Abstract
Kaposi sarcoma–associated (KS-associated) herpesvirus (KSHV) infection is linked to the development of both KS and several lymphoproliferative diseases. In all cases, the resulting tumor cells predominantly display latent viral infection. KS tumorigenesis requires ongoing lytic viral replication as well, however, for reasons that are unclear but have been suggested to involve the production of angiogenic or mitogenic factors by lytically infected cells. Here we demonstrate that proliferating cells infected with KSHV in vitro display a marked propensity to segregate latent viral genomes, with only a variable but small subpopulation being capable of stable episome maintenance. Stable maintenance is not due to the enhanced production of viral or host trans-acting factors, but is associated with cis-acting, epigenetic changes in the viral chromosome. These results indicate that acquisition of stable KSHV latency is a multistep process that proceeds with varying degrees of efficiency in different cell types. They also suggest an additional role for lytic replication in sustaining KS tumorigenesis: namely, the recruitment of new cells to latency to replace those that have segregated the viral episome.
Introduction
Kaposi sarcoma-associated herpesvirus (KSHV; also called human herpesvirus 8) is a novel human herpesvirus linked to the development of Kaposi sarcoma (KS). KS is a unique proliferative lesion with features that place it on the cusp between the benign and the malignant (1). At the center of KS pathology is the spindle cell, which is of endothelial origin. The proliferation of these cells is thought to be the driving force of KS histogenesis, but the lesion also contains two other hallmarks — a pronounced inflammatory infiltrate and a profusion of aberrant neovascular spaces. Spindle cells are the target of KSHV infection in vivo, with most cells being latently infected. Although these cells are considered the sine qua non of the lesion, they differ from standard tumor cells in many ways. First, unlike conventional tumor cells, they are often oligo- or polyclonal in KS lesions (2–4). KS spindle cells also lack the genetic instability of most tumor cells; they are typically diploid even in advanced lesions, and no characteristic chromosomal rearrangements have been found in the majority of KS tumors. Similarly, although spindle cells can be reproducibly grown in culture (5–10), they display few laboratory features commonly associated with transformed cells. For example, they remain heavily dependent upon exogenous growth factors, typically requiring cytokine-rich conditioned media from activated T cells for survival and proliferation (6). They do not form foci, grow in soft agar, or form tumors in nude mice (8, 9, 11, 12). These features have led to the proposal that the KS lesion results from reciprocal paracrine-signaling interactions between its various histologic components, no one component of which is fully autonomous. For example, spindle cells are known to produce proinflammatory and angiogenic factors, and inflammatory cells in turn produce cytokines that have been shown to sustain spindle cell survival and growth (see ref. 13 for review).
One of the most puzzling features of spindle cell biology has been the fact that every spindle cell line that has been derived from KS lesions over the years has been shown to lack the KSHV genome (7, 8, 14, 15), despite the fact that in advanced primary KS tumors nearly all spindle cells are latently infected. The reasons for this have been uncertain, but it is noteworthy that numerous cell lines have been derived from KSHV-associated primary effusion lymphomas (PELs) in which KSHV genomic persistence is readily demonstrable. This indicates that there are circumstances under which stable latency is achieved in vivo and that standard culture conditions are not incompatible with this state, at least in B cells. Because the failure of KS spindle cells to retain the genome in culture contravenes textbook notions of γ-herpesvirus latency, little attention has been paid to this phenomenon, which, where considered at all, has been brushed aside as a laboratory curiosity of doubtful relevance to KS biology.
Standard models of herpesviral oncogenesis generally ascribe to the latency program a primary role in driving tumor formation. Consistent with this, the KSHV latency program encodes several proteins with likely roles in the promotion of cell growth and extension of cell survival, including modulators of the p53-, Rb-, and β-catenin–signaling pathways (16–21). Latency alone, however, is not sufficient to sustain KS tumorigenesis in vivo. KS tumors also regularly display low levels of lytic viral replication (22, 23), and a recent clinical trial strongly indicates an important role for lytic infection in KS development (24). This trial showed that ganciclovir administration to severely immunodeficient AIDS patients who had been carrying KSHV for many years sharply inhibited the development of new KS lesions over the ensuing 6–12 months. Since ganciclovir is a specific inhibitor of lytic replication, this indicates that ongoing lytic KSHV replication is continuously required at all stages of the natural history of infection in order to promote KS tumorigenesis.
Several ideas have been put forward to explain how lytic infection might be important to KS tumor formation. Clearly, lytic replication is required early in the natural history of infection to allow spread from KSHV’s primary lymphoid reservoir to endothelial targets. In addition, the expression of several KSHV gene products with likely roles in angiogenesis and inflammation is restricted to the lytic cycle — for example, the virally encoded CC chemokines, which have both chemotactic and angiogenic activities (9, 25–30), and the viral G protein–coupled receptor, which can stimulate the release of VEGF from host cells and promote endothelial survival (31–35). Thus, ongoing lytic infection could be required to sustain the inflammatory and angiogenic components of a KS lesion.
Here we report another potential role for the lytic cycle in KS tumorigenesis. During experiments designed to study latent infection by KSHV, we observed that most proliferating cells in culture display marked instability of the latent phenotype — that is, as cell division proceeds, viral episomes are lost. This behavior, strikingly reminiscent of the behavior of authentic KS spindle cells following explantation, is regularly observed in every cell type we have examined. In some cell types, however, infrequent cells arise in which latency is stabilized, just as it is in PEL cells in vivo. Our data show that this stabilization is due not to host or viral mutations but to epigenetic changes operating on the viral genome in cis. These findings suggest that stable latency evolves via a multistep pathway and that cell types differ in the efficiency with which they support this pathway. Spindle cells appear to do so inefficiently, such that most newly latent cells lose the episome upon cell division. This finding suggests yet a third mechanism by which lytic replication could promote KS tumorigenesis: by sustaining the population of latently infected cells that otherwise would be diminished by segregation of latent viral episomes as spindle cells divide.
Methods
Plasmid constructs.
Plasmid pGFP and its derivatives pGTR4 and pGTR4:73 have been described (36). The vector backbone pGFP contains a GFP expression cassette driven by the CMV promoter; pGTR4 and pGTR8 were generated by assembling four or eight contiguous, head-to-tail–oriented units of the viral terminal repeats derived from pML1 (37) in a linker inserted into the backbone plasmid pGFP. pGTR4:73 was generated by inserting the CMV/ORF73 expression cassette from pCDNA3:ORF73 (38) into a polylinker upstream of the CMV promoter in pGTR4.
Cell lines and transfection procedures.
The KSHV-negative Burkitt’s lymphoma cell line BJAB was grown in RPMI-1640 medium supplemented with 10% FCS. The KSHV-positive PEL cell line BCBL-1 (39) was cultured in RPMI-1640 supplemented with 10% FCS, 0.005 mM 2-mercaptoethanol, 1 mM sodium pyruvate, and 2 mM L-glutamine. Telomerase immortalized dermal microvascular endothelial (TIME) cells (40) were maintained in EGM-2-MV medium from Cambrex Bio Science Rockland Inc. (Rockland Maine, USA). Primary human umbilical vein endothelial cells (HUVECs) were obtained from American Type Tissue Collection and maintained in EGM-2 medium (Cambrex Bio Science Rockland Inc.). Human foreskin fibroblasts (passage 16), the mouse hemangioendothelioma cell line, EOMA (41), human kidney (293 cells), and endothelial SLK cells (derived from a KS tumor) (10) were grown in DMEM H21 supplemented with 10% FCS.
