Abstract
Transport of macromolecules between the nucleus and the cytoplasm is an essential necessity in eukaryotic cells, since the nuclear envelope separates transcription from translation. In the past few years, an increasing number of components of the plant nuclear transport machinery have been characterised. This progress, although far from being completed, confirmed that the general characteristics of nuclear transport are conserved between plants and other organisms. However, plant-specific components were also identified. Interestingly, several mutants in genes encoding components of the plant nuclear transport machinery were investigated, revealing differential sensitivity of plant-specific pathways to impaired nuclear transport. These findings attracted attention towards plant-specific cargoes that are transported over the nuclear envelope, unravelling connections between nuclear transport and components of signalling and developmental pathways. The current state of research in plants is summarised in comparison to yeast and vertebrate systems, and special emphasis is given to plant nuclear transport mutants.
Keywords: Arabidopsis, Exportins, Importins, mRNA export, Nuclear transport, Transport mutants
Introduction
In eukaryotic cells, the nuclear envelope (NE) separates the cytoplasm, where protein synthesis occurs, from the nucleus, where RNA is produced by transcription of genomic DNA. Nuclear pore complexes (NPCs) span the double-membrane system of the NE and provide gateways for the exchange of proteins and macromolecular complexes between the cytoplasm and the nucleus (for recent reviews on plant NPC and NE components in comparison to animal and yeast systems see: Xu and Meier 2008; Meier and Brkljacic 2009). However, NPCs also create a permeability barrier that only nuclear transport receptors (NTRs) can pass efficiently (Frey et al. 2006; Frey and Görlich 2009). This set-up provides the basis for receptor-mediated nuclear transport, built on signals that reside on cargo molecules, which in turn are specifically recognised by adapter proteins and NTRs that are able to very efficiently carry cargo back and forth through the NPCs (Görlich and Kutay 1999; Pemberton and Paschal 2005; Cook et al. 2007). This system ensures a rapid exchange of material and information between the cytoplasm and the nucleus that is essential for a living cell. It works against concentration gradients, and it provides the possibility for the regulation of nuclear transport rates of cargoes in both directions that is often used as an additional control level for the regulation of signalling pathways that include an essential nuclear translocation step.
RanGTP-binding proteins and nuclear import and export
The majority of NTRs are Ran-binding proteins (RanBPs); more exactly, these proteins bind the small G protein Ran in its GTP-bound form (RanGTP). There is a gene family of at least 20 members encoding RanBPs in the human genome, 14 genes encoding different RanBP–NTRs are found in yeast, and 17 genes are found in the model plant Arabidopsis thaliana (Merkle 2003; Fig. 1a). The founding member was Importin beta (Görlich et al. 1995a), hence they are referred to as importin beta (or karyopherin beta) family of NTRs (Görlich et al. 1997). They display limited sequence similarity between each other, mainly in their Ran-binding domains in the N terminus of the proteins. These proteins contain HEAT repeats that greatly influence their structure, for instance a right-handed spiral in case of Transportin 1 (Pictures in Cell Biology 1999; Andrade et al. 2001; Cook et al. 2007). HEAT is an acronym derived from four proteins in which these repeats were identified: Huntington, Elongation Factor 3, the PR65/A subunit of Protein Phosphatase 2A, and the lipid kinase Target of Rapamycin (Andrade and Bork 1995). Importin beta-like NTRs bind to and transport very different classes of cargo across the NE and display differential cargo affinity, depending on whether or not they are associated with RanGTP (Görlich et al. 1996a). Importins among NTRs show high affinity to their cargo in the absence of RanGTP and are dissociated from their cargo as a consequence of RanGTP binding, once importin-cargo complexes reach the nuclear compartment (Fig. 2). In the case of Importin beta itself, cargo dissociation is an active displacement by RanGTP in the nucleus (Görlich and Kutay 1999; Cook et al. 2007). In contrast, exportins bind their cargo cooperatively with RanGTP and separate from their cargo and from the GTPase Ran after hydrolysis of GTP on Ran, once they encounter the cytoplasmic regulatory proteins of Ran, the GTPase-activating protein 1 (Ran GAP1) and Ran-binding protein 1 (RanBP1; Fig. 2). However, some NTRs may operate in both directions, as has been shown for Importin 13/RanBP13 (Mingot et al. 2001) and for Exportin 4 (Lipowsky et al. 2000; Gontan et al. 2009). The topological information provided by the steep gradient of RanGTP concentration over the NE that is created by the specific characteristics of the Ran GTPase cycle (Fig. 3) is exploited to impose directionality on nuclear transport processes mediated by NTRs of the importin beta family (Görlich et al. 1996a; Izaurralde et al. 1997). Input of metabolic energy into nuclear transport processes in the form of GTP is used for the recycling of NTRs rather than for the translocation of NTR-cargo complexes across the NPC, and the irreversible hydrolysis of GTP on Ran drives reactions to one side (Nakielny and Dreyfuss 1997; Ribbeck et al. 1999).
Fig. 1.
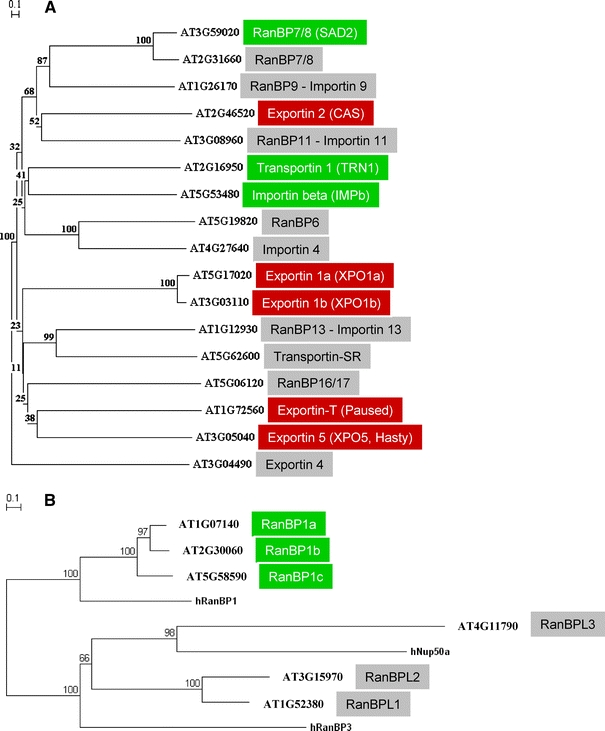
Ran-binding proteins (RanBPs) of Arabidopsis thaliana. a The phylogenetic tree shows all 17 Arabidopsis RanBPs of the Importin beta nuclear transport receptor family. AGI locus designations and Arabidopsis protein designations are given. If no specific name existed for a plant protein, the designation of the human protein that shows highest similarity is given. Plant importins that are functionally characterised are indicated by a green box, characterised plant exportins are labelled red. A grey box indicates that the protein has not been characterised in plants to date. b RanBP1-like proteins of Arabidopsis thaliana. The phylogenetic tree contains human (h) proteins for comparison. In contrast to the three very similar RanBP1 proteins, the three Arabidopsis RanBPL (Ran-binding protein 1-like) proteins that show similarity to human RanBP3 or NUP50 are not functionally characterised to date. Protein alignments were performed with full-length protein sequences using ClustalW2 (http:\\www.ebi.ac.uk/Tools/clustalw2), and the phylogenetic tree was constructed with TreeCon using Poisson correction and neighbour joining, taking insertions and deletions into account (Van de Peer and De Wachter 1997). Exportin 4 (a) or human NUP50a (b) were used to root the tree, respectively. Distance bars are given top left and bootstrap values are indicated at the nodes. Names and functions of animal and yeast homologs of plant NTRs are listed and compared in detail in Merkle (2003)
Fig. 2.
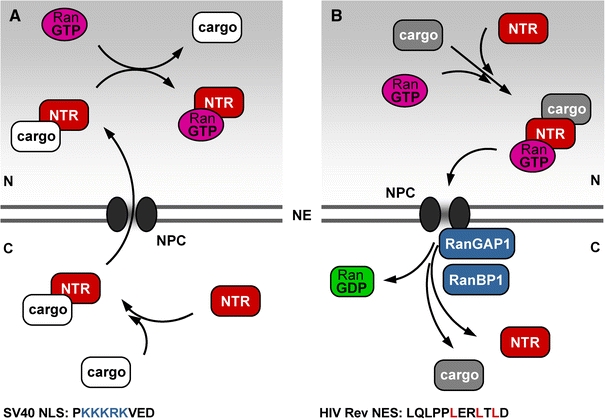
Simplified schemes of (a) nuclear import and (b) nuclear export facilitated by nuclear transport receptors (NTRs) that belong to the importin beta (or karyopherin beta) family. The nuclear pore complexes (NPC) that are embedded in the nuclear envelope (NE) provide the gateways for the exchange of molecules between the nucleus (N) and the cytoplasm (C). Simultaneously, NPCs create a diffusion barrier that can be overcome by NTRs, taking cargo with them through the NPCs. Importins among NTRs bind to their cargo in the absence of RanGTP in the cytoplasm, and dissociate from their cargo in the nucleus upon interaction with RanGTP, as shown in (a). The import cargo is released into the nucleoplasm and the importin travels back to the cytoplasm in complex with RanGTP, where the importin–RanGTP complex separates due to GTP hydrolysis on Ran (b). In contrast to importins, exportins among NTRs bind their cargo cooperatively with RanGTP in the nucleus (b). After translocation through the NPC, this triple complex is dissociated by GTP hydrolysis on Ran, catalysed by Ran-specific GTPase-activating protein 1 (RanGAP1) and Ran-binding protein 1 (RanBP1). The export cargo is released into the cytoplasm and the exportin recycles back to the nucleus on its own. The prototype of a basic nuclear localisation signal (NLS) from Simian Virus large T antigen and the prototype of a leucine-rich nuclear export signal (NES) from HIV Rev are given in (a) and (b) at the bottom, respectively. Amino acid residues that are important for NLS and NES function are given in colour
Fig. 3.
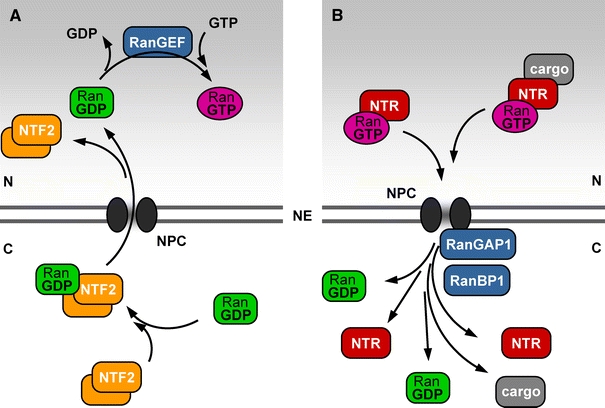
Simplified scheme of the Ran GTPase cycle in interphase. Like every G protein, Ran exists in two stable conformations, one bound to GTP and the other bound to GDP. Ran-specific regulatory proteins (given in blue colour) catalyse the interchange between these two conformations. a The Ran-specific guanine nucleotide exchange factor (RanGEF) is a chromatin-associated protein and catalyses the exchange of GDP for GTP in the nucleus. The RanGTP concentration therefore is very high in the nucleus, and thus RanGTP marks the position of the chromosomes, corresponding to the nuclear compartment in interphase. b RanGTP is exported with every recycling of an importin as NTR–RanGTP complex, or as part of nuclear export complexes. Two cytoplasmic regulatory proteins for Ran, Ran-specific GTPase-activating protein 1 (RanGAP1) and Ran-binding protein 1 (RanBP1), disassemble these NTR complexes by catalysing GTP hydrolysis on Ran once they reach the cytoplasm. As a consequence, RanGTP concentrations in the cytoplasm are very low. In order to compensate for the immense flux of Ran out of the nucleus, nuclear transport factor 2 (NTF2) acts as a nuclear import receptor for RanGDP, as shown in (a), and the cycle is completed. NE nuclear envelope; NPC nuclear pore complex; N nucleus; C cytoplasm
While many cargoes bind to their NTRs directly, other cargoes are linked to specific NTRs via adapter proteins (Table 1). A well-known adapter is Importin alpha (Fig. 4), identified in 1994 as the protein that is necessary for the first step of protein nuclear import (Görlich et al. 1994). Despite its similar name, it belongs to a different protein family than Importin beta. Importin alpha proteins contain armadillo (ARM) repeats and belong to a huge family of ARM repeat-containing proteins in plants (Mugdil et al. 2004). Importin alpha acts as a receptor for nuclear localisation signals (NLSs) on karyophilic proteins in the cytoplasm (Görlich et al. 1994, 1995a, b). These NLSs were known for quite some time (Görlich and Kutay 1999). They reside as permanent signals on karyophilic proteins and consist of a short stretch of basic amino acid residues (Fig. 2a) or two such motifs separated by a short spacer (bipartite NLS). Most karyophilic proteins contain such basic NLSs and are imported into the nucleus as a triple complex consisting of the cargo, the adapter Importin alpha, and the import receptor Importin beta (Görlich et al. 1995a, b). Upon binding of RanGTP to Importin beta in the nucleus, this complex is dissociated and the import cargo is released into the nucleoplasm (Görlich and Kutay 1999; Pemberton and Paschal 2005; Fig. 2a). However, it should be noted that basic NLSs are not the only nuclear import signals that exist, as there are additional nuclear import pathways that work independently of the Importin alpha/beta heterodimer (Görlich and Kutay 1999; Pemberton and Paschal 2005). An example for a nuclear import pathway that uses different import signals and is also characterised in plants depends on the NTR Transportin 1 (Ziemienowicz et al. 2003). On the other hand, the importance of the Importin alpha/beta nuclear import pathway for proteins may be reflected by the fact that one nuclear export receptor, Exportin 2 or CAS (cellular apoptosis susceptibility), has the one and only function to recycle Importin alpha adapters back to the cytoplasm, as known to date (Kutay et al. 1997; Haasen and Merkle 2002). In contrast to Importin alpha, the NTR Importin beta travels back to the cytoplasm on its own, in complex with RanGTP, and this complex is dissociated by GTP hydrolysis on Ran in the cytoplasm (Fig. 3b). Other adapter proteins for NTRs, their characteristics and their functions are listed in Table 1. The importance of the Importin alpha/beta nuclear import pathway may be also reflected by the fact that Importin alpha is encoded by a small gene family in humans as well as in Arabidopsis (Merkle 2001; Bhattacharjee et al. 2008; Mason et al. 2009; Fig. 4). In contrast, other adapters like snurportin 1 (Huber et al. 1998), NMD3 (Ho et al. 2000; Thomas and Kutay 2003; Trotta et al. 2003), or PHAX (Ohno et al. 2000) are encoded by single genes (Table 1). The advantage of adapters for NTRs may be that they constitute a hub to link unusual cargoes and/or many different cargo proteins containing slightly different signals to one NTR, while keeping the immediate NTR-adapter interaction relatively conserved. An example for the first scenario may be NMD3 that bridges the interaction between large ribosomal subunits and Exportin 1 (Ho et al. 2000; Thomas and Kutay 2003; Trotta et al. 2003), an example for the second scenario may be Importin alpha that links many karyophilic proteins with slightly different NLSs to Importin beta (Görlich and Kutay 1999) Adapters thus extend the cargo substrate range of specific NTRs.
