Abstract
Single nanopores attract a great deal of scientific interest as a basis for biosensors and as a system to study the interactions and behavior of molecules in a confined volume. Tuning the geometry and surface chemistry of nanopores helps create devices that control transport of ions and molecules in solution. Here, we present single conically shaped nanopores whose narrow opening of 8 or 12 nm is modified with single-stranded DNA molecules. We find that the DNA occludes the narrow opening of nanopores and that the blockade extent decreases with the ionic strength of the background electrolyte. The results are explained by the ionic strength dependence of the persistence length of DNA. At low KCl concentrations (10 mM) the molecules assume an extended and rigid conformation, thereby blocking the pore lumen and reducing the flow of ionic current to a greater extent than compacted DNA at high salt concentrations. Attaching flexible polymers to the pore walls hence creates a system with tunable opening diameters in order to regulate transport of both neutral and charged species.
Keywords: DNA strand, Nanopore, Ion transport
Introduction
Nanopores are powerful research tools to investigate the structural and dynamic properties of single biomolecules. Nanopores have inner diameters of a few nanometers, and following the size-exclusion principle, only individual DNA or protein molecules can fit into or pass through a pore. Single-particle translocation is conveniently detected via ionic current measurements through single pores. Importantly, a passing molecule blocks a pore, leading to a transient change of current. Parameters of the current blockade such as duration and amplitude can give information about the length and size of the molecule. One of the most widely published and scientifically attractive subjects is the sensing of DNA with nanopores. While an ultimate goal of these studies is to develop an inexpensive method to sequence DNA (Dekker 2007; Branton et al. 2008; Garaj et al. 2010; Schneider et al. 2010; Merchant et al. 2010), a large array of experiments have also shed light on the biophysics of DNA or RNA translocation through biological pores (Kasianowicz et al. 1996; reviewed in Branton et al. 2008; Wanunu et al. 2010; Healy 2007; Vercoutere and Akeson 2002; Deamer and Branton 2002; Marziali and Akeson 2001) as well as inorganic pores (reviewed in, e.g., Howorka and Siwy 2009; Healy et al. 2007; Dekker 2007). Examined aspects include (i) the frequency with which the strands thread into the pore (Henrickson et al. 2000; Meller and Branton 2002; Nakane et al. 2004; Maglia et al. 2008), (ii) the orientation of strands (Butler et al. 2006; Wang et al. 2004; Mathe et al. 2005; Wanunu et al. 2008; Wiggin et al. 2008; Li et al. 2003; Chen et al. 2004; Storm et al. 2005; Fologea et al. 2007; Steinbock et al. 2010), (iii) the speed of DNA transport (Meller et al. 2001; Wanunu et al. 2008), (iv) the influence of transmembrane potential (Aksimentiev and Schulten 2005; Mathe et al. 2005; Kathawalla et al. 1989; Heng et al. 2005; Keyser et al. 2006), (v) pore geometry (Howorka and Bayley 2002) as well as (vi) interaction with pore walls (Wiggin et al. 2008; Wanunu et al. 2008; Sigalov et al. 2008).
Covalently attaching DNA to the pore wall is an attractive variation in single-molecule research. In previous studies aimed at DNA sensing, one or multiple single stranded DNA (ssDNA) molecules were end-tethered to the pore wall of protein (Howorka et al. 2001a, 2001b) and inorganic nanopores (Iqbal et al. 2007; Harrell et al. 2004), respectively. Exposing the modified pores to a solution containing complementary or mismatched DNA molecules showed that DNA duplexes between the free and tethered strands form inside the nanopore. As the perfect and mismatched duplexes, respectively, had different lifetimes, the DNA-modified nanopores could be used as biosensor elements to distinguish nucleic acids with single-point mutations (Howorka et al. 2001a, b; Iqbal et al. 2007). The covalent attachment of DNA strands has also been exploited to control the electronic properties of a pore. In general, artificial pores that exhibit engineered properties such as voltage gating or ion selectivity are attractive model systems for biological voltage-gated ion channels (Hille 2001). Harrell et al. (2004) were the first to demonstrate that DNA-modified nanopores can function as ionic switches. A single gold nanotube carrying thiol-terminated DNA strands was shown to preferentially transport cations in one direction, while hindering transport in the other. The ion current rectification was explained by assuming that DNA molecules deflect by the external electric field and cause voltage-dependent pore opening. The attached DNA strands were not localized to a single subnanoscale position but covered the entire pore. Additionally, the length of the fully extended DNA molecules was smaller than the pore diameter; hence, it was not possible to completely block the current (e.g., 59-nm-diameter gold tube was modified with 30-mer DNA).