SLK cells were transfected with Fugene 6 (Roche Applied Science, Indianapolis, Indiana, USA), according to the manufacturer’s instructions. BJAB cells were transfected by electroporation (20 μg DNA and 1 × 107 to 2 × 107 cells per transfection) using a BioRad Gene Pulser II (0.4-cm gap electrode cuvettes; instrument settings at 250 V, 960 μF; Bio-Rad, Hercules, California, USA). For antibiotic selection, media were complemented with G418 (Invitrogen Corp., Carlsbad, California, USA) at final concentrations of 0.8 mg/ml (SLK) or 1.5 mg/ml (BJAB).
Isolation of single-cell clones from infected SLK mass cultures.
Tenfold dilutions of mass cultures of infected SLK cells were seeded into 96-well plates 65 days after the initial infection. Colonies in individual wells of plates in which approximately 40% of the total wells showed outgrowth after 2 weeks were considered to have arisen from a single cell clone. Cells from 106 individual wells were expanded and examined by immunofluorescence analysis (IFA) for latency-associated nuclear antigen (LANA) expression. The KSHV-negative (SLKN) and KSHV-positive cell lines (SLKP) were generated by pooling 14 LANA-negative (N01–N14) or seven LANA-positive (P01–P07) single-cell clones. Cell numbers of the individual single-cell clones were adjusted before pooling to ensure equal representation of each clone.
Preparation of viral stocks and in vitro infections.
BCBL-1 cells (density 4 × 105 cells/ml) were treated with 12-O-tetradecanoylphorbol-13-acetate (TPA; final concentration of 20 ng/ml) and ionomycin (500 ng/ml) to induce the lytic cycle of KSHV replication. Cells were washed and resuspended in fresh medium after 20 hours, and culture supernatants were harvested after 4 days. Supernatants were passed through a 0.45-μm filter, and virions were pelleted by centrifugation at 27,000 g for 2 hours. Virion pellets were subsequently resuspended in complete medium (1:100 of the volume of the original supernatants). Unconcentrated supernatants of recombinant KSHV-GFP virions (42) were generously provided by S.-J. Gao (The University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA).
For infection of adherent cell lines in vitro, cells at approximately 50% confluence were incubated with 500 μl of concentrated virion preparations diluted 1:10 in complete medium or 500 μl of unconcentrated recombinant KSHV-GFP supernatants supplemented with Polybrene at a final concentration of 8 μg/ml. After incubation at 37°C for 2 hours, cells were washed three times and analyzed for expression of viral gene products 48–72 hours later.
Gardella and Hirt analyses of episomal DNA.
Gardella gel analysis was performed as described (43). Briefly, the region above the wells of a horizontal 0.75% agarose gel in Tris-borate-EDTA (TBE) buffer was removed and replaced by a lysis gel (0.8% agarose in TBE, 2% SDS, 1 mg/ml pronase) (Calbiochem-Novabiochem Corp., San Diego, California, USA). Cells were washed twice in PBS and resuspended in loading buffer: TBE buffer containing 15% Ficoll, 100 μg/ml RNAse A (QIAGEN Inc., Valencia, California, USA), and 0.01% bromophenol blue. Wells were loaded with cell suspensions (2 × 106 cells per sample per well). To allow lysis of the cells in the wells, gels were run at 4°C for 2 hours at 40 V. Subsequently, gels were electrophoresed at 160 V for 12 hours. DNA was transferred to nitrocellulose membranes by Southern blotting and detected with 32P-labeled probes specific for ORF73 or GFP.
Extraction of episomal DNA was performed using a modified Hirt procedure as described (44). For detection of episomal reporter plasmids by PCR, DNA was extracted from 5 × 106 to 1 × 107 cells and resuspended in 50–100 μl of Tris-EDTA (TE) buffer at a relative concentration of 105 cells/μl. Two microliters of the eluate (corresponding to 2 × 105 cells) was subjected to PCR amplification (25 cycles) with primers specific for GFP. One-half of the reaction mixture was analyzed on an agarose gel.
IFA.
Cells seeded on chamber slides (Labtec, Campbell California, USA) were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. LANA was detected using polyclonal Ab’s (45) at a dilution of 1:500, and nuclei were stained with DAPI. For quantitation of LANA-positive cells, median percentage as well as standard deviation were calculated after counting LANA-positive and LANA-negative nuclei from microscopic images of at least five random fields of view (containing 50 to 400 cells each) per time point.
Results
Modeling KSHV latency with plasmids.
In previous work, viral factors required for persistence of latent KSHV genomes have been identified. By searching for viral cis-acting sequences that maintain recombinant plasmids in PEL cells, several groups have localized such sequences to the terminal repeats (TRs) of the viral genome and shown that two to four such repeats suffice to allow persistence of the plasmid in selected clones. The sole viral trans-acting factor required for such persistence is the LANA, the product of ORF73. In addition to mediating plasmid DNA replication (36, 46–48), LANA has been shown to bind to both TR DNA sequences (49–51) and to mitotic chromosomes (52, 53), suggesting that tethering to the latter might be one way to promote segregation of the viral genome to daughter cells during mitosis.
Accordingly, plasmids bearing two to four copies of the viral TR elements together with a LANA expression cassette would be expected to be stably maintained in host cells. Previous studies aiming at long-term persistence of artificial KSHV episomes were carried out under continuous drug selection after introducing TR-containing plasmids into cells stably expressing LANA in trans (54). To investigate the dynamics of stable episome establishment in a system more closely resembling authentic KSHV infection (i.e., provision of LANA in cis), we constructed the plasmid series shown in Figure 1a. Plasmid pGTR4:73 carries four copies of the TR sequence, a LANA (ORF73) gene driven by the strong CMV IE promoter, and a GFP cassette to allow easy scoring of plasmid-bearing cells. (The plasmid backbone also carries a selectable marker, neo, encoding G418 resistance.) Plasmid pGTR4 is an isogeneic plasmid lacking ORF73, and pGFP is the parental vector lacking both the cis- and trans-acting factors from KSHV.
Figure 1.
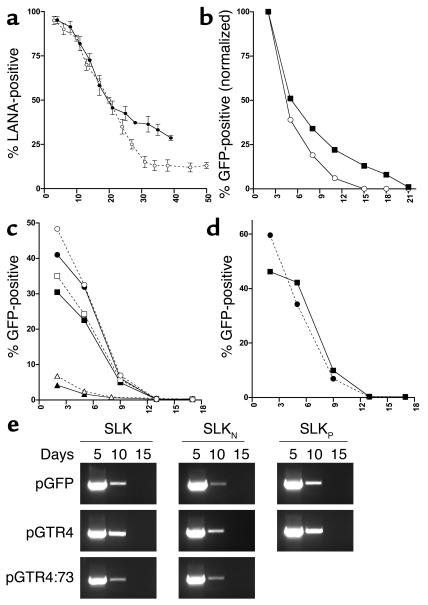
Loss of TR-containing reporter plasmids from transfected cells. (a) Functional elements of reporter plasmids. The vector backbone pGFP contains a GFP expression cassette driven by the CMV promoter. pGTR4 contains a GFP expression cassette as well as four contiguous units of the viral terminal repeats in authentic head-to-tail orientation. Construct pGTR4:73 contains, in addition to the GFP and TR elements, a CMV promoter–driven expression cassette for ORF73/LANA. (b–d) FACS analysis of cell lines transfected with the reporter plasmids described above. pGFP (open circles/dashed lines), pGTR4 (open squares/solid lines), or pGTR4:73 (filled triangles/solid lines) were introduced in SLK (b), BCBL-1 (c), or BJAB cells (d and e). BCBL-1 cells were only transfected with pGFP and pGTR4 because ORF73/LANA is provided in trans by endogenous KSHV episomes. The percentage of GFP-expressing cells was monitored over a period of 13–15 days after transfection by flow cytometry (FACS). (b–d) Analyses of transfected mass cultures. For the data shown in e, BJAB cells were FACS sorted 3 days after transfection for GFP expression in order to eliminate untransfected cells.
