Abstract
Helicobacter pylori is the etiological agent of diseases such as gastritis, gastric and duodenal ulcers, and two types of gastric cancers. While some insight has been gained into the etiology of these diverse manifestations, by and large, the reason that some individuals develop more severe disease remains elusive. Recent studies have focused on the roles of H. pylori toxins CagA and VacA on the disease process and have suggested that both toxins are intimately involved. Moreover, CagA and VacA are polymorphic within different H. pylori strains, and particular polymorphisms seem to show a correlation with the development of particular disease states. Among VacA polymorphisms, the intermediate region has recently been proposed to play a major role in disease outcome. In this article, we describe a detailed sequence analysis of the polymorphic intermediate region of vacA from strains obtained from a large South Korean population. We show that polymorphisms found at amino acid position 196 are associated with more severe disease manifestations. Additionally, polymorphisms found at amino acid position 231 are linked to disease in strains that carry the non-EPIYA-ABD allele of CagA. Collectively, these data help explain the impact of the VacA intermediate region on disease and lead to the hypothesis that there are allele-driven interactions between VacA and CagA.
The medically important microbe Helicobacter pylori colonizes the inhospitable niche of the gastric mucosa of over 50% of the world's population (34, 54). H. pylori is a spiral-shaped, microaerophilic, Gram-negative bacterium (33) that is the etiological agent of a multitude of diseases, including gastritis, peptic ulcers (both duodenal and gastric ulcers), as well as adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma (6, 12, 17, 18, 45). This class I carcinogen contributes to gastric cancer mortality, which is still one of the most common causes of mortality due to cancer (18, 42). This is especially true in East Asian countries, such as China, South Korea, and Japan (23).
Two H. pylori toxins that facilitate host cellular damage and directly interplay with the host immune system are the cytotoxin-associated gene A (CagA) and the vacuolating cytotoxin (VacA). A growing number of studies have begun to suggest that VacA and CagA interact in such a way as to affect disease severity (2, 27, 59, 62). VacA was identified within a few years of the discovery of H. pylori and appears to be produced and secreted by virtually all strains (4, 13). This toxin was initially identified and named for its ability to cause large cytoplasmic vacuoles in intoxicated host cells (14). However, VacA has subsequently been shown to have a multitude of functions. For instance, when inserted into the plasma membrane, VacA can act as an anion-selective channel (52), which may aid bacterial survival through leakage of host cytosolic anions that can be utilized by the bacterium (40). VacA also has the ability to induce apoptosis through permeabilization of the mitochondrial membrane, thereby causing cytochrome c release (32, 60). Furthermore, VacA can induce the autophagy pathway (53), disorganize the host cell cytoskeleton to cause spreading of host cells, inhibit T cell activation, and block T cell and B cell proliferation (20, 56).
CagA is directly injected into host cells through a type IV secretion apparatus (10) and is phosphorylated within host cells by host cell kinases. This phosphorylation event subsequently makes CagA competent for interaction with the Src homology 2 domain-containing protein tyrosine phosphatase (SHP-2) (23, 26). The downstream effects of this interaction include alterations in numerous host signaling pathways (22, 24-26, 41, 48, 58), which are believed to be responsible for the increased cancer risk associated with infection by strains that express CagA (7, 21). Phosphorylation of CagA occurs in the carboxy terminus of the protein at conserved tyrosine residues that exist as part of a repeated 5-amino-acid sequence (Glu-Pro-Ile-Tyr-Ala) referred to as the EPIYA repeat (25, 26). The numbers of these EPIYA repeats and the flanking amino acid regions surrounding these repeats vary dramatically across strains. On the basis of flanking sequences, four distinct EPIYA motifs have been identified (EPIYA-A, -B, -C, and -D) (25, 61). Strains that carry various combinations of these motifs can be divided into two main geographical distributions, which are hallmarked by differences in the primary phosphorylation sites, EPIYA-C and -D (25); East Asian strains contain EPIYA-ABD, whereas Western strains contain EPIYA-ABC and the EPIYA-C motif may be repeated up to five times (2, 25, 26, 43). These different EPIYA combinations have been suggested to impact disease progression (29, 61).
Similar to the polymorphic nature of CagA, VacA contains three distinct segments that exhibit variation starting within the amino terminus. These areas of variation are broadly defined as the signal (s), intermediate (i), and middle (m) regions, and two or more primary variants have been described for each region: s1 and s2 for the signal region, i1, i2, and i3 for the intermediate region, and m1 and m2 for the middle region (3, 11, 47). Various combinations of each s, i, and m region are then combined within each H. pylori strain to yield a particular vacA allele. The s region of VacA appears to influence the efficiency of anion channel formation based on the hydrophobicity of amino acid residues that are found near a proteolytic cleavage site found in this region (35, 46); the s1 form contains a hydrophobic region adjacent to the proteolytic cleavage site that increases membrane insertion and formation of membrane channels (30, 35). The m region affects host cell tropism (28); VacA toxins with the m1 region are toxic to a broader range of host cells (1, 44). The i region is positioned between the s and m regions and is the most recent region to be described. The i1 variants of VacA have been shown to have stronger vacuolating activity than toxins containing the i2 regions (47). Due to the increased anion channel formation capability, broader cell tropism, and enhanced vacuolating activity, individual associations between the s1, m1, and i1 types and more-severe forms of H. pylori-induced disease have been identified (5, 47). Furthermore, several studies have linked strains carrying the s1-m1 allele of the toxin to more-severe disease outcomes, since these strains show the strongest vacuolating activity to the broadest range of cells (31). However, recently, the i region has been suggested to be a better predictor of disease severity than either the s or m region, though the i region appears to covary with the s and m regions (47). This means that the more toxic i1 region is often associated with s1-m1 (47).
