Abstract
Phytochrome A (phyA) is the photolabile plant light receptor that mediates broad spectrum very low-fluence responses and high irradiance responses to continuous far-red light (FRc). An Arabidopsis mutant laf3-1 (long after far-red 3) was recovered from a screen for transposon-tagged mutants that exhibit reduced inhibition of hypocotyl elongation in FRc. The laf phenotype correlated well with a strongly attenuated disappearance of XTR7 transcript in FRc. The effects of laf3-1 on phyA-controlled CAB, CHS, and PET H expression were more subtle, and the mutation had no clear effects on PET E and ASN1 transcript levels in FRc. The use of two alternative transcription initiation sites in the LAF3 gene generates two isoforms that differ only at their N termini. Transcripts encoding both isoforms were induced during germination and were present at slightly higher levels in de-etiolated seedlings than in those grown in darkness. No significant differential regulation of the two isoforms was observed upon exposure to either FRc or continuous red light. Transcripts encoding the shorter isoform (LAF3ISF2) always appear to be more abundant than those encoding the longer isoform (LAF3ISF1). However, both isoforms were capable of full complementation of the laf3-1 hypocotyl phenotype in FRc. When fused to a yellow fluorescent protein, both isoforms localize to the perinuclear region, suggesting that LAF3 encodes a product that might regulate nucleo-cytoplasmic trafficking of an intermediate(s) involved in phyA signal transduction.
Phytochromes are soluble chromoproteins that regulate plant growth and development by their ability to interconvert between two stable spectral forms. Red light (R) converts the R-absorbing Pr form (the form synthesized in darkness) to the Pfr (far-red absorbing form) and far-red light (FR) reconverts Pfr to Pr. Two important features distinguish phytochrome A (phyA) from the other four phytochromes in Arabidopsis. First, although the Arabidopsis phytochromes phyB to phyE are activated exclusively by R and inactivated by irradiation with FR, phyA can be activated by FR and low fluences of R and blue light (B). Secondly, although phyA levels decrease rapidly after exposure to light as a result of both down-regulation of PHYA gene transcription (Cantón and Quail, 1999) and far greater photolability of phyA than other phytochromes (Sharrock and Clack, 2002), phyA is by far the most abundant phytochrome in etiolated seedlings. These features enable phyA to perform a seminal role in triggering the shift between skotomorphogenesis and photomorphogenesis. Together, they ensure that the inhibition of hypocotyl elongation, and the activation of physiological changes needed to ensure photosynthetic competence are already underway when seedlings emerge from the soil surface. Despite its lability in light-grown plants, the influence of phyA throughout the life cycle is evidenced by its involvement in sensing photoperiod to ensure that flowering is initiated at the proper time (Johnson et al., 1994; Yanovsky and Kay, 2002) and its requirement in mature plants for activation of the psbD promoter by B (Thum et al., 2001).
Extensive changes in gene expression underlie the dramatic shift between etiolated seedling growth and photomorphogenesis (Ma et al., 2001). Expression profiling also suggests that phyA initiates photomorphogenesis primarily by rapidly targeting the promoters of a set of transcription factors, which are in turn responsible for initiating a cascade of transcriptional induction or repression events (Tepperman et al., 2001). This conceivably enables orchestration of the expression of multiple downstream target genes in a highly branched signal transduction network that permits integration of phyA inputs with other environmental cues and endogenous developmental signals (Møller et al., 2002; Wang et al., 2002). Our current insight into the cell biology of phytochrome signaling (Møller et al., 2002) further validates the importance of regulated gene expression in phyA action. Although phyA appears to reside exclusively in the cytoplasm of seedlings grown in darkness, conditions known to activate phyA responses trigger a rapid translocation of some, but not all, phyA into the nucleus (Nagy and Schäfer, 2002). Considerable interest centers around the likely importance of the association of nuclear phyA with light-dependent transcriptional complexes (Fairchild et al., 2000), and it seems likely that the regulation of phyA localization may be critical to many aspects of phyA activity. The nuclear localization of COP1, a repressor of photomorphogenesis, also appears to be important for its role in targeting photomorphogenesis-promoting factors such as HY5 and LAF1 for degradation (von Arnim et al., 1997; Osterlund et al., 2000; Seo et al., 2003).
Although the majority of intermediates currently known to be essential for normal seedling responsiveness to activated phyA are predicted to localize to the nucleus (Hoecker et al., 1999; Hudson et al., 1999; Fairchild et al., 2000; Soh et al., 2000; Ballesteros et al., 2001; Dieterle et al., 2001; Wang and Deng, 2002), others are found exclusively in the cytoplasm (Bolle et al., 2000; Hsieh et al., 2000; Guo et al., 2001) or in both compartments (Choi et al., 1999; Desnos et al., 2001; Zeidler et al., 2001). Plastidic events (Møller et al., 2001) and peroxisomal processes (Hu et al., 2002) also contribute to an intact phyA signaling network. Most of the factors known to be essential for phyA signaling appear to be soluble proteins. There is presently little insight into the functional relationships between these signaling intermediates or the role of the majority of activated phyA that remains in the cytoplasm, even under conditions that are optimal for nuclear translocation (Nagy and Schäfer, 2002). Our insight into how these intermediates might relate to the apparent importance of phyA's protein kinase activity (Yeh and Lagarias, 1998; Fankhauser et al., 1999; Cólon-Carmona et al., 2000; Kim et al., 2002), the involvement of G proteins and fairly ubiquitous secondary messengers (Bowler et al., 1994; Guo et al., 2001; Kang et al., 2001), and the likely importance of protein degradation in regulating phyA-triggered photomorphogenesis (Dieterle et al., 2001) is also extremely limited. A more global view of the hierarchical arrangement of the intermediates and how closely they act within the phyA signaling network is likely to remain refractory until additional players in the phyA signaling pathway are identified.
Against this background, we have continued to search for mutants specifically compromised in the inhibition of hypocotyl elongation in continuous farred light (FRc), but which are unaffected in the phyA-mediated block of greening in white (W) light after pre-irradiation with FRc (Barnes et al., 1996a). Here, we report characterization of a laf (long after far-red) mutant defective in a protein (LAF3) that is located in the nuclear periphery. LAF3 encodes two isoforms with homology to a conserved family of bacterial proteins. The subcellular localization of LAF3 and fairly selective misregulation of phyA-controlled genes associated with its loss suggests that LAF3 participates in transmission of certain aspects of the phyA signal from the site of phyA activation in the cytoplasm of etiolated seedlings to the promoters of the genes that define certain downstream targets of activated phyA.
RESULTS
laf3-1 Is Impaired in phyA Signaling
A collection of Arabidopsis Ds-tagged lines (Sundaresan et al., 1995) was screened for reduced inhibition of hypocotyl elongation in FRc and one mutant, laf3-1 (long after far red 3), possessed longer hypocotyls under both low and high fluences of FRc, but no significant hypocotyl elongation phenotype in darkness or continuous red light (Rc), B, or W light (Fig. 1A). This indicates that the role of the LAF3 locus in inhibiting hypocotyl elongation is specific to the phyA photoreceptor (Fig. 1A). Under both fluences of FRc tested, laf3-1 hypocotyl lengths were always intermediate between those observed in WT and phyA null mutant seedlings, indicating a partial loss of responsiveness to FRc and no significant fluence dependence.