Each plasmid was transfected into the endothelial cell line SLK, and transfected cells were followed by examination for GFP fluorescence every 3–4 days (Figure 1b). As expected, GFP expression from pGFP and pGTR4 was rapidly lost from the culture. Surprisingly, however, the same was also true of pGTR4:73, which should express all functions thought to permit stable plasmid maintenance. (The initial level of pGTR4:73 transfection was lower than its sister plasmids, owing to its large size.) The terminal repeat sequences in the vectors were functional, since they mediated transient plasmid DNA replication in the presence of LANA as judged by the acquisition of Dpn1 resistance (36). Control experiments demonstrated that LANA was indeed efficiently expressed in the days following transfection; in fact, the decline in GFP-positive cells closely paralleled the loss of LANA-positive cells as judged by immunofluorescence (data not shown). Further proof that the instability of the TR-bearing plasmids is not simply attributable to inadequate LANA expression is shown in Figure 1c, in which control or pGTR4 plasmids have been introduced into BCBL-1 cells. BCBL-1 is a cell line derived from a KSHV-infected PEL. Like all PEL cell lines, it is latently infected by KSHV, expresses high levels of LANA, and stably maintains the viral genome as a closed circular nuclear episome. Nonetheless, when pGTR4 is introduced into such cells, GFP expression is lost nearly as rapidly as it is from cells transfected with plasmids (e.g., pGFP) lacking KSHV sequences altogether (Figure 1c).
Because this instability was unexpected, we wondered if SLK cells — or endothelial cells, generally — might have an unusual defect in supporting LANA function. The apparent instability of the pGTR4:73 replicon was equally apparent in transfected BJAB B cells (Figure 1d), however. To exclude the possibility that the culture was simply being overgrown by untransfected cells lacking LANA and GFP expression, we conducted an additional experiment in which transfected BJAB cells were first sorted by flow cytometry for high levels of GFP expression so that the starting cultures were 100% GFP positive. Nonetheless, GFP expression was rapidly lost from pGTR4:73-transfected cells (Figure 1e).
Figure 2 shows that the loss of GFP expression is, in fact, due to the loss of the plasmid genome and not simply the extinction of GFP expression. Figure 2a shows a Gardella gel analysis of the episome complement of the transfected, sorted BJAB cells of Figure 1e. In the Gardella protocol, cells are lysed in the well of the gel, and the electrophoresis conditions are such that chromosomal DNA remains trapped there; only episomal DNA is detected following electrophoresis, blotting, and hybridization to a GFP probe. As shown in Figure 2a, while pGTR4:73-transfected cultures harbored plasmid DNA on day 4 after FACS sorting (day 8 after transfection), they became negative soon thereafter. When analyzed by a more sensitive PCR technique applied to Hirt supernatant fractions, which are enriched for episomal DNA, plasmid DNA was detectable up to 6–10 days after sorting (10–14 days after transfection), but disappeared thereafter (Figure 2b). In Figure 2c we present an analysis of BJAB cells that were transfected with the plasmids of Figure 1a, then propagated in the presence of G418 to select for plasmid-bearing cells. Cells were examined every 5–6 days for episomal DNA by Gardella gel analysis. Even under these conditions, pGTR4:73 disappeared from the culture within 15 days. Notably, we could detect cells bearing stable levels of GFP fluorescence (as judged by FACS analysis) for many weeks. Such cells, however, lacked evidence of episomal DNA when examined in Gardella gels and presumably harbored only integrated plasmid sequences (data not shown). Thus, even in the presence of prolonged selection most cells still displayed loss of episomal KSHV replicons.
Figure 2.
Absence of stable episomal DNA from long-term cultures of cells transfected with TR reporter plasmids. (a) Aliquots of FACS-sorted BJAB cells transfected with the indicated reporter constructs were harvested at various time points after FACS sorting (indicated in days above the lanes) and subjected to Gardella gel analysis. Samples were derived from the same cultures shown in Figure 1e. As a control, untransfected BJAB cells were loaded in the lanes labeled BJAB. Plasmid DNA was detected with a probe specific for GFP. The band marked with an asterisk results from unspecific background hybridization, since it is also present in untransfected BJAB cells. (b) Episomal DNA was isolated from the same BJAB cultures described above by Hirt extraction and subjected to PCR amplification of the GFP cassette as described in Methods. (c) Gardella gel analysis of transfected BJAB cells cultures grown under antibiotic selection. BJAB cells were transfected with pGFP, pGTR4, or pGTR4:73 and cultured in the presence of G418. Aliquots of cells propagated for the time periods indicated above the lanes (in days) were examined for the presence of episomal DNA by Gardella analysis. Episomes were detected with a probe specific for the GFP cassette.
These results seem to contradict previous findings in which TR-containing plasmids were reported to persist in LANA-expressing cells over a period of several weeks (54). There are differences in the experimental design of the two studies, however, most notably the expression of LANA in cis instead of in trans and the absence of antibiotic drug selection in most of our studies. To investigate whether our reporter constructs would behave similarly under the conditions employed by Ballestas et al., we transfected pGTR4 as well as pGTR8 (which carries eight instead of four terminal repeat units) into SLK73 cells, a cell line stably expressing LANA. When these cultures were grown in the presence of neomycin, stable cultures of pGTR4- or pGTR8-transfected cells grew out more rapidly than those transfected with the vector backbone pGFP (data not shown), and at 2 weeks after transfection, approximately 90% of the pGTR4/pGTR8-transfected mass cultures were GFP positive, compared with about 35% GFP-expressing cells in the pGFP-transfected cultures (Figure 3a, left panel). PCR analysis of Hirt supernatants from the pGTR4/pGTR8-transfected cultures over a period of 2–5 weeks after transfection revealed the presence of extrachromosomal plasmid DNA, indicating that stable episomal maintenance had occurred exactly as observed by previous workers (Figure 3a, right panel). When transfected SLK73 cells were grown in the absence of G418 selection, however, GFP-expressing cells disappeared from the cultures after 3 weeks and episomal pGTR4/pGTR8 plasmids became undetectable by day 20 after transfection (Figure 3b).
Figure 3.
Efficient establishment of artificial KSHV episomes in vitro requires antibiotic selection and expression of LANA in trans. SLK73 cells were transfected with pGFP (open circles/dashed lines), pGTR4 (open squares/solid lines), or pGTR8 (filled squares/solid lines), and the transfected cultures were maintained in the presence (a) or absence (b) of G418. Cultures were analyzed at the indicated time points after transfection by FACS (left panels in a and b) or Hirt-PCR (right panels).
We do not fully understand why episomes are so readily lost under conditions where LANA is provided in cis and/or antibiotic selection is not applied, while they are more efficiently maintained if G418 selected in cells bearing a resident LANA gene. We suspect, however, that the immediate plasmid DNA amplification that occurs upon entry in the latter case may allow more cell generations to pass before loss of the incoming genomes; this allows more time to elapse during which secondary events that stabilize the episome can occur (see below).
The instability of authentic latent genomes.
