Abstract
Multidrug-resistant Acinetobacter baumannii is a worldwide nosocomial menace. We sought to better understand its behavior through studying the molecular epidemiology of this organism at the Royal Brisbane and Women's Hospital, Brisbane, Queensland, Australia, over a 10-year period. Multilocus sequence typing (MLST), semiautomated repetitive sequence-based PCR (rep-PCR), and pulsed-field gel electrophoresis (PFGE) were performed on a selection of 31 A. baumannii isolates collected over the 10-year period to determine their relationships to one another. MLST also allowed us to put this information in a global context. The presence or absence of blaOXA-23 was also established. The presence of blaOXA-23 closely correlated with carbapenem resistance in our collection. Sequence type 92 (ST92) was the dominant sequence type and was present in the hospital for 9 years. There was also evidence of the spread of ST69, ST73, and ST125 (novel) within the hospital, but this was not sustained over long periods. There were only single examples of the novel sequence types ST126 and ST127. The different typing methods clustered the isolates similarly; however, PFGE and rep-PCR were more discriminatory than MLST. Worldwide, ST92 and the associated clonal complex 92 represent the most sampled and widespread sequence type(s) and are also known as European clone 2 and worldwide clonal lineage 2. Antibiotic susceptibility within ST92 is variable, suggesting a role for mechanisms other than antibiotic resistance in its success.
Acinetobacter baumannii is a nonfermentative Gram-negative bacillus which is notable for its ability to acquire antibiotic resistance determinants and cause hospital outbreaks of infection (23). A. baumannii is an important pathogen of critically ill patients and can cause a range of infections, including ventilator-associated pneumonia, bloodstream infection, wound infection, and nosocomial meningitis (13, 15, 23). In many institutions, substantial difficulties arise because A. baumannii strains have become resistant to all beta-lactam antibiotics (including carbapenems), all fluoroquinolones, trimethoprim-sulfamethoxazole, and most, if not all, aminoglycosides (24). Thus, empirical treatment choices are extremely limited.
In order to better control multidrug-resistant A. baumannii (MRAB), an understanding of the molecular epidemiology of the infection is necessary. From a global perspective, it is known that A. baumannii is typically clonal in nature (11). Three clonal complexes (CCs) have predominated in Europe for more than a decade (6, 28); these clonal complexes have more recently also been documented in North America, Asia, Africa, and Australia (11). The precise origin of these clonal lineages will likely never be known. From this broader geographic perspective, it is remarkable how successful clones have spread, likely through the international transfer of patients (11, 22).
The molecular epidemiology of A. baumannii has typically been studied in the context of outbreaks of infection. However, an understanding of the epidemiology of the infection over longer time periods may allow new insights into the behavior of this emerging pathogen. In the study described in this paper, we have undertaken a longitudinal evaluation of the molecular epidemiology of multidrug-resistant A. baumannii in a single institution over a 10-year period.
MATERIALS AND METHODS
Setting.
The Royal Brisbane and Women's Hospital (RBWH) is a 900-bed teaching hospital of the University of Queensland. The hospital is a major referral center for trauma, burns, and hematologic transplantation. Within the hospital is a 19-bed long-stay intensive care unit (ICU), which has a bed occupancy of from 550 to 670 patient days per month.
Isolates and time period.
Isolates were selected from a collection of all A. calcoaceticus-A. baumannii complex strains with non-wild-type susceptibility profiles that had been cultured at RBWH between September 1998 and November 2008. A non-wild-type susceptibility profile was defined as resistance, by use of the Vitek GNI card, to trimethoprim-sulfamethoxazole, gentamicin, tobramycin, amikacin, ceftazidime, cefepime, ticarcillin-clavulanate (Timentin), piperacillin-tazobactam (Tazocin), ciprofloxacin, or meropenem for any isolate.
There were 483 isolates from unique patients available. The vast majority of isolates in the collection had been cultured from patients in the ICU or burns ward. The dominant phenotype was susceptibility only to amikacin or to amikacin and tobramycin (the isolates were not routinely tested for colistin and tigecycline susceptibility). More than half of the isolates collected over the 10-year period were from 2001-2002 (n = 204) and 2006 (n = 84); these periods corresponded to A. baumannii outbreaks in the hospital.