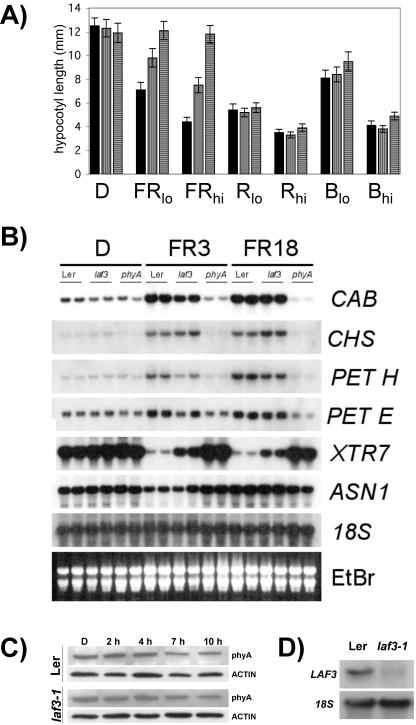
Figure 1.
Seedling phenotypes of laf3-1. A, Hypocotyl lengths of wild-type (WT; Landsberg erecta [Ler], solid bars), laf3-1 (vertical stripes), and phyA-201 (horizontal stripes) seedlings in darkness (D) and after irradiation with FRc (FRlo, 2.1 μmol m-2 s-1; FRhi, 6.3 μmol m-2 s-1), R (Rlo, 10.1 μmol m-2 s-1; Rhi, 27.4 μmol m-2 s-1), and B (Blo, 2.3 μmol m-2 s-1; Bhi, 9.4 μmol m-2 s-1) light. Hypocotyl lengths were measured after 4 d of growth under the indicated light condition. Values are means ± sd (n ≥ 80). All measurements were repeated at least three times with comparable results. B, Northern-blot analysis of phyA-regulated genes in WT (Ler), laf3-1, and phyA. Total RNA was harvested from 4-d-old seedlings grown in darkness, without exposure to FR (D) or after 3 h (FR3) or 18 h (FR18) irradiation with FRc (3.9 μmol m-2 s-1). Each lane contained 15 μg of total RNA. Duplicate samples for each treatment were from seedlings grown independently under identical conditions. The same blot was probed for transcripts encoding CAB, CHS, PET H, PET E, XTR7, and ASN1. Ethidium bromide staining and hybridization with an 18S rRNA probe are shown as loading controls. C, Western analysis of phyA abundance in 4-d-old etiolated laf3-1 and Ler seedlings without prior exposure to light (D) or after 2, 4, 7, or 10 h of exposure to FRc (5.3 μmol m-2 s-1). The mAA1 antibody did not detect any phyA protein (approximately 120 kD) in the phyA-201 null mutant (data not shown; Shinomura et al., 1996). A single band recognized by an actin antibody (approximately 45 kD) indicates comparable total protein levels. D, laf3-1 possesses severely reduced levels of LAF3 transcript. A 1.3-kb cDNA probe corresponding to the 3′ ends of both LAF3 isoforms was used to detect the combined levels of both transcripts in 10-d-old light-grown seedlings.
PCR analysis of a backcross-derived F2 population of 120 plants indicated that the laf3-1 hypocotyl phenotype cosegregated with kanamycin resistance conferred by a single Ds element and was inherited in a Mendelian fashion consistent with the presence of a single recessive mutation. The abundance of phyA is an important determinant of the sensitivity of seedling photomorphogenesis in FRc. Immunoblot analysis of WT and laf3-1 seedlings grown in darkness indicated that levels of phyA are not compromised in etiolated laf3-1 seedlings (Fig. 1C). Furthermore, the relatively slow disappearance of phyA in FRc to a steady-state level is comparable between laf3-1 and Ler (Fig. 1C). Therefore, the reduced responsiveness of laf3-1 to FRc does not result from a lower level of phyA in etiolated laf3-1 seedlings or from reduced rates of phyA degradation in FRc.
Arabidopsis seedlings with an intact phyA signaling pathway initiate chloroplast development in FRc but chloroplasts undergo photobleaching after subsequent W light treatment. In contrast, the phyA, fhy1/pat3, fhy3, and pat1 mutants are insensitive to this block of greening (Barnes et al., 1996a; Bolle et al., 2000). Germination and growth of laf3-1 and WT seedlings at very low fluences of FRc (0.4 μmol m-2 s-1) indicated that laf3-1 seedlings showed similar FR-induced photobleaching characteristics as WT seedlings (data not shown). Moreover, defects in apical hook opening, cotyledon expansion, and loss of negative gravitropism are not apparent in laf3-1, even at low fluences of FRc (data not shown).
Gene expression analysis indicated that of six phyA-regulated transcripts tested, laf3-1 was most strongly affected in FRc-mediated disappearance of a transcript encoding a xyloglucan endotransglycosylase-related protein (XTR7). However, XTR7 levels in FRc-irradiated laf3-1 seedlings were intermediate between those found in WT and phyA backgrounds (Fig. 1B). The effects of laf3-1 on FRc-stimulated increases in transcripts encoding CAB (chlorophyll a/b-binding) protein and ferredoxin: NADP(H) oxidoreductase (PET H) abundance were more subtle. Surprisingly, CHS (chalcone synthase) transcript levels in laf3-1 were slightly more elevated than in WT seedlings after both 3 and 18 h of exposure to FRc (Fig. 1B). The laf3-1 mutation had no clear effects on either plastocyanin (PET E) or Asn synthetase (ASN1) transcript levels in FRc.
The laf3-1 mutation does not appear to have any obvious developmental or morphological manifestations in mature plants. Under extended short-day conditions (8 h of high-intensity fluorescent light, 8 h of low-intensity incandescent light, and 8 h of darkness), the flowering time of laf3-1 plants and the number of leaves at the time of bolting were not significantly different from WT (data not shown). This was in sharp contrast to phyA mutants that showed delayed flowering under the same conditions (Johnson et al., 1994; Yanovsky and Kay, 2002).
LAF3 Encodes Two Isoforms with Homology to a Conserved Family of Bacterial Proteins
The flanking region of the gene disrupted by the Ds insertion in laf3-1 was cloned using inverse PCR. A database search using the TBLAST algorithm, and the tagged sequence as the query input indicated that the Ds element was inserted within an intron of a predicted open reading frame (ORF) near the bottom of Arabidopsis chromosome 3 (At3g55850 on bacterial artificial chromosome F27K19 in the vicinity of the marker SGCSNP86). Northern-blot analysis of laf3-1 and WT seedlings demonstrated substantially reduced levels of total LAF3 transcript (Fig. 1D). Thus far, no other mutations causing an FR-specific long-hypocotyl phenotype are found in the vicinity of the LAF3 locus. The only other locus on chromosome 3 implicated specifically in phyA signaling is FHY3 (Wang and Deng, 2002), found more than 12.9 Mb from LAF3. Screening of a flower cDNA library (Weigel et al., 1992) using this flanking region as a probe enabled isolation of a 1.3-kb cDNA that was complete at the 3′ end.
Interestingly, 5′-RACE analysis revealed that two isoforms of the LAF3 gene product are expressed in Arabidopsis seedlings. Neither isoform is identical to the gene product predicted by annotation of the Arabidopsis genome. Although the start codon of the first isoform (LAF3ISF1; 583 amino acids; accession no. AY295343) has been predicted correctly, the acceptor site of the second intron was predicted incorrectly, and the transcript encoding LAF3ISF1 contains 15 introns (Fig. 2A) instead of 14 introns. The transcription start site and start codon of the second isoform (LAF3ISF2; 576 amino acids; accession no. AY295344) both occur within the first intron of the ORF encoding LAF3ISF1 (Fig. 2A). Because there is an in-frame fusion of the first eight amino acids of LAF3ISF2 to the codon for Ala-16 of LAF3ISF1 and all other introns are identically spliced, the two isoforms differ only in the first few N-terminal amino acids. Transcription start sites for both isoforms are preceded by putative TATA boxes TATAAAT and TTATTT, which are found 43 and 101 bp upstream of the transcription start sites of LAF3ISF1 and LAF3ISF2, respectively. Database searches also indicated a single LAF3 cDNA in Arabidopsis (accession no. AY057597) that encodes LAF3ISF2. No coding regions are predicted to lie within the 3-kb upstream region of LAF3ISF1 or within the 3-kb downstream region of either isoform; moreover, no expressed sequence tags have been identified in these regions.
