Abstract
The redox-sensitive transcription factor SoxR in enteric bacteria senses and regulates the cellular response to superoxide and nitric oxide. In other bacterial groups, however, it may respond to redox-active small molecules, as demonstrated for pyocyanin sensing in pseudomonads. The antibiotic-producing soil bacterium Streptomyces coelicolor contains a gene for an SoxR homologue (SCO1697) whose DNA recognition helix is identical to that of Escherichia coli SoxR. Using the E. coli SoxR binding sequence, we predicted five candidate genes of the SoxR regulon and demonstrated that SoxR binds to their promoter regions and activates their expression concurrently with the production of the blue antibiotic actinorhodin (a benzoisochromanequinone). These genes encode a probable NADPH-dependent flavin reductase (SCO2478), an NADPH-dependent quinone reductase (SCO4266), an ABC transporter (SCO7008), a monooxygenase (SCO1909), and a hypothetical protein (SCO1178). Addition of actinorhodin to exponentially growing cells activated the expression of SoxR target genes in an SoxR-dependent manner. The secreted γ-actinorhodin was over 10-fold more effective in activation than the intracellular form of actinorhodin, suggesting that SoxR is specified to respond more to exogenous signals than to intracellular metabolites. The ΔsoxR mutant was not compromised in resistance against oxidants but was slow in forming aerial mycelium on R2YE medium with reduced sporulation, and its production of actinorhodin and undecylprodigiosin was lowered by about 50% and 30%, respectively, compared to that of the wild type. These results support the proposal that SoxR senses redox-active molecules, such as actinorhodin in S. coelicolor, and induces a protective function against them. It also functions to ensure that cells undergo optimal differentiation and secondary metabolite production.
Bacteria have evolved diverse ways of activating antioxidative defense genes to cope with harmful levels of endogenous and exogenous oxidants. One prominent and well-studied system is the SoxRS regulon in Escherichia coli, which responds to superoxide and nitric oxide through oxidation of the [2Fe-2S] cluster in SoxR; this leads to enhanced expression of SoxS, which then upregulates genes to remove oxidants and subsequent damage (14, 25, 32). However, the two-step regulatory mechanism involving SoxS as well as the antioxidative role of the SoxR regulon appears to be limited to enteric bacteria (11). Recent studies of SoxR regulons in pseudomonads indicated an alternative model to the E. coli SoxR-SoxS paradigm. This is based on the observations that (i) no SoxS homologue is found, (ii) the SoxR regulon does not include oxidative response genes, (iii) soxR mutations do not decrease resistance to oxidants, and (iv) the redox-active pigment pyocyanin can activate SoxR (10, 11, 19, 22, 23). SoxR homologues are found in a broad range of bacterial groups, including alpha-, beta-, delta-, and gammaproteobacteria and actinobacteria. Except in the enterobacterial branch of gammaproteobacteria (enteric bacteria), no SoxS homologues are found in bacterial genomes, and the predicted SoxR regulons include genes for putative efflux pumps, dehydrogenases, and monooxygenases (11).
In Pseudomonas aeruginosa SoxR responds to pyocyanin, an endogenous redox-active antibiotic, and activates transcription of two genes/operons encoding a probable efflux pump and a monooxygenase that might aid in phenazine transport and modification (10). In Streptomyces coelicolor, it has been proposed that two predicted SoxR regulon genes encoding putative oxidoreductases (SCO2478 and SCO4266) are expressed in cells that are capable of producing pigmented antibiotics but not in nonproducing cells (11). Considering that many of the SoxR regulon-containing bacteria also produce redox-active antibiotics (34), it has been hypothesized that SoxR may regulate transport and turnover of small redox-active molecules (11).
Streptomycetes produce a variety of secondary metabolites, including antibiotics. The best-characterized model organism S. coelicolor produces two pigmented antibiotics called actinorhodin and undecylprodigiosin (8). Actinorhodin, a benzoisochromanequinone, belongs to a class of aromatic polyketides and resembles the phenazine ring structure (21). It is a pH indicator that turns from red to blue at neutral to alkaline pH and hence is called a “blue” antibiotic (5). Its synthetic enzymes are encoded from a gene cluster (act genes, SCO5076 to SCO5092) (3) and are composed of a polyketide synthase complex that produce a 16-carbon polyketide backbone and a variety of modifying enzymes to produce the six-ringed actinorhodin molecules that accumulate in the cell. Intracellular actinorhodin is converted to a lactone form called γ-actinorhodin during or after export from the cell (6). Actinorhodin export is mediated through an efflux pump, ActA, whose expression is regulated by a TetR-like repressor, ActR, in response to the production of actinorhodin (6, 7, 29).
In this study, we identified five SoxR regulon genes, including the two previously predicted ones, and examined their expression in relation to actinorhodin. Whether the SoxR regulon responds to extracellular blue pigment, which is mostly γ-actinorhodin, as well as intracellular form of actinorhodin was also examined.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The S. coelicolor A3(2) M145 strain was used as the wild type and routinely grown in YEME liquid medium containing 10.3% sucrose (17). Solid and liquid R2YE media were used to grow cells for comparing phenotypes, isolation of RNAs, and measurement of antibiotic production. For PCR-targeted mutagenesis, E. coli BW25113 with the pIJ790 plasmid was used as recommended (15). E. coli ET12567 with the pUZ8002 plasmid was used for conjugal transfer (20).
Construction of ΔsoxR mutant and complementation.
