Abstract
To study the role of LecRK (lectin-like receptor kinase) genes in the legumerhizobia symbiosis, we have characterized the four Medicago truncatula Gaernt. LecRK genes that are most highly expressed in roots. Three of these genes, MtLecRK7;1, MtLecRK7;2, and MtLecRK7;3, encode proteins most closely related to the Class A LecRKs of Arabidopsis, whereas the protein encoded by the fourth gene, MtLecRK1;1, is most similar to a Class B Arabidopsis LecRK. All four genes show a strongly enhanced root expression, and detailed studies on MtLecRK1;1 and MtLecRK7;2 revealed that the levels of their mRNAs are increased by nitrogen starvation and transiently repressed after either rhizobial inoculation or addition of lipochitooligosaccharidic Nod factors. Studies of the MtLecRK1;1 and MtLecRK7;2 proteins, using green fluorescent protein fusions in transgenic M. truncatula roots, revealed that they are located in the plasma membrane and that their central transmembrane-spanning helix is required for correct sorting. Moreover, their lectin-like domains appear to be highly glycosylated. Of the four proteins, only MtLecRK1;1 shows a high conservation of key residues implicated in monosaccharide binding, and molecular modeling revealed that this protein may be capable of interacting with Nod factors. However, no increase in Nod factor binding was found in roots overexpressing a fusion in which the kinase domain of this protein had been replaced with green fluorescent protein. Roots expressing this fusion protein however showed an increase in nodule number, suggesting that expression of MtLecRK1;1 influences nodulation. The potential role of LecRKs in the legume-rhizobia symbiosis is discussed.
The lectin-like receptor kinases (LecRKs) are a class of proteins originally described from Arabidopsis (Hervé et al., 1996). They have a structure similar to other plant receptor-like kinases (RLKs; Shiu and Bleecker, 2001; Cock et al., 2002) with an N-terminal targeting signal, a presumably extracellular domain, a single transmembrane (TM)-spanning helix, and a cytosolic kinase domain. The Arabidopsis genome contains over 610 RLKs that have been shown to be monophylogenetic with respect to the kinase domain and most closely related to the fruitfly (Drosophila melanogaster) Pelle and related animal cytoplasmic kinases (Shiu and Bleecker, 2001). In cases in which they have been tested, these kinases have been shown to have specificity for phosphorylation on Ser/Thr residues (Shiu and Bleecker, 2001).
The plant RLKs can be grouped into more than 21 structural classes based on their extracellular domains (Shiu and Bleecker, 2001). The LecRKs represent one such class in which the extracellular domain is related to legume lectins. The Arabidopsis genome contains at least 42 LecRK sequences that have been grouped into three classes (A-C) plus an additional nine sequences of related soluble lectins (Barre et al., 2002). Searches in plant databases reveal that LecRK genes are widespread in higher plants, but apart from Arabidopsis (Hervé et al., 1996, 1999, Riou et al., 2002), LecRK genes have been studied only in lombardy poplar (Populus nigra var italica; Nishiguchi et al., 2002). RLK genes with legume lectin-like domains are not found in the complete genome sequences of yeast (Saccharomyces cerevisiae) or human (Homo sapiens).
The legume lectins are a class of proteins that are most abundant in the seeds of legumes (Brewin and Kardailsky, 1997). It is now clear that they are not legume specific because related genes have also been described in other plants and also in animals. A lectin may be defined as a protein, other than immunoglobulins, with at least one non-catalytic carbohydrate-binding domain, and they can be classified into a number of non-related families of which the legume lectins comprise one such class (Loris, 2002). Structural studies have been carried out on many legume lectins, and they are all calcium-/manganese-containing (glyco) proteins with a conserved tertiary structure composed of three β-sheets (Loris et al., 1998). They show variation in carbohydrate binding specificity due largely to variability around a conserved core of residues defining a monosaccharide-binding site. They are usually dimeric or tetrameric proteins, and for some of them (for example, the Dolichos biflorus lectin), oligomerization creates one or more binding sites for hydrophobic ligands such as adenine, related cytokinins, and auxins (Bouckaert et al., 1999; Hamelryck et al., 1999). In vivo, their actual ligands are not known but could include more complex glycans in addition to simple sugars or hydrophobic hormones. They have been implicated in such diverse physiological processes as protein storage, defense, recognition, protein sorting, embryogenesis, and development (Brewin and Kardailsky, 1997; Brill et al., 2001; Rudiger and Gabius, 2001), but whether they have a major biochemical function leading to these different physiological roles remains enigmatic.
Because of their homology to legume lectins, it seems reasonable to suppose that LecRKs could be involved in the recognition and transduction of saccharidic signals. However, sequence analysis and molecular modeling of Arabidopsis LecRKs has revealed a poor conservation of the residues involved in monosaccharide binding, whereas the hydrophobic-binding site appears to be better conserved (Hervé et al., 1999; Barre et al., 2002). Thus, these proteins are considered unlikely receptors for simple sugars but may be involved in recognition of either small hydrophobic hormones or more complex glycans (Barre et al., 2002). Plants recognize and respond to a variety of glycans of both endogenous (e.g. cell wall-derived molecules) and exogenous (e.g. from plant pathogens, predators, and symbionts) origin. These signals have been termed oligosaccharins and include the rhizobial lipochitooligosaccharides, pathogen-derived chitin fragments and glucan elicitors, and plant cell wall-derived pectin and xyloglucan oligosaccharides (Côté and Hahn, 1994; Creelman and Mullet, 1997). To date, the receptors of very few of these molecules have been cloned. Although there are as yet no functional studies on the role of LecRKs, detailed studies on the regulation of one of the Arabidopsis genes, Ath.lecRK-a1 (coding for protein At3g59700 in the Arabidopsis Information Resource [TAIR] database; Garcia-Hernandez et al., 2002), suggests that it is involved in plant development and also adaptive processes such as wounding (Riou et al., 2002). The poplar gene (PnLPK) is also up-regulated by wounding, but in contrast to Ath. lecRK-a1, it is more highly expressed in roots than other organs (Nishiguchi et al., 2002).
The structure of LecRKs has led to suggestions that in legumes they could be involved in the legumerhizobia symbiosis (Hervé et al., 1996; Hirsch, 1999). The establishment of this symbiosis requires at least two types of rhizobial saccharidic signals. First, lipochitooligosaccharidic Nod factors are obligatory for recognition, infection, and nodule organogenesis (Dénarié et al., 1996). These molecules elicit symbiotic responses at picomolar to micromolar concentrations and are considered to be perceived by plasma membrane (PM) receptors of high affinity and specificity, expressed in roots of legume plants (see Cullimore et al., 2001; Geurts and Bisseling, 2002). Using a biochemical approach, two different binding sites for these molecules have been characterized in Medicago spp. (Nod-factor binding sites 1 and 2, referred to as NFBS1 and NFBS2, respectively), but their role in nodulation is not clear (Bono et al., 1995; Gressent et al., 1999, 2002). A genetic approach has led recently to the cloning of a receptor kinase, with a novel extracellular domain, important for Nod factor responses (Endre et al., 2002; Stracke et al., 2002), but its dual role in establishment of a symbiosis with mycorrhizal fungi suggests that it may not be involved directly in Nod factor binding (Kistner and Parniske, 2002). The other type of saccharidic signals involved in the symbiosis are the rhizobial cell surface polysaccharides (exo-, lipo-, and capsular polysaccharides) and include small molecules derived from them that may play a signaling role (Niehaus and Becker, 1998). Generally, these saccharides appear to play a major role in infection rather than nodule organogenesis (Niehaus and Becker, 1998; Hirsch, 1999).
The other evidence that suggests that LecRKs could be involved in the symbiosis relies on the numerous studies on soluble legume lectins. In the 1970s, these studies culminated in the “lectin recognition hypothesis,” which proposed that plant lectins mediate specificity in the legume-rhizobia symbiosis (for review, see Brewin and Kardailsky, 1997; Hirsch, 1999). Since 1989, several studies using transgenic plants have lent support to this hypothesis because transgenic roots expressing heterologous lectins are capable of being nodulated by the heterologous rhizobial species (Diaz et al., 1989; Kijne et al., 1997; Hirsch, 1999). For this effect, the sugar-binding site of the lectin is required (Diaz et al., 2000; van Rhijn et al., 2001), and the latest results suggest that exopolysaccharide rather than Nod factors may be the critical rhizobial component for extension of the host range (van Rhijn et al., 2001). The lectins have been suggested to function as a glue, sticking the two partners together for Nod factors to operate, or as a cloaking device to avoid recognition as a pathogen, or as an element in signaling and friend recognition (Hirsch, 1999). For this latter role, LecRKs are suggested to provide a signal transduction mechanism through heterologously oligomerizing with the soluble lectins.
In this article, we describe the first study, to our knowledge, of LecRK genes in a legume, and we specifically address the question of their potential role in relation to the symbiosis with rhizobia. For this work, we have used the model legume Medicago truncatula Gaernt., in which a variety of genomic tools have been developed (Barker et al., 1990; Cook, 1999). This plant is nodulated by Sinorhizobium meliloti, the genome of which has been sequenced recently (Galibert et al., 2001). The major Nod factor produced by this species is described as NodSm-IV (Ac, S, C16:2) because it contains four N-acetyl glucosamine residues of which the reducing sugar is 6-O sulfated, whereas the terminal non-reducing sugar is 6-O acetylated and N-acylated with a C16:2 fatty acid.