From this collection, a total of 33 isolates were selected for study. Seventeen isolates were from outbreak periods. Of these, seven isolates had a pulsed-field gel electrophoresis (PFGE) profile demonstrating a close or possible relationship to the outbreak strain, six were different from the outbreak strain by PFGE, and four isolates were not typed by PFGE. Sixteen isolates were from sporadic cases. These isolates were selected because there was a point of difference in the antibiotic susceptibility profile or epidemiology (known international transfer or lack of association with the burns unit or ICU) or to allow representation of isolates from all years.
Species identification, antimicrobial susceptibility, and detection of carbapenemases.
Phenotypic identification and antibiotic susceptibility testing were performed by use of the Vitek or Vitek 2 system (bioMérieux, France), and the results were interpreted according to CLSI criteria (4). Genotypic identification as A. baumannii was confirmed by detection of blaOXA-51-like by a real-time PCR assay adapted from the gel-based method described previously (26). PCRs were performed using 20-μl reaction mixtures with 10 pmol each of forward and reverse primers, 7 μl of water, 10 μl of Platinum SYBR green quantitative PCR Supermix-UDG (Invitrogen, Carlsbad, CA), and 1 μl of extracted DNA. Amplification and detection were performed on a Rotor-Gene 6000 real-time system (Corbett Research, Sydney, Australia) under the following conditions: 94°C for 3 min and 35 cycles of 94°C for 45 s, 60°C for 45 s, and 72°C for 1 min, followed by a final extension of 72°C for 1 min.
Detection of blaOXA-23 (commonly associated with carbapenem resistance) was performed as described previously (2).
MLST.
Multilocus sequence typing (MLST) was performed as described previously (3), with the following exceptions: amplification of gyrB and gpi was performed with an annealing temperature of 50°C for some isolates, as product generation was inefficient at 55°C. The amplification product was purified with ExoSAP-IT reagent (Axygen, Union City, CA). Sequencing was performed with BigDye Terminator cycle sequencing kit premix version 3 and an ABI 3730 DNA analyzer (Applied Biosystems). For four isolates, sequencing of gdhB required use of diluted amplification primers, as sequencing was unsuccessful with the internal sequencing primers.
Editing and interpretation of electropherograms were performed visually and with the assistance of the following software: Finch TV (http://www.geospiza.com/Products/finchtv.shtml), Readseq (http://www.ebi.ac.uk/cgi-bin/readseq.cgi), and Clustal X (16). Analysis of allele sequences and sequence type (ST) assignment made use of the Oxford Acinetobacter baumannii MLST website (http://pubmlst.org/abaumannii/) (14). The eBURST diagram was constructed by V3 software (http://eburst.mlst.net/), using all available data from previous publications (3, 9, 12, 18, 21, 29) and unpublished data from the online database, where the submitter of the data consented to its use.
Typing using rep-PCR.
Repetitive sequence-based PCR (rep-PCR) was performed with the semiautomated Diversilab system (bioMerieux, Melbourne, Australia), according to the manufacturer's instructions. Diversilab fingerprints were analyzed with the Diversilab software using the Pearson correlation statistical method to determine clonal relationships.
PFGE.
PFGE was performed after digestion of genomic DNA with ApaI (New England Biolabs, Beverley, MA), as described previously (25), and the results were analyzed using Bionumerics software (Applied Maths, Belgium).
RESULTS
Of 33 isolates selected for analysis, 31 were blaOXA-51-like positive. These isolates that were confirmed to be A. baumannii underwent molecular typing and were examined for the presence of blaOXA-23.
MLST, antibiotic susceptibility, and blaOXA-23.
MLST findings and the associated antibiograms and blaOXA-23 results are summarized in Tables 1 and 2. It must be noted that many ST designations on the A. baumannii MLST website have changed since the publication of relevant studies (9, 10, 18, 20, 21), to avoid conflict with an earlier publication (3). For example, ST92 was previously referred to as ST22. Changes relevant to the current study are listed in Table 3.
TABLE 1.