Within the i region, three specific clusters have been identified as the main areas to contain polymorphisms, clusters A, B, and C. Of these three clusters, clusters B and C have been shown to impact the vacuolating activity of the toxin (47). Because of this link to toxin activity, researchers have sought to determine the roles of natural individual amino acid changes within these clusters in the ultimate outcome of disease. For example, variation in the ninth amino acid in cluster B (amino acid 231 of the protoxin) has been linked to disease development in the Taiwanese population (50). However, this amino acid appeared to have no impact in the South Korean population (27). Recently, we identified two additional positions in the VacA intermediate region that contained polymorphisms, positions 151 and 196 (27). While neither of these amino acids showed a statistically significant link to disease severity in the small population of samples that were examined, the distribution of amino acids found at position 196 displayed a trend toward significance (27). Given this, we sequenced the vacA intermediate region from 231 Korean isolates and then analyzed the distribution of polymorphisms across the entire region. Furthermore, we compared these polymorphic vacA distributions to the various cagA alleles carried by each strain as well as to the ultimate disease development. In this article, we present an expanded i1 and i2 consensus sequence, show that amino acid 196 impacts disease development, and show that amino acid 231 is important for disease development, but only within strains that carry a non-EPIYA-ABD CagA allele.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
This South Korean population of 260 strains has been described previously and includes 115 isolates from patients diagnosed with gastritis, 60 isolates from patients diagnosed with gastric ulcer, 55 isolates from patients diagnosed with duodenal ulcer, and 30 isolates from patients diagnosed with gastric cancer (27, 29). Isolates were preserved as stocks at −80°C and then grown and expanded on antibiotic-supplemented horse blood agar plates under microaerophilic conditions created by an Anoxomat evacuation/replacement system (Spiral Biotech, Norwood, MA) exactly as previously described (9, 29).
vacA i-region sequencing.
Chromosomal DNA from each of the 260 H. pylori strains was isolated using the Easy-DNA kit (Invitrogen, Carlsbad, CA). The vacA intermediate region was amplified and then Sanger dideoxy sequenced using the primers previously described by Rhead et al. (47), VacF1 (3′-GTTGGGATTGGGGGAATGCCG-5′) and VacR9 (3′-TGTTTATCGTGCTGTATGAAGG-5′). Sanger dideoxy sequencing was performed at both the Uniformed Services University of the Health Science Biomedical Instrumentation Center (Bethesda, MD) and Cosmo Genetech Co., Ltd. (Seoul, South Korea). The resulting DNA sequences were analyzed using Vector NTI version 9.1 (Invitrogen, Carlsbad, CA) and Sequencher 4.5 (Gene Codes Corp., Ann Arbor, MI). The amino acid numbering system used in this study is based on the VacA sequence of strain G27; the numbering begins at the translational start such that amino acid 1 is the first methionine of the translated protein.
Statistical analysis.
The Fisher exact test was used to analyze the association between the vacA allele, disease state, cagA allele, and specific amino acids within the intermediate region. Log linear modeling was used to assess higher-order associations that were significant at the 5% level. We fit a saturated model using categorical variables representing vacA genotype, cagA genotype, disease state, gender, and amino acids within the i region using a backward selection algorithm, which eliminates the least significant association at each step and then reforms the model to look for associations. Data were analyzed using SPSS version 16 software (SPSS Inc., Chicago, IL) or SAS version 9.1 software (SAS Institute Inc., Cary, NC).
Nucleotide sequence accession numbers.
The sequences for the i region of vacA from the 60 original strains analyzed (27) were previously deposited in GenBank under accession numbers GQ338184 to GQ338243, and the i-region sequences of the additional 171 strains have been deposited in GenBank under accession numbers HM047564 to HM047592 and HM047594 to HM047735 (see Table S1 in the supplemental material).
RESULTS
Sample acquisition and vacA i-region sequencing.
The strains used in this study have been previously characterized for distribution of both the cagA allele (27, 29) and the vacA allele (27). However, the previous study characterizing the vacA allele relied primarily on PCR-based typing methods and analyzed only the vacA intermediate region sequence from a subset of strains carrying the i1 allele, whereas in the current study, a detailed analysis of vacA sequences from the complete collection of South Korean strains was performed. The complete collection of 260 strains contained 254 strains for which we had complete epidemiological data. These 254 strains were obtained from patients with a mean age of 51 years and an age range of 14 to 86 years. These strains were evenly distributed by gender; 126 strains were from female patients with a mean age of 53 years and an age range of 21 to 86 years, and 128 strains were from male patients with a mean age of 50 years and an age range of 14 to 82 years. These strains were distributed across various H. pylori-induced disease states: 45% were from patients diagnosed with gastritis, 22% were patients diagnosed with gastric ulcers, 21% patients diagnosed with duodenal ulcers, and 12% patients diagnosed with gastric cancer (29).
Of the 260 strains in this entire collection, the vacA i regions in 231 strains were successfully sequenced, and we had complete epidemiological data and CagA and VacA genotypes for 222 of these strains (Fig. 1 and Table 1; see Table S1 in the supplemental material). These sequenced regions were from strains from patients with an age range of 14 to 86 years and a mean age of 50.3 years. Of the 222 isolates, 112 were from male patients aged 14 to 82 years with a mean age of 48.9 years, and 110 were from female patients aged 21 to 85 years with a mean age of 51.7 years. The sequenced strain distribution across disease states was similar to that of the overall population; 45.5% were from patients with gastritis, 41% were from patients suffering from ulcers (22.1% duodenal ulcers and 18.9% gastric ulcers), and 13.5% were from patients with cancer.
FIG. 1.
WebLogo image showing the VacA intermediate region's major polymorphic domains within the South Korean population. The WebLogo image was created by using the amino acid sequence from each of the three different intermediate alleles (220i1 sequences, 8 i2 sequences, and 3 i3 sequences) with the WebLogo generator as previously described (15, 49). The three primary regions of polymorphism (clusters A, B, and C) and the two amino acids shown to impact disease development (at positions 196 and 231) are indicated. The logo represents the alignment at each position by a stack of letters, where the height of each letter is proportional to the observed frequency of the corresponding amino acid, and the overall height of each stack is proportional to the sequence conservation, measured in bits, at that position (15).
TABLE 1.