RESULTS
Identification of Four M. truncatula LecRK (MtLecRK) Genes Expressed in Roots
To isolate clones related to LecRK genes potentially playing a role in the legume-rhizobia symbiosis, the following three root or nodule cDNA libraries were screened with a probe consisting of the kinase domain of the Arabidopsis Ath.lecRK-a1 gene: nitrogen-starved roots; roots following inoculation for 6, 24, and 48 h with S. meliloti; and 4-d-old nodules. Of the 35 kinase-containing clones isolated, six were related to either soluble or other receptor kinases. The rest of the clones were related to LecRKs, thus showing that the Ath.lecRK-a1 kinase probe is quite specific for LecRKs despite the large number of kinase genes in higher plants. Two, 25, and two clones were isolated from the three libraries, respectively (corresponding to an abundance of 1:165,000, 1:16,500, and 1:120,000). Restriction enzyme digestion and partial sequence analysis of the clones identified sequences related to four different genes. These genes were mapped on the molecular genetic linkage map of M. truncatula (Thoquet et al., 2002); three of them are located on linkage group 7, and the fourth is located on linkage group 1 (T. Huguet, personal communication). These genes have been named MtLecRK7;1 (GenBank accession no. AY358027), MtLecRK7;2 (GenBank accession no. AY358028), MtLecRK7;3 (GenBank accession no. AY358029), and MtLecRK1;1 (GenBank accession no. AY358030) according to their chromosomal location. The number of clones related to each of these MtLecRK genes was two, nine, nine, and nine, respectively. Recent screening of the M. truncatula expressed sequence tag (EST) banks (Quackenbush et al., 2001; Journet et al., 2002) revealed two, two, four, and two clones that are almost certainly derived from the MtLecRK7;1, MtLecRK7;2;MtLecRK7;3, and MtLecRK1;1 genes, respectively. Three ESTs (GenBank accession nos. AW692710, BF632073, and BE319405) represent three additional genes that are highly related to MtLecRK7;1/MtLecRK7;2, and a fourth EST (GenBank accession no. AW691413) represents a gene with high sequence similarity to MtLecRK7;3. The cluster MtD02854 with seven ESTs (http://medicago.toulouse.inra.fr/Mt/EST/DOC/MtB.html) also appears to encode an LecRK. Thus, we have so far identified nine LecRK genes in M. truncatula, and there are likely to be more.
Sequence and Phylogenetic Analysis of MtLecRKs
The longest clones related to each of the four genes were sequenced in their entirety, and for genes MtLecRK7;1, MtLecRK7;2, and MtLecRK7;3, the sequences were extended by 5′-RACE. Complete open reading frames (ORFs) were obtained for MtLecRK7;1, MtLecRK7;2, and MtLecRK1;1 (Fig. 1). For MtLecRK7;3, the initiating Met indicated in Figure 1 corresponds to the initiating Met of MtLecRK7;1 and 7;2, but it should be noted that we cannot exclude the possibility that the MtLecRK7;3 ORF starts further upstream. The MtLecRK7;1, MtLecRK7;2, MtLecRK7;3, and MtLecRK1;1 ORFs code for proteins of 659 (73.9 kD), 669 (74.9 kD), 682 (76.0 kD), and 678 (75.8 kD) amino acids, respectively, and these proteins are shown aligned with Ath.LecRK-a1 (At3g59700) in Figure 1. The M. truncatula proteins are more similar in their kinase domains (53%-87% similarity) than they are in their lectin-like domains (39%-83% similarity). MtLecRK7;1 and MtLecRK7;2 are the most closely related, followed by MtLecRK7;3, with MtLecRK1;1 being the most divergent. Structural predictions using the program THMHH suggest that all four proteins contain an N-terminal signal sequence and that they possess a TM-spanning helix of 23 residues situated between the lectin-like and kinase domains. Although there are some variation between the structural prediction programs, these proteins are generally predicted to be Type Ia PM proteins, with cytosolic kinase domains. The kinase domains of all four proteins, by alignment to the kinase domain of Ath.LecRK-a1, contain the 12 sub-domains and the two conserved sequence motifs in the catalytic regions of Ser/Thr kinases (Fig. 1). A region of high variability in these proteins is the carboxy-terminal region, which in protein Tyr kinases is implicated in regulatory functions (Hubbard and Till, 2000).
Figure 1.
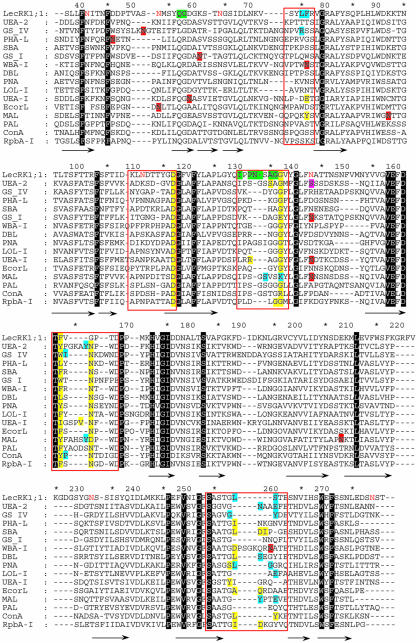
Alignment of the M. truncatula LecRKs and Ath.LecRK-a1. The position of the putative targeting sequences and the TM-spanning helices are marked by double underlining the corresponding regions in Ath.LecRK-a1. The lectin-like domains lie between the two marked sequences, and the kinase domains are C-terminal. The four starred residues in the lectin-like domain correspond to the position of the highly conserved residues involved in carbohydrate binding in soluble lectins (D, G, hydrophobic, and N, respectively). The four residues marked with crosses are putatively involved in cation binding. In the kinase domain, the position of the 12 sub-domains are marked, and the two motifs designating Ser/Thr specificity are singly underlined.
In the lectin-like domain, motif searches using ScanProsite revealed that MtLecRK1;1 contains the lectin_legume alpha and beta chain signatures that are not as highly conserved in the other three proteins. The lectin-like extracellular domains of MtLecRK7;1, MtLecRK7;2, MtLecRK7;3, and MtLecRK1;1 are predicted to be glycosylated with four, eight, seven, and eight N-glycosylation sites, respectively.
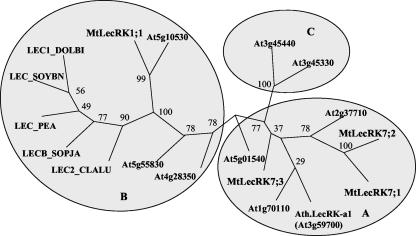
BLASTP comparisons with the National Center for Biotechnology Information nonredundant database (Altschul et al., 1997) or with TAIR Arabidopsis database (Garcia-Hernandez et al., 2002) were used to identify the sequences most closely related to either the lectin-like or the kinase domains of the four proteins. These analyses revealed that the closest relatives to each of the four MtLecRK kinase domains are the kinase domains of LecRKs from other higher plants (Arabidopsis, soybean [Glycine max], rice [Oryza sativa], and lombardy poplar). Similarly, the lectin-like domains of MtLecRK7;1, MtLecRK7;2, and MtLecRK7;3 have their closest current homologs in the lectin-like domains of higher plant LecRKs. The lectin-like domain of MtLecRK1;1, however, although clearly having a close homolog in Arabidopsis (At5g10530), is otherwise most similar to soluble legume lectins. A phylogenetic analysis was performed on the lectin-like domains of the four MtLec-RKs, nine representatives of the different classes (A-C) of Arabidopsis LecRKs (see Barre et al., 2002), and five soluble legume lectins that show closest homology to MtLecRK1;1 (Fig. 2). The results show that the lectin-like domains of MtLecRK7;1, MtLecRK7;2, and MtLecRK7;3 are most closely related to the Class A Arabidopsis LecRKs, whereas the lectin-like domain of MtLecRK1;1 falls into a clade containing the At5g10530 Class B protein and the soluble legume lectins.
Figure 2.
Phylogenetic analysis of the lectin-like domains of M. truncatula (Mt) and Arabidopsis (At) LecRKs and selected soluble legume lectins (LEC). The soluble lectins are from soybean (LEC_SOYBN), D. biflorus (LEC1_DOLB1), pea (Pisum sativum; LEC_PEA), Cladrastis lutea (LEC2_CLALU), and Sophora japonica (LECB_SOPJA). The chosen AtLecRKs have been classified as class A (At3g59700, At2g37710, At1g70110, and At5g01540), class B (At3g55830, At5g10530, and At4g28350), and class C (At3g45330 and At3g45440) by Barre et al. (2002). Alignment of the amino acid sequences was performed using ClustalX, distances between sequences were calculated using PROTDIST with 100 bootstrap analyses, and trees were constructed using BIONJ in the neighbor-joining programs of the PHYLIP packages. The bootstrap values are shown.
When compared with the sequences of soluble legume lectins of known structures, the four MtLecRK sequences appear to contain all conserved amino acids necessary for binding Ca2+ and Mn2+ (Fig. 1; Loris et al., 1998). However, the conservation of residues involved in carbohydrate binding is quite different (Fig. 3). Interaction of legume lectins with monosaccharides is mediated by key hydrogen bonds involving three extremely well-conserved residues: an Asp and an Asn that interact with the ligand by their side chain and a Gly that interacts by the backbone. In addition, stacking of the hydrophobic face of the sugar with aromatic amino acids is commonly observed. Of MtLecRK7;1, MtLecRK7;2, and MtLecRK7;3, the only residue conserved is the Asn in MtLecRK7;1 (Fig. 1). However, MtLecRK1;1 shows conservation of both Asp-118 and Gly-138, whereas the Asn in the triad is replaced by Gly at position 166. The aromatic amino acid Phe-164 is also in an appropriate position to participate in sugar binding (Figs. 1 and 3).
Figure 3.
Sequence alignment between the MtLecRK1;1 lectin-like domain and selected soluble legume lectin sequences corresponding to three-dimensional structures used in model building. Loops involved in ligand binding have been boxed in red. Amino acids involved in binding monosaccharides and oligosaccharides (or aglycons) have yellow and blue backgrounds, respectively. N-glycosylation sites confirmed by crystallography have a red background, whereas the O-GlcNAc site in UEA-2 has a pink background. Other conserved amino acids have a black background. The MtLecRK1;1 amino acids predicted to be involved in binding of the lipid moiety of Nod factors have a green background, and the ones that correspond to potential N-glycosylation site are colored in red. The alignment is based on conserved features in three-dimensional structures, and the extensions of the β-strands are indicated by arrows.
Molecular Modeling of the Lectin-Like Domain of MtLecRK1;1
Because MtLecRK1;1 is the only one of the four MtLecRK proteins that displays the required amino acids for binding carbohydrate, a three-dimensional model of the lectin-like domain of this protein was built. From the alignment with selected known three-dimensional structures (Fig. 3), a model of the region between amino acids 37 and 283 could be constructed, with the exception of the loop between Gly-221 and Gly-228 that corresponds to an insertion that is not present in known crystal structures. Because this loop is located at the opposite side to the monosaccharide-binding site, it should have no effect on the docking study, and no attempts to build it from ab initio were made.