The ΔsoxR mutant was constructed through PCR-targeted mutagenesis by replacing the entire coding sequence of SCO1697 with the apramycin resistance cassette [aac(3)IV]. The upstream forward primer used to create the ΔsoxR mutation contained the soxR gene sequences up to the start codon (bold) linked with aac(3)IV sequence (underlined) (5′-GTC CGC GCG GGC GAT CGG GTC GGT AGG GTT CGA GGG GTG ATT CCG GGG ATC CGT CGA CC-3′). The downstream reverse primer corresponds to the soxR stop codon (bold) and its downstream region connected with the aac(3)IV sequence (underlined) (5′-GCC ACG ACT GAC GAC CGG GCC AGG GTC ACC GCG CGG TCA TGT AGG CTG GAG CTG CTT C-3′). The purified PCR product was introduced by electroporation into E. coli BW25113 (9), a strain that harbors the λred recombination plasmid pIJ790 and a cosmid SC8F11 (a gift from the John Innes Centre) that contains the soxR gene. The resulting recombinant cosmid (SC8F11ΔsoxR::apr) recovered from the selected transformants was verified for its gene structure and introduced into E. coli ET12567 carrying pUZ8002, followed by conjugal transfer to S. coelicolor M145. Apramycin-resistant and kanamycin-sensitive exconjugants were selected, and we isolated three colonies that contained the expected gene structure as follows. Genomic PCR using forward primer 5′-GGT GTA CCC CAA ATG CTC GC-3′ (for the soxR upstream position) and reverse primer 5′-CCC GAG GTG CGA CGA CGG GT-3′ (in the soxR coding region) excluded colonies with the wild-type gene structure, and Southern hybridization of genomic DNA was performed with the aac(3)IV gene probe for the presence of the apramycin cassette. The three ΔsoxR isolates showed similar phenotypes, and we used one of the isolates for further experiments. To confirm mutation effects by complementation, the wild-type soxR gene was cloned in the pSET162 plasmid (18), a derivative of pSET152 (4) with a thiostrepton resistance marker, and the recombinant plasmid was introduced into the ΔsoxR mutant.
Overproduction and purification of S. coelicolor SoxR protein from E. coli.
The entire coding region of the soxR gene was amplified from cosmid SC8F11 using mutagenic primers SoxR-up (5′-GGT TCG AGC ATA TGC CTC AGA TTC-3′; an NdeI site is underlined) and SoxR-down (5′-GAC CGG GCC AGG ATC CCC GCG C-3′; a BamHI site is underlined). The 590-bp PCR product was digested with NdeI and BamHI and cloned into the pET15b vector (Novagen). The resulting recombinant plasmid (pET15b::soxR) was transformed into E. coli BL21(λDE3)/pLysS. To purify the SoxR protein, transformant cells grown in LB at 37°C to an optical density at 600 nm (OD600) of 0.5 were induced with 1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 30°C. His-tagged SoxR protein was purified through a nickel-charged nitrilotriacetic acid (NTA) column (Novagen) as recommended by the manufacturer. Following dialysis to remove imidazole and excess nickel, the SoxR sample was concentrated with a centrifugal filter device (Millipore; 3,000-molecular-weight cutoff) and further purified through a Superdex 75 column in a fast protein liquid chromatography (FPLC) system (ÄKTA standard; Amersham Biosciences). SoxR fractions were collected and further dialyzed against storage buffer (20 mM Tris-HCl [pH 7.8], 500 mM NaCl, 30% glycerol, and 2 mM dithiothreitol [DTT]). The concentration of purified SoxR protein was determined by the Bradford method, and the protein was stored at −80°C.
Electrophoretic mobility shift assay (EMSA) for SoxR-DNA binding.
DNA probes containing predicted SoxR binding sites were prepared by PCR using the following primer pairs: for SCO2478, GM143F (5′-GGT GAC CGG TGC CTC CGA AC-3′) and GM143R (5′-GGT GCG GTC GTC GTG TTC AC-3′); for SCO4266, GM127F (5′-CCT GAC GGC GGT ATC CCT CG-3′) and GM127R (5′-CAG TCG GAT GGC GTG CAT GG-3′); for SCO7008, GM131F (5′-GTG ACC GGC GCG CGG AAT CC-3′) and GM131R (5′-GCG TCC ACG CCG TGG TCT CC-3′); for SCO1909, GM144F (5′-CCC ATG TGT CTC CCC ACC GG-3′) and GM144R (5′-GTC GGG GTG GGT GTC GGT GC-3′); and for SCO1178, GM125F (5′-GAG TCC CGG TCC GTC TCG GTC-3′) and GM125R (5′-GGC TTC CAT GGG TGA GCC CTG-3′). Purified PCR products were labeled at the 5′ ends with [γ-32P]ATP using T4 polynucleotide kinase. The binding reaction was carried out by incubating approximately 2.2 fmol of labeled DNA and various amounts (0.5 to 5 pmol) of purified His-SoxR in 20 μl of reaction buffer [20 mM Tris-HCl (pH 7.8), 1 mM MgCl2, 40 mM KCl, 2 mM DTT, 0.1 mg/ml bovine serum albumin (BSA), 5% glycerol, and 0.1 μg poly (dI-dC)] for 20 min at room temperature. The binding mixture was subjected to electrophoresis on a 5% native polyacrylamide gel at 100 V in TBE running buffer (90 mM Tris-borate and 2 mM EDTA). The dried gels were exposed to imaging screens for quantification with a phosphorimage analyzer (FLA-2000; Fuji). For competition assay, either a specific competitor (unlabeled probe; 5- and 50-fold molar excesses) or a nonspecific competitor [plasmid pGEM3zf(+) digested with HpaII; 250- and 500-fold molar excesses] was added to the binding reaction mixture.
