ergosterol and tested for stimulation of in vitro kinase activity in the presence of increasing concentrations of ergosterol. Ypk1 activity from cells grown in the absence of ergosterol is significantly (and reproducibly) elevated in the presence of increasing amounts of ergosterol (Fig. 4B). Ypk1 activity was even higher when cells were grown in the presence of ergosterol and could be stimulated to a similar extent. Because Ypk1 was isolated from wild type cells which contain ergosterol, it might already contain bound ergosterol. We therefore purified Ypk1 protein from an erg4Δ strain which lacks ergosterol (Parks and Casey, 1995). Ypk1 protein levels were similar to preparations from wild type cells (Fig. 4C–a), however, Ypk1 activity was very low in erg4Δ strains (at least 5-fold lower) relative to wild type cells and could not be stimulated (Fig, 4B). These results demonstrate that ergosterol stimulates Ypk1 kinase activity. Since Ypk1 isolated from erg4Δ strains was low and could not be stimulated it is likely that some ergosterol must be present during Ypk1 synthesis and activation. Overall, these results demonstrate that ergosterol is critical for Ypk1 activity.
We also attempted to analyze the activity of Ssk22 (a kinase involved in osmosensing (Posas et al., 1996)) in wild type and erg4Δ cells. In multiple independent experiments we found that we could not purify Ssk22 from erg4Δ strains lacking ergosterol (Fig. 4C–a). Addition of exogenous ergosterol to the medium restored Ssk22 levels in the mutant strain. To explore this further, Ssk22 levels were examined for up to 7 hrs after expression was induced from a GAL promoter. As shown in Fig. 4B–b, copious amounts of Ssk22 protein are detected in wild type cells, but levels are substantially reduced (6 to 20-fold) in erg4Δ cells (Fig. 4C–b). Although the levels of Ssk22 were too low to measure kinase activity our results demonstrate that ergosterol is important for maintaining Ssk22 protein levels in wild type yeast.
Growth of strains deleted for Ypk1 and other ergosterol binding proteins is affected by ergosterol inhibitors
We next determined if ergosterol levels are important for the growth of yeast strains lacking ergosterol-binding kinases. Yeast strains lacking Ypk1 were shown previously to be sensitive to nystatin and fluconazole, two inhibitors of ergosterol biosynthesis (Gupta et al., 2003; Hillenmeyer et al., 2008). Six strains deleted for different ergosterol binding kinases as well as 4 strains deleted for kinases not found to bind ergosterol were grown in the presence of different concentrations of fluconazole and cell density determined. As shown in figure 4D, growth of ypk1Δ cells was inhibited by fluconazole, whereas ssk22Δ were resistant to fluconazole. Similarly, ypk1Δ cells were sensitive to nystatin (not shown). The mutants of other 4 ergosterol-binding protein kinases behaved similarly to the wild type cells and strains lacking non-ergosterol binding kinases. These results indicate that genetic interactions are evident between ergosterol-bound kinases and the ergosterol pathway.
An integrated global small metabolite-protein network
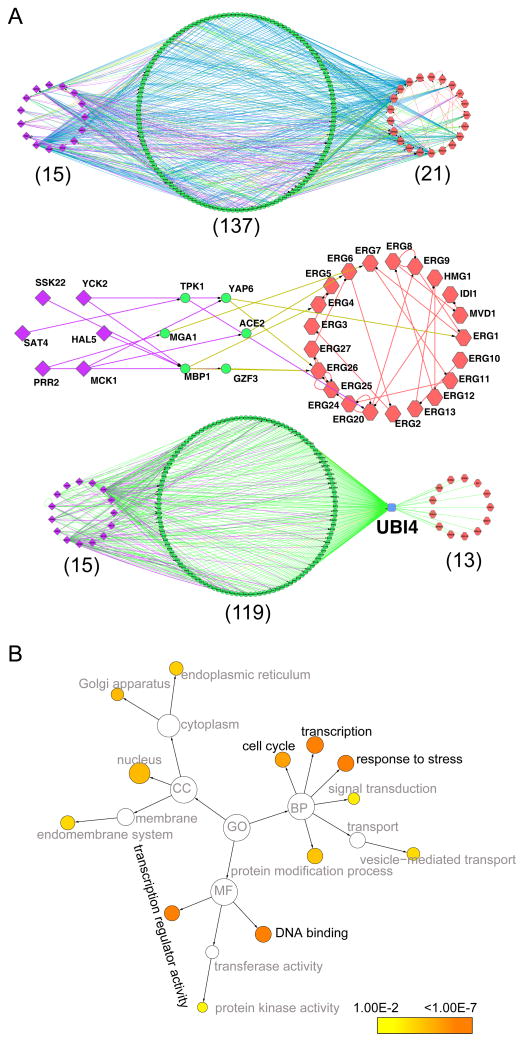
To better view how our results might be connected with other regulatory interactions, we next integrated the small metabolite-protein binding results with protein-protein interaction data, genetic interaction data and metabolite networks constructed by others. Our results suggest a highly connected network of interactions in which the small metabolites add an extra dimension of regulatory information. We found extensive interactions between the ergosterol-bound kinases, the ergosterol biosynthesis proteins and a wide variety of other cellular components. Simplification of the network to only those interactions that directly interact with the Erg pathway and ergosterol-bound kinases reveals a bipartite pattern (Fig. 5A). Interestingly, the kinases are connected to their interacting partners whereas ergosterol pathway proteins interact through genetic and phenotypic interactions. We suggest that Erg pathway components either often operate in the same pathways as the affected kinases and/or small metabolite products of the pathway affect kinase regulators and/or substrates. Overall, our results demonstrate close functional connections between ergosterol biosynthetic pathway components and ergosterol-bound kinases.