With the aim of achieving a greater pore blockade and ion current modulations, this study attempts to tune the ionic pore properties by restricting DNA immobilization to a small nanoscale region. Single conically shaped nanopores with an opening of 8 or 12 nm were selectively modified at the narrow section with ssDNA oligonucleotides (Fig. 1) with a length of 30 nucleotides. We compared the current–voltage curves of single nanopores before and after DNA modification at different KCl concentrations between 1 M and 10 mM. The experimental results indicate that the attached DNA either reduced or blocked the ionic current flow through the pore dependent on ionic strength, which also influenced the conformation of the DNA strands (Odijk 1977; Skolnick and Fixman 1977; Kaiser and Rant 2010). The pores hence show how DNA biophysics can influence ionic pore properties.
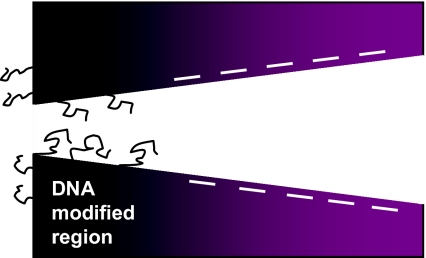
Fig. 1.
Scheme of the surface charge pattern obtained in the process of one-sided modification of a single conically shaped nanopore with point-tethered DNA molecules. We estimate the position of the junction between the DNA-modified zone and the zone with carboxyl groups to be several tens of nanometers from the small opening of the pore (Vlassiouk and Siwy 2007). Carboxyl groups are formed in the process of pore preparation
In addition, the DNA-modified conical nanopores presented here function as rectifiers. Rectifying systems have a preferential direction of mass flow and, in some cases, can entirely stop the flow in the opposite direction, forming a diode. Rectifiers and diodes based on surface charge patterns on the pore walls have previously been created, but these previous embodiments were able to switch the transport of just ions and charged species (Bassignana and Reiss 1983; Mafe and Ramirez 1997; Daiguji et al. 2005; Vlassiouk and Siwy 2007; Nguyen et al. 2010; Cheng and Guo 2009). As an example, an ionic bipolar diode was built based on nanopores that contained a junction between positive and negative charges on the pore walls (Vlassiouk and Siwy 2007; Cheng and Guo 2009). A unipolar diode is formed with a junction between positive (or negative) surface charges and a neutral zone (Karnik et al. 2007; Nguyen et al. 2010; Cheng and Guo 2009). By contrast, the DNA-modified nanopores presented here feature an opening diameter that is tuned by the operation conditions, e.g., KCl concentration and voltage. The rationally engineered pores can thus be applied to tune and switch the transport of both ionic and neutral species.
Materials and Methods
Preparation of Nanopores
Single conically shaped nanopores were prepared by the track-etching technique according to a published procedure (Fleischer et al. 1975; Spohr 1983). Briefly, 12-μm-thick films of polyethylene terephthalate (PET) were irradiated with single heavy ions (e.g., Au, U) with a kinetic energy of ~2 GeV. Irradiation was performed at the linear accelerator UNILAC at the Institute for Heavy Ions Research (GSI), Darmstadt, Germany (Spohr 1983). After tracking, foils were etched from one side in 9 M NaOH to form conically shaped nanopores (Apel et al. 2001). The other side of the membrane was protected against etching by an acidic stopping solution. The preparation was performed in a conductivity cell so that the pore opening could be observed electrochemically. Once an etching current of ~200 pA was recorded, the membrane was washed with the stopping medium and KCl solution. The transport properties of single PET nanopores were investigated in KCl solutions buffered to pH 8 with Tris buffer.
