Abstract
We recently demonstrated the existence of a previously uncharacterized subset of actomyosin fibers that form the perinuclear actin cap, a cytoskeletal structure that tightly wraps around the nucleus of a wide range of somatic cells. Fibers in the actin cap are distinct from well-characterized, conventional actin fibers at the basal and dorsal surfaces of adherent cells in their subcellular location, internal organization, dynamics, ability to generate contractile forces, response to cytoskeletal pharmacological treatments, response to biochemical stimuli, regulation by components of the linkers of nucleoskeleton and cytoskeleton (LINC) complexes, and response to disease-associated mutations in LMNA, the gene that encodes for the nuclear lamin component lamin A/C. The perinuclear actin cap precisely shapes the nucleus in interphase cells. The perinuclear actin cap may also be a mediator of microenvironment mechanosensing and mechanotransduction, as well as a regulator of cell motility, polarization and differentiation.
Key words: actin, nucleus, LINC complexes, nuclear lamina, cell shape, cancer, laminopathies
Introduction
We recently reported on the existence of a cytoskeletal structure containing a small number of thick actomyosin fibers that are highly organized at the apical surface of the interphase nucleus in a wide range of adherent cells (Fig. 1).1 These actin fibers form the perinuclear actin cap, which is physically connected to the nuclear envelope through linkers of nucleoskeleton and cytoskeleton (LINC) complexes. Actin filament bundles in the actin cap are distinct from well-characterized, conventional stress fibers located at the basal and dorsal surfaces of adherent cells in their subcellular location, internal organization, dynamics, response to cytoskeletal pharmacological treatments, response to biochemical stimuli, regulation by components of the LINC complexes, and response to disease-associated mutations in LMNA, the gene that encodes for the nuclear lamin component lamin A/C. The perinuclear actin cap shapes the nucleus of interphase cells, a function that is abrogated in cells deficient or mutated in LMNA, which lack an actin cap. Here, we aim to further highlight the functional and structural differences between conventional actin filament structures and the perinuclear actin cap in both physiological and pathological conditions. We also discuss other potential functions of the actin cap, including as a mediator of cellular mechanosensing and mechanotransduction, as well as a regulator of cell motility, polarization and differentiation.
Figure 1.
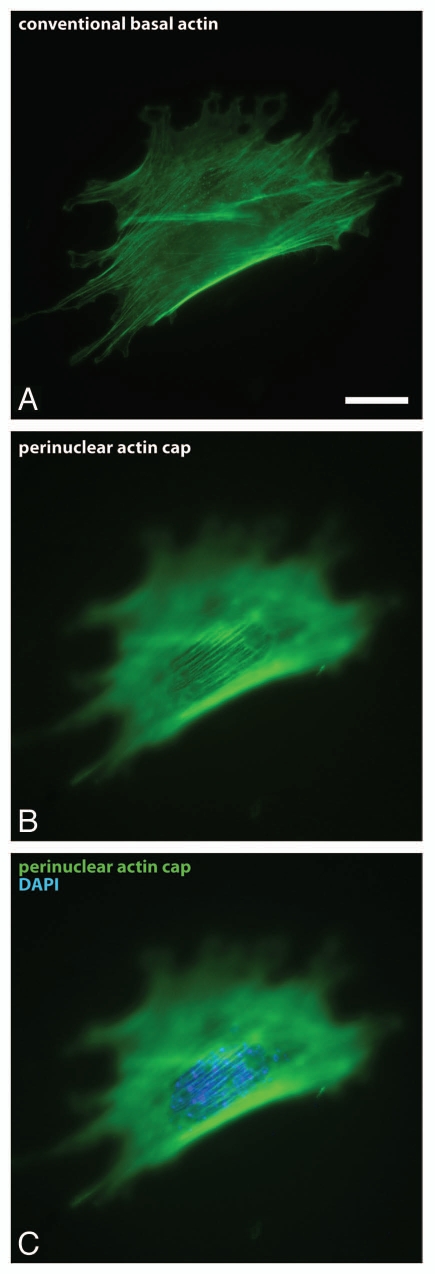
The perinuclear actin cap. (A–C) Fluorescence micrographs showing conventional basal stress fibers confined to the basal surface of the cell (A) and the perinuclear actin cap located at the apical surface of the nucleus (B and C). F-actin was stained using phalloidin (red); nuclear DNA was stained using DAPI (blue). Scale bar, 20 µm.
