Protein secretion in plant cells is generally thought to be achieved by vesicle-mediated traffic between the endoplasmic reticulum, Golgi apparatus, trans-Golgi network, and the plasma membrane. This study describes an unconventional secretory pathway involving a double-membrane exocyst-positive organelle that mediates exocytosis directly from the cytosol to cell wall.
Abstract
The exocyst protein complex mediates vesicle fusion with the plasma membrane. By expressing an (X)FP-tagged Arabidopsis thaliana homolog of the exocyst protein Exo70 in suspension-cultured Arabidopsis and tobacco (Nicotiana tabacum) BY-2 cells, and using antibodies specific for Exo70, we detected a compartment, which we term EXPO (for exocyst positive organelles). Standard markers for the Golgi apparatus, the trans-Golgi network/early endosome, and the multivesicular body/late endosome in plants do not colocalize with EXPO. Inhibitors of the secretory and endocytic pathways also do not affect EXPO. Exo70E2-(X)FP also locates to the plasma membrane (PM) as discrete punctae and is secreted outside of the cells. Immunogold labeling of sections cut from high-pressure frozen samples reveal EXPO to be spherical double membrane structures resembling autophagosomes. However, unlike autophagosomes, EXPOs are not induced by starvation and do not fuse with the lytic compartment or with endosomes. Instead, they fuse with the PM, releasing a single membrane vesicle into the cell wall. EXPOs are also found in other cell types, including root tips, root hair cells, and pollen grains. EXPOs therefore represent a form of unconventional secretion unique to plants.
INTRODUCTION
The exocyst is an octameric protein complex, first discovered in yeast (Novick et al., 1980; TerBush et al., 1996) and subsequently in mammals (Hsu et al., 1996), which mediates post Golgi vesicle fusion with the plasma membrane (PM) (Hsu et al., 2004; He and Guo, 2009). Of the eight proteins, seven (Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84) are associated with the vesicle membrane, whereas Sec3 and Exo70 are recruited to the PM via binding to phosphatidyl (4,5) biphosphate (He et al., 2007; Zhang et al., 2008). Exocyst assembly and function is mediated by the Rho family of GTPases (Hsu et al., 2004; Roumanie et al., 2005). Exocyst components are normally restricted to the tips of growing buds in yeast (TerBush and Novick, 1995) and in mammalian cells are recruited to regions of the PM where active exocytosis and membrane growth is occurring, such as the growing front of migrating cells (Zuo et al., 2006). Indeed, it has been proposed that the positioning of bound Sec3 and Exo70 mediates the targeting of secretory vesicles to particular domains of the PM (Orlando and Guo, 2009). However, in mammals, exocyst proteins have also been found associated with the trans-Golgi network (TGN) (Yeaman et al., 2001; Langevin et al., 2005) and with recycling endosomes (Prigent et al., 2003; Oztan et al., 2007).
Homologs to all eight exocyst proteins have been found in plants (Elias et al., 2003; Chong et al., 2010; Hála et al., 2008), but interestingly, there are over 23 paralogs of Exo70 in Arabidopsis thaliana, with Exo70A1 being the most abundant (Synek et al., 2006). It has recently been proposed by Zárský et al. (2009) that the large number of Exo70 proteins, together with the greater number of SNAREs and Rab GTPases (Rutherford and Moore, 2002; Lipka et al., 2007), reflect an increased variability in targeting domains for vesicle traffic in plant cells. Nevertheless, Arabidopsis Sec6, Sec8, and Exo70A1 have been localized to the tips of growing pollen tubes (Cole et al., 2005; Hála et al., 2008), and Sec3 and Exo70A1 appear to play crucial roles in polarized secretion in elongating root hairs (Wen et al., 2005) and in the stigma toward compatible pollen (Samuel et al., 2009).
In this article, we report on the distribution of one of the Arabidopsis Exo70 paralogs: Exo70E2. Not only does this exocyst protein locate to the PM in discrete punctate domains, but it is present in the unique double membrane structures that we term EXPO (for exocyst positive organelles). These structures are not labeled by any of the standard endomembrane markers used for identifying the Golgi apparatus, the TGN, or multivesicular body (MVB), nor do they become labeled with the endocytic tracer dye FM4-64 and do not colocalize with Atg8e, an autophagosome marker. They are also not affected by inhibitors of secretion (brefeldin A) or endocytosis (wortmannin). In high-pressure frozen/freeze-substituted samples of both Arabidopsis and tobacco (Nicotiana tabacum) BY-2 cells, we have been able to visualize stages in the fusion of EXPO with the PM, leading to the release of a cytosol-containing membrane vesicle outside of the PM. These structural observations are supported by following the fate of a cytosolically expressed protein, which associates with EXPO and subsequently is detectable outside of the cell. Per definition, EXPO is therefore an exosome but has a different origin to those described in mammalian cells (Denzer et al., 2000; Simons and Raposo, 2009).
RESULTS
Expression of (X)FP-Tagged Exo70 Paralogs in Arabidopsis Protoplasts
We prepared green fluorescent protein (GFP)- and red fluorescent protein (RFP)-tagged constructs of eight of the 23 Arabidopsis Exo70 paralogs and expressed them in protoplasts obtained from Arabidopsis suspension cultured cells under the control of the 35S promoter and the 3′ Nos terminator. Only three of these constructs, Exo70A1, Exo70B1, and Exo70E2, gave rise to punctate fluorescence located both at the PM and within the cytoplasm (see Supplemental Figures 1 and 2 online). All of the other constructs (Exo70B2, Exo70D1, Exo70D2, Exo70E1, and Exo70F1) lead to pronounced cytosolic signals (see Supplemental Figure 1 online). The punctate fluorescent signals produced by the coexpression of Exo70A1-GFP and Exo70E2-mRFP colocalized, as did the signals from the coexpression of Exo70B1-GFP and Exo70E2-mRFP (see Supplemental Figure 2A online). Because of the consistency and clarity of labeling, we restricted our observations to Exo70E2 for the rest of this investigation. Coexpression of different combinations of C- and N-terminally (X)FP-tagged Exo70E2 showed that neither the distribution nor the size of the fluorescent punctae is affected by the position or type of fluorescent tag (see Supplemental Figure 2B online).
Exo70E2 Labels the PM and Organelles That Do Not Lie on the Secretory or Endocytic Pathways
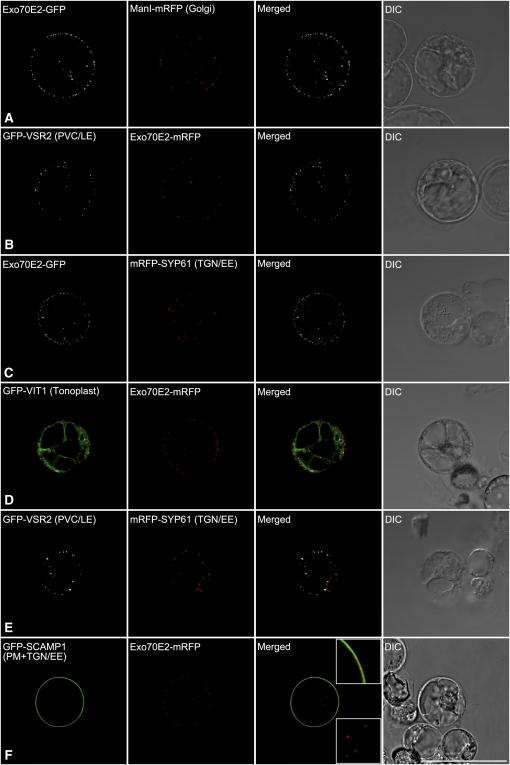
We coexpressed Exo70E2-(X)FP in Arabidopsis protoplasts with fluorescent marker proteins characteristic for the Golgi apparatus (ManI-RFP; Nebenführ et al., 1999; Tse et al., 2004), the prevacuolar compartment/late endosome (PVC/LE) (VSR2; Miao et al., 2006), the TGN/early endosome (EE) (SYP61 and SYP42; Sanderfoot et al., 2001; Uemura et al., 2004; Lam et al., 2007), the tonoplast (VIT1; Kim et al., 2006), and the PM. The cytosolic fluorescent punctae of Exo70E2 did not colocalize with any of the standard endomembrane markers (Figures 1A to 1E; see Supplemental Figure 3 online). However, a clear localization to the PM in the form of discrete punctae was observed (Figure 1F).
Figure 1.
Exo70E2 Localizes as Discrete Punctate Signals at the PM and in the Cytosol but Does Not Colocalize with Standard Organelle Markers.
Arabidopsis protoplasts were coelectroporated with Exo70E2-(X)FP, and the DNA of a single organelle marker as indicated. After 13 to 16 h of expression, the protoplasts were observed by CLSM. Bar = 50 μm.