Figure 5. Integrative interactomes of proteins and small metabolites.
(A) top, A protein-protein interaction network showing intermediate proteins (green circles) that link ergosterol-binding protein kinases (purple diamonds) from this study and ergosterol biosynthetic enzymes through known physical or genetic interactions or common phenotypes (red hexagons), The edge colors denote interaction types: green for physical, blue for genetic and phenotypic, purple for phosphorylation, red for metabolic, olive for transcription factor binding; Middle, A subnetwork showing a group of intermediate proteins phosphorylated by ergosterol-bound protein kinases and regulate ergosterol enzymes through transcription and phosphorylation; Bottom, A subnetwork showing only ubiquitin (Ubi4)-interacting intermediate proteins and ergosterol enzymes.
(B) Gene ontology (GO) enrichment analysis of the 137 intermediate proteins in A top (P value <0.01 for hypergeometric test against GOSlim_Yeast).
See also Table. S3.
Within the overall interaction network are a variety of interesting interactions. For example, Erg20, an essential enzyme for both isoprenoid and ergosterol (Daum et al., 1998) is indirectly phosphorylated by ergosterol-bound protein kinases Sat4 via Tpk1 whereas 5 other enzymes, Erg1, Erg4, Erg6, Erg7 and Erg26, are transcriptionally regulated by 5 proteins (in the middle circle, Fig. 5A). In addition, 13 of 21 enzymes (62%) in the ergosterol pathway and 3 ergosterol-binding kinases (Ypk1, Yak1, Mck1) have physical interactions with the ubiquitin Ubi4. This figure is statistically significant (p-value=1.5e−5 by Fisher’s exact test), as only 18% of all yeast proteins have physical interaction with Ubi4 (Fig 5A bottom). Perhaps many components involved in ergosterol biosynthesis are modified and/or degraded by the Ubi4-mediated ubiquitination pathway.
We next conducted gene function enrichment analyses for the 137 yeast genes known to interact physically, genetically, or phenotypically with both one of the 21 ergosterol biosynthetic proteins and one of the 15 ergosterol-bound protein kinases (Fig. 5A top). Several categories of cell division and growth are particularly overrepresented, such as cell cycle, stress responses, transcription, and general metabolism of lipids, vitamins and carbohydrates (Table S3 for KEGG pathway and Fig. 5B for gene ontology). These results further suggest that ergosterol can act through modulation of protein kinase activation as a general regulator in coordinating various cellular biological processes.
Discussion
A large number of metabolite-protein interactions exist in eukaryotes
Inside a cell, most proteins presumably encounter various small metabolites, which are in vast numerical excess as they constitute 5–8% of total cell weight (Alberts, 2002) and possess a small molecular weight (<1000 Da). Although these encounters may not always have functional consequences, it is likely that many of them will be important in modulation of protein/enzyme activity. Our study is the first systematic analysis of in vivo metabolite-protein interactions and has revealed many such interactions in a eukaryotic cell. Approximately 70% of ergosterol biosynthetic proteins and 20% of protein kinases were found to bind hydrophobic molecules. If a similar percentage exists for the entire proteome as found for protein kinases, then >1200 soluble yeast proteins would bind hydrophobic molecules. The analysis of hydrophilic small metabolites is likely to increase the fraction of metabolite-binding proteins even further. Thus, a substantial number of eukaryotic proteins are likely to bind metabolites.