Distribution of cagA and vacA alleles in patients with different diseases
| Allele and patient gender | Parameter | Value for parameter for patientsa |
||||
|---|---|---|---|---|---|---|
| All | Gastritis | Duodenal ulcers | Gastric ulcers | Gastric cancer | ||
| Overall | ||||||
| All patients | No. of patients | 222 | 101 | 49 | 42 | 30 |
| Age range (yr) | 14-86 | 19-82 | 14-72 | 34-84 | 37-86 | |
| Mean age (yr) | 50.3 | 48.3 | 45.1 | 55.1 | 56.6 | |
| Male | No. of patients | 112 | 34 | 30 | 32 | 16 |
| Age range (yr) | 14-82 | 19-76 | 14-70 | 34-82 | 38-70 | |
| Mean age (yr) | 48.9 | 46.6 | 41.3 | 54.5 | 55.1 | |
| Female | No. of patients | 110 | 67 | 19 | 10 | 14 |
| Age range (yr) | 21-86 | 21-82 | 31-72 | 46-84 | 37-86 | |
| Mean age (yr) | 51.7 | 49.2 | 51.1 | 57.1 | 61 | |
| cagA | ||||||
| EPIYA-ABD | ||||||
| All patients | No. of patients | 189 | 81 | 40 | 38 | 30 |
| Age range (yr) | 14-86 | 19-78 | 14-72 | 34-84 | 37-86 | |
| Mean age (yr) | 50.5 | 48.9 | 44.5 | 54.5 | 57.9 | |
| Male | No. of patients | 100 | 25 | 29 | 30 | 16 |
| Age range (yr) | 14-82 | 19-78 | 14-70 | 34-82 | 38-70 | |
| Mean age (yr) | 48.9 | 48.1 | 41.6 | 53.5 | 55.1 | |
| Female | No. of patients | 89 | 56 | 11 | 8 | 14 |
| Age range (yr) | 21-86 | 21-75 | 36-72 | 46-84 | 37-86 | |
| Mean age (yr) | 52.2 | 49.2 | 52.1 | 58.4 | 61 | |
| Otherb | ||||||
| All patients | No. of patients | 33 | 20 | 9 | 4 | 0 |
| Age range (yr) | 28-82 | 28-82 | 31-61 | 48-81 | N/A | |
| Mean age (yr) | 49.3 | 47.7 | 47.8 | 60.8 | N/A | |
| Male | No. of patients | 12 | 9 | 1 | 2 | 0 |
| Age range (yr) | 33-81 | 36-61 | 33 | 58-81 | N/A | |
| Mean age (yr) | 48.8 | 46 | N/A | 69.5 | N/A | |
| Female | No. of patients | 21 | 11 | 8 | 2 | 0 |
| Age range (yr) | 28-82 | 28-82 | 31-61 | 48-56 | N/A | |
| Mean age (yr) | 49.5 | 49 | 49.6 | 52 | N/A | |
| vacA | ||||||
| s1-i1-m1 | ||||||
| All patients | No. of patients | 200 | 92 | 41 | 40 | 27 |
| Age range (yr) | 14-84 | 19-78 | 14-70 | 34-84 | 37-78 | |
| Mean age (yr) | 49.8 | 48.5 | 43 | 54.9 | 57 | |
| Male | No. of patients | 104 | 32 | 27 | 32 | 15 |
| Age range (yr) | 14-82 | 19-78 | 14-70 | 34-82 | 38-70 | |
| Mean age (yr) | 50.1 | 48.2 | 40.3 | 54.5 | 55.9 | |
| Female | No. of patients | 94 | 60 | 14 | 8 | 12 |
| Age range (yr) | 21-84 | 21-75 | 31-61 | 46-84 | 37-78 | |
| Mean age (yr) | 50.5 | 48.7 | 48.3 | 56.5 | 58.3 | |
| s1-i1-m2 | ||||||
| All patients | No. of patients | 11 | 4 | 4 | 1 | 2 |
| Age range (yr) | 38-82 | 38-82 | 41-57 | 63 | 44-68 | |
| Mean age (yr) | 53.9 | 54 | 50.5 | N/A | 56 | |
| Male | No. of patients | 2 | 0 | 1 | 0 | 1 |
| Age range (yr) | 41-44 | N/A | 41 | N/A | 44 | |
| Mean age (yr) | 42.5 | N/A | N/A | N/A | N/A | |
| Female | No. of patients | 9 | 4 | 3 | 1 | 1 |
| Age range (yr) | 38-82 | 38-82 | 48-57 | 63 | 68 | |
| Mean age (yr) | 56.4 | 54 | 53.7 | N/A | N/A | |
| s1-i2-m2 | ||||||
| All patients | No. of patients | 8 | 4 | 3 | 1 | 0 |
| Age range (yr) | 38-72 | 38-68 | 61-72 | 56 | N/A | |
| Mean age (yr) | 56.6 | 50.8 | 64.7 | N/A | N/A | |
| Male | No. of patients | 2 | 1 | 1 | 0 | 0 |
| Age range (yr) | 43-61 | 43 | 61 | N/A | N/A | |
| Mean age (yr) | 52 | N/A | N/A | N/A | N/A | |
| Female | No. of patients | 6 | 3 | 2 | 1 | 0 |
| Age range (yr) | 38-72 | 38-68 | 61-72 | 56 | N/A | |
| Mean age (yr) | 58.2 | 53.3 | 66.5 | N/A | N/A | |
| s1-i3-m1 | ||||||
| All patients | No. of patients | 3 | 1 | 1 | 0 | 1 |
| Age range (yr) | 32-86 | 32 | 47 | N/A | 86 | |
| Mean age (yr) | 55 | N/A | N/A | N/A | N/A | |
| Male | No. of patients | 2 | 1 | 1 | 0 | 0 |
| Age range (yr) | 32-47 | 32 | 47 | N/A | N/A | |
| Mean age (yr) | 39.5 | N/A | N/A | N/A | N/A | |
| Female | No. of patients | 1 | 0 | 0 | 0 | 1 |
| Age range (yr) | 86 | N/A | N/A | N/A | 86 | |
| Mean age (yr) | N/A | N/A | N/A | N/A | N/A | |
N/A, not applicable.
Any genotype other than EPIYA-ABD, including Western strains and EPIYA-AABD, -BD, -BBD, -ABAB**D, and -AB**D where a mutation within the EPIYA-B motif is designated by two asterisks.
Vac i regions i1, i2, and i3.
The fact that the vacA i region shows polymorphism that may affect toxin activity was reported only recently. At that time, consensus sequences were identified for the i1 and i2 regions based on sequences from strains 60190 and Tx30a, respectively (Fig. 2) (47). Subsequently, the consensus sequences for the i1 and i2 regions were verified by analysis of 123 strains from four distinct populations by Chung et al., who also described an i3 region that appears to be a hybrid of the i1 and i2 sequences (11).
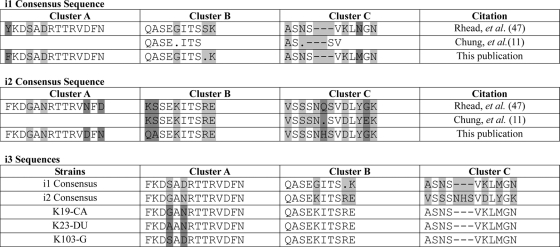
FIG. 2.
Consensus sequences for the vacA intermediate region. The consensus sequences of the i1 and i2 regions were previously defined by Rhead et al. (47) and Chung et al. (11) and are shown in comparison to sequences obtained in this study. Differences in the various i1 and i2 consensus sequences or differences in the i3 sequences are indicated by dark gray shading. Amino acids that are different in the i1 and i2 strains are indicated by light gray shading. A position where there is no consensus for the amino acid is indicated by a period. The points of insertion of additional amino acids that are found only in cluster C of the i2 allele are indicated by dashes.