Table 1.
Selected adapter proteins for nuclear transport receptors (NTRs)
| Adapter | Protein architecture | Link to NTR pathway | Recycling (NTR) | References |
|---|---|---|---|---|
| Importin alpha (protein family of ten members in Arabidopsis) | N-terminal IBB domain, 9-10 Armadillo (ARM) repeats; two Arabidopsis proteins differ from this architecture | Importin beta | Exportin 2/CAS | Görlich et al. (1994, 1995a, b, 1996b) |
| Snurportin 1 | N-terminal IBB domain, SPN or TMG-binding domain | Importin beta | Exportin 1 | Huber et al. (1998); Paraskeva et al. (1999) |
| NMD3 | NMD3 family domain, NLS, NES | Exportin 1 | Importin alpha/beta | Ho et al. (2000); Thomas and Kutay (2003); Trotta et al. (2003) |
| PHAX | NLS, NES, PHAX RNA-binding domain | Exportin 1 | Importin alpha/beta | Ohno et al. (2000) |
Only Importin alpha proteins have been characterised in plants to date. Importin alpha is the cytoplasmic receptor for most proteins containing a basic NLS. Other selected nuclear transport adapter proteins: Snurportin 1 is the import adapter for trimethylguanosine (TMG)-capped U snRNPs, NMD3 functions as export adapter for large ribosomal subunits, and PHAX (phosphorylated adapter for RNA export) functions in U snRNA export. In the Arabidopsis genome, a single gene encoding a protein with high sequence similarity to each of the human adapters Snurportin 1, NMD3, and PHAX exists
IBB Importin beta-binding; NLS nuclear localisation signal; NES nuclear export signal
Fig. 4.
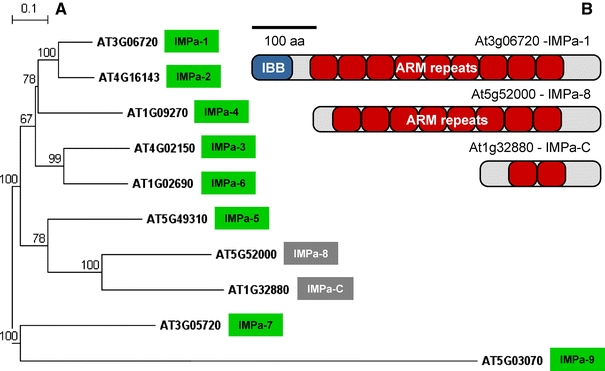
The Importin alpha family of Arabidopsis thaliana. a Phylogenetic tree of Importin alpha proteins and related proteins, b protein architecture of three selected family members. Protein names and AGI locus designations are given. Eight Importin alpha proteins (names in green boxes) meet the prototype architecture of Importin alpha 1 (IMPa-1). IMPa-1 contains an N-terminal Importin beta-binding (IBB) domain (b) that allows for interaction with Importin beta when an NLS substrate is bound to Importin alpha. Most of the remainder of the protein consists of 9–10 Armadillo (ARM) repeats that give the protein an elongated superhelical structure and form the NLS binding sites. Two related proteins are different (boxed grey). Importin alpha-like 8 (IMPa-8) lacks an IBB domain (b), and IMPa-C is a small protein with high similarity to the C terminus of Importin alpha proteins (b). The phylogenetic tree was constructed as described in the legend of Fig. 1. IMPa-9 was used to root the tree. Distance bar is given top left and bootstrap values are indicated at the nodes. Protein architectures in (b) are drawn to scale and a size bar is given top left
Seven of the plant Importin beta-like NTRs have been functionally characterised to date. These are the importins SAD2 (Zhao et al. 2007), Transportin 1 (Ziemienowicz et al. 2003), and Importin beta (Jiang et al. 1998), and the exportins Exportin 1/XPO1 (Haasen et al. 1999), Exportin 2/CAS (Haasen and Merkle 2002), Exportin-T (Paused; Hunter et al. 2003), and Exportin 5 (Hasty; Bollman et al. 2003). Although these NTRs transport plant-specific cargoes in and out of the nucleus, the general picture is a high functional conservation of importin beta-like proteins between vertebrates and plants. Transportin 1 imports RNA-binding proteins containing an M9 domain as import signal (Pollard et al. 1996; Bonfaci et al. 1997; Ziemienowicz et al. 2003), rice Importin beta binds to Importin alpha (Jiang et al. 1998; Görlich and Kutay 1999), Exportin 1, in humans termed CRM1 for Chromosomal Region Maintenance 1 (Adachi and Yanagida 1989), recognises proteins containing leucine-rich NESs (Fornerod et al. 1997, Fukuda et al. 1997; Ossareh-Nazari et al. 1997; Stade et al. 1997; Haasen et al. 1999), Exportin 2/CAS interacts with Importin alpha (Kutay et al. 1997; Haasen and Merkle 2002), Exportin-T/Paused complements a yeast los1 mutant deficient for the export receptor for tRNAs (Arts et al. 1998; Kutay et al. 1998; Hunter et al. 2003), and Exportin 5/Hasty binds to double-stranded RNAs (Bollman et al. 2003; Lund et al. 2004; Bohnsack et al. 2004; Park et al. 2005). In addition, the signals that confer nuclear transport are interchangeable between organisms, suggesting a high conservation of the basic principles of nuclear transport processes. Transportin 1 binds to the M9 domain of human hnRNP A1 and functions in nuclear import systems made from permeabilised HeLa cells (Ziemienowicz et al. 2003), Importin alpha 1 binds to all three types of NLS that have been identified in plants in vitro (Smith et al. 1997), the prototypical NLS of simian virus 40 large T antigen confers nuclear accumulation of proteins in plant cells and Arabidopsis XPO1 interacts with the NES derived from HIV protein Rev that also confers nuclear export of proteins in plant cells (Haasen et al. 1999).
RanBP1 represents another type of RanBPs. RanBP1-like proteins form a small family of six members in Arabidopsis and three in humans (Fig. 1b). These proteins contain a Ran-binding domain that is different from the one found in importin beta-like proteins (Görlich et al. 1997; Beddow et al. 1995). RanBP1-like proteins do not function as NTRs, however, they are important regulatory proteins for nuclear transport processes. RanBP1 is restricted in the cytoplasm due to a leucine-rich NES in the C terminus of the protein (Richards et al. 1996; Zolotukhin and Felber 1997; Haasen et al. 1999). RanBP1 cooperates with RanGAP1 to catalyse GTP hydrolysis on Ran in the cytoplasm (Bischoff et al. 1995). While RanBP2, a giant nucleoporin (Nup358) that contains Ran-binding domains of the RanBP1 type, zinc finger modules, and displays SUMO1 E3 ligase activity (Pichler et al. 2002), is missing in plant genomes, the Arabidopsis genome contains three genes encoding very similar RanBP1 proteins (Fig. 1b; Haasen et al. 1999). All three Arabidopsis proteins contain leucine-rich NESs (Haasen et al. 1999; Kim and Roux 2003), and display an exclusively cytoplasmic localisation (Haasen et al. 1999) RanBP1a was shown to specifically bind to RanGTP (Haizel et al. 1997) and RanBP1c acts as a co-activator of RanGAP1 in vitro (Kim and Roux 2003). There are three more Arabidopsis proteins containing Ran-binding domains of the RanBP1 type that have not been functionally characterised to date (Fig. 1b), designated RanBP1-like proteins (RanBPL1-3). RanBPL1 and RanBPL2 seem to be closer related to human RanBP3, and RanBPL3 groups with human Nup50 (Fig. 1b). However, these sequence similarities are insufficient to suggest putative functions of the uncharacterised Arabidopsis RanBPL proteins. Human RanBP3 shuttles between the nucleus and the cytoplasm and forms high affinity complexes with Exportin 1. However, RanBP3 is no export cargo but acts as a cofactor for the Exportin 1-dependent nuclear export pathway by influencing the relative affinity of Exportin 1 for different substrates in the nucleus (Englmeier et al. 2001; Lindsay et al. 2001) and by promoting efficient assembly of Exportin 1-containing export complexes (Nemergut et al. 2002). Human Nup50 (also called Npap60) also shuttles between the nucleus and the cytoplasm, and was reported to act as an import cofactor that accompanies Importin alpha/beta import complexes into the nucleus and stimulates nuclear import (Lindsay et al. 2002). Later, Matsuura and Stewart (2005) reported that Nup50 displaces NLS proteins from Importin alpha in the nucleus, thus acting in the final steps of nuclear import and in the recycling of Importin alpha. The finding that RanBPL proteins with high similarity to human RanBP3 and Nup50 exist in Arabidopsis suggests that these two important functions may be conserved in plants.
The Ran GTPase cycle
The small GTPase Ran has great impact on the regulation of nuclear transport processes. This function of Ran was established in 1993 (Moore and Blobel 1993). In Arabidopsis, three genes encode very similar Ran proteins, the Ran1 and Ran2 genes are arranged in tandem copies and show a much higher expression than Ran3 (Haizel et al. 1997). Arabidopsis, tomato and tobacco Ran proteins are functional in fission yeast (Ach and Gruissem 1994; Merkle et al. 1994; Haizel et al. 1997).
The extraordinary characteristics of the small GTPase Ran are: (1) Ran is a soluble small G protein, (2) Ran shuttles between the nucleus and the cytoplasm, and (3) the two different forms of Ran, RanGTP and RanGDP, are distributed asymmetrically over the NE (Moore and Blobel 1993; Görlich et al. 1996a; Izaurralde et al. 1997; Nachury and Weis 1999). The latter fact is a result of the asymmetric distribution of the major regulatory proteins for the Ran GTPase cycle. The Ran-specific Guanine nucleotide Exchange Factor (RanGEF, in humans called RCC1 for Regulator of Chromosome Condensation 1) that facilitates the dissociation of GDP from Ran so that GTP can bind (Fig. 3a; Bischoff and Ponstingl 1991a) is a chromatin-associated protein (Bischoff and Ponstingl 1991b). Hence, the concentration of RanGTP is very high in the nucleus (Fig. 3). RanGEF is not identified in plants to date. On the other hand, RanGAP1 and RanBP1 closely cooperate in the hydrolysis of GTP on Ran and are confined in the cytoplasm (Fig. 3b). RanBP1 binds to and releases RanGTP from complexes with RanBP–NTRs and presents RanGTP to RanGAP1, acting as a co-activator for RanGAP1 (Bischoff et al. 1995; Bischoff and Görlich 1997). In this way, RanGTP concentrations are kept very low in the cytoplasm. The resulting steep gradient of RanGTP over the NE is maintained in the cell and is used to impose directionality on nuclear transport processes by RanBP–NTRs (Izaurralde et al. 1997). Experiments to directly visualise the RanGTP gradient in living cells (Kalab et al. 2002, 2006) have not been performed in plant cells. However, most probably a very similar situation exists in plant cells as suggested by complementation experiments using plant proteins (Ach and Gruissem 1994; Merkle et al. 1994; Haizel et al. 1997; Pay et al. 2002), by the high degree of conservation between plant, yeast and vertebrate proteins, and by the cytoplasmic localisation of Arabidopsis RanBP1 (Haasen et al. 1999) and the cytoplasmic/NE localisation of Arabidopsis RanGAP1 (Jeong et al. 2005) in interphase. Another factor that is highly conserved between organisms is the nuclear import factor for RanGDP, Nuclear Transport Factor 2 (NTF2). NTF2 is a small protein that essentially consists of one domain, called NTF2-like domain. NTF2 forms homodimers and binds to RanGDP in the cytoplasm, where it also acts to inhibit GDP dissociation from Ran, and imports RanGDP into the nucleus (Fig. 3a; Ribbeck et al. 1998; Smith et al. 1998). Arabidopsis contains two genes encoding very similar proteins that are functional in yeast, designated NTF2a and NTF2b (Zhao et al. 2006). NTF2 is an example for an NTR that does not belong to the importin-beta family, and the NTF2-like domain confers interaction with phenylalanine-glycin (FG) repeat-containing nucleoporins (NUPs; Bayliss et al. 2002).