STs and allele numbers of Acinetobacter baumannii isolates, by year isolated
| Isolate no. | Yr isolated | ST | Allele no. |
||||||
|---|---|---|---|---|---|---|---|---|---|
| gltA | gyrB | gdhB | recA | cpn60 | gpi | rpoD | |||
| Q48, Q47, Q57 | 2000 | 92 | 1 | 3 | 3 | 2 | 2 | 7 | 3 |
| Q5, Q50, Q6, Q7 | 2001 | 92 | 1 | 3 | 3 | 2 | 2 | 7 | 3 |
| Q51, Q52, Q53 | 2002 | 92 | 1 | 3 | 3 | 2 | 2 | 7 | 3 |
| Q10, Q11 | 2004 | 92 | 1 | 3 | 3 | 2 | 2 | 7 | 3 |
| Q12 | 2005 | 92 | 1 | 3 | 3 | 2 | 2 | 7 | 3 |
| Q13, Q15, Q16, Q17, Q19, Q21 | 2006 | 92 | 1 | 3 | 3 | 2 | 2 | 7 | 3 |
| Q25, Q26, Q28 | 2008 | 92 | 1 | 3 | 3 | 2 | 2 | 7 | 3 |
| Q45 | 1998 | 69 | 1 | 46 | 3 | 2 | 2 | 58 | 3 |
| Q1 | 1999 | 69 | 1 | 46 | 3 | 2 | 2 | 58 | 3 |
| Q54 | 2002 | 69 | 1 | 46 | 3 | 2 | 2 | 58 | 3 |
| Q18, Q20 | 2006 | 73 | 1 | 47 | 53 | 1 | 1 | 59 | 32 |
| Q46, Q47 | 1999 | 125a | 1 | 52a | 59a | 12 | 1 | 18 | 44a |
| Q55 | 2003 | 126a | 10 | 53a | 4 | 11 | 4 | 64 | 5 |
| Q22 | 2007 | 127a | 1 | 33 | 57a | 11 | 26 | 11 | 6 |
Novel.
TABLE 2.
Antibiogram and blaOXA-23 result, by sequence type
| ST | No. of isolates | Antibiotic resistancea |
blaOXA-23 | ||||
|---|---|---|---|---|---|---|---|
| Gentamicin | Tobramycin | Amikacin | Ciprofloxacin | Meropenem | |||
| 92 | 22 | R (21/22) | R (12/22) | S (19/22) | R | R | Present |
| 69 | 3 | R (2/3) | R (2/3) | R | R | S | Absent |
| 73 | 2 | R | R | S | S | R | Present |
| 125 | 2 | R | R | R | R | R | Present |
| 126 | 1 | R | S | R | R | S | Absent |
| 127 | 1 | S | S | S | S | R | Present |
R, nonsusceptible (including resistant and intermediate); S, susceptible. Numbers in parentheses indicate the number of strains with the indicated susceptibility/total number of strains.
TABLE 3.
New designations for previously published sequence types discussed in the texta
| Current designation |
Previous designation |
|||
|---|---|---|---|---|
| ST | Allelic profile | ST | Allelic profile | Reference(s) |
| 92 | 1-3-3-2-2-7-3 | 22 | 1-3-3-2-2-7-3 | Park et al. (20), Mugnier et al. (18), Fu et al. (9), Ho et al. (12), Hamouda et al. (10) |
| 69 | 1-46-3-2-2-58-3 | 28 | 1-21-3-2-2-2-3 | Park et al. (20) |
| 73 | 1-47-53-1-1-59-32 | 32 | 1-22-19-1-1-23-18 | Park et al. (20 |
During December 2009/January 2010, changes were made to the MLST website (http://pubmlst.org/abaumannii/) to correct doubly allocated numbers to avoid conflict with an earlier publication (3). A complete list of changes is available at the website.