The Actin Cap in Health
The actin cap can readily be detected by staining cells for actin with phalloidin and conventional fluorescence microscopy at the top of the nucleus (Fig. 1). The actin cap is composed of thick, parallel actin filament bundles, which gently curve around the interphase nucleus (Fig. 1).1 In contrast, well-characterized stress fibers are confined to the basal and dorsal surfaces of the cell. We note that, unlike typically depicted in textbooks, the nucleus of adherent cells is not a spherical or an ellipsoid organelle; the nucleus forms a disk oriented parallel to the basal surface of the cell and is 15–25 µm in diameter and only 5–7 µm in thickness.1 This vertical confinement of the nucleus is mediated by the actin cap. Indeed, cell treatment with an extremely low dose of the actin depolymerizing drug latrunculin B (<60 nM), which only affects the actin cap, not conventional stress fibers, induces significant upward bulging of the nucleus.1 Moreover, movies of live cells at rest or under shear flow indicate that interphase nuclei of adherent cells undergo large rotational and translational excursions in the cytoplasm (detected by the movements of the nucleoli), which are confined to the same plane of focus, indicating no nuclear movements around a horizontal axis.2 These results suggest that another function of the actomyosin fibers in the actin cap in adherent cells is to pull the nucleus towards the cellular basal surface in a manner similar to ropes anchoring a hot-air balloon to the ground (Fig. 2).
Light microscopy of fixed and live specimens also reveals that actin filament bundles in the actin cap, whose number is of the order of ten (Fig. 1), tend to be highly parallel to one another within the actin cap, which is typically itself parallel to both the long axis of the nucleus and the long axis of the cell.1 In contrast actin filament bundles forming basal stress fibers located at the cortex and basal surface of the cell are much more numerous and display orientation correlation among themselves only locally within the cell (Fig. 1). Moreover, the orientation of basal stress fibers is uncorrelated with cellular and nuclear overall orientations.
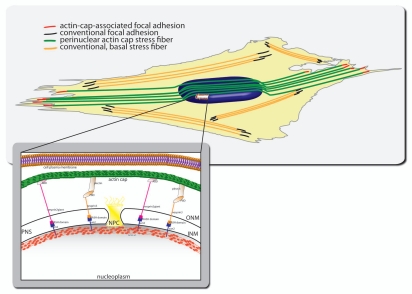
Our unpublished observations indicate that by progressively lowering the plane of focus of the confocal microscope of cells stained with phalloidin and antibodies against focal adhesion proteins vinculin, reveals that actin filament bundles of the actin cap are terminated at the basal surface of the cell by focal adhesions confined to the cell periphery (unpublished results, Kim D-H and Wirtz D) (Fig. 2). These actin cap-associated focal adhesions are larger and more elongated than conventional focal adhesions terminating stress fibers that lie entirely within the basal surface of the cell. Unlike actin-cap associated focal adhesions, conventional focal adhesions are distributed all over the basal surface.
Figure 2.
Schematic of subcellular organization of the perinuclear actin cap and associated focal adhesions. The actin cap is tightly connected to the apical surface of the nuclear envelope, not to the plasma membrane. Inset. Physical connections between the actin cap and the nucleus are mediated by components of the linker of nucleoskeleton and cytoskeleton (LINC) complexes, including Nesprin-3 and Nesprin-2giant, which are connected to the nuclear lamina through SUN proteins. Disruption of LINC complexes specifically disorganizes or eliminates the actin cap, without significantly affecting conventional stress fibers located at the basal surface of adherent cells. Similarly, treatment of cells with low dose (60 nM) of the F-actin-depolymerizing drug latrunculin B only affects the actin fibers of the actin cap because the actin cap is much more dynamic than conventional basal actin fibers. Actin filament bundles in the actin cap are terminated by specialized focal adhesions, which we name actin cap-associated focal adhesions. There are about ten actin cap-associated focal adhesions per cell. Actin cap-associated focal adhesions are restricted to the cell periphery and are larger and longer than conventional focal adhesions, which terminate conventional basal stress fibers.
Live-cell microscopy of cells transfected with EGFP-lifeact suggest that the actin cap is much more dynamic than conventional basal stress fibers.1 Unlike EGFP-actin, EGFP-lifeact allows us to monitor actin dynamics for long durations (>12 h) without significant photobleaching or interference with cell functions.3,4 The actin cap tends to continuously change its shape and can move large distances (>5 µm) over time scales of minutes, while stress fibers are immobile during these time intervals.1 The fast dynamics of the actin cap mirrors the movements of the nucleus, which are faster than the movements of the cell.2 This suggests that the actin cap directs the movements of the nucleus, although more work is required to determine cause and effect.