Figure 2.
A, Schematic representation of both coding regions of the LAF3 gene deduced from the comparison between the genomic sequence (accession no. AL163832) and cDNA sequences determined by RACE and reverse transcriptase (RT)-PCR. Black boxes, Exons encoding identical regions in isoforms. The transcription start site and start codon of LAF3ISF2 occur within the first intron of the primary LAF3ISF1 transcript, giving rise to isoforms differing in their N termini. The unique N termini of LAF3ISF1 and LAF3ISF2 are indicated by vertical and horizontal shading, respectively. The site of the transposon insertion in laf3-1 in the 14th intron of the LAF3ISF1 ORF and 13th intron of the LAF3ISF2 ORF is indicated by an arrow. B, Divergent N-terminal stretches of both LAF3 isoforms are boxed. Regions which are predicted to be transmembrane domains using five independent algorithms are underlined. Remarkably, the first portions of these regions span N-terminal stretches where the two isoforms are encoded by different DNA templates. Amino acids encoded by the same exon are in bold. Default parameters were used for the TMHMM (http://www.cbs.dtu.dk/services/TMHMM), PHDhtm, (http://www.embl-heidelberg.de/predictprotein), HMMtop (http://www.enzim.hu/hmmtop), TM-Pred (http://www.ch.embnet.org), and PSORT (http://psort.nibb.ac.jp) algorithms to predict transmembrane helices in the N termini of both LAF3 isoforms. Transmembrane segments predicted by the individual programs were considered overlapping if 12 or more amino acids were shared by all predicted segments. C, Alignment of two highly conserved regions in both LAF3 isoforms with homologous regions of putative proteins from rice (Oryza sativa), bacteria, and lower eukaryotes. Accession numbers of sequences and a complete alignment of LAF3ISF1 with 48 homologs are provided in Supplemental Data.
Sequence analysis did not reveal any extensive regions of similarity within either isoform with known protein domains, although several bacterial and archaeal hydrolases bear strong similarity only to an amino-terminal region of LAF3 indicated in Figure 2C. Both isoforms are predicted to contain a membrane-spanning region at their N termini (Fig. 2B). Interestingly, this is predicted to occur in the region where both proteins differ partly in their amino acid sequence. The LAF3 isoforms show closest similarity to a predicted rice gene product (554 amino acids; accession no. BAB64699) that is approximately 59% identical to the Arabidopsis LAF3ISF1 protein and, moreover, is predicted to contain a transmembrane helical region between Phe-7 and Leu-21. Curiously, the only other homologs in the database are from 45 different bacteria and the lower eukaryotes Neurospora crassa and Leishmania major. The only homolog for which any function is known is that from Pectobacterium carotovora, for which the LAF3 homolog is encoded by the aepA (activator of extracellular protein production A) locus. An aepA- knockout in this soft rot bacterium (previously in the genus Erwinia) was associated with reduced phytopathogenicity associated with coordinated down-regulation of genes encoding cell wall-degrading enzymes such as pectate lyase, polygalacturonase, cellulose, and protease (Liu et al., 1993). The molecular masses of these proteins range from 42 to 76 kD (both Arabidopsis LAF3 isoforms are around 63 kD) with an average of 59 kD. The estimated pI values of the 48 proteins range from 4.4 to 8.7 with an average pI of 5.9. The pIs of the Arabidopsis LAF3 isoforms are approximately 5.3. When compared with the LAF3ISF1 isoform, amino acid identities range between approximately 30% for the Deinococcus radiodurans, Novosphingobium aromaticivorans, Ralstonia metallidurans, Halobacterium sp. NRC-1, Thermoanaerobacter tengcongensis, Chloroflexus aurantiacus, and Bacillus halodurans homologs to around 17% for Aquifex aeolicus. There appear to be no homologs in animals. The homology between these proteins is likely to be significant because after alignment of all 48 homologs with LAF3ISF1, the LAF3ISF1 residues His-389, His-421, and Asp-485 displayed complete conservation; residues His-113, His-115, Gly-264, Gly-342, and Gly-542 were conserved in all but one of the homologs; and Pro-107, Pro-182, and Ala-550 were conserved in all but two homologs (all numbers pertain to LAF3ISF1; a complete alignment is provided as Supplemental Data available in the online version of this article at http://www.plantphysiol.org). An additional 11 residues of both Arabidopsis LAF3 isoforms were conserved in more than 85% of the 48 homologs, and another 20 residues displayed conservation in more than 70% of the proteins.
Genetic Complementation of laf3-1
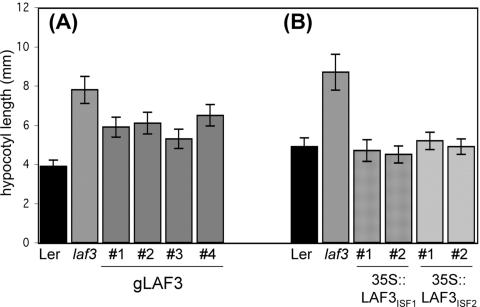
To confirm that the laf3-1 phenotype resulted from a single transposon insertion and to test whether only one or both of the LAF3 isoforms are required for an intact phyA signal transduction pathway, we tested the ability of three constructs to complement the laf3-1 hypocotyl phenotype in FRc. First, laf3-1 mutants were transformed with an 8,067-bp genomic fragment extending from 3,050 bp upstream of the start codon of LAF3ISF1 to 1,293 bp downstream of the stop codon of both isoforms. Of 23 T1 transformants, four transgenic lines displayed partial complementation (Fig. 3). The frequency of complementation was greater in laf3-1 mutants transformed with the 35S::LAF3ISF1 and 35S::LAF3ISF2 constructs, in which expression of both isoforms was under control of the CaMV 35S promoter. Of 65 35S::LAF3ISF1 transformants tested, 42 were indistinguishable from WT plants after 4 d of FRc light treatment, and of 63 35S::LAF3ISF2 transformants tested, 33 displayed clear complementation (Fig. 3). The greater efficacy of CaMV 35S-regulated transcription in complementing the mutant in comparison with the endogenous promoter suggests that possibly additional regulatory regions outside of the 3 kb upstream of the start codon or an additional factor(s), e.g. chromosomal modification of the LAF3 promoter, might normally regulate LAF3 expression.
Figure 3.
Expression of LAF3 restores the WT hypocotyl phenotype to laf3-1 mutant seedlings. A, Hypocotyl lengths of WT (Ler -0), laf3-1, and four independent transgenic laf3-1 lines (T3 generation) expressing the gLAF3 construct. The fluence of FRc was 5.7 μmol m-2 s-1. B, Hypocotyl lengths of WT (Ler -0), laf3-1, and transgenic laf3-1 lines (T2 generation) expressing either the LAF3ISF1 or LAF3ISF2 cDNA under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The fluence of FRc was 4.3 μmol m-2 s-1. For all experiments, values are means ± sd (n ≥ 80). Hypocotyl lengths were measured after 4 d of growth in darkness or FRc.