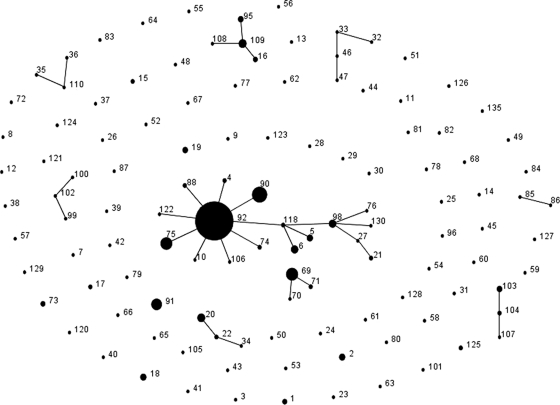
ST92 was the dominant sequence type (22 of 31 isolates) and was found in the hospital over the period from 2000 to 2008. ST92 is also the most frequently isolated and founding genotype of the largest and most widespread clonal complex (CC), CC92, of A. baumannii (Fig. 1). Within CC92, ST92 (18), ST6 (3), ST98 (10), and ST118 (18) have previously been shown to be representatives of European clone 2 (EU2) or worldwide clonal lineage 2 (WW2) by other typing methods. All of the isolates with this sequence type were carbapenem and ciprofloxacin resistant but had variable susceptibilities to aminoglycosides. All of the isolates were also blaOXA-23 positive.
FIG. 1.
eBURST population snapshot of A. baumannii constructed from previous publications where official sequence types have been assigned, plus unpublished data from the A. baumannii MLST website, where permission for use was granted by the submitter of the data. CC92 is currently the largest clonal complex. STs 19, 69, 9, 123, 28, 29, and 30 have double-locus-variant relationships with members of CC92 and are therefore closely related, although they are not considered members of the CC by conservative definition.
Three isolates were ST69. This is a double-locus variant (DLV) of multiple STs within CC92, implying a close relationship; however, an intervening single-locus variant (SLV) has not yet been identified, so it cannot be considered a member of CC92 by conservative definition (8). ST69 isolates were ciprofloxacin and amikacin resistant but carbapenem susceptible and blaOXA-23 negative. This phenotype was identified in only a small number of isolates, namely, isolates Q1 (isolated in 1998), Q45 (isolated in 1999), and Q54. Q54 was isolated on the day of admission from a patient transferred from Indonesia in 2002. In spite of the apparent repeated introduction of this ST, there was no evidence of persistence.
ST73 isolates were unique among local carbapenem-resistant strains; they possessed a distinct pulsotype and were ciprofloxacin susceptible. Only four isolates with this phenotype and pulsotype were detected, and all of these were isolated from patients over a 4-month period in 2006. The index ST73 isolate was cultured from a patient who had been transferred from Papua New Guinea. ST73 is a singleton by MLST and has previously been described in South Korea (20).
Three novel singleton STs were identified. ST125 was present in the hospital in 1999 only and was resistant to all aminoglycosides, ciprofloxacin, and meropenem. The source of the index case is unknown. The ST126 isolate (isolate Q55) had an antibiotic susceptibility profile similar to that of the ST69 isolates (i.e., it was resistant to amikacin and ciprofloxacin but susceptible to meropenem); however, these two STs were unrelated. The ST127 isolate (isolate Q22) was cultured from a patient 1 week after admission to the ICU; Q22 was resistant to meropenem only and possessed a blaOXA-23 resistance determinant.
Rep-PCR and PFGE.
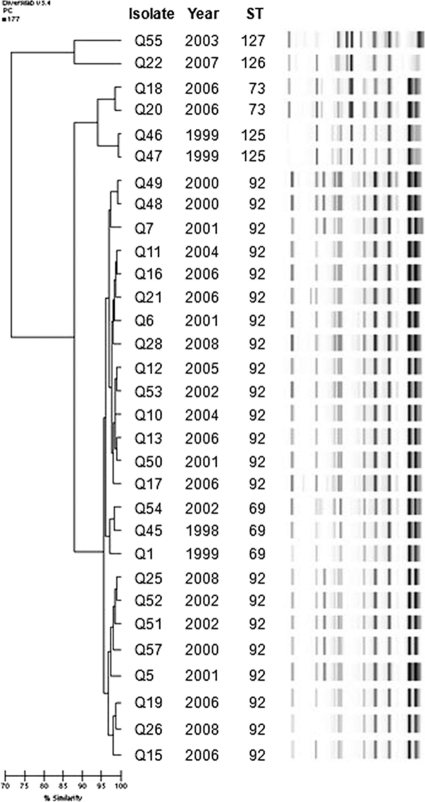
A dendrogram constructed from rep-PCR results is presented in Fig. 2. The results of the three typing methods were generally concordant. By rep-PCR, ST69 clustered >95% with ST92. By PFGE also, ST69 clustered among ST92 isolates (data not shown). This suggests that ST69 may belong to CC92, even though an SLV to link it to the complex by MLST has not yet been identified. Some isolates were indistinguishable by both rep-PCR and PFGE, despite being cultured from patients many years apart (e.g., 7 years for isolates Q6 and Q28 and 6 years for isolates Q28 and Q53, respectively).