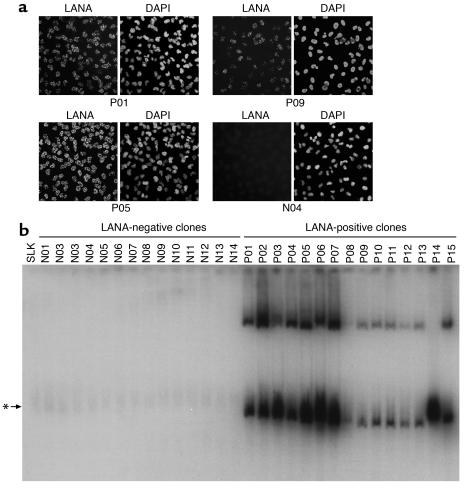
The inability of plasmids containing four copies of the TR to be maintained in KSHV-positive PEL cells (Figure 1c) raised the question of whether plasmid instability might simply be the result of inadequate TR copy numbers, since the authentic KSHV genome harbors over 30 copies of the TR (37). It is difficult to explore this issue with recombinant plasmids, since genomes bearing so many TRs are difficult to construct and would be expected to have a very low transfection efficiency. Instead, we opted to directly examine the stability of authentic KSHV genomes following their de novo introduction into cells through authentic viral infection. We have recently found that a wide variety of cultured adherent cell lines can be infected by KSHV, with latent infection being the result (55). Accordingly, we prepared a high-titer stock of KSHV from TPA-induced BCBL-1 cells and infected a variety of cell lines of endothelial (TIME, SLK, EOMA), epithelial (293 cells), and fibroblastic (HFF) origin. It was not possible to examine lymphoid cells in this manner, because neither we (55, 56) nor others (57) have been able to achieve de novo infection of established lymphoid cell lines. By 72 hours after infection, each of these cell lines displayed readily detectable LANA-positive cells; indeed, for HFF, TIME, and SLK, greater than 90% of the cells were initially positive for LANA expression (Figure 4). Nonetheless, in every case there was a rapid decline in the fraction of cells staining for LANA. We observed two patterns of LANA loss. In pattern 1, displayed by TIME, EOMA, and 293 cells, LANA-positive cells became virtually undetectable (less than 0.1% of the cells), while in pattern 2 (seen in SLK and HFF), after an initial steep decline, the percentage of LANA-positive cells stabilized at 10–20% of the total and remained fixed thereafter. We picked one representative of each pattern (TIME for pattern 1, SLK for pattern 2) for examination of viral episome content by Gardella gel analysis at varying times after infection. As expected from our earlier analysis of TIME cells (58), these cells displayed a rapid and progressive loss of latent viral episomes, which became undetectable by day 20 (Figure 5a). In sharp contrast, we observed a nearly constant signal of viral DNA in the infected SLK culture, irrespective of the time after infection at which the cells were assayed (Figure 5b). The explanation for this seemingly paradoxical result emerged when we conducted a detailed analysis of single-cell clones subcloned from the mass culture of infected SLK cells analyzed in Figure 4. As previously noted, at equilibrium about 80–90% of the cells in the mass culture were LANA negative and 10–20% were LANA positive. One hundred six single-cell clones were derived from the parental culture (at 65 days after infection) by limiting dilution. Consistent with the IFA of the parental culture, 91 of these clones were LANA negative and 15 were LANA positive. Interestingly, the LANA-positive clones fell into two classes: those with very strong LANA staining (as strong as seen in typical PEL lines) and a second class of less intensely positive cells. Although the latter clones all have typical punctate intranuclear dots of LANA, the intensity of the staining was reduced and the number of dots per nucleus was more variable. Figure 6a shows representative IFA images of each class of clones. Next we examined the 15 LANA-positive, as well as a collection of 14 LANA-negative, subclones (derived from the same mass culture) by Gardella gel analysis. As expected, all LANA-negative clones displayed no detectable KSHV genomes. By contrast, seven of seven strongly LANA-positive clones had high levels of KSHV DNA (comparable to that observed in uninduced BCBL-1 cells), while eight of eight LANA-intermediate clones showed intermediate levels of viral DNA (Figure 6b). When examined over a period of 2 months, a pool of the strongly LANA-positive clones (termed SLKP) maintained high expression levels of LANA in 100% of the cells (data not shown). Thus, the seeming stability of KSHV copy number in the mass culture (Figure 5b) is actually a composite of two processes: complete episome loss from the majority of cells combined with an amplification of genome copy number in the 10–15% of the cells in which stabilization of latency has occurred.
Figure 4.
Loss of LANA-positive cells in cultures infected with KSHV in vitro. The indicated cell lines were infected with viral supernatants from lytically induced BCBL-1 cells as described in Methods. Cells were cultured for 30–50 days after infection, and the percentage of LANA-positive cells was evaluated every 3–6 days by IFA.
Figure 5.
Gardella gel analysis of in vitro–infected SLK and TIME cells. Aliquots from KSHV-infected TIME (a) or SLK (b) cultures were evaluated by Gardella gel analysis at various time points after infection (indicated in days above the lanes) and KSHV episomes were detected using a LANA-specific probe. As a control, BCBL-1 cells were loaded in the leftmost and rightmost lanes in a and b, respectively. BCBL-1 cells were either untreated (lanes labeled BCBL-1) or treated with TPA and ionomycin (lanes labeled BCBL-1 ind.) for lytic cycle induction. The position of supercoiled episomes (upper band) and linear as well as nicked and partially degraded KSHV DNA (lower band) are marked by arrows labeled sup and lin, respectively. Note that the band marked with an asterisk in a, migrating slightly above the linear viral DNA, results from nonspecific background hybridization, because it is also present in the uninfected TIME cells loaded in lane 3. TPA/ionomycin-treated BCBL-1 samples show an increase in intensity of the lower (linear) band relative to uninduced BCBL-1 cells due to lytic replication and production of virus particles harboring linear KSHV genomes. (Note: Since spontaneous lytic replication does not occur in SLK [55], the linear DNA in b likely derives from fragmentation of circular KSHV genomes during handling and electrophoresis). infect., infected; ind., lytic cycle induction.
Figure 6.
KSHV-positive and KSHV-negative single cell clones derived from infected SLK mass cultures. SLK cells that had been cultured for 65 days following infection were subjected to single-cell cloning by limiting dilution. (a) Representative immunofluorescence pictures of three LANA-positive single-cell clones (P01, P05, and P09) as well as one LANA-negative clone (N04). LANA was detected with a polyclonal antiserum (left picture in each panel), and nuclei were stained with DAPI (right picture). (b) Gardella analysis of 14 LANA-negative (N01–N14) and 15 LANA-positive (P01–P15) single-cell clones derived from infected mass cultures. Uninfected SLK cells were loaded as a control in the leftmost lane. The arrow marked with an asterisk indicates the position of a background band that is also present in uninfected SLK control cells. KSHV episomes were detected using a probe specific for ORF73/LANA.
The availability of KSHV-positive and KSHV-negative derivatives of a once-infected SLK culture enabled us to ask additional questions about the nature of the changes that permit stable latency. First, we asked if pooled SLK clones that had become KSHV negative (termed SLKN) would be less susceptible to KSHV latency or segregate episomes more rapidly than the parental SLK population. Accordingly, 14 SLK-negative clones were pooled and reinfected with the same stock of KSHV used to create the original SLK infection. As shown in Figure 7a, LANA staining was lost at a rate that was similar to that of the original SLK infection of Figure 4 (also reproduced in Figure 7a to facilitate comparison). This suggests that these negative clones are no less permissive to latent genome maintenance than the starting cell line. In fact, stably latent cells arose at least as readily in this population as in the parental cell line. In agreement with this, Figure 7c shows that when the SLKN pool was transfected with pGTR4:73, GFP positivity was lost at a rate similar to that observed in similarly transfected parental SLK cells. To investigate whether SLKP are more susceptible for the establishment of latent episomes, we have examined the pool of strongly LANA-positive stable SLK clones (SLKP) for its ability to stably maintain a newly introduced KSHV replicon. SLKP cells were transfected with either pGFP or pGTR4, and the fraction of GFP-positive cells were recorded over time. Figure 7d shows that like BCBL-1 cells (Figure 1c), SLKP cells rapidly lose GFP positivity when transfected with TR-containing plasmids, despite their high levels of LANA expression and their high copy number of latent KSHV genomes. PCR analysis of the SLKN and SLKP transfectants (Figure 7e) confirmed that the incoming pGTR4:73 or pGTR4 plasmids were indeed lost from the culture by day 15. To confirm that these results can be generalized to the context of authentic viral infection, we examined the ability of SLKN and SLKP cells to support superinfection by a genetically marked KSHV virus. Recombinant KSHV-GFP virions (a generous gift of S.-J. Gao) were used to infect SLKN and SLKP cultures, and the fraction of initially infected (GFP positive) cells normalized to 100%. As can be seen in Figure 7b, over the next 3 weeks the superinfecting genomes were rapidly lost from the cultures, as manifested by the disappearance of GFP-positive cells. The fact that no stable GFP-positive clones were observed in these experiments (compared with the 10–20% stably infected cells in the long-term SLK cultures shown in Figure 4) is a likely result of the lower initial infection efficiencies obtained with the (unconcentrated) recombinant KSHV-GFP supernatants (2.9% and 4.2 % for SLKN and SLKP cells, respectively, compared with 95% in the experiments presented in Figure 4).