Because the lectin-like domain of MtLecRK1;1 is glycosylated in planta (see later), the seven putative N-glycosylated sites of the 37 to 283 domain were compared with the ones that have been proven to be occupied in related legume lectins (Fig. 3). N-glycosylation in legume lectins is common but, with the exception of one site, does not display a conserved scheme. The N-glycans are known to affect the oligomerization mode of some legume lectins, but not their carbohydrate binding. In the known structures, only the region close to amino acids 100 to 120 (Asn-113 in Maackia amurensis lectin) contains a partially conserved N-glycosylation site, which corresponds to Asn-144 in the modeled lectin-like domain of MtLecRK1;1. This loop is close to the monosaccharide-binding site, but, in comparison with known structures displaying glycosylation in this position, the N-glycan is predicted to be orientated such that it does not interact at this site. At present, it is difficult to predict if other sites are glycosylated and whether this would affect ligand binding.
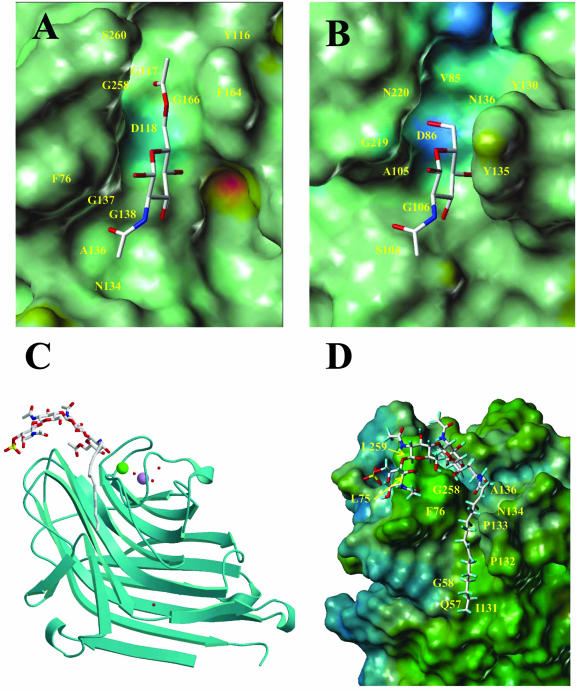
The major Nod factor of S. meliloti, NodSm-IV (Ac, S, C16:2), was docked into the protein as follows. A 6-O-acetyl GlcNAc residue was docked into the binding site of MtLecRK1;1 by comparison with the binding of GlcNAc by Ulex europaeus isolectin II (UEA-2; Loris et al., 2000; Fig. 4, A and B). The binding site of MtLecRK1;1 is less deep than that of UEA-2. This latter lectin appears to have one additional aromatic amino acid (Tyr-135) that creates a wall on one side of the binding site. Only two hydrogen bonds are established in the MtLecRK1;1 site: Asp-118 receives one from hydroxyl O4, whereas Gly-138 donates one to O3. The acetate group at position 6 fits nicely into a cavity formed by the presence of two very small amino acids (Gly-117 and Gly-166). From the alignment, it appears that only MtLecRK1;1 presents Gly residues at these two positions and, thus, would have the potential to accommodate an acetate at position 6.
Figure 4.
Molecular modeling of the lectin-like moiety of MtLecRK1;1. A, Model of the complex between 6-O-acetyl-GlcNAc and the carbohydrate recognition domain of MtLecRK1;1. The protein moiety is represented by its Connolly surface color coded according to the electrostatic potential (from blue for negative to red for positive). B, Crystal structure of the complex between lectin II of U. europaeus and GlcNAc (pdb code 1QOO), with the same color coding as in A. C, Ribbon representation of the model of the lectin-like domain of MtLecRK1;1 complexed with Nod factor. The ligand and water molecules are represented by sticks, and the cations are represented by space fill. D, Blow-up of the binding site of MtLecRK1;1 with Connolly surface of the molecule color coded according to the hydrophobicity potential (from blue for hydrophilic to brown for hydrophobic).
Three additional GlcNAc residues were added to form a tetrameric Nod factor backbone while performing a systematic conformational search. Several conformations could be generated, but the one retained in the model is the one that presents most interactions between the oligosaccharide and the protein surface (Fig. 4, C and D). No additional hydrogen bonds are observed because all of the three residues interact through their hydrophobic face with a hydrophobic patch of amino acids at the protein surface (Leu-75, Phe-76, and Leu-259). The sulfate group does not make any direct contact with the protein, but it lies not far from a basic residue, Arg-77, that is specific to MtLecRK1;1 and, thus, can participate in electrostatic stabilization.
The C16:2 lipid moiety of S. meliloti Nod factors was built into the model from the N-acetyl group of the first docked GlcNAc residue. Different conformations were tested, and the one that results in the best fit between the chain and the protein surface consists simply of an all trans-chain, with expected kinks at the two cis-linkages. In fact, the presence of these two cis-bridges is the factor that brings the chain exactly into a deep hydrophobic crevasse at the surface of the protein (Fig. 4D). The lipid makes most interaction with a strand of hydrophobic amino acids (Ile-131-Pro-Pro-133) that does not exist in the other lectins.
Taken together, the model suggests that the lectin-like domain of MtLecRK1;1 may adopt a structure similar to soluble legume lectins and that the major S. meliloti Nod factor could be a possible ligand with its terminal reducing sugar inserted into the position of the putative monosaccharide-binding site.
Expression of the MtLecRK Genes
As an initial approach to study the physiological role of the MtLecRKs, the expression patterns of the four MtLecRK genes were analyzed. This was done by northern analysis using probes to their lectin domains. In Southern analyses, the probes of MtLecRK1;1 and MtLecRK7;3 did not cross-hybridize with the other genes, but a low degree of cross-hybridization was observed between MtLecRK7;1 and 7;2. The abundance of mRNA related to the MtLecRK7;2, MtLecRK7;3, and MtLecRK1;1 probes was found to be higher in roots than in the other plant organs tested (nodules at different stages, leaves, stems, flowers, and shoot apices). Moreover, the mRNA levels of all three genes showed a clear increase in roots following nitrogen starvation (Fig. 5A). The three genes also appeared to be expressed in a cell culture line of M. varia, in which a high-affinity Nod factor-binding site (NFBS2) has been characterized. The MtLecRK7;3 gene seems to be expressed also in leaves, and mRNA related to both MtLecRK7;2 and MtLecRK7;3 could be detected in stems. MtLecRK1;1 seemed to show the highest organ-specific expression, being highly specific for roots but with detectable mRNA levels also in 8-d-old nodules and in stems. The probe to MtLecRK7;1 hybridized very weakly to the blots in Figure 5, suggesting that this gene is expressed only weakly in the plant. The MtLecRK1;1 and MtLecRK7;2 genes were selected for further detailed study based on their expression levels, their sequence divergence, and their different chromosomal locations.
Figure 5.
Expression of the MtLecRK genes by northern analysis. A, Different organs of plants grown in aeroponic chambers. Roots were from 2-week-old plants grown on 5 mm NH4NO3 and following nitrogen starvation for 4 d (Root-N). Nodules (Nods) were harvested at different times following inoculation. Cell cultures are of Medicago varia. B, Roots following inoculation (at T = 0) with S. meliloti wild type (Wt) or a non-nodulating mutant (nodA). M. truncatula Jemalong A17 or the hypernodulating skl (sickle) mutant were used. C, Roots following addition at T = 0 of 10-9 m NodSm factors. The specific lectin-like domains of the MtLecRK1;1 (1;1), MtLecRK7;2 (7;2), and MtLecRK7;3 (7;3) genes were used as probes. The expression of MtENOD11 and histone was used as controls. A rRNA probe was used to check RNA loading.
To study the expression of the MtLecRK7;2 and MtLecRK1;1 proteins in M. truncatula, antisera were raised to the lectin-like domains of the two proteins. These proteins were expressed as glutathione-S-transferase fusions in Escherichia coli. The lectin fusion proteins were found to be largely insoluble when the bacteria were grown at room temperature, but by expressing the proteins at 4°C, enough soluble protein was obtained to immunize rabbits. The resulting antisera were tested in western blots with the E. coli-expressed proteins and were found to recognize their own protein but not to cross-react with the other lectin domain (data not shown). These antisera were used in western blots of crude and membrane extracts of M. truncatula roots, but no specific protein corresponding to the endogenous LecRKs could be identified (Fig. 6B). Because the antisera recognize the lectin-like domains when overexpressed in transgenic roots (see below), it seems that the endogenous proteins are of too low an abundance in the root tissues to be detected by western analysis. Therefore, further studies on the regulation of expression of MtLecRK1;1 and MtLecRK7;2 were carried out by northern analysis of their mRNAs.
Figure 6.
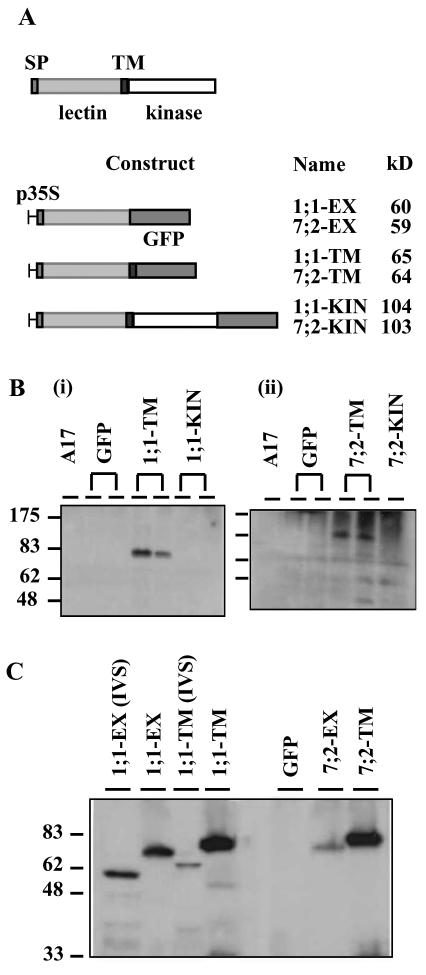
Expression of MtLecRK1;1 and MtLecRK7;2 green fluorescent protein (GFP) fusion proteins in transgenic roots of M. truncatula. A, Schematic structure of the LecRK proteins and constructs. SP, Signal peptide. p35S, Cauliflower mosaic virus (CaMV) 35S promoter. The protein sizes are predicted from the amino acid sequences. B, Western analysis of roots of wild-type plants (A17) and of different transgenic lines expressing the GFP fusion proteins, using antisera to the lectin-like domains of MtLecRK1;1 (i) and MtLecRK7;2 (ii). C, Western analysis using anti-GFP detection to compare the size of the fusion proteins in roots either from aeroponically grown plants or from in vitro “hairy” root cultures with those of the fusions expressed in an E. coli in vitro synthesis (IVS) system. Note that the plant expressed proteins are considerably larger than those expressed in the E. coli in vitro system, probably reflecting glycosylation of the lectin domains. Constructs with MtLecRK1;1 and MtLecRK7;2 are designated as 1;1 and 7;2, respectively, as shown in A.