FIG. 2.
Dendrogram of A. baumannii rep-PCR patterns by the Pearson correlation method. Isolate number, year of isolation, and ST are also shown.
DISCUSSION
In this study we have examined the epidemiology of multidrug-resistant A. baumannii at a single institution longitudinally over 10 years. We found that in spite of six different STs being represented by non-wild-type A. baumannii isolates tested, ST92 dominated heavily and was isolated from 2000 to 2008. Within ST92 there were diverse, but related, pulsotypes and rep-PCR patterns. The only previous comparison of a large group of A. baumannii isolates by PFGE and MLST showed an unexpected divergence of sequence types by PFGE for a geographically diverse collection of isolates (10). In this study, the sequence types cluster together. These differences likely reflect the sources of the strains comprising the two collections. The only discrepant result between the typing methods was that ST69 isolates clustered among ST92 isolates by PFGE and rep-PCR. However, these ST69 isolates had a distinct epidemiology and phenotype consistent with the differentiation by MLST.
Our analysis identified isolates with indistinguishable pulsotypes or rep-PCR patterns separated by 6 and 7 years, respectively. This suggests clonal spread of a successful A. baumannii strain. In contrast, although there was introduction and transmission of ST69 (on two occasions), ST73, and ST125, we did not find evidence of the persistence of any of these STs for more than 1 year. Possible explanations for the repeated isolation of ST92 for nearly 10 years include readmission of previously colonized local patients (the potential duration of carriage is currently uncertain), reintroduction from interhospital transfers, and long-term persistence in the hospital environment, in spite of intensive infection control efforts. Our results and the known ability of this organism to resist desiccation (23) support the latter possibility. Previous studies using an alternative MLST scheme have also shown repeated isolation of ST2 isolates, which are also representatives of EU2, including isolates that directly correspond to ST92 (18), in a single location (5) or geographic region (7) over long periods of time, but without the continuity at one site presented here.
On a global scale, ST92 is the predicted founder of CC92, the largest and most geographically diverse clonal complex by MLST. The results presented here confirm that our facility in Australia is included in this global epidemic. CC92 corresponds to EU2/WW2 on the basis of previous typing results by other methods for members of the clonal complex, including ST92 and ST118 (18). Further, an EU2 reference strain, RUH 134, has been reported in previous publications both to be ST6 (3) and to have an allelic profile corresponding to that of ST98 (10) by this MLST scheme. It is unclear why the reference strain has given two STs, but ST6 and ST98 are SLVs that differ by only one nucleotide in the gyrB locus. Our rep-PCR results also show similarities between ST92 and WW2 (11), though this is an inferior method for comparison between laboratories.
Although ST92 is heavily represented in the current MLST data set, we cannot rule out the possibility that it may not be truly representative of the diversity and relative abundance of A. baumannii STs. There is presumably a bias toward investigation and typing of strains resistant to multiple antimicrobials. Indeed, this is the case for our collection of isolates, as the antibiotic-resistant strains were those that had been stored for potential future study, and half the strains that we tested were isolated during outbreaks. We attempted to overcome this by including a number of diverse sporadic strains in our analyses; over half of these were, in fact, ST92. We note also that we tested only a representative subset of the total number of A. baumannii isolates collected from 1998 to 2008, and thus, it is possible that some important clones were missed. Globally, there is also likely to be temporal and geographic bias related to the availability of sequencing technology. For example, eBURST analysis of European strains in the database identifies ST98 as the founder of the dominant carbapenem-resistant clonal complex, whereas ST92 is identified as the founder if Asia/Oceania strains are analyzed. The diversity within clonal complexes and the designation of putative founders may change with the collection of more sequence typing data.