Figure 7.
Analysis of SLKP and SLKN cells. (a) SLKN cells were infected with viral supernatants from lytically induced BCBL-1 cells. The percentage of LANA-positive cells was evaluated over a period of 40 days by IFA (solid line). The curve obtained from the initial infection of the parental SLK mass cultures (see Figure 4) is shown for comparison (dashed line). (b) SLKN (open circles) or SLKP cells (filled squares) were infected with recombinant KSHV-GFP supernatants, and the percentage of GFP-positive cells was analyzed by FACS over a period of 3 weeks. Shown are normalized percentages relative to the initial infection level (absolute infection efficiencies were 2.9% and 4.2% for SLKN and SLKP cells, respectively). (c) SLKN cells (solid lines, filled symbols) or uninfected SLK cells (dashed lines, open symbols) were transfected with the reporter constructs pGFP (circles), pGTR4 (squares), or pGTR4:73 (triangles). The percentage of GFP-expressing cells was monitored over a period of 17 days after transfection by FACS. (d) SLKP cells were transfected with pGFP (circles) or pGTR4 (squares) and analyzed by FACS over a period of 17 days. (e) PCR analysis of the transfected SLK, SLKN, and SLKP cultures described above. Episomal DNA was isolated at the time points indicated (in days) above the lanes by Hirt extraction and subjected to PCR amplification of the GFP cassette as described in Methods.
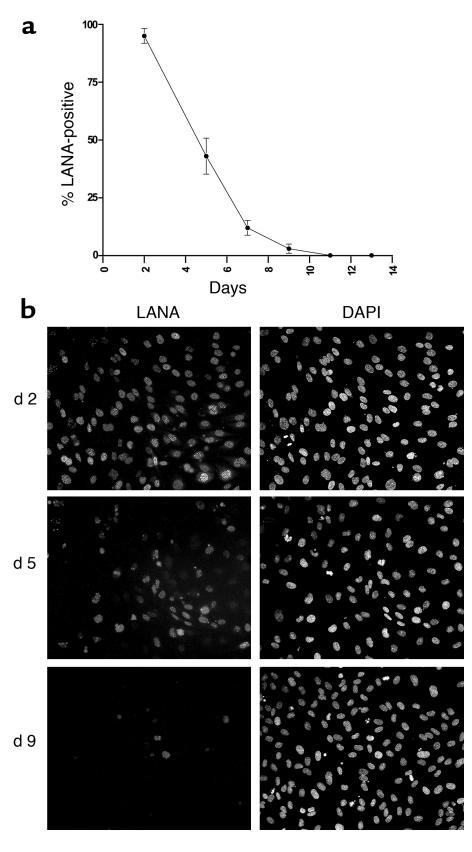
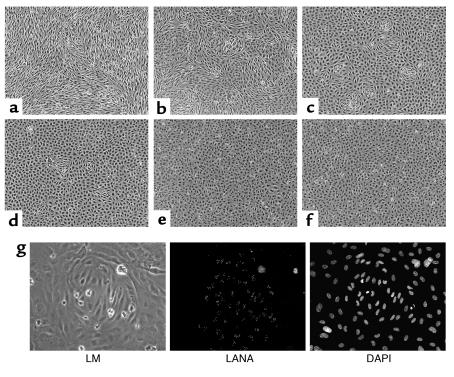
All of the above experiments were conducted in established cell lines, many of which are immortalized, transformed, or derived from tissues that are not normally hosts for KSHV. To study episome maintenance in a system more directly relevant to KS pathogenesis, we examined the fate of the KSHV genome in newly infected primary endothelial cells. Hayward and colleagues (59) have previously shown that such cells can be latently infected by KSHV, whereupon they undergo morphologic changes that are strikingly reminiscent of those displayed by KS spindle cells in vivo. Accordingly, primary HUVECs were exposed to KSHV on passage 2 in vitro. LANA staining revealed that initially approximately 95% of the cells were latently infected (Figure 8, a and b). When such cultures were grown to confluence, nearly all cells displayed the characteristic elongation and spindling induced by KSHV (Figure 9a). In continuously growing cultures (passaged every 2 to 3 days), however, virtually all the cells had lost LANA staining after five passages (11 days after infection; Figure 8, a and b) and, in concert with this, the culture was once again devoid of spindlelike cells (Figure 9e). IFA confirmed that the clusters of spindle-shaped cells in confluent cultures derived from intermediate passages (Figure 9, b–d) were indeed infected with KSHV; in contrast, the surrounding, cobblestone-shaped cells were KSHV negative (Figure 9g).
Figure 8.
Loss of KSHV-positive cells in continuously growing HUVEC cultures. HUVEC cells were infected with KSHV in vitro and passaged every 2 to 3 days for a period of 2 weeks. (a) Graph showing the percentage of LANA-positive cells at each passage as judged by IFA. (b) Representative immunofluorescence pictures of cultures at 2, 5, or 9 days after infection stained for LANA (left picture in each complement) or nuclei (right pictures).
Figure 9.
Loss of spindle cell morphology correlates with loss of KSHV episomes in KSHV-infected HUVEC cultures. At each passage, aliquots of the continuously growing KSHV-infected HUVEC cultures shown in Figure 6a were allowed to reach confluence and maintained for an additional 4 days to allow for complete conversion of infected cells to spindle cell morphology. Shown are cultures seeded at passage 1 (day 2 after infection (a), passage 2 (day 5 after infection) (b), passage 3 (day 7 after infection) (c), passage 4 (day 9 after infection) (d), and passage 5 (day 13 after infection) (e). An image of a mock-infected culture propagated in parallel is shown (f). (g) Cluster of spindle-shaped cells from a culture seeded at day 7 after infection analyzed by light microscopy (LM, left) and immunofluorescence staining for LANA (center). Nuclei were stained with DAPI (right).