Fluorescence recovery after photobleaching (FRAP) analysis of cells transfected with EGFP-actin indicates that exchange dynamics between F-actin in the actin cap and monomeric actin (G-actin) in the cytoplasm is much faster that actin exchange dynamics in basal stress fibers (unpublished data, Khatau SB and Wirtz D). Moreover, the actin cap in cells treated with low dose of latrunculin B, which sequesters G-actin away from the polymerizable pool of actin, disappears quickly, while conventional stress fibers remain intact for long periods of time.1 Together these results indicate that the actin cap is a much more dynamic structure than conventional basal stress fibers.
A key function of the perinuclear actin cap is to regulate the shape of the interphase nucleus.1 Early work by Champy and Carleton suggested a correlation between the overall shape of cells of different origin (including animal and plant cells) and the shape of their nucleus.5 We first tested whether this strong correlation between cellular shape and nuclear shape still held in a single type of cells, a hypothesis that, surprisingly, had never previously been tested. This is despite the predictive power of nuclear shape in cancer staging and in a wide range of other human diseases, including muscular dystrophy and accelerated aging.6–9 We found that nuclear shape and cellular shape strongly correlated within a single type of cell (here MEFs),1 suggesting the existence of physical connections between cell periphery and the nuclear envelope. Because the shape of cells is highly variable for cells plated on culture dishes, cells were positioned on adhesive fibronectin-coated microstripes of width between 10 and 50 µm, i.e., respectively smaller and larger than the natural size of the cell (∼40 µm) on non-patterned substrates to control overall cell shape.1 These adhesive stripes were flanked by non-adhesive polyethylene glycol (PEG)-coated stripes to confine the cells to the adhesive stripes. Remarkably, cell adhesion to stripes wider than the size of cells on unpatterned surfaces induced rounder nuclei than in cells on non-patterned surfaces, while cells on narrow stripes showed highly elongated nuclei.1 This result is not a priori obvious as nuclei in cells on narrow stripes could simply bulge out from their basal confinement and, therefore, remain relatively round. We predicted the existence of a structure that was keeping nuclei highly confined not only laterally, but also vertically, promoting a round nuclear shape in cells on wide stripes and an elongated nuclear shape in cells on thin stripes. Confocal microscopy revealed that this cytoplasmic structure confining the nucleus was indeed the perinuclear actin cap.1
To demonstrate that cell shape controlled nuclear shape through the actin cap, cells were treated with low dose of latrunculin B.1 The shape of nuclei in latrunculin B-treated cells did not respond to changes in the width of the underlying adhesive stripe. As already mentioned above, latrunculin B does not affect conventional stress fibers as quickly as it does the actin cap. Cells treatment with low doses of drugs inhibiting myosin II/ROCK-based actomyosin contractility10,11 affected phosho-MLC content in the actin cap, not in conventional stress fibers, but they did not affect regulation of nuclear shape by cell shape on narrow stripes.1 In contrast these inhibitors eliminated the ability of the actin cap to uniformly stretch the nucleus beyond its spontaneous size and roundness.1 These results suggest that the cell shape controls nuclear shape through the contractile actin filament fibers of perinuclear actin cap.
Micropipette manipulation is routinely used to probe the mechanical properties of the nucleus. This method suggests that lamin A/C greatly contributes to nuclear mechanics.12 As assessed by immunofluorescence microscopy, F-actin disassembly and the associated dismantlement of the actin cap does not significantly affect the organization of the major nuclear lamina component lamin A/C.1 This important result suggests that the inability of nuclei of treated cells to respond to forced cell shape changes is not due mechanical weakening of the nuclear lamina, per se. Rather, this result suggests both that the nuclear lamina is necessary but not sufficient to shape the nucleus and that the shape of the nucleus depends on the actin cap. Unfortunately, to allow for close contact with the nucleus, micropipette manipulation requires pre-treatment of cells using a high dose (>1 µM) of F-actin depolymerizing drug (e.g., cytochalasin D or latrunculin B), which, among other things, eliminates the actin cap. Hence micropipette manipulation probes the intrinsic mechanical properties of the nucleus, not the mechanical properties of the nucleus in the larger context of nucleus-cytoplasm interconnections mediated by the actin cap.13 One way to reconcile these seemingly contradictory results—nuclear shaping by the actin cap and nuclear mechanics dominated by lamin A/C—is a model where lamin A/C determines the intrinsic rigidity of the nuclear cortex, while the actin cap, which we discuss below, is directly connected to the nuclear lamina though specific connecting proteins, controls the shape of the lamin A/C-rich stiff nucleus. We note that other biophysical methods preserving the integrity of nucleus-cytoskeleton connections, including particle tracking microrheology or magnetic tweezers,14–16 have been used to directly probe the mechanical properties of the nucleus in living cells.