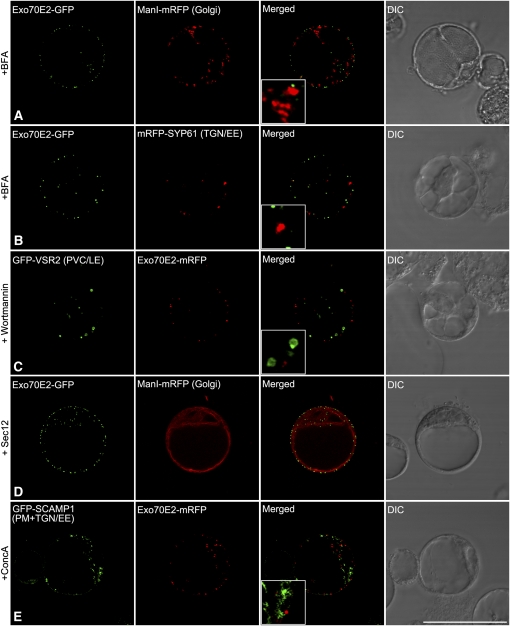
We examined this novel expression pattern by applying known inhibitors of the secretory and endocytic pathways in plants (Robinson et al., 2008a, 2008b). We first applied brefeldin A (BFA), which blocks the function of guanine-nucleotide exchange factors for ADP-ribosylation factor GTPases and interferes with vesicle trafficking (Anders and Jürgens, 2008). However, since the Arabidopsis protoplasts were prepared from a suspension culture originally derived from roots, BFA did not cause the cis-Golgi marker ManI-mRFP to redistribute into the endoplasmic reticulum (ER) (Figure 2A). This is because Arabidopsis root cells, unlike tobacco cells, have a Golgi-localized BFA-resistant guanine-nucleotide exchange factor for ADP-ribosylation factor GTPases (Richter et al., 2007; Teh and Moore, 2007). Nevertheless, a slight aggregation of the Golgi signal was registered, although this effect was not shared by the Exo70E2-GFP signal. Similarly, a small enlargement of the TGN signal (from mRFP-SYP61) resulted from BFA treatment, but again the Exo70E2-GFP was unaffected (Figure 2B). We then tried wortmannnin, which is known to block transport to the vacuole (daSilva et al., 2005) and characteristically causes the PVC/LE to dilate (Tse et al., 2004). Such an enlargement of the PVC marker GFP-VSR2 was recorded for Arabidopsis protoplasts upon treatment with wortmannin, but this had no effect on the Exo70E2-GFP signal (Figure 2C). The punctate Exo70E2-GFP signals were also completely unaffected by the overexpression of Sec12, which inhibits COPII vesicle formation at the ER (Phillipson et al., 2001) and consequently leads to the accumulation of Golgi enzymes (e.g., ManI-mRFP) in the ER (Figure 2D). Disruption of the TGN, caused by treatment with the V-ATPase inhibitor concanamycin A (ConcA; Dettmer et al., 2006), was visualized using the endocytic cargo molecule SCAMP1 as a marker (Lam et al., 2007; Lam et al., 2008), but again this treatment also had no effect on the punctate Exo70E2-mRFP signal (Figure 2E).
Figure 2.
EXPOs Are Not Affected by Secretory and Endocytosis Inhibitors in Protoplasts.
Arabidopsis protoplasts were coelectroporated with Exo70E2-(X)FP and marker organelle DNA as indicated and treated with either BFA, wortmannin (1 h), or ConcA (13 h) before observing by CLSM. For inhibition of ER export, protoplasts expressing both Exo70E2-GFP and ManI-mRFP were additionally electroporated with Sec12. DIC, differential interference contrast. Bar = 50 μm.
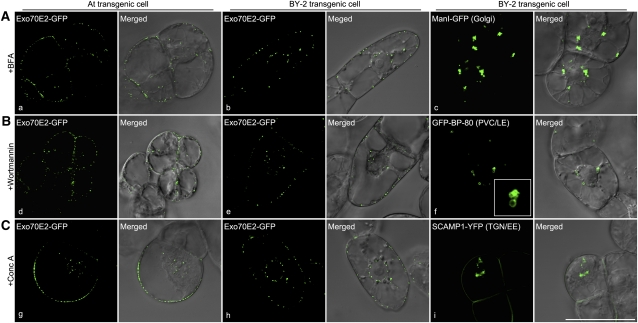
Since the experiments just described were performed on protoplasts transiently expressing Exo70E2 plus marker constructs, as a precautionary measure, we decided to repeat them on stable cell lines of suspension-cultured Arabidopsis and tobacco BY-2 cells expressing Arabidopsis Exo70E2-GFP. Essentially, we obtained the same results. Immunofluorescent signals obtained with antibodies against ManI, VSR2, and SYP61 did not colocalize with the Exo70E2 signals in either cell line (see Supplemental Figure 4 online). We were also able to confirm that BFA did not affect the Exo70E2-GFP signals, neither in Arabidopsis (Figure 3A, a) nor in BY-2 cells (Figure 3A, b), but caused ManI-GFP–marked Golgi to form aggregates in BY-2 cells (Figure 3A, c). Wortmannin was also without affect in both cell lines expressing Exo70E2-GFP (Figure 3B, d and e) but elicited the typical PVC dilations (Figure 3B, f). The same was the case with ConcA (Figure 3C, g and h) but also causing TGN aggregation (Figure 3C, i).
Figure 3.
EXPOs Are Not Affected by Secretory and Endocytosis Inhibitors in Transgenic Arabidopsis and BY-2 Cells.
Transgenic Arabidopsis (a, d, and g) and BY-2 (b, e, and h) cells expressing Exo70E2-GFP were treated with either BFA (A), wortmannin (1 h) (B), or ConcA (2 h) (C) before observing by CLSM. As controls, transgenic BY-2 cell lines expressing either the Golgi marker ManI-GFP (c), the PVC/LE marker GFP-BP-80 (f), or the TGN/EE marker SCAMP1-YFP (i) were used in treatments with these three drugs, respectively. Bar = 50 μm.
[See online article for color version of this figure.]
Arabidopsis Exo70E2-Specific Antibodies Confirm Identity of EXPO
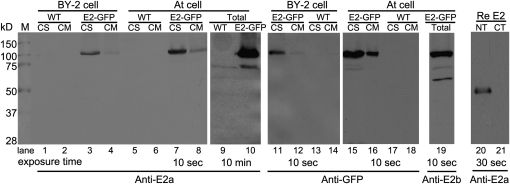
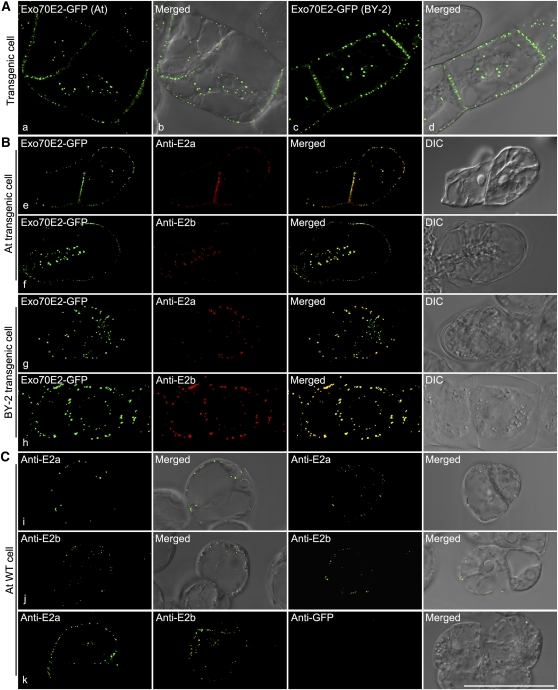
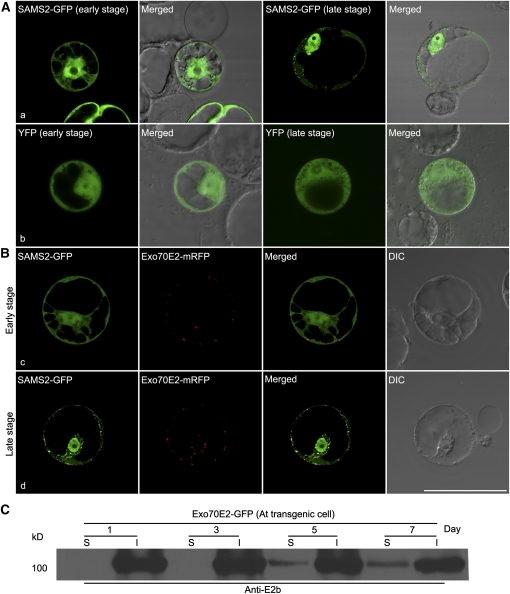
We generated two sets of antibodies (E2a and E2b) against either an N-terminal synthetic peptide or a recombinant 300–amino acid N terminus of Exo70E2, respectively. In immunoblots, these antibodies recognized this domain but not the C terminus of E. coli–derived recombinant proteins (Figure 4, lanes 20 and 21). In addition, we used either E2a or a GFP antibody that in immunoblots was able to recognize a protein of the expected molecular mass (100 kD) for Exo70E2-GFP fusion in the transformed BY-2 and Arabidopsis cell lines with a more prominent signal in the cytosolic than in the membrane fractions (Figure 4). A similar prominent distribution of Exo70E2 proteins in the cell-soluble fraction was also observed in wild-type Arabidopsis cells (see Supplemental Figure 5B online). One can expect to detect Exo70E2-GFP in both cell soluble and cell membrane fractions as it is a cytosolic soluble protein located in both cytosol and EXPO. The E2a antibodies were also able to detect endogenous Exo70E2 (~75 kD) in the total protein fraction in both the wild-type (lane 9) and transgenic (lane 10) Arabidopsis cell culture but only after prolonged exposure periods (10 min) (Figure 4). This was not possible with wild-type tobacco BY-2 cells. In addition, E2b antibodies also detected both Exo70E2-GFP and endogenous Exo70E2 as well as a smaller protein at ~70 kD in transgenic Arabidopsis cells (Figure 4, lane 19). This 70-kD protein band was also detected by E2b antibodies in wild-type Arabidopsis cells (see Supplemental Figure 5A online). Thus, to avoid the possible unspecific labeling by E2b antibodies, we mainly used E2a for immunofluorescence and immunogold electron microscope (EM) labeling in this study, while anti-E2b was used as supporting antibodies.
Figure 4.
Immunoblot Characterization of Transgenic Arabidopsis or Tobacco BY-2 Cell Lines Expressing Exo70E2-GFP and Exo70E2.