Both Isoforms of LAF3 Localize in the Perinuclear Region
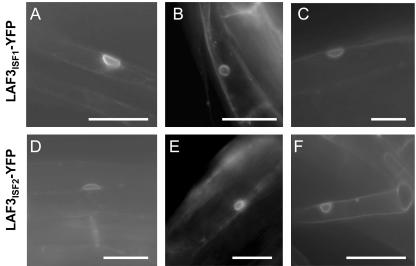
Preliminary studies involving transient expression of LAF3ISF1-yellow fluorescent protein (YFP) and LAF3ISF2-YFP fusions in onion (Allium cepa) epidermal cells indicated that both isoforms were targeted to the nuclear periphery with no significant fluorescence within the cytoplasm, nucleus, or cell periphery (data not shown). The same preferential perinuclear distribution was observed when LAF3ISF1-YFP and LAF3ISF2-YFP were constitutively expressed under the control of CaMV 35S in laf3-1 seedlings (Fig. 4). There was no evidence of fluorescence in the cortical endoplasmic reticulum, and visualization with both cyan fluorescent protein and YFP filters suggested that any signal associated with the plasma membrane was artifactual. Irradiation with light did not affect the subcellular distribution of either LAF3 fusion in etiolated seedlings. Fusion of either isoform to the N terminus of YFP did not significantly affect the ability of both isoforms to complement the laf3-1 phenotype (data not shown).
Figure 4.
LAF3-YFP localizes to the nuclear periphery in stably transformed laf3-1 seedlings. A signal with significant enrichment in the nuclear periphery was observed in several independent lines expressing 35S::LAF3ISF1-YFP in hypocotyl (A and B) or root (C) cells. The same distribution of signal was observed in lines expressing 35S::LAF3ISF2-YFP in hypocotyl (D and E) or root (F) cells. Scale bars = 20 μm.
Expression of LAF3 Transcripts
Preliminary northern-blot analysis using a probe incapable of distinguishing between both isoforms suggested that an approximately 2-kb LAF3 transcript was detected at highly reduced levels in laf3-1 compared with WT seedlings (Fig. 1D). Examination of the tissue-specific regulation of LAF3 transcripts using the same probe indicated a comparable low level of expression in seedlings grown in W light, rosette leaves, stems, flowers, and siliques, with an approximately 2- to 3-fold higher abundance in roots (data not shown). Examination of the light regulation of LAF3 expression in WT plants using this probe suggested a slight and relatively slow increase in LAF3 transcript abundance after irradiation of etiolated seedlings with FRc (data not shown).
The demonstration that LAF3 encodes two protein isoforms with distinct N termini introduces the interesting question of whether the choice of LAF3 transcript initiation site has important regulatory implications for LAF3 function. To validate the northern-blot analyses and reveal any differential regulation of both isoforms, we employed mimic-controlled RT-PCR analysis. Oligonucleotides were designed to amplify regions encoding the divergent N termini of both isoforms and the first 73 amino acids common to both LAF3ISF1 and LAF3ISF2. The two exogenous standards used in competitive PCR had the same oligonucleotide templates as the target cDNA but generated smaller PCR products than the target DNA. Depending on the relative ratio of mimic to target, one or the other will be amplified preferentially.
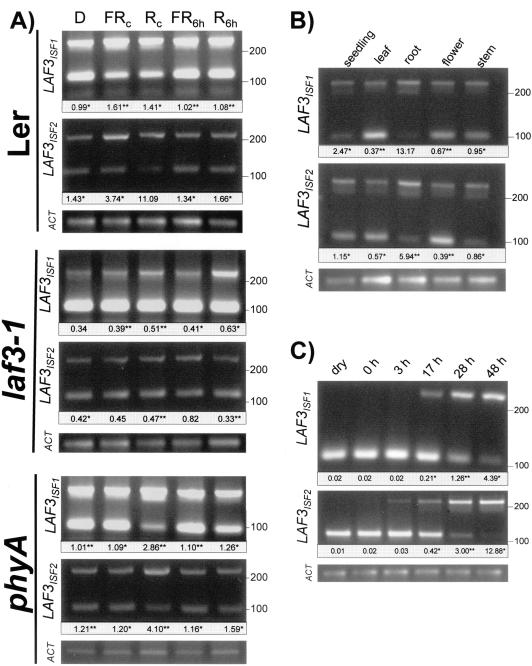
Comparison of LAF3ISF1 and LAF3ISF2 transcript levels in 4-d-old seedlings indicated that transcripts encoding both isoforms were at least 2- to 3-fold higher in seedlings grown in Rc and FRc than in etiolated seedlings but that 6 h of irradiation with light of either wavelength was inadequate to induce accumulation of either transcript to the level observed in seedlings germinated and grown for 4 d under either monochromatic wavelengths (Fig. 5A). Not surprisingly, induction of accumulation of both transcripts by FRc displayed an absolute dependence on phyA, although a detectable increase in both LAF3ISF1 and LAF3ISF2 transcripts by Rc treatments indicates that activation of other phytochromes is also adequate to induce accumulation of LAF3 transcripts (Fig. 5A). Nonetheless, phyA does play a significant role in the early phases of induction of LAF3 transcript accumulation by Rc. Inclusion of laf3-1 seedlings in this experiment enabled confirmation by RT-PCR that both LAF3ISF1 and LAF3ISF2 transcript levels are significantly lower in laf3-1 than in WT seedlings (Fig. 5A). RT-PCR analysis of LAF3ISF1 and LAF3ISF2 transcript levels in 4-d-old seedlings germinated in W light and roots, rosette leaves, stems, and flowers of mature plants indicated that both transcripts were expressed at the highest levels in roots, with comparable levels in leaves, flowers, and stems and slightly higher levels in seedlings (Fig. 5B). Based on the different concentrations of mimics used for the amplification reactions for each isoform, it appears that LAF3ISF1 transcript levels are at least an order of magnitude lower than those of LAF3ISF2 in all of the tissue types examined (Fig. 5B).
Figure 5.
Transcriptional regulation of the LAF3 gene. Fragments specific to LAF3 isoforms are both approximately 250 bp, whereas fragments arising from amplification of mimics are 110 bp. Values below each lane indicate the mean ratio (determined from three independent PCRs) of signal arising from amplification of LAF3 transcript to that arising from amplification of the mimic target. **, sd < 5% of mean ratio; *, sd<10% of mean ratio. For each RT product, amplification of a fragment specific to ACTIN2 transcript (ACT) confirmed comparable amounts of starting RNA and comparable RT efficiency within all comparisons. A, Relative levels of the LAF3ISF1 and LAF3ISF2 transcripts in 4-d-old Ler, laf3-1, and phyA seedlings after continuous growth in darkness (D), FRc of intensity 4.7 μmol m-1 s-1, Rc of intensity 5.1 μmol m-1 s-1, growth for 4 d in darkness before a 6-h pulse with 4.7 μmol m-1 s-1 FRc (FR6 h), or growth for 4 d in darkness before a 6 h pulse with 5.1 μmol m-1 s-1 Rc (R6 h). The results indicated were obtained after 22 cycles of PCR containing 10-7 pmol LAF3ISF1 mimic or 10-6 pmol LAF3ISF2 mimic. B, Both LAF3 transcripts are present at higher levels in roots of mature plants than in 4-d-old seedlings germinated and grown in W light or leaves, flowers, and stems of adult plants. The results indicated were obtained after 20 cycles of PCR containing either 5× 10-8 pmol LAF3ISF1 mimic or 10-7 pmol LAF3ISF2 mimic. C, Relative levels of the LAF3ISF1 and LAF3ISF2 transcripts in dry Ler seeds (dry), seeds imbibed at 4°C for 4 d (0 h), and throughout the process of germination (times indicate hours after transfer of imbibed seeds imbibed to continuous W light at 22°C). The intensity of W light was 16 μmol m-1 s-1. For most seeds, radicle emergence was evident by 24 h and seedling cotyledon opening, and greening was evident by 48 h. The results indicated were obtained after 20 cycles of PCR containing 5× 10-8 pmol LAF3ISF1 mimic or 10-7 pmol LAF3ISF2 mimic. No amplification was obtained for either of the LAF3ISF1 and LAF3ISF2-specific fragments even after 35 cycles of PCR in the absence of a mimic fragment.