In addition to MEFs, for which most of the work on the actin cap has been conducted so far, the actin cap has been detected in a wide range of somatic cells, including human foreskin fibroblasts, human lung fibroblasts, human umbilical vein endothelial cells, as well as Swiss 3T3 mouse fibroblasts.1 However, the actin cap in normal human breast epithelial (MCF-10A) cells appeared on top of the nucleus as short actin filament bundles that formed a reticulated network, as opposed to parallel bundles (unpublished results, Khatau SB and Wirtz D). Moreover, actin caps are absent in HeLa cells, a human cervical cancer cell line. Why actin caps are absent in these cells remains and whether the absence of actin caps is a general feature of transformed cells remain to be explored.
One may wonder whether the actin cap is an artifact of conventional cell culture using two-dimensional (2-D) substrates. Clearly, endothelial and epithelial cells form multicellular structures in vivo that are essentially locally 2-D. In this case, the actin cap may play an important role in vivo, as described above. However, cells such as fibroblasts reside in an in vivo milieu that is three-dimensional. In unpublished studies, we investigated the organization of the actin cap in cells that were fully embedded inside a three-dimensional (3-D) collagen I matrix. We found that the equivalent structure of the actin cap is topologically different from that in cells in two-dimensional (2-D) cultures, but functionally similar. In the 3-D case, the actin cap seems to be composed of thick actin filament bundles that radiate from the perinuclear region and impinge on the plasma membrane to form highly polarized pseudopodial protrusions, which themselves drive cell motility (unpublished results, Bloom RJ and Wirtz D).17 Similar to the 2-D case, the elimination of the 3-D actin cap prevents cells from shaping their nucleus. As discussed in more details below, the specific elimination of the actin cap in cells embedded in a 3-D matrix, dramatically reduce cell speed and persistence of migration, while this has little effect on single-cell motility on 2-D substrates.
The Actin Cap in Disease
Defects in nuclear shape are commonly used in clinical setting as markers of disease and differentiation in human cells and tissues.6 In particular, laminopathic cells harvested from patients and animal models of a wide range of diseases stemming from mutations in LMNA are characterized by ill-shaped nuclei.7–9 We investigated the status of the actin cap in MEFs lacking LMNA from a mouse model for muscular dystrophy.9 Fluorescence microscopy detects no significant difference in actin filament network organization and focal adhesions between Lmna+/+ and Lmna−/− cells.18 However, while a majority of Lmna+/+ cells show a prominent actin cap, a majority of Lmna−/− cells display either no actin cap or a disorganized actin cap.1 Moreover, Lmnal530p/l530p cells from other mouse models of laminopathies, including cells from mice displaying symptoms of accelerated aging (progeria),19 tend to show even fewer caps than Lmna-/- cells.1 Lmna−/− and Lmnal530p/l530p cells both show a cytoplasm that is significantly softer than the cytoplasm of wild-type cells, another consequence of the critical importance of functional linkages between nucleus and cytoplasm.18,20,21
LINC complexes connect lamin A/C to the cytoskeleton, span the nuclear envelope, and mediate physical connections between the nuclear lamina and the cytoskeleton.22 LINC complex proteins Sun1 and Sun2 recruit KASH-domain-containing proteins Nesprin-2giant and Nesprin-3 to the outer nuclear membrane through Sun-KASH interactions.23–28 Lmna−/− and Lmnal530p/l530p cells both display disrupted LINC complexes at the nuclear envelope. Forced disruption of the LINC complexes using an EGFP-KASH2 construct,29 which prevents binding between Sun proteins and nesprins, recapitulates the inability of Lmna−/− and Lmnal530p/l530p cells to shape their nucleus. KASH2-transfected cells also show significantly fewer actin caps than control cells. Importantly, similarly to low-dose treatment of latrunculin B, the disruption of LINC complexes does not significantly affect the organization of basal stress fibers organization and associated focal adhesions and specifically affects the actin cap.1,18 This suggests that the actin cap could be topologically disconnected from the rest of the actin cytoskeleton.