Different plant-bacterial couples were used to look further at the regulation of MtLecRK7;2 and MtLecRK1;1 during the establishment of the legumerhizobia symbiosis (Fig. 5B). Gene expression during the essentially wild-type symbiosis between M. truncatula cv Jemalong A17 and S. meliloti was compared with expression during either a non-nodulating interaction (wild-type M. truncatula with S. meliloti nodA) or a hypernodulating interaction (between the M. truncatula ethylene-insensitive skl mutant [Penmetsa and Cook, 1997] and wild-type S. meliloti). MtENOD11 (Journet et al., 2001), histone, and rRNA probes were used as controls for symbiotic and non-symbiotic regulation and for RNA loading, respectively. As previously shown (Navarro-Gochicoa et al., 2003), MtENOD11 is more highly induced in the skl mutant than in wild-type M. truncatula, and this induction is specific for the nodulating interaction (Fig. 5B). Hybridization with the MtLecRK probes suggests that rhizobial inoculation leads to a rapid (within 3 h) and transient (largely restored within 48 h) decrease in the root mRNA levels of MtLecRK7;2 and MtLecRK1;1. Moreover, the decrease seemed to be partially dependent on a successful nodulation because a lesser decrease was observed with the nodA S. meliloti mutant, but it is notable in this case that a reduction still occurred. Because this mutant does not produce Nod factors, experiments were performed to examine the effect of addition of these signals on the expression of the genes. The results (Fig. 5C) show that Nod factor addition does lead to a partial and transient reduction in the abundance of the mRNA of the two MtLecRK genes, whereas there is no great effect on histone mRNA.
Localization and Targeting of the LecRKs to the PM
To investigate the subcellular localization of the MtLecRK1;1 and 7;2 proteins, constructs of each gene were prepared with the GFP marker. Three constructs were prepared for each gene with the GFP tag fused as a C-terminal domain after either the predicted extracellular domain (constructs EX), the TM-spanning helix (constructs TM), or the kinase domain (constructs KIN). These protein fusions (Fig. 6A) were expressed from the CaMV 35S promoter in roots of M. truncatula after Agrobacterium rhizogenes transformation (Boisson-Dernier et al., 2001). Many fluorescent lines of the EX and TM constructs were obtained. However, very few lines were obtained with the KIN constructs, and those that did thrive were only weakly fluorescent. Some of the lines were tested for expression of the proteins by western analysis using antisera raised to the lectin-like domains. These antibodies recognized the overexpressed EX and TM (Fig. 6B) fusion proteins, thus suggesting that the antisera are capable of recognizing the plant-expressed proteins. No specific protein, however, was detected in non-transformed roots (A17). In roots containing the KIN constructs, no fusion proteins were observed, thus confirming the poor expression of these proteins. Using antibodies to GFP, it was notable that some cleavage of the EX and TM fusion proteins occurred in the fluorescent transgenic lines because an approximate 25-kD GFP protein was also observed by western analysis (data not shown).
By western analysis of the transgenic roots, the size of the EX and TM fusions seemed to be bigger than predicted from the sequences. To investigate this further, the 1;1-EX and 1;1-TM constructs were expressed in an E. coli in vitro transcription/translation system, and their sizes compared with the same protein constructs expressed in the M. truncatula roots (Fig. 6C). The two 1;1 fusion proteins appear to be about 14 kD bigger when expressed in the plant compared with the in vitro system. The 7;2 proteins seemed to be similar in size to the corresponding 1;1 fusion proteins. These results suggest that the lectin-like domains of both proteins are probably heavily glycosylated.
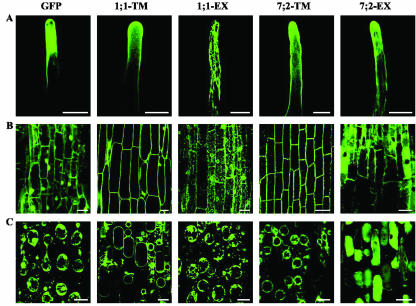
The subcellular locations of the fusion proteins were investigated by confocal microscopy (Fig. 7). The 35S promoter gave a characteristically high expression in the vasculature, but particular attention was paid to the epidermis (Fig. 7A) and cortex (Fig. 7B), where the MtLecRKs are more likely to play a symbiotic role. Plasmolysis was used (Fig. 7C) to distinguish between fluorescence in the cytosol, PM, and wall at the periphery of the cells. The GFP-alone construct showed the typical nuclear and cytosolic localization previously shown for this protein. Both TM fusion proteins (1;1-TM and 7;2-TM) were found to be located primarily at the periphery of the cell. After plasmolysis, the GFP fluorescence was found to be located in the PM and not in the cell wall. With the two EX fusions, the GFP fluorescence showed a complex localization within the cell, with the 1;1-EX protein seeming to be mainly located in the endoplasmic reticulum (ER) and the 7;2-EX fusion in the vacuole and cytosol (Fig. 7).
Figure 7.
Confocal microscopy showing the localization of MtLecRK1;1 and MtLecRK7;2 GFP fusion proteins in transgenic roots. A, Root hairs; B, cortical cells; and C, plasmolyzed cortical cells from the GFP, 1:1-TM, 1;1-EX, 7;2-TM, and 7;2-EX constructs. Note the PM localization of the two TM-GFP fusion proteins.
Functional Analysis of the MtLecRKs in Nod Factor Binding and Nodulation
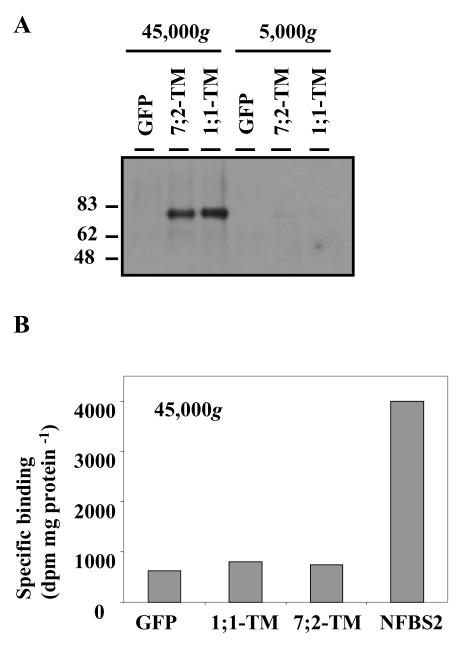
Because the TM fusions were well expressed with the fusion proteins appearing to be glycosylated and targeted to the PM (Figs. 6 and 7), these transgenic hairy roots lines were used in experiments to address the function of the MtLecRK1;1 and MtLecRK7;2 proteins. First, these lines were used to examine whether Nod factors bind specifically to the EX + TM domains of the proteins. A GFP-expressing line was used as a control for either endogenous or nonspecific binding. Cell-free extracts of the roots were prepared and centrifuged sequentially at 5,000g and then 45,000g to obtain different membrane fractions. As expected for PM proteins, the fusion proteins were observed by western analysis in the 45,000g fraction (Fig. 8A). This fraction, from the three extracts, was used in binding experiments with a radio-labeled ligand corresponding to the major Nod factor of S. meliloti (NodSm-IV, Ac, 35S, C16:2). The 45,000g fraction of a M. varia cell culture line, in which NFBS2 has been characterized previously (Gressent et al., 1999), was used as a positive control. The results (Fig. 8B) show a similar low specific binding of the Nod factor in all three transgenic root lines, whereas the specific binding to NFBS2 was 4-fold higher. Thus, the EX + TM domains of the two MtLecRK proteins do not appear to bind Nod factors at high affinity in the conditions examined.
Figure 8.
Analysis of Nod factor binding in extracts of roots expressing the MtLecRK1;1-TM and MtLecRK7;2-TM constructs. A, Western analysis, using anti-GFP antibodies, showing the distribution of LecRK-related fusion proteins in the pellets of extracts centrifuged at 5,000g and 45,000g from 1;1-TM, 7;2-TM, and GFP constructs. Note the presence of the MtLecRK-TM fusion proteins in the 45,000g (membrane) fraction. B, Nod factor binding to the above 45,000g extracts using, as ligand, 1 nm radiolabeled NodSm-IV (Ac, 35S, C16:2). Extracts of M. varia cell cultures containing NFBS2 were used as a positive control. Nonspecific binding was measured in the presence of 2 μm unlabeled Nod factor.
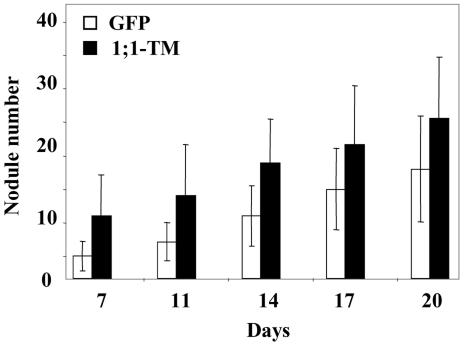
In the second approach, the effect of overexpression of the TM constructs on nodulation was examined. The GFP construct was again used as a control. Chimeric plants with transgenic roots of the three lines were transferred to growth pouches, inoculated with S. meliloti, and nodule number was counted at various times. A pilot experiment and two larger scale experiments were performed. Figure 9 shows the results of the second larger scale experiments for the GFP and 1;1-TM constructs. Although there was considerable variation between the numbers of nodules per plant, the 1;1-TM construct led to an increase in the mean number of nodules per plant compared with the GFP control, starting by 7 d after inoculation. By 14 d, the increase compared with the control was 2.8- and 1.9-fold in the two larger scale experiments. Statistical analysis using the Student's t test showed that the differences from the control were highly significant with P values of less than 0.005 for all the time points in the first experiment and with P values generally of less than 0.005 for the second experiment (Fig. 9). The 7;2-TM construct led to an increase in nodule number in one of the experiments, but no difference was found in the second experiment using the same statistical criteria. For each construct, nodules from several transgenic lines were stained with methylene blue and observed by microscopy. No differences could be observed in nodule structure or infection of the LecRK-expressing roots compared with those expressing the GFP alone.