S1 nuclease mapping analysis.
To prepare RNA, wild-type (M145) and ΔsoxR and M511 (ΔactII-ORF4) (12) mutant cells were grown for various lengths of time on cellulose membrane-covered R2YE solid medium or in liquid YEME medium containing 10.3% sucrose to exponential phase at an OD600 of 0.3 to 0.4. RNAs were purified by using acidic phenol after fixation with RNAprotect bacterial reagent (Qiagen). Gene-specific S1 probes for soxR, actII-ORF4, actR, actA, SCO2478, SCO4266, SCO7008, SCO1909, and SCO1178 transcripts were generated by PCR using S. coelicolor M145 genomic DNA as a template. The probes spanned the following nucleotide (nt) positions relative to the start codon: for soxR, −202 (upstream) to +132 (downstream); for actII-ORF4, −92 to +47; for actR, −105 to +54; for actA, −114 to +69; for SCO2478, −177 to +100; for SCO4266, −174 to +132; for SCO7008, −162 to +138; for SCO1909, −152 to +72; and for SCO1178, −168 to +73. The forward primers for actA (5′-CGG ACG GAT CCT CAT CGT ATG G-3′) and actR (5′-GGA GT G GAT CCT CGA CTA TTG G-3′) were designed to include mutagenized residues (underlined) to inhibit antisense hybridization and to create BamHI sites (bold). The probe DNA fragments were labeled with [γ-32P]ATP and T4 polynucleotide kinase. Hybridization and S1 nuclease mapping were carried out according to standard procedures (17). The protected DNA fragments were loaded on 6% (wt/vol) polyacrylamide gels containing 7 M urea. The dried gels were exposed to a phosphorimage screen (FLA-2000; Fuji).
Quantification of actinorhodin and undecylprodigiosin.
About 1 × 108 spores each from the wild-type (M145+pSET162) and ΔsoxR+pSET162, ΔsoxR+pSET162::soxR, and ΔactII-ORF4 (M511) mutant strains were inoculated and grown to an OD600 of 0.5 to 0.6 in YEME medium for seed culture. The cultured cells were inoculated to 1% (vol/vol) in R2YE liquid medium and incubated at 30°C with shaking at 180 rpm. Cells were harvested at specified time points (19, 29, 42, 52, 66, 76, 90, 100, 114, 124, and 138 h after inoculation) to measure the contents of actinorhodin and undecylprodigiosin as described previously (6, 17, 27, 31, 33). For actinorhodin, each 1-ml sample was first treated with 50 μl of 10 N NaOH, gently vortexed, and allowed to stand for 5 min, followed by centrifugation at 4,000 × g for 5 min. The absorption spectra of supernatants were monitored with a UV/visible spectrophotometer (Shimadzu model UV-1650PC). The absorbance at 608 nm was used to calculate the concentration of actinorhodin based on the extinction coefficient of 25,320 M−1 cm−1 (6). Undecylprodigiosin was extracted similarly except that the cell mass was resuspended with 100% methanol and acidified with 10 N HCl. The absorbance at 530 nm was used to calculate its concentration based on the extinction coefficient of 100,500 M−1 cm−1 (33). In order to prepare partially pure γ-actinorhodin solution, we collected 2.5 ml blue extracellular droplets from 15 R2YE plates densely inoculated with M145 spores at 4 days after inoculation by inverting the plates. The collected blue sap was filtered through a 0.2-μm filter set and centrifuged at 12,000 rpm for 1 h, followed by heating at 70°C for 1 h. UV/visible absorption spectra indicated that the primary component of the blue sap is γ-actinorhodin with absorption peaks at 542 nm and 583 nm (6), with little contamination of intracellular actinorhodin (absorption peak at 640 nm) or its intermediates, such as (S)-DNPA (peak at 410 nm) and (R)-DNPA (peak at 434 nm) (28). The concentration of γ-actinorhodin was calculated to be 12.5 μM.
RESULTS
Prediction of SoxR target genes in S. coelicolor.
Genome sequence information for S. coelicolor strain M145 revealed the presence of a putative gene (SCO1697) for SoxR (ScSoxR; 175 amino acids [aa]) that shares 60% and 69% identical and similar amino acids, respectively, with E. coli SoxR (EcSoxR; 154 aa). Strikingly, the amino acid sequence in the DNA recognition helix of the helix-turn-helix motif is identical to that of EcSoxR. We therefore utilized the target sequence of EcSoxR, the inverted repeat of 18 bp (CCTCAAGTTAACTTGAGG) in the E. coli soxS promoter (16), as a query to search for ScSoxR target genes with the RSAT (Regulatory Sequence Analysis Tool; http://rsat.bigre.ulb.ac.be/rsat/) program, allowing up to seven mismatches. Among 1,746 candidates, 85 sites were within 200 nt from the coding regions, and 5 sites overlapped with the consensus −35 promoter element in a similar way as in the E. coli soxS promoter. These five candidates included the two previously predicted genes SCO2478 and SCO4266, which encode a probable NADPH-dependent flavin reductase and a probable quinone reductase, respectively, and three novel genes: SCO7008, encoding an ABC-type transporter; SCO1909, encoding a monooxygenase likely to be involved in antibiotic production; and SCO1178, of nonpredictable function (Table 1). As expected for a MerR-type regulator, the predicted binding sites all resided between −35 and −10 putative promoter elements that are spaced by 19 nt. Gene expression of SCO2478 and SCO4266 has been previously demonstrated to be elevated in wild-type S. coelicolor that produces pigmented antibiotics compared with the nonproducing mutants (11). The SCO2478-2477-2476 cluster is conserved in all Streptomyces genomes sequenced (KEGG gene cluster; http://www.genome.jp/kegg), suggesting that they form an operon. SCO2477 and SCO2476 encode a probable short-chain dehydrogenase/reductase similar to the FabG 3-ketoacyl reductase. The SCO4266-4265-4264 cluster is conserved in Streptomyces avermitilis and is likely to be an operon as well. These genes encode a probable NAPDH:quinone reductase (SCO4266), an efflux pump (SCO4265), and a phosphotransferase (SCO4264), suggesting that they may participate in modification and transport of redox-active metabolites.
