Abstract
DET1 is a pleiotropic regulator of Arabidopsis development and controls the expression of many light-regulated genes. To gain a better understanding of the mechanism by which DET1 controls transcription from light-regulated promoters, we identified elements in the chlorophyll a/b-binding protein 2 (CAB2) promoter that are required for DET1-mediated expression. Using a series of reporter constructs in which the luciferase gene is controlled by CAB2 promoter fragments, we defined two DET1-responsive elements in the CAB2 promoter that are essential for proper CAB2 transcription. A 40-bp DET1 dark-response element (DtRE) is required for both dark and root-specific repression of CAB2, whereas the known CAB upstream factor-1 element is required for DET1 activation-associated effects in the light and repression in the roots. HY5, a factor that binds CAB upstream factor-1, is also required for DET1 effects in the light. DtRE binds two distinct activities in Arabidopsis seedling extracts: a novel activity with binding site CAAAACGC that we have named CAB2 DET1-associated factor 1 plus an activity that is likely to be the myb transcription factor Circadian Clock-Associated 1. Both activities are altered in dark-grown det1 extracts as compared with wild type, correlating a change in extractable DNA binding activity with a major change in CAB2 expression. We conclude that DET1 represses the CAB2 promoter in the dark by regulating the binding of two factors, CAB2 DET1-associated factor 1 and Circadian Clock-Associated 1, to the DtRE.
Plants respond to their ambient light environment via sets of photoreceptors that generate a complex web of integrated signals to control growth. Accurate and coordinated responses downstream of these primary light receptors are crucial because plants are sessile and dependent upon light as an energy source. Photomorphogenetic growth is controlled by two major types of photoreceptors, the phytochromes, which respond to red/far-red light (PHYA–E) and the cryptochromes, which respond to blue/UV-A light (CRY1/2; Briggs and Olney, 2001; Fankhauser, 2001). Excitation of these photoreceptors by light ultimately leads to selective alterations in the transcription of genes involved in growth and development (Kuno and Furuya, 2000; Ma et al., 2001; Tepperman et al., 2001). Genetic screens for mutants with altered light responses have been used successfully to identify signaling components downstream of the photoreceptors (Hudson, 2000; Neff et al., 2000). On the basis the results of genetic and biochemical studies, a preliminary model for light signaling has emerged in which the phytochrome- and cryptochrome-signaling pathways have both unique and shared components (Chory and Wu, 2001; Fankhauser, 2001; Quail, 2002a, 2002b; Schäfer and Bowler, 2002).
One such class of shared components is defined by the Arabidopsis DET/COP/FUS class of mutants (11 recessive loci). These mutants display constitutive light signaling in the absence of light and are defined by a de-etiolated, or light-grown, morphology accompanied by an increase in light-regulated gene expression in dark-grown seedlings (Hardtke and Deng, 2000; Schwechheimer and Deng, 2000). The phenotypes of mutants in this DET/COP/FUS class are pleiotropic, and epistasis studies indicate that these loci act genetically downstream of the three major photoreceptors (PHYA, PHYB, and CRY1), suggesting that the products of these genes are convergence points for the integration of many signals or are involved in processes fundamental to general signal transduction in plants (Chory, 1992; Kwok et al., 1996). One of these signaling processes appears to involve COP1/9-mediated turnover of transcription factors such as HY5 (Hardtke and Deng, 2000). However, additional mechanisms by which light signals arising from the phytochromes and cryptochromes are integrated with each other and with the DET/COP/FUS class to produce specific seedling morphology and gene expression profiles remain elusive.
One major output of light signaling is the regulated expression of genes (Kuno and Furuya, 2000; Ma et al., 2001; Tepperman et al., 2001). To affect developmental processes such as de-etiolation, changes in both nuclear and chloroplastic gene expression must occur. Changes in the abundance of more than 800 mRNAs can be observed in response to light (Ma et al., 2001; Tepperman et al., 2001; Schroeder et al., 2002). Ultimately, light signals lead to the regulation of transcription factors that bind to elements within the promoters of light-regulated genes (Terzaghi and Cashmore, 1995). Several elements necessary for promoter activity in the light are commonly found multiple times in light-regulated promoters (Kehoe et al., 1994; Terzaghi and Cashmore, 1995). Thus far, no single light-regulated element (LRE) has been found to be sufficient for light responsiveness; rather, pairings of elements appear to be required for light-regulation of a promoter (Puente et al., 1996; Chattopadhyay et al., 1998).
The nuclear encoded chlorophyll a/b-binding protein (CAB or Lhcb) gene promoters are strongly induced by light while repressed in the dark. A number of LREs have been defined by promoter deletion studies, and the factors that bind these elements have been characterized (Giuliano et al., 1988; Perisic and Lam, 1992; Williams et al., 1992; Anderson et al., 1994; Carré and Kay, 1995; Degenhardt and Tobin, 1996). A region of the CAB1 (Lhcb1*3) promoter necessary for phytochrome regulation was found to bind the MYB-related transcription factor Circadian Clock-Associated 1 (CCA1; Sun et al., 1993; Kenigsbuch and Tobin, 1995; Wang et al., 1997). CCA1 overexpression results in plants with long hypocotyls, a delayed flowering time, and abolished circadian rhythms (Wang and Tobin, 1998). CCA1 null mutants have a wild-type morphology but altered circadian rhythms (Green and Tobin, 1999). Other MYB-related factors that appear to have a role in circadian regulation have been cloned such as Late Elongated Hypocotyl, REVEILLE1, and REVEILLE2. These factors also alter circadian rhythms when overexpressed and are able to bind the same elements as CCA1 (Schaffer et al., 1998; C Andersson and S. Kay, unpublished data).
Two other elements defined as LREs have been found in the CAB2 (Lhcb1*1) promoter. The CAB2 GATA factor 1 (hereafter CGF-1/GT-1) element contributes both to the acute peak of light induction and the absolute level of induction (Anderson and Kay, 1995; Anderson et al., 1997). This element, consisting of the sequence GATAN2GATTN6GATA, is bound by tobacco CGF-1 and a closely related factor from Arabidopsis extracts, GT-1 (Anderson et al., 1994; Hiratsuka et al., 1994; Teakle and Kay, 1995). GT-1 has been shown to undergo Ca2+-dependent phosphorylation in response to light signals (Marechal et al., 1999). The CAB upstream factor 1 (CUF-1) element contains an ACGT G-box core and contributes to high levels of CAB2 expression but is not required for phytochrome or circadian regulation (Anderson et al., 1994; Anderson and Kay, 1995). Interestingly, the CUF-1 element can be bound by HY5 (Chattopadhyay et al., 1998a) a basic Leu zipper transcription factor discovered in a mutant screen for seedlings with reduced response to light (Oyama et al., 1997). Mutations in HY5 cause seedlings to be deficient in hypocotyl growth inhibition in response to red, far-red, and blue light (Chory, 1992). Mutations in HY5 are also able to partially suppress the deetiolated morphology of both the det1 and cop1 photomorphogenetic mutants (Ang and Deng, 1994; Pepper and Chory, 1997).
The mutant det1 grows as a light-grown plant in the dark with a short hypocotyl, open and expanded cotyledons, partial chloroplast development, and expression of light-regulated genes encoded by both the nuclear and chloroplastic genomes (Chory et al., 1989). Recently, genechip experiments have shown that more than half of early light-induced genes are overexpressed in dark-grown det1 further supporting a role for DET1 in the transcriptional response to light (Hu et al., 2002; Schroeder et al., 2002). Some of the genes overexpressed in the dark are those for CAB, chalcone synthase (CHS), the small subunit of ribulose bisphosphate carboxylase, nitrate reductase, and LEAFY. DET1 is thus thought to be a signal transduction component linking the perception of light to a switch in developmental program. DET1 also plays a role in the light, as evidenced by its light-grown phenotypes in both seedling and adult stages. Light-grown det1 is a pale dwarf with reduced apical dominance, reduced fertility, and ectopic expression of both CHS and CAB (Chory and Peto, 1990). The ectopic expression of CAB that occurs in the roots is associated with chloroplast development and can be seen as a more rapid greening of the root pericycle relative to wild type (Chory and Peto, 1990). Consistent with the pale leaves and cotyledons of det1, CAB is underexpressed in shoots of light-grown det1 when compared with wild type. This is in contrast with a high level of overexpression of CAB in dark-grown det1 seedlings as compared with wild type. Thus, DET1 contributes to CAB gene repression in the dark but activation in the light. Other light-related phenotypes of the det1 mutant include a shortened circadian rhythm of CAB expression and early flowering in short-day photoperiods (Millar et al., 1995; Pepper and Chory, 1997). However, DET1 regulation is not restricted to light-related genes because other gene sets are also mis-regulated in det1 under both light and dark growth conditions (Mayer et al., 1996; Schroeder et al., 2002).