Figure 9.
Number of nodules at different times after rhizobial inoculation of composite M. truncatula plants with transgenic roots expressing either the GFP or the MtLecRK1;1-TM construct. The composite plants with transgenic roots were transferred to growth pouches for 4 d before inoculation with S. meliloti. The data show the mean number of nodules per plant and the associated se. Using the Student's t test, the P values for a difference between the MtLecRK1;1-TM and the GFP construct were 0.0029, 0.0052, 0.0007, 0.0036, and 0.010 for the 7-, 11-, 14-, 17-, and 20-d time points, respectively.
DISCUSSION
In this paper, we have tested the hypothesis (Hervé et al., 1996; Hirsch, 1999) that LecRKs may play a role in the legume-rhizobia symbiosis. Screening of cDNA libraries and examination of M. truncatula EST banks has shown that the model legume contains at least nine LecRK genes. The four genes studied here (MtLecRK1;1, MtLecRK7;1, MtLecRK7;2, and MtLecRK7;3) are the most highly expressed LecRK genes in roots (as judged by their clone abundance in root and nodule cDNA libraries) and, hence, are the most likely LecRK genes to play a role in the symbiosis. Although their mRNAs could be easily monitored by northern analysis, antisera raised against the lectin-like domains of MtLecRK1;1 and MtLecRK7;2 failed to detect their endogenous proteins by western analysis, suggesting that these receptors are low-abundance proteins.
The four genes encode proteins with a classic LecRK structure, with an N-terminal signal peptide, a lectin-like domain, a single TM-spanning helix, and a kinase domain (Fig. 1). They are most likely PM proteins as fusions in which the kinase domains of MtLecRK7;2 and MtLecRK1;1 were replaced by GFP localized to the PM in transgenic roots (Fig. 7). For this localization, the internal TM helix is required because without it, the MtLecRK1;1 fusion became stuck in the ER, and the MtLecRK7;2 fusion located mainly to the vacuole. These localizations are surprising because the default destination of soluble proteins in the secretory pathway is secretion (Barrieu and Chrispeels, 1999; Hadlington and Denecke, 2000), thus suggesting that the lectin-like domain fusions contain information for retention in the ER (for MtLecRK1;1) or sorting to the vacuole (for MtLecRK7;2). However, such sorting or retention signals could not be identified in the sequences.
The above localizations suggest that the TM helices of the two LecRKs are required not only to anchor the LecRK proteins in a membrane but also as a positive sorting signal for the PM. In studies in yeast (see Rayner and Pelham, 1997) and more recently in plants (Brandizzi et al., 2002), the length of the TM helix has been shown to play a major role in protein sorting because the helix needs to be at least 23 amino acids to span and to permit sorting to the PM. The predicted TM helices of MtLecRK1;1 and MtLecRK7;2 are 23 amino acids in length, thus fulfilling this sorting requirement.
The lectin-like domains of MtLecRK1;1 and MtLecRK7;2 appear to be heavily glycosylated because the sizes of the EX and TM GFP fusion proteins were about 14 kD greater than the proteins expressed in an E. coli-based in vitro protein production system (Fig. 6). The size of the Ath.LecRK-a1 protein in PM extracts of Arabidopsis suggests that this protein is also highly glycosylated (Hervé et al., 1999). Assuming a molecular mass of plant N-glycans of between 1,200 and 2,000 D, the molecular masses of the plant-expressed MtLecRK1;1 and MtLecRK7;2 fusion proteins suggest that many of the predicted eight N-glycosylation sites in the lectin domains may be used. Whether the lectin-like domain glycans are involved in protein sorting requires further experimentation. Studies, mainly on animal cells and yeast (Helenius and Aebi, 2001) but also on plant-expressed proteins (Bardor et al., 1999), suggest that N-linked glycans may play a variety of roles including protein folding, oligomerization, quality control, sorting, and transport.
The four MtLecRKs are expected to be active in protein phosphorylation because their kinase domains show a high conservation of the sub-domains, residues, and motifs required for Ser/Thr protein kinase activity (Fig. 1). To date, the kinase domains of only two LecRKs have been tested for functionality (Ath.LecRK-a1 and PnLPK), and both have been shown to be active in autophosphorylation (Hervé et al., 1996; Nishiguchi et al., 2002); however, their cellular targets remain unknown.
The lectin-like domains of the four MtLecRKs were found be very divergent; hence, clones of these different genes probably would not have been isolated using probes from the lectin-like domain of Ath.lecRK-a1. Thus, the isolation of such divergent MtLecRK genes relied on the strategy of using the kinase domain as a probe. The success of this strategy in isolating mainly LecRK genes, coupled with subsequent sequence analysis, suggest that the kinase domain attached to the variant lectin-like domains have a similar phylogenetic origin, in agreement with studies by Shiu and Bleecker (2001). The greater divergence of the lectin-like domains suggests that they evolve more quickly than the kinase domains, although it cannot be excluded that some of the LecRK genes may have picked up new lectin-like domains from soluble lectins. Comparative analysis of the four M. truncatula LecRKs with the 42 Arabidopsis LecRKs suggests that at least the Class A and B subfamilies arose before the speciation of the higher plants. Analysis of the complete genome of M. truncatula is probably required before direct orthologs can be assigned between the two species. However, by similarity, the MtLecRK1;1 gene may be the ortholog of the Class B gene encoding At5g10530. The other genes studied in this article (MtLecRK7;1, 7;2, and 7;3) are all members of Class A, are clustered on the same chromosome, and, hence, probably arose from more recent duplications.
An obvious question on LecRKs is whether the lectin-like domains are true lectins and bind carbohydrates, perhaps as ligands, to activate the kinase domain. Careful analysis of this domain revealed that the residues shown to be involved in sugar binding in soluble legume lectins are more highly conserved in MtLecRK1;1 (three of four) than any of the other MtLecRKs so far identified. Moreover, of the 42 AtLecRKs, the conservation of the D, G, F/Y, and N residues are shown in two, six, four, and 11 proteins, respectively, with only two proteins showing conservation of three of these residues (Barre et al., 2002). To date, no binding studies have been done on any of the Arabidopsis LecRKs, but sequence analysis and molecular modeling of Ath.LecRK-a1 suggest that it is unlikely to bind monosaccharides (Hervé et al., 1999). The lectin-like domain of the poplar LecRK was incubated with erythrocytes as a test for carbohydrate binding, but no hemagglutination activity was observed (Nishiguchi et al., 2002). The much higher conservation of a hydrophobic cavity rather than the monosaccharide-binding site in AtLecRKs may suggest that their ligands are small hydrophobic hormones such as auxins and cytokinins (Barre et al., 2002). However, some of the large range of oligosaccharin signaling molecules that have yet unknown receptors (Côté and Hahn, 1994) could be good candidates for LecRK ligands (Barre et al., 2002).
Due to the conservation of key sugar-binding residues in MtLecRK1;1, molecular modeling was performed on this protein using the known structures of soluble legume lectins (Figs. 3 and 4). Because Nod factors are a key signaling molecule in root symbioses and the principle target of our research, the terminal non-reducing sugar of the major Nod factor of S. meliloti was docked into the monosaccharide-binding site, and the Nod factor was fitted into the protein. The results suggest that the two Nod factor substitutions on the terminal non-reducing sugar (the O-acetate on C6 and the C16:2 fatty acyl chain on C2) could fit nicely into cavities on the MtLecRK1;1 protein with the other sugars extending to the surface of the protein. The number of predicted hydrogen bonds between the protein and the Nod factor ligand is low (only two); however, it should be noted that sugars are amphiphilic molecules, and evidence suggests that contact between the hydrophobic face of the glycan and hydrophobic patches on the protein surface are of primary importance for both the affinity and the specificity of binding.
Such an interaction between Nod factors and the lectin-like domains would not be predicted for the other three MtLecRK proteins or for soluble legume lectins. Thus, the structural model predicts that MtLecRK1;1 could be a specific receptor for Nod factors. However, when this hypothesis was tested, no increase in Nod factor-specific binding was found in root extracts overexpressing the MtLecRK1;1-TM fusion in comparison with roots expressing either a similar construction with MtLecRK7;2 or GFP alone (Fig. 8). Thus, no evidence has been obtained that these proteins do bind Nod factors. However, the technique used to measure binding is designed for high-affinity interactions and, thus, we cannot exclude the possibility that Nod factors do bind to this protein but with a dissociation constant (KD) in the micromolar to millimolar rather than the nanomolar range. Moreover, ligand binding could require the activity of the kinase domain, although this is not a general requirement for ligand binding to RLKs (Cock et al., 2002). In our constructs, this kinase domain was replaced by GFP to detect the expressed proteins and because the complete LecRK fusion proteins (KIN constructs) could not be overexpressed.
The lack of many transformants with the KIN constructs and the poor fusion protein expression observed in the few transgenic roots that grew could suggest that overexpression of the LecRK genes is detrimental to root development. Riou et al. (2002) have shown that two Arabidopsis LecRK genes are regulated developmentally and have suggested that they play an important role in both the developmental program and adaptive processes (e.g. wounding) in this species. In a similar way, the two MtLecRK genes studied here could be playing an essential role in roots that needs to be carefully regulated. An alternative explanation for the poor expression of the KIN constructs could be that the sequences of the kinase domain or the functional LecRK proteins could be involved in a negative regulation of their expression.
The high expression of the four MtLecRK genes in roots compared with other organs (Fig. 5) suggests roles for these genes predominantly in this organ. Thus, they are predicted to have a different role than two LecRK genes studied in Arabidopsis that are expressed predominantly in aerial tissues (Hervé et al., 1996, 1999; Riou et al., 2002). Interestingly, an LecRK gene from poplar is also predominantly expressed in roots (Nishiguchi et al., 2002). The three MtLecRK genes tested were induced in roots by nitrogen starvation, which is consistent with a role in the legume-rhizobia symbiosis because a combined nitrogen source inhibits nodulation. Moreover, further studies on MtLecRK1;1 and MtLecRK7;2 showed that the levels of their mRNAs were transiently decreased by addition of rhizobia or Nod factors (Fig. 5), indicating a regulation during the symbiosis. The similar regulation of these two genes is somewhat surprising in view of their phylogenetic divergence; nevertheless, these studies provide circumstantial evidence for a role of both these MtLecRK genes in the legume-rhizobia symbiosis.