Functions of the Actin Cap
We have already identified a major function for the actin cap: it shapes the interphase nucleus. Given its location, one can reason that another function of the actin cap could be position the nucleus within the cytoplasm and mediate its translocation. Recent work by our group and Gundersen and coworkers suggest that the position of the nucleus in the cell is not random and is mediated by actin.2,30 Given the fact that only actin-cap fibers are physically attached to the nucleus, it is likely that nuclear motion and positioning are mediated by the actin cap, although a direct demonstration is still missing.
Actin filament bundles that make up the perinuclear actin cap are terminated by a subset of focal adhesions, which we name actin-cap associated focal adhesions. It is tempting to speculate that actin-cap associated focal adhesions play a critical role in cellular mechanosensing and mechanotransduction, i.e., the ability of cells to sense and respond to changes in substrate rigidity and externally applied forces, respectively.31 Indeed, the discovery of the actin cap and its connections to the nuclear lamina through the LINC complexes identifies for the first time an entirely physical (as opposed to biochemical) pathway directly connecting the extracellular microenvironment to the genome.32 This pathway contains the following components: (1) actin-cap associated focal adhesions which connect the extracellular matrix to F-actin bundles in the actin cap; (2) the actin cap bundles, which bind Nesprins at the nuclear envelope through either multi-domain actin-binding protein plectin which binds Nesprin-3,33,34 or the actin-binding domain of Nesprin-2giant;35,36 (3) Nesprins, which bind SUN proteins through KASH-SUN interactions;22,29 (4) SUN proteins, which bind lamin A/C;29 and (5) finally lamin A/C, which directly or indirectly interacts with DNA.37 Therefore, it is plausible that cellular response to changes in substrate rigidity or to externally applied forces are dominated by the actin cap as opposed to conventional stress fibers, which are not connected to the nucleus, although much more work is required to directly test this hypothesis. Recent FRAP analysis indicate that LINC complex components are highly dynamic.38 Therefore, the physical connections between the LINC complexes and F-actin in the actin cap, which is itself highly dynamic, are not permanent, presumably allowing for rapid response and remodeling of the actin cap and the genome to extracellular stimuli, such as changes in the physical and biochemical properties of the microenvironment.
Lmna−/− cells show impaired cell migration in the wound-healing assay.18 The wound-healing assay not only involves intrinsic motility, but also intercellular contacts, which can affect migration. Indeed, single cell motility parameters of single Lmna+/+ and Lmna−/− cells on flat substrates, including cell speed and persistence, are similar. However, Lmna−/− cells completely embedded inside a 3-D matrix show a severe cell motility phenotype when compared to Lmna+/+ cells (unpublished results, Bloom RJ, Khatau SB and Wirtz D). Lamin-deficient cells show few actin filament bundles that can produce protrusions, suggesting the elimination of the 3-D analog of the actin cap observed in 2-D cell culture. This combined motility/protrusion defect is rescued in Lmna−/− cells transfected with EGFP-lamin A/C, for which actin filament bundles protruding from the nuclear region are restored, and is recapitulated in cells transfected with EGFP-KASH2 for such actin filament bundles are eliminated (unpublished results, Bloom RJ, Khatau SB and Wirtz D). These observations suggest the importance of nuclear-actin-cap connections in mediating motility and protrusion activity for cells in a 3D matrix through the regulation of the organization of the 3D actin cap.
The above results suggest a remarkably wide range of critically important functions for the actin cap, functions mediated by the actin cap as opposed to other actin structures in the cell, such as dorsal or basal actin. The above studies indicate that the perinuclear actin cap is different from conventional stress fibers confined to the basal cellular surface in terms of organization, orientation, subcellular position, dynamics and functions. However, future studies will need to identify molecular markers that are specific to the actin cap, not basal stress fibers, and actin-cap associated focal adhesions, not conventional focal adhesions, both in 2-D and 3-D micro-environments.39 Moreover, how the actin cap may affect chromosomal organization through its connections to the lamina remains to be studied.