On the basis of this new (i1, i2, and i3) nomenclature, our South Korean population contained four different vacA alleles, s1-i1-m1 (200 isolates), s1-i1-m2 (11 isolates), s1-i2-m2 (eight isolates), and s1-i3-m1 (three isolates) (Table 1). All three of the isolates defined as having an i3 region contained an i2 cluster B consensus sequence and an i1 cluster C consensus sequence (Fig. 1 and 2). Of the s1-i1-m1 strains, 92 were from gastritis patients, 41 were from duodenal ulcer patients, 40 were from gastric ulcer patients, and 27 were from gastric cancer patients. Four of the s1-i1-m2 strains were from gastritis patients, four were from duodenal ulcer patients, one was from a gastric ulcer patient, and two were from cancer patients. Of the eight s1-i2-m2 strains, four were from gastritis patients, three were from duodenal ulcer patients, and one from a gastric ulcer patient. The three s1-i3-m1 strains were from one gastritis patient, one duodenal ulcer patient, and one cancer patient (Table 1). There was no statistical association between the distribution of the vacA alleles and the different disease states (P = 0.6865).
Given the fact that we were able to determine the i-region sequence from 231 strains, we next examined the i-region sequences to identify amino acids that predominated in East Asian strains, as well as to determine the amino acid differences between the i1 and i2 vacA alleles that might differ from the defined consensus sequence (47). For this analysis, we defined the consensus sequence in our South Korean population as the amino acids encoded for by genes from at least 85% of all strains. Comparison of our consensus sequence to that defined by Rhead et al. (47) revealed the following. In cluster A, we found the following. (i) There was a phenylalanine instead of tyrosine at the first amino acid for the i1 allele. (ii) The 12th and 14th amino acids of the i2 allele were reversed (asparagine, phenylalanine, and aspartic acid versus aspartic acid, phenylalanine, and asparagine in our population). (iii) The main difference between the i1 and i2 vacA alleles in our population were the fourth and sixth amino acids (i1: SAD and i2: GAN) (Fig. 2). In cluster B, we found the following. (i) Our population displayed variability at the ninth amino acid of the i1 allele compared to the previously described serine. (ii) The first two amino acids of the i2 alleles in our population were similar to the first two amino acids of the i1 sequence and contained glutamine followed by alanine rather than the lysine followed by serine found in other populations. (iii) The major differences in cluster B sequences in i1 and i2 alleles in our population were the fifth, ninth, and tenth amino acids (Fig. 2) (11, 47). In cluster C, we found the following. (i) There was a methionine instead of an asparagine at the eighth amino acid of the i1 allele in our population. (ii) There was a histidine instead of a glutamine at the sixth amino acid of the i2 allele. (iii) The major differences in cluster C sequences in the i1 and i2 alleles were the insertion of three additional amino acids in the i2 allele, an asparagine, histidine, and serine, after the fourth amino acid and then subsequent differences in the 1st, 3rd, 6th, 8th, and 10th amino acids of the i1 allele (correlating with the 1st, 3rd, 9th, 11th, and 13th amino acids of the i2 allele, respectively) (Fig. 2) (11, 47).
The newest allele of the intermediate region, the i3 region has been defined as the region that contains a cluster B sequence from either the i1 or i2 allele and a cluster C sequence from the other allele (11). This South Korean population contained three i3 strains. All of these strains contained a cluster B sequence from an i2 allele and a cluster C sequence from an i1 allele. The differences in the cluster A sequence in this population between an i1 allele and an i2 allele are the fourth and sixth amino acids. It is noteworthy that the three i3 sequences showed no consensus for these amino acids; one i3 sequence was identical to the i1 consensus sequence, another was identical to the i2 consensus sequence, and one contained two different amino acids (Fig. 2).
Amino acid 151.
In-depth sequence analysis of the entire i region revealed that, as previously described (27), amino acid 151 showed polymorphism. Strains contained either a tyrosine (91 isolates [41%]) or phenylalanine (131 isolates [59%]) at this position. The distribution of amino acids at this position was not associated with gender (P = 0.1749); the distribution between males and females was fairly even (Table 2 ). In agreement with the smaller subset of isolates we previously analyzed (27), there was no association with variation at this residue and the CagA allele (P = 0.4433). Variation was also not associated with the overall VacA allele (P = 0.2177) or with either the i (P = 0.1692) or m (P = 0.8097) subregions of the VacA allele. Polymorphisms at this amino acid also did not impact disease state, even when individual disease states were compared directly to each other (Table 3).
TABLE 2.
Distribution of amino acids at polymorphic positions within the intermediate region
| Amino acid and patient gender | Parameter | Value for parameter for patientsa |
||||
|---|---|---|---|---|---|---|
| All | Gastritis | Duodenal ulcers | Gastric ulcers | Gastric cancer | ||
| Amino acid 151 | ||||||
| Tyrosine | ||||||
| All patients | No. of patients | 91 | 41 | 15 | 20 | 15 |
| Age range (yr) | 14-84 | 25-74 | 14-70 | 34-84 | 38-78 | |
| Mean age (yr) | 49.8 | 46.7 | 45.5 | 54.3 | 56.4 | |
| Male | No. of patients | 51 | 17 | 10 | 17 | 7 |
| Age range (yr) | 32-81 | 28-74 | 14-70 | 34-81 | 38-64 | |
| Mean age (yr) | 45.9 | 45.1 | 42.4 | 52.7 | 51.1 | |
| Female | No. of patients | 40 | 24 | 5 | 3 | 8 |
| Age range (yr) | 37-84 | 25-74 | 41-57 | 48-84 | 41-78 | |
| Mean age (yr) | 49 | 47.9 | 51.6 | 63 | 61 | |
| Phenylalanine | ||||||
| All patients | No. of patients | 131 | 60 | 34 | 22 | 15 |
| Age range (yr) | 19-86 | 19-82 | 20-72 | 41-82 | 37-86 | |
| Mean age (yr) | 50.7 | 49.9 | 44.9 | 55.9 | 59.3 | |
| Male | No. of patients | 61 | 17 | 20 | 15 | 9 |