Immunoblot analysis of proteins isolated from either transgenic cell lines (Arabidopsis and BY-2) expressing Exo70E2-GFP (E2-GFP) or their wild-type (WT) cells using E2a/b or GFP antibodies as indicated, showing the specificity of rabbit polyclonal anti-E2a/b antibodies generated against the N-terminal synthetic peptide or recombinant 300–amino acid N terminus of At Exo70E2, respectively. The predicted molecular masses for Exo70E2-GFP and endogenous At Exo70E2 are ~100 and 75 kD, respectively. Both anti-E2a/b and anti-GFP recognized the full-length Exo70E2-GFP fusion proteins in both transgenic cell lines (lanes 3 and 4, 7 and 8, 11 and 12, 15 and 16, and 19). Anti-Ea/b also detected the endogenous At Exo70E2 proteins in both wild-type (lane 9) and transgenic (lanes 10 and 19) Arabidopsis cells. CS, cell-soluble proteins; CM, cell membrane proteins; total, total proteins. NT, N terminus of recombinant Exo70E2; CT, C terminus of recombinant Exo70E2.
We performed immunofluorescence on the two transgenic cell lines with the Exo70E2a/b antibodies and obtained a very good colocalization between the GFP and Alexa Fluor signals (Figure 5B, e to h). We also performed immunofluorescence with the Exo70E2a/b antibodies on wild-type cells of Arabidopsis. Endogenous Exo70E2 was detected both at the PM and as discrete punctae in the cytoplasm, albeit at lower levels than in the transgenics (Figure 5C, i to k). Anti-GFP labeling did not give any signal in wild-type Arabidopsis cells (Figure 5C, k). The average number of EXPOs detected in transgenic Arabidopsis Exo70E2-GFP cells (60 EXPOs per cell) was slightly higher than that detected by the anti-E2a antibodies in wild-type Arabidopsis cells (45 EXPOs per cell). This could be due to overexpression of Exo70E2-GFP in transgenic cells or to the insufficiency of detection by the E2a antibodies in wild-type Arabidopsis cells.
Figure 5.
Confocal Characterization of Transgenic Arabidopsis or Tobacco BY-2 Cell Lines Expressing Exo70E2-GFP and Exo70E2.
(A) The expression patterns of Exo70E2-GFP in both cell lines are similar to that seen by transient expression in protoplasts. DIC, differential interference contrast.
(B) At Exo70E2a/b antibodies specifically recognize the Exo70E2-GFP signals in both transgenic Arabidopsis and BY-2 cell lines.
(C) At Exo70E2a/b antibodies recognize the endogenous At Ex070E2 proteins in wild-type Arabidopsis cells with similar patterns as transgenic cells expressing Exo70E2-GFP. Bar = 50 μm.
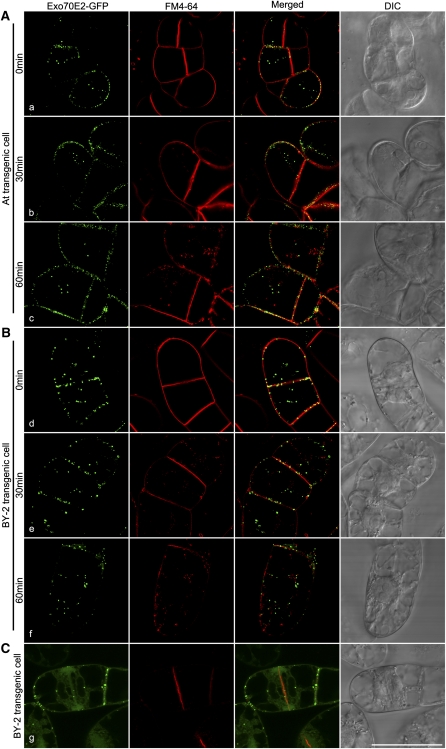
EXPOs Do Not Label with FM4-64
To determine whether EXPOs were endocytic organelles that we had not screened for, we performed uptake experiments with the styryl dye FM4-64. This dye initially stains the PM before being internalized, first to the TGN/EE, then the PVC/LE before reaching the tonoplast (Bolte et al., 2004). Over a 60-min time course, FM4-64 was taken up by both transgenic cell lines and gave rise to small fluorescent punctae. At no stage during the uptake period with either cell line was a colocalization with EXPO detected (Figures 6A and 6B, a to f). Moreover, although FM4-64 is known to accumulate in the growing cell plate during cytokinesis (Lam et al., 2008; Toyooka et al., 2009), this labeling pattern was not shared by EXPO (Figure 6C, g).
Figure 6.
Time Course of FM4-64 Uptake in Transgenic Arabidopsis and BY-2 Cell Lines Stably Expressing Exo70E2-GFP.
FM4-64 (red) uptake study was performed in transgenic Arabidopsis or BY-2 cells expressing Exo70E2-GFP (green), followed by confocal image collection at indicated times. At no time is there a colocalization to be seen between internalized FM4-64 and the Exo70E2-GFP signals in Arabidopsis (A) or BY-2 (B). During cytokinesis, whereas FM4-64 intensively labels the developing cell plate, an equivalent distribution of Exo70E2 signal is not observed (C). Arabidopsis cell line (A); tobacco BY-2 cell lines ([B] and [C]). Bar = 50 μm.
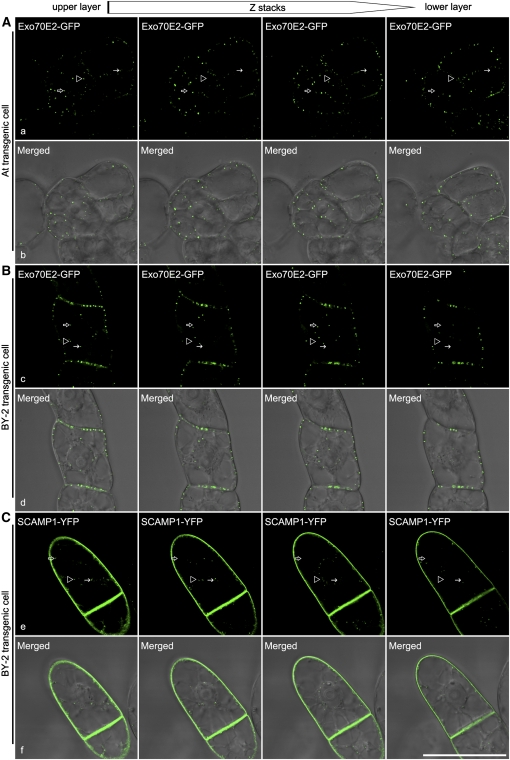
EXPOs Are True Organelles in the Cytoplasm
Since many of the sectioned EXPOs were found in close proximity to the PM, there was a possibility that some were attached to the PM in planes above or below the section. However, serial optical sections in the confocal laser scanning microscopy (CLSM) confirmed the bona fide particulate nature of the EXPO (Figure 7). This was supported by three-dimensional reconstructions (see Supplemental Movie 1 online), as well as by live time imaging revealing the mobile nature of EXPOs (see Supplemental Movie 2 online). The most convincing evidence, however, was provided by a three-dimensional projection (using Imaris software) of multiple optical sections at 0.52 μm per step of a transgenic BY-2 cell expressing Exo70E2-GFP, which had been exposed to FM4-64 for 15 min. This enabled both FM4-64 carrying endocytic structures (TGN) and EXPO to be resolved as structures separate from the PM (see Supplemental Movie 3 online).
Figure 7.
EXPOs Are Cytosolic and Not Associated with the PM.
Shown are examples of series optical sections of confocal images from top to bottom of transgenic Arabidopsis (A) or tobacco BY-2 cells (B) expressing Exo70E2-GFP or BY-2 cells (C) expressing rice (Oryza sativa) SCAMP1-YFP (Lam et al., 2007), which is located in TGN and PM, as a control. Arrows and arrowheads indicated examples of EXPO not associated with the PM. Bar = 50 μm.
[See online article for color version of this figure.]
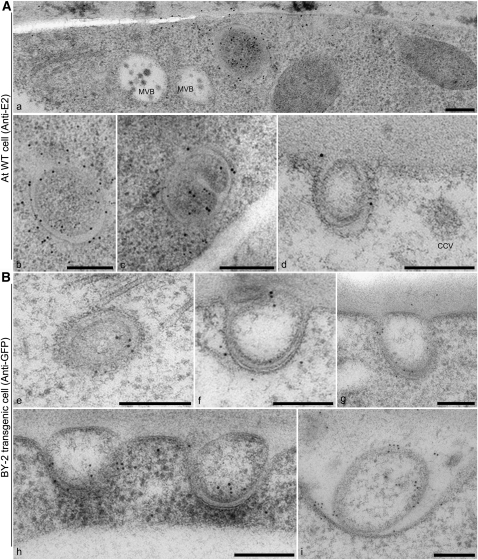
EXPOs Are Exocytic Double Membrane Organelles
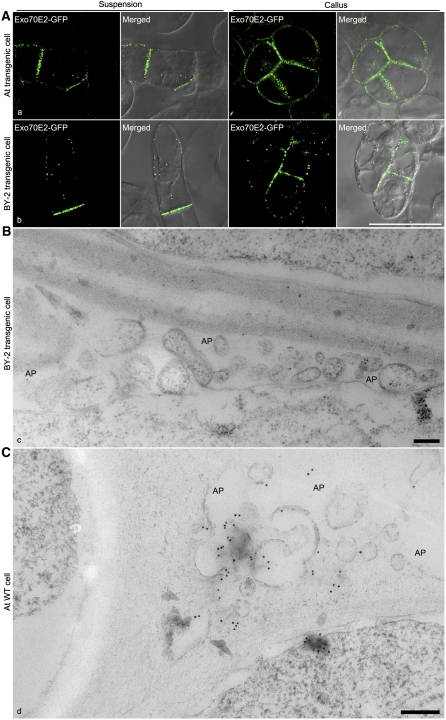
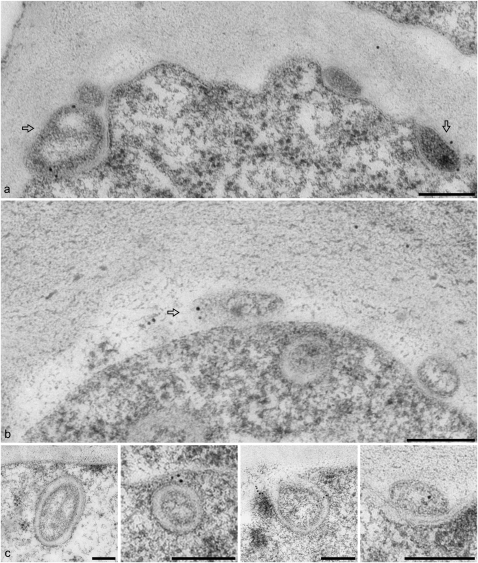
The ultrastructure of EXPO and the PM-localized Exo70E2-positive punctae was revealed by immunogold labeling of thin sections cut from high-pressure frozen/freeze-substituted samples of wild-type Arabidopsis cells and transgenic BY-2 cells expressing Exo70E2-GFP using E2a and GFP antibodies, respectively. Labeling on both cell types was very specific, with gold particles being found over two types of structure. One was a double membrane-bound structure similar in size to MVB/LE (Figure 8A, a to c, and 8B, e), the contents of which was very similar to the surrounding cytoplasm. Gold label was present on both the inner and outer membranes. We interpret these structures as representing the EXPO in the CLSM. The other structure was an unusual fusion-like profile at the PM (Figure 8A, d, and 8B, f to i; see Supplemental Figures 6 and 7 online), which we presume corresponds to the Exo70E2-positive punctae at the PM seen in the CLSM (Figures 1 to 3; see Supplemental Figures 2 and 3 online). Since identical EXPO fusion profiles were detected in wild-type Arabidopsis cells (see Supplemental Figures 6 and 7A online) and transgenic BY-2 cells expressing Exo70E2-GFP (see Supplemental Figure 7B online), we can discount their formation being solely the result of Exo70E2 overexpression. We also detected EXPO-like structures in a number of other plant cells (see Supplemental Figure 7C, panels c and d, online).