In the A. baumannii isolates examined here, similar antibiotic resistance profiles were seen within STs. The presence of blaOXA-23 correlated closely with carbapenem resistance, as has previously been the case where Australian strains have been tested (17, 27), including a single ST92 isolate (16). However, antibiotic resistance profiles are known to be an inaccurate predictor of clonality for A. baumannii (1, 6, 11, 13, 19), and there is also diversity of antibiotic resistance determinants within strains that are related by MLST. For example, carbapenem-susceptible ST92 isolates are widespread in China (9), and carbapenem-resistant ST69 isolates have been identified in South Korea (21), in contrast to the antibiotic susceptibility patterns of our isolates. The large number of carbapenem-susceptible ST92 isolates in China supports the notion that antibiotic resistance may not be the primary determinant of hospital adaptiveness for this clone.
For A. baumannii of CC92/WW2/EU2, the ability to persist for over 9 years in a single hospital and the variable antibiotic susceptibility of CC92 (and within ST92) suggest that adaptation to the hospital environment, as well as antibiotic resistance, may be important for the success of A. baumannii as a nosocomial pathogen. This raises the question of why this clonal complex has been so successful. With the progressive accumulation of global data, MLST is proving to be a powerful tool for the study of A. baumannii epidemiology, and the addition of Australian data adds new evidence for the intercontinental spread of the most successful clonal complex. Colistin-resistant isolates of ST92 have recently been described in South Korea (21), a further step toward a truly panresistant epidemic A. baumannii clone. In order to fully understand and combat multidrug-resistant nosocomial A. baumannii, the mechanisms of hospital adaptiveness beyond antibiotic resistance demand more attention.
Acknowledgments
Funding for this study was provided by Pathology Queensland's Study, Education and Research Trust Fund. The Diversilab equipment used in this study was loaned by bioMérieux, Melbourne, Australia.
We are grateful to the following people who allowed use of their unpublished data available on the A. baumannii MLST website for the construction of the eBURST diagram: Mari Matsui, Fernando Chavez, Nuno Mendonca, Guo-Bao Tian, Yohei Doi, Liulin Luo, P. Ravasi, A. Mussi, A. Limansky, A. Viale, and Jose Luis Juan-Banon. We also thank the curator of the database, Sergio Bartual, who facilitated this. The technical assistance provided by the following people is also gratefully acknowledged: Narelle George, Jacqueline Schooneveldt, Haakon Bergh, Fatimah Haslina, Tarrant Hansen, and Angela Duffy.
Footnotes
Published ahead of print on 25 August 2010.
REFERENCES
- 1.Abbo, A., S. Navon-Venezia, O. Hammer-Muntz, T. Krichali, Y. Siegman-Igra, and Y. Carmeli. 2005. Multidrug-resistant Acinetobacter baumannii. Emerg. Infect. Dis. 11:22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams-Haduch, J. M., D. L. Paterson, H. E. Sidjabat, A. W. Pasculle, B. A. Potoski, C. A. Muto, L. H. Harrison, and Y. Doi. 2008. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob. Agents Chemother. 52:3837-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartual, S. G., H. Seifert, C. Hippler, M. A. Luzon, H. Wisplinghoff, and F. Rodriguez-Valera. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Diancourt, L., V. Passet, A. Nemec, L. Dijkshoorn, and S. Brisse. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. [DOI] [PMC free article] [PubMed]
- 6.Dijkshoorn, L., H. Aucken, P. Gerner-Smidt, P. Janssen, M. E. Kaufmann, J. Garaizar, J. Ursing, and T. L. Pitt. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Popolo, A., M. Giannouli, M. Triassi, S. Brisse, and R. Zarrilli. 28 April 2010. Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin. Microbiol. Infect. [Epub ahead of print.] [DOI] [PubMed]