| Age range (yr) | 19-82 | 19-78 | 20-70 | 41-80 | 44-70 | |
| Mean age (yr) | 49.8 | 50 | 40.7 | 56.5 | 58.2 | |
| Female | No. of patients | 70 | 43 | 14 | 7 | 6 |
| Age range (yr) | 21-86 | 21-82 | 31-72 | 46-63 | 37-86 | |
| Mean age (yr) | 51.5 | 49.9 | 50.9 | 54.6 | 61 | |
| Amino acid 231 | ||||||
| Glycine | ||||||
| All patients | No. of patients | 153 | 67 | 34 | 30 | 22 |
| Age range (yr) | 14-84 | 19-82 | 14-70 | 34-82 | 37-78 | |
| Mean age (yr) | 50.3 | 50.2 | 42.6 | 54.6 | 57 | |
| Male | No. of patients | 83 | 25 | 23 | 24 | 11 |
| Age range (yr) | 14-84 | 19-78 | 14-70 | 34-82 | 38-68 | |
| Mean age (yr) | 48.6 | 49.3 | 40.7 | 53 | 54.5 | |
| Female | No. of patients | 70 | 42 | 11 | 6 | 11 |
| Age range (yr) | 25-84 | 25-82 | 31-61 | 48-84 | 37-78 | |
| Mean age (yr) | 52.3 | 50.7 | 46.8 | 60.7 | 59.5 | |
| Otherb | ||||||
| All patients | No. of patients | 69 | 34 | 15 | 12 | 8 |
| Age range (yr) | 21-86 | 27-74 | 31-72 | 41-81 | 44-86 | |
| Mean age (yr) | 50.4 | 45.5 | 50.5 | 57.3 | 60.3 | |
| Male | No. of patients | 29 | 9 | 7 | 8 | 5 |
| Age range (yr) | 29-81 | 29-61 | 31-61 | 41-81 | 44-70 | |
| Mean age (yr) | 50 | 42.6 | 43.3 | 60.1 | 47.6 | |
| Female | No. of patients | 40 | 25 | 8 | 4 | 3 |
| Age range (yr) | 21-86 | 21-74 | 37-72 | 46-57 | 48-86 | |
| Mean age (yr) | 50.7 | 46.6 | 56.9 | 51.8 | 66.7 | |
| Amino acid 196 | ||||||
| Serine | ||||||
| All patients | No. of patients | 148 | 64 | 28 | 31 | 25 |
| Age range (yr) | 19-84 | 19-82 | 23-70 | 34-84 | 37-78 | |
| Mean age (yr) | 50.9 | 48.8 | 43.6 | 56.7 | 57.4 | |
| Male | No. of patients | 76 | 22 | 18 | 24 | 12 |
| Age range (yr) | 19-82 | 19-76 | 23-70 | 34-82 | 38-68 | |
| Mean age (yr) | 49.6 | 46.6 | 40.7 | 56.1 | 55.5 | |
| Female | No. of patients | 72 | 42 | 10 | 7 | 13 |
| Age range (yr) | 21-84 | 21-82 | 31-61 | 48-84 | 37-78 | |
| Mean age (yr) | 52.3 | 48.4 | 48.7 | 58.9 | 59.1 | |
| Leucine | ||||||
| All patients | No. of patients | 74 | 37 | 21 | 11 | 5 |
| Age range (yr) | 14-86 | 24-78 | 14-72 | 38-69 | 44-86 | |
| Mean age (yr) | 49.1 | 48.4 | 47 | 50.5 | 60.4 | |
| Male | No. of patients | 36 | 12 | 12 | 8 | 4 |
| Age range (yr) | 14-78 | 28-78 | 14-70 | 38-69 | 44-70 | |
| Mean age (yr) | 48.8 | 49.2 | 42.1 | 49.6 | 54 | |
| Female | No. of patients | 38 | 25 | 9 | 3 | 1 |
| Age range (yr) | 24-86 | 24-74 | 39-72 | 46-57 | 86 | |
| Mean age (yr) | 50.7 | 48 | 53.7 | 53 | N/A | |
N/A, not applicable.
Any amino acid other than a glycine at this position.
TABLE 3.
P values of the distribution of amino acids at polymorphic positions in patients with various diseases
| Comparison of the distribution of amino acids in patients with different diseasesa |
P valuea for the distribution of amino acids at position: |
||
|---|---|---|---|
| 151 | 231 | 196 | |
| Across all diseases | 0.2639 | 0.8914 | 0.0624 |
| Gastritis vs all other diseases (duodenal ulcers, gastric ulcers, and gastric cancer) | 1.0000 | 0.4694 | 0.3916 |
| Gastritis vs peptic ulcers (both duodenal and gastric ulcers) | 0.7700 | 0.6418 | 0.8808 |
| Gastritis vs duodenal ulcers | 0.2818 | 0.8529 | 0.4796 |
| Gastritis vs gastric ulcers | 0.4627 | 0.6946 | 0.2500 |
| Gastritis vs gastric cancer | 0.4044 | 0.5132 | 0.0460 |
| Duodenal ulcers vs all other diseases | 0.1025 | 1.0000 | 0.1237 |
| Duodenal ulcers vs gastric ulcers | 0.1306 | 1.0000 | 0.1250 |
| Duodenal ulcers vs gastric cancer | 0.0994 | 0.8015 | 0.0254 |
| Gastric ulcers vs all other diseases | 0.3847 | 0.8533 | 0.3636 |
| Gastric ulcers vs gastric cancer | 1.0000 | 1.0000 | 0.3994 |
| Gastric cancer vs all other gastric diseases | 0.3207 | 0.6732 | 0.0389 |
| Gastric cancer vs peptic ulcers | 0.2910 | 0.8201 | 0.0689 |
Associations that were statistically significant and the corresponding P values are in boldface type.
Amino acid 231.
Amino acid 231, which is the ninth amino acid in cluster B, was previously shown to contain amino acid polymorphisms that are important for disease development in a Taiwanese population (50). In our South Korean population, we identified five different amino acids at this position: glycine, serine, aspartic acid, asparagine, and arginine. To determine whether the residue at this position was important, statistical associations were analyzed across all the different amino acids and for glycine versus all other amino acids combined; a previous study identified the presence of the glycine residue as important for the progression to more severe disease (50). Since we identified no difference between which associations were statistically significant and since the previous literature assessed the glycine residue versus any other amino acid (50), the numbers we present compare glycine versus all other amino acids found at position 231. The amino acid at position 231 was not associated with gender (P = 0.1109) (Table 2). The strong association we previously identified using a subset of South Korean isolates (27) between the distribution of amino acid polymorphisms at this position and the distribution of the CagA allele was maintained in this larger population (P = 0.0081). More specifically, among EPIYA-ABD strains, glycine was more prevalent at this position (72.5%) than among strains carrying a non-EPIYA-ABD cagA allele (48.5%). Polymorphisms at this site were also associated with the overall VacA allele (P < 0.0001), every subregion of the VacA allele (i region, P < 0.0001; m region, P = 0.0168), and every combination of subregions (s and i regions, P < 0.0001; m and i regions, P < 0.0001; s and m regions, P = 0.0168). However, even though glycine at this position was previously identified as important for the progression to more severe disease (50), we found that polymorphisms at this position did not impact the disease state. Additionally, there was no significance when comparing across any individual disease, whether individual disease states were compared alone or whether a combination of disease states was analyzed (Table 3). These data suggest that this amino acid is not important for disease progress or that other factors are masking the contribution of this amino acid to disease state in the South Korean population.