In vertebrates, a fraction of RanGAP1 is sumoylated and associated with the cytoplasmic side of the NPC in close interaction with RanBP2 (Matunis et al. 1996; Mahajan et al. 1997; Saitoh et al. 1997). RanBP1 function is also provided by RanBP2/Nup358 that is required for efficient nuclear transport processes in vertebrates (Bernad et al. 2004; Hutten et al. 2008). As stated above, RanBP2 is missing in plants, and RanGAP1 is anchored to the NPC in a completely different way than in vertebrates (Rose and Meier 2001; Xu et al. 2007a, b). Arabidopsis contains two RanGAP genes that complemented the yeast RanGAP mutant rna1 (Pay et al. 2002). Arabidopsis RanGAP proteins share a plant-specific domain located in the N terminus, termed WPP domain that is necessary and sufficient to target RanGAP1 to the NPC (Rose and Meier 2001). Two families of NE-localised proteins were shown to confer NPC targeting of Arabidopsis RanGAP1, the WIPs (WPP domain-interacting proteins; Xu et al. 2007a, b) and WITs (WPP domain-interacting tail-anchored proteins; Zhao et al. 2008). Both protein classes are plant-specific and share additional common features. They contain a coiled-coil domain next to a putative transmembrane domain, which is why they were designated CC-TMD proteins (for coiled-coil-transmembrane domain; Zhao et al. 2008). These findings suggest that RanGAP1 targeting to the NPC has evolved at least twice, and differently in plants and vertebrates. The importance of RanGAP1 function for nuclear transport, however, seems to be conserved.
The small GTPase Ran, together with its regulatory proteins RCC1 and RanGAP1, has multiple functions. Ran is not only the key regulator for directionality of nuclear transport involving RanBP–NTRs, but also regulates key aspects of mitosis, like spindle formation, kinetochore attachment, spindle checkpoint control, and post-mitotic re-formation of the nuclear envelope and NPC assembly (reviewed in: Joseph 2006; Clarke and Zhang 2008). Also, nucleo-cytoplasmic transport components are involved in these processes, including Importin alpha, Importin beta and Exportin 1 (reviewed in: Harel and Forbes 2004; Clarke and Zhang 2008). The finding that Arabidopsis RanGAP1 labels the preprophase band and remains associated with it during mitosis and cytokinesis (Xu et al. 2008) strongly suggests that at least parts of the regulatory role of Ran and RanGAP1 during cell division may be conserved in plants as well. Further suggesting a role of Ran in mitosis and plant cell division control, inducible depletion of RanGAP1 leads to misplaced cell walls similar to other mutants with division plane defects (Xu et al. 2008).
Nuclear export of mRNA in plants
Nuclear export of mRNA is a highly complex and regulated process that is intertwined with mRNA maturation (Cullen 2003; Sommer and Nehrbass 2005; Carmody and Wente 2009). Specific nuclear export receptors as well as many other proteins are necessary for bulk mRNA export (Vinciguerra and Stutz 2004). Nuclear export of mRNA in vertebrates and in yeast is facilitated by specific NTRs that, like the nuclear import receptor for RanGDP, NTF2, are unrelated to importin beta-like RanBPs. They were designated Tip-Associated Protein (TAP) or Nuclear RNA Export Factor 1 (NXF1) in humans, and Mex67p and in yeast (Cullen 2003; Rodriguez et al. 2004). TAP/NXF1 and Mex67p function in complex with a small protein termed p15 or NTF2-like Export Factor 1 (NXT1) in humans and mRNA Transport Regulator 2 (Mtr2p) in yeast (Cullen 2003; Stutz and Izaurralde 2003). Interestingly, human p15/NXT1 and yeast Mtr2p share no sequence similarity, but are functional homologs (Fribourg and Conti 2003). The common feature of TAP, p15 and related proteins seems to be the conserved NTF2-like domain that is primarily important for protein–protein interaction (Suyama et al. 2000) and for interaction with FG repeat-containing NUPs (Fribourg et al. 2001). The p15, like NTF2 itself is a small protein that consists of an NTF2-like domain, only. However, in contrast to NTF2, p15 does not form homodimers and does not interact with RanGDP. Instead, it binds to TAP, a protein containing several specific domains, including an NTF2-like domain as well, a non-canonical RNA recognition motif (RRM), leucine-rich repeats (LRRs), and the so-called TAP-C domain (Katahira et al. 2002; Wiegand et al. 2002). The TAP-C domain is located at the very C terminus of the protein, belongs to the superfamily of UBA-like domains, and together with the NTF2-like domain is important for binding to FG repeat-containing NUPs (Fribourg et al. 2001; Levesque et al. 2006). The TAP/p15 heterodimer does not make direct use of the topological information provided by the RanGTP gradient across the nuclear envelope. Directionality of nuclear export of mRNA has thus to be achieved by other means. However, the RanGTP gradient is needed for the re-import of proteins involved in mRNA export after they dissociate from their cargo (Izaurralde et al. 1997). While there is only one protein in yeast, Mex67p, a small gene family encoding TAP-like proteins is present in humans (Herold et al. 2000).
In plants, much less is known about mRNA export and the regulatory proteins involved. Genes encoding homologs of human TAP/NXF1 or yeast Mex67p are missing in plant genomes. This situation leaves two possibilities: either plant genes that encode proteins with functions equivalent to human TAP or yeast Mex67p exist but are structurally unrelated and/or plants use different components for mRNA export than yeast or humans. However, there is a candidate for an Arabidopsis homolog of vertebrate p15. Zhao et al. (2006) characterised NTF2a and NTF2b as the Arabidopsis homologs for the nuclear import receptor for RanGDP. Unlike NTF2a and NTF2b, a third related protein termed NTL for NTF2-like could not functionally replace the yeast NTF2 gene. Its function remains uncharacterised to date.
Some plant genes encoding proteins with similarity to vertebrate or yeast proteins that function in mRNA export are characterised by mutants that show accumulation of poly(A)+ RNA in nuclei, indicating a role in mRNA export in plants. Concomitantly, these mutants show pleiotropic phenotypes, including altered development, altered responses to phytohormones and environmental stresses, impaired disease resistance, and early flowering. Many genes encode Arabidopsis NUPs, like AtTPR/AtNUA (Jacob et al. 2007; Xu et al. 2007a, b), and AtNUP160/SAR1 (Dong et al. 2006; Parry et al. 2006), AtNUP96/SAR3/MOS3 (Parry et al. 2006; Zhang and Li 2005), and AtNUP1 (Lu et al. 2010). NUPs are discussed in detail below. Of specific interest, however, is the Arabidopsis LOS4 gene (Gong et al. 2005) since it encodes the most likely candidate for a plant ortholog of yeast Dbp5p to date. Since human TAP and yeast Mex67p do not belong to the importin beta NTR family (Stutz and Izaurralde 2003), directionality of transport through the nuclear pore complexes mediated by these proteins has to be achieved independently of the Ran GTPase system. In yeast, Dbp5p associates with mRNA early in the nucleus and accompanies it to the cytoplasmic side of the NPC where it concentrates by binding to NPC filaments. Dbp5p is a nucleo-cytoplasmic shuttle protein. Its ATPase activity is activated by interaction with Gle1p, an RNA export factor that is also associated with the cytoplasmic side of the NPC, and inositol polyphosphate IP6. The activity of the DEAD box helicase Dbp5 leads to ATP-dependent remodelling of mRNPs at the cytoplasmic side of the NPC (Cole and Scarcelli 2006; Stewart 2007; Tran et al. 2007). This is discussed as the crucial step for dissociation of specific factors and as a way to impose directionality on mRNP export by dissociating factors that would be needed for the way back (Stewart 2007; Carmody and Wente 2009). The phenotype of los4 mutants in Arabidopsis and the nuclear rim localisation of LOS4-GFP thus suggest the existence of a similar mechanism to achieve directionality of nuclear export of mRNAs in plants.
Other plant proteins that show high similarity to vertebrate and/or yeast proteins that contribute to mRNA export are the proteins of the cap-binding complex (CBC). After capping of the 5′ end of mRNA in the nucleus, CBC forms at the cap structure and contributes to mRNA maturation, splicing, and nuclear export. Although nuclear at steady state, the CBC accompanies ribonucleoprotein particles containing mRNA (mRNPs) to the cytoplasm, where cap-binding proteins (CBPs) are stripped off and re-imported into the nucleus (Lewis and Izaurralde 1997). Arabidopsis contains two single genes that encode homologs of the large and the small subunit of the CBC, termed CBP80 and CBP20, respectively (Kmieciak et al. 2002). Furthermore, poly(A)-binding proteins (PABPs) are major constituents of mRNPs in the nucleus and in the cytoplasm. PABPs with a predominantly cytoplasmic localisation (PABPCs) are also important for mRNA export. While yeast contains one gene, Arabidopsis contains a small gene family of eight members (Belostotsky 2003). Yeast Pab1p is essential, shuttles between the nucleus and the cytoplasm, contains an NES in its N terminus (Brune et al. 2005), and is implicated in mRNA biogenesis and export, regulation of mRNA turnover, and initiation of translation (Kühn and Wahle 2004). Cross-species complementation experiments revealed differential functions of Arabidopsis PABPCs. Arabidopsis PAB2 restored viability of a pab1 mutant strain and functioned in many of the post-transcriptional processes that were characterised for yeast Pab1p (Palanivelu et al. 2000). This included poly(A) shortening by interaction with the yeast poly(A) nuclease complex, translation initiation, and the restoration of the linkage between deadenylation, decapping and mRNA decay. Arabidopsis PAB3 or PAB5 also rescued the lethal phenotype of yeast strains that lack Pab1p (Belostotsky and Meagher 1996; Chekanova et al. 2001). PAB3, like yeast Pab1p, directly interacted with target mRNA, was localised in the yeast nucleus, and acted in mRNA export to the cytoplasm, as was shown by genetic interactions with Gle1 and Nab2 (Chekanova and Belostotsky 2003). Taken together, some PABPC functions are highly conserved between yeast and plants. PAB2 came closest to yeast Pab1p of all tested Arabidopsis PABPCs (Palanivelu et al. 2000). Arabidopsis PABPCs may recruit several other proteins to the mRNP, including other RNA-binding proteins (Bravo et al. 2005). Serine/arginine-rich (SR) proteins are splicing regulators and have a function in mRNA export as well (Huang and Steitz 2005). The Arabidopsis SR protein RSZp22 shows high intra-nuclear dynamics and contains an NES and shuttles between the nucleus and the cytoplasm (Tillemans et al. 2006). Proteomic analysis of the Arabidopsis nucleolus identified SR proteins and many components of the post-splicing exon-junction complex that functions in mRNA export and surveillance (Pendle et al. 2005). These findings suggest that plant nucleoli are implicated in these processes. Arabidopsis also contains four homologs of vertebrate ALY/REF proteins (Uhrig et al. 2004). They are highly conserved RNA-binding proteins that function in mRNA export as adapters to recruit other proteins to the mRNP in vertebrates, most notably TAP/p15 (Stutz et al. 2000; Stutz and Izaurralde 2003).
Nucleo-cytoplasmic transport and its implications for plant homeostasis and development
The mRNA has to be exported from the nucleus in order to be translated in the cytoplasm as well as other RNA species that have different functions in the cytoplasm. In contrast, many karyophilic proteins have to be imported into the nucleus to gain access to the genome in order to be able to fulfil their regulatory or enzymatic functions. The possibility to regulate the flow of specific macromolecules over the NPCs creates a control level that is unique to eukaryotic cells. There are numerous examples in vertebrates and in yeast that demonstrate that nucleo-cytoplasmic partitioning of specific proteins is exploited as a regulatory mechanism to control signalling to and from the nucleus. The classic examples are the transcription factors NF-kappaB in humans and Pho4 in yeast (reviewed in: Kaffman and O’Shea 1999; Turpin et al. 1999). The term “nucleo-cytoplasmic partitioning of proteins” refers to the phenomenon that the ratio of the concentration of a protein in the nucleus versus the cytoplasm differs between different states of a cell. If this is achieved by regulation of the nuclear import and/or nuclear export rates of the protein, this change in steady state localisation may be fully reversible (shuttling). Nucleo-cytoplasmic shuttling of proteins can be visualised in vivo in real time using specific fluorescence microscopy techniques (Köster et al. 2005) or photoswitchable or photoactivatable fluorescent proteins (Martini et al. 2007). However, nucleo-cytoplasmic partitioning may also involve selective and regulated degradation of proteins in one or the other cellular compartment. In plants, nucleo-cytoplasmic partitioning of specific proteins is also used to regulate developmental and signalling pathways (Merkle 2003). This includes regulation of temperature stress (heat and cold stress), light signalling, phytohormone (cytokinin, gibberellin, brassinosteroid) signalling, the self-incompatibility response and disease resistance (reviewed in Merkle 2003; Table 2). An illustrative example for nucleo-cytoplasmic partitioning of a regulatory plant protein is Arabidopsis Brassinazole Resistant 1 (BZR1; Ryu et al. 2007) and its rice ortholog (Bai et al. 2007). Brassinosteroids (BRs) are a group of phytohormones that are implicated in the regulation of plant growth and development. BRs are perceived by a receptor kinase complex in the plasma membrane. Binding of BR to the receptor Brassinosteroid Insensitive 1 (BRI1) results in dissociation of a repressor and in complex formation of BRI1 with a receptor kinase-like protein. A downstream signal transduction cascade is then activated that modulates the activity of two related proteins that are the key transcription factors of BR signalling, and BZR1 is one of them (Ryu et al. 2007). The nucleo-cytoplasmic localisation and the phosphorylation status of BZR1 is regulated in a BR-dependent manner. BR induces rapid dephosphorylation of BZR1 in the cytoplasm by a plant-specific phosphatase named BRI1 Suppressor 1 (BSU1). This correlates with nuclear accumulation of BZR1, where it acts as a transcriptional repressor (Ryu et al. 2007). Nuclear import of BZR1 most likely occurs via Importin alpha/beta heterodimers. In contrast, phosphorylation of BZR1 by the nuclear kinase Brassinosteroid Insensitive 2 (BIN2) directly inhibits DNA binding, induces nuclear export by XPO1 and cytosolic retention of BZR1 (Ryu et al. 2007). Interestingly, 14-3-3 proteins interact with BZR1 in a phosphorylation-dependent manner and are implicated in nuclear export and/or cytosolic retention of BZR1 (Gampala et al. 2007; Ryu et al. 2007). In sum, BR regulates the transcriptional activity of BZR1 by controlling its nucleo-cytoplasmic partitioning by a switch involving the opposing actions of the nuclear kinase BIN2 and the cytoplasmic phosphatase BSU1. Such a mechanism allows for quick responses to environmental and developmental signals without de novo synthesis of BZR1.