Further evidence for such a role for MtLecRK1;1 was obtained by studies of nodulation in roots expressing the TM constructs. Western analysis suggests that the MtLecRK1;1 and MtLecRK7;2 TM constructs were expressed at least 100-fold higher than the endogenous proteins (Fig. 8), and good expression was seen in the epidermal cells that are in direct contact with symbiotic bacteria (Fig. 7). For both constructs, plants produced normal looking nodules that were infected with rhizobia. Thus, expression of these catalytically defective proteins does not interfere with either nodule development or infection. However, roots expressing the MtLecRK1;1-TM construct developed a greater number of nodules than roots expressing the GFP construct alone (Fig. 9).
One explanation for this result could be that the endogenous MtLecRK1;1 gene plays a negative role in controlling nodule number and that overexpressing a catalytically inactive protein acts as a dominant negative mutation inhibiting the normal functioning of the endogenous gene (perhaps by soaking up the normal ligands). Nodule number is known to be controlled by an autoregulatory response involving a shoot-effective gene that controls the number of root and nodule meristems (Caetana-Anolles and Gresshoff, 1991). Recently, this gene has been cloned from Lotus japonicus (Krusell et al., 2002; Nishimura et al., 2002) and soybean (Searle et al., 2003) and shown to be a CLAVATA1-like receptor kinase with a predominantly Leu-rich repeat extracellular domain. The autoregulation of nodulation is proposed to occur via a root-derived signal that is produced by nodulation and that is perceived by the CLAVATA1-like receptor in the shoot. In turn, this receptor would lead to the production of an inhibitor that represses further nodule development in the root (Caetana-Anolles and Gresshoff, 1991; Downie and Parniske, 2002). Nonfunctioning of the hypothetical proteins involved in either production of the root-derived signal or responses to the shoot-derived signal would, like the CLAVATA1-like gene, be predicted to lead to an increase in nodulation. Our work raises the question of whether MtLecRK1;1 could be one of these root-acting proteins.
A second explanation for the increase in nodule number is that overexpression of the MtLecRK1; 1-TM construct could have a positive effect on the establishment of the symbiosis, which is independent of the kinase domain and is perhaps due to the lectin activity of the protein. MtLecRK1;1 would be predicted to have a greater effect than MtLecRK7;2 because it is predicted to be a better lectin. In a manner similar to that proposed for overexpression of soluble lectins (Hirsch, 1999), the lectin domain on the exterior of the epidermal cells could serve to increase attachment of rhizobia via their polysaccharide coats, thus facilitating signal exchange and nodulation. The question remains, however, of whether this is a normal physiological function of endogenous MtLecRK1;1 or is a function derived from overexpression of a “sticky” lectin.
Clearly, further experimentation is required to understand the potential and rather enigmatic role of the MtLecRK1;1 gene in the control of nodule number. Although the rapid hairy root system has proven invaluable as an expression system to study the structure and location of these proteins, a more detailed functional study of this gene may require stably transformed plant lines in which the plant-to-plant variability is considerably less than in the independently transformed roots of the hairy root system. Moreover, although ectopic overexpression of functional or nonfunctional proteins can lead to indications of the physiological role of a protein, this strategy suffers from the problem that the overexpressed protein may function in a way different to its endogenous homolog. Thus, such studies need to be complemented by strategies designed to inhibit functioning of the endogenous gene either by isolating specific gene mutants or by using antisense or RNA interference strategies.
In conclusion, we have shown that M. truncatula contains a family of at least nine LecRK genes. The four genes studied here are the most highly expressed MtLecRK genes in roots. Three of them encode proteins related to the Class A Arabidopsis LecRKs, whereas the fourth gene (MtLecRK1;1) encodes a proteins that is more closely related to a Class B LecRK. The considerable divergence in the sequence of their lectin-like domains suggests that these putative receptors may perceive different ligands and perform different physiological functions. The structure of the MtLecRK1;1 lectin-like domain suggests that this protein is a good candidate for interacting with saccharidic ligands, and studies with transgenic roots suggest that its expression may influence the regulation of nodulation, although probably not through Nod factor binding. Clearly, the identification of the ligands for these proteins, coupled with studies of stably transformed transgenic plants modified in their expression, may help to determine the physiological role of these intriguing receptors in both symbiotic and non-symbiotic conditions.
MATERIALS AND METHODS
Growth Conditions and Plant Material
All plant lines and bacterial strains are listed in Table IV of Navarro-Gochicoa et al. (2003). Medicago truncatula Gaernt. cv Jemalong lines A17 or J5 were grown in aeroponic chambers (unless otherwise indicated) in a 16-h photoperiod on medium containing 5 mm NH4NO3 for 2 weeks, essentially as described by Journet et al. (2001). Different organs were harvested from such plants. For nodulation, the media was replaced by a similar one lacking a nitrogen source, and the plants were then starved of nitrogen for 4 d before either inoculation with Sinorhizobium meliloti or treatment with 10-9 m NodSm factors. Several whole-root systems or nodules from several plants were then harvested at different times, frozen in liquid nitrogen, and stored at -80°C before RNA or protein extraction.
For the experiments described in Figures 6 and 9, composite plants with transgenic roots (Boisson-Dernier et al., 2001) were transferred to growth pouches (four plants per pouch) and grown for 6 d using the above media lacking a nitrogen source before harvesting or inoculation with S. meliloti. For some of the experiments described in Figures 6 and 8, transgenic “hairy root” cultures were established (Boisson-Dernier et al., 2001), and roots were harvested from solid media.
Isolation, Characterization, and Sequencing of LecRK cDNAs of M. truncatula
About 980,000 clones were screened essentially as described by Gamas et al. (1996) from the following three M. truncatula cDNA libraries prepared in Lambda ZapII (Stratagene, Stratagene, La Jolla, CA): about 240,000 plaque-forming units (pfu) of a cDNA library prepared from 4-d-old nodules (Gamas et al., 1996); about 412,000 pfu from a cDNA library from roots harvested 6, 24, and 48 h after rhizobial inoculation (Szybiak-Strozycka et al., 1995); and about 330,000 pfu from a library prepared from roots following inoculation for 3, 6, 9, and 24 h with S. meliloti nodA, GMI 5382 (T. Arcondeguy and P. Gamas, unpublished data). The filters were hybridized at low stringency (hybridization: 40% [v/v] formamide, 5× sodium chloride/sodium phosphate/EDTA buffer, 5× Denhardts, 0.1% [w/v] SDS, and 100 μg mL-1 denatured salmon sperm DNA at 37°C for 20 h; and wash: 6× sodium chloride/sodium phosphate/EDTA buffer and 0.1% [w/v] SDS at 48°C) using a 32P-labeled probe consisting of a 988-bp (AvaII/HpaI) fragment of the Ath.lecRK-a1 kinase domain (Hervé et al., 1996). Hybridizing clones were plaque purified, converted to plasmid clones in the vector pBluescript SK- (Stratagene), and classified by restriction mapping and DNA sequencing. The longest cDNAs of each class were sequenced on both strands with specific primers using a 373A automatic sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA). Clones corresponding to MtLecRK7;1, 7;2, and 7;3 were extended by 5′-RACE and sequenced.
Sequence and Phylogenetic Analyses
MtLecRK ESTs were identified by using the four full-length MtLecRK cDNAs in BLAST analyses with the GenBank EST database (Altschul et al., 1997), the M. truncatula Gene Index Release 4.0 database at The Institute for Genomic Research (http://www.tigr.org/tdb/tgi/mtgi/; Quackenbush et al., 2001) and the M. truncatula EST database (January 2003 version) at the Institut National de la Recherche Agronomique (http://medicago.toulouse.inra.fr/Mt/EST/DOC/MtB.html; Journet et al. 2002). Sequence data were analyzed using the GCG software (Genetics Computer Group, Madison, WI). Derived protein sequences were analyzed using the SwissProt Expasy server and the following programs: PROTPARAMS, PSORT, ScanProsite, and TMHMM2.0. Sequences showing homology were identified using either the Arabidopsis TAIR server (Garcia-Hernandez et al., 2002) or the National Center for Biotechnology Information BLAST server (Altschul et al., 1997) and recovered from either the GenBank or SwissProt databases. The following soluble lectin sequences (with their SwissProt accession nos.) were used in phylogenetic analyses: LEC_SOYBN (P05046), LEC1_DOLBI (P05045), LEC_PEA (P02867), LEC2_CLALU (Q39529), and LECB_SOPJA (P93538).
For the alignment shown in Figure 1 and for the phylogenetic analysis in Figure 2, ClustalX (Thompson et al. 1997) was used to align the sequences. Minor adjustments were made either to improve the alignments or to delete the N-terminal targeting sequences and, where appropriate, the C-terminal TM and kinase domains using GeneDoc (Nicholas and Nicholas, 1997). Phylogenetic analyses were then performed on the 272-amino acid sequence alignment of the putative lectin domains using the PHYLIP package of programs (Felsenstein, 1993) on the Pasteur Institute Server (http://bioweb.pasteur.fr/seqanal/phylogeny/). PROTDIST with 100 bootstrap analyses was used to calculate the distance between the sequences before analysis with the neighbor-joining program BIONJ and calculation of the consensus tree using CONSENSE. Trees were viewed and edited using TREEVIEW (Page, 1996).
Molecular Modeling
The N-terminal 300-amino acid sequence of MtLecRK1;1 was aligned with the sequences of 15 legume lectins selected among the ones with known structures that are available in the Protein Data Bank (Berman et al., 2000). The homology modeling COMPOSER program (Blundell et al., 1988) of the Sybyl software (Tripos Associates, St. Louis) was used to build the structurally conserved regions of the lectin model. Loops were built from a library containing all legume lectins structures, and their geometries were optimized. The stereochemistry of the final model was checked using the PROCHECK program (Laskowski et al., 1993). Hydrogen atoms were added on all atoms, and partial atomic charges were derived using the Pullman procedure. Calcium and manganese cations were inserted in a model with a partial charge of 2. Five water molecules that are observed in most crystal structure of legume lectins and that play a role in cation coordination and loop architecture (Loris et al., 1994) have also been included.