Discussion
These data show that latent KSHV episomes tend to be lost from a wide variety of dividing cells, despite the presence of both cis- and trans-acting components previously shown to be required for episome maintenance. Several lines of evidence suggest that the accumulation of LANA-negative cells in these experiments reflects episome loss from infected cells rather than the selective overgrowth of uninfected or untransfected cells (such as might occur if, for example, LANA expression conferred a growth disadvantage). First, the loss was observed even when genetically marked infected cells were sorted (Figure 1e) or selected (Figure 2c) to eliminate untransfected cells. Second, during KSHV infection of SLK, stable latent clones accumulated with elevated levels of LANA, hardly the result one would anticipate if LANA were in some way inhibitory to growth. Third, TR-containing genomes were unstable even in BCBL-1 or SLKP cells that continuously express high levels of LANA. Finally, we have directly searched for effects of LANA on cell growth and viability by performing colony suppression assays. In this assay, cells are cultured under antibiotic selection after transfection with expression constructs conferring drug resistance. We have observed no significant differences in the number of drug-resistant colonies arising from cultures receiving a LANA expression construct or the empty control vector (data not shown). Likewise, when SLK or BJAB were transfected with an expression vector for LANA and cultured under antibiotic selection, we obtained mass cultures in which approximately 40–50% of the cells expressed LANA (as judged by immunofluorescence). The proportion of LANA-expressing cells remained stable when these cultures were monitored over a period of several weeks (data not shown). Detrimental effects of LANA on cell growth, in contrast, would be expected to result in declining levels of LANA-positive cells over time.
In accord with earlier findings of others (54), we were able to derive stable lines that display efficient long-term maintenance of episomal KSHV TR plasmids (Figure 3a). This outcome required two features, however: drug selection and introduction of a TR-containing episome into a cell already expressing LANA. Interestingly, neither one of these factors alone is sufficient in culture (Figures 1, 2, 3b, and 8), and neither feature is typically present in a natural infection with KSHV in vivo. In fact, in most cases in which a TR-bearing plasmid or viral genome is presented to a dividing cell without drug selection, the element is lost from the majority of cells, often extremely rapidly. Even in the absence of selection, however, rare cells can be identified in which authentic viral latency is stably maintained. The frequency with which such cells arise varies from cell line to cell line and ranges from less than 0.1% to 0.2% (so-called pattern 1) to more than 10% (pattern 2). Lines in which stable episomes arise have not undergone heritable genetic changes that render them more competent to stably replicate viral episomes; when new viral episomes are presented to them, either by transfection or infection, they are lost from these cells as rapidly as from KSHV-naive cell lines. This indicates that changes have occurred to resident viral genomes in cis. In principle, such changes could be genetic or epigenetic. If the former, we would expect virus derived from a stably latent cell line (e.g., BCBL-1) to engender stable latency upon infection of a naive cell. That is not what is observed, however: even virus derived from BCBL-1 is initially unstable in newly infected cells (Figures 4 and 8). Moreover, we have directly looked for genetic rearrangements in our SLKP clones using both Southern blot analysis and PCR and have found little evidence for such changes (data not shown; available as supplemental information on http://itsa.ucsf.edu/∼micro/Faculty/ganem_folder/data/suppdata.html). Thus, the cis-acting changes in stably latent genomes are most likely epigenetic. Latency establishment must therefore be a multistep process involving not only the expression of LANA and its action to promote viral DNA replication, but also one or more epigenetic modifications of the latent viral genome.
Our ability to isolate SLK clones in which episome stabilization has occurred now opens this phenomenon to experimental scrutiny. Given that the responsible changes are epigenetic, several biochemical processes can be considered, for example, changes in chromatin structure, DNA methylation, or histone modification. Such changes could, for instance, alter the ability of the TR to be segregated by allowing the viral chromosome access to specialized subnuclear compartments that favor stable maintenance. It is even possible that the initial replication of the viral DNA directed by LANA might be a signal that recruits the modification machinery (DNA methyltansferases, histone methylases) to the viral chromatin; if so, this might explain, in part, the increased stability of TR plasmids transfected into cells already expressing LANA. By comparing the chromatin structure or methylation state of viral genomes in stable SLKP cells with those in their unstable progenitors, we should be able to develop a more precise biochemical understanding of the determinants of episome stabilization.
All of our experiments have of necessity been conducted in cultured cells, so it could be argued that the results might not be applicable to spindle cells in vivo. For example, spindle cells or endothelial cells in general might express factors that facilitate episomal maintenance in vivo, and such factors might not be expressed in cell lines cultured in vitro. There are strong reasons to believe that our conclusions do apply to authentic KS spindle cells as well, however. First, we note that even primary or secondary endothelial cultures, which retain differentiated endothelial function and faithfully reproduce the spindle-cell phenotype upon KSHV infection, display unstable latency following infection. Second, this behavior exactly mirrors that of authentic spindle cells explanted directly from KS biopsies, which rapidly segregate the genome when placed in conditions supporting cell proliferation (7, 8, 14, 15). Moreover, the kinetics of this loss strikingly parallels that observed in our experiments, resembling that referred to above as pattern 1, that is, rapid and complete loss. This suggests that most latently infected spindle cells in vivo have not undergone whatever epigenetic changes are necessary to stabilize latency. There clearly are some KSHV-infected cells in vivo that have done so, however, best exemplified by the tumorous B cells of KSHV-associated PELs. These cells are uniformly LANA positive in vivo, grow readily in culture, and maintain viral episomes at substantial copy number indefinitely in vitro. Thus, all of the latency phenotypes we observe in vitro have their counterparts in cells that exist in vivo.
In fact, the view of latency we present here violates no experimentally validated fact about γ-herpesviral latency. First, despite the widespread impression that EBV-based plasmids bearing oriP and EBNA-1 efficiently achieve stable episomal maintenance, Sugden and colleagues (60) have recently reported (and we have confirmed; A. Grundhoff and D. Ganem, unpublished data) that such plasmids are, in fact, frequently lost from most proliferating cells, with only a small subpopulation entering stable latency. Thus, even for EBV, the prevailing notions about efficient latency induction rest upon very insecure foundations. It is true that in EBV biology virtually all B cell tumors explanted from patients display stable episome maintenance. To our knowledge, no classical EBV-induced lymphoma displays the kind of instability we observe in KS tumors. But there are many examples of explants from nasopharyngeal carcinoma in which loss of the EBV episome has occurred, and only a few NPC lines exist in which the EBV genome has been retained (61, 62).
The seemingly paradoxical existence of a multistep (and often inefficient) pathway to latency in both KSHV and EBV may, however, be rationalized by viewing them from an evolutionary perspective. The natural target cell of both KSHV and EBV latency is the B cell. Although not well studied for KSHV, the natural history of B cell latency in vivo has been extensively examined in EBV infection (63–67). In the peripheral blood of healthy carriers, the virus is restricted to latently infected, memory B cells that are not proliferating, but rather are resting in the G0 phase of the cell cycle. These cells represent a major site of long-term persistent infection. They express few EBV genes and do not express the full repertoire of EBV latency that is expressed in proliferating lymphoblastoid cell lines in vitro. In vivo, the latter program is largely restricted to newly infected naive B cells and triggers a growth program resulting in B cell activation. Although the duration of this proliferative state is unknown, by analogy with antigen-induced activation (which it somewhat resembles), it is likely to be brief; these cells then differentiate into long-lived memory B cells, most likely in germinal centers. If this view is correct, then the relative brevity of virus-induced B cell proliferation would not require a highly efficient plasmid maintenance machinery to allow long-term persistence. This implies that there would have been little selective pressure to drive the evolution of a mechanism to ensure the efficient stabilization of latency in cells undergoing sustained proliferation.
While not important in infrequently dividing cells, the relative instability of newly established KSHV latency in actively proliferating cells has important potential implications for the pathogenesis of KS. Since KS is characterized by sustained endothelial proliferation, such instability would be expected to lead to the loss of KSHV infection over time, just as is seen in newly infected primary endothelial cells or in explanted KS spindle cells. If our findings are relevant in vivo, one might expect to find many uninfected spindle cells in KS lesions. In fact, in early KS lesions, this is precisely what is observed (68). In more advanced lesions, however, KS spindle cells display high levels of latent infection. How are such high levels maintained? We propose that lytically infected cells (both within the lesion and elsewhere) produce virus that can infect (or reinfect) such cells, restoring them to latency. (Horizontal spread did not occur in our in vitro experiments. While a small percentage of lytically infected cells was detectable immediately after the initial infection [data not shown], spontaneous lytic reactivation ceased generally within 3 to 5 days after infection in all continuously growing cultures.)
