Abstract
Rop small GTPases are plant-specific signaling proteins with roles in pollen and vegetative cell growth, abscisic acid signal transduction, stress responses, and pathogen resistance. We have characterized the rop family in the monocots maize (Zea mays) and rice (Oryza sativa). The maize genome contains at least nine expressed rops, and the fully sequenced rice genome has seven. Based on phylogenetic analyses of all available Rops, the family can be subdivided into four groups that predate the divergence of monocots and dicots; at least three have been maintained in both lineages. However, the Rop family has evolved differently in the two lineages, with each exhibiting apparent expansion in different groups. These analyses, together with genetic mapping and identification of conserved non-coding sequences, predict orthology for specific rice and maize rops. We also identified consensus protein sequence elements specific to each Rop group. A survey of ROP-mRNA expression in maize, based on multiplex reverse transcriptase-polymerase chain reaction and a massively parallel signature sequencing database, showed significant spatial and temporal overlap of the nine transcripts, with high levels of all nine in tissues in which cells are actively dividing and expanding. However, only a subset of rops was highly expressed in mature leaves and pollen. Intriguingly, the grouping of maize rops based on hierarchical clustering of expression profiles was remarkably similar to that obtained by phylogenetic analysis. We hypothesize that the Rop groups represent classes with distinct functions, which are specified by the unique protein sequence elements in each group and by their distinct expression patterns.
Rho family GTPases are well-characterized regulators of cellular morphogenesis in fungal, insect, and mammalian cells (Lu and Settleman, 1999; Hall and Nobes, 2000; Settleman, 2001). A plant-specific family of Rho homologs, known as the Rop family (Rho-related protein from plants), has important roles in plant development (Li et al., 2001; Yang, 2002). Rops have been linked to the regulation of pollen tube and root hair growth, vegetative cell expansion, cell wall synthesis, and cell proliferation in the meristem (Valster et al., 2000; Zheng and Yang, 2000; Fu and Yang, 2001). They carry out at least some of their developmental functions through F-actin (Kost et al., 1999a; Fu et al., 2001, 2002), a crucial component in plant cell morphogenesis (Fowler and Quatrano, 1997; Kost et al., 1999b). In addition to roles in development, Rops may have significant roles in signaling pathways through which plants respond to their environment. For example, Rop plays a role in abscisic acid signaling (Lemichez et al., 2001; Zheng et al., 2002) and in tolerance to oxygen deprivation (Baxter-Burrell et al., 2002). Furthermore, overexpression of a constitutively active form of the rice (Oryza sativa) Rop OsRac1 promotes both the generation of reactive oxygen species and a pathogen-induced cell death response (Kawasaki et al., 1999; Ono et al., 2001). Similarly, maize (Zea mays) ROPs can induce the formation of reactive oxygen species when expressed heterologously in mammalian cells (Hassanain et al., 2000).
Rops form a multigene family in all plants characterized to date, and thus far, a precise picture of Rop function in higher plants is lacking. Although there are some hints that specific Rops are uniquely associated with specific pathways (e.g. AtROP10 with abscisic acid signaling; Zheng et al., 2002), it is not known to what extent certain Rops have unique functions in specific developmental processes or stress responses and to what extent redundancy exists between different Rops. Many current functional studies have used overexpression of dominant negative Rop mutants, which could have promiscuous inhibitory effects on more than one Rop, and thus do not definitively answer these questions. The importance of Rop signaling activity in the growth of the dicot pollen tube, root hair, and leaf cell has been established (Yang, 2002), but questions remain about the roles of specific Rops throughout the plant life cycle, particularly in monocots. As a prelude to genetically dissecting the functions of individual Rops in development and in response to stress, we have undertaken a molecular description of rop genes in the monocot maize and have used the recently completed genome sequence of rice, as well as other genome project databases, to place this information within the context of the angiosperm Rop family. Such analyses of sequence and of mRNA expression patterns can generate more specific, testable hypotheses for the functions of individual rops.
Phylogenetic analysis can also assist in generating hypotheses for gene function. One likely result of gene duplication and divergence is that two genes that were derived more recently from a common ancestor via duplication are more likely to have similar functions than are a pair of genes that were derived from a common ancestor longer ago. If this holds true during the evolution of a gene family, then genes of similar function are likely to group together in clades when arranged in a phylogenetic tree, in what has been termed a “phylogeny of function” (Pereira-Leal and Seabra, 2001). A well-supported phylogeny can also assist in defining subgroup-specific sequence elements that are highly conserved. Such elements are potential functional determinants for each Rop subgroup. Previous analyses of the Rop family have delineated either two (Winge et al., 2000) or four (Zheng and Yang, 2000) such clades, or subgroups, within the Rop family. However, these analyses included relatively few monocot sequences, leaving it unclear as to whether the defined Rop subgroups include all monocot Rops or whether monocot-specific groups exist. Due to the recent availability of the rice genome sequence and the dramatic increase in expressed sequence tag (EST) data, as well as the development of new techniques for determining phylogeny (Huelsenbeck et al., 2001), we decided to revisit the Rop phylogeny.
Our interest in plant cell signaling during development and defense response led us to characterize the Rop family in the monocot species maize and rice. We have completed the identification of the full complement of rop genes—seven—in rice and have isolated nine rop genes in maize. Analysis of gene structure, conserved non-coding sequences (CNSs), genetic mapping, and comparison of Rop coding sequences provided insight into the evolutionary relationships among plant Rops, particularly in monocots. On the basis of our analyses, we propose a modified delineation of four Rop subgroups, which are present in multiple angiosperm species. These groupings will guide the testing of Rop functions by providing a framework for assessing whether closely related Rops have similar functions in different species. Our analysis also illustrates the utility of CNSs (Kaplinsky et al., 2002; Guo and Moose, 2003) in defining orthology among monocot genes.
In addition, we have used developmental expression profiles for the nine maize rops to help identify tissues that express multiple Rops and thus have a high likelihood of overlapping Rop functions, as well as tissues that are associated with transcription of specific subsets of family members. These profiles also allowed us to address whether similar rop expression patterns correlated with close phylogenetic relationships. A survey of mRNA from various tissues indicated that maize rops were differentially expressed, with the highest and most widespread levels of expression in vegetative tissues in which cells were actively dividing and/or expanding. In contrast, we found that only a subset of maize Rops were highly expressed in pollen and mature leaf tissues.
RESULTS
Identification of Monocot Rops
We used two approaches to isolate full-length monocot Rho family cDNA sequences. First, we screened at low stringency a maize shoot apical meristem library using a probe from a highly conserved region of two rice rop EST clones (corresponding to the OsRac1 and OsRac2 genes; Kawasaki et al., 1999). Full-length cDNAs for rop5, rop6, and rop7 were recovered. Second, we searched both public and private EST databases for Rho family GTPases. We identified and sequenced cDNAs for several novel monocot genes in the Pioneer Hi-Bred International proprietary maize EST database (racA-D; Hassanain et al., 2000), in the public ZmDB maize EST database (rop8 and rop9), and in the public Rice Genome Research Program (RGP) database (OsRop4, OsRop5). No other genes in the Rho family were detected in current (April 2003) EST databases for maize or rice. On the basis of recent analyses indicating that all plant Rho genes are part of a unique, plant-specific family designated ROP (Winge et al., 2000; Yang, 2002), we propose to change the names of maize racA to racD to rop1 to rop4.
Sequencing of the identified cDNA clones, as well as of the genomic regions of two pairs of closely related rops (rop2 and rop9, rop6 and rop7), established that the nine maize genes corresponded to bona fide unique rop genes (Supplemental Fig. 1). Sequence identity among maize rops ranged from 97% (rop2 and rop9) to 72% (rop3 and rop4) at the nucleotide level, and 99.5% (ROP2 and ROP9) to 75% (ROP2 and ROP3) at the amino acid level. This range of conservation is similar to that seen in Arabidopsis ROPs (Winge et al., 2000).
Our two additions to the rice Rop family, along with the previously described rice Rops (Kawasaki et al., 1999), bring the number of expressed rice genes to seven (Fig. 1). BLAST searches of both the Syngenta rice genomic sequence database (Goff et al., 2002) at the Torrey Mesa Research Institute (TMRI; http://portal.tmri.org/rice/) and rice genomic sequence in GenBank with each of the seven known rice genes (OsRac1, Rac2, Rac3, RacB, RacD, Rop4, and Rop5) accounted for each of these identified cDNAs and did not reveal any other full-length rop genes. One other genomic rice Rop sequence was identified in both databases (TMRI accession no. CL000714.398; GenBank accession no. AP003516) but appeared to be a pseudogene. The putative pseudogene was missing a detectable final exon, had a nonsense mutation in the sixth exon, and had numerous amino acid changes, including a five amino acid deletion in the first GTP-binding domain (data not shown). These changes were unlikely to be sequencing artifacts, because the independent GenBank and TMRI sequences showed identical changes. Thus the number of expressed rice Rops (seven) is similar to the number of ROPs in Arabidopsis (11).
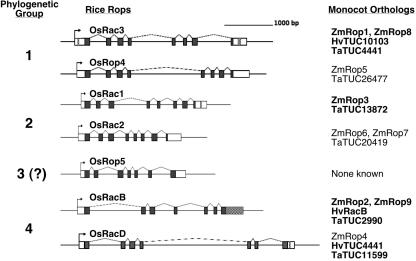
Figure 1.
Genomic structures of rice Rops. Coding regions are in dark gray, untranslated regions (UTRs; based on known cDNA sequences) are in white, and blocks of CNSs are checkered. The Rop phylogenetic group to which each gene belongs (see Fig. 2) is noted to the left, and other grass species orthologs are identified to the right. Orthologs that share the identified CNS sequences are in bold. Groups 1 and 2 have eight exons, and groups 3 and 4 have seven exons; exon/intron junction sites are conserved among Rops for which the genomic sequence is known.