Given the central role played by phyA in regulating germination and early seedling development, we investigated LAF3 transcript levels in dry seeds, seeds that had been imbibed in darkness at 4°C for 4 d (0-h time point denotes extracts prepared immediately after the end of stratification), and then at four time points after exposure of stratified seeds to continuous W light at 22°C. These spanned the time of radicle emergence (around 24 h) and cotyledon expansion and greening (complete within 48 h of growth in continuous W light at 22°C). In the absence of mimic fragments, neither transcript was detectable in dry seeds or seeds imbibed in darkness, although both transcripts were present within 3 h after transfer of stratified seeds to germination-promoting conditions (data not shown). As indicated in Figure 5C, levels of both transcripts increased progressively throughout the first 2 d after transfer to continuous W light at 22°C with lower levels of LAF3ISF1 than LAF3ISF2.
DISCUSSION
Molecular events that signal the availability of light play a central role during plant growth and development. As one of the most critical developmental transitions in the life cycle of flowering plants, seedling photomorphogenesis provides an excellent experimental system to elucidate the components required for light signal transduction. Activation of phyA in etiolated seedlings causes cotyledon expansion and greening and inhibition of hypocotyl cell elongation. Despite much recent progress, the molecular basis of these morphological manifestations is still largely unknown. This arises primarily from the difficulty in establishing how known intermediates in phyA signaling interact with one another in the context of a transduction network.
The Contribution of LAF3 to the phyA Signaling Network
The specific hyposensitivity of laf3-1 hypocotyl elongation to FR in comparison with R and B implicates the requirement of LAF3 for complete responsiveness to activated phyA. Defective LAF3 expression does not affect the inhibition of hypocotyl elongation triggered by activation of any of the other known photoreceptors in Arabidopsis. Clearly, LAF3 is essential for full response capacity to activated phyA, although the map position of LAF3 excludes the possibility of allelism with any other FR-specific long hypocotyl mutants. Like all known mutants defective in phyA signaling intermediates, significantly reduced levels of LAF3 transcripts do not completely eliminate any of the FRc-induced seedling responses tested in this study. Furthermore, the absence of any effects of the laf3-1 mutation on FRc-regulated hook opening and cotyledon expansion, hypocotyl gravitropism, greening in W light after a prolonged FRc treatment, PET E and ASN1 regulation, or phyA-mediated photoperiod sensing in mature plants indicates that the LAF3 locus may normally modulate a more discrete subset of phyA-regulated responses than are affected in most mutants deficient in positively acting intermediates in phyA signaling (Hudson et al., 1999; Bolle et al., 2000; Fairchild et al., 2000; Fankhauser and Chory, 2000; Hsieh et al., 2000; Soh et al., 2000; Ballesteros et al., 2001; Møller et al., 2001; Wang and Deng, 2002; Wang et al., 2002).
Analysis of LAF3 transcript levels (Fig. 5) and failure to discern any LAF3 overexpression phenotype strongly suggest that LAF3 action is unlikely to be significantly regulated at the transcriptional level. However, we cannot eliminate the possibility that both LAF3 isoforms may be differentially regulated by light posttranscriptionally. The ubiquitous expression of LAF3 and the lack of specificity for FR wavelengths to modulate LAF3 levels might be interpreted as indicating that LAF3 normally plays a permissive role in phyA signaling, although the apparent developmental specificity of the laf3-1 mutant phenotype argues against a role for LAF3 in regulating basic housekeeping functions. Although LAF3 transcripts are higher in roots than in other tissues, and root elongation in FR is inhibited in phyA seedlings (Büche et al., 2000), we failed to observe significant effects of the laf3 mutation on root elongation in 4-d-old seedlings irradiated with FRc or on the frequency of lateral root initiation in 14-d-old seedlings grown in continuous W light (data not shown). The use of the CaMV 35S promoter in complementation studies may have obscured a role for one of the two LAF3 isoforms in mediating phyA responses. We cannot eliminate the possibility that when levels of an isoform normally not involved in phyA signaling are considerably elevated, it might be capable of fulfilling a role normally attributable to the other isoform. Further investigation of whether such genetic redundancy might occur would require knowledge of the endogenous levels of both isoforms in seedlings. Therefore, the biological significance of the use of alternative transcription initiation sites to generate two isoforms that differ only in their N termini remains enigmatic.
LAF3 Displays a Perinuclear Distribution
Insight into the relationship between LAF3 and known intermediates in phyA signaling might benefit from knowledge of its subcellular localization. It is accepted that phyA responses are regulated primarily at the level of altered gene expression. In accordance, two key events involving the movement of signaling components across the nuclear envelope appear to be essential for complete responsiveness to activated phyA. The first is the relatively rapid translocation of phyA from the cytoplasm to the nucleus triggered by the same light conditions required for phyA activation (Nagy and Schäfer, 2002). Here, phyA activates waves of transcriptional activation or repression to control the downstream target genes that define discrete branches of the phyA-regulated transcriptional network (Tepperman et al., 2001). The second event is the simultaneous derepression of gene expression that appears to be mediated at least in part by the relatively slow movement of COP1 out of the nucleus (von Arnim et al., 1997). The FR-induced nuclear depletion of COP1 is dependent on phyA (Osterlund and Deng, 1998). In addition to these two processes, phyA signaling in Arabidopsis is also likely to mediate the light-dependent nuclear translocation of transcription factors that are cytoplasmic in darkness (Kircher et al., 1999).
Although we cannot formally exclude the possibility that expression of LAF3 proteins under the control of the CaMV 35S promoter might cause them to be targeted to a subcellular site different from that of the naturally expressed proteins, our characterization of LAF3 reiterates the apparent importance of physical separation of cellular processes as a control point in phyA signaling (Møller et al., 2002; Nagy and Schäfer, 2002). Intriguingly, the significant enrichment in the abundance of LAF3 at the nuclear rim closely resembles that reported for SUB1, a cytoplasmic calcium-binding protein that negatively regulates both cryptochrome and phyA responses (Guo et al., 2001), although unlike sub1, laf3 does not display a dramatic hypocotyl elongation phenotype in B (Fig. 1A). The laf3-1 hypocotyl phenotype also appears to display far less fluence dependence than that of sub1 (Guo et al., 2001). Neither of the LAF3 isoforms nor SUB1 possess the recently identified WPP domain that appears to be a novel plant motif necessary and sufficient for targeting plant RanGAPs and the plant-specific nuclear envelope-associated MAF1 protein to the nuclear rim (Rose and Meier, 2001). The role of the putative transmembrane domains of both LAF3 isoforms remains unclear.
It is tempting to speculate that LAF3 may participate in the phyA-dependent translocation of a latent factor(s) across the nuclear envelope from its subcellular location in darkness to a site where it triggers phyA responses in the light. Spatial confinement is an excellent mechanism to regulate gene expression by controlled movement of signaling intermediates across the nuclear envelope (Amador et al., 2001; Heerklotz et al., 2001; Igarashi et al., 2001; Carmo-Fonseca, 2002; Stone et al., 2003). Regulated subcellular compartmentation may also be essential for the regulation of posttranslational modifications such as the addition or removal of ligands [e.g. (de)phosphorylation, (de)acetylation, and (de)conjugation with ubiquitin or ubiquitin-like tags] and/or regulation of the stability of gene products. The combined regulation of ligand modification and subcellular localization potentially confers considerable complexity on the regulation of a signaling intermediate. For instance, four different species, each with a distinct function, might be envisaged for a protein that displays nucleocytoplasmic shuttling and can be reversibly conjugated to a regulatory ligand.