Figure 8.
EXPOs Have Two Membranes and Fuse with the PM Expelling a Single Membrane Vesicle into the Apoplast.
Immunogold labeling of sections cut from high-pressure frozen/freeze-substituted samples of wild-type Arabidopsis ([A], a to d), and transgenic tobacco BY-2 ([B], e to i) cells. Bars = 200 nm.
(A) EXPOs are distinct in morphology from MVBs, which do not label with Exo70E2a antibodies. Label is distributed over both membranes of the EXPO.
(B) Gallery of fusion profiles. Label is found on the inner surface of the inner vesicle and is also present in the neighboring vicinity, suggesting that the released vesicle ultimately bursts, releasing Exo70E2 into the cell wall.
The fusion of the outer membrane of the EXPO with the PM leads to the expulsion of a single membrane-bound structure into the apoplast (Figure 8B, f to i; see Supplemental Figures 6 and 7 online). The inner surface of this membrane is also labeled (Figure 8) and confirms that Exo70E2 is attached to both the inner and outer membranes of the EXPO. Gold label was also found in the cell wall in the vicinity of the EXPO fusion profiles (Figure 8B, i), suggesting that Exo70E2 detaches from the surface of the inner membrane after its release into the apoplast and bursts.
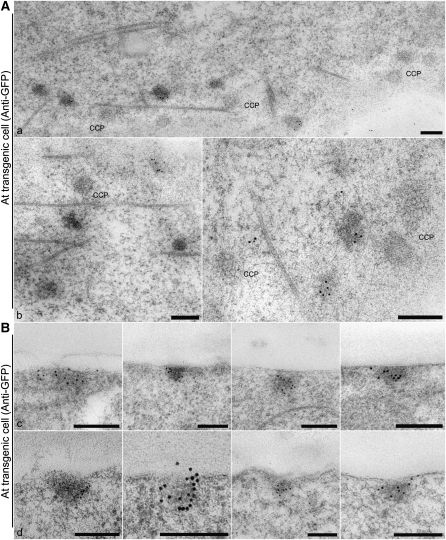
Interestingly, in both transgenic Arabidopsis and tobacco BY-2 cells expressing Exo70E2-GFP, stronger GFP signals representing punctate EXPO are commonly visible in PM/cell wall areas connecting two cells compared with other areas facing directly against the media (Figure 9A, a and b). Indeed, in the EM, single-membrane EXPOs labeled by either GFP or ExoE2 antibodies are commonly found at this location in both transgenic BY-2 and wild-type Arabidopsis cells (Figures 9B and 9C). Glancing sections through the PM allowed for a differentiation between clathrin-coated pits and similarly sized electron-opaque Exo70E2-positive patches in transgenic Arabidopsis cells expressing Exo70E2-GFP using GFP antibodies (Figure 10A, a and b). We managed to visualize these structures in cross section but were not able to ascertain an inwardly directed membrane fold (Figure 10B, c and d). In addition, in all the immunogold EM labeling, control experiments such as secondary antibodies did not result in any specific labeling (see Supplemental Figure 8 online).
Figure 9.
Likely Nature of EXPO Release in Plant Cells.
(A) GFP-positive EXPOs are more abundant in PM/cell wall areas connecting two cells in transgenic Arabidopsis and BY-2 cells expressing Exo70E2-GFP. Bar = 50 μm.
(B) Immuno-EM detection of released EXPOs in the apoplasts (AP; as indicated) of adjacent transgenic BY-2 cells expressing Exo70E2-GFP using high-pressure freezing. Bar = 200 nm.
(C) Immuno-EM detection of released EXPOs in the apoplasts of adjacent wild-type Arabidopsis cells using high-pressure freezing. Bar = 200 nm.
[See online article for color version of this figure.]
Figure 10.
Immunogold Labeling of EXPO/PM Fusions.
(A) Tangential sections through the PM–cell wall interface showing that clathrin-coated pits (CCPs) are separate from GFP antibody–labeled electron opaque patches.
(B) Cross sections through the PM–cell wall interface. The Exo70E2-positive patches do not reveal clearly fusion profiles. We interpret these as being the remains of fusions, where exocyst molecules are still attached to the PM.
Bars = 200 nm.
Antibodies directed against other exocyst proteins (Exo70A1, Sec6, and Sec8; Hála et al., 2008) were also employed to prove that the Exo70E2 labeling represented the exocyst complex and not just the presence of a single protein. Of the three antibodies, Exo70A1 gave a similar labeling pattern to that of Exo70E2 (Figure 11). Sec6 produced high background labeling, but EXPOs were also clearly labeled. Sec8 gave only a very low labeling, but EXPOs were nonetheless still marked (see Supplemental Figure 9, panels a and b, online). When immunofluorescence was performed using these antibodies in transgenic Arabidopsis cells expressing Exo70E2-GFP, once again the Exo70A1 antibodies largely colocalized with Exo70E2-GFP, whereas both Sec6 and Sec 8 showed unspecific labeling (see Supplemental Figure 10 online). This result is consistent with the immuno-EM labeling and the documented specificity of these antibodies (Hála et al., 2008).
Figure 11.
EXPOs Are Also Labeled by Exo70A1 Antibodies in Wild-Type Arabidopsis Cells.
Immunogold labeling of sections cut from high-pressure frozen/freeze-substituted samples of Arabidopsis cells using Exo70A1 antibodies, with similar labeling patterns as that of Exo70E2a. Bars = 200 nm.
EXPOs Are Not Autophagosomes but Sequester Cytosolic Proteins to Release Them into the Apoplast
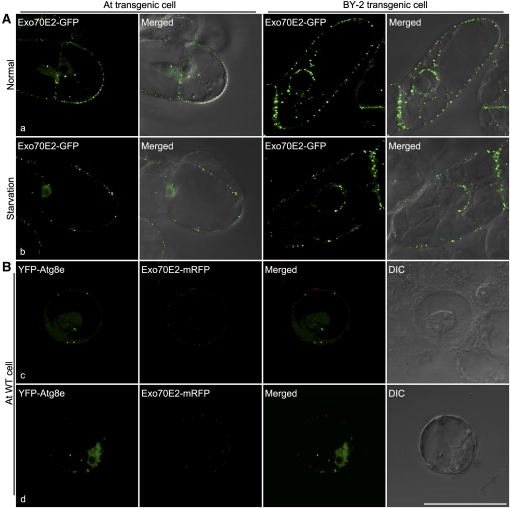
Because of their double membrane appearance, EXPOs are morphologically similar to autophagosomes (Baba et al., 1994, 1995; Klionsky, 2007). We decided therefore to examine the possibility that EXPOs are related to autophagosomes. First, we observed no increase in numbers of EXPO numbers in transgenic Arabidopsis cells and BY-2 cells expressing Exo70E2-GFP as a consequence of sucrose starvation (Figure 12A, a and b). On the contrary, the numbers of EXPO decreased under C-starvation by 20 to 30%. Second, as seen in Figure 12B, c and d, Exo70E2 does not colocalize with Atg8e, a standard marker for the autophagosome membrane (Yoshimoto et al., 2004; Contento et al., 2005). Nevertheless, on the basis of expression experiments conducted with the Arabidopsis protein S-adenosylmethionine synthetase 2 (SAMS2) (AT4G01850), it appears that cytosolic proteins can be sequestered into the interior of the EXPO, presumably as the membranes close. Based on software analysis, SAMS2 lacks a signal peptide and is predicted to locate to the cytoplasm. Interestingly, although being unable to enter the secretory pathway, it has been claimed that SAMS2 is present in the cell wall of Arabidopsis based on proteome analysis (Bayer et al., 2006). This is similar to SAMS3, one of the four SAMS enzymes that convert Met to S-adenosyl Met, which has been shown to be the major substrate for lignin methylation in the cell wall (Shen et al., 2002). We expressed a SAMS2-GFP construct in Arabidopsis cells and followed the pattern of its expression over time (Figures 13A and 13B). At early expression times (24 h after transformation), the fluorescent signal is distributed throughout the cytoplasm but is also visible in the matrix of the nucleus. At later expression times (48 h after transformation), small fluorescent punctae become visible in the the cytoplasm. No such punctate structures were visible in protoplasts expressing as a control cytosolic yellow fluorescent protein (YFP) at these two stages (Figure 13A, b). When coexpressed with Exo70E2-mRFP, SAMS2-GFP remained cytosolic at early stages but showed punctate at later stages, and these punctae colocalized with Exo70E2-mRFP (Figure 13B).