We annotated the rice genomic sequence to determine the structure of the rice Rops (Fig. 1). These structures were compared with both the known maize rop structures (rop2, rop6, rop7, and rop9) and to the rop structures in Arabidopsis (Winge et al., 2000). The comparisons revealed an almost complete conservation of rop gene exon/intron structure among rice, maize, and Arabidopsis, including the presence of an additional exon at the 3′ end of a subset of plant Rops (e.g. rice OsRac1, maize rop6, and Arabidopsis ROP9), designated type II Rops (Winge et al., 2000; Lavy et al., 2002). Only the final exon of rice OsRop4 and maize rop5 varied, in that the splice acceptor sites of both were shifted toward the 5′ end by 6 bp.
Phylogenetic Analyses Place Rops within Distinct Subgroups
Previous analyses placed members of the plant Rho family into a monophyletic group distinct from the Rho, Rac, and Cdc42 families of animals and fungi (Winge et al., 1997, 2000; Li et al., 1998; Vernoud et al., 2003). Furthermore, analyses of the dicot rop genes revealed phylogenetic subgroups, some of which contained members that were expressed in similar patterns. These subgroups have been hypothesized to provide distinct biological functions (Li et al., 1998; Yang, 2002). To determine whether the monocot rop genes were distributed within the same subgroups identified using primarily dicot sequences, we analyzed 82 full-length Rop-like nucleotide sequences from GenBank, The Institute for Genomic Research (TIGR) Gene Indices (GI), and the Plant Genome Database (PlantGDB) EST Clusters, including gene sequences from nonangiosperm plants (moss, pine, and spruce). Phylogenetic trees were generated by three methods: Bayesian inference, maximum parsimony, and maximum likelihood. The Bayesian approach is a recent development that incorporates evolutionary models for DNA substitution and is thought to provide better resolution of proposed relationships among deeply diverging sequences, as well as a more accurate estimation of the confidence in these relationships (Huelsenbeck et al., 2001). All three sets of analyses produced tree topologies that were largely congruent with one another; therefore we present the tree derived by Bayesian inference (Fig. 2) and discuss the other results.
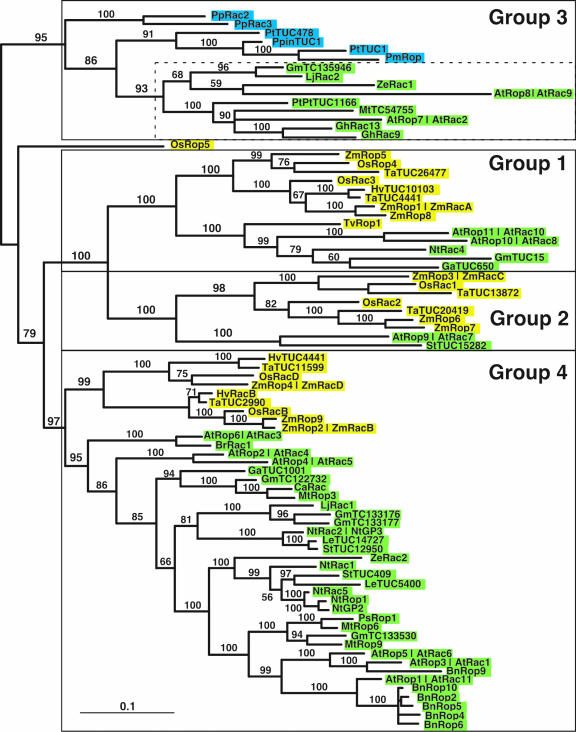
Figure 2.
Midpoint-rooted phylogram of Rop genes, based on Bayesian analysis of the 82 publicly available plant Rho family nucleotide sequences (mean log-likelihood = -17,930.4; variance = 100.6). Monocot Rops are highlighted in yellow, dicot Rops in green, and nonangiosperm sequences in blue. Four phylogenetically distinct groups are boxed; angiosperm sequences in group 3 (all dicots) are outlined by a dotted line. Sequences identified as tentative consensus sequences (TC; TIGR) and tentative unique contig sequences (TUCs; PlantGDB) refer to tentative cDNA consensus sequences based on multiple ESTs. Clade credibility values (Bayesian posterior probabilities) are shown in bold with each branch. At, Arabidopsis; Bn, oilseed rape (Brassica napus); Br, turnip (Brassica rapa); Ca, chickpea (Cicer arietinum); Ga, tree cotton (Gossypium arboreum); Gh, upland cotton (Gossypium hirsutum); Gm, soybean (Glycine max); Hv, barley (Hordeum vulgare) barley; Le, tomato (Lycopersicon esculentum); Lj, Lotus japonicus; Mt, barrel medic (Medicago truncatula); Nt, tobacco (Nicotiana tabacum); Os, rice; Pm, black spruce (Picea mariana); Pp, moss (Physcomitrella patens); Ppin, maritime pine (Pinus pinaster); Ps, pea (Pisum sativum); Pt, loblolly pine (Pinus taeda); PtPt, European aspen (Populus tremula × Populus temuloides); St, potato (Solanum tuberosum); Ta, wheat (Triticum aestivum); Tv, spiderwort (Tradescantia virginiana); Ze, zinnia (Zinnia elegans); Zm, maize. All identification numbers (GenBank accession, GDB TUC, and TIGR-GI TC) for the Rop nucleotide sequences used in our phylogenetic analyses are listed in Supplemental Table III.
The Bayesian and maximum likelihood methods both identified four phylogenetically related groups defined by basal nodes on the midpoint-rooted tree (Fig. 2), designated groups 1, 2, 3, and 4 for consistency with earlier studies (Zheng and Yang, 2000). These same groupings were also identified by maximum parsimony analysis, although it placed group 3 within, and toward the distal tip of, group 4. Except for OsRop5, the constituent members of these groups did not differ in the three types of analyses. OsRop5 was placed either adjacent to group 3 (Bayesian; Fig. 2), in group 3 (likelihood), or adjacent to group 4 (parsimony) and thus remained an enigmatic sequence. All other monocot Rop sequences fell within the previously defined monophyletic Rop family, and were distributed among groups 1, 2, and 4. Our more extensive analysis generally corroborated the results of an earlier study defining four Rop groups based primarily on dicot sequences (Zheng and Yang, 2000), although with some significant modifications. First, two earlier studies (Winge et al., 2000; Zheng and Yang, 2000) suggested that AtROP8/AtRAC9 constitutes a group of its own, whereas our trees indicated that it was a highly divergent member of group 3. This discrepancy is probably due to the greater number of sequences in our analyses, because increasing the sample size more accurately predicts the placement of such divergent (“long branch”) sequences. Second, our analysis suggested that Rop groups 1 and 2 should be delineated separately, rather than combined into a single class (previously referred to as group II). Third, Bayesian posterior probabilities indicated strong support (≥95%) for basal branches separating the four groups, whereas most basal branches were not well supported in previous analyses, which primarily employed neighbor-joining and parsimony methods.
Groups 1, 2, and 4 contained at least one monocot and one dicot sequence, and within each group, the most basal node separated the monocot from the dicot Rops, with the exception of the monocot TvRop1 in group 1. Except for OsRop5, the entire complement of rice Rops fell into one of these three groups, alongside dicot Rops. The enigmatic OsRop5 could be excluded from groups 1 and 2, because it had only seven exons (see below), and its placement in the Bayesian and maximum likelihood trees suggested that it could be a divergent member of group 3. Group 3 also contained all six nonangiosperm Rop sequences (from moss, pine, and spruce). Taken together, our analyses indicated that all four groups originated before the monocot/dicot split. One hypothesis to account for these observations is that four ancestral Rop paralogs were present in a progenitor of monocots and dicots; each was maintained in both monocots and dicots and gave rise to the four extant Rop groups.
The Rops in groups 1 and 2 are distinct from typical Rho GTPases in that their carboxy-terminal sequences have diverged (Ivanchenko et al., 2000; Winge et al., 2000; Lavy et al., 2002). Genomic sequence for Rops in groups 1 and 2 revealed that each gene contained an additional eighth intron and exon just 3′ to the typical Rho translation stop (Fig. 1; Supplemental Fig. 1). The initial divergence of Rop groups 1 and 2 from the typical Rho sequence can thus be traced to the addition of this exon early in angiosperm evolution. Additional sampling of Rop sequences from species basal to angiosperms will be required to determine whether this event occurred only in the angiosperm lineage.
CNSs and Genetic Mapping Independently Verify rop Relationships between Maize and Rice
As expected based on the relatively close evolutionary relationship among the grass species (Gale and Devos, 1998) and the duplication that produced the modern maize genome (Helentjaris et al., 1988; Wilson et al., 1999), most of the monocot sequences in the phylogenetic tree were arranged in tight groups at the branch tips (e.g. one rice Rop closely related to one or two maize Rops). These distal clades are predicted to contain orthologs. To find evidence that independently verified the Rop phylogeny, we searched for CNSs in the monocot rop UTRs and used these as phylogenetic “markers” for orthologous genes in the monocot rops. CNSs in grass species have been defined by previous studies (Kaplinsky et al., 2002; Guo and Moose, 2003), although their function is unknown. Earlier studies focused primarily on CNSs in promoters and introns and the possibility that they encode sites for gene regulatory elements. We searched instead for CNSs in the rop transcript UTRs, given that these sequences were widely available in EST databases.