One important advantage of an unbiased and systematic analysis of in vivo protein-metabolite interactions is that many unexpected interactions are revealed, such as interactions of protein kinases with sterols, the binding of ergosterol biosynthetic proteins to different ergosterol-related metabolites and pentaporphyrin, a heme-related compound that presumably binds noncovalently to proteins. In many cases the results can explain previous unexplained observations. For example, yeast strains defective in heme biosynthesis fail to grow in the absence of ergosterol (Parks and Casey, 1995). Likewise, the finding that erg7 mutants fail to grow in the absence of ergosterol may be due to our observation that lanosterol binds and likely regulates many key steps in the Erg pathway. Finally, the observation that ypk1 mutants fail to grow in the presence of ergosterol inhibitors is consistent with a role for ergosterol in Ypk1 pathway or related pathway function. Thus, the advantage of an unbiased screen reveals many novel interactions that appear to be functional in vivo, and provides potential insights into previously unexplained mutant phenotypes.
An assay for in vivo protein-small metabolite interactions
Investigation of protein-small metabolite interactions is difficult for two reasons: First, sufficient quantities of proteins are required for detection, and many proteins can be difficult to purify. Second, small metabolites vary in chemical properties, which prohibits a universal detection method for untargeted profiling. We overcame these challenges by using an epitope-tagged protein expression system for systematic analysis of large numbers of proteins in yeast (Gelperin et al., 2005) and sensitive MS to detect trace amounts of small metabolites at picomole scales. We further customized our MS approach to target non-polar hydrophobic small metabolites because of the concern of losing hydrophilic metabolites during protein purification and aqueous washes (Morozov et al., 2003). The assay we developed will not detect non-volatile compounds and may miss many phospholipids (albeit see Carrier et al., 2000) and perhaps other compounds as well. Nonetheless, it is capable of detecting many diverse compounds (Tables 1,S2) and yields not only reproducible and simple mass spectra amenable for further interpretation (Fig. 2–3), but also meaningful results that could be validated using other assays (Fig. 4). With modification, this method may also be used for profiling protein-bound hydrophilic metabolites.
Our method differs significantly from several studies (Maynard et al., 2009; Morozov et al., 2003; Tagore et al., 2008) in several important aspects: 1) we detect in vivo bound small metabolites, which is analogous to co-immunoprecipitation, a gold standard method for detecting in vivo protein-protein interactions; 2) the metabolomic composition exposed to each protein of study is at its in vivo physiological state, thus avoiding bias due to overloading and/or disproportional composition of small molecules that can occur in in vitro studies. 3) Finally the procedure we have established is highly scalable and can be used to analyze large number of proteins.
Our current method cannot distinguish between physical and indirect association of protein and small metabolites, as the metabolites we identify may purify with associated proteins. Thus, the results are analogous to those for protein-protein interaction studies that use affinity purification or two hybrid methods. Since the purified protein preparations typically have a single overproduced peptide it is likely that most interactions are direct, but indirect interactions are possible. In some cases the interaction might be suggestive of a membrane association; for the cases of ergosterol-bound protein kinases none have transmembrane domains and only one, Yck2, has been shown to be membrane-associated through palmitoylation (Babu et al., 2004). Other limitations of our assay are: 1) transient interactions, as might be expected in interactions with substrates and products, may be difficult to observe. Those that are detected may prove to be important limiting steps that control biochemical flux through pathways; 2) interactions not normally present within the cell might be detected because the proteins are overproduced; 3) false negative and false positive are difficult to assess. False negatives may be transient interactions, have high off rate in aqueous phase or require particular interaction conditions (e.g. protein modifications or interacting partners); apparent false positives may be bona fide events that occur under conditions that have not been assessed previously or due to nonspecific interactions.