Amino acid 196.
As we previously reported for a subset of South Korean strains (27), amino acid 196 was identified as a position that contained amino acid polymorphisms. In this larger population, either a serine (148 isolates [67%]) or a leucine (74 isolates [33%]) was found at this position. The distribution of amino acids at this position was not associated with gender (P = 0.7762) and was fairly evenly distributed between males and females (Table 2). Additionally, there was still no association with the CagA allele (P = 0.6928). However, polymorphisms at this residue were associated with the VacA allele (P = 0.0003). This association was present for the i region of the VacA allele (P = 0.0001) or any combination that contained the i region (s and i regions, P = 0.0001; m and i regions, P = 0.0003). However, there was no association when the m region was assessed alone (P = 0.7600) or when the combination of regions did not include the i region (s and m regions, P = 0.0760). In fact, all of the i2 strains contain a leucine at this position (Fig. 1).
Polymorphisms at this amino acid did not impact disease state (P = 0.0624) as a whole (Table 3). Additionally, this position was not significant when gastritis was compared to any noncancer disease state: gastritis versus all other disease states (duodenal ulcers, gastric ulcers, and gastric cancers) (P = 0.3916), gastritis versus peptic ulcers (both duodenal and gastric ulcers) (P = 0.8808), gastritis versus duodenal ulcers (P = 0.4796), or gastritis versus gastric ulcers (P = 0.2500). It was also not significant when peptic ulcers were compared to noncancer disease states: duodenal ulcers versus all other disease states (P = 0.1237), duodenal ulcers versus gastric ulcers (P = 0.1250), or gastric ulcers versus all other disease states (P = 0.3636). There was also no association with the amino acid at position 196 with gastric cancers versus peptic ulcers (P = 0.0689). However, we did find an association between the amino acid at this position and the development of gastric cancer to development of all other disease states (P = 0.0389), versus duodenal ulcers alone (P = 0.0254), and versus gastritis alone (P = 0.0460). Additionally, there was a statistical association between polymorphisms at position 196 and more-severe disease manifestations (gastric ulcers and gastric cancer) versus less-severe disease manifestations (gastritis and duodenal ulcers) (P = 0.0155) (Table 3). While the presence of a serine at this position was more prevalent in all patients, patients suffering from gastric cancer were five times more likely than other patients to carry a serine at this location. These data suggest that position 196 does impact disease development but that the overall impact is masked by the progression of this disease through gastric ulcers.
Higher-order associations.
Log linear modeling using a combination of available data revealed two direct three-way associations: disease state, cagA allele, and variation at amino acid 231 (P = 0.012) and disease state and variation at amino acids 196 and 231 (P = 0.029). As mentioned above, we found no direct association with variation of amino acid 231 and disease state in our population (P = 0.8914). However, we noted that the two direct three-way associations described above involved disease state and amino acid 231, or disease state, amino acid 231, and the cagA allele. We therefore asked whether the presence of a Western or East Asian CagA allele affected the ability of variation at residue 231 to be associated with disease progression. We found that a two-way association did exist between disease state and amino acid 231 but only within the non-EPIYA-ABD population (P = 0.0367). This again suggests that the effects of different virulence factors or polymorphisms within these virulence factors may be masked when the CagA allele is present.
DISCUSSION
Polymorphisms within vacA have been studied for several years but have primarily focused on the s and m regions. However, the newest identified polymorphic region of vacA, the i region, has been suggested to be a determinant of vacuolating activity, as well as the best indicator of disease pathology, at least in Western strains (2, 16, 47). Within this region, three clusters of polymorphisms have been reported: cluster A, B, and C (47). In fact, two different amino acid substitutions within i1 clusters have been identified as potential markers for Taiwanese VacA (50). However, as previously suggested by our group (27) both of these amino acid substitutions are conserved within the South Korean population: we found a phenylalanine instead of a tyrosine for the first amino acid in cluster A (94.4%) and a methionine for the second asparagine within cluster C (99.5%). This suggests that these amino acid substitutions are not limited to the Taiwanese population but could serve as a general marker for East Asian VacA.
Further analysis of the amino acid consensus sequences of clusters A, B, and C revealed several differences from the consensus sequence previously reported by Rhead et al. (47) and Chung et al. (11). This South Korean population provides the largest number of VacA i sequences analyzed so far, allowing us to better identify amino acids that differ between the i1 and i2 alleles (Fig. 2). Cluster A was well conserved in strains, and the i1 and i2 consensus sequences were very similar to each other. Interestingly, each of the three i3 strains that we identified showed differences in cluster A; one of the strains contained the exact i1 consensus sequence, one contained the exact i2 consensus sequence, and one was different (Fig. 2), indicating that these strains may be in the process of evolving from one allele to the other allele.
Within cluster B, the fifth, ninth, and tenth amino acids are the main differences between the i1 and i2 alleles. The ninth amino acid (residue 231) was previously shown to have a role in disease (50). We found this residue was not variable in the small subset of i2 strains within our population, though it was variable within the i1 strains. Moreover, this variability at amino acid 231 does impact disease development but only within strains containing a non-EPIYA-ABD CagA. This finding suggests that within strains carrying the most virulent CagA allele, this residue is less important.
The differences between the i1 and i2 alleles were most pronounced in cluster C. This is primarily due to the addition of three polar amino acids, asparagine, histidine, and serine, but there were an additional five amino acids that are different in the i1 and i2 alleles (Fig. 2). Since both clusters B and C have been suggested to affect toxin activity, studies examining the specific roles of these amino acids in vacuolating activity would be of interest. Creation and examination of the activity of isogenic toxin derivatives in which only the residue of interest was different would aid in this effort. Furthermore, given that the i3 strains appear to be hybrids of the i1 and i2 alleles at clusters B and C, it would be of significant interest to determine whether there is a functional difference between the vacuolating activity of i1, i2, and i3 toxins. To further investigate the roles of these clusters, toxin activity could be assessed among the three i3 strains we identified and strains that contain an i1 cluster B and i2 cluster C. Finally, even though it has not been suggested to be important for activity, it would also be interesting to assess the sequences from cluster A to analyze the variance of amino acids four and six, potentially providing insight into VacA evolution.