DET1 encodes a nuclear-localized protein that is a component of a 350-kD complex of unknown biochemical function (Schroeder et al., 2002). The level of DET1 mRNA is constant throughout early development and does not increase with light treatment (Pepper et al., 1994; Schroeder et al., 2002). DET1 by itself does not have any detectable DNA-binding activity in vitro. However, DET1 purifies from plants as a complex with Damaged DNA-Binding Protein-1, a protein implicated in chromatin modification possibly via recruitment of histone acetyltransferase (Schroeder et al., 2002). Also, the tomato (Lycopersicon esculentum) homolog of DET1 (tDET1) has been shown to interact with the core histone H2B both in vitro and in vivo (Benvenuto et al., 2002), and the fruitfly (Drosophila melanogaster) homolog of DET1, ABO, has been shown to associate with chromatin (Berloco et al., 2001). Although specific DET1 action is yet to be determined, a mechanistic understanding of DET1 function that involves chromatin remodeling is beginning to emerge.
The CAB2 promoter is mis-regulated in four distinct developmental scenarios in det1 mutants. It is de-repressed in the dark, has reduced activation in the light, is expressed ectopically in roots, and has a shortened circadian period (Chory et al., 1990; Millar et al., 1995). Thus the CAB2 promoter is ideal for studying the transcriptional effects of DET1 on a light-regulated promoter. By defining how DET1 controls expression of the CAB2 promoter, we may begin to understand how DET1 controls light-regulated gene expression in general.
In this study, a series of CAB2 promoter fragments were fused to the luciferase reporter gene (CAB2::LUC), and their activities were compared between wild-type and det1-1 genetic backgrounds. Two elements in the CAB2 promoter were found to be required for DET1 signal transduction: a new 40-bp element, DET1 dark response element (DtRE), required for dark repression of CAB2, and the G-box element CUF-1 and its b-ZIP-binding factor HY5 for full expression of CAB2 in the light. In roots, both the DtRE and the CUF-1 element are required for DET1 effects on transcription. We characterized the factors that bind to the DtRE by using electrophoretic mobility shift analysis (EMSA) and identified a new activity, CAB2 DET1-associated factor 1 (CDA-1), that binds to the CAB2 promoter. CDA-1-binding activity is increased by light, and this increase is dependent upon the myb-transcription factor CCA1, which may also bind the DtRE. We also found that HY5 and an element to which HY5 can bind, CUF-1, are required for DET1 effects in light-grown conditions. Our data provide a framework for further exploration of DET1-signaling events at the promoter level.
RESULTS
Control of CAB2::LUC by DET1 Is Not Dependent on De-Etiolation
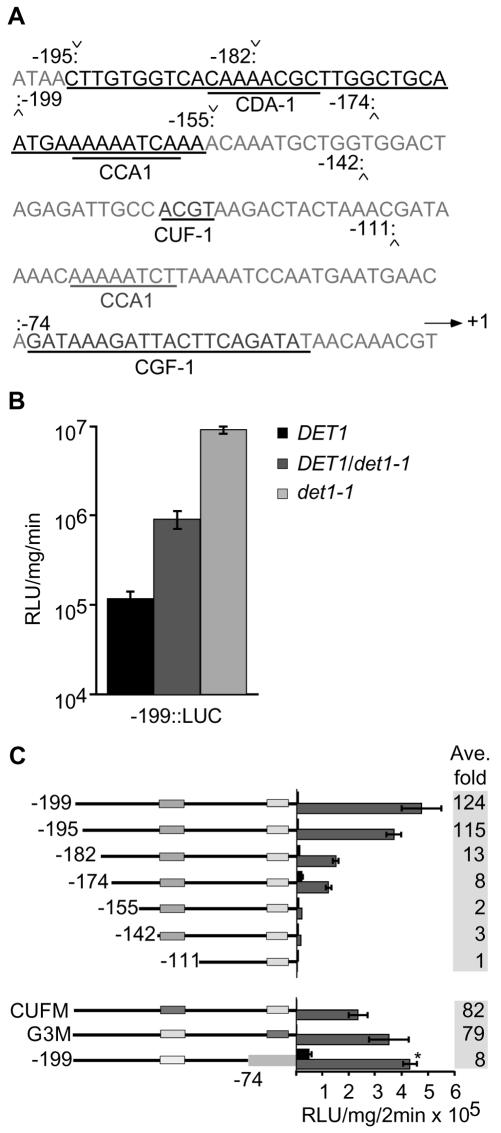
The CAB2::LUC reporter is de-repressed in dark-grown det1-1 mutants. To define promoter elements involved in DET1 repression of the CAB2 promoter, we measured the activities of a series of truncated and mutated CAB2::LUC reporters in the det1-1 background as compared with wild type. Previous studies have shown that a -199 CAB2C::LUC construct recapitulated the native promoter (Anderson et al., 1994). We measured luciferase activity from the -199CAB2::LUC (-199::LUC; consisting of -199 to +1 of the CAB2 promoter fused to luciferase) and a number of 5′ deletions: -195::LUC, -182::LUC, -174::LUC, -155::LUC, -142::LUC, and -111::LUC, as well as a -199 construct containing mutant LREs (CUFM::LUC and G3M::LUC; Anderson et al., 1994; Anderson and Kay, 1995; this work). Ten homozygous lines for each construct were selected, and two representative lines were chosen for further analysis. The selected lines were crossed into the det1-1 intermediate strength allele to locate DET1-responsive elements. The det1-1 allele contains 2% of wild-type DET1 RNA levels due to a splicing defect (Pepper et al., 1994; Pepper and Chory, 1997). A diagram of the CAB2 promoter with the positions of the truncations and relevant cis-elements is shown in Figure 1A.
Figure 1.
Regulation of the CAB2 promoter by DET1 in dark-grown seedlings is primarily dependent upon a 40-bp region between -195 and -155 and is not dependent upon a de-etiolated morphology. A, Diagram of the CAB2 promoter with the positions of the truncations used in luciferase reporter fusions indicated by chevrons. The 40-bp DtRE at -195 to -155 is indicated by a single underline. The binding sites of the two activities within the DtRE, CDA-1 and CCA1, are indicated by double underline. The core of the CUF-1 site, the entire CGF-1/GT-1 site, and a second CCA1 site is also indicated by a single underline. B, Expression of the -199::LUC transgene in different genetic backgrounds; wild type, heterozygous det1, and homozygous det1. Bars represent se. C, Expression of the CAB2::LUC constructs in dark-grown wild-type and det1 seedlings. Results from one representative experiment for each construct is shown as a bar graph (± se) with the average for three experiments expressed as the average-fold difference between wild type (black) and det1 (gray) adjacent to the graph. For -195::LUC through -155::LUC, two independent lines were used in luciferase assays. For all other constructs, one representative line was tested. *, Scale for this line is relative light units (RLU) mg-1 2 min-1 × 107.
Although det1 mutations have been described in the past as recessive, we had some indications that, at the level of gene expression, det1-1 (hereafter det1) might be semidominant (J. Chory, unpublished data). Activity of the -199::LUC reporter construct was examined in det1 homozygotes, heterozygotes, and wild-type seedlings grown in the dark. Figure 1B shows that reporter activity in dark-grown heterozygous DET1/det1 seedlings, which exhibit a wild-type etiolated morphology, is 10-fold higher than wild type. det1 thus affects CAB2::LUC transcription, but not seedling morphology, in a semidominant manner. This indicates that increased CAB2::LUC transcription in det1 mutants is not simply an indirect effect of a general pattern of de-etiolated growth.
A 40-bp Element, DtRE, Is Required for Expression of CAB2::LUC in Dark-Grown det1
Luciferase expression from the CAB2::LUC reporter series in 7-d-old dark-grown wild-type and det1(homozygous) seedlings was compared, and the results are shown in Figure 1C. The activity measured for -195::LUC was, on average, approximately 100-fold higher in dark-grown det1 than in wild type, whereas for -155::LUC, there was only a 2- to 3-fold difference in luciferase activity between det1 and wild type (P < 0.002). These results indicate that the major promoter region required for overexpression of CAB2::LUC in dark-grown det1 lies between -195 and -155 bp 5′ of the CAB2 transcriptional start site. We have designated this region the DtRE. One short truncation within the 40-bp DtRE resulted in a large loss of regulation. Specifically, a 13-bp deletion from -195 to -182 resulted in the loss of overexpression of CAB2::LUC in the det1 background from 100- to 13-fold over wild type (Fig. 1C). Further deletion of an additional 28 bp (to -155) resulted in only a 2-fold difference in expression between det1 and wild type.