The metabolite-protein interactions are likely to be specific for several reasons. First, only a limited number of metabolites were found to be bound to any protein and distinct metabolites are bound to a particular protein and even between closely related proteins. In fact, the majority of yeast proteins do not bind ergosterol or other metabolites, even though most are just as abundant in our preparations as the metabolite binding proteins (Fig. S3D). Second, the stoichiometry of the protein-small metabolite interaction is usually on the order of 1:1 (or 1:2 for Erg27), suggestive of specific interaction. Third, for most of the interactions the signal intensities are highly reproducible between experiments. These results are consistent with specific interactions rather than nonspecific adsorption to protein surfaces. Thus, although some of the interactions we detect may be nonspecific, our data indicate that many of them are specific.
Small metabolites as general regulators of cellular processes
As demonstrated in this study, ergosterol, an important building material for new cell membranes, regulates the activity of Ypk1 (Schmelzle et al., 2002) and the levels of Ssk22. Ypk1 is important for stress response, cell wall integrity, lipid uptake and budding (Jacquier and Schneiter, 2010) and Ssk22 is involved in the Hog1-mediated osmosensory pathway (Posas et al., 1996). Since even low ergosterol concentrations can stimulate Ypk1 activity it is possible that Ypk1 is a biomass sensor for ergosterol levels and help coordinate cell wall synthesis and budding. The positions of both ergosterol and Ypk1 in the integrated network (Fig. 5) are consistent with key roles in coordinating these processes along with cell cycle control and other related biological processes (Table S3). Furthermore, the role of ergosterol as a coordinator of many molecular and cellular processes has general similarities to other key molecular regulators such as cAMP and phospholipid derivatives (Alberts, 2002). Since ergosterol is a critical component of cell membranes it is ideally suited to coordinate membrane synthesis with related biological processes that depend upon membrane and cell integrity such as the stress response, endocytosis, budding, cell division. The regulation by ergosterol may differ from classical signalling regulators in terms of magnitude and selectivity, because as a structural component of membranes, ergosterol serves more diverse general roles than small molecules made specifically for signalling. As such ergosterol is well situated for simultaneously monitoring cell integrity, intracellular processes and environmental interactions. In addition to possible roles as a structural component and signalling molecule, ergosterol might also serve as a facilitator of protein folding as cells grown in the absence of ergosterol have low Ypk1 kinase activity and low levels of Ssk22. Perhaps ergosterol is important for the proper folding of these enzymes and homeostasis for Ssk22.
Small metabolites and human disease
The interplay of metabolites and proteins may have profound importance in human health. Strong associations between urine metabolites and human diseases have been documented (e.g. Nicholson et al., 2008). Thus, the knowledge gained from this study may pave the way, not only to more fully understand the molecular basis of disease, but to potentially modulate specific biological pathways by manipulating the metabolite concentration through nutrient uptake or intervention. Consistent with the latter possibility, many key enzymatic steps are controlled by pharmaceuticals that are analogs of cellular metabolites; these could be used to modulate pathways regulated by metabolite interactions.
Experimental Procedures
Yeast strains and media
Yeast MORF strains expressing tagged proteins were grown and processed as described previously (Gelperin et al., 2005) Yeast knockout strains were from Yeast Genome Deletion Project. Y258 is MATa pep4-3, his4-580, ura3-53, leu2-3,112. The Y258 strain used as negative control in this study was integrated with a URA3 gene from pRS426 vector. For yeast transformation, MORF plasmids were rescued from yeast MORF strains and then introduced into the yeast using standard methods (Gietz and Woods, 2002). Standard YPAD or SC-URA media supplemented with glucose, raffinose, or galactose were used. For ergosterol treatments, a suspension at 0.05g/ml in ethanol was added to 300 volumes of a yeast raffinose culture.
Affinity purification and metabolite extraction
Frozen yeast cell pellets from 150 ml cultures were resuspended in 1ml lysis solution (200 mM NH4Ac, 1x Complete Protease Inhibitor Cocktail (Roche), 1 mM EGTA). One volume of Zirconia silica beads was added to facilitate cell lysis using a FastPrep 24 machine (MP Biomedicals) and a setting of 60 s for 3 times at 6.5 m/s. The lysis was repeated once with another 1 ml of lysis solution. The combined supernatants were then incubated with 50 μl of rabbit IgG-crosslinked M-270 epoxy Dynabeads (Invitrogen) for 2 h at 4°C. The beads were washed once in 300 mM NH4Ac and once in 150 mM NH4Ac; each wash lasted 5 min. To extract the protein-bound metabolites, 60 μl of pure methanol was added to the beads and incubated at RT for 10 min. The methanol extract was then immediately transferred to a Waters max recovery glass vial and analyzed by a mass spectrometer the same day. The beads were then boiled in 30 μl 2x SDS sample buffer for 10 min, 15 μl of the supernatant were loaded on a 4–15% SDS-PAGE gel for separation and stained by ProtoBlue Safe reagent (National Diagnostics).