Characterization of the Pore Opening
An approximate diameter of the small opening of the conical pore was determined using conductance measurements as described previously (Apel et al. 2001). In brief, both sides of a single nanopore membrane were placed in contact with 1 M KCl. Ag/AgCl electrodes were immersed into each solution, and a current–voltage curve was recorded for voltages between +100 and −100 mV. In this voltage regime the nanopores produce a linear current–voltage curve whose slope provides the ionic resistance (R
p) of the nanopore. The tip diameter (d) is related to R
p via 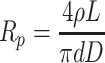 , where L is the membrane thickness, ρ is the specific ionic resistance of the electrolyte and D is the base (big opening of the cone) diameter. The diameter D was determined from the so-called bulk-etch rate of a polymer material. For PET and etching conditions of 20°C and 9 M NaOH, the bulk-etch rate is equal to 2.13 nm/min (Apel et al. 2001). The value of D was then determined from the formula D = 2 × 2.13t (nm), where t is time of etching in minutes. The validity of this relation was confirmed by scanning electron microscopy studies (Wharton et al. 2007).
, where L is the membrane thickness, ρ is the specific ionic resistance of the electrolyte and D is the base (big opening of the cone) diameter. The diameter D was determined from the so-called bulk-etch rate of a polymer material. For PET and etching conditions of 20°C and 9 M NaOH, the bulk-etch rate is equal to 2.13 nm/min (Apel et al. 2001). The value of D was then determined from the formula D = 2 × 2.13t (nm), where t is time of etching in minutes. The validity of this relation was confirmed by scanning electron microscopy studies (Wharton et al. 2007).
Modification with DNA
Pore walls and the surface of track-etched PET films contain carboxyl groups at a density of ~1 group/nm2 (Wolf-Reber 2002). The carboxyl groups were the attachment points to covalently couple amino-modified DNA using a procedure which had been previously applied for other surface modification purposes. In the procedure, carboxyl groups are activated with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and then coupled to the amine, forming an amide (Vlassiouk and Siwy 2007). A DNA oligonucleotide with a 5′-terminal amino C12 spacer and the sequence 5′-CGC GAG AAGTTA CAT GAC CTG TAG ACG ATC-3′ was purchased from Integrated DNA Technologies (Coralville, IA). The received amount of 1,267 nM was dissolved in 500 μl of 0.1 M MES buffer, pH 5.5. In the amidation protocol, the DNA and EDC solution was placed in contact with the small opening of the pore, while the large opening of the pore was in contact with 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) solution, pH 5.5, for 24-h incubation. Placing the modifying agent only on one side of the membrane is known to create a very nonhomogeneous distribution of the chemicals along the pore axis (Vlassiouk and Siwy 2007). Due to the conical pore shape, the concentrations of EDC and DNA were high at the first tens of nanometers at the narrow tip and decayed hyperbolically along the pore axis. Consequently, DNA attachment was expected to occur only at the region close to the small opening where the reagents’ concentration was sufficiently high. It is expected that our nanopore system has a surface chemistry as schematically shown in Fig. 1. Four DNA-modified nanopores with opening diameters between 4 and 12 nm were studied.
Recordings of Current–Voltage Curves
The ionic properties of the nanopores were studied in the same conductivity cell in which the etching was performed. Two Ag/AgCl electrodes were used to apply the transmembrane potential and to measure the ion current. Ag/AgCl electrodes are very stable and largely nonpolarizable. We used a two-electrode setup in which the electrode placed at the narrow end of a conical nanopore was grounded, while the other electrode next to the big pore opening was used to apply a given potential difference with respect to the ground electrode. The voltage was swept between −3 and +3 V for 1 M KCl, and between −4 and +4 V for lower concentrations, with a voltage step of 100 mV. The presented current–voltage curves are averages of typically three sweeps, unless otherwise stated (Figs. 3c, d, 5). The current variations in the averaged signals typically did not exceed 8% (the current variations were the strongest for negative voltages above 2 V).
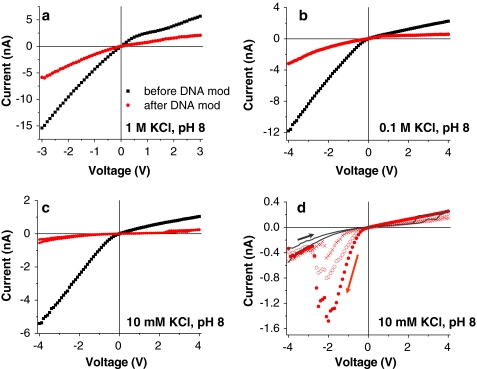
Fig. 3.