Acknowledgements
This work was supported by NIH grants GM084204 and CA143868. S.B.K. was partially supported by an NSF-IGERT graduate training program in the Johns Hopkins Institute for NanoBioTechnology. We thank members of the Wirtz group for sharing their unpublished results.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/12331
References
- 1.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, et al. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JS, Chang MI, Tseng Y, Wirtz D. Cdc42 mediates nucleus movement and MTOC polarization in Swiss 3T3 fibroblasts under mechanical shear stress. Mol Biol Cell. 2005;16:871–880. doi: 10.1091/mbc.E03-12-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wildt B, Wirtz D, Searson PC. Programmed subcellular release for studying the dynamics of cell detachment. Nat Methods. 2009;6:211–213. doi: 10.1038/nmeth.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champy C, Carleton HM. Memoirs: observations on the shape of the nucleus and its determination. J Cell Sci. 1921;65:589–625. [Google Scholar]
- 6.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 7.Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 8.Mounkes L, Kozlov S, Burke B, Stewart CL. The laminopathies: nuclear structure meets disease. Curr Opin Genet Dev. 2003;13:223–230. doi: 10.1016/s0959-437x(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JS, Panorchan P, Hale CM, Khatau SB, Kole TP, Tseng Y, et al. Ballistic intracellular nanorheology reveals ROCK-hard cytoplasmic stiffening response to fluid flow. J Cell Sci. 2006;119:1760–1768. doi: 10.1242/jcs.02899. [DOI] [PubMed] [Google Scholar]
- 11.Panorchan P, Lee JS, Kole TP, Tseng Y, Wirtz D. Microrheology and ROCK signaling of human endothelial cells embedded in a 3D matrix. Biophys J. 2006;91:3499–3507. doi: 10.1529/biophysj.106.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher D. E. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng Y, Lee JS, Kole TP, Jiang I, Wirtz D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J Cell Sci. 2004;117:2159–2167. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 14.Wirtz D. Particle-tracking microrheology of living cells: principles and applications. Annu Rev Biophys. 2009;38:301–326. doi: 10.1146/annurev.biophys.050708.133724. [DOI] [PubMed] [Google Scholar]
- 15.Celedon A, Nodelman IM, Wildt B, Dewan R, Searson P, Wirtz D, et al. Magnetic tweezers measurement of single molecule torque. Nano Lett. 2009;9:1720–1725. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haber C, Wirtz D. Magnetic tweezers for DNA micromanipulation. Rev Sci Instrum. 2000;71:4561–4570. [Google Scholar]
- 17.Bloom RJ, George JP, Celedon A, Sun SX, Wirtz D. Mapping local matrix remodeling induced by a migrating tumor cell using three-dimensional multiple-particle tracking. Biophys J. 2008;95:4077–4088. doi: 10.1529/biophysj.108.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, et al. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J. 2008;95:5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mounkes LC, Kozlov S, Hernandez L, Sullivan T, Stewart CL. A progeroid syndrome in mice is caused by defects in A-type lamins. Nature. 2003;423:298–301. doi: 10.1038/nature01631. [DOI] [PubMed] [Google Scholar]
- 20.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holaska JM, Wilson KL, Mansharamani M. The nuclear envelope, lamins and nuclear assembly. Curr Opin Cell Biol. 2002;14:357–364. doi: 10.1016/s0955-0674(02)00329-0. [DOI] [PubMed] [Google Scholar]
- 22.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starr DA, Han M. ANChors away: an actin based mechanism of nuclear positioning. J Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 24.Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, et al. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- 25.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 27.Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly. 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- 28.Technau M, Roth S. The Drosophila KASH domain proteins Msp-300 and Klarsicht and the SUN domain protein klaroid have no essential function during oogenesis. Fly. 2008;2 doi: 10.4161/fly.6288. [DOI] [PubMed] [Google Scholar]
- 29.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Vogel V, Sheetz MP. Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Curr Opin Cell Biol. 2009;21:38–46. doi: 10.1016/j.ceb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ketema M, Wilhelmsen K, Kuikman I, Janssen H, Hodzic D, Sonnenberg A. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J Cell Sci. 2007;120:3384–3394. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- 34.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, Lu W, et al. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 37.Shoeman RL, Traub P. The in vitro DNA-binding properties of purified nuclear lamin proteins and vimentin. J Biol Chem. 1990;265:9055–9061. [PubMed] [Google Scholar]
- 38.Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraley SI, Feng F, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]