Figure 12.
EXPOs Are Not Induced by Sucrose Starvation and Are Distinct from an Autophagy Marker.
(A) Shown are representative examples of transgenic Arabidopsis and BY-2 cells expressing Exo70E2-GFP under normal and starvation cultured conditions after 24 h.
(B) Separation of Exo70E2-mRFP from the autophagy marker YFP-Atg8e in wild-type Arabidopsis protoplasts. Bar = 50 μm.
Figure 13.
EXPO Sequesters Cytosolic SAMS2-GFP in Arabidopsis Protoplasts.
(A) When transiently expressed in Arabidopsis protoplasts, SAMS2-GFP showed cytosol and nucleus patterns at early stages (24 h) but became punctate at later stages (a; 48 h). By contrast, YFP remained cytosolic in both stages (b).
(B) When coexpressed with Exo70E2-mRFP in Arabidopsis protoplasts, SAMS2-GFP showed cytosol and nucleus patterns at early stages (c), but at later stages, SAMS2-GFP became colocalized with Exo70E2-mRFP in EXPO (d). Bar = 50 μm.
(C) Immunoblot detection of Exo70E2-GFP proteins in the culture medium of transgenic Arabidopsis cells expressing Exo70E2-GFP. S, secretion medium proteins; I, intracellular proteins; day, days after subculture.
We have not been able to detect SAMS2 extracellularly via transient expression in protoplasts. This could either be due to low expression levels or to the low transformation efficiency of protoplasts. Rapid degradation of SAMS2 in the medium can also not be excluded. However, we have obtained evidence for the presence of Exo70E2 in the culture medium. Samples were taken from Arabidopsis cell cultures expressing Exo70E2-GFP over a 7-d period, and the cells separated from the medium by centrifugation. Immunoblots were made of intracellular proteins (I) and medium secretion (S) proteins using the E2a antibodies. As seen in Figure 13C, a band for Exo70E2-GFP at ~100 kD became visible in the medium extracts after 5 d of culture, and its intensity increased at 7 d (Figure 13C). The detection of Exo70E2-GFP in the media is not due to broken cells because the other antibodies tested (anti-Man1, anti-TUB, and anti-VSR) did not detect any corresponding proteins in the culture medium of day 7 Arabidopsis Exo70E2-GFP cells (see Supplemental Figure 11 online). Tubulin (TUB) was used as a control for cytosolic proteins because it is a major cytosolic protein whose assembly into microtubules is critical to many cellular processes (Yaffe et al., 1992). In addition, when identical experiments were performed for transgenic BY-2 cells expressing SCAMP-YFP or GFP-BP-80 cells (Lam et al., 2007), no GFP fusions were detected in the culture media (data not shown).
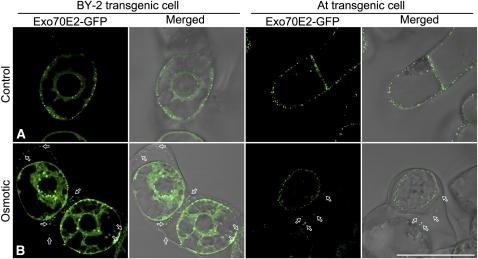
To determine if EXPO reaches the outside of the cells after fusion and release from the PM, we performed an experiment in which day 3 transgenic tobacco BY-2 or Arabidopsis cells expressing Exo70E2-GFP were first subjected to an osmotic treatment with 250 mM NaCl solution for 10 min so that the PM would retract from the cell wall, followed by confocal imaging. As shown in Figure 14, many small punctate fluorescent structures were seen close to the cell wall but clearly separated from the PM (Figure 14B, indicated by arrows). We believe that they represent released single membrane–bound EXPO that are in the process of bursting because the intensity of the fluorescent signals was generally weaker than the EXPO inside the cells or those remaining PM associated. This result demonstrated that EXPO did reach outside of the PM, which is different from transgenic BY-2 cells expressing the PM/TGN-localized SCAMP1-GFP, where this GFP fusion remained in the PM after similar osmotic treatment (Lam et al., 2007). In addition, when an identical experiment was performed for transgenic BY-2 cells expressing the Golgi marker Man1-GFP, no punctate GFP signal representing Golgi organelle was observed outside of the PM (data not shown), again indicating that the observed secretion of EXPO is specific for Exo70E2-GFP.
Figure 14.
EXPOs Are Detected Outside the PM in Transgenic Cells.
Transgenic tobacco BY-2 or Arabidopsis cells expressing Exo70E2-GFP were first subjected to osmotic treatment with 250 mM NaCl for 10 min, followed by confocal imaging. Untreated cells were included as controls. Bars = 50 μm.
[See online article for color version of this figure.]
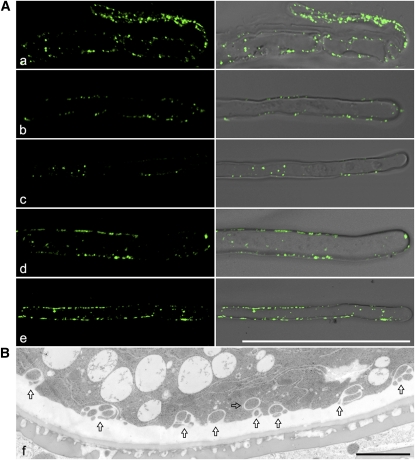
Since Exo70E2 is highly expressed in suspension culture cells based on microarray data analysis (Synek et al., 2006), it is thus not surprising that EXPOs are commonly found in both BY-2 and Arabidopsis culture cells. EXPOs are also found in other cell types, as seen from immune-EM and structural EM studies (see Supplemental Figure 7 online). To have a better picture of the existence and abundance of EXPO in other cell types, we also performed confocal immunofluorescent labeling with Exo70E2a antibodies in Arabidopsis root tip cells. As shown in Figure 15, E2a antibodies labeled many EXPOs in both Arabidopsis root tip cells (Figure 15A) and root hair cells (Figures 15B to 15E), with almost identical punctate patterns in both cytosol and close to PM as seen in suspension culture cells. In addition, many EXPOs in the process of fusing with the PM are also clearly found in tobacco pollen grains (Figure 15F). These results demonstrate that EXPOs are not only present in culture cells but are also abundantly seen in other plant cell types, including root tip cells, root hair cells, and pollen grains. Taken together, our results support a model of double-membrane EXPO formation in the cytosol, fusion with the PM, and release of single-membrane EXPOs outside of the cell. Bursting of single-membrane EXPOs in the harsh environment completes the transport or secretion of cytosolic contents into cell exterior (Figure 16).
Figure 15.
Immunofluorescent and Structural EM Detection of EXPO in Other Cell Types.
(A) Shown are examples of EXPO detected by anti-Exo70E2a antibodies in wild-type Arabidopsis root tip cells (a) and root hair cells (b to e). Bars = 50 μm.
(B) Structural EM detection of EXPO in an ultrathin section prepared from high-pressure freezing/frozen-substituted wild-type tobacco pollen grains. Arrows indicated examples of EXPO. Bars = 2 μm.
[See online article for color version of this figure.]
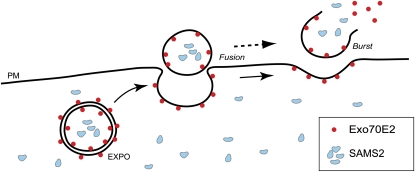
Figure 16.
Working Model of EXPO in Plant Cells.
Illustration depicting the relationship between Exo70E2, and double-membrane EXPO, their fusion with the PM, and release and bursting of the single-membrane EXPO in the harsh environment to complete the unconventional secretion. Meanwhile, cytosolic protein SAMS2 is sequestered by EXPO and finally reaches the cell wall.
[See online article for color version of this figure.]
DISCUSSION
Exocyst and EXPO
Together with COG, GARP, HOPS, and TRAPP, the exocyst belongs to the subgroup of vesicle tethering factors that are multimeric protein complexes (Sztul and Lupashin, 2006; Cai et al., 2007). Since these tethering complexes are evolutionary conserved (Koumandou et al., 2007), their presence in plants should not come as a surprise. Indeed, considerable data have accumulated over the last 5 years that confirm the existence of the exocyst in plants and its importance for polarized plant growth (Zárský et al., 2009). Not only has in silico research resulted in the identification of homologs of all exocyst subunits (Elias et al., 2003; Synek et al., 2006; Chong et al., 2010), but a 900-kD complex containing seven of the eight exocyst subunits has recently been successfully identified from an Arabidopsis cell extract (Hála et al., 2008). Moreover, a number of studies have described the deleterious effects on growth, especially localized tip growth in pollen tubes and root hairs in plants expressing mutant exocyst proteins (Cole et al., 2005; Wen et al., 2005; Synek et al., 2006; Hála et al., 2008; Samuel et al., 2009).
The subcellular localization of exocyst subunit proteins in plants has been performed on two previous occasions using fluorescence microscopy. Hála et al. (2008) showed by immunofluorescence microscopy that Arabidopsis Sec6, Sec8, and Exo70A1-positive structures accumulated at the tips of growing pollen tubes. Discrete punctae at the PM were also reported but not shown. Most recently, Chong et al. (2010) looked at the short-term (6 h) expression of GFP-tagged (N-terminal) Arabidopsis Sec5a, Sec6, Sec8, Sec15a, Sec15b, and Exo84b in tobacco BY-2 cells. While Sec6 and Sec8 gave rise to cytosolic signals, Sec5a, Sec15a, Sec15b, and Exo84b localized to punctae within the cytosol. These authors also performed immunofluorescence on cells coexpressing GFP-tagged markers for the TGN and PVC (Syp21, Syp42, and Syp52), together with myc-tagged Sec15b and Exo70E2. Based on the images presented, they claimed to have visualized “significant overlapping patterns of localization” between the exocyst proteins and the endosomal markers. Curiously, no labeling of the PM was seen.