Current–voltage curves of the same nanopore studied in Fig. 2 after modification with 30-nucleotide ssDNA for a 1 M KCl, b 0.1 M KCl and c 10 mM KCl (two sweeps from −4 to +4 V are shown for the DNA-modified nanopore). All recordings were performed at pH 8. Red curves in d present three consecutive reverse sweeps of voltages from +4 to −4 V, showing a big variation between the sweeps and hysteresis compared to the forward sweeps from −4 to +4 V (shown in gray). In 1 and 0.1 M KCl, no differences between currents recorded for forward and reverse voltage sweeps were observed
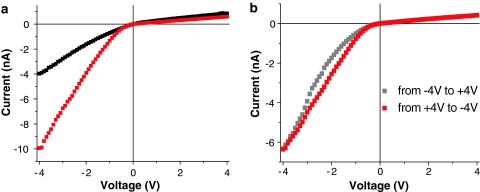
Fig. 5.
a Current–voltage curves before and after modification with 30-mer ssDNA of a single conically shaped nanopore with a narrow opening of 12 nm. Recordings were performed in 0.1 M KCl, pH 8. b Current–voltage curves of 12-nm DNA-modified nanopore in 10 mM KCl, pH 8, for forward bias (gray squares) and reverse bias (red squares)
Results and Discussion
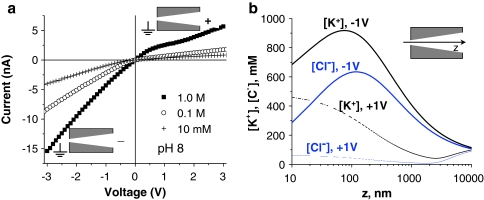
We first analyzed a pore that had not been modified with DNA. Figure 2a presents a set of current–voltage curves of a single conically shaped nanopore with small and big openings of 8 and 400 nm, respectively. A positive voltage in our electrode configuration corresponds to a positively biased electrode placed at the big opening of a conical nanopore, and vice versa for a negative voltage. Similar to previous reports, the conical nanopore rectified the ion current due to breaking of the symmetry of the electric potential inside the pore (Siwy and Fulinski 2002; Hanggi and Marchesoni 2009; Cervera et al. 2005). In particular, the broken symmetry affected the interactions between K+ ions and the negatively charged carboxyl groups on the pore walls. The rectification was quantified by calculating the so-called rectification degree, which is a ratio of currents for voltages of the same absolute value but of opposite polarity, e.g., I(−3 V)/I(+3 V). The negatively charged nanopores have transference numbers found from the reversal potential (Hille 2001) of at least 0.8, which indicates that ~80% of the current is carried by potassium ions (Cervera et al. 2007; Gillespie et al. 2008). The larger currents for negative voltages correspond to cations moving from the small opening to the large opening of the pore. The experimentally found ion current rectification and selectivity properties of conically shaped nanopores were modeled using the Poisson-Nernst-Planck (PNP) equations (Cervera et al. 2005, 2006, 2007; White and Bund 2008) as well as Monte Carlo (He et al. 2009) and molecular dynamics (Cruz-Chu et al. 2009) approaches. Figure 2b presents average ionic concentrations along the pore axis for +1 and −1 V, 0.1 M KCl, obtained bynumerically solving the PNP equations, as described (Vlassiouk et al. 2008a, b; Powell et al. 2009). At negative voltages, ionic concentrations of both ions, potassium and chloride, are significantly higher than at the corresponding positive voltage, thereby validating that the negative voltage corresponds to the high conductance state. Given that the concentration of cations is higher than that of anions for negative and positive potentials, the plot in Fig. 2b also confirms the cation selectivity of our pore. The modeling furthermore revealed that in the case of conically shaped nanopores, the quantitative estimation of cation selectivity (based on the reversal potential measurements) holds true only for low voltages. At high negative potentials, the cation selectivity becomes less pronounced (Fig. 2b), indicated by an increase in the concentration of both potassium and chloride ions within the pore (Cervera et al. 2006; Powell et al. 2009).