Table 2.
Plant proteins that show nucleo-cytoplasmic partitioning, including cargos for Exportin 1
| Protein | Locus (organism) | Pathway | References, comments |
|---|---|---|---|
| RanBP1a, b, c | AT1G07140 | Ran-dependent nucleo-cytoplasmic transport | Haasen et al. (1999); Kim and Roux (2003), interaction with XPO1 ensures cytoplasmic localisation |
| AT2G30060 | |||
| AT5G58590 | |||
| bZIP transcriptional repressor BZR1 | AT1G75080 | Brassinosteroid signal transduction | Ryu et al. (2007), cargo for XPO1 |
| Rice bZIP transcription factor RSG | OS12G06520 | Gibberellin signal transduction | Igarashi et al. (2001), cargo for XPO1 |
| Rice DELLA transcription factor SLR1 | OS03G49990 | Gibberellin signal transduction | Itoh et al. (2002), regulated nuclear import, degradation |
| PHOR1 | Potato | Gibberellin signal transduction | Amador et al. (2001), regulated nuclear import (nuclear export?) |
| AHP1, AHP2 | AT3G21510, AT3G29350 | Cytokinin signal transduction | Hwang and Sheen (2001); Punwani et al. (2010), nucleo-cytoplasmic distribution by balanced nuclear import and export |
| bZIP10 | AT4G02640 | Basal defence and cell death | Kaminaka et al. (2006), cargo for XPO1 |
| High mobility group type B protein (HMGB) | AT5G23405 | Unknown | Grasser et al. (2006), cargo for XPO1 |
| Cyclin-dependent kinase inhibitor ICK1/KRP1 | AT2G23430 | Cell cycle regulation | Jacoby et al. (2006), cargo for XPO1 |
| Heat shock transcription factor Hsf2a | tomato | Heat stress response | Heerklotz et al. (2001), cargo for XPO1 |
| RING finger protein HOS1 | AT2G39810 | Cold signalling | Ishitani et al. (1998); Lee et al. (2001), regulated nuclear export (nuclear export?; degradation?) |
| Nucleosome assembly proteins (NAP1) | Tomato, rice | Nucleosome assembly, chromatin remodelling | Dong et al. (2005), cargo for XPO1 |
| SR splicing factor RSZp22 | AT4G31580 | mRNA processing and export | Tillemans et al. 2006, cargo for XPO1 |
| Ribonuclease III-like protein RTL2 | AT3G20420 | RNA metabolism (unknown) | Comella et al. (2007), cargo for XPO1 |
| ARM repeat protein ARC1 | Brassica | Self-incompatibility response | Stone et al. (2003), cargo for XPO1 |
| Phytochromes phyA, phyB | AT1G09570, AT2G18790 | Light signal transduction | Kircher et al. (1999a); Yamaguchi et al. (1999); Gil et al. (2000); Kim et al. (2000), light-induced nuclear import, nucleo-cytoplasmic partitioning |
| Cryptochrome cry1 | AT4G08920 | Light signal transduction | Cashmore et al. (1999); Guo et al. (1999); Kleiner et al. (1999); Yang et al. (2000), cry1 and cry2 are predominantly nuclear protein, evidence for shuttling of cry1 |
| FHY1 | AT2G37680 | Light signal transduction | Zeidler et al. (2004), cargo for XPO1; Hiltbrunner et al. (2005, 2006), required for phyA nuclear import |
| bZIP transcription factor CPRF2 | Parsley | Light signal transduction | Kircher et al. (1999b) |
| bZIP transcription factor GBF2 | AT4G01120 | Light signal transduction | Terzaghi et al. (1997) |
| COP1 | AT2G32950 | Light signal transduction | Von Arnim and Deng (1994); von Arnim et al. (1997); Stacey et al. (1999, 2000), nuclear in darkness, strong reduction of protein levels in light |
| Chaperone DjC6 | AT5G06910 | Protein folding | Suo and Miernyk (2004), cargo for XPO1 |
| LHY/CCA1-like 1 (LCL1) | AT5G02840 | Circadian clock | Martini et al. (2007), cargo for XPO1 |
| ERM kinase | Parsley | Desease resistance and defence | Ligterink et al. (1997), elicitor-induced nuclear translocation |
| NPR1 | AT1G64280 | Systemic acquired resistance | Kinkema et al. (2000), salicylic acid-induced nuclear translocation |
Partitioning of proteins between the nucleus and the cytoplasm may not only be due to a regulated change of the rates of nuclear import versus nuclear export. Instead or in addition, it may involve regulated selective protein degradation in one or the other compartment. For a more detailed description of pathways see Merkle (2003)
Mutations in plant genes that cause impaired nucleo-cytoplasmic transport
In the past few years, mutations in many plant genes that encode NUPs, NTRs, nuclear transport adapters, or other proteins of the nuclear transport machinery have been investigated. In some cases, gene expression has been experimentally manipulated by (conditional) over-expression or by applying antisense technology. The consequences of these de-regulations of nucleo-cytoplasmic transport for plant development and homeostasis are discussed in this chapter and are summarised in Table 3.
Table 3.
Plant pathways that are affected by mutations in or de-regulations of genes encoding proteins that function in nucleo-cytoplasmic transport of macromolecules
| Genes | Comments | Pathways affected by mutation or de-regulation | References |
|---|---|---|---|
| MOS6 (IMPa-3) | Encodes an adapter for nuclear import of NLS proteins | Immune response | Palma et al. (2005) |
| IMPa-4 | Encodes an adapter for nuclear import of NLS proteins | Agrobacterium tumefaciens-mediated transformation | Bhattacharjee et al. (2008) |
| SAD2 | Encodes an NTR of the importin beta family (RanBP7/8 ortholog) | ABA response, cold stress, osmotic stress, UV-B response | Zhao et al. (2007); Verslues et al. (2006) |
| XPO1A, XPO1B | Encode NTRs of the importin beta family | Gametophyte development | Blanvillain et al. (2008) |
| Hasty/Exportin 5 | Encodes an NTR of the importin beta family | Pre-miRNA biogenesis/export defect, many developmental pathways incl. flowering time | Bollman et al. (2003); Park et al. (2005) |
| Paused/Exportin-T | Encodes an NTR of the importin beta family | tRNA export defect, many developmental pathways incl. flowering time | Hunter et al. (2003); Li and Chen (2003); Park et al. (2005) |
| Ran1, Ran2 | Encode GTPase Ran (overexpession in Arabidopsis and tobacco) | Auxin response, cell division, osmotic and salt stress | Wang et al. (2006); Zang et al. (2010) |
| RanBP1c | Encodes a regulatory protein of the GTPase Ran (antisense overexpession) | Auxin response | Kim et al. (2001) |
| RanGAP1 | Regulatory protein of the GTPase Ran (sense overexpession) | Cell division, cytokinesis | Xu et al. (2008) |
| LOS4/Cryophyte | Encodes a DEAD box RNA helicase, putative yeast Dbp5 ortholog | mRNA export defect, cold stress | Gong et al. (2002, 2005) |
| CBP20, ABH1/CBP80 | Encode proteins of the nuclear Cap-binding complex (CBC) | ABA response, flowering time | Hugouvieux et al. (2002); Bezerra et al. (2004); Papp et al. (2004a, b) |
| Rae1 (Nicotiana benthamiana) | Encodes a putative ortholog of the yeast RNA export factor Rae1p (RNAi-mediated silencing) | mRNA export defect, growth defects, cell division | Lee et al. (2009) |
| AtTPR/AtNUA | Encodes a nucleoporin | mRNA export defect, flowering time | Jacob et al. (2007); Xu et al. (2007a, b) |
| Nup85 (Lotus japonicus) | Encodes a nucleoporin | Bacterial and fungal symbiosis | Saito et al. (2007) |
| Nup133 (Lotus japonicus) | Encodes a nucleoporin | Bacterial and fungal symbiosis | Kanamori et al. (2006) |
| MOS7 (AtNup88) | Encodes a nucleoporin | Immune response | Cheng et al. (2009) |
| MOS3/SAR3 (AtNup96) | Encodes a nucleoporin | mRNA export defect, immune response, auxin response, flowering time | Parry et al. (2006); Zhang and Li (2005) |
| SAR1 (AtNup160) | Encodes a nucleoporin | mRNA export defect, auxin response, flowering time, cold stress | Dong et al. (2006); Parry et al. (2006) |
| AtNup1 | Encodes a nucleoporin | mRNA export defect, many developmental pathways | Lu et al. (2010) |
| AtTHP1 | Encodes a homolog of a component of the yeast TREX-2 complex | mRNA export defect, many developmental pathways | Lu et al. (2010) |
If not stated otherwise, the genes are from Arabidopsis thaliana
ABA abscisic acid; NTR nuclear transport receptor
Arabidopsis Exportin 5 and Exportin-T genes are not essential but indispensable for normal plant development
The Hasty (HST) gene was identified in a genetic screen for Arabidopsis developmental mutants that showed an accelerated change from the juvenile to the adult phase (Telfer and Poethig 1998). HST encodes a protein with high sequence similarity to human Exportin 5 (Bollman et al. 2003). The hst mutant plants show a pleiotropic phenotype with defects in many different processes in Arabidopsis development, including the size of the shoot apical meristem, accelerated vegetative phase change, the transition to flowering, disruption of the phyllotaxis of the inflorescence and reduced fertility. In addition, hst seedlings have an abnormally short hypocotyl and primary root. Cargo substrates for human Exportin 5 are eEF1A via tRNA (Bohnsack et al. 2002; Calado et al. 2002), small RNAs that contain a double-stranded mini-helix domain that are associated with RNA-binding proteins (Gwizdek et al. 2003, 2004), and precursor microRNAs (pre-miRNAs; Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). These findings suggested that Arabidopsis Exportin 5/HST may also act in miRNA biogenesis in plants, which was verified by Park et al. (2005). However, there are major differences between plants and metazoa regarding miRNA biogenesis. Two RNase III-like activities are involved in metazoan miRNA biogenesis, Dicer and Drosha, whereas DCL1 is the major RNase III-like protein that is responsible for miRNA biogenesis in Arabidopsis (Jones-Rhoades et al. 2006). DCL1 is responsible for both processing steps, and since DCL1 is localised predominantly in the nucleus, plant pre-miRNAs are very short-lived intermediates and mature single-stranded miRNAs are already produced in the plant nucleus (Papp et al. 2003; Park et al. 2005). It is unclear to date whether HST binds to and facilitates nuclear export of single-stranded miRNAs and/or of the miRNA:miRNA* duplexes (Park et al. 2005). Interestingly, leaves of hst plants are curled upwards, and the abaxial layer of leaf mesophyll cells resembles the adaxial layer in the hst plants. This defect in organ polarity was also obvious in carpels (Bollman et al. 2003). This phenotype is reminiscent of the polarity defects in leaves caused by de-regulation of the transcript levels of class III homeodomain-leucine zipper transcription factors that are controlled by miR165/miR166 (Mallory et al. 2004). Similarly, many of the defects of hst plants may be direct or indirect consequences of impaired miRNA biogenesis.
Paused (PSD) (Hunter et la. 2003) encodes the ortholog of the export receptor for tRNAs, termed Exportin-T in vertebrates (Kutay et al. 1998) and Los1p in yeast (Hellmuth et al. 1998). It was identified in several genetic screens. One screen was designed to identify mutations that affect meristem initiation during embryogenesis, a second screen was designed to identify mutations resulting in adult characteristics in the first two normally juvenile leaves (Hunter et al. 2003). Finally, the third screen was a genetic modifier screen for enhancers of the weak class C loss-of-function phenotype of hua1-1 hua2-1 double-mutant Arabidopsis plants (Li and Chen 2003). In line with its identification in three different genetic screens, psd mutants, like hst mutants, show a pleiotropic phenotype. In psd mutants phase change is also affected, the plants display adult characteristics on leaves that are juvenile in wild-type plants. They also show defects in the shoot apical meristem that were already detectable at embryo stages, and in the phyllotaxis of the inflorescence. They are delayed in the transition to reproductive development, in root growth, lateral root initiation, and they also show significantly reduced fertility (Hunter et al. 2003). The psd plants are fully viable, which is unexpected for a defect in an export pathway for an important bulk cargo like tRNA. Recently, a T-DNA insertion line for the rice PSD ortholog was isolated and evidence for the involvement of rice PSD in nuclear export of tRNA was reported (Yao et al. 2008). The mutant displayed severe morphological defects including retarded development and low fertility compared with wild-type rice.