TABLE 1.
S. coelicolor genes with SoxR binding sequences
| Probable function | Gene | No. of mismatches | Predicted SoxR binding sequencea | Distance/translational start codonb |
|---|---|---|---|---|
| E. coli soxSp | 0 | TTTACCTCAAGTTAACTTGAGGAATTATACT | ||
| Flavin reductase | SCO2478 | 2 | TTGACCTCAAGCAAACTTGAGGTACCAGGCT | 52/GTG |
| Quinone reductase | SC04266 | 4 | TTGACCTCAAGCAGGCTTGAGGTCGTTGAGT | 40/ATG |
| ABC transporter | SCO7008 | 5 | TTGACCTCAAGGTTGGTCGAGGTTCTACGGT | 13/ATG |
| Monooxygenase | SCO1909 | 6 | TTGACCTCAACCTTGGTCGAGGTCGGAGGGT | 40/ATG |
| Unknown | SCO1178 | 7 | TTGACCTCAACCGTGGTCGAGGTACCACGGT | 6/ATG |
The boldface in the E. coli SoxSp sequence indicates the 18-bp inverted repeat that we used as a query to screen putative SoxR target genes by using the RSAT program (http://rsat.bigre.ulb.ac.be/rsat/). The predicted −10 and −35 promoter elements with 19-nt spacing are underlined. The mismatches with respect to the E. coli SoxS promoter sequence are in italic.
Distance between the −10 promoter and translational start codon (ATG or GTG).
SoxR-dependent expression of predicted target genes in actinorhodin-producing cells.
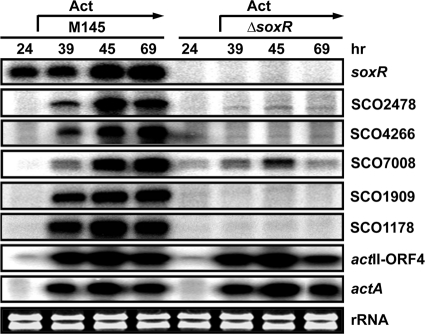
We examined whether the predicted five genes are indeed under the control of SoxR in vivo. For this purpose, RNAs prepared from the wild-type and ΔsoxR null mutant cells grown for various lengths of time on solid R2YE medium were analyzed by S1 mapping. On cellophane-covered R2YE plates, production of blue pigment (actinorhodin) was observed at 39 h and after longer culture following spore inoculation. As demonstrated in Fig. 1, we found that transcripts from the five candidate genes of the SoxR regulon appeared concurrently with the presence of blue pigment in the wild-type cells (after 39 h), whereas soxR gene expression itself is independent of growth stages. In the ΔsoxR mutant, however, none of these transcripts were produced. As indicators of actinorhodin-related gene expression, we examined the presence of transcripts from the actII-ORF4 gene, encoding a pathway-specific regulator for actinorhodin biosynthesis (2, 13), and the actA gene, encoding an actinorhodin exporter induced by actinorhodin (1, 7, 29). Production of actII-ORF4 and actA transcripts correlated with the appearance of blue pigments, as expected, and was independent of SoxR. These results clearly indicate that SCO2478, -4266, -7008, -1909, and -1178 are indeed regulated by SoxR and that SoxR-mediated induction occurs in accordance with actinorhodin production. Since the soxR gene is transcribed even in the absence of actinorhodin production, actinorhodin appears to exert posttranscriptional modulation, most likely at the level of SoxR activity.
FIG. 1.
SoxR-dependent induction of predicted SoxR target genes during the antibiotic-producing period. Transcripts from soxR, predicted soxR target genes, actII-ORF4, and actA genes were analyzed by S1 mapping. Wild-type and ΔsoxR cells were surface cultured on R2YE for various lengths of time from 24 to 69 h to prepare RNA. Blue actinorhodin was clearly visible from 39 h of culture.
Direct binding of SoxR to its target genes.
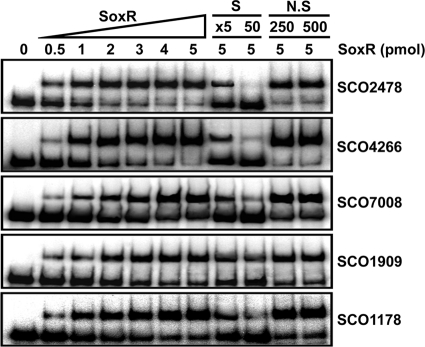
Whether SoxR directly recognizes its predicted binding sites was examined through DNA binding assays with purified ScSoxR protein and PCR-generated gene probes. Increasing amounts of His-tagged ScSoxR protein prepared from E. coli (0 to 5 pmol) were incubated with radiolabeled DNA probes and subjected to gel electrophoresis. The specificity of interaction was evaluated by adding either specific unlabeled probes (5-fold or 50-fold molar excess) or nonspecific DNA fragments [HpaII digest of plasmid pGEM3zf (+); 250- or 500-fold molar excess] to the binding reaction mixture. The results in Fig. 2 indicate that all the candidate genes bind SoxR protein specifically in vitro, confirming that these genes indeed are direct target genes of SoxR, qualifying as genuine SoxR regulon members.