Our fluorescence microscopy data on the localization of Exo70E2 is at considerable variance to the results of Chong et al. (2010). Not only were we able to visualize Exo70E2 as discrete punctae in cells expressing (X)FP-tagged constructs, but a punctate distribution was also revealed in immunofluorescence. Significantly, Exo70E2 was also present at the PM, again in the form of discrete punctae. Our studies with standard endomembrane markers also ruled out the localization of Exo70E2 to the Golgi apparatus, the TGN/EE, and the PVC/LE. Significantly, the Exo70E2-positive structures, which we termed EXPOs, did not label with the endocytic dye FM4-64 and did not respond to inhibitors of secretion or endocytosis. However, these unusual features conform with the novel ultrastructure of EXPO and EXPO fusion profiles as revealed in immunogold labeled sections taken from high-pressure frozen samples.
We wondered why EXPO-like structures have not previously been reported in the plant literature, even though high-pressure freezing/freeze substitution is no longer that uncommon. However, we would like to point to two early articles where freeze fracturing was performed after rapid freezing (both propane jet and high-pressure freezing) of suspension-cultured cells (Staehelin and Chapman, 1987) and root tips (Craig and Staehelin, 1988). In both cell types, novel circular, sometimes horseshoe like, indentations in the PM were visualized. These structures were clearly different from clathrin-coated pits. We think that these might possibly represent images of EXPOs fusing with the PM. Although there are several reports that exocyst proteins localize to the TGN and recycling endosomes (see Introduction), the great majority of the articles dealing with exocyst in yeast and mammalian cells places this tethering factor at the PM (He and Guo, 2009; Orlando and Guo, 2009). In keeping with this, we would maintain that plant EXPOs are also principally exocytic in function. Our study also shows that, in addition to suspension culture cells, EXPOs are also present in pollen, root tip, and root hair cells.
Are EXPOs Related to Autophagosomes?
Double-membraned structures encapsulating cytosol and organelles are well known as autophagosomes that develop in mammalian cells in response to starvation (Xie and Klionsky, 2007; Kirkin et al., 2009). Much smaller structures are also present as cytoplasm-to-vacuole-targeting vesicles in wild-type yeast cells (Baba et al., 1997) but enlarge to form autophagosomes upon nutrient depletion (Kraft et al., 2009). Autophagosomes have also been reported in plants (Bassham, 2009). Their origin has been something of a mystery for a number of years (for review, see Reggiori, 2006; Yoshimori and Noda, 2008), but recent EM tomography studies on mammalian cells have now provided conclusive evidence for a connection to the ER (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009). Usually, autophagosomes ultimately fuse with a lytic compartment: either the lysosome or the vacuole. However, two recent articles reported on a novel autophagosome-mediated secretion of acyl-CoA binding protein, which occurs when yeast or Dictyostelium cells are starved (Duran et al., 2010; Manjithaya et al., 2010). These autophagosomes fuse first with recycling endosomes in a GRASP (Golgi reassembly and stacking protein) dependent manner, and the resulting multivesicular carriers then fuse with the PM.
EXPOs are also structures with two membranes, but they do not fuse with the tonoplast. Furthermore, they do not colocalize with the phagophore marker Atg8e and do not label with FM4-64 unlike yeast autophagosomes (Journo et al., 2008). Since EXPOs do not alter in number when the cells are subjected to starvation, we do not consider them to be autophagic in character. On the other hand, the presence of Exo70E2 on both the inner and outer membranes of the EXPO suggests that EXPO develops like an autophagosome by starting out as a phagophore-like structure. Such a structure is exposed to the cytosol on both membrane surfaces, to which Exo70E2 would attach. Upon closure of the phagophore, an autophagosome-like structure would be formed with Exo70E2 on both the cytosolic surface of the outer membrane as well as the inner surface of the inner membrane (Figure 16).
EXPOs and Exosomes: Agents of Unconventional Secretion
Vesicles released from cells are termed exosomes. Usually, they are expelled to the cell surface through the fusion of multivesicular endosomes with the PM, a phenomenon well described in mammalian cells (Février and Raposo, 2004; Simons and Raposo, 2009), but also reported in plants (An et al., 2007). Whereas clear functions have been ascribed to this form of unconventional secretion in mammals, such as recycling of transferrin and its receptor (Harding et al., 1984; Pan et al., 1985) and antigen presentation (Denzer et al., 2000; Van Niel et al., 2003), the nature of the molecules released through multivesicular body-PM fusion in plants in still unclear, although there seems to be a connection to the plant pathogen response (An et al., 2007). In this regard, EXPO-mediated exocytosis might be responsible for the secretion of apoplastic mannitol dehydrogenase upon salicylic acid treatment in plant cells (Cheng et al., 2009).
EXPO is distinct from a multivesicular body and does not lie on the endocytic pathway, but broadly speaking, the internal vesicle of the EXPO when released into the apoplast through fusion of the EXPO with the PM is also an exosome. Our demonstration that a soluble protein without a signal peptide (SAMS2) is sequestered by EXPO suggests that EXPO serves to release proteins into the apoplast that cannot enter the secretory pathway. The physiological significance of this unconventional delivery mechanism remains to be ascertained. Purification and proteomic analysis of isolated EXPO from plant cells will allow the identification and characterization of EXPO proteins in the future, which will certainly shed light on the biological significance of EXPO-mediated secretion in plants.
METHODS
General methods for the construction and characterization of recombinant plasmids, maintenance of tobacco (Nicotiana tabacum) BY-2 and Arabidopsis thaliana suspension culture cells, and preparation and characterization of antibodies have been described previously (Jiang and Rogers, 1998; Jiang et al., 2000, 2001; Tse et al., 2004).
Plasmid Construction
The full-length At Exo70E2 cDNA was amplified by PCR using the cDNA obtained from RIKEN Genomic Science Center (Seki et al., 1998, 2002) as template and the primer sets listed in Supplemental Table 1 online. The amplified Exo70E2 cDNA fragments were inserted into a premade GFP vector in pBI221 via XbaI and XhoI restriction enzyme sites to generate AtExo70E2-GFP and GFP-AtExo70E2, respectively, and used for transient expression. Monomeric RFP (mRFP) versions of the AtExo70E2 construct were generated similarly. For Agrobacterium tumefaciens–mediated transformation of tobacco BY-2 and Arabidopsis suspension culture cells, the Exo70E2-GFP cDNA fragment was cut out using ScaI and EcoRI restriction enzymes and cloned into the binary vector pBI121 (Chen et al., 2003) via the same restriction sites.
Cloning and Screening of Different Predicted Members of Arabidopsis Exo70s
The full-length cDNAs of Arabidopsis Exo70A1, Exo70B1, Exo70B2, Exo70D1, Exo70D2, Exo70E1, and Exo70F1 were amplified using their corresponding cDNAs obtained from RIKEN Genomic Science Center (Seki et al., 1998, 2002) as templates and the primer sets listed in Supplemental Table 1 online. The amplified fragments were cloned into the pBI221-GFP vector using specific restriction cutting sites to generate various Exo70s-GFP fusion constructs for transient expression.
Transient Expression and Transformation of Tobacco BY-2 and Arabidopsis Cells
Transient expression using Arabidopsis protoplasts has been described previously (Miao and Jiang, 2007). Transformation and generation of transgenic tobacco BY-2 cells have also been described (Tse et al., 2004; Lam et al., 2007). For the generation of transgenic Arabidopsis cells, Arabidopsis cells were maintained in Murashige and Skoog (MS; Sigma-Aldrich) medium at 22°C in an orbital shaker set at 130 rpm and subcultured once a week. Agrobacterium-transfected Arabidopsis cells were transferred into a CELLSTAR six-well cell culture plate (Greiner Bio-One) and shaken at 25°C in an orbital shaker setting at 130 rpm for 2 d. After washing with fresh MS medium three times, the transfected cells were transferred onto MS agar (1% agar, w/v) plates containing kanamycin (100 μg/mL) and cefotaxime sodium (250 μg/mL) for 3 to 4 weeks until transformed colonies formed. Subsequent screening and maintenance of transformed colonies were as previously described (Tse et al., 2004; Lam et al., 2007).
Antibodies
A synthetic peptide (CQHKRHVQPDYLAVSSRRKD) within the N terminus of Arabidopsis Exo70E2 was synthesized (GenScript) and used as antigen to raise antibodies as described previously (Rogers et al., 1997; Tse et al., 2004; Lam et al., 2007). Similarly, the Escherichia coli–derived recombinant N terminus and C terminus of Exo70E2 were also used as antigens for antibody generation. Only affinity-purified E2 antibodies were used in this study. Polyclonal antibodies against VSRAt-1 and ManI were described previously (Tse et al., 2004). The Arabidopsis SYP61 antibodies were kindly provided by Natasha Raikhel (University of California, Riverside, CA). The Arabidopsis Exo70A1, Sec6, and Sec8 antibodies were kindly provided by Viktor Zarsky (Prague, Czech Republic). For immunoblotting and confocal immunofluorescent analysis, antibodies were used at 4 μg/mL (Wang et al., 2007, 2009). Polyclonal Alexa Fluor-568 anti-rabbit antibodies (Molecular Probes) were used as secondary antibodies for immunofluorescent detection.
Immunofluorescent Studies
Fixation and preparation of tobacco BY-2, Arabidopsis cells, and Arabidopsis seedlings and their labeling and analysis by confocal immunofluorescence have been described previously (Jiang and Rogers, 1998; Jiang et al., 2000; Ritzenthaler et al., 2002; Tse et al., 2004; Lam et al., 2007). Confocal fluorescent images were collected using an Olympus FluoView FV1000 confocal microscope with ×60 water lens or Leica TCS SP5 II confocal microscope with ×63 water lens. The settings for collecting confocal images and processing of images using Adobe Photoshop software were described previously (Jiang and Rogers, 1998).