Since tRNAs may also be exported from the nucleus by human Exportin 5 (Bohnsack et al. 2002; Calado et al. 2002), double-mutant plants were generated. Arabidopsis psd hst plants are also viable, show a combination of the hst and psd mutant phenotypes, and are further decreased in size as compared to plants carrying a single mutation (Hunter et al. 2003). The authors speculate that this may indicate the existence of another nuclear export pathway for tRNA that is unknown to date. However, tRNAs may also leave the nucleus by passive diffusion, and this very inefficient process may be enough for survival of single and double-mutant plants. Park et al. (2005) tested whether PSD may partially compensate for an impaired HST function. They found that PSD does not transport miRNAs, since miRNA biogenesis is not affected in psd mutants, whereas the accumulation of most miRNAs is reduced in hst mutants. On the other hand, in psd mutants, the biogenesis of a tRNA was impaired, but this pathway was not affected in hst plants (Park et al. 2005). The most likely explanation for the psd phenotype is reduced supply of tRNAs and hence, reduced supply of proteins. A defect in a pathway that efficiently supplies the cytosol with an important substrate like tRNA may influence all aspects of plant growth and development. The phenotypes observed in psd mutants may thus hint to developmental processes that are especially sensitive for an under-representation of specific protein factors. On the other hand, the most likely explanation for the hst phenotype is an under-representation of most miRNAs. Both scenarios explain the pleiotropic phenotypes of both mutants, since many cellular processes are affected. However, miRNAs are still produced in hst plants, albeit less efficiently than in wild-type plants. The miRNAs may exit the nucleus in association with proteins, in complex with mRNPs, or even in complex with their mRNA targets, as proposed by Park et al. (2005). It is also unknown whether or not plant miRNAs are already associated with components of the RNA-induced silencing complex in the nucleus or when transported to the cytoplasm. Park et al. (2005), however, found indirect evidence for nuclear activity of miRNA-dependent mRNA cleavage. Since so many different pathways are potentially impaired, however, conclusions are difficult.
XPO1 function is essential, and mutant xpo1 plants show severe gametophytic defects
Arabidopsis contains two genes encoding very similar XPO1 proteins (Haasen et al. 1999; Merkle 2003; Blanvillain et al. 2008). T-DNA insertion lines for both loci, XPO1A and XPO1B, were isolated and single homozygous plants appeared phenotypically normal, indicating that each paralogous gene could functionally replace the other (Blanvillain et al. 2008). However, homozygous double-mutant plants carrying insertions in both loci could not be recovered. Co-transmission of mutant alleles through the gametes was impaired, as indicated by a distorted segregation of genotypes originating from crosses between xpo1A and xpo1B mutants. Inspection of mature siliques for the presence of aborted ovules indicated a female gametophyte lethal phenotype if two mutant xpo1 alleles were co-inherited. Siliques of double-heterozygous (xpo1A/+ xpo1B/+) plants contained about 25% aborted ovules, in siliques of homozygous–heterozygous (xpo1A/xpo1A xpo1B/+ and xpo1A/+ xpo1B/xpo1B) plants this number was about 50% (Blanvillain et al. 2008). Reciprocal crosses using wild-type pollen and homozygous–heterozygous recipients and vice versa showed that xpo1A and xpo1B alleles could not be co-transmitted through the female, whereas co-transmission through the male was reduced to 5% (Blanvillain et al. 2008). As a conclusion, Exportin 1 is essential for gametophyte function in Arabidopsis, and the double-mutant pollen was partially functional, but the female double-mutant embryo sac was not. The xpo1 mutations affected pollen germination and pollen tube growth. The female gametophyte was affected much more severely. In addition, Blanvillain et al. (2008) reported a maternal effect, since xpo1A and xpo1B alleles were not equivalent when maternally inherited in the mutant background of the paralogous gene. xpo1A/+ xpo1B/xpo1B plants produced double-mutant female gametophytes that failed to undergo all the mitotic divisions or failed to complete embryo sac maturation. In contrast, double-mutant female gametophytes of xpo1A/xpo1A xpo1B/+ plants had normal mitotic divisions, fertilisation occurred, and in most of these embryo sacs the endosperm started to divide but an embryo failed to develop. This finding suggests that XPO1B plays a greater role in female gametophyte development although its expression in pistils is lower than the expression of XPO1A.
The defects in Arabidopsis xpo1 mutants could be due to the lack of export of specific factors that are normally excluded from the nucleus or to the shortage of specific proteins or RNAs in the cytoplasm that are essential for gametophyte development and function. Since CRM1/XPO1 in complex with RanGTP is also involved in mitotic spindle assembly and kinetochore attachment in mammalian somatic cells (reviewed in: Arnaoutov and Dasso 2005; Budhu and Wang 2005) and is linked to the spindle pole body in yeast (Neuber et al. 2008), there may be a third possibility to explain gametophyte defects in xpo1 plants. In fact, Crm1/Xpo1 was originally identified in a genetic screen for cold-sensitive mutants in fission yeast with deformed chromosome domains (Adachi and Yanagida 1989). If XPO1 were involved in similar processes in plants as well, a lack of XPO1 function would most probably affect cell division. However, involvement of XPO1 in these cellular processes has not been shown in plants to date. Due to the very early gametophytic defects of xpo1 mutant plants, involvement of XPO1 in other signalling pathways could not be investigated.
Putative mRNA export factors and mutations that cause nuclear accumulation of poly(A)+ RNA
Arabidopsis contains two single genes that encode homologs of the large and the small subunit of the nuclear CBC, termed CBP80 and CBP20, respectively, which are highly conserved between organisms (Kmieciak et al. 2002). Arabidopsis CBP80/ABH1 (for ABA Hypersensitive 1) was identified in a genetic screen, and the abh1 mutation confers abscisic acid (ABA)-hypersensitive regulation of seed germination, stomatal closure and cytosolic calcium increase in guard cells (Hugouvieux et al. 2001). The authors concluded that mRNA processing factors act as negative regulators for ABA signalling. ABH1 is mainly localised in the nucleus at steady state, but a cytosolic localisation was also detected, indicating partitioning between the nucleus and the cytoplasm (Hugouvieux et al. 2002). In addition, an abh1 mutant suppressed the Frigida-mediated delay in flowering (Bezerra et al. 2004). The authors showed that this phenotype was caused by the inability of Frigida to increase mRNA levels of the floral repressor Flowering Locus C (FLC) in the abh1 mutant. Interestingly, mutations in genes encoding Arabidopsis NUPs also show flowering phenotypes (see below). A mutation in the Arabidopsis CBP20 gene was also characterised phenotypically (Papp et al. 2004a, b). The cbp20 mutant shows phenotypic characteristics that are very similar to those of the abh1 mutant, confers drought tolerance and shows defects in ABA signalling.
Several mutations in Arabidopsis genes encoding NUPs have been described. The vertebrate NUP Tpr (for Translocated promoter region) is located at the filaments of the nuclear basket of the NPC and serves as docking site for mRNPs. The Arabidopsis gene AtTPR was identified in a screen for suppressors of the floral repressor FLC (Jacob et al. 2007). The attpr mutants are characterised by an eightfold increase of poly(A)+ RNA in the nucleus. In addition, microarray analyses showed that homeostasis between nuclear and cytoplasmic RNA was disturbed, as revealed by a loss in correlation of transcript abundance, but not transcript composition, of the nuclear versus the total RNA pool. Furthermore, a pleiotropic phenotype indicated that several signalling pathways were affected, including the flowering pathway, according to the design of the screen. The attpr mutants are early flowering, and show defects in small RNA abundance and in signalling of the phytohormone auxin (Jacob et al. 2007). Interestingly, attpr and hst mutants have similar negative effects on the abundance of many miRNAs, whereas siRNAs are not affected. This finding suggests that HST-dependent nuclear export of miRNAs needs functional AtTPR and the export complex interacts with this NUP. Xu et al. (2007a, b) also described the characterisation of a mutation in the same gene, which they named Nuclear Pore Anchor (NUA). Accumulation of poly(A)+ RNA and an early flowering phenotype was also described for nua mutants. NUA was localised to the inner surface of the nuclear envelope. Since nua mutants phenocopy the effects of a mutation in the gene Early In Short Days 4 (ESD4) that encodes a SUMO protease, interaction of the two proteins was tested and verified in yeast two-hybrid experiments (Xu et al. 2007a, b). This result is supported by findings that yeast Mlp1p/Mlp2p (Myosin-like protein 1/2) proteins and mammalian Tpr, homologs of Arabidopsis TPR/NUA, also bind a SUMO protease and suggests a role for ESD4 in Arabidopsis mRNA export as well.
Like in attpr mutant plants (Jacob et al. 2007), defects in auxin signalling were also described in the sar3 mutant (Parry et al. 2006). This gene was characterised as Suppressor of Auxin Resistance 3, and encodes another Arabidopsis NUP, the homolog of mammalian NUP96. Both sar3 and attpr mutants are strong suppressors of the auxin-resistant 1 (axr1) mutant that lacks auxin sensitivity and shows an overall reduction in auxin responses (Lincoln et al. 1990; Leyser et al. 1993). Also similar to attpr mutants, sar3 mutants are early flowering. Yet another Arabidopsis mutant shows accumulation of poly(A)+ RNA in the nucleus. The atnup160-1 mutant was identified in a screen for mutations that impair cold-induced transcription of a reporter gene (Dong et al. 2006). The atnup160-1 mutation renders plants more sensitive to chilling stress. It encodes the Arabidopsis homolog of mammalian NUP160, and was also isolated as sar1 in a screen for suppressors of auxin resistance conferred by the axr1 mutation (Parry et al. 2006). The sar1 and sar3 mutants showed pleiotropic growth defects and nuclear accumulation of poly(A)+ RNA (Parry et al. 2006), like the atnup160-1 mutant (Dong et al. 2006). This result and nuclear rim localisation of NUP160/SAR1 (Dong et al. 2006) link this Arabidopsis NUP with the export of mRNA. However, not only mRNA export was affected. Both sar1 and sar3 mutations also affect the nucleo-cytoplasmic localisation of the transcriptional repressor AXR3/IAA17 (Indole Acetic Acid 17), most probably the nuclear import (Parry et al. 2006). This finding provides a very likely explanation for the suppression of the axr1 phenotype. Again, the flowering pathway was also affected since atnup160-1/sar1 mutants are early flowering.
Interestingly, in two more Arabidopsis mutants, export of poly(A)+ RNA from the nucleus is impaired. The cryophyte/los4-2 mutation was identified as a mutation that confers low expression of osmotically sensitive genes and, like mutations in the Arabidopsis NUP160/SAR1 gene, shows a defect in cold signalling. It confers cold and freezing tolerance to plants, but renders them more sensitive to heat stress (Gong et al. 2005). In addition, los4-2 mutants are hypersensitive to ABA, which is reminiscent to mutations in the ABH1/CBP80 gene. The mutation is allelic with the los4-1 mutation that was identified earlier (Gong et al. 2002). However, los4-1 mutants show an opposite phenotype since this mutation renders the plants more cold-sensitive. The los4-1 mutants show accumulation of poly(A)+ RNA at low and high temperatures, whereas in los4-2 mutants RNA export is impaired at warm and high temperatures, only (Gong et al. 2005). This suggests that los4-2 is a temperature-sensitive allele of the DEAD box RNA helicase that is encoded by this gene. The los4 phenotypes again show that temperature and phytohormone signalling are especially sensitive to impaired mRNA export. As discussed above, LOS4 is the most likely candidate for an Arabidopsis ortholog of yeast Dbp5p that is involved in remodelling of mRNPs and thus conferring directionality for mRNP export (Cole and Scarcelli 2006; Stewart 2007). Dbp5p is highly conserved between organisms, and, although Arabidopsis contains a multigene family encoding highly related DEAD box RNA helicases, LOS4 is most similar to yeast Dbp5p. In addition, Gong et al. (2005) demonstrated nuclear rim localisation of LOS4-GFP.
Accumulation of poly(A)+ RNA in the nucleus was also reported recently for plants that carry mutations in the AtNUP1 gene that encodes the Arabidopsis homolog of yeast Nup1p/vertebrate Nup153, and for mutants of the AtTHP1 gene encoding a yeast Thp1p homolog (Lu et al. 2010), a component of the yeast TREX-2 (transcription-coupled export) complex that is anchored to the NE via Nup1p (Köhler and Hurt 2007). thp1 mutants showed diverse developmental defects, including smaller plants, shorter roots, fewer lateral roots, and curly leaves. THP1-YFP fusion proteins were localised to the nucleus and to the nuclear rim in root cells. Surprisingly, mutants of other putative Arabidopsis TREX-2 components that were also isolated did not result in nuclear accumulation of poly(A)+ RNA (Lu et al. 2010). In addition to the defect in mRNA export, nup1 mutant plants revealed diverse developmental defects, including fewer rosette leaves and reduced fertility. There is evidence from one mutant nup1 line that NUP1 may be an essential gene. NUP1–YFP fusion proteins localised to the nuclear rim in root cells (Lu et al. 2010).