Expression of CAB2::LUC in Dark-Grown det1 Does Not Require the CUF-1 or CGF-1/GT-1 Elements
Because the CUF-1 and CGF-1/GT-1 elements are known to mediate light regulation of CAB2, we examined their possible roles in DET1-mediated CAB2 regulation. The -142::LUC and -155::LUC reporter fusions retain a minimal ability to be regulated by DET1 (P < 0.001 and P < 0.002, respectively; Fig. 1C). The activity of the -142::LUC construct in the det1 background was similar to -155::LUC, and luciferase activity was the same in both det1 and wild type when the promoter was truncated to -111 (P > 0.15). Within the -142/+1 promoter region, there is the CUF-1 G-box element. The CUF-1 element contributes to a high level of CAB2 expression in light-grown conditions (Anderson and Kay, 1995) and thus may be a target of DET1 repression in the dark. To test whether this LRE was required for DET1-mediated repression in the dark, we assayed luciferase activity in the CUFM::LUC reporter line in which the CUF-1-binding site was mutated from a core G-box sequence of ACGT to AATT in the context of -199/+1. The difference in activity between det1 and wild type in the CUFM line (82-fold) was similar to that in the -199::LUC line (124-fold). Thus CUF-1 was not absolutely required for response to DET1 in the dark but may contribute to the response to some small extent. Within the -111/+1 promoter region, there is one strong CCA1-binding site and one CGF-1/GT-1 site. Although both are known to mediate light induction of the CAB2 promoter (Anderson and Kay, 1995; Anderson et al., 1997; Wang et al., 1997), in the context of -111/+1, they are not sufficient to mediate DET1 regulation. In support of these data, the mutant reporter line G3M that contains a disruption of the CGF-1/GT-1-binding site in the context of the wild-type -199 promoter shows a 79-fold increase in activity in a det1 background as compared with wild type. Again, this is similar to the difference in activity between det1 and wild type in the -199::LUC line (124-fold). In addition, a strong heterologous promoter -199 to -74CAB2::-9035S::LUC lacking CGF-1/GT-1 was repressed in the wild type, again showing that CGF-1/GT-1 is not required for DET1 to affect CAB2::LUC activity in the dark.
CCA1 is known to be involved in light signaling, and there are four CCA1-binding sites present in the CAB2 promoter (Wang et al., 1997; Green and Tobin, 1999; C. Andersson and S. Kay, unpublished data). det1 lines carrying a -199 CAB2::LUC reporter in which all four CCA1-binding sites were mutated still overexpressed the LUC reporter by more than 100-fold in the dark compared with wild type (data not shown). This indicates that these CCA1-binding sites are not required for DET1-mediated CAB2 regulation. Also, fusion of the DtRE to either -90 35S::LUC or -111::LUC did not result in increased LUC activity in the context of the det1 mutant (data not shown). This may indicate that sequences in addition to DtRE, other than those for CUF-1, CGF-1/GT-1, and CCA1, are required for DET1-mediated regulation of the CAB2 promoter in the dark.
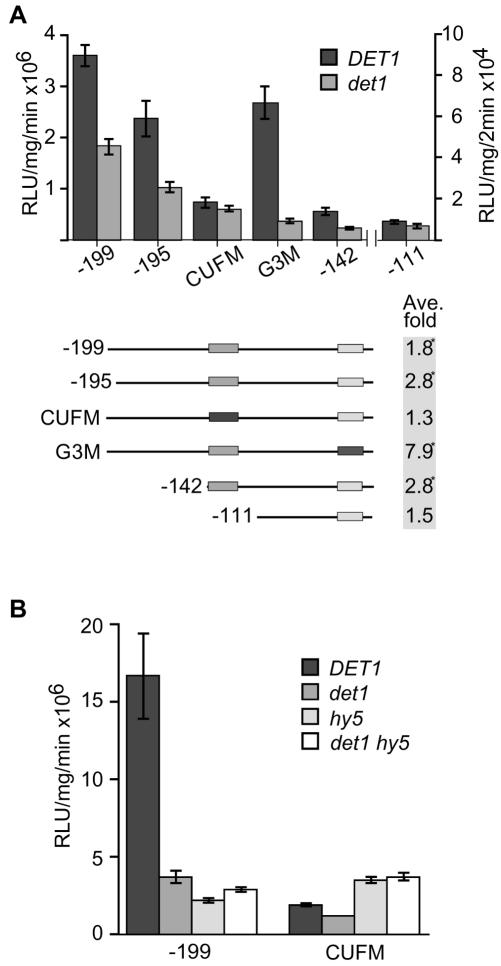
The CUF-1 Element, But Not the DtRE, Is Required for DET1 Regulation of CAB2 in the Light
CAB mRNA is underexpressed in light-grown det1 compared with wild type, which is consistent with the pale phenotype of both det1 seedlings and adults (Chory et al., 1989; Chory and Peto, 1990). The CAB2 promoter truncations were assayed in light-grown seedlings and luciferase activity compared between det1 and wild type. The results are shown in Figure 2A. Luciferase activity was 2-fold lower in light-grown det1 compared with wild type for the -199::LUC reporter (P < 0.001). The -142::LUC construct lacks the DtRE but still behaves the same with a 2.8-fold difference in expression between wild type and det1 (P < 0.001). Therefore, the DtRE is not required for DET1-mediated effects in light-grown seedlings. However, when the CUF-1 element was mutated in the context of -199/+1 (CUFM::LUC) or removed (-111::LUC), no significant difference in expression was seen between det1 and wild type (P > 0.25). Thus the CUF-1 element is required for DET1-mediated effects in the light. Previous studies have shown that this element is required for high levels of CAB2 expression (Anderson et al., 1994); in addition, this element has been shown to bind the bZIP transcription factor HY5, which promotes photomorphogenetic phenotypes (Chory, 1992; Oyama et al., 1997; Chattopadhyay et al., 1998a; C. Fankhauser and J. Chory, unpublished data). Mutation of the GT-1-binding element CGF-1/GT1 (G3M::LUC) results in an enhanced effect of the det1 mutation on CAB2::LUC expression (Fig. 2A).
Figure 2.
HY5 and DET1 are in the same pathway leading to CAB2 activation via CUF-1 in light-grown seedlings. A, Representative experiments for each reporter line are shown as a bar graph ± se. The average fold difference between light-grown wild type and det1 for three experiments is shown for each reporter construct below the graph. *, Statistically significant difference P ≤ 0.05. B, Luciferase activity driven by the -199::LUC and CUFM::LUC reporters in light-grown det1, hy5, and det1 hy5 double mutant as compared with wild type.
hy5 mutants also underexpress -199::LUC by about 2- to 3-fold compared with wild type (P < 0.001) and an intact CUF-1 element is required to generate this difference (Anderson et al., 1997; this work). We therefore tested hy5 det1 double mutants to determine the effect on CAB2 expression. Figure 2B shows that hy5 det1 double mutants have approximately the same amount of LUC expression as the single mutants for both the -199::LUC or the CUFM::LUC reporters. That is, the effects of the det1 and hy5 mutations on CAB2 expression are not additive. This suggests that HY5 and DET1 act in the same pathway leading to expression of CAB2 in the light.
Both the CUF-1 Element and the DtRE Are Required for DET1-Mediated CAB2 Regulation in Roots
Previous studies have shown that Arabidopsis roots have low levels of CAB mRNA and contain amyloplasts rather than chloroplasts. In contrast, det1 mutants that are grown on plates in the light have green roots. Although wild-type roots may develop chloroplasts after extended growth in the light, det1 roots turn green faster and in response to lower levels of light (Chory and Peto, 1990). In addition, multiple CAB promoters are overexpressed in det1 roots. To determine the elements that are required for CAB2 overexpression in det1 roots, we assayed excised roots for CAB::LUC reporter expression. Luciferase activity in the -199::LUC lines was 5-fold higher in det1 roots as compared with wild-type roots as shown in Figure 3 (P < 0.01). Mutation of the CUF-1 element (CUFM::LUC) or removal of the DtRE (-155::LUC) resulted in suppression of CAB2 overexpression in det1 roots (P > 0.7 and P > 0.3, respectively). These data show that both the CUF-1 element and the DtRE are required for this DET1 effect and that each of these elements alone is not sufficient to drive overexpression in det1 roots.
Figure 3.
Both the CUF-1 element and the DtRE (-195 to -155) are required for DET1 regulation of CAB2 in root tissue. Comparison of luciferase activity in excised roots from wild-type and det1 seedlings grown for 2.5 weeks in white light. The average reported is for three experiments, whereas the bar graph shown is for a single experiment with se bars.