If, as is generally believed, latency produces a cell-autonomous growth or survival advantage in spindle cells, then loss of latency would result in loss of said advantage. (The disappearance of the spindling phenotype in HUVEC cultures is the in vitro correlate of this.) For a KS tumor mass to expand under such conditions, continuous (re)infection of such KSHV-negative cells by infectious virus produced by lytic replication would be required to sustain high levels of latency in the tumor. This is consistent with clinical studies showing that progression to advanced KS is linked to high KSHV viral loads (69–71). Our findings encourage the view that a principal role of HIV-induced immune depletion in KS pathogenesis may be to cause the host to lose control of lytic KSHV replication.
Of course, the instability of newly established latency does not mean that every pathologic process linked to infection will display continuous dependence on lytic replication. This is certainly not the case for KSHV-induced PEL, for example, or EBV-related Burkitt’s lymphoma. Presumably, during the evolution of these classical clonal malignancies, one of the (many) changes that is selected for is epigenetic stabilization of the viral episome. The more rapid and autonomous the proliferation of the clone, the stronger such a selection is likely to be. One of the unsolved puzzles of KS is why such stabilization has not been regularly selected for in this process. The relative indolence of the proliferation in vivo, taken together with a high episomal copy number, could be factors in this regard.
We emphasize that our findings do not exclude other contributions of lytic replication to the pathogenesis of KS, such as the production of paracrine factors that promote proliferation and angiogenesis (see ref. 32 for review). Indeed, we favor the view that both contributions of the lytic cycle are important and regard them as complementary: one allows the lytic cycle to participate directly in KS histogenesis, the other acts by supporting the contributions of the latency program to the process. Taken together, they provide a strong rationalization for the many disparate clinical and biological observations linking lytic KSHV replication to this remarkable neoplasm.
Footnotes
See the related Commentary beginning on page 21.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Kaposi sarcoma-associated herpesvirus (KSHV); Kaposi sarcoma (KS); primary effusion lymphoma (PEL); telomerase immortalized microvascular endothelial cells (TIME); human umbilical vein endothelial cells (HUVEC); hemangioendothelioma (EOMA); latency-associated nuclear antigen (LANA); KSHV-negative cell line (SLKN); KSHV-positive cell line (SLKP); 12-O-tetradecanoylphorbol-13-acetate (TPA); Tris-borate-EDTA (TBE); terminal repeat (TR); immunofluorescence analysis (IFA).
References
- 1.Herndier B, Ganem D. The biology of Kaposi’s sarcoma. Cancer Treat. Res. 2001;104:89–126. doi: 10.1007/978-1-4615-1601-9_4. [DOI] [PubMed] [Google Scholar]
- 2.Delabesse E, et al. Molecular analysis of clonality in Kaposi’s sarcoma. J. Clin. Pathol. 1997;50:664–668. doi: 10.1136/jcp.50.8.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill PS, et al. Evidence for multiclonality in multicentric Kaposi’s sarcoma. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8257–8261. doi: 10.1073/pnas.95.14.8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Judde JG, et al. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi’s sarcoma and other diseases. J. Natl. Cancer Inst. 2000;92:729–736. doi: 10.1093/jnci/92.9.729. [DOI] [PubMed] [Google Scholar]
- 5.Corbeil J, Evans LA, Vasak E, Cooper DA, Penny R. Culture and properties of cells derived from Kaposi sarcoma. J. Immunol. 1991;146:2972–2976. [PubMed] [Google Scholar]
- 6.Ensoli B, et al. AIDS-Kaposi’s sarcoma-derived cells express cytokines with autocrine and paracrine growth effects. Science. 1989;243:223–226. doi: 10.1126/science.2643161. [DOI] [PubMed] [Google Scholar]
- 7.Aluigi MG, et al. KSHV sequences in biopsies and cultured spindle cells of epidemic, iatrogenic and Mediterranean forms of Kaposi’s sarcoma. Res. Virol. 1996;147:267–275. doi: 10.1016/0923-2516(96)82285-0. [DOI] [PubMed] [Google Scholar]
- 8.Lebbe C, et al. Characterization of in vitro culture of HIV-negative Kaposi’s sarcoma-derived cells. In vitro responses to alfa interferon. Arch. Dermatol. Res. 1997;289:421–428. doi: 10.1007/s004030050215. [DOI] [PubMed] [Google Scholar]
- 9.Salahuddin SZ, et al. Angiogenic properties of Kaposi’s sarcoma-derived cells after long-term culture in vitro. Science. 1988;242:430–433. doi: 10.1126/science.2459779. [DOI] [PubMed] [Google Scholar]
- 10.Herndier BG, et al. Characterization of a human Kaposi’s sarcoma cell line that induces angiogenic tumors in animals. AIDS. 1994;8:575–581. doi: 10.1097/00002030-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Fiorelli V, Gendelman R, Samaniego F, Markham PD, Ensoli B. Cytokines from activated T cells induce normal endothelial cells to acquire the phenotypic and functional features of AIDS-Kaposi’s sarcoma spindle cells. J. Clin. Invest. 1995;95:1723–1734. doi: 10.1172/JCI117849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensoli B, et al. Block of AIDS-Kaposi’s sarcoma (KS) cell growth, angiogenesis, and lesion formation in nude mice by antisense oligonucleotide targeting basic fibroblast growth factor. A novel strategy for the therapy of KS. J. Clin. Invest. 1994;94:1736–1746. doi: 10.1172/JCI117521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ensoli B, Sturzl M. Kaposi’s sarcoma: a result of the interplay among inflammatory cytokines, angiogenic factors and viral agents. Cytokine Growth Factor Rev. 1998;9:63–83. doi: 10.1016/s1359-6101(97)00037-3. [DOI] [PubMed] [Google Scholar]
- 14.Flamand L, Zeman RA, Bryant JL, Lunardi-Iskandar Y, Gallo RC. Absence of human herpesvirus 8 DNA sequences in neoplastic Kaposi’s sarcoma cell lines. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1996;13:194–197. doi: 10.1097/00042560-199610010-00011. [DOI] [PubMed] [Google Scholar]
- 15.Dictor M, Rambech E, Way D, Witte M, Bendsoe N. Human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) DNA in Kaposi’s sarcoma lesions, AIDS Kaposi’s sarcoma cell lines, endothelial Kaposi’s sarcoma simulators, and the skin of immunosuppressed patients. Am. J. Pathol. 1996;148:2009–2016. [PMC free article] [PubMed] [Google Scholar]
- 16.Cesarman E, et al. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J. Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y, et al. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. (Letter). doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 18.Li M, et al. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. J. Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 20.Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y. Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 2001;75:429–438. doi: 10.1128/JVI.75.1.429-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimuro M, et al. A novel viral mechanism for dysregulation of beta-catenin in Kaposi’s sarcoma-associated herpesvirus latency. Nat. Med. 2003;9:300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- 22.Orenstein JM, et al. Visualization of human herpesvirus type 8 in Kaposi’s sarcoma by light and transmission electron microscopy. AIDS. 1997;11:F35–F45. doi: 10.1097/00002030-199705000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Staskus KA, et al. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin DF, et al. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N. Engl. J. Med. 1999;340:1063–1070. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 25.Benelli R, et al. Distinct chemotactic and angiogenic activities of peptides derived from Kaposi’s sarcoma virus encoded chemokines. Int. J. Oncol. 2000;17:75–81. doi: 10.3892/ijo.17.1.75. [DOI] [PubMed] [Google Scholar]