Using stringent criteria (at least 70% identity in a window of at least 20 bp) defined by Guo and Moose (2003), we identified four unique CNS blocks in three orthologous rop groups defined by the rice genes OsRac1, OsRac3, and OsRacD (Fig. 1; Supplemental Fig. 2). In addition, we identified a remarkable fifth CNS block, encompassing the entire 3′-UTR of the orthologous group defined by the OsRacB gene (Fig. 3). None of the identified CNS blocks were related to other sequences in the rice genome, by BLAST, and thus served as unique phylogenetic footprints that confirmed orthology in these groups of grass rops. Except for maize rop4, every monocot gene identified as an ortholog of these four rice genes by phylogenetic analysis of coding sequence (Fig. 1) shared the corresponding CNSs.
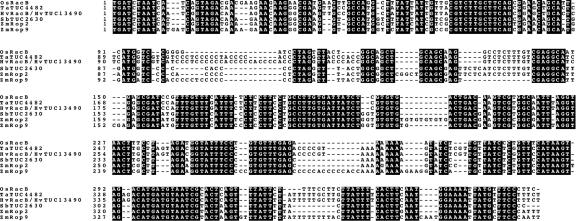
Figure 3.
Alignment of the 3′-UTRs of monocot OsRacB orthologs demonstrates an extraordinarily long CNS block. Nucleotides that are identical in all five species (wheat-Ta, rice-Os, barley-Hv, sorghum-Sb, and maize-Zm) are highlighted in black. Numbering begins at the stop codon (TGA) in each gene; according to available cDNA sequences, the poly(A) tail is added immediately following the final base pair in the alignment, except in HvRacB, whose poly(A) tail addition site is 50 bp further downstream. The HvRacB 3′-UTR is from a full-length PlantGDB TUC identified using the incomplete HvRacB sequence in GenBank.
The conservation of the 3′-UTR in the OsRacB group was remarkable, and it allowed us to identify an additional EST contig from sorghum, with only partial coding sequence, as a likely ortholog (Fig. 3). The nucleotide identity across the entire approximately 350- to 400-bp UTR ranged from a high of 98% (TaTUC4482 versus HvRacB) to a low of 83% (OsRacB versus maize rop2 or rop9), and most of the identity was present in all six sequences. Thus, the entire 3′-UTR could be considered a CNS, making it the longest grass CNS yet identified. The shorter OsRacD CNS resembled a portion of the OsRacB 3′-UTR; both regions were located 40 to 50 bp 3′ to the stop codon. Similarly, the 3′ OsRac3 CNS showed some similarity to sequences at a corresponding position in the more distant monocot TvRop1 (Supplemental Fig. 2).
In addition to the CNS blocks, we genetically mapped most of the maize and rice rop genes to determine whether the putative orthologs were in syntenous positions (Table I). Maize has retained pairs of chromosomal regions that are similar in sequence and gene content, indicating that they were derived via genome duplication. These maize chromosome segments have been mapped relative to each other and to syntenous regions in other grass species (Gale and Devos, 1998; Wilson et al., 1999). Pairs of genes resulting from genome duplication or species divergence often map to such regions of synteny.
Table I.
Monocot rop map positions nd, Not done.
| Map Position
|
Map Position
|
||||
|---|---|---|---|---|---|
| Rice Gene | Chromosome | CentiMorgans | Maize Ortholog(s) | Chromosome | Bin |
| OsRac1 | 1 | 36.9 | rop3 (racC) | 6 | 6.04 |
| OsRac2 | 5 | 104-105 | rop6 | 6 | 6.06 |
| rop7 | 8 | 8.05 | |||
| OsRac3 | 2 | Near 131 | rop1 (racA) | 5 | 5.07 |
| rop8 | nd | ||||
| OsRop4 | 6 | Near 38.3 | rop5 | nd | |
| OsRop5 | 2 | Near 53.5 | None found | ||
| OsRacB | 2 | 4.8 | rop2 (racB) | 4 | 4.11 |
| rop9 | nd | ||||
| OsRacD | 2 | Near 157.9 | rop4 (racD) | 5 | 5.08 |
One pair of maize rops mapped to duplicated regions: rop6 (bin 6.06) and rop7 (bin 8.05). In conjunction with the phylogenetic analysis, these data argued that rop6 and rop7 were created by genome duplication. On the basis of the predicted phylogeny, two other gene pairs (rop1/rop8 and rop2/rop9) may also have been created by genome duplication. However, we were unable to find suitable polymorphisms to map three of the maize rops, and thus could not address this possibility based on map position. Of the five maize/rice pairs for which data were available, four were in syntenous regions, based on current comparative genome maps (Ahn and Tanksley, 1993; Wilson et al., 1999): rop2 (maize 4L) and OsRacB (rice 2S); rop6 (maize 6L)/rop7 (maize 8L) and OsRac2 (rice 5L); rop1 and rop4 (both on maize 5L); and OsRac3 and OsRacD (both on rice 2L). In total, the CNS (Fig. 1) and map data (Table I) confirm several predictions of our tree (Fig. 2), thus providing strong independent support for the accuracy of the phylogenetic hypothesis.
Identification of Rop Subfamily-Specific Protein Sequences
In animals, the Rho GTPase family has diverged into three major subfamilies, Rac, Rho, and Cdc42; each has specific signaling functions that are conserved across species boundaries (Hall, 1998; Johnson, 1999; Settleman, 2001). Similarly, the Rab GTPases have diverged into subfamilies, each with specialized roles in endomembrane trafficking; these subfamilies are conserved among all eukaryotes (Pereira-Leal and Seabra, 2001; Vernoud et al., 2003). The specialized functions of each subfamily are thought to be conferred by the specific binding of member GTPases to interacting proteins, either upstream activators or downstream effectors. Analysis of Rab sequences has revealed “subfamily regions” called RabSF, which contain sequence elements that are conserved among members of the same subfamily, but differ between subfamilies (Pereira-Leal and Seabra, 2000). These elements are hypothesized to help control the unique functions of each Rab subfamily by providing structural determinants that specify binding to the requisite interactors. We wanted to determine whether analogous sequence patterns were present in the four Rop groups and were conserved in both monocots and dicots.
Because elements at the Rop C terminus had been characterized previously (Li et al., 1999; Ivanchenko et al., 2000; Lavy et al., 2002), we focused on the N-terminal GTPase domain and devised consensus sequences for each of the four phylogenetically defined groups (Fig. 4). Our consensus sequences identified amino acid positions (marked by *) that contained highly conserved differences between at least two groups, as well as amino acid positions (marked by ‡) that were highly conserved in one or more groups, but less highly conserved in others. These positions differentiated the groups from each other and defined amino acids that were potentially under distinct selective pressures. Two clusters of group-specific amino acids were apparent, designated RopSF1 and RopSF2. When mapped with respect to the presumed secondary structure of the ROP protein (based on the conserved small GTPase structure; Vetter and Wittinghofer, 2001), RopSF1 was associated with the α3/β5 turn region, and RopSF2 was associated with α3b. These regions present surfaces that can mediate interactions between a small GTPase and an interacting protein (Ostermeier and Brunger, 1999; Lapouge et al., 2000).
Figure 4.
Group consensus amino acid sequences identify conserved regions that differentiate angiosperm Rop groups. Two clusters of group-specific residues are designated RopSF1 and RopSF2. Each position in which at least two groups have a different highly conserved amino acid is marked by *; each position with a given highly conserved amino acid in one or more groups, but less highly conserved residues in other groups is marked by ‡. The secondary structure of human Rac1 (Hirshberg et al., 1997) is shown mapped above corresponding residues in ROP: α-Helices are horizontally striped; β-sheets are diagonally hatched; switch regions (those that change conformation in the GDP-versus GTP-bound protein) are unfilled. The consensus sequences are based on manual inspection of all angiosperm ROP sequences, except the divergent AtROP8 and OsROP5 (74 total). In the ROP family consensus (top line), near identity in the family (at least 95% identity at a position) is capitalized, and highly conserved amino acids (85%-95% identity) are lowercase. Within a Rop group, highly conserved amino acids (at least 85% identity and present in both monocots and dicots) are capitalized. Lowercase letters in the Rop groups indicate an amino acid present in at least one-half of the members and in both monocots and dicots; dots indicate the lack of a predominant amino acid. Complete identity in the ROP family or in a group is marked by an underline. (Note that group 3d contains only dicot sequences.) Number of sequences in each group: group 1, 14; group 2, 9; group 3d, 8; group 4, 43.
To test the predictive utility of these group-specific sequence patterns, we used the consensus sequence for each group from the combined RopSF1/SF2 region to search the PlantGDB database. The top BLAST hits other than those from the original analysis were: group 1, tomato TUC LEtuc02-10-21.3073; group 2, Sorghum propinquum TUC SPtuc02-10-22.2133 and soybean TUC GMtuc03-04-25.17388; group 3, soybean TUC GMtuc03-04-25.5521; and group 4, winter rye (Secale cereale) EST WHE503_E02_J03ZR. Two criteria indicated that the genes corresponding to these sequences belonged in the designated groups: (a) Conceptual translations showed that, where sequence was available, the translations matched the Rop group consensus sequences in Figure 4; and (b) inclusion of these sequences in additional phylogenetic analyses supported their placement in the identified groups (data not shown). Because these new sequences were not full-length, confirmation that they represent a Rop in each group will need additional data. However, this test suggested that the RopSF1/SF2 sequences were good predictors of group membership.