Given the comparatively weak phenotype of laf3-1, we do not suspect that LAF3 normally acts at the level of the phyA photoreceptor itself. The phyA-specific signaling intermediate FHY1/PAT3 also occurs in both nuclear and cytoplasmic compartments (Desnos et al., 2001; Zeidler et al., 2001), although a significant role for FHY1 in regulating CHS and plastocyanin (PET E) but not CAB transcript abundance in FRc (Barnes et al., 1996b) contrasts with the role of LAF3 in the regulation of these genes (Fig. 1B). Therefore, it is unlikely that LAF3 acts at the level of FHY1 action. The likelihood of a direct effect of LAF3 on COP1 subcellular localization is decreased by the specificity of the hypocotyl phenotype to FR wavelengths. Moreover, the nuclear depletion of COP1 is much slower than the time taken for many of the light-induced changes in photoresponsive gene expression that are compromised in laf3-1 (von Arnim et al., 1997).
LAF3 Appears to Be an Ortholog of a Ubiquitous Bacterial Protein
Bacteria that contain putative gene products homologous to LAF3 are represented in the Proteobacteria (purple, non-sulfur bacteria), Archaebacteria, class Firmicutes, Actinobacteria and the Aquificae, Chloroflexi, Fusobacteria, and Thermotogae. Only two of the non-plant LAF3 homologs are predicted to contain transmembrane domains, although with a lower probability than was predicted for the Arabidopsis or rice gene products. In both Pseudomonas aeruginosa and Ralstonia solanacearum, these predicted regions were found within the first 30 amino acids at the N termini of the proteins.
It is intriguing that LAF3 is homologous to aepA, an Erwinia sp. gene product that is responsible for regulating enzymes capable of depolymerizing the cell wall and cell wall components. These enzymes facilitate the maceration of host plant tissues and the liberation of nutrients. The aepA gene product is believed to regulate the timely induction of these exoenzymes by responding to levels of specific components of plant extracts to ensure optimal destruction of plant tissues before initiation of the host's defense response (Barras et al., 1994). The homology of LAF3 with this regulatory gene product is consistent with its role in coordinated induction and repression of several phyA-regulated mRNAs (Fig. 1B). Of the phyA-regulated transcripts tested, XTR7 levels clearly display the strongest regulation by LAF3 (Fig. 1B). XTR7 transcript abundance correlates well with the extent of inhibition of hypocotyl elongation in different genetic backgrounds and under different light conditions (Fig. 1B). Although like aepA, the XTR7 gene product is also involved in loosening the network of cellulose and xyloglucan fibers in plant cell walls, the reduced sensitivity of laf3 to FR-induction of genes unrelated to cell wall loosening (Fig. 1B) argues against a role restricted to regulation cell elongation. It is proposed that coordinated activation of the aepABH (activator of extracellular protein production) operon down-regulates expression of the rsmA repressor of the synthesis of cell wall-degrading enzymes (Barras et al., 1994), although the precise role of aepA action in this process does not appear to have elucidated. Certainly, the ubiquitous presence of aepA and rsmA homologs throughout microbial kingdoms but not in animals suggests that their roles might extend beyond mediating pathogenicity. It appears that like the phytochromes and His kinase receptors for ethylene and cytokinin, the ancestry of plant LAF3s can be traced back to their prokaryotic heritage and that the functions of LAF3 might have been modified in different species to enhance integration with diverse signaling mechanisms. Notably, N. crassa and at least 12 of the bacteria that are predicted to have LAF3 homologs also express putative bacteriophytochrome photoreceptors (Montgomery and Lagarias, 2002). This raises the intriguing possibility that the relationship between LAF3 and phyA signaling may have fairly ancient evolutionary origins. Nonetheless, it is difficult to reconcile the apparently constitutive perinuclear localization of both Arabidopsis LAF3 isoforms with the proposal that aepA activates transcription of a pectate lyase gene from P. carotovorum (Liu et al., 1993). We cannot eliminate the possibility that the mode of action of LAF3 homologs in bacteria and fungi is distinct from their function(s) in plants.
The use of alternative transcription initiation sites (e.g. Tamaoki et al., 1995), intron splicing sites (Eckardt, 2002), or translational start sites (Chabregas et al., 2003) have all been shown previously to generate isoform diversity in plants. However, pretranslational mechanisms of increasing the flexibility of gene expression by generating alternative products with separable functions do not appear to be used as widely in plants as they are in animal systems. In a time when the availability of genome sequences has led to a strong reliance on the use of predicted ORFs, our unanticipated discovery of two LAF3 isoforms emphasizes the importance of confirming transcript initiation and termination sites to assess possible sources of the fine tuning of biological responses. Although the exact significance of the evolution of distinct transcription initiation sites in the LAF3 gene is not clear, identification of this locus provides a further contribution to ongoing efforts to interpret plant physiological responses to light in the context of a molecular framework.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Genetic Analysis
The laf3-1 and phyA-201 mutants are in the Arabidopsis Ler background and were compared with WT Ler-O in all analyses. The laf3-1 mutant corresponds to Ds-tagged line GT3069 (Sundaresan et al., 1995). Unless otherwise stated, plant growth conditions and light sources were identical to those described by Bolle et al. (2000). Procedures for mutant screening and PCR-mediated linkage analysis were identical to those described previously (Møller et al., 2001). Cosegregation of the mutant phenotype with transposon-borne genes was confirmed in 120 F2 seedlings. Seedlings with the laf phenotype were rescued from the block in chloroplast development in greening observed in WT plants after transfer from FRc to W by incubation for 2 d in darkness on Murashige and Skoog medium supplemented with 3% (w/v) Suc before exposure to W.
Molecular Characterization of laf3-1
The sequence flanking the transposon in laf3-1 was cloned by inverse PCR. A 519-bp fragment was amplified using the oligonucleotides 5′-GGTCGGTACGGGATTTTCCC-3′ and 5′-CTAAAAAGTGAAAAGGATCATGGC-3′. To verify that the cloned genomic region was adjacent to the Ds element, a combination of two Ds oligonucleotides and two LAF3 oligonucleotides was used to amplify four fragments spanning the left and right Ds borders. Sequence analysis of these fragments confirmed the predicted site of Ds integration. The flanking sequence isolated by inverse PCR was used as a probe to screen a flower cDNA library (Weigel et al., 1992) obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). For western analysis, seedlings were ground with a pestle and mortar under liquid nitrogen, and the powder was suspended in 2 volumes of SDS-PAGE sample buffer followed by incubation at 95°C for 5 min. After 10 min of centrifugation, the protein concentrations of the supernatant solutions were quantified using the RC DC protein assay (Bio-Rad, Hercules, CA), and 40 μg of total protein was resolved on 10% (w/v) SDS-PAGE gels before transfer to Immun-Blot PVDF membranes (Bio-Rad). Western blotting was according to standard procedures using the phyA monoclonal antibody mAA1 (Shinomura et al., 1996) and monoclonal anti-actin clone C4 (ICN Biochemicals Inc, Aurora, OH). Levels of phyA and actin were assessed using the ECL PLUS chemiluminescent detection system (Amersham, Buckinghamshire, UK). Northern-blot analysis was conducted as described previously (Møller et al., 2001) using CHS, CAB, PET E, PET H, and XTR7 probes identical to those described elsewhere (Ballesteros et al., 2001; Møller et al., 2001). The 730-bp ASN1 probe was obtained from a cDNA library by PCR-mediated amplification using the oligonucleotides 5′-GAGGCACAGAGGACCTGACTGG-3′ and 5′-GAGCCCTCAAGACCAACGCAAAAGG-3′. Comparable RNA loading was verified by ethidium bromide staining and by hybridization with an Arabidopsis 18S rRNA probe.