Silencing of Nicotiana benthamiana Rae1, a homolog of the metazoan Rae1 and yeast Gle2p RNA export factor, resulted in growth defects and abnormal leaf development (Lee et al. 2009). In addition, nuclear accumulation of poly(A)+ RNA was reported, and depletion of NbRae1 also led to reduced mitotic activities, disrupted mitotic spindle organisation and chromosomal missegregation. The protein was localised mainly at the nuclear rim during interphase and found associated with mitotic microtubules during mitosis (Lee et al. 2009). Rae1 is involved in the delivery of poly(A)+ RNA and TAP/NXF1 to the NPC by direct binding to Nup98 (Blevins et al. 2003). In addition, it is a microtubule-associated protein that is required for spindle assembly (Blower et al. 2005), and is an essential mitotic checkpoint regulator (Babu et al. 2003).
Mutations in the SAD2 gene define a novel plant nuclear import pathway
In a genetic screen for sad (super sensitive to ABA and drought) mutations, a mutation was isolated that harbours a T-DNA insertion in a gene encoding an importin beta-like protein (Verslues et al. 2006). SAD2 is one of the Arabidopsis homologs of mammalian RanBP7/8, and Arabidopsis, like humans, contains two genes that encode very closely related proteins (Fig. 1). SAD2 is expressed at low levels in all tissues except flowers in wild-type plants, and was not inducible by ABA treatment. The sad2 mutants showed higher levels of transcripts of several ABA- and stress-responsive genes, and are characterised by ABA hypersensitivity in seed germination and seedling growth (Verslues et al. 2006). A T-DNA insertion mutant of the closely related gene At2G31660 did not phenocopy the sad2 mutant, indicating different functions of these two genes in Arabidopsis. A SAD2–GFP fusion protein localised predominantly to the nucleus, however, a cytoplasmic localisation was clearly detectable as well (Verslues et al. 2006). Interestingly, sad2 abh1 double mutants showed additive effects in responses to ABA and were much more sensitive to drought and salt stress than any of the single mutants (Verslues et al. 2006). The sad2 mutants were more tolerant to UV-B radiation than wild-type plants (Zhao et al. 2007). They accumulated fewer cyclobutane pyrimidine dimers than wild-type, but wild-type and sad2 seedlings reacted equally when exposed to genotoxic stress. As a conclusion, SAD2 functions in UV-B protection rather than in DNA damage repair (Zhao et al. 2007). MYB4 is an R2R3-type MYB transcription factor that negatively regulates the Cinnamate-4-Hydroxylase (C4H) gene and thus the expression of sinapoyl esters and flavonoids that are discussed as UV protecting compounds (Jin et al. 2000; Bieza and Lois 2001). Transcription levels of MYB4 and C4H were significantly higher in sad2 than in wild-type seedlings, as was the level of UV-absorbing compounds (Zhao et al. 2007). MYB4 did not localise to the nucleus in sad2, and SAD2 and MYB4 were co-immunoprecipitated, suggesting that SAD2 is the nuclear import receptor for MYB4. MYB4 and its mRNA are part of a negative feedback loop that is altered in sad2 due to lack of nuclear localised MYB4 (Zhao et al. 2007). As a consequence, C4H (and MYB4) is expressed constitutively, leading to increased levels of UV-protecting pigments, thus explaining the sad2 phenotype (Zhao et al. 2007). The nuclear import signal on MYB4 that is recognised by SAD2 is not known. SAD2 is involved in yet another pathway (Gao et al. 2008). The sad2 mutants had fewer trichomes than wild-type plants, but there was no difference in trichome development or in the number and development of root hairs between sad2 and wild-type seedlings (Gao et al. 2008). In shoots of sad2 mutants, the expression levels of six transcription factors are de-regulated that are involved in Arabidopsis trichome initiation and development (Gao et al. 2008). The molecular basis of this de-regulation, however, remains unclear.
Nucleo-cytoplasmic transport and immune response
The SNC1 gene (Suppressor of npr1-1, Constitutive 1) is a resistance gene (R-gene) that encodes a protein of the Toll Interleukin 1 (TIR) receptor family (Li et al. 1999). Besides the N-terminal Toll Interleukin 1 receptor domain, the SNC1 protein contains a central nucleotide-binding site and a C-terminal leucine-rich repeat domain. These domains are shared among many R-proteins (Ellis et al. 2000; Meyers et al. 2003). The Arabidopsis gain-of-function mutant snc1 is characterised by constitutive expression of pathogenesis-related (PR) genes and shows constitutive pathogen resistance (Li et al. 2001; Zhang et al. 2003). Two genes are essential for the activation of downstream signalling in snc1, EDS1 (Enhanced Disease Susceptibility 1) and PAD4 (Phytoalexin Deficient 4) (Li et al. 2001; Zhang et al. 2003). Both genes encode lipase-like proteins that interact with each other (Feys et al. 2001). Other proteins are important regulators of R-gene function as well, but are not essential for signalling in snc1 (Zhang and Li 2005). In order to identify additional components required for snc1 signalling, a genetic screen for mutations that suppress the phenotypes of snc1 was performed. Several mutants were obtained and analysed in detail. Among them are modifier of snc1, 3 (mos3; Zhang and Li 2005), mos6 (Palma et al. 2005), and mos7 (Cheng et al. 2009). MOS3 and MOS7 encode NUPs, NUP96 and NUP88, respectively, whereas MOS6 encodes one of the Arabidopsis importin alpha import adapter proteins, Importin alpha 3 (IMPa-3). Interestingly, three mutant alleles of PAD4 were isolated in this screen as well (Zhang and Li 2005). The mos3 is allelic to sar3 mutants isolated in a genetic screen for suppressor mutations of the axr1 phenotype (Parry et al. 2006).
The mos mutants were analysed in the snc1 background. mos3 snc1 and mos6 snc1 double-mutant plants lacked PR gene expression that is high in snc1, mos7 snc1 plants showed a strong reduction of PR gene expression. Suppression of the elevated levels of salicylic acid in snc1 was verified in all double mutants, suppression of pathogen resistance was found in mos3 snc1 and in mos7 snc1, in mos6 snc1 the suppression was partial (Palma et al. 2005; Zhang and Li 2005; Cheng et al. 2009). Intracellular localisation of the different MOS proteins was assayed as GFP fusion proteins that were able to complement the phenotype when expressed in the respective double mutant. MOS3-GFP and MOS7-GFP localised to the nuclear rim, as expected for NUPs (Zhang and Li 2005; Cheng et al. 2009). MOS6-GFP showed a nuclear localisation (Palma et al. 2005). Importin alpha proteins show a predominant nuclear localisation at steady state (Smith et al. 1997; Haasen and Merkle 2002). The phenotypes of mos single mutants are of particular interest for their role in nucleo-cytoplasmic traffic. The mos3 and mos7 plants were compromised in basal and R-gene-mediated resistance, whereas mos6 single mutants showed enhanced susceptibility to a virulent oomycete pathogen but not to a bacterial pathogen (Palma et al. 2005; Zhang and Li 2005; Cheng et al. 2009). The mos7 plants were also analysed for possible defects in abiotic stress and phytohormone signalling, since sar3 and sar1 mutants of Arabidopsis NUP96 and NUP160 genes, respectively, showed altered responses to chilling stress and auxin (Dong et al. 2006; Parry et al. 2006). No altered responses to salt stress or altered ethylene or auxin responses were found in mos7-1 mutants (Cheng et al. 2009). However, mos7 null alleles are lethal, and no information was given for flowering time, responses to temperature stress, and possible accumulation of poly(A)+ RNA.
The mos phenotypes and the nature of the proteins encoded by the MOS genes indicate that nucleo-cytoplasmic partitioning of regulatory proteins plays an important role in disease resistance against pathogens. Since the MOS6 gene encodes IMPa-3, the most direct explanation for mos6 phenotypes is that nuclear import of (an) essential regulator(s) is necessary to trigger immune responses. Possible candidates are EDS1, PAD4, and NPR1 (Non-Expressor of PR Genes 1). NPR1 is required for the establishment of systemic acquired resistance (SAR) in Arabidopsis (Cao et al. 1994) and is involved in the regulation of specific binding of basic leucine zipper transcription factors to the promoter of the PR-1 gene (Johnson et al. 2003). NPR1 is localised in the cytoplasm in uninduced cells and is imported into the nucleus after induction by salicylic acid (Kinkema et al. 2000; Mou et al. 2003). However, analyses in mos7 mutants revealed additional aspects. Using a GFP-based nuclear export/import assay system (Haasen et al. 1999), Cheng et al. (2009) reported that NES-mediated nuclear export is generally enhanced in mos7-1 plants. At first sight, this is not easy to understand for plants impaired in NUP88 function. For comparison, in human cells, Nup88 and Nup214 are localised in a complex at the NPC, Nup214 provides binding sites for CRM1-containing export complexes, and both NUPs are essential for NES-mediated nuclear protein export, as shown by depletion of Nup214 (Hutten and Kehlenbach 2006). On the other hand, there are reports that, in Drosophila, mutations in the gene encoding Nup214 (Xylourgidis et al. 2006) or down-regulation of the Nup88 homolog leads to changes in the nucleo-cytoplasmic partitioning of selected proteins that may involve enhanced export (Uv et al. 2000; Roth et al. 2003). Irrespective of the question of whether nuclear import rates are attenuated or nuclear export rates are enhanced for specific cargoes in mos7-1, the importance of nucleo-cytoplasmic partitioning of regulatory proteins for plant innate immunity is further underlined by the findings that nuclear accumulation of snc1-GFP, NPR1-GFP, and EDS1 was reduced in mos7-1, as compared to the wild-type situation.
De-regulation of other components of the plant nuclear transport machinery
Down-regulation of the Arabidopsis transcripts encoding RanBP1c using antisense technology rendered roots of transgenic plants hypersensitive to auxin and caused altered auxin-induced root growth and development by arresting mitotic progress (Kim et al. 2001). An auxin hypersensitive phenotype was also reported in transgenic Arabidopsis plants over-expressing wheat RAN1 (Wang et al. 2006). Interestingly, the over-expression of rice RAN2 renders Arabidopsis hypersensitive to ABA, salinity and osmotic stress (Zang et al. 2010). The molecular basis of these results and their possible connection to nucleo-cytoplasmic traffic is not known. However, although these results are very difficult to interpret, the GTPase Ran is involved in several regulatory steps of mitosis in animals (reviewed in: Joseph 2006, Clarke and Zhang 2008), and auxin is essential for many aspects of plant development, including meristem activity, the initiation of primordia and the control of mitotic activity in plants (reviewed in: Perrot-Rechenmann 2010), to name a few. For these reasons, mitotic defects and de-regulation of auxin signalling could be expected among the phenotypes of plants with de-regulated expression of Ran and RanBP1 genes.
Bhattacharjee et al. (2008) reported that Arabidopsis Importin alpha isoform IMPa-4 is preferentially involved in Agrobacterium-mediated plant transformation. In an earlier report, Ballas and Citovsky (1997) showed that IMPa-1 specifically interacted with the Agrobacterium tumefaciens virulence protein VirD2. They thus claimed that IMPa-1 plays a pivotal role in Agrobacterium-mediated transformation. However, they could not show interaction of VirE2 with IMPa-1. Interestingly, Bhattacharjee et al. (2008) showed that only depletion of IMPa-4 rendered the mutant plants resistant to Agrobacterium-mediated transformation, although all tested members of the Arabidopsis Importin alpha family could interact with VirD2 and VirE2. Over-expression of six Arabidopsis Importin alpha isoforms including IMPa-4 rescued the transformation-resistant phenotype in the impa-4 mutant background. The findings of Bhattacharjee et al. (2008) suggest differential functionality for at least Importin alpha isoform IMPa-4. However, this functionality is not exclusive to IMPa-4 but is also found to a lesser extend in other Importin alpha family members. Hence differential function of IMPa-4 could only be detected under normal expression conditions, but not if other Importin alpha members were over-expressed.
Conclusions
In the past few years, the effects of mutations in genes that encode NUPs, NTRs, nuclear transport adapters, or other regulatory proteins of the nuclear transport machinery have been investigated in Arabidopsis thaliana and in other plants. In most cases, these mutations resulted in a range of morphological and developmental phenotypes. These pleiotropic phenotypes are not surprising given the fact that nuclear transport processes are elementary cellular activities that affect most, if not all, cellular pathways to different extents. Often, these phenotypes are not easy to interpret, but at the same time they offer a very interesting means to assess sensitivity of specific pathways to impaired nuclear transport processes. In some cases, mutations were identified in genetic screens and the resultant phenotypes were interpreted in a way that a given nuclear transport component would act specifically in the pathway under investigation, according to the design of the genetic screen. Such statements most certainly are over-interpretations, because other defects were not investigated and/or have not been identified under the given experimental conditions. An example for this point of view is the fact that several mutations in nuclear transport components have been identified in quite different genetic screens, like mutations in the genes encoding Exportin-T (Paused; Hunter et al. 2003; Li and Chen 2003), NUP96 (Zhang and Li 2005; Parry et al. 2006), or NUP160 (Dong et al. 2006; Parry et al. 2006). In addition, if mutations result in defects very early in development, defects in other pathways may not be discovered. As a consequence, investigations on defects in plants with impaired nucleo-cytoplasmic transport provide valuable experimental evidence for signalling pathways that include an essential nuclear translocation step. However, not all pathways may be discovered in this way.