- 26.Boshoff C, et al. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 27.Haque NS, Fallon JT, Taubman MB, Harpel PC. The chemokine receptor CCR8 mediates human endothelial cell chemotaxis induced by I-309 and Kaposi sarcoma herpesvirus-encoded vMIP-I and by lipoprotein(a)-stimulated endothelial cell conditioned medium. Blood. 2001;97:39–45. doi: 10.1182/blood.v97.1.39. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Okruzhnov Y, Li H, Nicholas J. Human herpesvirus 8 (HHV-8)-encoded cytokines induce expression of and autocrine signaling by vascular endothelial growth factor (VEGF) in HHV-8-infected primary-effusion lymphoma cell lines and mediate VEGF-independent antiapoptotic effects. J. Virol. 2001;75:10933–10940. doi: 10.1128/JVI.75.22.10933-10940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sozzani S, et al. The viral chemokine macrophage inflammatory protein-II is a selective Th2 chemoattractant. Blood. 1998;92:4036–4039. [PubMed] [Google Scholar]
- 30.Stine JT, et al. KSHV-encoded CC chemokine vMIP-III is a CCR4 agonist, stimulates angiogenesis, and selectively chemoattracts TH2 cells. Blood. 2000;95:1151–1157. [PubMed] [Google Scholar]
- 31.Bais C, et al. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 32.Cesarman E, Mesri EA, Gershengorn MC. Viral G protein-coupled receptor and Kaposi’s sarcoma: a model of paracrine neoplasia? J. Exp. Med. 2000;191:417–422. doi: 10.1084/jem.191.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirshner JR, Staskus K, Haase A, Lagunoff M, Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi’s sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J. Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smit MJ, et al. Kaposi’s sarcoma-associated herpesvirus-encoded G protein-coupled receptor ORF74 constitutively activates p44/p42 MAPK and Akt via G(i) and phospholipase C-dependent signaling pathways. J. Virol. 2002;76:1744–1752. doi: 10.1128/JVI.76.4.1744-1752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang TY, et al. Transgenic expression of the chemokine receptor encoded by human herpesvirus 8 induces an angioproliferative disease resembling Kaposi’s sarcoma. J. Exp. Med. 2000;191:445–454. doi: 10.1084/jem.191.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grundhoff A, Ganem D. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 2003;77:2779–2783. doi: 10.1128/JVI.77.4.2779-2783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus. Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 38.Renne R, et al. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renne R, et al. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 40.Venetsanakos E, et al. Induction of tubulogenesis in telomerase-immortalized human microvascular endothelial cells by glioblastoma cells. Exp. Cell Res. 2002;273:21–33. doi: 10.1006/excr.2001.5424. [DOI] [PubMed] [Google Scholar]
- 41.Obeso J, Weber J, Auerbach R. A hemangioendothelioma-derived cell line: its use as a model for the study of endothelial cell biology. Lab. Invest. 1990;63:259–269. [PubMed] [Google Scholar]
- 42.Zhou FC, et al. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 2002;76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arad U. Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques. 1998;24:760–762. doi: 10.2144/98245bm14. [DOI] [PubMed] [Google Scholar]
- 45.Polson AG, et al. Kaposi’s sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 2001;75:3175–3184. doi: 10.1128/JVI.75.7.3175-3184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garber AC, Hu J, Renne R. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 2002;277:27401–27411. doi: 10.1074/jbc.M203489200. [DOI] [PubMed] [Google Scholar]
- 47.Hu J, Garber AC, Renne R. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 2002;76:11677–11687. doi: 10.1128/JVI.76.22.11677-11687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim C, Sohn H, Lee D, Gwack Y, Choe J. Functional dissection of latency-associated nuclear antigen 1 of Kaposi’s sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 2002;76:10320–10331. doi: 10.1128/JVI.76.20.10320-10331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ballestas ME, Kaye KM. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 2001;75:3250–3258. doi: 10.1128/JVI.75.7.3250-3258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cotter MA, II, Subramanian C, Robertson ES. The Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology. 2001;291:241–259. doi: 10.1006/viro.2001.1202. [DOI] [PubMed] [Google Scholar]
- 51.Garber AC, Shu MA, Hu J, Renne R. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2001;75:7882–7892. doi: 10.1128/JVI.75.17.7882-7892.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cotter MA, II, Robertson ES. The latency-associated nuclear antigen tethers the Kaposi’s sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 53.Piolot T, Tramier M, Coppey M, Nicolas JC, Marechal V. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 2001;75:3948–3959. doi: 10.1128/JVI.75.8.3948-3959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 55.Bechtel JT, Liang Y, Hvidding J, Ganem D. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J. Virol. 2003;77:6474–6481. doi: 10.1128/JVI.77.11.6474-6481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J. Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blackbourn DJ, et al. The restricted cellular host range of human herpesvirus 8. AIDS. 2000;14:1123–1133. doi: 10.1097/00002030-200006160-00009. [DOI] [PubMed] [Google Scholar]
- 58.Lagunoff M, et al. De novo infection and serial transmission of Kaposi’s sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol. 2002;76:2440–2448. doi: 10.1128/jvi.76.5.2440-2448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ciufo DM, et al. Spindle cell conversion by Kaposi’s sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 2001;75:5614–5626. doi: 10.1128/JVI.75.12.5614-5626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leight ER, Sugden B. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol. Cell Biol. 2001;21:4149–4161. doi: 10.1128/MCB.21.13.4149-4161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheung ST, et al. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer. 1999;83:121–126. doi: 10.1002/(sici)1097-0215(19990924)83:1<121::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 62.Hui AB, Cheung ST, Fong Y, Lo KW, Huang DP. Characterization of a new EBV-associated nasopharyngeal carcinoma cell line. Cancer Genet. Cytogenet. 1998;101:83–88. doi: 10.1016/s0165-4608(97)00231-8. [DOI] [PubMed] [Google Scholar]
- 63.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 64.Babcock GJ, Decker LL, Freeman RB, Thorley-Lawson DA. Epstein-Barr virus-infected resting memory B cells, not proliferating lymphoblasts, accumulate in the peripheral blood of immunosuppressed patients. J. Exp. Med. 1999;190:567–576. doi: 10.1084/jem.190.4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Babcock GJ, Hochberg D, Thorley-Lawson AD. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity. 2000;13:497–506. doi: 10.1016/s1074-7613(00)00049-2. [DOI] [PubMed] [Google Scholar]
- 66.Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thorley-Lawson DA, Babcock GJ. A model for persistent infection with Epstein-Barr virus: the stealth virus of human B cells. Life Sci. 1999;65:1433–1453. doi: 10.1016/s0024-3205(99)00214-3. [DOI] [PubMed] [Google Scholar]
- 68.Dupin N, et al. Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boivin G, Gaudreau A, Routy JP. Evaluation of the human herpesvirus 8 DNA load in blood and Kaposi’s sarcoma skin lesions from AIDS patients on highly active antiretroviral therapy. AIDS. 2000;14:1907–1910. doi: 10.1097/00002030-200009080-00004. [DOI] [PubMed] [Google Scholar]
- 70.Campbell TB, et al. Relationship of human herpesvirus 8 peripheral blood virus load and Kaposi’s sarcoma clinical stage. AIDS. 2000;14:2109–2116. doi: 10.1097/00002030-200009290-00006. [DOI] [PubMed] [Google Scholar]
- 71.Quinlivan EB, et al. Elevated virus loads of Kaposi’s sarcoma-associated human herpesvirus 8 predict Kaposi’s sarcoma disease progression, but elevated levels of human immunodeficiency virus type 1 do not. J. Infect. Dis. 2002;185:1736–1744. doi: 10.1086/340652. [DOI] [PubMed] [Google Scholar]