A GlcNAc residue was docked in the primary binding site with the same location and orientation as in the crystalline complex between UEA-II and GlcNAc (Loris et al., 2000). From this starting point, the Nod factor was graphically built step by step with a systematic conformational search at each linkage. In each case, the lowest energy conformer that brings the molecule in close contact with the protein surface was selected. All energy calculations were performed with the Tripos force-field (Clark et al., 1989) using energy parameters previously derived for carbohydrates (Imberty et al., 1999). Connolly surface of the protein was calculated using the MOLCAD program (Waldherr-Teschner et al., 1992).
RNA Isolation and Northern Analysis
The techniques for isolating and determining the abundance of specific RNAs by northern analysis are described by Navarro-Gochicoa et al. (2003). cDNA fragments corresponding to the lectin-like domains were used to produce specific MtLecRK probes. Probes were also prepared from cDNA fragments of MtENOD11 (Journet et al., 2001), histone, and rRNA genes, as described by Navarro-Gochicoa et al. (2003).
Production of Antisera to MtLecRK1;1 and MtLecRK7;2 Lectin-Like Domains
The glutathione-S-transferase gene fusion system was used for MtLecRK1;1 and MtLecRK7;2 lectin-like domain purification (Amersham Biosciences, Buckinghamshire, UK). The lectin-like domains corresponding to amino acids 35 to 293 and 19 to 254 for MtLecRK1;1 and MtLecRK7;2, respectively, were cloned into the pGEX 6P-1 plasmid in Escherichia coli strain BL21 DE3. The bacteria were grown at 20°C and induced at 4°C using 0.1 mm isopropylthio-β-galactoside-containing media. In these conditions, some of the MtLecRK1;1 fusion protein was obtained in a soluble form. The MtLecRK7;2 fusion protein was solubilized from the insoluble fraction in buffer containing 1.5% (w/v) sarkosyl and 2% (w/v) triton, followed by keeping 0.5% (w/v) triton in all the washing steps. The lectin-like proteins were purified on a glutathione Sepharose4B matrix, followed by enzymatic cleavage using the PreScission protease enzyme (Amersham Biosciences). The samples were then run on an SDS-PAGE (10% [w/v] acrylamide) gel, stained with Coomassie Blue, and the acrylamide fragments corresponding to the MtLecRK1;1 and 7;2 lectin-like domains were cut out and used for antibody production in rabbits (four injections with 100 μg of antigen per injection, performed by Eurogentec Bel S.A., Seraing, Belgium).
Protein Extraction and Western Blotting
Frozen plant material and E. coli samples were crushed in a mortar and pestle and extracted by boiling in a 2× concentrated SDS-PAGE loading buffer. After SDS-PAGE on 10% (w/v) acrylamide gels, the proteins were transferred into Hybond-P membrane (Amersham Biosciences) in Laemmli buffer containing 10% (v/v) methanol and 0.02% (w/v) SDS. The protein marker used was a broad range prestained protein marker (New England Biolabs, Beverly, MA). The membrane was pre-incubated in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TTBS) and 5% (w/v) nonfat dried milk. The incubation steps with the appropriate antibody dilutions were performed in TTBS buffer with 1% (w/v) nonfat dry milk and the washing steps in TTBS buffer. The secondary antibody used was an anti-rabbit IgG:horseradish peroxidase-linked whole antibody from donkey, and the western blotting detection was by chemiluminescence (using the ECL Chemiluminescent kit, Amersham Biosciences). In some experiments, membranes were stripped using 100 mm 2-mercaptoethanol, 2% (w/v) SDS, and 62.5 mm Tris-HCl (pH 6.8) before reuse.
Construction and Expression of GFP Fusions in Transgenic Plants and in an in Vitro System
Constructs were prepared in the binary vector pGreen0029 that confers resistance to kanamycin in the transformed plants (Hellens et al., 2000). The CaMV 2x35S promoter and tobacco mosaic virus Ω 5′ untranslated region were cloned from pBE113-GUS into the HindIII site of the vector, and the CaMV terminator region was cloned into the StuI site near the right border. In this way, a plasmid was created (termed pGr29-35SOMCaMVT) that contains sites for XbaI, BamHI, SmaI, SacI, and EcoRI for gene expression from the 35S promoter. The enhanced GFP sequence from plasmid pEGFP-N2 (CLONTECH Laboratories, Palo Alto, CA) was cloned into the SmaI and SacI sites to create plasmid pGr29-35SOM-EGFP-CaMVT that contains sites for XbaI, BamHI, SmaI, BamHI, and NcoI upstream of the GFP. Using specific primers and PCR, XbaI and NcoI sites were created at the start of the cDNA of MtLecRK1;1 so that the NcoI site encodes the start codon and an SmaI site was introduced either at amino acid 297 (1;1-EX construct), 342 (1;1-TM construct), and 678 (1;1-KIN construct). A similar strategy was used for MtLecRK7;2 except that a BamHI site was created just before the start codon and the SmaI sites were at positions 278 (7;2-EX construct), 326 (7;2-TM construct), and 669 (7;2-KIN construct). The sequence changes were verified by DNA sequencing, and the LecRK-containing fragments were then cloned just upstream of the GFP in pGr29-35SOM-EGFP-CaMVT to produce in-frame LecRK-GFP fusions. The construct-containing plasmids were cotransformed into Agrobacterium rhizogenes ARqua1 (Quandt et al., 1993) with pSoup (Hellens et al., 2000), and the transformants were verified by PCR. Transformation of M. truncatula to produce composite plants with transgenic roots was carried out essentially as described by Boisson-Dernier et al. (2001). Only one transgenic root on each transformed plant was allowed to grow, and this root was either harvested and used directly in western analyses, or a portion of the root was excised and maintained in in vitro culture on solid media (Boisson-Dernier et al., 2001). In some experiments, the composite plants were used in nodulation tests (see below).
The MtLecRK1;1 EX- and TM-GFP-fusions were also cloned into vector pIVEX2.3 and expressed from the T7 promoter in the in vitro cell-free protein expression system, termed the Rapid Translation System, of Roche Applied Science (Mannheim, Germany). After expression, the reaction mixture was used directly for western analysis.
Confocal Microscopy
Transformed roots growing on agar were covered with a gas-permeable plastic foil (bioFolie 25, Sartorius AG, Vivascience Support Center, Göttingen, Germany) and observed with a TCS SP2 confocal microscope (Leica Microsystems, Wetzlar, Germany) using an HCX Plan Apo 63× 1.2-W objective. Optical sections were made with a separating distance of 1 μm between subsequent sections. Image projections were made with the Leica confocal software, and these were processed with Image-Pro plus (Media Cybernetics, Silver Spring, MD). To distinguish between labeling of the PM and the cell wall, root epidermal cells were plasmolyzed by incubation in a 10% (w/v) KNO3 solution.
Nod Factor-Binding Assay
Transgenic root material from in vitro hairy root cultures was extracted as described by Gressent et al. (1999) and centrifuged sequentially at 5,000g and 45,000g. The pellets were resuspended in binding buffer with 50% (w/v) glycerol. Nod factor binding was carried out using 1 nm NodSm-IV (Ac, 35S, C16:2) as described by Gressent et al. (1999). Nonspecific binding was determined using 2 μm unlabeled ligand.
Analysis of Nodulation
A. rhizogenes-transformed composite plants were transferred to growth pouches for 3 to 4 d using media lacking a nitrogen source before inoculation with S. meliloti, essentially as described by Vernoud et al. (1999). The number of developing nodules was counted at different times. At the end of the experiment, the nodulated root systems were fixed, stained with methylene blue, and observed by bright-field microscopy. The probability (P value) of the plants with a lectin construct producing a different number of nodules than the GFP control was analyzed by a Welch-modified Student's t test, which takes into account the difference in variability of the two samples. Splus software (version 3.4, release 1 for Sun SPARC, MathSoft, Inc., Cambridge, MA) was used for these analyses.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
NOTE ADDED IN PROOF
Map-based cloning of M. truncatula and Lotus japonicus genes involved in Nod factor responses has revealed RLKs with LysM domains as putative Nod factor receptors (Limpens E., Franken C, Smit P, Willemse J, Bisseling T, Geurts R [2003] LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630-633; Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S. Sandal N, Stougaard J (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585-592).
Acknowledgments
We thank Brigitte Mangin (Institut National de la Recherche Agronomique, Toulouse, France) for help with statistical analysis and Prof. Douglas Cook (University of California, Davis) for sending us the M. truncatula skl mutant.