Maize rop Genes Are Developmentally Regulated
We were interested in whether the maize rops were expressed during growth and development and if so, whether the different genes were expressed in distinct patterns. The high degree of conservation among the rop mRNAs made northern-blot hybridization problematic. Therefore, we used Multiplex Titration reverse transcriptase (RT)-PCR (MTRP; Nebenführ and Lomax, 1998) to assay the relative abundance of rop transcripts, in relation to an internal control (either actin1 or EF-1α). MTRP allows estimation of relative transcript abundance by determining the step in a serial dilution of cDNA in which a specific template (i.e. a reverse-transcribed mRNA species) becomes limiting for amplification. The more highly a given rop is expressed, the more dilute its cDNA template can be made while still allowing amplification and visualization of a band. Thus, we avoid some concerns regarding the potentially differential amplification efficiencies of distinct primer pairs (Nebenführ and Lomax, 1998), because the determination of relative expression levels depends only on the dilution step at which the band is no longer visible and not on the intensity of particular bands.
We designed gene-specific primers (GSPs) to regions of greatest divergence for all nine maize rops (Supplemental Table I) and empirically optimized the MTRP assay on three reaction mixes to achieve specificity, as well as a nearly identical “amplification response” (i.e. the inability to amplify a product at a specific dilution step), for each rop (Supplemental Fig. 3). We used these primer mixes to determine the relative expression level of the nine rops using cDNA samples generated from W22 inbred tissue samples (Fig. 5). We sampled four vegetative tissues: root tip (encompassing the root apical meristem and the region of active cell division), root shank (a region of active cell expansion), shoot apex (including the shoot apical meristem and several primordial leaves), and the fully differentiated mature leaf. We also assayed expression in mature maize pollen, as several Arabidopsis ROPs are highly expressed in pollen (Li et al., 1998). Internal control primers corresponded either to maize actin1 in vegetative samples or to EF-1α in pollen samples (due to the low expression of actin1 in pollen).
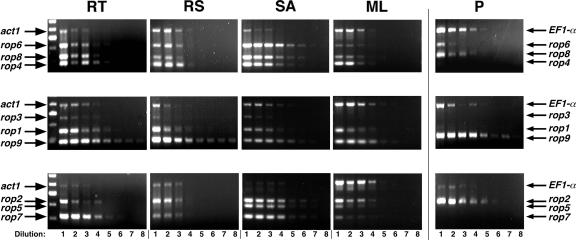
Figure 5.
Agarose gel electrophoresis of MTRP amplification products shows a high degree of overlap in individual maize rop expression patterns in vegetative tissues. PCR templates are 4-fold serial dilutions of cDNA made from root tip (RT), root shank (RS), shoot apex (SA), mature leaf (ML), and pollen (P) RNA samples. Lanes correspond to the dilution steps taken from the original cDNA (i.e. the template used in the lane 1 reaction was the initial 4-fold dilution of cDNA). Amplification of the actin1 gene is an internal control for the vegetative tissue samples; Elongation Factor1-α is the internal control for the pollen samples. The left-most lane in the RT panel shows the Mr standard (100-bp markers; the 500 bp marker is the brightest band). Similar results for the relative expression levels of each rop were observed in three independent experiments.
Our data indicated that all nine rops were widely expressed in different maize organs, with some developmentally regulated differences. Notably, mRNA levels of most of the rops were significantly down-regulated in mature leaf tissue, compared with shoot apex: bands corresponding to the rop3, rop5, rop6, rop7, and rop8 genes showed at least a two dilution (approximately 16-fold) difference in amplification response, and two other genes (rop2 and rop4) consistently displayed at least a one dilution (approximately 4-fold) difference in mature leaf versus shoot apex. In contrast, rop1 and rop9 were highly expressed in all vegetative samples tested, with no consistent differences in expression level detectable by MTRP between mature and developing cells. This pattern of high rop expression in dividing and/or differentiating cells and lower expression in mature cells is consistent with a role for rop genes in maize development.
The most striking example of differential expression was evident in pollen: mRNAs for five of the nine rops were undetectable or barely detectable using MTRP in mature pollen, and rop6 mRNA was expressed at a relatively low level. In contrast, three maize rops were detected at relatively high levels in pollen: the duplicate group 4 genes rop2 and rop9, and the group 1 gene rop8. Thus, maize rop2 and rop9 appeared similar to the AtROP1 gene in Rop group 4, which is crucial for pollen tube growth (Li et al., 1999). However, no data yet address whether group 1 Rops (e.g. maize rop8) are important for pollen development.
Definition of rop Expression Profiles Using a High-Throughput Expression Database
We confirmed and extended our RT-PCR observations by analyzing maize expression data from a large proprietary database, which was generated using massively parallel signature sequencing (MPSS; Brenner et al., 2000a). MPSS quantitates how many times a gene-specific 17-bp signature sequence is represented in a population of 2 × 105 to 2 × 106 cDNAs derived from an RNA sample. Because such 17-bp sequences almost always correspond to unique cDNAs, the “count” of each signature (normalized in parts per million) effectively measures the abundance of the corresponding mRNA in a sample. In addition, statistical tests can determine the significance of differences in transcript abundance (Audic and Claverie, 1997).
We compiled the MPSS values for the nine maize rop genes in 57 RNA samples from a broad spectrum of maize tissues and developmental stages (supplemental data). We then asked three questions: (a) Do the MPSS data confirm the trends observed by MTRP? (b) Do the MPSS data help identify additional trends in rop expression, e.g. patterns that are unique to specific genes or groups of genes? (c) How similar are rop expression patterns, as assayed by MPSS expression profiling, to one another, particularly for those rops that are predicted by our phylogenetic analysis to be most closely related (Fig. 2)?
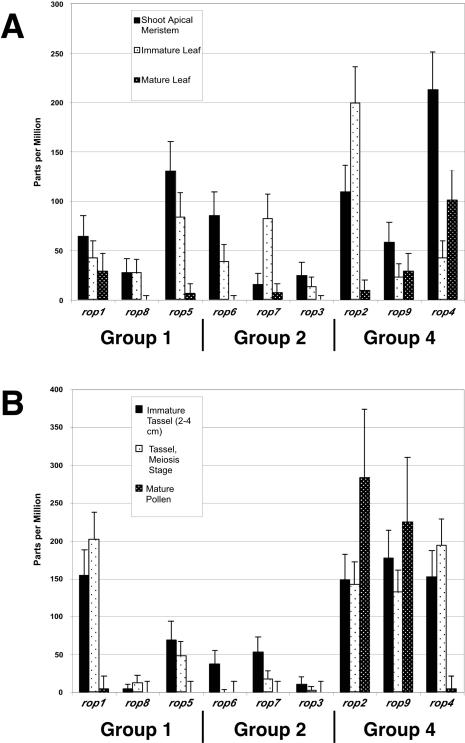
Initially, we compared MPSS values from samples that were most similar to those in our MTRP experiments, i.e. those from vegetative meristems, immature and mature leaves, and immature tassels and mature pollen (Fig. 6). The MPSS data generally agreed with our MTRP observations. For example, almost all rops were expressed, and at relatively high levels, in the more actively dividing and/or expanding tissues (shoot apical meristem, immature leaf, and immature tassel). In addition, all rops showed a statistically significant decrease in expression in mature leaf compared with vegetative meristem and/or immature leaf. However, expression of rop1, rop4, and rop9 decreased by only approximately 2-fold, whereas the expression of all other rops was either undetectable or was dramatically reduced. In mature pollen, MPSS confirmed that rop2 and rop9 were highly expressed, but it failed to detect rop8. Because RT-PCR directed specifically at rop8 transcript confirmed the MTRP results (data not shown), we believe that the MPSS value for rop8 in pollen is artifactually low. Transcript-specific features (e.g. relative distance of the signature sequence from the poly(A) tail) can affect the ability of MPSS to detect a given transcript, and rop8 could be recalcitrant to MPSS detection. This is consistent with the relatively low values for rop8 throughout the data set. In addition, the number of signatures gathered for the pollen sample (3 × 105) was lower than that for most of the other experiments.
Figure 6.
MPSS expression values for maize rops in tissues also assayed by MTRP. MPSS values for each experimental sample are normalized to parts per million; error bars represent 95% confidence intervals for experiments sampling identical numbers of signatures (Audic and Claverie, 1997), also normalized to parts per million. Expression values shown in A are for rops in the shoot apex (shoot apical meristem, immature leaf) and mature leaf (compare with SA and ML in Fig. 6), and in B, in the tassel and mature pollen (compare with P in Fig. 6). Total number of signatures determined in each experiment: shoot apical meristem, 1,243,089; immature leaf, 1,191,133; mature leaf, 923,217; immature tassel, 1,082,651; tassel, meiosis stage, 1,325,062; mature pollen, 280,724.
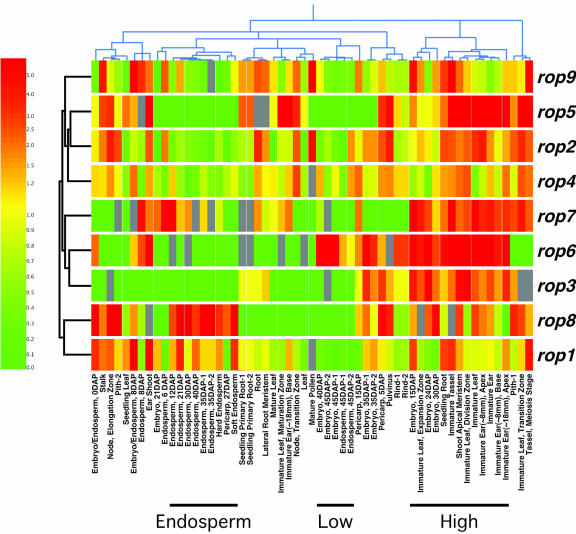
To conduct a global analysis of rop expression, we used GeneSpring software for hierarchical clustering of all samples in the data set (see “Materials and Methods”), which facilitates visualization of experimental samples and genes with similar expression profiles (Eisen et al., 1998; Perou et al., 1999). To better identify trends in rop expression (relative increases versus decreases) and to help control for differences in the ability of MPSS to detect each transcript, MPSS values for each gene were normalized to that gene's median expression value in the entire data set before clustering. The relative level of expression for each gene was then represented using a color scale, from red (representing a 5-fold or greater increase over median) to yellow (median level) to green (a 5-fold or greater decrease from the median; Fig. 7). The validity of the clustering algorithm for identifying samples with similar expression patterns was supported by the distribution of the seven pairs of duplicate samples present in the data set. Six of the seven clustered very close to each other, as expected; only one duplicate pair (Pith-1 and Pith-2) did not. This clustering of pairs supported the idea that MPSS generates reproducible results and that the clustering algorithm recognizes samples with similar rop expression.