FIG. 2.
Electrophoretic mobility shift assay of binding of purified SoxR protein to target promoters. Various amounts of purified His-tagged SoxR protein (from 0 to 5 pmol) were incubated with 32P-labeled probe DNAs (2.2 fmol) as described in the text. To verify the specificity of binding, either specific (S) (unlabeled probe DNA) or nonspecific (N.S) (HpaII digest of pGEM-3Zf plasmid) competitors were added in a 5- to 50-fold or 250- to 500-fold molar excess, respectively, to the binding reaction mixture.
Actinorhodin production is essential for SoxR-mediated induction of its target genes.
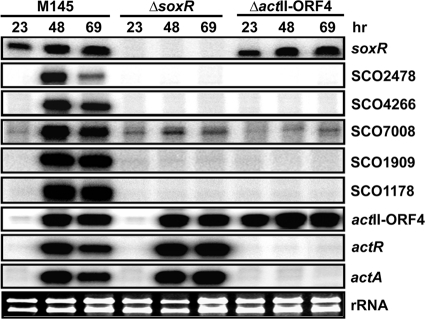
We examined whether actinorhodin production is essential for SoxR-mediated induction of its target genes by examining the effect of the actII-ORF4 mutation, which blocks actinorhodin production. For this purpose, strain M511, which lacks 178 internal amino acids in the 255-aa-long ActII-ORF4 (12), was examined for the expression of SoxR regulon genes. Transcripts from the remaining portion of actII-ORF4, along with actA and actR, which encodes the TetR-like repressor ActR for actA transcription, were examined in parallel. The results in Fig. 3 demonstrate that in the actII-ORF4 mutant, all SoxR target genes were not expressed, even though soxR transcription was unaffected. The actR and actA transcripts were independent of SoxR but dependent on actinorhodin. Therefore, ActII-ORF4, and possibly the production of actinorhodin, is required not only for ActR-mediated induction of the actR-actA gene pair but also for SoxR-mediated induction of its target genes. Unexpectedly, we found that actII-ORF4 transcripts are produced constitutively in the absence of functional ActII-ORF4 protein, suggesting its negative autoregulatory role in its own expression.
FIG. 3.
Actinorhodin-dependent induction of the SoxR regulon. Transcripts from soxR, SoxR regulon genes, actII-ORF4, actA, and actR were analyzed by S1 mapping in wild-type and ΔsoxR and ΔactII-ORF4 mutant cells grown on R2YE solid medium for 23, 48, and 69 h.
Induction of SoxR regulon genes by extracellular γ-actinorhodin.
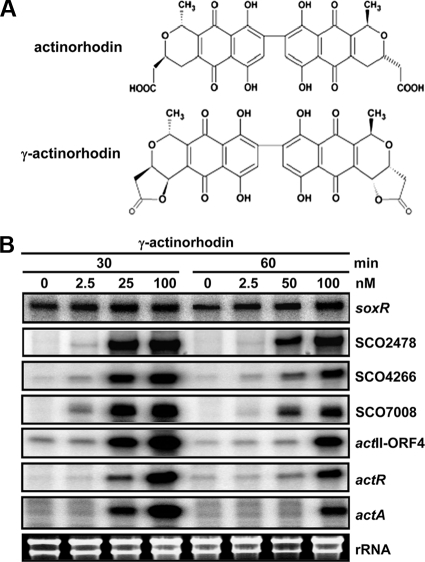
To examine whether actinorhodin serves as a signal molecule to activate SoxR, we monitored the effect of secreted blue pigment on the expression of soxR, its target genes, and some act genes. For this purpose, we collected blue secreted droplets from densely sporulated wild-type M145 colonies on R2YE plates at 96 h after inoculation. The primary component of the excreted blue pigment has been determined to be a lactone form of actinorhodin called γ-actinorhodin (6). We confirmed its predominant presence by the UV/visible absorption spectral profile (data not shown).
M145 wild-type cells were grown to mid-exponential phase (OD600 of 0.3 to 0.4) in YEME liquid medium, and then various amounts of γ-actinorhodin were added and left for 30 or 60 min before cell harvest. S1 mapping analysis demonstrated that treatment with more than 25 nM γ-actinorhodin for 30 min induced expression of all SoxR regulon genes as well as actR and actA, even though actR and actA induction is slightly less sensitive to γ-actinorhodin than that of SoxR target genes (Fig. 4). Longer incubation, for 60 min, diminished gene induction, which is likely to be due to increased export. These results clearly demonstrate that extracellular γ-actinorhodin can serve as a signal molecule to activate the SoxR regulon. We also found that γ-actinorhodin can induce transcription from actII-ORF4.
FIG. 4.
Activating effect of secreted actinorhodin (γ-actinorhodin). (A) Chemical structures of actinorhodin and γ-actinorhodin. (B) S1 mapping analysis. Transcripts were analyzed by S1 nuclease mapping in wild-type cells grown to an OD600 of 0.3 in YEME liquid medium, followed by treatment with secreted blue pigment (γ-actinorhodin) at 2.5, 25, and 100 nM (final concentration) for 30 and 60 min or by no treatment.