UPLC-coupled APCI mass spectrometry
The mass spectrometry system used in this study was comprised of an Acquity UPLC system and a Micromass Q-TOF mass spectrometer equipped with an APCI probe (Waters Co., Milford MA). The operating software was MassLynx v4.1 (Waters Co., Milford, MA). For each run, 10 μl metabolite extract was loaded on an Acquity UPLC BEH C18 column protected by a VanGuard pre-column using a binary solvent gradient of 0 to 100% methanol in water for 10 min and 100% methanol for another 10 min. The collection mass range was 100–1500 m/z in profile scan mode to avoid missing uncommon mass adducts. The probe and source temperatures were 500 and 130 °C, respectively.
Mass spectrometry assay development and validation
Pure standards of highest available purity were prepared in methanol to 10 μM final concentration. Five μl of the solution was loaded on the same LC system using identical gradient settings. The results were then compared with the mass peaks from real samples and other known compound standards for matched retention time and monoisotopic exact mass patterns. Most identifications were performed using MarkerLynx or XCMS. For ergosterol, lanosterol and pentoporphyrin, standards were also analyzed by LC-MS (Figs. 2,3,S1).
In vitro small metabolite-protein binding assay
Purified protein kinases (in 50 mM Hepes pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 30% glycerol) were either kept on ice (native) or boiled for 10 min (denatured) before distribution in 10 μl aliquots. One μl of ergosterol stock (in 10% DMSO) or pentaporphyrin stock (in methanol) was added from a 2-fold dilution series. After incubation for 15 min at 25°C, the mixture was loaded to Zeba desalting microcolumn (Pierce) for 2 min. Ninety μl of methanol was then added to the flowthrough and vortexed briefly before loading on mass spec for quantification. A standard curve was constructed using the loading standards diluted 1000 fold in methanol. The quantification method was optimized with 10 μM ergosterol or pentaporphyrin on a TSQ Vantage triple quad (Thermo) run on SRM mode. The monitored reaction transition was 379 m/z to 239 m/z for ergosterol and 331 m/z to 282 m/z for pentaporphyrin, respectively.
In vitro Kinase activity assay
Kinase activity assay was performed on Corning 384-well plates using Z’-lyte Kinase Assay Kit-Ser/Thr 6 Peptide according to manual instruction (Invitrogen). The incubation time was 2 h at 25°C. Protein and ATP concentrations (1 mM) were determined empirically by titration according to manufacturer’s instruction.
Data analyses
The mass spectra were first analyzed in MarkerLynx SCN639 to generate a table with peak intensity, peak elemental composition, and associated proteins. Further analyses were conducted in Microsoft Excel for peak intensity averages and the two-tailed t-test comparison assuming unequal variance. Stringent thresholds were used (see Fig. S1D for the MarkerLynx settings; thresholds are in Fig. S3C legend and Table 1). The initial cut-off value of signal to noise ratio was set to ≥ 5. For XCMS analysis (Smith et al., 2006), mass spectra were first centroided and converted to netCDF format in MassLynx. UPLC and Q-TOF specific peak calling and grouping parameters were used.
Construction of interaction network
All networks were visualized using Cytoscape (Shannon et al., 2003). The raw interaction network was created by including all genes that interact with one of the ergosterol-bound kinases and one of the proteins in the Erg pathway. The interactions were gathered from BioGRID (Breitkreutz et al., 2008), transcription factor binding data (Teichmann and Babu, 2004) and phosphorylome data (Ptacek et al., 2005).
Gene function enrichment analyses
The GO enrichment graph was created using the raw interaction network after filtering phosphorylation and transcription factor binding data. The genes in the middle layer of the resulting network were analyzed by BiNGO (Maere et al., 2005).
Supplementary Material
Acknowledgments
We thank E. Davidov and X. Zhou for technical assistance, and Drs. H. Im, M. Bruno and L. Wu for comments on the manuscript. This work was supported by grants from the NIH (M.S. and M.G.).