Figure 7.
Hierarchical cluster diagram for all nine maize rops and all 57 MPSS experimental samples. Two dendograms are shown, clustering both the RNA samples (above the diagram) and the genes (to the left of the diagram) based on similarity of expression profiles. Gene names are to the right of the diagram, and short descriptive titles for each experiment are below. MPSS values were normalized (see “Materials and Methods”), and the relative level of expression for each gene is represented using a log-based color scale (left), from red (representing a 5-fold or greater increase over median) to yellow (median level = 1) to green (a 5-fold or greater decrease from the median). Genes are represented by a row of colored boxes; experimental samples are represented as columns. Grays represent MPSS values between 5 and 0, which are not significantly different from zero, at a 95% confidence level; these values were ignored by the clustering algorithms. Six of seven pairs of duplicate samples cluster in close proximity, from left to right: endosperm, 35 d after pollination (DAP); seedling primary root; embryo, 45 DAP; endosperm, 45 DAP; embryo, 35 DAP; and rind. The three sample clusters discussed in the text are noted by bars below the experiment titles.
Three clusters were noteworthy: high, low, and endosperm (Fig. 7). Samples in the high group were characterized by high relative expression of most or all of the nine rops. This cluster included the meristematic/immature leaf samples mentioned earlier (Fig. 6), along with other dividing and expanding tissues, most notably several immature ear samples and embryos at 15, 24, and 30 DAP. In contrast, the Low group included samples with relatively low overall rop expression, and consisted of embryo and endosperm samples approaching quiescence (40 and 45 DAP). Intriguingly, rop6 appears to be uniquely expressed at a high level in these samples.
In addition to identifying samples that expressed the entire rop family at high or low levels, the cluster analysis also suggested certain sample groupings are associated with high expression of specific rops or sets of rops. The most obvious example of this was the endosperm cluster, in which eight of nine adjacent samples spanned the period of endosperm development from 12 to 40 DAP. In the endosperm group, rop8, and to a lesser extent rop1, showed high relative levels of expression, in contrast to the other rops, which were relatively low. We also assembled a table of the samples that showed the highest relative levels of expression for each rop. This confirmed that endosperm was among the samples in which rop8 was most highly induced (Table II). Another example of preferential high expression was in 15- to 24-DAP embryo samples, which ranked among the highest expression values for all three of the group 2 rops (rop3, rop6, and rop7; Table II). In contrast, rop4 was expressed at relatively constant levels in almost all of the samples: expression in none of the samples was induced by more than 3-fold above the median, and only four samples showed more than a 3-fold decrease from the median. Thus, despite apparent coordinate high expression of all nine rops in certain tissue types (the High cluster), each family member did show a unique developmental expression profile, which could be important for any rop gene-specific functions.
Table II.
Tissue samples with the highest MPSS values for each maize rop
| Phylogenetic Group | Gene | Median Expression Value | Tissue Samples with Highest Expression | -Fold Induction over Median |
|---|---|---|---|---|
| 1 | rop 1 (racA) | 45 | Embryo/endosperm, 8 DAP | 10.5 |
| Ear shoot | 5.3 | |||
| Tassel, meiosis stage | 4.5 | |||
| rop8 | 9 | Endosoperm, 35 DAP-2 | 10.1 | |
| Pith-2 | 9.7 | |||
| Soft endosperm | 8.1 | |||
| rop5 | 6 | Ear shoot | 27.3 | |
| Immature leaf, division zone | 25.7 | |||
| Shoot apical meristem | 21.7 | |||
| 2 | rop3 (racC) | 5a | Embryo, 24 DAP | 8.0 |
| Immature leaf, division zone | 5.6 | |||
| Shoot apical meristem | 4.8 | |||
| Embryo, 15 DAP | 4.4 | |||
| rop6 | 5a | Embryo, 24 DAP | 19.6 | |
| Immature ear (∼18mm), apex | 18.6 | |||
| Embryo, 15 DAP | 18.4 | |||
| Shoot apical meristem | 17.0 | |||
| rop7 | 7 | Embryo/endosperm, 6 DAP | 12.6 | |
| Embryo, 15 DAP | 11.7 | |||
| Immature leaf | 11.7 | |||
| 4 | rop2 (racB) | 58 | Mature pollen | 4.9 |
| Node, elongation zone | 4.0 | |||
| Immature ear (∼8mm), apex | 4.0 | |||
| Pulvinus | 4.0 | |||
| rop9 | 26 | Embryo/endosperm, 8 DAP | 10.5 | |
| Mature pollen | 8.7 | |||
| Immature tassel | 6.8 | |||
| rop4 (racD) | 94 | None above three times induced | - |
Median values for rop3 and rop6 were set at 5, the smallest value significantly different from 0 (95% confidence interval). This was due to overall low MPSS values for these genes in the experimental samples. The actual median for rop3 was 2; for rop6, 4. All values in this table are significantly different from the median, with a greater than 99.9% confidence interval.
Finally, we used Spearman rank-order correlation and hierarchical clustering to determine whether rop expression patterns were similar across the 57 samples (Fig. 7; Supplemental Table II). Perhaps surprisingly, the most similar pairs of genes, based on expression pattern, were not the three duplicate pairs identified by phylogenetic analysis, but rather two group 2 genes: rop3 and rop6 (rs = 0.731). The only other correlations with rs greater than 0.5 were rop2 with rop5 (rs = 0.570), with rop4 (rs = 0.529), and with rop3 (rs = 0.502). These data suggest that, despite the high conservation of coding sequences, the expression patterns of the duplicates have been significantly altered through evolution. However, despite the low absolute correlation values among the rops, the clustering algorithm produced a tree that was remarkably similar to the predicted gene phylogeny (Fig. 7). The most closely related clusters of genes, based on expression pattern similarity, included: rop3, rop6, and rop7 (phylogenetic group 2); rop2, rop5, rop4, and rop9 (all in phylogenetic group 4, except rop5); and rop1 and rop8 (a duplicate pair in phylogenetic group 1).
DISCUSSION
We have undertaken an initial investigation of the ROP GTPase family in monocots, using genomic resources to identify and classify rop genes in several grass crops, most notably, rice and maize. Following EST searches we sequenced two new rop mRNAs in rice, and identified their cognate genes, bringing the total to seven. This completes the rop family in rice, because our searches of rice genomic sequence identified only one other rop-like sequence, an apparent pseudogene. In comparison, the dicot Arabidopsis has 11 Rop genes (Winge et al., 2000; Vernoud et al., 2003). Additional screening and bioinformatics approaches identified six tentative unique rops in wheat, three in barley, and nine confirmed rop genes in maize.
Comparison of the maize and rice rop family suggests the existence of least one other maize rop, an ortholog of OsRop5 (Fig. 2; Table I). Also, duplicate rops originating from the ancestral maize genome tetraploidization (Gale and Devos, 1998) have not been identified for maize rop3, rop4, or rop5. If any of these maize rops exist, they are probably expressed at low levels or in a limited number of cells, because they are absent from current maize EST databases. An analysis of the knox homeobox gene family has shown that only two of the seven knox group I genes in rice have duplicated orthologs in maize (Sentoku et al., 1999). Thus, maize genes from the ancient tetraploidization may be lost at a high frequency (Freeling, 2001), suggesting the possibility that the “missing” rop duplicates have been eliminated from the genome.
An Evolutionary Framework for the Rop GTPase Family
A robust hypothesis describing the evolutionary history of a gene family can assist in relating experimental results across species boundaries, and in focusing experimental analyses on groups of closely related genes. Our phylogenetic analyses extend and refine previous analyses (Winge et al., 2000; Zheng and Yang, 2000), both by adding new monocot sequences and several new EST-derived dicot sequences, and by using Bayesian inference to evaluate more confidently the earliest duplication and divergence events in Rop family evolution. The inferred phylogeny allows us to define more clearly the four Rop groups that are present in angiosperms (Fig. 2). The simplest explanation for the genesis of these groups is that the common ancestor of monocots and dicots possessed at least four Rop paralogs, which were the progenitors for each of the modern Rop groups. Our phylogenetic analysis suggests two major modifications to previously published trees (Winge et al., 2000; Zheng and Yang, 2000): (a) It separates the group previously designated as Rop group II into groups 1 and 2; and (b) it suggests that AtROP8 is a highly divergent member of group 3, rather than a member of the separate group (previously designated group 2). The long branch on which AtROP8 is placed suggests that it is evolving more rapidly than other Rops, and the lack of highly conserved orthologs for this gene in other dicot sequence databases suggests it may have a function unique to Arabidopsis. Our alternative, strongly supported placement of AtROP8 illustrates the advantages of using gene sequences from multiple species (i.e. increasing taxon sampling) in phylogenetic analyses, which helps to overcome difficulties in correctly placing such divergent sequences.