Comparison of the effect of purified actinorhodin with that of γ-actinorhodin.
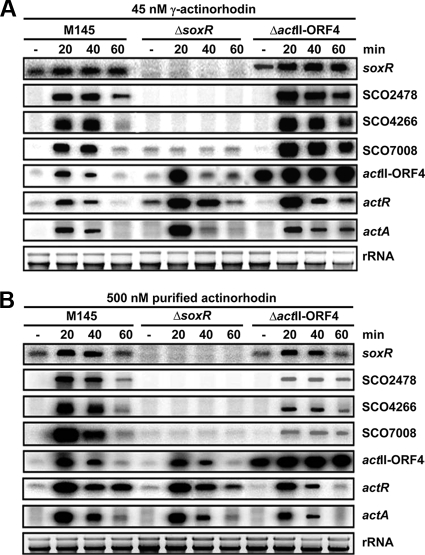
We then compared the effect of extracellular γ-actinorhodin with that of purified intracellular actinorhodin (a gift from Koji Ichinose, Musashino University). As demonstrated in Fig. 5, SoxR regulon genes were not induced in the ΔsoxR mutant in the presence of added γ-actinorhodin at 45 nM (final concentration), confirming that actinorhodin signaling is indeed mediated through SoxR. In the ΔactII-ORF4 mutant, γ-actinorhodin enhanced expression of SoxR regulon genes, and the induction was sustained for even longer than in the wild type. This suggests that defects in some metabolic functions in the ΔactII-ORF4 mutant might have facilitated the maintenance of signaling molecules. In order to produce a similar stimulatory effect in the wild type, we needed to add more than a 10-fold-larger amount of purified actinorhodin than of γ-actinorhodin. In the ΔactII-ORF4 mutant, purified actinorhodin was even less effective than in the wild type. These results indicate that the secreted form of actinorhodin is more effective than the intracellular form in activating the SoxR regulon, implying that the SoxR system may be more specified toward signals from outside.
FIG. 5.
Effect of the intracellular form of actinorhodin in comparison with extracellular γ-actinorhodin in various genetic backgrounds. (A) Transcripts for wild-type and ΔsoxR and ΔactII-ORF4 mutant cells grown and treated with 45 nM extracellular γ-actinorhodin as described for Fig. 4 were analyzed. (B) Effect of purified actinorhodin. Samples were prepared as for panel A except that 500 nM (final concentration) of the purified intracellular form of actinorhodin was added to exponentially growing cells.
Slower differentiation and decreased production of antibiotics in the ΔsoxR mutant.
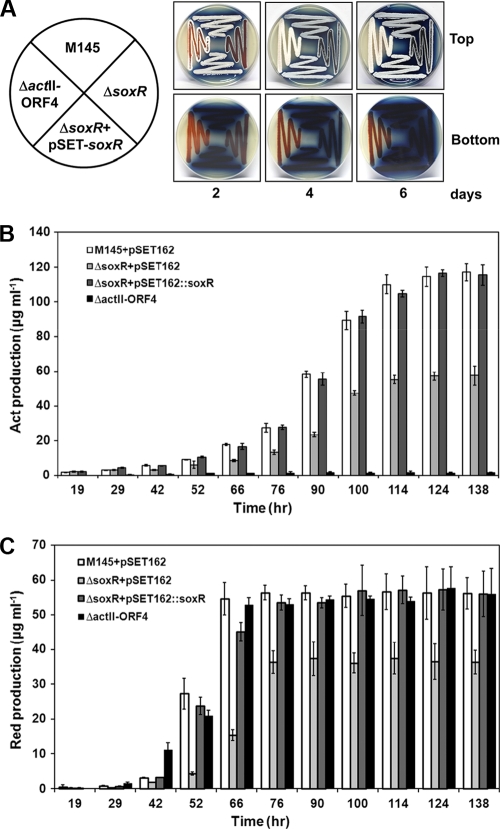
When plated on solid R2YE, the ΔsoxR mutant was delayed in forming aerial mycelia and pigmented antibiotics by about 2 days (Fig. 6A). The amounts of sporulation as well as red and blue pigments were also reduced in the mutant. This phenomenon is reproducibly observed on R2YE plates but not as much on other media, such as NA, MM, DNB, NMP, SMMS, R2, and SFM (data not shown). The defective phenotypes in the ΔsoxR mutant were restored to the wild-type level by introducing the soxR gene through plasmid pSET162 (Fig. 6). We measured the amount of pigmented antibiotics in cells grown in R2YE liquid medium. Total amounts of actinorhodin and undecylprodigiosin in the wild type (pSET162) and in the ΔsoxR (pSET162), ΔsoxR (pSET162::soxR), and ΔactII-ORF4 mutants were determined at specified growth time points (19, 29, 42, 52, 66, 76, 90, 100, 114, 124, and 138 h). The quantification demonstrated that actinorhodin production was delayed and that the maximal level was decreased to about 50% of the wild-type level (Fig. 6B). The undecylprodigiosin production was also delayed and the maximal level was reduced by about 30% in the ΔsoxR mutant compared with the wild type and the ΔactII-ORF4 mutant (Fig. 6C). Cell growth of the mutant in liquid medium was not affected, as monitored by total cell mass (wet weight).
FIG. 6.