Two Distinct Activities Bind the DtRE
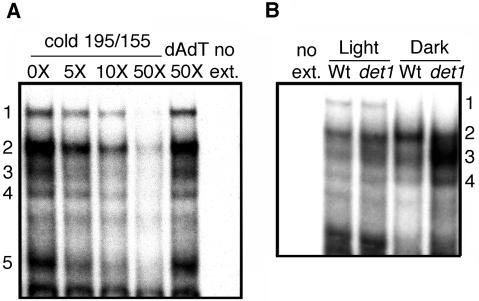
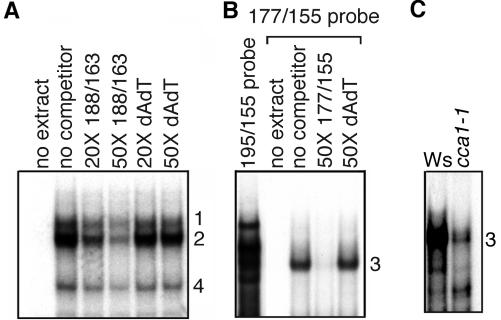
To determine how many biochemically distinct activities bind the DtRE, we first performed EMSAs using the entire 40-bp element as the probe (-195/-155). The EMSA in Figure 4A shows five activities present in wild-type light-grown extracts with specific binding to the DtRE probe. These activities were competed away by DtRE but not by a large excess of nonspecific DNA (dAdT). Binding activity 5 subsequently showed inconsistent behavior and was not followed further. Division of the DtRE probe into shorter sections resulted in the separation of activities 1, 2, and 4 (hereafter 1/2/4) from activity 3. As shown in Figure 5A, binding activities 1/2/4 bound the short probe -188/-163. Activity 3 alone bound the short probe -177/-155 as shown in Figure 5B. The short probes (-188/-163 and -177/-155) could specifically compete with the long probe (-195/-155) for a subset of activities, either 1/2/4 or 3, respectively. We therefore concluded that the activities binding to the shorter probes were the same as those binding the longer probe (data not shown). Thus there are at least two separable activities present that can bind within the DtRE.
Figure 4.
Gel-shift activities that bind specifically to the DtRE are altered in dark-grown det1 extracts versus wild type. A, EMSA using crude nuclear extracts made from wild-type seedlings grown in the light and the DtRE as a probe. Amount of cold competitor indicated along the top is a molar ratio. Numbers indicate individual activities with specificity to the DtRE. no ext., No extract. B, EMSA comparing DtRE-binding activities in wild-type and det1 extracts. Seedlings were grown under the same light or dark conditions as used for the reporter expression assays shown in Figures 1 and 2. An equal amount, 2 μg, of extracted protein was used in each binding reaction. Activities are labeled as they correspond to those in A.
Figure 5.
At least two separable activities, 1/2/4 and 3, bind the DtRE; activity 3 is greatly reduced in cca1-1 null extracts. A, EMSA using extracts from wild-type seedlings grown in the light and a probe, 188/163, covering a 26-bp region of the DtRE. The type and amount, as a molar ratio, of cold competitor is indicated above the lanes. Activities are labeled as they correspond to those in Figure 1A. B, EMSA using extracts from light-grown wild-type seedlings and 177/155 as a probe, covering a 23-bp region of the DtRE. Binding to the entire DtRE, 195/155 is shown on the left for comparison. C, EMSA comparing binding to the 177/155 probe in extracts made from CCA1 null seedlings (cca1-1) and the corresponding wild-type ecotype (Ws) grown in the light. An equal amount of extracted protein was used in each binding reaction.
DtRE-Binding Activities Are Altered in det1 Mutants
To investigate the effect of the det1 mutation on binding of activities 1/2/4 and 3 to the DtRE, we compared extracts prepared from wild-type and det1 seedlings grown in the dark. Activities 3 and 4 showed an increase and activity 2 showed a decrease in binding when extracts from dark-grown det1 were compared with wild type as shown in Figure 4B. In contrast, no significant difference was detected between wild type and det1 for light-grown seedling extracts. The slight difference in band 3 seen between light-grown det1 and wild type in Figure 4B was variable between replicates, whereas the other differences described above were consistently repeated. This is consistent with previous results showing no role for DtRE in DET1-dependent transcription of CAB2::LUC in the light (Fig. 2A). This result also demonstrates that changes in DtRE binding in det1 are specific to dark-grown conditions and are not a general effect of the det1 mutation. These changes in DtRE binding correlate with a approximately 50-fold change in CAB2::LUC expression in the dark (see Fig. 1C, compare -199::LUC with -155::LUC). The binding pattern of extracts prepared from dark-grown det1 seedlings does not mimic that of light-grown wild-type extracts. This suggests that the mechanism behind expression of CAB2 in dark-grown det1 is not the same as in light-grown wild-type seedlings. Intriguingly, binding activity 1 is light specific but is not affected in det1 mutants (Fig. 4B).
CCA1 Binds to the DtRE and Is Necessary But Not Sufficient for Overexpression of CAB2::LUC in det1
The CAB2 promoter contains four CCA1-binding sites: two strong and two weak (C. Andersson and S. Kay, unpublished data). One of the strong binding sites is contained within the DtRE; -164: AAAAATCA:-157 (see Fig. 1A). This CCA1-binding site is found within the -177/-155 probe that exclusively binds activity 3. Accordingly, we found that recombinant CCA1 is able to bind a -177/-155 probe with a mobility shift similar to that of activity 3 (data not shown). There are elevated levels of both CCA1 mRNA and protein in dark-grown det1 (Z. Wang and E. Tobin, personal communication). We hypothesized that binding activity 3 was the transcription factor CCA1. We therefore tested cca1-1 null mutant extracts for changes in this activity. As shown in Figure 5C, there was a dramatic decrease in the binding of activity 3 in cca1-1 null extracts as compared with the wild type (Ws ecotype). The residual banding may be due to other myb-like factors, such as Late Elongated Hypocotyl (LHY), present in the extracts. However, extracts from light-grown CCA1-OX lines showed the same banding pattern as wild type (data not shown). To further test whether activity 3 behaved like CCA1, we used cold competitors that contained mutations known to affect the binding affinity of recombinant CCA1 (C. Andersson and S. Kay, unpublished data) in EMSA. These mutants competed for activity 3 binding with the same reduced efficiency as they did for CCA1 (data not shown). Taken together, these data suggest that binding activity 3 is CCA1. Alternately, this activity is closely related to CCA1 in its binding characteristics and is dependent upon the presence of CCA1 for binding or expression.
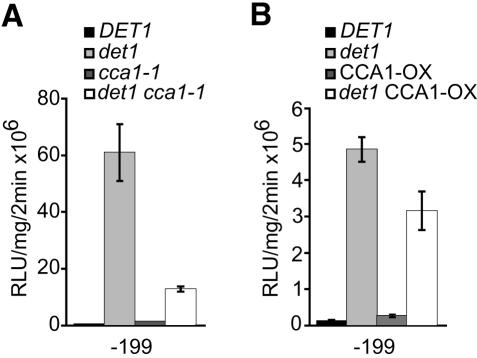
Binding activity 3 is increased in dark-grown det1 extracts, and behaves like CCA1 in EMSAs. We therefore tested cca1-1 null mutants for suppression of, and CCA1-OX for enhancement of, CAB2::LUC expression in the det1 background using the -199::LUC reporter. The cca1-1 det1 double mutant reporter lines overexpress -199 CAB2::LUC by only 10-fold as compared with the approximately 100-fold overexpression in det1 as shown in Figure 6. This shows that CCA1 is at least partially required for CAB2 overexpression in dark-grown det1. However, overexpression of CCA1 in a det1 background did not enhance CAB2::LUC overexpression (compare det1 and CCA1-OX det1 lines in Fig. 6B). Also, CAB2::LUC expression in dark-grown CCA1-OX/DET1 seedlings was the same as in wild type. Thus, CCA1 is necessary, but not sufficient, for dramatic overexpression of CAB2 in det1. Also, in a wild-type background, CCA1 overexpression alone is not sufficient to cause up-regulation of CAB2 in the dark. These data are consistent with the results from the EMSA experiments that show no difference in banding patterns between CCA1-OX and wild-type extracts (data not shown). Taken together, these data imply that the increase in activity 3 binding seen in dark-grown det1 extracts does not likely arise from CCA1 overexpression alone but is dependent upon additional effects of the det1 mutation.
Figure 6.
Overexpression of -199::LUC in det1 is dependent upon CCA1 but is not enhanced by overexpression of CCA1. A, Luciferase activity in det1, cca1-1, det1 cca1-1, and wild-type lines containing the -199::LUC reporter. B, Luciferase activity in det1, CCA1-OX, det1 CCA1-OX, and wild-type lines containing the -199::LUC reporter. Sample number (n) was ≥ 10 for each line. se bars are shown.