Fig. 2.
a Current–voltage curves of a single conically shaped nanopore with openings of ~8 and 400 nm, recorded at pH 8 for three KCl concentrations as indicated. Insets show the position of the electrical ground and external voltage relative to the two pore openings. b Numerical solutions of the PNP equation for a conically shaped nanopore with surface charge density of 0.5 e/nm2 showing average ionic concentrations along the pore axis. Modeling was performed using software Comsol as described (Vlassiouk et al. 2008a, b; Powell et al. 2009). Opening diameters of the modeled nanopore were set at 5 and 500 nm
It is well known that the pores rectify only if the walls have excess surface charge and when the pore diameter is comparable to the thickness of the electrical double-layer (Siwy and Fulinski 2002; Cervera et al. 2006). As a consequence, the rectification degree of the conically shaped nanopores in 1 M KCl was lower than that in 0.1 M and 10 mM KCl. The values at 3 V are (Fig. 2a) 2.7 (|−15.4| nA, 5.68 nA) in 1 M, 4.7 (|−8.32| nA, 1.75 nA) in 0.1 M KCl and 4.9 (|−4.08| nA, 0.828 nA) in 10 mM KCl. The small difference in rectification degrees between 0.1 M and 10 mM KCl is in agreement with predictions based on the PNP equations, which point to a nonlinear dependence of the rectification degree on KCl concentration (Cervera et al. 2006). The modeling also showed the optimal ratio between the pore diameter and the screening length that produces the highest rectification degree. For a 6-nm-diameter conical pore, the concentration range at which the pore was predicted to rectify most was between ~0.12 and 0.40 M KCl (Cervera et al. 2006). In this concentration range, the rectification degree was shown to change very little. For wider pores, the optimal concentration range is shifted to more diluted solutions; and in the 8-nm pore studied here, at 10 mM and 0.1 M KCl the pore rectified almost equally well.
A very different behavior was observed after modifying the tip of the same nanopore with 30-nucleotide-long ssDNA. Figure 3 compares current–voltage curves of the pore before and after DNA modification for 1 M, 0.1 M and 10 mM KCl. For all these concentrations, the DNA modification caused a significant decrease of the current, which suggests that the long DNA molecules physically occluded the pore and diminished its effective opening diameter.
The experimental results suggest that occlusion of the pore by DNA becomes more pronounced with lowering the ionic strength. At 1 M KCl, attachment of DNA caused a decrease of ~2.5-fold of ion current compared to a non-modified pore (Fig. 3a), while at 10 mM KCl, the DNA-modified nanopore exhibited at least five times lower current (Fig. 3c). Any explanation of the observed current change has to consider two effects which were initially used to describe experiments performed with non-attached pore-translocating DNA strands (Smeets et al. 2006). Firstly, each DNA molecule excludes a certain volume in the pore so that fewer ions are available for transport. Secondly, DNA molecules are heavily charged; thus, their presence brings more counter-ions to the pore lumen. Insertion of DNA molecules in the pore can therefore cause a decrease or an increase of the current, depending on whether the number of excluded ions is larger or smaller than the number of additionally introduced DNA counterions. Both effects have been observed in systematic experiments on threading single DNA molecules through unmodified nanopores (Smeets et al. 2006). In dependence of the concentration of the background electrolyte, the pore-threaded DNA molecules caused a positive or negative spike of the current. In all reported cases, lowering the KCl concentration of the background electrolyte caused the DNA counter-ions to become dominant.
Our observation that a decrease in KCl concentration leads to an increase of the pore blockage suggests that a simple explanation based on volume exclusion and electrostatic effects is insufficient. It is more likely that the transmembrane current is additionally influenced by changes in the conformation of the DNA molecules. This effect has recently been observed with DNA molecules attached to gold surfaces (Kaiser and Rant 2010). In the experiments, ssDNA molecules in high KCl concentrations assumed a random coil and compact configuration. Decreasing the background electrolyte concentration caused the DNA molecules to be more extended, and in concentrations ~1 mM the molecules became entirely stretched. The length of the DNA strand was determined by applying a negative potential to the gold surface and determining the quenching of a fluorophore which was attached to the other DNA end.