Differentiation and antibiotic-producing phenotypes of ΔsoxR. (A) Differentiation progress of wild-type (M145 with pSET162), ΔsoxR with pSET162, ΔsoxR complemented with pSET162::soxR, and ΔactII-ORF4 (M511) strains on R2YE solid plates. Formation of aerial mycelia, spores, and pigmented antibiotics was examined visually by taking photographs at 2, 4, and 6 days after inoculation. (B) Quantification of actinorhodin (Act) production. Cells from the M145 and ΔsoxR strains with the parental pSET162 plasmid, the ΔsoxR mutant complemented with pSET162::soxR, and M511 were grown in R2YE liquid medium for various lengths of time from 19 to 138 h. Average values and standard deviations from triplicate cultures are presented. (C) Quantification of undecylprodigiosin (Red) in cells prepared as for panel B.
DISCUSSION
In this study, we demonstrated that the previously predicted SoxR homologue (SCO1697) in S. coelicolor indeed positively regulates its direct target genes in response to actinorhodin, especially the secreted form. The SoxR regulon genes include two putative operons (SCO2478 to -2476 and SCO4266 to -4264) encoding oxidoreductases (SCO2478 to -2476 and -4266), an efflux pump (SCO4265), and a phosphotransferase (SCO4264) and three monocistronic genes for an ABC-type transporter (SCO7008), a monooxygenase (SCO1909), and a hypothetical protein (SCO1178). The predicted functions of SoxR regulon products suggest that they most likely reduce, oxidize, or modify redox-active compounds, such as quinones and keto-acids, and pump them out. The lack of antisuperoxide or anti-nitric oxide functions coincides with observations in pseudomonads, supporting the proposal that the SoxR regulon in nonenteric proteobacteria and actinobacteria is specified for reacting toward redox-active toxic compounds rather than superoxides or nitric oxides (11). This also coincides with the lack of SoxR-inducing effects of various oxidants, such as paraquat, menadione, hydrogen peroxide, and cumene hydroperoxide, in S. coelicolor (data not shown).
The preference for the secreted form (γ-actinorhodin) over intracellular actinorhodin to induce SoxR activation is intriguing. This contrasts with the preference observed for another actinorhodin-sensing regulator, ActR, in S. coelicolor. ActR, which is a repressor of its divergently neighboring gene actA, encoding an efflux pump for actinorhodin within the act gene cluster, responds to intermediates in the actinorhodin biosynthetic pathway in addition to actinorhodin (29). It actually binds more tightly to three-ringed intermediate than to the six-ringed actinorhodin (30). Therefore, ActR-ActA is regarded as a system that detects the intracellular production of actinorhodin and confers self-resistance by elevating the pumping function. In comparison with the ActR system, which intimately coordinates its activity with the intracellular production of actinorhodin, SoxR may preferentially sense chemical signals that enter the cell from outside. Although it has not been experimentally demonstrated that ScSoxR contains [2Fe-2S] and senses redox-active chemicals through oxidizing this cofactor in S. coelicolor, conservation of [2Fe-2S] binding residues and overall amino acid sequences suggest that this is very likely to be true. The redox-active chemicals to which SoxR responds could be both endogenous and exogenous. Whether SoxR responds to a broader list of heterocyclic redox-active compounds needs be investigated in the future. For actinorhodin, our study indicates that S. coelicolor utilizes two separate pathways: one for protecting cells from intracellular antibiotic as soon as it is made and the other for protecting cells from incoming antibiotic. There could be some overlap between these two pathways, depending on the flexibility in the ligand specificities of these two regulators. Considering the determined structure of SoxR from E. coli (36), where the [2Fe-2S] cluster is exposed at the surface of the protein, it is likely that SoxR responds to a broader range of molecules than ActR. For bacteria with diverse antibiotic-producing capacities such as streptomycetes, utilization of multiple sensor-exporter systems for small toxic molecules could have provided survival advantages.
Regulation of various genes in accordance with antibiotic production continues to be an interesting subject to investigate further. One of the unexpected observations in our study suggests negative autoregulation of actII-ORF4 gene by its own product. The gene for ActII-ORF4 is known to be regulated by several transcription factors in response to nutritional starvation and perturbation. Direct transcriptional regulators include the GntR family regulator DasR (26) and the TetR-family regulator AtrA (35). Even though additional factors are reported to bind to the promoter regions of actII-ORF4 in vitro (24), their role in vivo has not been determined. In addition to these regulators, ActII-ORF4 may add a negative feedback loop. How this effect occurs needs further investigation.
Further studies on the spectrum of SoxR-activating compounds, its sensing mechanism (which most likely involves [2Fe-2S]), and characterization of SoxR regulon gene products are expected to reveal functions of the SoxR system in S. coelicolor and will shed light on its general role in actinobacteria as well as in other bacterial groups.
Acknowledgments
We thank Koji Ichinose (Research Institute of Pharmaceutical Science, Musashino University, Japan) for purified actinorhodin and Andy Hesketh (John Innes Centre) for strain M511.
This work was supported by a grant from the National Research Laboratory for Molecular Microbiology to J.-H. Roe (NRF-2010-0000802). J.-H. Shin was supported by a BK21 postdoctoral fellowship for Life Sciences at SNU. A. K. Singh was supported as a Korean government scholarship grantee by the Ministry of Education, Science and Technology.