Interestingly, five possible different amino acids were found to occur at amino acid 231 (the ninth amino acid of cluster B): glycine, serine, arginine, asparagine, and aspartic acid. Of these five amino acids, glycine is the only nonpolar amino acid, suggesting that perhaps this residue is important for the conformation or folding of VacA. Indeed, this could explain the statistical association between the distribution of amino acids at this position with every region of VacA. Previous work showed that a glycine found at amino acid 231 was linked to disease development within the Taiwanese population (50). However, in our South Korean population, the distribution of amino acids found at this position had no overall impact on disease development. Conversely, there was a strong association with variation at this residue and the cagA allele and two direct three-way associations between disease state, the distribution of amino acids at this position, and either cagA allele or amino acid 196, both of which affect the development of cancer in this population (Table 3) (27, 29). Given the association with cagA, the impact of this amino acid across East Asian and Western CagA alleles was examined, and a direct two-way association between amino acid 231 and disease state was found among the strains that carry non-EPIYA-ABD CagA alleles. This emphasizes the importance of the i region when carried within the context of Western strains and suggests that there may be other factors that are more important in disease development or that mask the importance of the i region in the development of severe disease among East Asian strains. Indeed, even among Western strains, the CagA allele was the most important virulence factor for development of gastric cancer, whereas the VacA i region was the best indicator for development of peptic ulcer disease (5). Further exploration of differences between Western and East Asian strains may help to explain the exact mechanism of interaction of CagA and VacA.
Variation at amino acid 196 was statistically linked only to the vacA i region, indicating that it may be a true indicator of i-region-associated impacts. While overall this amino acid was not linked to disease state and there was not an association between the less severe disease states and distribution of amino acids at position 196, there was a statistical difference in the distribution of amino acids at this position when gastric cancer was compared to any other disease state, gastritis, or duodenal ulcers. However, there was no statistical difference in the distribution of amino acids at this position between gastric cancer and gastric ulcers. This may be due to the fact that gastric ulcers can be a precursor to gastric cancer (19, 36). Given this, more-severe disease states (gastric cancer and ulcers) were next compared to less-severe disease manifestations (duodenal ulcers and gastritis), and a statistical association was identified. This suggests that variation at residue 196 is important for progression to severe disease and that the overall significance of the amino acids found at this position is probably masked by the lack of association between gastric cancer and gastric ulcers.
While a serine at this position was more prevalent across the overall population, patients suffering from gastric cancer were five times more likely to carry a serine at this location. However, all of the i2 alleles carried a leucine at this position, and if this trend holds true for a larger population of i2 strains, then its contribution to disease development may become more evident. Since the distribution of amino acids at this position was linked not only to the overall Vac allele but also specifically to the i region, the different prevalences of serine found at this position may explain why a previous study concluded that the i region was the best predictor of disease (47).
Since both CagA and VacA polymorphisms influence disease development, perhaps it is not surprising that within a population of isolates from a country with one of the highest rates of H. pylori colonization and gastric cancer (21, 51, 55), the majority of strains carry genes that encode the most toxic form of both CagA and VacA. While polymorphisms important for disease severity have been identified within both CagA and VacA individually, strains that are CagA positive and carry the VacA s1-m1 allele have been shown to induce highly active corpus gastritis, which has been associated with the progression to gastric cancer (37-39). Additionally, we recently presented evidence that within this high-risk population for gastric cancer development, there is a significant interaction between the VacA allele, CagA allele, and disease state (27). However, the reason only a small percentage of the population develops cancer is still unclear. Although it is evident that host, environmental, and bacterial factors play a role in H. pylori-induced disease (reviewed in references 8 and 57), additional studies are required to determine the contribution of all of these factors, both individually and in conjunction with each other, to the development of H. pylori-induced gastric cancer.
Supplementary Material
Acknowledgments
We thank Jeannette Whitmire and Beth Carpenter for critical reading of the manuscript.
This work was supported by the Basic Science Research Program through the National Research Foundation of South Korea (NRF) funded by the Ministry of Education, Science, and Technology (R13-2003-013-04002-0) (to J.-H.C.).
The contents of this article are the sole responsibility of the authors and do not necessarily represent the official views of the Department of Defense.
Footnotes
Published ahead of print on 17 November 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Amieva, M. R., and E. M. El-Omar. 2008. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 134:306-323. [DOI] [PubMed] [Google Scholar]
- 2.Argent, R. H., et al. 2008. Functional association between the Helicobacter pylori virulence factors VacA and CagA. J. Med. Microbiol. 57:145-150. [DOI] [PubMed] [Google Scholar]
- 3.Atherton, J. C., et al. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 4.Atherton, J. C., et al. 1999. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J. Clin. Microbiol. 37:2979-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basso, D., et al. 2008. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 135:91-99. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1998. Helicobacter pylori and gastric diseases. BMJ 316:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser, M. J., et al. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 8.Brenner, H., D. Rothenbacher, and V. Arndt. 2009. Epidemiology of stomach cancer. Methods Mol. Biol. 472:467-477. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, B. M., et al. 2007. Expanding the Helicobacter pylori genetic toolbox: modification of an endogenous plasmid for use as a transcriptional reporter and complementation vector. Appl. Environ. Microbiol. 73:7506-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Censini, S., et al. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. U. S. A. 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, C., et al. 2010. Diversity of VacA intermediate region among Helicobacter pylori strains from several regions of the world. J. Clin. Microbiol. 48:690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 13.Cover, T. L., and S. R. Blanke. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320-332. [DOI] [PubMed] [Google Scholar]
- 14.Cover, T. L., S. A. Halter, and M. J. Blaser. 1992. Characterization of HeLa cell vacuoles induced by Helicobacter pylori broth culture supernatant. Hum. Pathol. 23:1004-1010. [DOI] [PubMed] [Google Scholar]