Cloning the LAF3 Gene and Complementation Analysis
A bacteriophage-based Arabidopsis genomic library (CLONTECH Laboratories, Palo Alto, CA) from the Columbia ecotype was screened using a 1.3-kb LAF3 cDNA probe. An 8,067-bp genomic fragment, beginning 3,050 bp upstream of the start codon of the LAF3ISF1 ORF and encoding 1,293 bp downstream of the stop codon of both LAF3 isoforms, was excised from one of the isolates by digestion with XhoI and SalI and cloned into a promoterless binary plant transformation vector (Møller et al., 2001) to generate the construct gLAF3. The entire fragment was sequenced.
To identify the transcription start sites of the LAF3 gene, 5′-RACE analysis was performed using an Arabidopsis cDNA library generated from 4-d-old etiolated Columbia seedlings using the Marathon cDNA Amplification Kit (CLONTECH Laboratories). Nested gene-specific oligonucleotides used were 5′-GCGTTCAGATGGAATCCAAGCG-3′ and 5′-GGGAGGTATTCTTTTAACCGCGG-3′. Three independent fragments corresponding to the 5′ termini of LAF3ISF1 were recovered, and eight independent fragments corresponding to the 5′ termini of LAF3ISF2 were recovered. To test the ability of 35S::LAF3ISF1 and 35S::LAF3ISF2 to complement laf3-1, full-length LAF3ISF1 and LAF3ISF2 cDNAs were amplified by RT-PCR using total RNA isolated from 4-d-old FRc-irradiated Ler seedlings. First strand cDNA was prepared using RNA from etiolated seedlings and the oligonucleotide 5′-GTAAAGTTTGTTTCTTAATTAGATAAAGACTGCATCATC-3′ specific to the 3′-untranslated region of both isoforms and M-MLV RT (Invitrogen, Carlsbad, CA) as recommended by the manufacturer. Oligonucleotides used to amplify full-length LAF3ISF1 from first strand product were 5′-TACTCGAGATGACCGGTTGGTATGAGTTTCC-3′ and 5′-GACTAGTCTCATGGATACAATTGCTTTCCTCC-3′. Oligonucleotides used to amplify LAF3ISF2 were 5′-TACTCGAGATGAACCTCTTCGTCAGCGTTTCAGC-3′ and 5′-GACTAGTCTCATGGATACAATTGCTTTCCTCC-3′. The PCR products were sequenced and cloned into the binary transformation vector pBA002 (Kost et al., 1998) under the control of the CaMV 35S promoter. The resulting constructs were transformed into laf3-1 plants using the Agrobacterium tumefaciens-mediated floral dip method and transformants were selected on media containing glufosinate ammonium (BASTA, Crescent Chemical Co., Inc., New York). The progeny from 23 gLAF3 transformants, 65 35S::LAF3ISF1 transformants, and 63 35S::LAF3ISF2 transformants were analyzed for restoration of a WT phenotype.
Subcellular Localization of LAF3 Isoforms
A binary vector YFP-pBA was constructed containing a multicloning site and a full-length YFP cDNA ligated into the MluI and SpeI sites of pBA002 (Kost et al., 1998). This introduced unique XhoI, AscI, MluI, PacI, AvrII, BamHI, XmaI/SmaI, and AatII sites between the CaMV 35S promoter and YFP in pBA002. Full-length cDNAs encoding LAF3ISF1 and LAF3ISF2 were amplified using oligonucleotides that removed the stop codons and cloned as translational fusions to YFP in YFP-pBA. laf3-1 plants were transformed with the resulting 35S::LAF3ISF1-YFP and 35S::LAF3ISF2-YFP constructs as described previously, and primary transformants were selected by their resistance to glufosinate ammonium (BASTA, Crescent Chemical Co., Inc.). The progeny of these lines (T2 generation) was used to assess complementation of the laf3-1 hypocotyl phenotype in FRc and to visualize the subcellular localization of LAF3ISF1-YFP and LAF3ISF2-YFP in root and hypocotyl cells of etiolated seedlings using an inverted TE2000 epifluorescence microscope (Nikon, Tokyo) and Openlab imaging software (Improvision, Coventry, UK).
Mimic-Controlled RT-PCR
The oligonucleotides 5′-AAGTTCAGCATTGTATCAACTTCCGG-3′ and 5′-ATGAACCTCTTCGTCATCGTTTCAGCT-3′ were both used in conjunction with 5′-CTTGAGAGTGGCAAAGCTTCCAAC-3′ to quantify levels of the LAF3ISF1 and LAF3ISF2 transcripts, respectively. The LAF3ISF1-specific oligonucleotide recognizes part of the 5′-untranslated region of the LAF3ISF1 transcript beginning 28 nucleotides upstream of the start codon of LAF3ISF1. The LAF3ISF2-specific oligonucleotide is complementary to the region encoding the first nine amino acids of the LAF3ISF2 isoform that are spliced out of the primary transcript encoding LAF3ISF1. The reverse oligonucleotide common to both RT-PCRs recognizes the sequence encoding the last eight amino acids encoded by the third exon of LAF3ISF1 (i.e. the second exon of LAF3ISF2). Both fragments indicating LAF3ISF1 and LAF3ISF2 levels span an intron to eliminate the possibility that genomic DNA contamination interfered with the competition between the target and mimic templates.
The mimics used for LAF3ISF1 and LAF3ISF2 transcript quantification were the oligos 5′-AAGTTCAGCATTGTATCAACTTCCGGGTAACGTTGCGTTCCAGGGTTGTCACACTGTCTTCTCAGAGCAGATAGTTTGACTGACAGGTTGGAAGCTTTGCCACTCTCAAG-3′ and 5′-ATGAACCTCTTCGTCATCGTTTCAGCGGTGGGACGAAATCCGTCATACTGTAGTTGTTGTCGTCTCTGTCGTGGTTGGAAGCTTTGCCACTCTCAAG-3′, respectively. Because the mimic fragments contain the recognition sequences for the amplification oligonucleotides, they compete with target LAF3ISF1 and LAF3ISF2 cDNAs during amplification. Fragments specific to both LAF3ISF1 and LAF3ISF2 are both approximately 250 bp, whereas fragments arising from amplification of mimics are 110 bp. Artifactual genomic DNA amplifications are predicted to be approximately 400 (for LAF3ISF1) and 340 bp (for LAF3ISF2) bp.
With the exception of RNA extraction from the seeds (extracted according to Vicient and Delseny, 1999), RNA was extracted using the RNeasy Plant Minikit (Qiagen USA, Valencia, CA). Total RNA samples were treated with RNase-free DNase I (Invitrogen) to eliminate contaminating genomic DNA. After confirmation of RNA integrity by analysis using an Agilent 2100 Bioanalyzer, 3 μg of RNA from each sample was combined with 10 pmol oligo 5′-CTTGAGAGTGGCAAAGCTTCCAAC-3′ in a total volume of 10 μL, and the oligo was annealed by heating the sample to 72°C for 2 min before transferring immediately to ice for 2 min. The RT reaction was performed by adding 2 μL of dithiothreitol, 2 μL of dNTP mix (10 mm each), 2 μL of M-MLV RT (Invitrogen), and 4 μL of 5× buffer provided by the supplier of the enzyme followed by incubation for 1 h at 42°C.