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Ahn, S. K., K. Tahlan, Z. Yu, and J. Nodwell. 2007. Investigation of transcription repression and small-molecule responsiveness by TetR-like transcription factors using a heterologous Escherichia coli-based assay. J. Bacteriol. 189:6655-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, P., M. A. Fernández-Moreno, and F. Malpartida. 1999. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J. Bacteriol. 181:6958-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 5.Brockmann, H., and E. Hieronimus. 1955. Uber Actinomycetenfarbstoffe, V. Mitteil.: Zur Konstitution des Actinorhodins, III. Mitteil. Chem. Ber. 88:1379-1390. [Google Scholar]
- 6.Bystrykh, L. V., M. A. Fernandez-Moreno, J. K. Herrema, F. Malpartida, D. A. Hopwood, and L. Dijkhuizen. 1996. Production of actinorhodin-related blue pigments by Streptomyces coelicolor A3(2). J. Bacteriol. 178:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caballero, J. L., F. Malpartida, and D. A. Hopwood. 1991. Transcriptional organization and regulation of an antibiotic export complex in the producing Streptomyces culture. Mol. Gen. Genet. 228:372-380. [DOI] [PubMed] [Google Scholar]
- 8.Chater, K. F., and D. A. Hopwood. 1993. Streptomyces, p. 83-89. In J. P. Sonenshein (ed.), Bacillus subtilis and other gram-positive bacteria. Biochemistry, physiology and molecular genetics. ASM Press, Washington, DC.
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich, L. E. P., A. Price-Whelan, A. Petersen, M. Whiteley, and D. K. Newman. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308-1321. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich, L. E. P., T. K. Teal, A. Price-Whelan, and D. K. Newman. 2008. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321:1203-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Floriano, B., and M. J. Bibb. 1996. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 21:385-396. [DOI] [PubMed] [Google Scholar]
- 13.Gramajo, H. C., E. Takano, and M. J. Bibb. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7:837-845. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg, J. T., P. Monach, J. H. Chou, P. D. Josephy, and B. Demple. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidalgo, E., and B. Demple. 1994. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 13:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 18.Kim, I. K., C. J. Lee, M. K. Kim, J. M. Kim, J. H. Kim, H. S. Yim, S. S. Cha, and S. O. Kang. 2006. Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2). Mol. Microbiol. 60:1179-1193. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, K., and S. Tagawa. 2004. Activation of SoxR-dependent transcription in Pseudomonas aeruginosa. J. Biochem. 136:607-615. [DOI] [PubMed] [Google Scholar]
- 20.Mazodier, P., R. Petter, and C. Thompson. 1989. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 171:3583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto, S., T. Taguchi, K. Ochi, and K. Ichinose. 2009. Biosynthesis of actinorhodin and related antibiotics: discovery of alternative routes for quinone formation encoded in the act gene cluster. Chem. Biol. 16:226-236. [DOI] [PubMed] [Google Scholar]
- 22.Palma, M., J. Zurita, J. A. Ferreras, S. Worgall, D. H. Larone, L. Shi, F. Campagne, and L. E. N. Quadri. 2005. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect. Immun. 73:2958-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, W., S. Peña-Llopis, Y. H. Lee, and B. Demple. 2006. Regulation of superoxide stress in Pseudomonas putida KT2440 is different from the SoxR paradigm in Escherichia coli. Biochem. Biophys. Res. Commun. 341:51-56. [DOI] [PubMed] [Google Scholar]
- 24.Park, S. S., Y. H. Yang, E. Song, E. J. Kim, W. S. Kim, J. K. Sohng, H. C. Lee, K. K. Liou, and B. G. Kim. 2009. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 36:1073-1083. [DOI] [PubMed] [Google Scholar]
- 25.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 26.Rigali, S., F. Titgemeyer, S. Barends, S. Mulder, A. W. Thomae, D. A. Hopwood, and G. P. van Wezel. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO reports. 9:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strauch, E., E. Takano, H. A. Baylis, and M. J. Bibb. 1991. The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 5:289-298. [DOI] [PubMed] [Google Scholar]
- 28.Taguchi, T., Y. Ebizuka, D. A. Hopwood, and K. Ichinose. 2001. A new mode of stereochemical control revealed by analysis of the biosynthesis of dihydrogranaticin in Streptomyces violaceoruber Tu22. J. Am. Chem. Soc. 123:11376-11380. [DOI] [PubMed] [Google Scholar]
- 29.Tahlan, K., S. K. Ahn, A. Sing, T. D. Bodnaruk, A. R. Willems, A. R. Davidson, and J. R. Nodwell. 2007. Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol. 63:951-961. [DOI] [PubMed] [Google Scholar]
- 30.Tahlan, K., Z. Yu, Y. Xu, A. R. Davidson, and J. R. Nodwell. 2008. Ligand recognition by ActR, a TetR-like regulator of actinorhodin export. J. Mol. Biol. 383:753-761. [DOI] [PubMed] [Google Scholar]
- 31.Takano, E., H. C. Gramajo, E. Strauch, N. Andres, J. White, and M. J. Bibb. 1992. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6:2797-2804. [DOI] [PubMed] [Google Scholar]
- 32.Tsaneva, I. R., and B. Weiss. 1990. SoxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol. 172:4197-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsao, S. W., B. A. Rudd, X. G. He, C. J. Chang, and H. G. Floss. 1985. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. 38:128-131. [DOI] [PubMed] [Google Scholar]
- 34.Turner, J. M., and A. J. Messenger. 1986. Occurrence, biochemistry and physiology of phenazine pigment production. Adv. Microb. Physiol. 27:211-275. [DOI] [PubMed] [Google Scholar]
- 35.Uguru, G. C., K. E. Stephens, J. A. Stead, J. E. Towle, S. Baumberg, and K. J. McDowall. 2005. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol. Microbiol. 58:131-150. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe, S., A. Kita, K. Kobayashi, and K. Miki. 2008. Crystral structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc. Natl. Acad. Sci. U. S. A. 105:4121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]