- 15.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douraghi, M., et al. 2009. Multiple gene status in Helicobacter pylori strains and risk of gastric cancer development. Digestion 80:200-207. [DOI] [PubMed] [Google Scholar]
- 17.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. [DOI] [PubMed] [Google Scholar]
- 19.Furuta, T., et al. 1998. Effect of Helicobacter pylori infection on gastric juice pH. Scand. J. Gastroenterol. 33:357-363. [DOI] [PubMed] [Google Scholar]
- 20.Gebert, B., W. Fischer, E. Weiss, R. Hoffmann, and R. Haas. 2003. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science 301:1099-1102. [DOI] [PubMed] [Google Scholar]
- 21.Gwack, J., et al. 2006. CagA-producing Helicobacter pylori and increased risk of gastric cancer: a nested case-control study in Korea. Br. J. Cancer 95:639-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatakeyama, M. 2008. Linking epithelial polarity and carcinogenesis by multitasking Helicobacter pylori virulence factor CagA. Oncogene 27:7047-7054. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama, M. 2004. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 4:688-694. [DOI] [PubMed] [Google Scholar]
- 24.Higashi, H., et al. 2004. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J. Biol. Chem. 279:17205-17216. [DOI] [PubMed] [Google Scholar]
- 25.Higashi, H., et al. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. U. S. A. 99:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higashi, H., et al. 2002. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295:683-686. [DOI] [PubMed] [Google Scholar]
- 27.Jang, S., et al. 2010. Epidemiological link between gastric disease and polymorphisms in VacA and CagA. J. Clin. Microbiol. 48:559-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji, X., et al. 2000. Cell specificity of Helicobacter pylori cytotoxin is determined by a short region in the polymorphic midregion. Infect. Immun. 68:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, K. R., et al. 2009. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J. Clin. Microbiol. 47:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letley, D. P., and J. C. Atherton. 2000. Natural diversity in the N terminus of the mature vacuolating cytotoxin of Helicobacter pylori determines cytotoxin activity. J. Bacteriol. 182:3278-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letley, D. P., J. L. Rhead, R. J. Twells, B. Dove, and J. C. Atherton. 2003. Determinants of non-toxicity in the gastric pathogen Helicobacter pylori. J. Biol. Chem. 278:26734-26741. [DOI] [PubMed] [Google Scholar]
- 32.Manente, L., et al. 2008. The Helicobacter pylori's protein VacA has direct effects on the regulation of cell cycle and apoptosis in gastric epithelial cells. J. Cell. Physiol. 214:582-587. [DOI] [PubMed] [Google Scholar]
- 33.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed] [Google Scholar]
- 34.Matysiak-Budnik, T., and F. Megraud. 1997. Epidemiology of Helicobacter pylori infection with special reference to professional risk. J. Physiol. Pharmacol. 48(Suppl. 4):3-17. [PubMed] [Google Scholar]
- 35.McClain, M. S., et al. 2001. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J. Bacteriol. 183:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McColl, K. E., and E. el-Omar. 1996. Helicobacter pylori and disturbance of gastric function associated with duodenal ulcer disease and gastric cancer. Scand. J. Gastroenterol. Suppl. 215:32-37. [DOI] [PubMed] [Google Scholar]
- 37.Meining, A., et al. 1997. Differing degree and distribution of gastritis in Helicobacter pylori-associated diseases. Virchows Arch. 431:11-15. [DOI] [PubMed] [Google Scholar]
- 38.Miehlke, S., et al. 1998. Severe expression of corpus gastritis is characteristic in gastric cancer patients infected with Helicobacter pylori. Br. J. Cancer 78:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miehlke, S., et al. 2000. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int. J. Cancer 87:322-327. [PubMed] [Google Scholar]
- 40.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell Biol. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 41.Neel, B. G., H. Gu, and L. Pao. 2003. The ′Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 28:284-293. [DOI] [PubMed] [Google Scholar]
- 42.Neugut, A. I., M. Hayek, and G. Howe. 1996. Epidemiology of gastric cancer. Semin. Oncol. 23:281-291. [PubMed] [Google Scholar]
- 43.Nguyen, L. T., T. Uchida, K. Murakami, T. Fujioka, and M. Moriyama. 2008. Helicobacter pylori virulence and the diversity of gastric cancer in Asia. J. Med. Microbiol. 57:1445-1453. [DOI] [PubMed] [Google Scholar]
- 44.Pagliaccia, C., et al. 1998. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc. Natl. Acad. Sci. U. S. A. 95:10212-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsonnet, J., et al. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 46.Pugsley, A. P. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhead, J. L., et al. 2007. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 133:926-936. [DOI] [PubMed] [Google Scholar]
- 48.Roovers, K., and R. K. Assoian. 2000. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays 22:818-826. [DOI] [PubMed] [Google Scholar]
- 49.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheu, S. M., K. H. Hung, B. S. Sheu, H. B. Yang, and J. J. Wu. 2009. Association of nonsynonymous substitutions in the intermediate region of the vacA gene of Helicobacter pylori with gastric diseases in Taiwan. J. Clin. Microbiol. 47:249-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin, A., et al. 2005. A nested case-control study of the association of Helicobacter pylori infection with gastric adenocarcinoma in Korea. Br. J. Cancer 92:1273-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szabo, I., et al. 1999. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 18:5517-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terebiznik, M. R., et al. 2009. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy 5:370-379. [DOI] [PubMed] [Google Scholar]
- 54.The EUROGAS Study Group. 1993. Epidemiology of, and risk factors for, Helicobacter pylori infection among 3194 asymptomatic subjects in 17 populations. Gut 34:1672-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tokudome, S., et al. 2007. Are there any real Helicobacter pylori infection-negative gastric cancers in Asia? Asian Pac. J. Cancer Prev. 8:462-463. [PubMed] [Google Scholar]
- 56.Torres, V. J., S. E. VanCompernolle, M. S. Sundrud, D. Unutmaz, and T. L. Cover. 2007. Helicobacter pylori vacuolating cytotoxin inhibits activation-induced proliferation of human T and B lymphocyte subsets. J. Immunol. 179:5433-5440. [DOI] [PubMed] [Google Scholar]
- 57.Tsugane, S., and S. Sasazuki. 2007. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 10:75-83. [DOI] [PubMed] [Google Scholar]
- 58.Tsutsumi, R., A. Takahashi, T. Azuma, H. Higashi, and M. Hatakeyama. 2006. Focal adhesion kinase is a substrate and downstream effector of SHP-2 complexed with Helicobacter pylori CagA. Mol. Cell. Biol. 26:261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Doorn, L. J., et al. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116:823-830. [DOI] [PubMed] [Google Scholar]
- 60.Willhite, D. C., and S. R. Blanke. 2004. Helicobacter pylori vacuolating cytotoxin enters cells, localizes to the mitochondria, and induces mitochondrial membrane permeability changes correlated to toxin channel activity. Cell. Microbiol. 6:143-154. [DOI] [PubMed] [Google Scholar]
- 61.Yamaoka, Y., T. Kodama, K. Kashima, D. Y. Graham, and A. R. Sepulveda. 1998. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J. Clin. Microbiol. 36:2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamazaki, S., et al. 2005. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J. Clin. Microbiol. 43:3906-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.