This work was supported by the European Community's Framework 5 Human Potential Program (Marie Curie Fellowship to M.-T.N.G., contract no. HPMF-CT-1999-00073, and Research Training Network on Oligosaccharide Signaling in Plants, contract no. HPRN-CT-2002-00251 and by the Région Midi-Pyrénées, France.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027680.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardor M, Faye L, Lerouge P (1999) Analysis of the N-glycosylation of recombinant glycoproteins produced in transgenic plants. Trends Plant Sci 4: 376-380 [DOI] [PubMed] [Google Scholar]
- Barker D, Bianchi S, Blondon F, Dattée Y, Duc G, Essad S, Flament T, Gallusci T, Genier G, Guy P (1990) Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep 8: 40-49 [Google Scholar]
- Barre A, Hervé C, Lescure B, Rougé P (2002) Lectin receptor kinases in plants. Crit Rev Plant Sci 21: 379-399 [Google Scholar]
- Barrieu F, Chrispeels MJ (1999) Delivery of a secreted soluble protein to the vacuole via a membrane anchor. Plant Physiol 120: 961-968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28: 235-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell T, Carney D, Gardner S, Hayes F, Howlin B, Hubbard T, Overington J, Singh DA, Sibanda BL, Sutcliffe M (1988) 18th Sir Hans Krebs lecture: knowledge-based protein modelling and design. Eur J Biochem 172: 513-520 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant-Microbe Interact 14: 695-700 [DOI] [PubMed] [Google Scholar]
- Bono JJ, Riond J, Nicolaou KC, Bockovich NJ, Estevez VA, Cullimore JV, Ranjeva R (1995) Characterization of a binding site for chemically synthesized lipo-oligosaccharidic NodRm factors in particulate fractions prepared from roots. Plant J 7: 253-260 [DOI] [PubMed] [Google Scholar]
- Bouckaert J, Hamelryck T, Wyns L, Loris R (1999) Novel structures of plant lectins and their complexes with carbohydrates. Curr Opin Struct Biol 9: 572-577 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077-1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin NJ, Kardailsky IV (1997) Legume lectins and nodulation by Rhizobium. Trends Plant Sci 2: 92-98 [Google Scholar]
- Brill LM, Evans CJ, Hirsch AM (2001) Expression of MsLEC1- and MsLEC2-antisense genes in alfalfa plant lines causes severe embryogenic, developmental and reproductive abnormalities. Plant J 25: 453-461 [DOI] [PubMed] [Google Scholar]
- Caetana-Anolles G, Gresshoff PM (1991) Plant genetic control of nodulation. Annu Rev Microbiol 45: 345-382 [DOI] [PubMed] [Google Scholar]
- Clark M, Cramer RDI, van den Opdenbosch N (1989) Validation of the general purpose Tripos 5.2 force field. J Comput Chem 10: 982-1012 [Google Scholar]
- Cock JM, Vanoosthuyse V, Gaude T (2002) Receptor kinase signalling in plants and animals: distinct molecular systems with mechanistic similarities. Curr Opin Cell Biol 14: 230-236 [DOI] [PubMed] [Google Scholar]
- Cook D (1999) Medicago truncatula: a model in the making! Curr Opin Plant Biol 2: 301-304 [DOI] [PubMed] [Google Scholar]
- Côté F, Hahn M (1994) Oligosaccharins: structures and signal transduction. Plant Mol Biol 26: 1379-1411 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1997) Oligosaccharins, brassinolides, and jasmonates: nontraditional regulators of plant growth, development, and gene expression. Plant Cell 9: 1211-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore JV, Ranjeva R, Bono JJ (2001) Perception of lipochitooligosaccharidic Nod factors in legume. Trends Plant Sci 6: 24-30 [DOI] [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Promé JC (1996) Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65: 503-535 [DOI] [PubMed] [Google Scholar]
- Diaz CL, Melchers LS, Hooykass PJJ, Lugtenberg BJJ, Kijne JW (1989) Root lectin as a determinant of host-plant specificity in the Rhizobium-legume symbiosis. Nature 338: 579-581 [Google Scholar]
- Diaz CL, Spaink HP, Kijne JW (2000) Heterologous Rhizobial lipochitin oligosaccharides and chitin oligomers induce cortical cell divisions in red clover roots, transformed with the pea lectin gene. Mol Plant-Microbe Interact 13: 268-276 [DOI] [PubMed] [Google Scholar]
- Downie JA, Parniske M (2002) Fixation with regulation. Nature 420: 369-370 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevel Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962-966 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1993) PHYLIP (Phylogeny Inference Package) Version 3.5c. Department of Genetics, University of Washington, Seattle
- Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P et al. (2001) The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293: 668-672 [DOI] [PubMed] [Google Scholar]
- Gamas P, de Carvalho-Niebel F, Lescure N, Cullimore JV (1996) Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant-Microbe Interact 9: 233-242 [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez M, Berardini TZ, Chen G, Crist D, Doyle A, Huala E, Knee E, Lambrecht M, Miller N, Mueller LA et al. (2002) TAIR: a resource for integrated Arabidopsis data. Funct Integr Genomics 2: 239-253 [DOI] [PubMed] [Google Scholar]
- Geurts R, Bisseling T (2002) Rhizobium Nod factor perception and signalling. Plant Cell S239-S249 [DOI] [PMC free article] [PubMed]
- Gressent F, Drouillard S, Mantegazza N, Samain E, Geremia RA, Canut H, Niebel A, Driguez H, Ranjeva R, Cullimore JV et al. (1999) Ligand specificity of a high-affinity binding site for lipo-chitooligosaccharidic Nod factors in Medicago cell suspension cultures. Proc Natl Acad Sci USA 96: 4704-4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressent F, Mantegazza N, Cullimore JV, Driguez H, Ranjeva R, Bono JJ (2002) High-affinity Nod factor binding site from Phaseolus vulgaris cell suspension cultures. Mol Plant-Microbe Interact 15: 834-839 [DOI] [PubMed] [Google Scholar]
- Hadlington JL, Denecke J (2000) Sorting of soluble proteins in the secretory pathway of plants. Curr Opin Plant Biol 3: 461-468 [DOI] [PubMed] [Google Scholar]
- Hamelryck TW, Loris R, Bouckaert J, Dao-Thi MH, Strecker G, Imberty A, Fernandez E, Wyns L, Etzler ME (1999) Carbohydrate binding, a novel quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus. J Mol Biol 286: 1161-1177 [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M (2001) Intracellular functions of N-linked glycans. Science 291: 2364-2369 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819-832 [DOI] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Galaud JP, Rougé P, Lescure B (1996) Characterization of an Arabidopsis thaliana gene that defines a new class of putative plant receptor kinases with an extracellular lectin-like domain. J Mol Biol 258: 778-788 [DOI] [PubMed] [Google Scholar]
- Hervé C, Serres J, Dabos P, Canut H, Barre A, Rougé P, Lescure B (1999) Characterization of the Arabidopsis lecRK-a genes: members of a superfamily encoding putative receptors with an extracellular domain homologous to legume lectins. Plant Mol Biol 39: 671-682 [DOI] [PubMed] [Google Scholar]
- Hirsch AM (1999) Role of lectins (and rhizobial exopolysaccharides) in legume nodulation. Curr Opin Plant Biol 2: 320-326 [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Till JH (2000) Protein tyrosine kinase structure and function. Annu Rev Biochem 69: 373-398 [DOI] [PubMed] [Google Scholar]
- Imberty A, Bettler E, Karababa M, Mazeau K, Petrova P, Pérez S (1999) Building sugars: the sweet part of structural biology. In M Vijayan, N Yathindra, AS Kolaskar, eds, Perspectives in Structural Biology. Indian Academy of Sciences and Universities Press, Hyderabad, India, pp 392-409
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscular-containing cells. Mol Plant Microbe Interact 14: 737-748 [DOI] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O et al. (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579-5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijne JW, Bauchrowitz MA, Diaz CL (1997) Root lectins and rhizobia. Plant Physiol 115: 869-873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C, Parniske M (2002) Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci 7: 511-518 [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F et al. (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422-426 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283-291 [Google Scholar]
- Loris R (2002) Principles of structures of animal and plant lectins. Biochim Biophys Acta 1572: 198-208 [DOI] [PubMed] [Google Scholar]
- Loris R, De Greve H, Dao-Thi MH, Messens J, Imberty A, Wyns L (2000) Structural basis of carbohydrate recognition by lectin II from Ulex europaeus, a protein with a promiscuous carbohydrate binding site. J Mol Biol 301: 987-1002 [DOI] [PubMed] [Google Scholar]
- Loris R, Hamelryck T, Bouckaert J, Wyns L (1998) Legume lectin structure. Biochim Biophys Acta 1383: 9-36 [DOI] [PubMed] [Google Scholar]
- Loris R, Stas PP, Wyns L (1994) Conserved waters in legume lectin crystal structures: the importance of bound water for the sequence-structure relationship within the legume lectin family. J Biol Chem 269: 26722-26733 [PubMed] [Google Scholar]
- Navarro-Gochicoa MT, Camut S, Niebel A, Cullimore JV (2003) Expression of the apyrase-like APY1 genes in roots of Medicago truncatula is induced rapidly and transiently by stress and not by Sinorhizobium meliloti or Nod factors. Plant Physiol 131: 1124-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. Available at http://www.psc.edu/biomed/genedoc/
- Niehaus K, Becker A (1998) The role of microbial surface polysaccharides in the Rhizobium-legume interaction. Subcell Biochem 29: 73-116 [DOI] [PubMed] [Google Scholar]
- Nishiguchi M, Yoshida K, Sumizono T, Tazaki K (2002) A receptor-like protein kinase with a lectin-like domain from lombardy poplar: gene expression in response to wounding and characterization of phosphorylation activity. Mol Genet Genomics 267: 506-514 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M et al. (2002) HAR1 mediates systemic regulation of symbiotic organ development. Nature 420: 426-429 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357-358 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook D (1997) A legume ethylene-insensitive mutant hyper-infected by its rhizobial symbiont. Science 275: 527-530 [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt HJ, Puhler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant-Microbe Interact 6: 699-706 [Google Scholar]
- Rayner JC, Pelham HRB (1997) Transmembrane domain-dependent sorting of proteins to the ER and Plasma membrane in yeast. EMBO J 16: 1832-1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, Hervé C, Pacquit V, Dabos P, Lescure B (2002) Expression of an Arabidopsis lectin kinase receptor gene, lecRK-a1, is induced during senescence, wounding and in response to oligogalacturonic acids. Plant Physiol Biochem 40: 431-438 [Google Scholar]
- Rudiger H, Gabius HJ (2001) Plant lectins: occurrence, biochemistry, functions and applications. Glycoconj J 18: 589-613 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109-112 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763-10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959-962 [DOI] [PubMed] [Google Scholar]
- Szybiak-Strozycka U, Lescure N, Cullimore JV, Gamas P (1995) A cDNA encoding a PR-1-like protein in the model legume Medicago truncatula. Plant Physiol 107: 273-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins D (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoquet P, Gherardi M, Journet EP, Kereszt A, Ane JM, Prosperi JM, Huguet T (2002) The molecular genetic linkage map of the model legume Medicago truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BioMed Central Plant Biol 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rhijn P, Fujishige NA, Lim PO, Hirsch AM (2001) Sugar-binding activity of pea lectin enhances heterologous infection of transgenic alfalfa plants by Rhizobium leguminosarum biovar viciae. Plant Physiol 126: 133-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Journet EP, Barker DG (1999) MtENOD20, a Nod factor-inducible molecular marker for root cortical activation. Mol Plant-Microbe Interact 12: 604-614 [Google Scholar]
- Waldherr-Teschner M, Goetze T, Heiden W, Knoblauch M, Vollhardt H, Brickmann J (1992) MOLCAD: computer aided visualization and manipulation of models in molecular science. In FH Post, AJS Hin, eds, Advances in Scientific Visualization. Springer, Heidelberg, pp 58-67