There is strong support for regarding groups 1 and 2 as sister clades, originating by gene duplication before the monocot/dicot divergence. Proteins in these groups have a C-terminal extension, compared with conventional Rho GTPases (Ivanchenko et al., 2000; Winge et al., 2000), due to the addition of an intron/exon pair at the rop 3′ end. Intriguingly, our more extensive data bear out an earlier suggestion (Winge et al., 2000) that groups 1 and 2 have expanded to a much greater extent in monocots (four genes in rice, at least four in wheat, and six in maize) than in dicots (only three in Arabidopsis, poor representation in dicot EST databases). In contrast, expansion of group 4 is greater in dicots (six genes in Arabidopsis, three each identified in barrel medic, soybean, and tobacco) than in monocots (two genes in rice, two identified in barley and wheat, and three identified in maize). Thus, the Rop gene family has evolved in distinct evolutionary patterns in monocots and dicots. Whether these contrasting trends in Rop family evolution have any functional consequence is unknown. However, assigning specific functions to monocot Rops based on orthology to Arabidopsis Rops may be problematic, because the duplicated Rops in each lineage are likely to have acquired different functions during evolution.
It is intriguing that the six nonangiosperm plant sequences (four gymnosperm and two moss) occupy basal branches in group 3, which is the most basally placed group on the midpoint-rooted tree. At face value, the placement of these sequences suggests that the root of the Rop tree may actually be at the base of group 3, i.e. it may represent the more ancestral group of the four detected. However, few Rop genes have been isolated from nonangiosperm plants, and additional sampling from such species will be needed to clarify whether nonangiosperm members of groups 1, 2, and 4 exist and where the root of the four angiosperm ROP groups should be placed. No unequivocal monocot members of Rop group 3 were identified in public sequence databases, although OsRop5 is a candidate. However, the inconsistent placement of OsRop5 by the different phylogenetic methods and the low posterior probability (79) supporting its current position in the Bayesian tree indicate that other possibilities have not been ruled out. For example, OsRop5 could be a highly diverged, rice-specific member of group 4; if this is the case, then group 3 Rops would appear to have been lost during monocot evolution. Identifying monocot orthologs of OsRop5 should help resolve this issue.
Our well-supported phylogeny also provides a useful test for the suggestion that CNSs could serve as phylogenetic footprints to assist in identifying orthologs among similar genes in a gene family (Kaplinsky et al., 2002; Guo and Moose, 2003). Although examining CNSs alone predicts orthology for a smaller number of genes than the more comprehensive alignment-based phylogenetic analysis, the two methods give exactly the same answer where CNSs exist. Thus, our analysis supports the idea that the presence of a certain CNS in a grass gene can be taken as strong evidence of orthology; however, the absence of such a CNS (e.g. in ZmRop4) does not rule out orthology. That we were able to find CNSs in the 3′-UTRs of four of six identified orthologous groups of grass rops suggests that these will be quite useful general tools for determining orthology among grass genes, given the rapid increase in large-scale EST projects, and the bias toward 3′-UTR sequence versus 5′-UTR sequence due to truncated cDNA clones.
We examined all ROPs to define consensus amino acid sequences in the N-terminal GTPase domain that could differentiate among the four Rop groups, and showed that group consensus sequences in the regions designated RopSF1 and RopSF2 are useful for classifying novel Rop ESTs by group. Although only a limited number of changes differentiate groups 3 and 4, longer sequence elements, primarily RopSF1 and RopSF2, differentiate among groups 1, 2, and 3/4. Other, smaller clusters of Rop group-specific amino acids are located at the N terminus, the α1/switch I region, and the β2/β3 region. Similar analyses have defined subfamily-specific sequence elements in Rab GTPase proteins, which have been mapped onto the conserved GTPase structure in positions similar to those of some of the Rop group-specific motifs (Pereira-Leal and Seabra, 2000, 2001). These Rab sequence elements have been proposed to help mediate interactions with protein interactors specific for each Rab subfamily (Pereira-Leal and Seabra, 2000). The capacity of human GTP-bound Cdc42 and RhoA to bind to certain downstream effectors is also determined by elements in some of these same regions. For example, the β2/β3 turn contains residues important for determining the specificity for GTPase binding to certain Cdc42/Rac-Interactive Binding motif proteins (Abdul-Manan et al., 1999; Mott et al., 1999; Owen et al., 2000). The elements identified by the Rop group consensus sequences may serve similar functions. For example, differences among Rops in these regions could help distinguish among the ROP-Interacting Cdc42/Rac-Interactive Binding-containing proteins, a class of Rop targets whose members show differing affinities for Arabidopsis ROP1 (Wu et al., 2001).
Developmental Regulation of Maize rop Genes
To determine whether maize rop expression is upor down-regulated during development, we used MTRP and the high-throughput technique, MPSS, to survey rop expression levels in different maize tissues. Both techniques insured that cross-hybridization among members of the highly conserved rops did not interfere with collection and interpretation of the data. High expression levels for all nine rops are correlated with tissues undergoing rapid cell division and/or expansion (e.g. the shoot apex). Six of the nine rops assayed are expressed at much lower levels in mature leaves; only three rops (rop1, rop4, and rop9) are not dramatically reduced, relative to shoot apical meristem or immature leaf levels. This pattern is consistent with a role for Rop in maize vegetative development and cell morphogenesis, and with reduced requirements for most ROPs in fully mature cells.
Our experiments also reveal a high degree of overlap in the expression patterns of the nine rops, because relatively few experimental samples show preferential expression for one or two rops (e.g. rop8 and rop1 in endosperm). However, neither assay can address whether expression of specific rop genes is restricted to distinct cell types within these tissues. Our results, although much more extensive, match the general pattern seen for ROP expression in Arabidopsis: transcripts and protein are detectable in all tissues, with considerable overlap among genes (Winge et al., 1997; Li et al., 1998). However, the extensive rop expression during embryogenesis (from 15 to 30 DAP) and the differences in expression between undifferentiated and mature tissues had not been noted previously, and may hint at specific gene function. For example, the maintenance of rop1, rop4, and rop9 expression in mature leaf and in most vegetative tissues assayed suggests that these genes could be needed in sporophytic cells in a constitutive, “housekeeping” capacity. Because Rops are thought to participate in pathogen and stress responses in mature plants (Yang, 2002), our data suggest that these three genes are more likely to have important roles in such responses than the rops that are strongly down-regulated in mature tissue. This supposition is consistent with the finding that ROP1 and ROP4 are more potent than ROP2 and ROP3 in inducing superoxide production (a likely intermediate in pathogen response) when expressed heterologously in mammalian cells (Hassanain et al., 2000).
One of the most striking rop expression patterns was in maize pollen. In contrast to the overlapping rop expression in vegetative tissues, only rop2, rop8, and rop9 are expressed at high levels in pollen. This expression pattern, as well as their conserved sequence, suggests that rop2 and rop9 are functionally analogous to the Rop1-related group from dicots, which is required for pollen tube growth and polarity (Lin and Yang, 1997; Kost et al., 1999a; Li et al., 1999). The three Arabidopsis genes AtROP1, AtROP3, and AtROP5 are all expressed in pollen (Li et al., 1998) and may have arisen from relatively recent gene duplications (Winge et al., 2000). Although AtROP3 and AtROP5 are expressed both in vegetative tissue and, more weakly, in pollen, expression of AtROP1 is limited primarily to pollen. In contrast, maize rop2 and rop9 are expressed at high levels in both vegetative tissue and pollen. These observations suggest that, in Arabidopsis, duplication has allowed AtROP1 to specialize as a pollen-specific gene, whereas the maize genes have retained the ancestral vegetative and pollen expression patterns.
The Rop evolutionary framework provides a starting point for comparing gene expression profiles in the rop family among ancient duplicates and across species boundaries. The observation that more closely related genes also tend to cluster together based on expression profiling (Fig. 7) argues that these genes have at least the potential to carry out more closely related functions in the plant. It is also clear that expression patterns, even between maintained duplicate genes, do diverge over time, although the divergence can be minor (e.g. rop6 and rop7) or can constitute a more substantial change (rop5 versus rop1/rop8). As more high-throughput expression data for both Arabidopsis and rice become available, our rop MPSS expression database should become useful for addressing questions regarding the maintenance and divergence of specific gene expression patterns over the course of evolution. The robust Rop evolutionary framework we have proposed insures that comparisons of such expression data, as well as functional data, are made among the most suitable sets of genes.
CONCLUSIONS
We propose that angiosperm Rops be classified into four distinct groups (designated groups 1, 2, 3, and 4), based on phylogeny and conserved amino acid sequence elements. Each of the four primary groups is present in at least two widely divergent species (soybean and Arabidopsis), and Rops in at least three of the four groups have been identified in four other species (cotton, rice, wheat, and maize). These observations suggest that each Rop group provides some selective advantage that prevents its loss due to mutation. We believe our data further support and refine the hypothesis (Li et al., 1998; Yang, 2002) that each Rop group provides some unique function that has been maintained during evolution. We suggest that Rop family proteins in the different groups could assume specific functions based on (a) the identified distinct sequence elements in their GTPase domains, which help determine protein-protein interactions in vivo, and (b) differences in their C termini, which target Rops to distinct subcellular sites (Li et al., 1999; Ivanchenko et al., 2000; Lavy et al., 2002). In addition, rop expression profiles suggest that some Rop gene function is developmentally specialized. However, there exists the potential for genetic redundancy among Rops within a group, due to gene duplication, overlapping expression patterns and highly conserved sequences. Therefore, testing the hypothesis that group functions are distinct will likely require the construction of genetic stocks that are mutant for several rops, with the goal of eliminating all gene function within each group.