These results were explained by considering the dependence of the mechanical rigidity of ssDNA on the ionic strength of the environment. Theories of Odijk (1977) as well as Skolnick and Fixman (1977) distinguish bare (l
0) and electrostatic (l
e) contributions to the total persistence length of a charged molecule l
p: 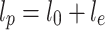 . The bare persistence length of ssDNA is known to be only 2–3 nm (Tinland et al. 1997), while the value of l
e is ionic strength–dependent and can be calculated as
. The bare persistence length of ssDNA is known to be only 2–3 nm (Tinland et al. 1997), while the value of l
e is ionic strength–dependent and can be calculated as 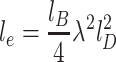 , where
, where 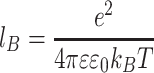 is the Bjerum length, λ is the linear density of the polymer and l
D stands for the screening length given by the Debye-Hueckel theory. The parameters k
b and T have their usual meaning of the Boltzmann constant and temperature, respectively.
is the Bjerum length, λ is the linear density of the polymer and l
D stands for the screening length given by the Debye-Hueckel theory. The parameters k
b and T have their usual meaning of the Boltzmann constant and temperature, respectively.
The length l e becomes important only at lower concentrations; e.g., at 10 mM KCl it equals ~10 nm. Applied to our DNA-modified pore, this high length obviously increases the volume of the DNA strands and could thereby help explain the high level of occlusion observed for the nanopore which measures 8 nm in diameter (Fig. 2c).
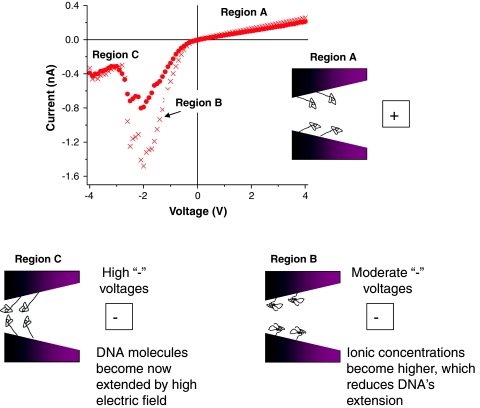
It is also important to mention that for the lowest studied KCl concentration of 10 mM, the current–voltage curves exhibited a strong hysteresis, especially for negative voltages (Fig. 3d). When the voltage sweep started at negative voltages, the currents were typically significantly lower than in the case when the sweep started from positive voltages. We think that the hysteresis is caused by the voltage-induced movement of the attached DNA strands as well as voltage-dependent ionic concentrations, as discussed above. Figure 4 summarizes a model which accounts for the observed hysteresis and high currents for a sweep from positive to negative transmembrane potentials. When a positively biased electrode is placed at the wide opening of the pore, the DNA molecules are expected to be deflected away from the narrow opening and toward the wide opening, which does not present any significant steric hindrance for the strands (Fig. 4, region A). Due to the conical shape of the pore, this voltage polarity constitutes the lower conductance state of the device. Reducing the positive voltage and switching it to the negative values forces the DNA strands to deflect toward the small opening of the pore (Fig. 4, region B). It is recalled that at negative voltages concentrations of potassium and chloride ions in the region close to the small pore opening increase at least several times compared to the bulk concentration (Fig. 2b). This region with elevated ionic concentration is rather wide and reaches several hundreds of nanometers away from the narrow opening. Consequently, the DNA strands in the modified pore are in contact with high concentrations of potassium and chloride ions, which in turn likely causes DNA to assume a more compact form (Fig. 4, region B)—in line with results from Kaiser and Rant (2010). At moderate negative voltages the currents through the pore are large, leading to high rectification degrees. At negative voltages above a threshold value of ~−2 V (Figs. 3d, 4), DNA molecules might become extended due to the electric field (Heng et al. 2005; Randall et al. 2006) so that the nanopore is again more blocked (Fig. 4, region C).
Fig. 4.