FM4-64 Uptake Study
The two transgenic Arabidopsis and BY-2 cell lines expressing Exo70E2-GFP were used in the FM4-64 uptake study. FM4-64 uptake experiments were performed as previously described (Tse et al., 2004; Lam et al., 2007, 2008). Images were collected at different time points as indicated.
Expression and Purification of Recombinant Protein
Exo70E2 was split into two parts: the N terminus (first 300 amino acids from N terminus) and the C terminus (the remaining protein sequence). Both the N and C termini of Exo70E2 were amplified by PCR using Exo70E2-GFP as template and the primer sets listed in Supplemental Table 1 online. These amplified fragments were inserted into pET30a (+) vector via the EcoRI and XhoI restriction sites and then transferred into E. coli strain BL21 for expression. The expressed recombinant proteins were harvested by sonicating the cell pellets in cell lysis buffer (20 mM sodium phosphate, 0.5 M NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.1% Nonidet P-40, 1 mM PMSF, and 1 mM TCEP, pH 7.0) for 10 min and centrifuged at 12,000g for 15 min to collect the supernatants. The supernatants were then purified using a HiTrap IMAC HP column (GE Healthcare) coated with nickel(II) sulfate hexahydrate.
Drug Treatments
BFA and wortmannin treatment experiments were performed as previously described (Tse et al., 2004; Miao et al., 2006; Lam et al., 2007; Wang et al., 2010). For ConcA experiments, aliquots of ConcA (stock solution at 1 mM in DMSO) solution were added to Arabidopsis protoplasts to give the final concentration of 2 μM after electroporation and protoplasts incubation. These drug-treated cells were then removed from the cultures at indicated times for direct confocal imaging. Similar BFA and wortmannin treatments were done for transgenic cell lines, but cells were with ConcA at 2 μM for 2 h.
Osmotic Treatments of Culture Cells
Day 3 transgenic tobacco BY-2 or Arabidopsis cells expressing Exo70E2-GFP were first washed with fresh culture medium (pH 7.5) twice, followed by culture in fresh medium for 2 h, prior to osmotic treatment with 250 mM NaCl solution for 10 min and confocal imaging.
EM of Resin-Embedded Cells
The general procedures for transmission electron microscopy sample preparation and thin sectioning of samples of BY-2 and Arabidopsis cells were performed essentially as described previously (Ritzenthaler et al., 2002; Tse et al., 2004; Lam et al., 2007). For high-pressure freezing, BY-2 and Arabidopsis cells were harvested by filtering and immediately frozen in a high-pressure freezing apparatus (Leica EM Part2 and Bal-Tec HPM010). Immunolabeling on HM20 sections was done using standard procedures as described previously (Tse et al., 2004) with Exo70E2a antibodies at 65 μg/mL and gold-coupled secondary antibodies at 1:50 dilution. Sections were poststained with aqueous uranyl acetate/lead citrate and then examined with a either a Hitachi H7650 transmission electron microscope (Hitachi High-Technologies) with MacroFire monochrome CCD camera (Optronics) operating at 80 kV described previously (Tse et al., 2004; Wang et al., 2007) or a JEM1400 (JEOL) with images recorded with a FastScan F214 digital camera (TVIPS).
Confocal Imaging for Three-Dimensional Projection Using Imaris
Transgenic BY-2 cells expressing Exo70E2-GFP were stained with FM4-64 for 15 min and washed twice with fresh medium before confocal imaging of multiple optical sections at 0.52 μm per step at z axis and used for three-dimensional movie construction using Imaris software (Bitplane).
Secretion Assay and Immunoblot Analysis
Secretion assay was performed as described previously (Suen et al., 2010). Intracellular and medium proteins were prepared from different subculture days of transgenic Arabidopsis cells expressing Exo70E2-GFP, followed by protein separation via SDS-PAGE and immunoblot analysis using either GFP or E2 or other control antibodies as indicated. For regular immunoblot analysis of cells, cell-soluble and cell membrane protein fractions were prepared as previously described (Jiang and Rogers, 1998), followed by protein separation via SDS-PAGE and immunoblot analysis using the appropriate antibodies at 4 μg/mL.
Accession Numbers
Sequence data for different At Exo70s cDNAs can be found in the GenBank/EMBL data libraries under following accession numbers. Arabidopsis Genome Initiative codes for Arabidopsis loci are also provided in parentheses: At Exo70A1, NM_120434 (At5g03540), At Exo70B1 NM_125229 (At5g58430), At Exo70B2 NM_100573 (At1g07000), At Exo70D1 NM_105906 (At1g72470), At Exo70D2 NM_104286 (At1g54090), At Exo70E1 NM_113866 (At3g29400), At Exo70E2 NM_001037039 (At5g61010) and At Exo70F1 NM_124420 (At5g50380), as well as At SAM2 (AT4G01850).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transient Expression of Various GFP Fusions with Selected Members of Exo70.
Supplemental Figure 2. Colocalization of Pairs of Exo70-XFP Fusions.
Supplemental Figure 3. The TGN Marker SYP42 Is Separated from Exo70E2-mRFP in Arabidopsis Protoplasts.
Supplemental Figure 4. Exo70-GFP–Positive Organelles Are Distinct from Known Secretory Compartments.
Supplemental Figure 5. Immunoblot Characterization of Endogenous Exo70E2 Proteins in Wild-Type Arabidopsis Cells.
Supplemental Figure 6. Ultrastructure and Fusion Nature of EXPO in Wild-Type Arabidopsis Suspension Culture Cells.
Supplemental Figure 7. Ultrastructure of EXPO in Plant Cells.
Supplemental Figure 8. Immuno-EM Controls.
Supplemental Figure 9. EXPOs Are Also Labeled by Sec6 and Sec8 Antibodies in Arabidopsis Cells.
Supplemental Figure 10. Immunofluorescence of Exo70E2-GFP Arabidopsis Transgenic Cells Using Different Exocyst Subunit Antibodies.
Supplemental Figure 11. Immunoblot Detection of Endogenous Arabidopsis Proteins.
Supplemental Table 1. Primers Used in This Study.
Supplemental Movie 1. Three-Dimensional Image of a Transgenic Tobacco BY-2 Cell Expressing Exo70E2-GFP, Showing the Existence of Two Populations of GFP-Positive EXPOs in the Cytosol and Close to the Plasma Membrane.
Supplemental Movie 2. Dynamics of Exo70E2-GFP–Marked EXPO in Transgenic BY-2 Cells.
Supplemental Movie 3. Three-Dimensional Image Animation of a Transgenic Arabidopsis Cell Expressing Exo70E2-GFP, Showing the Cytosolic Organelle Nature of Some Internal GFP-Positive EXPOs.
Supplementary Material
Acknowledgments
We thank Viktor Zarsky (Charles University, Pragus, Czech Republic) for sending us Exo70A1, Sec5, and Sec8 antibodies. This work was supported by grants from the Research Grants Council of Hong Kong (CUHK488707, CUHK465708, CUHK466309, CUHK466610, and HKUST6/CRF/08), Chinese University of Hong Kong (CUHK) Schemes B and C, University Grants Council/Areas of Excellence to L.J., as well as from the German Research Council (DFG) to D.G.R.