MATERIALS AND METHODS
Plasmids and Sequencing
Plasmids with inserts corresponding to the maize (Zea mays) rop5, rop6, and rop7 cDNAs were isolated from a Lambda ZAP maize shoot apical meristem cDNA library (provided by S. Hake and L. Smith, U.S. Department of Agriculture-Plant Gene Expression Center, Albany, CA) using standard low-stringency radioactive screening methods (Sambrook et al., 1989). The probe used to recover these clones was amplified from the highly conserved region (corresponding to amino acids 14-69 of maize ROP1) of two rice (Oryza sativa) EST clones (GenBank accession nos. D23963 and D41794) identified by the RGP of the Japanese Ministry of Agriculture. Sequenced cDNA clones for racA, racB, racC, and racD were identified by BLAST searches of the Pioneer Hi-bred International EST database (Hassanain et al., 2000). These genes will be referred to as rop1 through rop4. EST clones corresponding to rop8 (GenBank accession no. AI964615) and rop9 (GenBank accession nos. AW506911 and AI668259) were identified by searching the ZmDB maize EST database (Dong et al., 2003); similarly, rice EST clones generated by the RGP, corresponding to the previously uncharacterized rice genes designated OsRop4 (GenBank accession nos. C26233 and AU077893) and OsRop5 (GenBank accession no. C74803), were identified. These clones were requested and sequenced in full. All sequencing used standard automated protocols, and was done at the University of North Carolina DNA Sequencing Facility, at the Oregon State University Center for Gene Research and Biotechnology Central Services Lab, or at Pioneer Hi-Bred, International. Sequences were analyzed using the Wisconsin Package v10.3 (Accelrys Inc., San Diego) and Web-based BLAST (Altschul et al., 1997).
Genomic sequences for maize rop2, rop6, rop7, and rop9 were obtained by PCR amplification using GSPs, followed by direct sequencing or sequencing of the cloned product. Certain amplified fragments were cloned with either the pPCR-Script (+) kit (Stratagene, La Jolla, CA) or the TA Cloning kit (Invitrogen, Carlsbad, CA) using the manufacturer's protocol. Rice rop genomic sequences were identified in GenBank by BLAST, and annotated using GenePalette (http://www.genepalette.org). Intron/exon junctions were determined by comparing cDNA and genomic sequence, assisted by the Splice Predictor application (Usuka et al., 2000). Because neither rop9 cDNA was full-length, we used a GSP in the 5′-UTR of rop2, in combination with a rop9 intron-specific primer, to amplify the 5′ end of the rop9 coding region. Thus, the initial 126 bp of the rop9 coding region was derived from genomic sequence.
Phylogenetic Analysis
Phylogenetic trees were generated from nucleotide sequences using the software packages PAUP* 4.0b10 (Phylogenetic Analysis Using Parsimony, Sinaur Associates, Sunderland, MA; Unix) for maximum parsimony and maximum likelihood trees, and MrBayes v2.01 (Macintosh) for Bayesian inference of phylogeny (Huelsenbeck and Ronquist, 2001). All publicly available Rop gene sequences were retrieved from the January 2003 versions of GenBank. In addition, the TIGR Gene Indices (http://www.tigr.org/tdb/tgi.shtml; Quackenbush et al., 2001) and the PlantGDB EST Clusters (http://www.plantgdb.org/) were searched by BLAST for Rop sequences from barley (Hordeum vulgare), wheat (Triticum aestivum), sorghum, tomato (Lycopersicon esculentum), potato (Solanum tuberosum), soybean (Glycine max), tree cotton (Gossypium arboreum), barrel medic (Medicago truncatula), European aspen (Populus tremula × Populus temuloides), and loblolly pine (Pinus taeda) by BLAST, and unique contigs were retrieved. Two TUCs, PtTUC1 (loblolly pine) and PpinTUC1 (maritime pine [Pinus pinaster]), were assembled from downloaded GenBank ESTs by Phrap. The reference numbers for these sequences are in Supplemental Table III. Only sequences containing the full-length coding region were used in the analysis, and TIGR-GI TC and PlantGDB TUC sequences were used only if they were derived from at least three ESTs. No outgroup sequences were included, because (a) the evolutionary distance between the plant and non-plant Rho sequences prevented a confident alignment of a significant portion of coding sequence, and (b) trees generated using the more limited alignment possible with outgroup sequences indicated that sequence divergence was too great to provide a useful and well-supported root for the Rop tree (data not shown). Due to high conservation among Rop proteins, nucleotide sequences were used to allow the maximum phylogenetic resolution. The nucleotide alignment was generated using GCG Pileup and ClustalX and adjusted manually with guidance from the corresponding amino acid alignment. Due to the presence of numerous insertions and/or deletions and repeated codons, Rop sequences near the 3′ end of the coding sequence (noted on the amino acid alignment) were not included in the data set used for analysis, because they could not be aligned with confidence. Use of longer data sets including a “best guess” alignment of this 3′ end region did not markedly alter the resulting tree topology or increase the support for the branches (data not shown). Both alignments are available at http://oregonstate.edu/~fowlerjo/RopAlign/.
For parsimony analyses in PAUP*, gaps in the nucleotide sequence were treated as a fifth character state (coded for by an “I”), and heuristic searches were conducted employing 500 replicates of stepwise random sequence addition and tree bisection-reconnection branch swapping. Two hundred replicates of non-parametric bootstrapping (Felsenstein, 1985) with the same settings as the maximum parsimony analyses, but on informative characters only, were used to estimate branch support. Sequence gaps were treated as missing data in both maximum likelihood and Bayesian analyses. For maximum likelihood analyses in PAUP*, heuristic replicates were conducted using a general time-reversible evolutionary model with estimated base frequencies and rate variation across sites modeled by codon position. For phylogeny inference in MrBayes, a general time reversible evolutionary model was used with estimated base frequencies and rate variation across sites modeled by codon position, with each position having its own γ-distribution. Posterior probabilities were calculated after a run of 1,000,000 generations, with the first 100,000 generations as the “burnin” period. Posterior probabilities were interpreted as estimates of branch support. Settings for all phylogenetic analyses are included in the executable block at the end of the sequence alignment file on the Web site. Tree files generated by PAUP* or MrBayes were rooted using midpoint rooting, and then imported into Treeview (Page, 1996) for formatting into figures.
CNSs and Genetic Mapping
CNSs were identified in the UTRs of the monocot Rop transcripts using BLAST 2 Sequences (Tatusova and Madden, 1999), ClustalX, and the parameters defined by Kaplinsky et al. (2002) and Guo and Moose (2003).
Maize mapping was done using RFLP or CAPS markers for each gene in sets of recombinant inbred lines established by Pioneer Hi-Bred (rop1-4), or Brookhaven National Labs (rop6 and rop7). Rice rops were mapped by BLAST of cDNA sequence versus GenBank rice genomic sequence produced by the International Rice Genome Sequencing Program. International Rice Genome Sequencing Program bacterial artificial chromosome clones are genetically mapped, providing an approximate map position for the rop.
RNA Isolation and Multiplex RT-PCR
Corn seeds (W22 inbred) were grown on moist paper towels for 2 weeks at room temperature. RNA was extracted from: (a) root tip, the last 5 mm of the primary and adventitious seminal roots; (b) root shank, tissue basal to the root tip, including the zone of elongation, with few or no visible root hairs; (c) shoot apex, dissected shoot apices, 5 mm long, including the apical meristem and immature leaves, but lacking the coleoptile; and (d) mature leaf, from juvenile leaf blades of 2-week-old seedlings, sampled 10 mm from the leaf tip. Pollen from newly exerted anthers of W22 plants was collected over a 2-h period. Samples of approximately 100 mg were ground following freezing in liquid nitrogen using RNase-free pestles, and RNA was extracted from homogenized tissues using Trizol reagent (Invitrogen). RNA concentrations were determined by spectrophotometry, and cDNA was generated from 5 μg of total RNA using oligo-d(T) primers and the SuperScript cDNA Synthesis kit (Invitrogen).
To determine relative expression levels of the rop genes, we used MTRP (Nebenführ and Lomax, 1998) with each of the three primer sets. PCR primers specific to each maize rop gene (Supplemental Table I) were designed to minimize cross-amplification by selecting diverged sequences as targets. The primers generated amplified products of 280 to 600 bp; this narrow size range minimized preferential amplification of shorter fragments. Two control primer sets amplified either maize actin1 (Mac1; GenBank accession no. J01238) or a widely expressed Elongation Factor1-α gene (Fernandes et al., 2002; PlantGDB ZMtuc03-04-07.21400). Three sets of primers (set 1, primers for rop4, 6, and 8; set 2, rop1, 3, and 9; set 3, rop2, 5, and 7) were used for multiplex PCR. PCR parameters (Supplemental Table IV) were optimized for each reaction set using an equimolar mix of all nine rop cDNAs (in plasmids) as a positive control. Amplification conditions (35 cycles of 95°C for 45 s, 63°C for 1 min, and 72°C for 1 min) were chosen as near-optimal for all three sets of primers. Eight serial dilutions (1:4) of each cDNA sample were used as templates for separate PCR reactions, and products were separated using 1.5% to 2% (w/v) agarose. All MTRP experiments were repeated in triplicate, starting with RNA isolation from three independently frozen aliquots of each tissue.