It is intriguing to see that, as an overarching theme to effects of mutations in different nuclear transport components that are analysed to date, signalling of abiotic stress, ABA, auxin and flowering time control are very often affected. This may indicate that these pathways are especially sensitive to nucleo-cytoplasmic partitioning of signalling components and/or to impaired supply of such components. In humans, it is well established that impaired nucleo-cytoplasmic partitioning of signalling components may have important consequences for developmental pathways, including disease and oncogenesis. The cargoes that need to be translocated over the NE carry the specificity for proper function of signalling pathways. With the uncovering of more and more cargoes that show partitioning between the nucleus and the cytoplasm in the near future, signal transduction research in plants enters a novel and exciting era. In addition, it will be exciting to learn about more plant-specific aspects and components of nucleo-cytoplasmic transport.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Ach RA, Gruissem W. A small nuclear GTP-binding protein from tomato suppresses a Schizosaccharomyces pombe cell cycle mutant. Proc Natl Acad Sci USA. 1994;91:5863–5867. doi: 10.1073/pnas.91.13.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kD protein preferentially localized in the nucleus and at its periphery. J Cell Biol. 1989;108:1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador V, Monte E, Garcia-Martinez J-L, Prat S. Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell. 2001;106:343–354. doi: 10.1016/s0092-8674(01)00445-7. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Bork P. HEAT repeats in the Huntington’s disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O’Donoghue SI, Müller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A, Dasso M. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 2005;4:1161–1165. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R, Leung SW, Baker RP, Quimby BB, Corbett AH, Stewart M. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 2002;21:2843–2853. doi: 10.1093/emboj/cdf305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddow AL, Richards SA, Orem NR, Macara IG. The Ran/TC4 GTPase-binding domain: identification by expression cloning and characterization of a conserved sequence motif. Proc Natl Acad Sci USA. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky BA. Unexpected complexity of poly(A) binding protein gene families in flowering plants: three conserved lineages that are at least 200 million years old and possible auto- and cross-regulation. Genetics. 2003;163:311–319. doi: 10.1093/genetics/163.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky DA, Meagher RB. A pollen- ovule-, and early embryo-specific poly(A) binding protein from Arabidopsis complements essential functions in yeast. Plant Cell. 1996;8:1261–1275. doi: 10.1105/tpc.8.8.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R, van der Velde H, Fornerod M, Pickersgill H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol. 2004;24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra IC, Michaels SD, Schomburg FM, Amasino RM. Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 2004;40:112–119. doi: 10.1111/j.1365-313X.2004.02194.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Lee L-Y, Oltmanns H, Cao H, Veena Cuperus J, Gelvin SB. IMPa-4, and Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell. 2008;20:2661–2680. doi: 10.1105/tpc.108.060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieza K, Lois R. An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol. 2001;126:1105–1115. doi: 10.1104/pp.126.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Görlich D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain R, Boavida LC, McCormick S, Ow DW. Exportin 1 genes are essential for development and function of the gametophytes in Arabidopsis thaliana. Genetics. 2008;180:1493–1500. doi: 10.1534/genetics.108.094896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem. 2003;278:20979–20988. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Görlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman KM, Aukerman MJ, Park M-Y, Hunter C, Berardini TZ, Poethig RS. HASTY, the Arabidopsis orthologs of exportin 5/MSN5, regulates phase change and morphogenesis. Development. 2003;130:1493–1504. doi: 10.1242/dev.00362. [DOI] [PubMed] [Google Scholar]
- Bonfaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta 2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Aguilar-Henonin L, Olmedo G, Guzmán P. Four distinct classes of proteins as interaction partners of the PABC domain of Arabidopsis thaliana Poly(A)-binding proteins. Mol Genet Genomics. 2005;272:651–665. doi: 10.1007/s00438-004-1090-9. [DOI] [PubMed] [Google Scholar]
- Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhu AS, Wang XW. Loading and unloading: orchestrating centrosome duplication and spindle assembly by Ran/Crm1. Cell Cycle. 2005;4:1510–1514. doi: 10.4161/cc.4.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Treichel N, Müller E-C, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: Blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Belostotsky DA. Evidence that poly(A) binding protein has an evolutionary conserved function in facilitating mRNA biogenesis and export. RNA. 2003;9:1476–1490. doi: 10.1261/rna.5128903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Shaw RJ, Belostotsky DA. Analysis of an essential requirement for the poly(A)-binding protein function using cross-species complementation. Curr Biol. 2001;11:1207–1214. doi: 10.1016/s0960-9822(01)00371-2. [DOI] [PubMed] [Google Scholar]
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, Garcia AV, Wirthmueller L, Despres C, Parker JE, Zhang Y, Li X. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Cole CN, Scarcelli JJ. Transport of messenger RNA from the nucleus to the cytoplasm. Curr Opin Cell Biol. 2006;18:299–306. doi: 10.1016/j.ceb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Comella P, Pontvianne F, Lahmy S, Vignols F, Barbezier N, DeBures A, Jobet E, Brugidou E, Echeverria M, Saez-Vasquez J. Characterization of a ribonuclease III-like protein required for the cleavage of the pre-rRNA in the 3′ETS in Arabidopsis. Nucleic Acids Res. 2007;36:1163–1175. doi: 10.1093/nar/gkm1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Nuclear RNA export. J Cell Sci. 2003;116:587–597. doi: 10.1242/jcs.00268. [DOI] [PubMed] [Google Scholar]
- Dong A, Liu Z, Yu F, Li Z, Cao K, Shen WH. Interacting proteins and differences in nuclear transport reveal specific functions for the NAP1 family proteins in plants. Plant Physiol. 2005;138:1446–1456. doi: 10.1104/pp.105.060509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Hu X, Tang W, Zheng X, Kim YS, Lee BH, Zhu JK. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol Cell Biol. 2006;26:9533–9543. doi: 10.1128/MCB.01063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T. Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol. 2000;3:278–284. doi: 10.1016/s1369-5266(00)00080-7. [DOI] [PubMed] [Google Scholar]
- Englmeier L, Fornerod M, Bischoff FR, Petosa C, Mattaj IW, Kutay U. RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBP Rep. 2001;2:926–932. doi: 10.1093/embo-reports/kve200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Frey S, Görlich D. FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 2009;28:2554–2567. doi: 10.1038/emboj.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Richter RP, Görlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- Fribourg S, Conti E. Structural similarity in the absence of sequence homology of the messenger RNA export factors Mtr2 and p15. EMBO Rep. 2003;4:699–703. doi: 10.1038/sj.embor.embor883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S, Braun IC, Izaurralde E, Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export receptor. Mol Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, Chen H, Shibagaki N, Ferl RJ, Ehrhardt D, Chong K, Burlingame AL, Wang ZY. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Gong X, Cao W, Zhao J, Fu L, Wang X, Schumaker KS, Guo Y. SAD2 in Arabidopsis functions in trichome initiation through mediating GL3 function and regulating GL1, TTG1 and GL2 expression. J Integr Plant Biol. 2008;50:906–917. doi: 10.1111/j.1744-7909.2008.00695.x. [DOI] [PubMed] [Google Scholar]
- Gil P, Kircher S, Adam E, Bury E, Kozma-Bognar L, Schäfer E, Nagy F. Photocontrol of subcellular partitioning of phytochrome-B:GFP fusion protein in tobacco seedlings. Plant J. 2000;22:135–145. doi: 10.1046/j.1365-313x.2000.00730.x. [DOI] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA. 2002;99:11507–11512. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005;17:256–267. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C, Güttler T, Engelen E, Demmers J, Formerod M, Grosveld FG, Tibboel D, Görlich D, Poot RA, Rottier RJ. Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J Cell Biol. 2009;185:27–34. doi: 10.1083/jcb.200810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser M, Lentz A, Lichota J, Merkle T, Grasser KD. The Arabidopsis genome encodes structurally and functionally diverse HMGB-type proteins. J Mol Biol. 2006;358:654–664. doi: 10.1016/j.jmb.2006.02.068. [DOI] [PubMed] [Google Scholar]
- Guo H, Duong H, Ma N, Lin C. The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J. 1999;19:279–287. doi: 10.1046/j.1365-313x.1999.00525.x. [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Ossareh-Nazari B, Brownawell AM, Doglio A, Bertrand E, Macara IG, Dargemont C. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J Biol Chem. 2003;278:5505–5508. doi: 10.1074/jbc.C200668200. [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Ossareh-Nazari B, Brownawell AM, Evers S, Macara IG, Dargemont C. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem. 2004;279:884–891. doi: 10.1074/jbc.M306808200. [DOI] [PubMed] [Google Scholar]
- Haasen D, Merkle T. Characterization of an Arabidopsis homologue of the nuclear export receptor CAS by its interaction with Importin alpha. Plant Biol. 2002;4:432–439. [Google Scholar]
- Haasen D, Köhler C, Neuhaus G, Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999;20:695–705. doi: 10.1046/j.1365-313x.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- Haizel T, Merkle T, Pay A, Fejes E, Nagy F. Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 1997;11:93–103. doi: 10.1046/j.1365-313x.1997.11010093.x. [DOI] [PubMed] [Google Scholar]
- Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress trasncription factor HsfA2. Mol Cell Biol. 2001;21:1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Künzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A, Viczian A, Bury E, Tscheuschler A, Kircher S, Toth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol. 2005;15:2125–2130. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Tscheuschler A, Viczian A, Kunkel T, Kircher S, Schäfer E. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47:1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Lührmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Murata Y, Young JJ, Kwak JM, Mackesy DZ, Schroeder JI. Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol. 2002;130:1276–1287. doi: 10.1104/pp.009480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CA, Aukerman MJ, Sun H, Fokina M, Poethig RS. PAUSED encodes the Arabidopsis Exportin-t orthologs. Plant Physiol. 2003;132:2135–2143. doi: 10.1104/pp.103.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Kehlenbach RH. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol Cell Biol. 2006;26:6772–6785. doi: 10.1128/MCB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Flotho A, Melchior F, Kehlenbach RH. The Nup358–-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol Biol Cell. 2008;19:2300–2310. doi: 10.1091/mbc.E07-12-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell. 2001;13:2483–2497. doi: 10.1105/tpc.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Lee H, Stevenson B, Zhu J-K. HOS1, a genetic locus involved in cold responsive gene expression in Arabidopsis thaliana. Plant Cell. 1998;10:1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Mongkolsiriwatana C, Veley KM, Kim SY, Michaels SD. The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 2007;144:1383–1890. doi: 10.1104/pp.107.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby MJ, Weinl C, Pusch S, Kuijt SJH, Merkle T, Dissmeyer N, Schnittger A. Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol. 2006;141:1293–1305. doi: 10.1104/pp.106.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Joseph J, Dasso M, Meier I. Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J. 2005;42:270–282. doi: 10.1111/j.1365-313X.2005.02368.x. [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Imamoto N, Matsuki R, Yoneda Y, Yamamoto N. In vitro characterization of rice importin beta 1: molecular interaction with nuclear transport factors and mediation of nuclear protein import. FEBS Lett. 1998;437:127–130. doi: 10.1016/s0014-5793(98)01207-1. [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell. 2003;15:1846–1858. doi: 10.1105/tpc.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Joseph J. Ran at a glance. J Cell Sci. 2006;119:3481–3484. doi: 10.1242/jcs.03071. [DOI] [PubMed] [Google Scholar]
- Kaffman A, O′Shea EK. Regulation of nuclear localisation: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a RanGTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kaminaka H, Näke C, Epple P, Dittgen J, Schütze K, Chaban C, Holt BF, III, Merkle T, Schäfer E, Harter K, Dangl JL. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006;25:4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, Jensen TH, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA. 2006;103:359–364. doi: 10.1073/pnas.0508883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Straesser K, Saiwaki T, Yoneda Y, Hurt E. Complex formation between TAP and p15 affects binding to FG-repeat nucleoporins and nucleocytoplasmic shuttling. J Biol Chem. 2002;277:9242–9246. doi: 10.1074/jbc.M110007200. [DOI] [PubMed] [Google Scholar]
- Kim SH, Roux SJ. An Arabidopsis Ran-binding protein, AtRanBP1c, is a co-activator of Ran GTPase-activating protein and requires the C-terminus for its cytoplasmic localization. Planta. 2003;216:1047–1052. doi: 10.1007/s00425-002-0959-2. [DOI] [PubMed] [Google Scholar]
- Kim L, Kircher S, Toth R, Adam E, Schäfer E, Nagy F. Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 2000;22:125–134. doi: 10.1046/j.1365-313x.2000.00729.x. [DOI] [PubMed] [Google Scholar]
- Kim SH, Arnold D, Lloyd A, Roux SJ. Antisense expression of an Arabidopsis Ran binding protein renders transgenic roots hypersensitive to auxin and alters auxin-induced root growth and development by arresting mitotic progress. Plant Cell. 2001;13:2619–2630. doi: 10.1105/tpc.010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12:2339–2350. doi: 10.1105/tpc.12.12.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schäfer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome-A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Wellmer F, Nick P, Rugner A, Schäfer E, Harter K. Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J Cell Biol. 1999;144:201–211. doi: 10.1083/jcb.144.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner O, Kircher S, Harter K, Batschauer A. Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J. 1999;19:289–296. doi: 10.1046/j.1365-313x.1999.00535.x. [DOI] [PubMed] [Google Scholar]