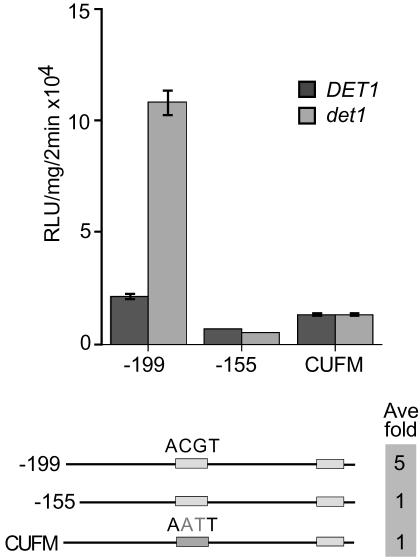
A Novel Activity, CDA-1, Binds the DtRE at CAAAACGC
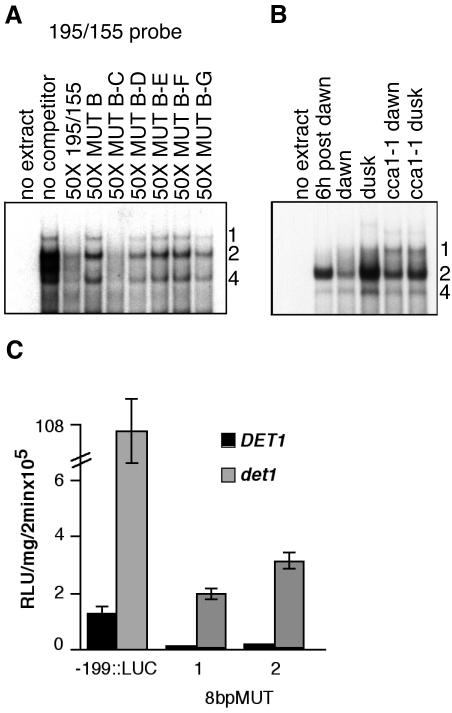
As described above, activities 1/2/4 represent a group of binding activities that are separate from binding activity 3 (CCA1). The binding of this potential complex is disrupted by mutation in a broad 10-bp region called “B” as shown in Figure 7A. We further defined the nucleotides required for binding of this activity by making a series of 2-bp mutations within region B in the context of the entire 40-bp DtRE. These mutants were used as cold competitors in EMSAs with -195/-155 as the probe. This experiment defined an 8-bp binding site CAAAACGC with a core of AAAC for activities 1/2/4 (Fig. 7A). This element, CAAAACGC, is missing in the reporter -182::LUC, which shows only approximately 10-fold overexpression of CAB2::LUC in the det1 background as compared with the 100-fold difference seen in the det1 lines harboring a -195::LUC reporter (see Fig. 1, A and C). We have named this novel binding activity CDA-1. To test whether mutation of the CDA-1-binding site could affect the ability of the CAB2 promoter to be regulated by DET1, we constructed a CAB2::LUC reporter with the 8-bp binding site mutated (8bpMUT::LUC) in the context of the -199/+1 promoter fragment. As shown in Figure 7C, mutation in the 8-bp CDA-1-binding site results in an approximate 20-fold difference in CAB2::LUC expression in the det1 background versus wild type. This is similar to the 13-fold difference between det1 and wild type seen for the -182::LUC reporter in which the CDA-1-binding site is disrupted.
Figure 7.
CDA-1 binds the site CAAAACGC, is light or circadian dependent, and is also CCA1 dependent. Mutation of the CDA-1-binding site greatly suppresses CAB2 overexpression in det1 mutants. A, EMSA using the 195/155 DtRE probe and cold competitors with 2-bp mutations along region “B” (-187 to -178). Extracts were from light-grown wild-type seedlings. Sequences of the cold competitors were: 195/155, cttgtggtcacaaaacgcttggctgcaatgaaaaaatcaaa; MUT B, cttgtggtacaccccatattggctgcaatgaaaaaatcaaa; MUT B-C, cttgtggtaccaaaacgcttggctgcaatgaaaaaatcaaa; MUT B-D, cttgtggtcaacaaacgcttggctgcaatgaaaaaatcaaa; MUT B-E, cttgtggtcacaccacgcttggctgcaatgaaaaaatcaaa; MUT B-F, cttgtggtcacaaacagcttggctgcaatgaaaaaatcaaa; and MUT B-G, cttgtggtcacaaaactattggctgcaatgaaaaaatcaaa. B, EMSA using the probe 188/163 and extracts made from wild-type or cca1-1 seedlings grown in a 12-h-light/12-h-dark cycle and collected just after lights-on (dawn), 6 h after lights-on (6 h post dawn), and just after lights-off (dusk). C, Comparison of luciferase activity in det1 versus wild-type seedlings for reporter lines containing a wild-type (-199::LUC) or mutant version (8bpMUT::LUC) of the CDA-1-binding site. Results for two independent lines carrying the mutant reporter (line 1 and 2) are shown. The CDA-1-binding site sequence was mutated to ACCCCATA from CAAAACGC.
Interestingly, CDA-1 binding increases from dawn to 6 h after dawn, then to a peak around dusk, indicating a possible light-regulation or circadian rhythm of activity as shown in Figure 7B. This change in activity is partially dependent upon CCA1 because the difference between dawn and dusk is almost entirely lost when extracts from cca1-1 are used. This dusk peak in binding activity may be in opposition to the binding of CCA1, whose expression is highest at dawn and whose binding site is only 13 bp downstream. Thus CDA-1, like CCA1, may be a light-regulated circadian clock-associated transcription factor that displays altered binding activity in det1 mutants.
DISCUSSION
As suggested by the pleiotropic phenotype of det1 mutants, DET1 regulation of gene transcription is likely to be complex. Although the CAB2 promoter has previously been well studied, we have uncovered a new element in this promoter important for DET1-mediated transcriptional regulation. By using a combination of in vivo and in vitro assays, we have identified targets for DET1 repressive effects in etiolated seedlings. These include a novel cis-element, DtRE, and its associated binding activity, CDA-1. The bulk of CAB2 regulation in the dark by DET1 is accomplished through the DtRE. This element is bound by the previously characterized myb-transcription factor CCA1, as well as the novel activity CDA-1. The CDA-1-binding site and the CCA1 protein are both partially required for overexpression of CAB2 in dark-grown det1 (Figs. 7C and 6A). In addition, changes in extractable CDA-1 activity over the day are partially dependent upon CCA1. It is interesting that these two components required for DET1 signaling bind to the CAB2 promoter within only 13 bp of each other. Sequences containing the CDA-1-binding site alone could compete for activity3/CCA1 binding (B. Maxwell and J. Chory, unpublished data). This hints that CDA-1 may be able to recruit CCA1 to DNA. Taken together, these data suggest that CDA-1 and CCA1 may interact on the promoter and that this interaction is involved in DET1 regulation of CAB2. This interaction would explain why a promoter lacking all four CCA1-binding sites is still able to be overexpressed in a det1 background (data not shown) contrary to the genetic evidence that shows that CCA1 is required (see Fig. 6A). CDA-1 may be able to recruit CCA1 to a promoter that lacks CCA1-binding sites allowing overexpression of this mutant promoter in a det1 background. In a cca1-1 null background, no CCA1 is available either to bind directly or to be recruited by CDA-1. It is also possible that CCA1 indirectly affects CDA-1 activity.
The DtRE Does Not Require the GATA Element CGF-1/GT-1 for Function in the Dark
Previous data have shown that synthetic promoters containing a G-box-GATA pair mimic native promoters under certain conditions and can be regulated by DET1 (Chattopadhyay et al., 1998b). Our data show that the G-box-GATA pair composed of CUF-1 and CGF-1/GT-1 in the CAB2::LUC reporters account for only a 2- to 3-fold difference between det1 and wild type, whereas the DtRE has a much more profound effect. In addition, the GATA-type element CGF-1/GT-1 was shown to be dispensable by two reporter-based assays for constructs G3M::LUC and -199/-74::-9035S::LUC. These constructs contain an I-box at position -109 to -106 (Carré and Kay, 1995). This I-box binds factors distinct from CGF-1/GT-1 and thus is unlikely to compensate for the G3M mutation or for the removal of CGF-1/GT-1 in -199/-74::-9035S::LUC (Carré and Kay, 1995). Thus the GATA repeat, CGF-1/GT-1, is not required for DtRE function. The DtRE fused to the CGF-1/GT-1-containing -111::LUC reporter does not confer DET1-mediated regulation (data not shown), thus the DtRE alone is not sufficient to confer DET1 regulation of CAB2. Alternatively, the distance between the DtRE and the basal transcription machinery, which is altered in the DtRE::-111::LUC construct, may be important. It should be noted that these assays reflect regulation by DET1 and not necessarily by light, and the paradigm of paired elements conferring light-dependent regulation may not hold true for DET1-mediated regulation.