References
- An Q., van Bel A.J., Hückelhoven R. (2007). Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2: 4–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders N., Jürgens G. (2008). Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65: 3433–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Osumi M., Ohsumi Y. (1995). Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 20: 465–471 [DOI] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S.V., Klionsky D.J., Ohsumi Y. (1997). Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139: 1687–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Takeshige K., Baba N., Ohsumi Y. (1994). Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124: 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C. (2009). Function and regulation of macroautophagy in plants. Biochim. Biophys. Acta 1793: 1397–1403 [DOI] [PubMed] [Google Scholar]
- Bayer E.M., Bottrill A.R., Walshaw J., Vigouroux M., Naldrett M.J., Thomas C.L., Maule A.J. (2006). Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6: 301–311 [DOI] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Cai H.Q., Reinisch K., Ferro-Novick S. (2007). Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12: 671–682 [DOI] [PubMed] [Google Scholar]
- Chen P.Y., Wang C.K., Soong S.C., To K.Y. (2003). Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breed. 11: 287–293 [Google Scholar]
- Cheng F.Y., Zamski E., Guo W.W., Pharr D.M., Williamson J.D. (2009). Salicylic acid stimulates secretion of the normally symplastic enzyme mannitol dehydrogenase: A possible defense against mannitol-secreting fungal pathogens. Planta 230: 1093–1103 [DOI] [PubMed] [Google Scholar]
- Chong Y.T., Gidda S.K., Sanford C., Parkinson J., Mullen R.T., Goring D.R. (2010). Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol. 185: 401–419 [DOI] [PubMed] [Google Scholar]
- Cole R.A., Synek L., Zarsky V., Fowler J.E. (2005). SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol. 138: 2005–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Craig S., Staehelin L.A. (1988). High pressure freezing of intact plant tissues. Evaluation and characterization of novel features of the endoplasmic reticulum and associated membrane systems. Eur. J. Cell Biol. 46: 81–93 [PubMed] [Google Scholar]
- daSilva L.L.P., Taylor J.P., Hadlington J.L., Hanton S.L., Snowden C.J., Fox S.J., Foresti O., Brandizzi F., Denecke J. (2005). Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. Plant Cell 17: 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzer K., Kleijmeer M.J., Heijnen H.F., Stoorvogel W., Geuze H.J. (2000). Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 113: 3365–3374 [DOI] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran J.M., Anjard C., Stefan C., Loomis W.F., Malhotra V. (2010). Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M., Drdova E., Ziak D., Bavlnka B., Hala M., Cvrckova F., Soukupova H., Zarsky V. (2003). The exocyst complex in plants. Cell Biol. Int. 27: 199–201 [DOI] [PubMed] [Google Scholar]
- Février B., Raposo G. (2004). Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 16: 415–421 [DOI] [PubMed] [Google Scholar]
- Hála M., Cole R., Synek L., Drdová E., Pecenková T., Nordheim A., Lamkemeyer T., Madlung J., Hochholdinger F., Fowler J.E., Zárský V. (2008). An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell 20: 1330–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C., Heuser J., Stahl P. (1984). Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: Demonstration of a pathway for receptor shedding. Eur. J. Cell Biol. 35: 256–263 [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- He B., Guo W. (2009). The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 21: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Xi F., Zhang X., Zhang J., Guo W. (2007). Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 26: 4053–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.C., TerBush D., Abraham M., Guo W. (2004). The exocyst complex in polarized exocytosis. Int. Rev. Cytol. 233: 243–265 [DOI] [PubMed] [Google Scholar]
- Hsu S.C., Ting A.E., Hazuka C.D., Davanger S., Kenny J.W., Kee Y., Scheller R.H. (1996). The mammalian brain rsec6/8 complex. Neuron 17: 1209–1219 [DOI] [PubMed] [Google Scholar]
- Jiang L., Phillips T.E., Hamm C.A., Drozdowicz Y.M., Rea P.A., Maeshima M., Rogers S.W., Rogers J.C. (2001). The protein storage vacuole: a unique compound organelle. J. Cell Biol. 155: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Phillips T.E., Rogers S.W., Rogers J.C. (2000). Biogenesis of the protein storage vacuole crystalloid. J. Cell Biol. 150: 755–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Rogers J.C. (1998). Integral membrane protein sorting to vacuoles in plant cells: Evidence for two pathways. J. Cell Biol. 143: 1183–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journo D., Winter G., Abeliovich H. (2008). Monitoring autophagy in yeast using FM 4-64 fluorescence. Methods Enzymol. 451: 79–88 [DOI] [PubMed] [Google Scholar]
- Kim S.A., Punshon T., Lanzirotti A., Li L., Alonso J.M., Ecker J.R., Kaplan J., Guerinot M.L. (2006). Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Kirkin V., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J. (2007). Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8: 931–937 [DOI] [PubMed] [Google Scholar]
- Koumandou V.L., Dacks J.B., Coulson R.M.R., Field M.C. (2007). Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol. Biol. 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Reggiori F., Peter M. (2009). Selective types of autophagy in yeast. Biochim. Biophys. Acta 1793: 1404–1412 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Cai Y., Hillmer S., Robinson D.G., Jiang L.W. (2008). SCAMPs highlight the developing cell plate during cytokinesis in tobacco BY-2 cells. Plant Physiol. 147: 1637–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.K., Siu C.L., Hillmer S., Jang S., An G., Robinson D.G., Jiang L. (2007). Rice SCAMP1 defines clathrin-coated, trans-golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19: 296–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin J., Morgan M.J., Sibarita J.B., Aresta S., Murthy M., Schwarz T., Camonis J., Bellaïche Y. (2005). Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev. Cell 9: 365–376 [DOI] [PubMed] [Google Scholar]
- Lipka V., Kwon C., Panstruga R. (2007). SNARE-ware: The role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23: 147–174 [DOI] [PubMed] [Google Scholar]
- Manjithaya R., Anjard C., Loomis W.F., Subramani S. (2010). Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 188: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Jiang L. (2007). Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Miao Y., Yan P.K., Kim H., Hwang I., Jiang L. (2006). Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiol. 142: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A., Gallagher L.A., Dunahay T.G., Frohlick J.A., Mazurkiewicz A.M., Meehl J.B., Staehelin L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. (1980). Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21: 205–215 [DOI] [PubMed] [Google Scholar]
- Orlando K., Guo W. (2009). Membrane organization and dynamics in cell polarity. Cold Spring Harb. Perspect. Biol. 1: a001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oztan A., Silvis M., Weisz O.A., Bradbury N.A., Hsu S.C., Goldenring J.R., Yeaman C., Apodaca G. (2007). Exocyst requirement for endocytic traffic directed toward the apical and basolateral poles of polarized MDCK cells. Mol. Biol. Cell 18: 3978–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B.T., Teng K., Wu C., Adam M., Johnstone R.M. (1985). Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 101: 942–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillipson B.A., Pimpl P., daSilva L.L.P., Crofts A.J., Taylor J.P., Movafeghi A., Robinson D.G., Denecke J. (2001). Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell 13: 2005–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent M., Dubois T., Raposo G., Derrien V., Tenza D., Rossé C., Camonis J., Chavrier P. (2003). ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J. Cell Biol. 163: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F. (2006). 1. Membrane origin for autophagy. Curr. Top. Dev. Biol. 74: 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Geldner N., Schrader J., Wolters H., Stierhof Y.D., Rios G., Koncz C., Robinson D.G., Jürgens G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492 [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C., Nebenführ A., Movafeghi A., Stussi-Garaud C., Behnia L., Pimpl P., Staehelin L.A., Robinson D.G. (2002). Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14: 237–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Jiang L., Schumacher K. (2008a). The endosomal system of plants: Charting new and familiar territories. Plant Physiol. 147: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Langhans M., Saint-Jore-Dupas C., Hawes C. (2008b). BFA effects are tissue and not just plant specific. Trends Plant Sci. 13: 405–408 [DOI] [PubMed] [Google Scholar]
- Rogers S.W., Burks M., Rogers J.C. (1997). Monoclonal antibodies to barley aleurain and homologs from other plants. Plant J. 11: 1359–1368 [DOI] [PubMed] [Google Scholar]
- Roumanie O., Wu H., Molk J.N., Rossi G., Bloom K., Brennwald P. (2005). Rho GTPase regulation of exocytosis in yeast is independent of GTP hydrolysis and polarization of the exocyst complex. J. Cell Biol. 170: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S., Moore I. (2002). The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 5: 518–528 [DOI] [PubMed] [Google Scholar]
- Samuel M.A., Chong Y.T., Haasen K.E., Aldea-Brydges M.G., Stone S.L., Goring D.R. (2009). Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Kovaleva V., Bassham D.C., Raikhel N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12: 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Carninci P., Nishiyama Y., Hayashizaki Y., Shinozaki K. (1998). High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15: 707–720 [DOI] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Shen B., Li C., Tarczynski M.C. (2002). High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-L-methionine synthetase 3 gene. Plant J. 29: 371–380 [DOI] [PubMed] [Google Scholar]
- Simons M., Raposo G. (2009). Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21: 575–581 [DOI] [PubMed] [Google Scholar]
- Staehelin L.A., Chapman R.L. (1987). Secretion and membrane tecycling in plant cells: Novel intermediary structures visualized in ultrarapidly frozen sycamore and carrot suspension-culture cells. Planta 171: 43–57 [DOI] [PubMed] [Google Scholar]
- Suen P.K., Shen J.B., Sun S.S.M., Jiang L.W. (2010). Expression and characterization of two functional vacuolar sorting receptor (VSR) proteins, BP-80 and AtVSR4 from culture media of transgenic tobacco BY-2 cells. Plant Sci. 179: 68–76 [Google Scholar]
- Synek L., Schlager N., Eliás M., Quentin M., Hauser M.T., Zárský V. (2006). AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 48: 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul E., Lupashin V. (2006). Role of tethering factors in secretory membrane traffic. Am. J. Physiol. Cell Physiol. 290: C11–C26 [DOI] [PubMed] [Google Scholar]
- Teh O.K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496 [DOI] [PubMed] [Google Scholar]
- TerBush D.R., Maurice T., Roth D., Novick P. (1996). The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 15: 6483–6494 [PMC free article] [PubMed] [Google Scholar]
- TerBush D.R., Novick P. (1995). Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J. Cell Biol. 130: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K., Goto Y., Asatsuma S., Koizumi M., Mitsui T., Matsuoka K. (2009). A mobile secretory vesicle cluster involved in mass transport from the Golgi to the plant cell exterior. Plant Cell 21: 1212–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Van Niel G., Mallegol J., Bevilacqua C., Candalh C., Brugière S., Tomaskovic-Crook E., Heath J.K., Cerf-Bensussan N., Heyman M. (2003). Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut 52: 1690–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Tse Y.C., Law A.H., Sun S.S., Sun Y.B., Xu Z.F., Hillmer S., Robinson D.G., Jiang L. (2010). Vacuolar sorting receptors (VSRs) and secretory carrier membrane proteins (SCAMPs) are essential for pollen tube growth. Plant J. 61: 826–838 [DOI] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li Y., Lo S.W., Hillmer S., Sun S.S., Robinson D.G., Jiang L. (2007). Protein mobilization in germinating mung bean seeds involves vacuolar sorting receptors and multivesicular bodies. Plant Physiol. 143: 1628–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T.J., Hochholdinger F., Sauer M., Bruce W., Schnable P.S. (2005). The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol. 138: 1637–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. (2007). Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Yaffe M.B., Farr G.W., Miklos D., Horwich A.L., Sternlicht M.L., Sternlicht H. (1992). TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature 358: 245–248 [DOI] [PubMed] [Google Scholar]
- Yeaman C., Grindstaff K.K., Wright J.R., Nelson W.J. (2001). Sec6/8 complexes on trans-Golgi network and plasma membrane regulate late stages of exocytosis in mammalian cells. J. Cell Biol. 155: 593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. (2009). 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185 [DOI] [PubMed] [Google Scholar]
- Yoshimori T., Noda T. (2008). Toward unraveling membrane biogenesis in mammalian autophagy. Curr. Opin. Cell Biol. 20: 401–407 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zárský V., Cvrcková F., Potocký M., Hála M. (2009). Exocytosis and cell polarity in plants - Exocyst and recycling domains. New Phytol. 183: 255–272 [DOI] [PubMed] [Google Scholar]
- Zhang X., Orlando K., He B., Xi F., Zhang J., Zajac A., Guo W. (2008). Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J. Cell Biol. 180: 145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X., Zhang J., Zhang Y., Hsu S.C., Zhou D., Guo W. (2006). Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat. Cell Biol. 8: 1383–1388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.