Standard 50-μL PCRs contained 2 μL of first strand reaction product, 50 pmol of each amplification oligo, and the appropriate mimic at a predetermined concentration shown to not have reached the limits of amplification after the number of PCR cycles used for the particular set of RNA samples being compared. The RT-PCR runs varied between 18 and 23 cycles (each 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s), depending on the exponential range of amplification. In all cases, identical samples subjected to an additional five cycles were run in parallel to ensure that amplification of the mimic was still in the exponential phase at the time when the sample was withdrawn for use in comparative analyses. Comparable results were obtained when the PCR reaction was repeated once with the same reverse-transcribed sample and twice with an independent reverse-transcribed sample from the same preparations of total RNA. For all experiments, fragments arising from amplification of targets were pooled and sequenced to confirm their identity.
The intensities of bands arising from amplification of LAF3 transcripts and those arising from amplification of mimics were determined using Quantity One Version 4 image quantification software (Bio-Rad) after image acquisition using a Gel Doc 2000 gel documentation system (Bio-Rad). Comparison of the ratios of the intensities of bands arising from LAF3 transcripts to bands arising from mimics enabled assessment of the relative levels of the expression of both isoforms.
RT-PCR with the oligonucleotides 5′-TTGCCATTCAGGCCGTTCTTTCT-3′ and 5′-ACCCGCAAGATCAAGACGAAGGA-3′ was used to ensure comparable starting concentrations of RNA and RT efficiency in all comparisons. These oligonucleotides recognize sites that span the second intron in the ACTIN2 gene (At5g09810) to amplify a 147-bp fragment from correctly spliced mRNA transcripts. First strand cDNA was synthesized by priming using the latter oligonucleotide.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
We are grateful to Venkatesan Sundaresan for providing Ds-tagged lines, Qi-Wen Niu for mutant screening, Mathias Zeidler for the cDNA used for 5′-RACE analysis, and Akira Nagatani for phyA antibody.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.028480
This work was supported by NIH grant GM 44640, a JST (CREST) grant, a grant from the Biotechnology and Biological Science Research Council (S.G.M.), and by the South African National Research Foundation (postdoctoral fellowship to P.D.H.).
The online version of this article contains Web-only data.
References
- Amador V, Monte E, García-Martínez J-L, Prat S (2001) Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106: 343-354 [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada J-P, Grossniklaus U, Chua N-H (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613-2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua N-H (1996a) Far red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Quaggio RB, Whitelam GC, Chua N-H (1996b) fhy1 defines a branch point in phytochrome A signal transduction pathways for gene expression. Plant J 10: 1155-1161 [DOI] [PubMed] [Google Scholar]
- Barras F, van Gijsegem F, Chatterjee AK (1994) Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol 32: 201-234 [Google Scholar]
- Bolle C, Koncz C, Chua N-H (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14: 1269-1278 [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua N-H (1994) Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 77: 73-81 [DOI] [PubMed] [Google Scholar]
- Büche C, Poppe C, Schäfer E, Kretsch T (2000) eid1: a new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell 12: 547-558 [PMC free article] [PubMed] [Google Scholar]
- Cantón FR, Quail PH (1999) Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol 121: 1207-1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M (2002) The contribution of nuclear compartmentation to gene regulation. Cell 108: 513-521 [DOI] [PubMed] [Google Scholar]
- Chabregas SM, Luche DD, van Sluys M-A, Menck CFM, Silva-Filho MC (2003) Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1. J Cell Sci 116: 285-291 [DOI] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon Y-K, Sho MS, Shin B, Luka Z, Hahn T-R, Song P-S (1999) Phytochrome signaling is mediated through nucleoside diphosphate kinase 2. Nature 401: 610-613 [DOI] [PubMed] [Google Scholar]
- Cólon-Carmona A, Chen DL, Yeh K-C, Abel S (2000) Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol 124: 1728-1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15: 2980-2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou Y-C, Schäfer E, Funk M, Kretsch T (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt NA (2002) Alternative splicing and the control of flowering time. Plant Cell 14: 743-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377-2391 [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J (2000) RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol 124: 39-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh K-C, Lagarias JC, Zhang H, Elich TD, Chory J (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539-1541 [DOI] [PubMed] [Google Scholar]
- Guo H, Mockler T, Duong H, Lin C (2001) SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome action. Science 291: 487-490 [DOI] [PubMed] [Google Scholar]
- Heerklotz D, Doring P, Bonzelius F, Winkelhaus S, Nover L (2001) The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol Cell Biol 21: 1759-1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH (1999) SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496-499 [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng X-W (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14: 1958-1970 [PMC free article] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405-409 [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13: 2017-2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483-2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC (1994) Photoresponses of light-grown phyA mutants of Arabidopsis: phytochrome A is required for the perception of daylength extensions. Plant Physiol 105: 141-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-G, Ju Y, Kim D-H, Chung K-S, Fujioka S, Kim J-I, Dae H-W, Yoshida S, Takatsuto S, Song P-S et al. (2001) Light and brassinosteroid signals are integrated via a dark-induced small G-protein in etiolated seedling growth. Cell 105: 625-636 [DOI] [PubMed] [Google Scholar]
- Kim D-H, Kang J-G, Yang S-S, Chung K-S, Song P-S, Park C-M (2002) A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14: 3043-3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Wellmer F, Nick P, Rügner A, Schäfer E, Harter K (1999) Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J Cell Biol 144: 201-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua N-H (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393-401 [DOI] [PubMed] [Google Scholar]
- Liu Y, Murata H, Chatterjee A, Chatterjee AK (1993) Characterization of a novel regulatory gene aepA that controls extracellular enzyme production in the phytopathogenic bacterium Erwinia carotovora subsp. carotovora. Mol Plant-Microbe Interact 6: 299-308 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng X-W (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589-2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller SG, Ingles PJ, Whitelam GC (2002) The cell biology of phytochrome signaling. New Phytol 154: 553-590 [DOI] [PubMed] [Google Scholar]
- Møller SG, Kunkel T, Chua N-H (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev 15: 90-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BL, Lagarias JC (2002) Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci 7: 357-366 [DOI] [PubMed] [Google Scholar]
- Nagy F, Schäfer E (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol 53: 329-355 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Deng X-W (1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J 16: 201-208 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng X-W (2000) Targeted destabilization of HY5 in light development of Arabidopsis. Nature 405: 462-466 [DOI] [PubMed] [Google Scholar]
- Rose A, Meier I (2001) A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA 98: 15377-15382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang J-Y, Ishikawa M, Bolle C, Ballesteros ML, Chua N-H (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995-999 [DOI] [PubMed] [Google Scholar]
- Sharrock RA, Clack T (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130: 442-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M (1996) Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 8129-8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh M-S, Kim Y-M, Han S-J, Song P-S (2000) REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12: 2061-2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797-1810 [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Tsugawa H, Minami E, Kayano T, Yamamoto N, Kano-Murakami Y, Matsuoka M (1995) Alternative RNA products from a rice homeobox gene. Plant J 7: 927-938 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang H-S, Wang X, Quail PH (2001) Multiple transcription factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437-9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum KE, Kim M, Christopher DA, Mullet JE (2001) Crytochrome 1, cryptochrome 2, and phytochrome A co-activate the chloroplast psbD blue light-responsive promoter. Plant Cell 13: 2747-2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Delseny M (1999) Isolation of total RNA from Arabidopsis thaliana seeds. Anal Biochem 268: 412-413 [DOI] [PubMed] [Google Scholar]
- von Arnim A, Osterlund MT, Kwok SF, Deng X-W (1997) Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol 114: 779-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339-1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma L, Habashi J, Li J, Zhao H, Deng X-W (2002) Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J 32: 723-733 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843-859 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308-312 [DOI] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC (1998) Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA 95: 13976-13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M, Bolle C, Chua N-H (2001) The phytochrome A specific component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol 42: 1193-1200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.