- Kmieciak M, Simpson CG, Lewandowska D, Brown JW, Jarmolowski A. Cloning and characterization of two subunits of Arabidopsis thaliana nuclear cap-binding complex. Gene. 2002;283:171–183. doi: 10.1016/s0378-1119(01)00859-9. [DOI] [PubMed] [Google Scholar]
- Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Köster M, Frahm T, Hauser H. Nucleocytoplasmic shuttling revealed by FRAP and FLIP technologies. Curr Opin Biotechnol. 2005;16:28–34. doi: 10.1016/j.copbio.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kühn U, Wahle E. Structure and function of poly(A)-binding proteins. Biochim Biophys Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu J-K. The ArabidopsisHOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Lee HS, Wi SJ, Park KY, Schmit AC, Pai HS. Dual functions of Nicotiana benthamiana Rae1 in interphase and mitosis. Plant J. 2009;59:278–291. doi: 10.1111/j.1365-313X.2009.03869.x. [DOI] [PubMed] [Google Scholar]
- Levesque L, Bor Y-C, Matzat LH, Jin L, Berberoglu S, Rekosh D, Hammarskjöld M-L, Paschal MB. Mutations in TAP uncouple RNA export activity from translocation through the nuclear pore complex. Mol Biol Cell. 2006;17:931–943. doi: 10.1091/mbc.E04-07-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Li J, Chen X. PAUSED, a putative Exportin-t, acts pleiotropically in Arabidopsis development but is dispensable for viability. Plant Physiol. 2003;132:1913–1924. doi: 10.1104/pp.103.023291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang Y, Clarke JD, Li Y, Dong X. Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell. 1999;98:329–339. doi: 10.1016/s0092-8674(00)81962-5. [DOI] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang Y, Dong X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- Ligterink W, Kroj T, zur Nieden U, Hirt H, Scheel D. Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science. 1997;276:2054–2057. doi: 10.1126/science.276.5321.2054. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Holaska JM, Welch K, Paschal BM, Macara IG. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol. 2001;153:1391–1402. doi: 10.1083/jcb.153.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Plafker K, Smith AE, Clurman BE, Macara IG. Npap60/Nup50 is a tri-stable switch that stimulates Importin-alpha:beta-mediated nuclear protein import. Cell. 2002;110:349–360. doi: 10.1016/s0092-8674(02)00836-x. [DOI] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Görlich D. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 2000;19:4362–4371. doi: 10.1093/emboj/19.16.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Xurong T, Gang T, Fang W, Kede L, Vi N, Kohalmi SE, Keller WA, Tsang EWT, Harada JJ, Rothstein SJ, Cui Y. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010;61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini J, Schmied K, Palmisano R, Toensing K, Anselmetti D, Merkle T. Multifocal two-photon laser scanning microscopy combined with photo-activatable GFP for in vivo monitoring of intracellular protein dynamics in real time. J Struct Biol. 2007;158:401–409. doi: 10.1016/j.jsb.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Mason DA, Stage DE, Goldfarb DS. Evolution of the metazoan-specific importin alpha gene family. J Mol Evol. 2009;68:351–365. doi: 10.1007/s00239-009-9215-8. [DOI] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24:3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Brkljacic J. Adding pieces to the puzzling plant nuclear envelope. Curr Opin Plant Biol. 2009;12:752–759. doi: 10.1016/j.pbi.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Merkle T. Nuclear import and export of proteins in plants: a tool for the regulation of signalling. Planta. 2001;213:499–517. doi: 10.1007/s004250100621. [DOI] [PubMed] [Google Scholar]
- Merkle T. Nucleo-cytoplasmic partitioning in plants: implications for the regulation of environmental and developmental signalling. Curr Genet. 2003;44:231–260. doi: 10.1007/s00294-003-0444-x. [DOI] [PubMed] [Google Scholar]
- Merkle T, Haizel T, Matsumoto T, Harter K, Dallmann G, Nagy F. Phenotype of the fission yeast cell cycle regulatory mutant pim1–46 is suppressed by a tobacco cDNA encoding a small, Ran-like GTP-binding protein. Plant J. 1994;6:555–565. doi: 10.1046/j.1365-313x.1994.6040555.x. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot J-M, Kostka S, Kraft R, Hartmann E, Görlich D. Importin 13: a novel mediator of nuclear import and nuclear export. EMBO J. 2001;20:3685–3694. doi: 10.1093/emboj/20.14.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Mugdil Y, Shiu S-H, Stone SL, Salt JN, Goring DR. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004;134:59–66. doi: 10.1104/pp.103.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr Biol. 1997;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- Nemergut ME, Lindsay ME, Brownawell AM, Macara IG. Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J Biol Chem. 2002;277:17385–17388. doi: 10.1074/jbc.C100620200. [DOI] [PubMed] [Google Scholar]
- Neuber A, Franke J, Wittstruck A, Schlenstedt G, Sommer T, Stade K. Nuclear export receptor Xpo1/Crm1 is physically and functionally linked to the spindle pole body in budding yeast. Mol Cell Biol. 2008;28:5348–5358. doi: 10.1128/MCB.02043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachlerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Belostotsky DA, Meagher RB. Arabidopsis thaliana poly (A) binding protein 2 (PAB2) functions in yeast translational and mRNA decay processes. Plant J. 2000;22:187–198. doi: 10.1046/j.1365-313x.2000.00721.x. [DOI] [PubMed] [Google Scholar]
- Palma K, Zhang Y, Li X. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol. 2005;15:1129–1135. doi: 10.1016/j.cub.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJM. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol Biol. 2004;55(5):679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol Biol. 2004;55:679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmann E, Lührmann R, Görlich D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M-Y, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell. 2006;18:1590–1603. doi: 10.1105/tpc.106.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pay A, Resch K, Frohnmeyer H, Fejes E, Nagy F, Nick P. Plant RanGAPs are localised at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J. 2002;30:699–709. doi: 10.1046/j.1365-313x.2002.01324.x. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JWS, Shaw PJ. Poteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell. 2005;16:260–269. doi: 10.1091/mbc.E04-09-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol. 2010;2:1–10. doi: 10.1101/cshperspect.a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- Pictures in Cell Biology Structures of nuclear transport components. Trends Cell Biol. 1999;9:310–311. doi: 10.1016/s0962-8924(99)01610-4. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J. 2010;62:473–482. doi: 10.1111/j.1365-313X.2010.04165.x. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Görlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Kutay U, Paraskeva E, Görlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr Biol. 1999;9:47–50. doi: 10.1016/s0960-9822(99)80046-3. [DOI] [PubMed] [Google Scholar]
- Richards SA, Llounsbury KM, Carey KL, Macara IG. A nuclear export signal is essential for the cytosolic localization of Ran binding protein RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Stutz F. Nuclear export of RNA. Biol Cell. 2004;96:639–655. doi: 10.1016/j.biolcel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA. 2001;98:15377–15382. doi: 10.1073/pnas.261459698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth P, Xylourgidis N, Sabri N, Uv AE, Fornerod M, Samakovlis C. The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J Cell Sci. 2003;163:701–706. doi: 10.1083/jcb.200304046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yoshikawa M, Yano K, Miwa H, Uchida H, Asamizu E, Sato S, Tabata S, Imaizumi-Anraku H, Umehara Y, Kouchi H, Murooka Y, Szczyglowski K, Downie JA, Parniske M, Hayashi M, Kawaguchi M. NUCLEOPORIN 85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell. 2007;19:610–624. doi: 10.1105/tpc.106.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Pu R, Cavenagh M, Dasso M. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc Natl Acad Sci USA. 1997;94:3736–3741. doi: 10.1073/pnas.94.8.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM, Hicks GR, Raikhel NV. Importin alpha from Arabidopsis thaliana is a nuclear import receptor that recognizes three classes of import signals. Plant Physiol. 1997;114:411–417. doi: 10.1104/pp.114.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Brownawell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- Sommer P, Nehrbass U. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr Opin Cell Biol. 2005;17:294–301. doi: 10.1016/j.ceb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Stacey MG, Hicks SN, von Arnim AG. Discrete domains mediate the light-responsive nuclear and cytoplasmic localisation of Arabidopsis COP1. Plant Cell. 1999;11:349–363. doi: 10.1105/tpc.11.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Kopp OR, Kim TH, von Arnim AG. Modular domain structure of Arabidopsis COP1. Reconstitution of activity by fragment complementation and mutational analysis of a nuclear localisation signal in planta. Plant Physiol. 2000;124:979–989. doi: 10.1104/pp.124.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell. 2003;15:885–898. doi: 10.1105/tpc.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Izaurralde E. The interplay of nuclear mRNP assembl, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo Y, Miernyk JA. Regulation of nucleocytoplasmic localization of the AtDjC6 chaperone protein. Protoplasma. 2004;224:79–89. doi: 10.1007/s00709-004-0050-x. [DOI] [PubMed] [Google Scholar]
- Suyama M, Doerks T, Braun IC, Sattler M, Izaurralde E, Bork P. Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep. 2000;1:53–58. doi: 10.1093/embo-reports/kvd009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Poethig RS. HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development. 1998;125:1889–1898. doi: 10.1242/dev.125.10.1889. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Bertekap RL, Cashmore AR. Intracellular localization of GBF proteins and blue light-induced import of GBF2 fusion proteins into the nucleus of cultured Arabidopsis and soybean cells. Plant J. 1997;11:967–982. doi: 10.1046/j.1365-313x.1997.11050967.x. [DOI] [PubMed] [Google Scholar]
- Thomas F, Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- Tillemans V, Leponce I, Rausin G, Dispa L, Motte P. Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell. 2006;18:3218–3234. doi: 10.1105/tpc.106.044529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22:2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin P, Ossareh-Nazari B, Dargemont C. Nuclear transport and transcriptional regulation. FEBS Lett. 1999;452:82–86. doi: 10.1016/s0014-5793(99)00533-5. [DOI] [PubMed] [Google Scholar]
- Uhrig JF, Canto T, Marshall D, MacFarlane SA. Relocalization of nuclear ALY proteins to the cytoplasm by the tomato Bushy stunt virus p19 pathogenicity protein. Plant Phys. 2004;135:2411–2423. doi: 10.1104/pp.104.046086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uv AE, Roth P, Xylourgidis N, Wickberg A, Cantera R, Samakovlis C. Members only encodes a Drosophila nucleoporin required for rel protein import and immune response activation. Genes Dev. 2000;14:1945–1957. [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 1997;13:227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- Verslues PE, Guo Y, Dong CH, Ma W, Zhu JK. Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant J. 2006;47:776–787. doi: 10.1111/j.1365-313X.2006.02833.x. [DOI] [PubMed] [Google Scholar]
- Vinciguerra P, Stutz F. mRNA export: an assembly line from genes to nuclear pores. Curr Opin Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell type specific modulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Osterlund MT, Kwok SF, Deng X-W. Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 1997;114:779–788. doi: 10.1104/pp.114.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xu Y, Han Y, Bao S, Du J, Yuan M, Xu Z, Chong K. Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol. 2006;140:91–101. doi: 10.1104/pp.105.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Coburn GA, Zeng Y, Kang Y, Bogerd HP, Cullen BR. Formation of TAP/NXT1 heterodimers activates TAP-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol Cell Biol. 2002;22:245–256. doi: 10.1128/MCB.22.1.245-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MX, Meier I. The nuclear pore comes to the fore. Trends Plant Sci. 2008;13:20–27. doi: 10.1016/j.tplants.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol. 2007;17:1157–1163. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19:1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Zhao Q, Rodrigo-Peiris T, Brkljacic J, He CS, Muller S, Meier I. RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc Natl Acad Sci USA. 2008;105:18637–18642. doi: 10.1073/pnas.0806157105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C. The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFkappaB activation in Drosophila. J Cell Sci. 2006;119:4409–4419. doi: 10.1242/jcs.03201. [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR. The C termini of Arabidopsis cryptochromes mediate a constititive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- Yao C, Ge L, Li W, Zhao B, Li C, Ruan K, Lin H, Lin Y. Expression and characterization of rice putative PAUSED gene. Acta Biochim Biophys Sin. 2008;40:893–900. [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang A, Xu X, Neill S, Cai W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J Exp Bot. 2010;61:777–789. doi: 10.1093/jxb/erp341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M, Zhou Q, Sarda X, Yau CP, Chua NH. The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J. 2004;40:355–365. doi: 10.1111/j.1365-313X.2004.02212.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li X. A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Leung S, Corbett AH, Meier I. Identification and characterization of the Arabidopsis orthologs of Nuclear Transport Factor 2, the nuclear import factor of Ran. Plant Phys. 2006;140:869–878. doi: 10.1104/pp.105.075499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y. SAD2, an importin-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell. 2007;19:3805–3818. doi: 10.1105/tpc.106.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Brkljacic J, Meier I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell. 2008;20:1639–1651. doi: 10.1105/tpc.108.059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemienowicz A, Haasen D, Staiger D, Merkle T. Arabidopsis Transportin is the nuclear import receptor for the circadian clock-regulated RNA-binding protein AtGRP7. Plant Mol Biol. 2003;53:201–212. doi: 10.1023/B:PLAN.0000009288.46713.1f. [DOI] [PubMed] [Google Scholar]
- Zolotukhin AS, Felber BK. Mutations in the nuclear export signal of human Ran-binding protein RanBP1 block the Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Biol Chem. 1997;272:11356–11360. doi: 10.1074/jbc.272.17.11356. [DOI] [PubMed] [Google Scholar]