Two current–voltage curves (the same as shown in Fig. 3d) for an 8-nm-diameter nanopore modified with DNA, recorded when the voltage was changed from +4 to −4 V. Insets schematically indicate possible configurations of the attached DNA molecules as a function of applied voltage
By contrast, when the voltage is swept from negative to positive values, DNA molecules have to deflect from the small opening toward the large opening of the pore. We think that this process occurs with significantly more severe steric hindrance because lowering the negative applied voltage decreases the ionic concentrations in the pore, causing the DNA to maintain a more elongated form induced by the high voltage, thereby leading to a greater pore blockade.
In order to support our general hypothesis on the conformation dependent occlusion of nanopores by DNA molecules, we studied a wider nanopore with a narrow diameter of 12 nm. We expected that in this case the DNA would lead to a less extensive pore blockade based on geometric grounds. Figure 5 shows the current–voltage curves of the 12-nm pore in 0.1 M KCl, pH 8, before and after DNA attachment. In contrast to the data obtained with the 8-nm pore (Fig. 3), the wider structure is characterized by not only a less extensive blockade but also higher negative currents after the DNA modification. The larger current after DNA modification suggests, according to Smeets et al. (2006), that the number of DNA counter-ions exceeds the number of ions that are excluded from the nanopore. Most likely, DNA attachment increases the local surface charge density and consequently enhances ionic concentrations at the narrow opening.
The wider nanopore also exhibited an ion current rectification degree in 0.1 M KCl, 4 V equal to almost 17, which is three times higher than the rectification degree of the 8-nm DNA-modified pore (Fig. 3b, red curve). This observation was at first puzzling to us as one would expect a higher rectification degree for a narrower pore. One possible explanation is that the DNA conformational change makes the pore structure less asymmetric and less rectifying. A wider pore would be less affected by this phenomenon. Another explanation takes into account the dependence of the rectification degree of a conical nanopore on ionic strength, which we already discussed above. There is an optimal ratio of pore diameter and screening length for a maximal rectification degree (Cervera et al. 2006). For concentrations that are lower than the optimum for a given pore diameter, the pores rectify less. With DNA, the nanopore is effectively smaller so that the pore might be outside its optimal rectifying regime.
As expected, for the wider conical nanopore in 10 mM KCl, the hysteresis of ion currents recorded for forward and reverse voltage sweeps was much less pronounced compared to the 8-nm nanopore (see Figs. 3d, 5b).
Conclusions
We have prepared an ionic rectifying system based on single conically shaped nanopores whose small opening was modified with ssDNA. In contrast to previously designed bipolar and unipolar diodes (Vlassiouk and Siwy 2007; Karnik et al. 2007) containing stationary surface charges, the attached DNA molecules are flexible and assume different configurations depending on the operation conditions such as KCl concentration and voltage. Our experimental results provide evidence that the level of nanopore blockage and, most likely, the persistence length of tethered DNA increase with lower KCl concentration. Due to the salt-dependent DNA volume, the nanopore changes its effective diameter and thus can be applicable in building systems to control transport of both charged and neutral species. In future research, we will systematically study the transport properties of DNA-modified pores as a function of DNA length. We expect to be able to tune the rectification properties of the pores as well as the effective pore diameter as a function of external voltage. We will also try to identify a particular ratio of DNA length and pore diameter under which DNA-modified nanopores exhibit spontaneous openings and closings of the pore similar to biological voltage-gated channels.