DET1 Has a Complex Relationship with the CGF-1/GT-1 Element in the Light
In light-grown seedlings, the CGF-1/GT-1 element is not required for, but appears to play a complex role in, DET1-mediated regulation of the CAB2 promoter. The G3M mutant construct shows enhanced underexpression in det1 of about 7-fold as compared with the 2-fold underexpression of the wild-type promoter -199::LUC in the light (Fig. 2A). Put another way, an intact CGF-1/GT-1 element partially compensates for the det1 mutation. One possible explanation for this is that DET1 and CGF-1/GT-1 support the same interaction on the promoter but in different ways. HY5 binding at CUF-1 causes DNA bending (Q. Zhu, R. Larkin, and J. Chory, unpublished data), which is a process known to be involved in transcription factor recruitment to promoters (Pérez-Martín and de Lorenzo, 1997). CGF-1/GT-1 is required for acute induction of CAB2 by red light and binds the transcription factor GT-1, which interacts with the TFIIATBP-TATA complex (Teakle and Kay, 1995; Anderson et al., 1997; Le Gourrierec et al., 1999; Zhou, 1999). Thus, GT-1 interactions on the promoter are likely to be important for light-dependent CAB2 expression. The role of DET1 may be to allow recruitment of GT-1 to the promoter via HY5-mediated DNA bending effects, where it binds to the element CGF-1/GT-1 and associates with the basal transcription machinery. An intact CGF-1/GT-1 element would partially compensate for a lack of GT-1 recruitment in a det1 mutant background.
DET1 Requires the CUF-1 Element and HY5 for Action in the Light But Not the Dark
Although the bulk of CAB2 repression in the dark by DET1 is accomplished through the DtRE, a construct lacking the DtRE (-155::LUC) is still overexpressed by 2- to 3-fold in dark-grown det1. There may be additional targets of DET1 regulation in the CAB2 promoter between -155 and -111. This region contains the CUF-1 element. Although CUF-1 may be a target of DET1 regulation, the DtRE does not require CUF-1 for function. The DtRE may require a functional G-box but does not specifically require the CUF-1 element or the G-box/CUF-1-binding factor HY5 for function in dark-grown seedlings. The CUFM::LUC reporter line retains nearly a full response to DET1 despite a 2-bp mutation in the CUF-1 element. However, the CUFM::LUC reporter contains an intact ACGT G-box core at -48 to -45, which may partially compensate for the loss of CUF-1 (Carré and Kay, 1995). The possibility that a G-box is required is supported by the mutual requirement for DtRE and CUF-1 in roots. More detailed studies are needed to determine whether DtRE requires a G-box to be present on the promoter for function. It would be of interest to see if a synthetic G-box promoter would gain sensitivity to DET1 if DtRE, rather than GATA, were added.
HY5 binds and activates at CUF-1, is genetically downstream of DET1, and is overexpressed in det1 (C. Fankhauser and J. Chory, unpublished data; Oyama et al., 1997; Pepper and Chory, 1997; Osterlund et al., 2000). However, hy5 does not suppress the CAB overexpression phenotype and yet does suppress both the short hypocotyl and the CHS overexpression phenotypes of dark-grown det1 (Pepper and Chory, 1997). The semidominant effects of DET1 on CAB2 gene expression but not hypocotyl length imply different mechanisms behind DET1 control of these phenotypes (see Fig. 1B). Variable mechanisms for DET1 control of light-regulated gene expression in dark-grown seedlings must also exist because CHS expression appears to be controlled via HY5 whereas CAB2 is not. Interestingly, overexpression of HY5 alone affects hypocotyl length only under light-grown conditions (Ang et al., 1998). Other factors in addition to HY5 must be required for hypocotyl inhibition in the dark. Thus, the overexpression of HY5 in dark-grown det1 seedlings contributes to, but is not the sole cause of, a short hypocotyl in det1.
HY5 also mediates DET1 effects in the light. DET1 contributes to activation of the CAB2 promoter in the light via HY5 and the CUF-1 element (see Fig. 2, A and B). The lack of a simple additive effect of the det1 and hy5 mutations on the expression of CAB2 implies that HY5 and DET1 are in the same pathway. In addition, the mutual use of the CUF-1 element suggests that DET1 affects transcription by regulating HY5, which then binds and activates at CUF-1. Oddly, light-grown det1-8 mutants have been shown to accumulate more HY5 protein than wild type, which should theoretically lead to overexpression of CAB2 in the light rather than underexpression (Osterlund et al., 2000). Either another factor is limiting, or HY5 requires some sort of DET1-dependent activation. Because DET1 has been shown to bind nonacetylated H2B, a possibility is that DET1 allows HY5 access to the CUF-1-binding site by facilitating acetylation of H2B. It is also possible that while det1-8 null mutants accumulate more HY5 relative to wild type, the det1-1 mutants do not. HY5 has been shown to be phosphorylated by CK2, which results in higher binding affinity both for DNA and COP1 (Hardtke et al., 2000). DET1 could affect this CK2-dependent modification thus allowing HY5 activation. Alternatively, the det1-1 mutation could result in the down-regulation of a gene encoding a factor that limits HY5 activity. Interestingly, the phytochrome interacting factor PIF3 is able to bind the same type of G-box as HY5, and thus the CUF-1 site on the CAB2 promoter potentially represents a common site of regulation by phytochrome, PIF3, HY5, and DET1 (Martinez-Garcia et al., 2000).
The DtRE and CCA1
The altered CCA1 binding (activity 3), as well as CDA-1 binding, in det1 extracts correlates with CAB2 overexpression in the dark. Overexpression of CCA1 alone is insufficient for CAB2 overexpression in a wild-type context; the det1 mutation is required (see Fig. 6). Either another factor is limiting, or a post-translational change to CCA1 is required for activity. One, or possibly both, of these two conditions is met in the context of the det1 mutant. Because EMSAs do not reflect the effects of histone regulation, the change in binding of activities from det1 extracts does not reflect a change in DNA accessibility. CCA1 has been shown to be phosphorylated by CK2; however, CK2 phosphorylation alone is not sufficient for an increase in CCA1 binding to DNA (Sugano et al., 1998). An additional factor present in extracts is required. In det1 mutants, overexpression or modification of another factor, which allows CCA1 binding, may occur. One interesting possibility is that CDA-1 is the factor required. It is also possible that activity 3 is not CCA1 itself but a closely related factor dependent upon CCA1 for activity.
DET1 and Chromatin
DET1 requires DtRE in the dark but not the light and the CUF-1 element in the light but not the dark. This is not surprising because DET1 represses CAB2 in the dark but contributes to its activation in the light. The mechanisms behind these opposing actions are likely to be different. Requirement for both DtRE and CUF-1 for de-repression of CAB2::LUC in det1 mutant roots implies interdependence of the two elements downstream of DET1, the significance of which is unclear. However, it has been shown that HY5 mRNA is overexpressed in det1-1 roots (Oyama et al., 1997). DET1 may control the expression or activity state of both HY5 and factors that bind the DtRE in root tissue, which are dependent upon each other for activation. Alternatively, this interdependence may be based on HY5 bending of DNA allowing factors bound at the DtRE to associate with factors more proximal to the transcriptional start site. Data showing enhancement of the det1 defect when the binding element CGF-1/GT-1 is mutated in the light point to a role for DET1 in factor recruitment or activation status rather than DNA availability.