Footnotes
Author contributions The conceptual design for the project was developed by X.L. and M.S. Laboratory experiments were performed by X.L. Data analyses were performed by X.L., T.G., K.Y. and M.G. The manuscript was prepared by X.L. and M.S.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberts B. Molecular biology of the cell. 4. New York: Garland Science; 2002. [Google Scholar]
- Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- Babu P, Deschenes RJ, Robinson LC. Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J Biol Chem. 2004;279:27138–27147. doi: 10.1074/jbc.M403071200. [DOI] [PubMed] [Google Scholar]
- Beloqui A, Guazzaroni ME, Pazos F, Vieites JM, Godoy M, Golyshina OV, Chernikova TN, Waliczek A, Silva-Rocha R, Al-ramahi Y, et al. Reactome Array: Forging a Link Between Metabolome and Genome. Science. 2009;326:252–257. doi: 10.1126/science.1174094. [DOI] [PubMed] [Google Scholar]
- Brazil DP, Hemmings BA. Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem Sci. 2001;26:657–664. doi: 10.1016/s0968-0004(01)01958-2. [DOI] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier A, Parent J, Dupuis S. Quantitation and characterization of phospholipids in pharmaceutical formulations by liquid chromatography-mass spectrometry. J Chromatogr A. 2000;876:97–109. doi: 10.1016/s0021-9673(00)00148-5. [DOI] [PubMed] [Google Scholar]
- Cascante M, Marin S. Metabolomics and fluxomics approaches. Essays Biochem. 2008;45:67–81. doi: 10.1042/BSE0450067. [DOI] [PubMed] [Google Scholar]
- Caydasi AK, Pereira G. Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev Cell. 2009;16:146–156. doi: 10.1016/j.devcel.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gupta SS, Ton VK, Beaudry V, Rulli S, Cunningham K, Rao R. Antifungal activity of amiodarone is mediated by disruption of calcium homeostasis. J Biol Chem. 2003;278:28831–28839. doi: 10.1074/jbc.M303300200. [DOI] [PubMed] [Google Scholar]
- Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Bluthgen N, Borger S, Costenoble R, Heinemann M, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak CE, Snyder M. Global analysis of gene expression in yeast. Funct Integr Genomics. 2002;2:171–180. doi: 10.1007/s10142-002-0065-3. [DOI] [PubMed] [Google Scholar]
- Hunter T, Plowman GD. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Schneiter R. Ypk1, the yeast orthologue of the human serum- and glucocorticoid-induced kinase, is required for efficient uptake of fatty acids. J Cell Sci. 2010;123:2218–2227. doi: 10.1242/jcs.063073. [DOI] [PubMed] [Google Scholar]
- Karst F, Lacroute F. Ertosterol biosynthesis in Saccharomyces cerevisiae: mutants deficient in the early steps of the pathway. Mol Gen Genet. 1977;154:269–277. doi: 10.1007/BF00571282. [DOI] [PubMed] [Google Scholar]
- Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Maynard JA, Lindquist NC, Sutherland JN, Lesuffleur A, Warrington AE, Rodriguez M, Oh SH. Surface plasmon resonance for high-throughput ligand screening of membrane-bound proteins. Biotechnol J. 2009;4:1542–1558. doi: 10.1002/biot.200900195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov VN, Morozova TY, Johnson KL, Naylor S. Parallel determination of multiple protein metabolite interactions using cell extract, protein microarrays and mass spectrometric detection. Rapid Commun Mass Spectrom. 2003;17:2430–2438. doi: 10.1002/rcm.1213. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Elliott P. The metabolome-wide association study: a new look at human disease risk factors. J Proteome Res. 2008;7:3637–3638. doi: 10.1021/pr8005099. [DOI] [PubMed] [Google Scholar]
- Parks LW, Casey WM. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Helliwell SB, Hall MN. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol Cell Biol. 2002;22:1329–1339. doi: 10.1128/mcb.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Snyder M, Gallagher JE. Systems biology from a yeast omics perspective. FEBS Lett. 2009;583:3895–3899. doi: 10.1016/j.febslet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagore R, Thomas HR, Homan EA, Munawar A, Saghatelian A. A global metabolite profiling approach to identify protein-metabolite interactions. J Am Chem Soc. 2008;130:14111–14113. doi: 10.1021/ja806463c. [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36:492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- Yano K, Fukasawa T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:1721–1726. doi: 10.1073/pnas.94.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.