- 8.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu, Y., J. Zhou, H. Zhou, Q. Yang, Z. Wei, Y. Yu, and L. Li. 2010. Wide dissemination of OXA-23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J. Antimicrob. Chemother. 65:644-650. [DOI] [PubMed] [Google Scholar]
- 10.Hamouda, A., B. A. Evans, K. J. Towner, and S. G. Amyes. 2010. Characterisation of epidemiologically unrelated Acinetobacter baumannii isolates from four continents using multilocus sequence typing, pulsed-field gel electrophoresis and sequence-based typing of blaOXA-51-like genes. J. Clin. Microbiol. 48:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins, P. G., C. Dammhayn, M. Hackel, and H. Seifert. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:233-238. [DOI] [PubMed] [Google Scholar]
- 12.Ho, P. L., A. Y. Ho, K. H. Chow, E. L. Lai, P. Ching, and W. H. Seto. 2009. Epidemiology and clonality of multidrug-resistant Acinetobacter baumannii from a healthcare region in Hong Kong. J. Hosp. Infect. 74:358-364. [DOI] [PubMed] [Google Scholar]
- 13.Hujer, K. M., A. M. Hujer, E. A. Hulten, S. Bajaksouzian, J. M. Adams, C. J. Donskey, D. J. Ecker, C. Massire, M. W. Eshoo, R. Sampath, J. M. Thomson, P. N. Rather, D. W. Craft, J. T. Fishbain, A. J. Ewell, M. R. Jacobs, D. L. Paterson, and R. A. Bonomo. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50:4114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet-distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, B. N., A. Y. Peleg, T. P. Lodise, J. Lipman, J. Li, R. Nation, and D. L. Paterson. 2009. Management of meningitis due to antibiotic-resistant Acinetobacter species. Lancet Infect. Dis. 9:245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 17.Mak, J. K., M. J. Kim, J. Pham, J. Tapsall, and P. A. White. 2009. Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 63:47-54. [DOI] [PubMed] [Google Scholar]
- 18.Mugnier, P. D., L. Poirel, T. Naas, and P. Nordmann. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemec, A., L. Krizova, M. Maixnerova, L. Diancourt, T. J. van der Reijden, S. Brisse, P. van den Broek, and L. Dijkshoorn. 2008. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 62:484-489. [DOI] [PubMed] [Google Scholar]
- 20.Park, Y. K., J. Y. Choi, S. I. Jung, K. H. Park, H. Lee, D. S. Jung, S. T. Heo, S. W. Kim, H. H. Chang, H. S. Cheong, D. R. Chung, K. R. Peck, J. H. Song, and K. S. Ko. 2009. Two distinct clones of carbapenem-resistant Acinetobacter baumannii isolates from Korean hospitals. Diagn. Microbiol. Infect. Dis. 64:389-395. [DOI] [PubMed] [Google Scholar]
- 21.Park, Y. K., S. I. Jung, K. H. Park, H. S. Cheong, K. R. Peck, J. H. Song, and K. S. Ko. 2009. Independent emergence of colistin-resistant Acinetobacter spp. isolates from Korea. Diagn. Microbiol. Infect. Dis. 64:43-51. [DOI] [PubMed] [Google Scholar]
- 22.Peleg, A. Y., J. M. Bell, A. Hofmeyr, and P. Wiese. 2006. Inter-country transfer of Gram-negative organisms carrying the VIM-4 and OXA-58 carbapenem-hydrolysing enzymes. J. Antimicrob. Chemother. 57:794-795. [DOI] [PubMed] [Google Scholar]
- 23.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seifert, H., L. Dolzani, R. Bressan, T. van der Reijden, B. van Strijen, D. Stefanik, H. Heersma, and L. Dijkshoorn. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turton, J. F., N. Woodford, J. Glover, S. Yarde, M. E. Kaufmann, and T. L. Pitt. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valenzuela, J. K., L. Thomas, S. R. Partridge, T. van der Reijden, L. Dijkshoorn, and J. Iredell. 2007. Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J. Clin. Microbiol. 45:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dessel, H., L. Dijkshoorn, T. van der Reijden, N. Bakker, A. Paauw, P. van den Broek, J. Verhoef, and S. Brisse. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105-112. [DOI] [PubMed] [Google Scholar]
- 29.Wisplinghoff, H., C. Hippler, S. G. Bartual, C. Haefs, D. Stefanik, P. G. Higgins, and H. Seifert. 2008. Molecular epidemiology of clinical Acinetobacter baumannii and Acinetobacter genomic species 13TU isolates using a multilocus sequencing typing scheme. Clin. Microbiol. Infect. 14:708-715. [DOI] [PubMed] [Google Scholar]