Because many genes are both overexpressed and underexpressed in det1 mutants (Schroeder et al., 2002), DET1 must simultaneously negatively and positively control transcription. One possibility is that DET1 controls the transcription of multiple genes by regulating chromatin conformation. The CDA-1 element may be used to designate those areas that are under the transcriptional control of DET1. The fruitfly homolog to DET1, ABO (25% identity, 52% similarity), has been shown to repress the transcription of specific histones during oogenesis by association with histone promoters (Berloco et al., 2001). Tomato DET1 has been shown to interact with the nonacetylated tail of histone H2B (Benvenuto et al., 2002). Arabidopsis DET1 interacts both genetically and biochemically with Damaged DNA-Binding Protein-1 whose mammalian homolog is known to interact with histone acetyltransferases (Schroeder et al., 2002). Possibly, DET1 controls chromatin conformation in Arabidopsis thereby affecting the expression of many genes, resulting in a pleiotropic mutant phenotype. In this scenario, DET1 may mediate the recruitment of complexes to the CAB2 promoter via the nonacetylated H2B tail, which has been proposed to be a “platform” for histone regulatory activity (Schreiber and Bernstein, 2002). Depending on the promoter context, light conditions, and time of day, DET1 may facilitate the assembly of histone-associated complexes involved in repression or activation. This type of reversible system would be well adapted to produce both activation and repression effects at the same binding site on the DNA, such as is the case for CUF-1. Much work remains to be done to uncover the mechanism behind DET1 control of transcription. These experiments provide a framework for further exploration of DET1-signaling events at the promoter level.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Transgenic Arabidopsis lines containing a fusion of the reporter gene firefly luciferase to the CAB2 gene promoter (CAB2::LUC) were crossed to det1-1 mutants. F3 plants homozygous for both the transgene and the det1 mutation were selected for resistance to kanamycin (50 μg mL-1) and morphological phenotype, respectively. The CAB2::LUC reporter fusions -199::LUC, -142::LUC, and -111::LUC have been previously described (Anderson et al., 1994). The CUFM::LUC and G3M::LUC constructs have been described by Anderson and Kay (1995). In brief, the CUFM::LUC line contains a mutation in the CUF-1 element that converts the core ACGT to AATT, and the G3M::LUC line contains mutations in the element GATAN2GATTN6GATA, which has been converted to CATAN2CATTN6CATA. The -199/-74::35S::LUC construct has been described by Carré and Kay (1995). The promoter deletions -195::LUC, -182::LUC, -174::LUC, and -155::LUC were created by using PCR primers that amplified the corresponding region of the CAB2 promoter to +1 and introduced a 5′ BamHI site and a 3′ HindIII site for cloning into the binary vector Vip11 in front of the luciferase reporter gene. The pMON721 based vector from which Vip11 derives is described by Millar et al. (1992). The sequence immediately 5′ of each deletion construct is CCCGGGGATCC. Arabidopsis transformation was done by the Agrobacterium sp. floral dip method (Clough and Bent, 1998). Lines segregating the transgene 3:1 in the T2 generation were carried to the T3 generation. Approximately 10 homozygous transgenic lines for each construct were tested for luciferase expression in constant light and constant dark. Two lines were then chosen to be crossed to det1, one with average and one with high expression levels of the transgene. Pair wise comparisons for each reporter line were made between det1 and wild type. All plants transformed were of the Columbia-0 ecotype.
Seeds were surface sterilized by shaking for 10 min in 33% (v/v) sodium hypochlorite and 0.1% (v/v) Tween 20. They were then rinsed in sterile dH2O, stratified in 0.1% (w/v) phytagar for 4 d at 4°C, and plated on 1× Murashige and Skoog medium (Sigma-Aldrich, St. Louis) with 1% (w/v) Suc and 0.6% (w/v) phytagar. Dark-grown seedlings were given a 2- to 12-h light treatment before being wrapped in aluminum foil and maintained in a growth chamber at 20°C for 7 d. Light-grown seedlings were grown under a 12-h-light/12-h-dark photoperiod in a growth chamber at 20°C at a fluence rate of 250 μE m-2 s-1 white fluorescent light. Light-grown seedlings were collected on d 7 at 6 h after lights-on when the peak of CAB2::LUC expression occurs in both wild type and det1 (B. Maxwell, unpublished data). For populations of seedlings germinated and grown in the dark, CAB2::LUC expression is not significantly affected by circadian timing. Seedlings from which root tissue was harvested were grown in constant white fluorescent light at 250 μE m-2 s-1 for 2.5 weeks.
For the luciferase activity assayed in DET1/det1-1 (Fig. 1B), crosses were made between DET1/DET1 and det1/det1 lines homozygous for the -199::LUC transgene. The resulting DET1/det1 seeds were germinated and grown in the dark for 7 d alongside the parental lines, and the amount of luciferase activity was determined for each.
Crosses for the double reporter mutants with CCA1-overexpressing lines (CCA1-OX) and cca1-1 were screened for the overexpression construct or the null mutation by PCR. A primer based on the Feldman T-DNA left border (FELDLFT) with a forward primer from the third exon of CCA1 (exon3 FOR) produced a 450-bp band, indicating the presence of the T-DNA insertion. The same exon3 FOR primer paired with primer exon5 REV gave a 600-bp band for the wild-type copy of CCA1 and a 133-bp band when 35S:CCA1 cDNA was present. Exon3 FOR, 5′-aaagcaacgtgaaaggtggtggactga-3′; Exon5 REV, 5′-cttaggccgtggaggaggaatag-3′; and FELDLFT, 5′-gatgcactcgaaatcagccaattttagac-3′. Crosses for the double reporter mutants with hy5 were screened by the intermediate phenotype of the double. Both light- and dark-grown hy5 det1 seedlings are intermediate in height between the two single mutants.
The reporter construct 8bpMUT was made by site-directed mutagenesis of the -199/+1 CAB2 promoter fragment in the pGEM-T vector. Primers 8bpMUT1 (5′-attaacttgtggtcaaccccatattggctgcaatgaaaa-3′) and 8bpMUT2 (5′-ttttcattgcagccaatatggggttgaccacaagttaatc-3′) were used in separate single-strand extension reactions for two cycles with an annealing temperature of 47°C. The two reactions were then mixed, and 15 more cycles of PCR were carried out. The products were digested with DpnI overnight at 37°C and transformed into Escherichia coli. The -199/+1 promoter region carrying the CDA-1-binding site mutation was then subcloned into the Vip11 binary vector, described above, for transformation into Arabidopsis.
In Vivo Promoter Analysis
Ten light-grown seedlings were collected per sample for both det1 and wild type. For dark-grown seedlings, 10 det1 or 60 wild-type etiolated seedlings were collected per sample. Sample size (n) was generally 10 for each treatment in each experiment (range 5–20). Luciferase was extracted and quantified following the manufacturer's instructions (luciferase assay system E1500, Promega, Madison, WI). Root tissue is more acidic and brings with it more fluid from the plate media. Thus, for root tissue, the amount of Reporter Lysis buffer was increased to 200 μL (versus 150 μL) per sample containing an equivalent amount of tissue. Total protein in these extracts was assayed in duplicate by the BCA system (Pierce, Rockford, IL) according to the manufacturer's instructions for the microtiter plate method. Light emission as measured by a Berthold Microplate Luminometer (LB96V, PerkinElmer Life Sciences, Boston) was expressed as RLU per milligram of total protein. Absolute RLU per milligram per minute values obtained for a given line varied between experiments (luciferase activity measured is temperature dependent and also declines linearly with time after protein extraction). Thus, comparisons between treatments/lines were only made within an experiment where temperature and time between extraction and measurement were the same. For differences between category means less than 5-fold, significance was tested for by two-tailed t tests and a P value of ≤0.05 taken to be significant (Glantz, 1997).
Nuclear Extracts
The following procedure for the enrichment of plant nuclei was adapted from Dignam et al. (1983). Seven-day-old seedlings were frozen in liquid nitrogen, placed in a ceramic mortar, coated with one-third volume of NH1 buffer (50 mm HEPES-NaOH, pH 7.9, 1.1 m Suc, 25 mm NaCl, 25 mm EDTA, 1.1 mm spermine, 1.6 mm spermidine, 5 mm dithiothreitol [DTT], 2 mm phenylmethylsulfonyl fluoride [PMSF], 1 μm leupeptin, 0.4 mm Pefablock, 0.2% [v/v] Triton X-100, 3.2% [w/v] Dextran T500, and 0.6% [w/v] polyvinylpolypyrrolidone), allowed to partially thaw, and then ground gently but completely. This lysate was divided among 1.5-mL microcentrifuge tubes at 400 μL each and spun at 6,000g for 3 min at 4°C. The supernatant was discarded, and the pellet was resuspended with a soft brush in an equal volume of NH2 buffer (50 mm HEPES-NaOH, pH 7.9, 1.1 m Suc, 25 mm NaCl, 25 mm EDTA, 1.1 mm spermine, 1.6 mm spermidine, 5 mm DTT, 2 mm PMSF, 1 μm leupeptin, and 0.4 mm Pefablock). Resuspended material was spun as before. The pellet was resuspended in an equal volume of 2× NE buffer (40 mm HEPES-NaOH pH 7.9, 0.8 m NaCl, 3 mm MgCl, 0.4 mm EDTA, 1 mm DTT, 0.1 PMSF, and 40% [v/v] glycerol) and incubated on ice for 30 min with gentle mixing every 5 min. The nuclear lysate was spun at 20,000g for 5 min saving the supernatant. A final 180,000g spin for 90 min at 4°C was performed, and the cleared solution of nuclear proteins was dialyzed against two changes of the following buffer for 1 h at 4°C using a Slide-ALyzer 2K (Pierce): 25 mm HEPES-KOH, pH 7.5, 50 mm KCl, 20% (v/v) glycerol, 0.1 mm PMSF, 0.4 mm Pefablock, 0.1 mm EDTA, and 1 mm DTT. These extracts were frozen in liquid nitrogen and stored at -80°C.