Generation and Analysis of MPSS Data
The expression data for all nine maize rop genes were extracted from a large proprietary database of MPSS experiments. This database was created using the MPSS methodology as described (Brenner et al., 2000a, 2000b). In brief, poly(A) RNA was extracted from various maize tissues, reverse transcribed, and “cloned” onto microbeads. Between 2 × 105 and 2 × 106 individual cDNAs in each sample were assayed for 17-mer signature sequences, and the total number of each signature sequence in these samples was tabulated and normalized to parts per million to provide an estimate of transcript abundance that could be compared across experiments. The 5′ end of each signature is defined by the DpnII (GATC) site nearest the poly(A) tail. Signature sequences for each of the nine maize rops, each of which uniquely corresponds to one rop, were determined based on cDNA sequence, and MPSS values corresponding to these signatures were extracted from the larger database and assigned to the correct rop in an Excel spreadsheet (supplemental data). Values for a given transcript in two different experiments can be compared using statistics derived from the Poisson distribution (Audic and Claverie, 1997), and 95% confidence intervals were calculated based on the raw signature count and signature sample size to evaluate whether observed differences were statistically significant (Fig. 7).
MPSS values for all nine rops from 57 different experiments covering a representative spectrum of maize tissues and developmental stages were imported into GeneSpring 5.1 (Silicon Genetics, Redwood City, CA) for a more comprehensive analysis. To provide a measure of the relative level of transcription of each rop, MPSS values were normalized with respect to the median value for each rop across this set of experiments. For rop3 and rop6, which were associated with low expression values throughout the data set, we normalized with respect to 5, because this is the value that is significantly different from 0, using a 95% confidence interval. Any positive MPSS values less than five were ignored in the subsequent analyses. The normalized expression values were then used to hierarchically cluster the gene expression patterns (Eisen et al., 1998), using Spearman rank-order correlation, which tests for similarity when values may not be normally distributed. Hierarchical clustering was also used to arrange the different experiments (developmental stages and tissues) into groups of similar rop expression patterns. Relative expression values were displayed graphically using a color scale. The samples in which each given gene was the most highly expressed were identified (Table II), and the statistical significance (99.9% confidence interval) of the difference between the highly expressed sample and the median sample for each gene was confirmed.
Distribution of Materials
Novel materials described in this publication may be available for noncommercial research purposes upon acceptance and signing of a material transfer agreement. In some cases, such materials may contain or be derived from materials obtained from a third party. In such cases, distribution of material will be subject to the requisite permission from any third-party owners, licensors, or controllers of all or parts of the material. Obtaining any permissions will be the sole responsibility of the requestor.
Supplementary Material
Acknowledgments
We thank D. Braun, R. Cole, M. Foss, C. Lawrence, and C. Rivin for useful critiques of the manuscript. We also appreciate the assistance of C. Simmons (Pioneer Hi-Bred) with the interpretation of the MPSS data. We acknowledge both the RGP of the Japanese Ministry of Agriculture (http://rgp.dna.affrc.go.jp/) and the National Science Foundation-sponsored Maize Gene Discovery project (http://www.zmdb.iastate.edu/) for providing plasmids from their EST sequencing projects. We acknowledge the Oregon State University Center for Gene Research and Biotechnology Central Services Lab and the Pioneer DNA Sequencing Facility for sequencing.
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 98-35304-6670 to J.E.F.) and by the National Science Foundation (grant no. IBN-0111078 to J.E.F.); the project was initiated by J.E.F. in the lab of R.S.Q.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.029900.
References
- Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Siminovitch KA, Rosen MK (1999) Structure of Cdc42 in complex with the GTPase-binding domain of the “Wiskott-Aldrich syndrome” protein. Nature 399: 379-383 [DOI] [PubMed] [Google Scholar]
- Ahn S, Tanksley SD (1993) Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA 90: 7980-7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7: 986-995 [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J (2002) RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296: 2026-2028 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M et al. (2000a) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630-634 [DOI] [PubMed] [Google Scholar]
- Brenner S, Williams SR, Vermaas EH, Storck T, Moon K, McCollum C, Mao JI, Luo S, Kirchner JJ, Eletr S et al. (2000b) In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc Natl Acad Sci USA 97: 1665-1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Roy L, Freeling M, Walbot V, Brendel V (2003) ZmDB, an integrated database for maize genome research. Nucleic Acids Res 31: 244-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791 [DOI] [PubMed] [Google Scholar]
- Fernandes J, Brendel V, Gai X, Lal S, Chandler VL, Elumalai RP, Galbraith DW, Pierson EA, Walbot V (2002) Comparison of RNA expression profiles based on maize expressed sequence tag frequency analysis and micro-array hybridization. Plant Physiol 128: 896-910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler JE, Quatrano RS (1997) Plant cell morphogenesis: plasma membrane interactions with the cytoskeleton and cell wall. Annu Rev Cell Dev Biol 13: 697-743 [DOI] [PubMed] [Google Scholar]
- Freeling M (2001) Grasses as a single genetic system: reassessment 2001. Plant Physiol 125: 1191-1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wu G, Yang Z (2001) Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J Cell Biol 152: 1019-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yang Z (2001) Rop GTPase: a master switch of cell polarity development in plants. Trends Plant Sci 6: 545-547 [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95: 1971-1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92-100 [DOI] [PubMed] [Google Scholar]
- Guo H, Moose SP (2003) Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15: 1143-1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A (1998) Rho GTPases and the actin cytoskeleton. Science 279: 509-514 [DOI] [PubMed] [Google Scholar]
- Hall A, Nobes CD (2000) Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos Trans R Soc Lond B Biol Sci 355: 965-970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanain HH, Sharma YK, Moldovan L, Khramtsov V, Berliner LJ, Duvick JP, Goldschmidt-Clermont PJ (2000) Plant rac proteins induce superoxide production in mammalian cells. Biochem Biophys Res Commun 272: 783-788 [DOI] [PubMed] [Google Scholar]
- Helentjaris T, Weber D, Wright S (1988) Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118: 353-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshberg M, Stockley RW, Dodson G, Webb MR (1997) The crystal structure of human rac1, a member of the rho-family complexed with a GTP analogue. Nat Struct Biol 4: 147-152 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754-755 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294: 2310-2314 [DOI] [PubMed] [Google Scholar]
- Ivanchenko M, Vejlupkova Z, Quatrano RS, Fowler JE (2000) Maize ROP7 GTPase contains a unique, CaaX box-independent plasma membrane targeting signal. Plant J 24: 79-90 [DOI] [PubMed] [Google Scholar]
- Johnson DI (1999) Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev 63: 54-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinsky NJ, Braun DM, Penterman J, Goff SA, Freeling M (2002) Utility and distribution of conserved noncoding sequences in the grasses. Proc Natl Acad Sci USA 99: 6147-6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K (1999) The small GTP-binding protein Rac is a regulator of cell death in plants. Proc Natl Acad Sci USA 96: 10922-10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua N-H (1999a) Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol 145: 317-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Mathur J, Chua NH (1999b) Cytoskeleton in plant development. Curr Opin Plant Biol 2: 462-470 [DOI] [PubMed] [Google Scholar]
- Lapouge K, Smith SJ, Walker PA, Gamblin SJ, Smerdon SJ, Rittinger K (2000) Structure of the TPR domain of p67phox in complex with Rac GTP. Mol Cell 6: 899-907 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S (2002) A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431-2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemichez E, Wu Y, Sanchez JP, Mettouchi A, Mathur J, Chua NH (2001) Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev 15: 1808-1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z (1999) Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11: 1731-1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol 126: 670-684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wu G, Ware D, Davis KR, Yang Z (1998) Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiol 118: 407-417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Yang Z (1997) Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell 9: 1647-1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Settleman J (1999) The role of rho family GTPases in development: lessons from Drosophila melanogaster. Mol Cell Biol Res Commun 1: 87-94 [DOI] [PubMed] [Google Scholar]
- Mott HR, Owen D, Nietlispach D, Lowe PN, Manser E, Lim L, Laue ED (1999) Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK. Nature 399: 384-388 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Lomax TL (1998) Multiplex titration RT-PCR: rapid determination of gene expression patterns for a large number of genes. Plant Mol Biol Rep 16: 323-339 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759-764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier C, Brunger AT (1999) Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell 96: 363-374 [DOI] [PubMed] [Google Scholar]
- Owen D, Mott HR, Laue ED, Lowe PN (2000) Residues in Cdc42 that specify binding to individual CRIB effector proteins. Biochemistry 39: 1243-1250 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357-358 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC (2000) The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol 301: 1077-1087 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC (2001) Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313: 889-901 [DOI] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC et al. (1999) Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA 96: 9212-9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, Matsuoka M (1999) Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11: 1651-1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settleman J (2001) Rac 'n Rho: the music that shapes a developing embryo. Dev Cell 1: 321-331 [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL (1999) BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174: 247-250 [DOI] [PubMed] [Google Scholar]
- Usuka J, Zhu W, Brendel V (2000) Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics 16: 203-211 [DOI] [PubMed] [Google Scholar]
- Valster AH, Hepler PK, Chernoff J (2000) Plant GTPases: the rhos in bloom. Trends Cell Biol 10: 141-146 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131: 1191-1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299-1304 [DOI] [PubMed] [Google Scholar]
- Wilson WA, Harrington SE, Woodman WL, Lee M, Sorrells ME, McCouch SR (1999) Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics 153: 453-473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge P, Brembu T, Bones AM (1997) Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol Biol 35: 483-495 [DOI] [PubMed] [Google Scholar]
- Winge P, Brembu T, Kristensen R, Bones AM (2000) Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics 156: 1959-1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Gu Y, Li S, Yang Z (2001) A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13: 2841-2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (2002) Small GTPases: versatile signaling switches in plants. Plant Cell Suppl 14: S375-S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14: 2787-2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Yang Z (2000) The Rop GTPase: an emerging signaling switch in plants. Plant Mol Biol 44: 1-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.