EMSAs
Oligonucleotides for 195/155 probe construction were annealed in 50 mm NaCl and 10 mm Tris-HCl, pH 7.5, at a concentration of 9.4 mm using a PCR machine as follows; 3 min at 95°C, decrease to 86°C at 1°C min-1, decrease to 76°C at 0.1°C min-1, decrease to 20°C at 2°C min-1. Oligo sequences were 5′-cttgtggtcacaaaacgcttggctgcaatgaaaaaatcaaa-3′ and 5′-tttgattttttcattgcagccaagcgttttgtg-3′ leaving an 8-bp 3′ underhang for fill-in. Endfill reactions contained 100 ng of annealed oligo, 3 pmol of 3,000 Ci mmol-1 [32P]dCTP, 0.25 mm dA/G/TP mix, and 10 units of Klenow (exo-; New England Biolabs, Beverly, MA) and was followed by a cold chase with 0.1 mm dNTPs. Sodium chloride was added to 50 mm and then Klenow was heat inactivated for 20 min at 75°C, and the reaction was subsequently cooled to 20°C at a rate of 1°C min-1. Free [32P]dCTP was removed using a 1-mL Sephadex G-25 column. Per gel shift reaction, 100,000 cpm was used, representing approximately 0.1 ng of endfilled probe. Each 20-μL reaction contained 900 ng of double-stranded poly(dA-dT), 0.1 ng of probe between 2 μg and 4.4 μg of nuclear proteins in 25 mm HEPES-KOH, pH 7.5, 10% (v/v) glycerol, 50 mm KCl, and 1 mm DTT. After 15 min of incubation on ice, the samples were loaded onto a 5% (w/v) acrylamide gel (29:1) made with 0.5× Tris-borate/EDTA buffer containing 2% (v/v) glycerol and run for 90 min at 10 W of constant power in a 4°C cold room. Gel was pre-run at 10 W during the sample incubation time. Oligos used in cold competition and the shorter probes were annealed using the same method as for the 195/155 probe. When cold competitor was added to an assay, a pre-incubation period of 15 min on ice was included before the addition of labeled probe. EMSAs were repeated with two to three independent extracts to confirm results.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Jianping Hu and Dana Schroeder for comments on the manuscript; Stacey Harmer, Rob Larkin, and Zhi-Yong Wang for sharing materials and experimental results; and Pablo Cerdan for experimental collaboration.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025114.
This work was supported by the National Science Foundation (grant no. MCB96–31390 to J.C.) and by the Howard Hughes Medical Institute. B.B.M. was partially supported by the National Institutes of Health (training grant no. HD 07495).
References
- Anderson SL, Kay SA (1995) Functional dissection of circadian clock- and phytochrome-regulated transcription of the Arabidopsis CAB2 gene. Proc Natl Acad Sci USA 92: 1500-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Somers DE, Millar AJ, Hanson K, Chory J, Kay SA (1997) Attenuation of phytochrome A and B signaling pathways by the Arabidopsis circadian clock. Plant Cell 9: 1727-1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Teakle GR, Martino-Catt SJ, Kay SA (1994) Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J 6: 457-470 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213-222 [DOI] [PubMed] [Google Scholar]
- Ang LH, Deng XW (1994) Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6: 613-628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C (2002) The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr Biol 12: 1529-1534 [DOI] [PubMed] [Google Scholar]
- Berloco M, Fanti L, Breling A, Orlando V, Pimpinelli S (2001) The maternal effect gene, abnormal oocyte (abo), of Drosophila melanogaster encodes a specific negative regulator of histones. Proc Natl Acad Sci USA 98: 12126-12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs W, Olney M (2001) Photoreceptors in plant photomorphogenesis to date: five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol 125: 85-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré IA, Kay SA (1995) Multiple DNA-protein complexes at a circadian-regulated promoter element. Plant Cell 7: 2039-2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998a) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng XW, Wei N (1998b) Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J 15: 69-77 [DOI] [PubMed] [Google Scholar]
- Chory J (1992) A genetic model for light-regulated seedling development in Arabidopsis. Development 115: 337-354 [Google Scholar]
- Chory J, Peto CA (1990) Mutations in the DET1 gene affect cell-type-specific expression of light regulated genes and chloroplast development in Arabidopsis. Proc Natl Acad Sci USA 87: 8776-8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991-999 [DOI] [PubMed] [Google Scholar]
- Chory J, Wu D (2001) Weaving the complex web of signal transduction. Plant Physiol 125: 77-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Tobin EM (1996) A DNA binding activity for one of two closely defined phytochrome regulatory elements in an Lhcb promoter is more abundant in etiolated than in green plants. Plant Cell 8: 31-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C (2001) The phytochromes, a family of red/far-red absorbing photoreceptors. J Biol Chem 276: 11453-11456 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik US, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089-7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz SA (1997) Primer of Biostatistics. McGraw-Hill, New York
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96: 4176-4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Deng XW (2000) The cell biology of the COP/DET/FUS proteins: regulating proteolysis and beyond? Plant Physiol 124: 1548-1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19: 4997-5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka K, Wu X, Fukuzawa H, Chua N-H (1994) Molecular dissection of GT-1 from Arabidopsis. Plant Cell 6: 1805-1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405-409 [DOI] [PubMed] [Google Scholar]
- Hudson ME (2000) The genetics of phytochrome signalling in Arabidopsis. Semin Cell Dev Biol 11: 475-483 [DOI] [PubMed] [Google Scholar]
- Kehoe DM, Degenhardt J, Winicov I, Tobin EM (1994) Two 10-bp regions are critical for phytochrome regulation of a Lemna gibba Lhcb gene promoter. Plant Cell 6: 1123-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenigsbuch D, Tobin EM (1995) A region of the Arabidopsis Lhcb1*3 promoter that binds to CA-1 activity is essential for high expression and phytochrome regulation. Plant Physiol 108: 1023-1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno N, Furuya M (2000) Phytochrome regulation of nuclear gene expression in plants. Semin Cell Dev Biol 11: 485-493 [DOI] [PubMed] [Google Scholar]
- Kwok SF, Piekos B, Misera S, Deng XW (1996) A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol 110: 731-742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gourrierec J, Li YF, Zhou DX (1999) Transcriptional activation by Arabidopsis GT-1 may be through interaction with TFIIA-TBP-TATA complex. Plant J 18: 663-668 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589-2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal E, Hiratsuka K, Delgado J, Nairn A, Qin J, Chait BT, Chua N-H (1999) Modulation of GT-1 DNA-binding activity by calcium-dependent phosphorylation. Plant Mol Biol 40: 373-386 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859-863 [DOI] [PubMed] [Google Scholar]
- Mayer R, Raventos D, Chua N-H (1996) det1, cop1, and cop9 mutations cause inappropriate expression of several gene sets. Plant Cell 8: 1951-1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Hiratsuka K, Chua N-H, Kay SA (1992) Firefly luciferase as a reporter of regulated gene expression in higher plants. Plant Mol Biol Rep 10: 324-337 [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua N-H, Kay SA (1995) The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163-1166 [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257-271 [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462-466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983-2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J (1994) DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78: 109-116 [DOI] [PubMed] [Google Scholar]
- Pepper AE, Chory J (1997) Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics 145: 1125-1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J, de Lorenzo V (1997) Clues and consequences of DNA bending in transcription. Annu Rev Microbiol 51: 593-628 [DOI] [PubMed] [Google Scholar]
- Perisic O, Lam E (1992) A tobacco DNA binding protein that interacts with a light-responsive box II element. Plant Cell 4: 831-838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW (1996) Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J 15: 3732-3743 [PMC free article] [PubMed] [Google Scholar]
- Quail PH (2002a) Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Cell Biol 14: 180-188 [DOI] [PubMed] [Google Scholar]
- Quail PH (2002b) Phytochrome photosensory signalling networks. Nat Rev 3: 85-93 [DOI] [PubMed] [Google Scholar]
- Schäfer E, Bowler C (2002) Phytochrome-mediated photoperception and signal transduction in higher plants. EMBO Rep 3: 1042-1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219-1229 [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Bernstein BE (2002) Signaling network model of chromatin. Cell 111: 771-778 [DOI] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J (2002) De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol 12: 1462-1472 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Deng XW (2000) The COP/DET/FUS proteins: regulators of eukaryotic growth and development. Semin Cell Dev Biol 11: 495-503 [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM (1998) Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA 95: 11020-11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Doxsee RA, Harel E, Tobin EM (1993) CA-1, a novel phosphoprotein, interacts with the promoter of the cab140 gene in Arabidopsis and is undetectable in det1 mutant seedlings. Plant Cell 5: 109-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teakle GR, Kay SA (1995) The GATA-binding protein CGF-1 is closely related to GT-1. Plant Mol Biol 29: 1253-1266 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437-9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Light regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46: 445-474 [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491-507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207-1217 [DOI] [PubMed] [Google Scholar]
- Williams ME, Foster R, Chua N-H (1992) Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell 4: 485-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D-X (1999) Regulatory mechanisms of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci 4: 210-214 [DOI] [PubMed] [Google Scholar]