Acknowledgments
This work was supported in part by the National Science Foundation (CHE 0747237), and the Royal Society of Chemistry’s Journals Grants for International Authors programme. The authors are grateful to Dr. Christina Trautmann and Dr. Eugenia Tomil-Molares (GSI Darmstadt) for providing us with irradiated polymer films. We are grateful to Dr. Ivan Vlassiouk for numerically solving the PNP equations.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aksimentiev A, Schulten K. Imaging alpha-hemolysin with molecular dynamics: ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys J. 2005;88:3745–3761. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel PY, Korchev YE, Siwy Z, Spohr R, Yoshida M. Diode-like single ion-track membrane prepared by electro-stopping. Nucl Instrum Methods Phys Res B. 2001;184:337–346. doi: 10.1016/S0168-583X(01)00722-4. [DOI] [Google Scholar]
- Bassignana IC, Reiss H. Ion transport and water dissociation in bipolar ion exchange membranes. J Membr Sci. 1983;15:27–41. doi: 10.1016/S0376-7388(00)81360-5. [DOI] [Google Scholar]
- Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler TZ, Gundlach JH, Troll MA. Determination of RNA orientation during translocation through a biological nanopore. Biophys J. 2006;90:190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera J, Schiedt B, Ramirez P. A Poisson/Nernst-Planck model for ionic transport through synthetic conical nanopores. Europhys Lett. 2005;71:35–41. doi: 10.1209/epl/i2005-10054-x. [DOI] [Google Scholar]
- Cervera J, Schiedt B, Neumann R, Mafe S, Ramirez P. Ionic conduction, rectification, and selectivity in single conical nanopores. J Chem Phys. 2006;124:104706. doi: 10.1063/1.2179797. [DOI] [PubMed] [Google Scholar]
- Cervera J, Alcaraz A, Schiedt B, Neumann R, Ramirez P. Asymmetric selectivity of synthetic conical nanopores probed by reversal potential measurements. J Phys Chem C. 2007;111:12265–12273. doi: 10.1021/jp071884c. [DOI] [Google Scholar]
- Chen P, Gu JJ, Brandin E, Kim YR, Wang D, Branton D. Probing single DNA molecule transport using fabricated nanopores. Nano Lett. 2004;4:2293–2298. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L-J, Guo LJ. Ionic current rectification, breakdown, and switching in heterogeneous oxide nanofluidic devices. ACS Nano. 2009;3:575–584. doi: 10.1021/nn8007542. [DOI] [PubMed] [Google Scholar]
- Cruz-Chu E, Ritz T, Siwy ZS, Schulten K. Molecular control of ionic conduction in polymer nanopores. Faraday Discuss. 2009;143:47–62. doi: 10.1039/b906279n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiguji H, Oka Y, Shirono K. Nanofluidic diode and bipolar transistor. Nano Lett. 2005;5:2274–2280. doi: 10.1021/nl051646y. [DOI] [PubMed] [Google Scholar]
- Deamer DW, Branton D. Characterization of nucleic acids by nanopore analysis. Acc Chem Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- Fleischer RL, Price PB, Walker RM. Nuclear tracks in solids. Principles and applications. Berkeley: University of California Press; 1975. [Google Scholar]
- Fologea D, Brandin E, Uplinger J, Branton D. DNA conformation and base number simultaneously determined in a nanopore. Electrophoresis. 2007;28:3186–3192. doi: 10.1002/elps.200700047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467:190–194. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D, Boda D, He Y, Apel P, Siwy ZS. Synthetic nanopores as a test case for ion channel theories: the anomalous mole fraction effect. Biophys J. 2008;95:609–619. doi: 10.1529/biophysj.107.127985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanggi P, Marchesoni F. Artificial Brownian motors: controlling transport on the nanoscale. Rev Mod Phys. 2009;81:387–442. doi: 10.1103/RevModPhys.81.387. [DOI] [Google Scholar]
- Harrell CC, Kohli P, Siwy Z, Martin CR. DNA-nanotube artificial ion channels. J Am Chem Soc. 2004;126:15646–15647. doi: 10.1021/ja044948v. [DOI] [PubMed] [Google Scholar]
- He Y, Gillespie D, Boda D, Vlassiouk I, Eisenberg RS, Siwy ZS. Tuning transport properties of nanofluidic devices with local charge inversion. J Am Chem Soc. 2009;131:5194–5202. doi: 10.1021/ja808717u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy K. Nanopore-based single-molecule DNA analysis. Nanomedicine. 2007;2:459–481. doi: 10.2217/17435889.2.4.459. [DOI] [PubMed] [Google Scholar]
- Healy K, Schiedt S, Morrison AP. Solid-state nanopore technologies for nanopore-based DNA analysis. Nanomedicine. 2007;2:875–879. doi: 10.2217/17435889.2.6.875. [DOI] [PubMed] [Google Scholar]
- Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. Stretching DNA using the electric field in a synthetic nanopore. Nano Lett. 2005;5:1883–1888. doi: 10.1021/nl0510816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Driven DNA transport into an asymmetric nanometer-scale pore. Phys Rev Lett. 2000;85:3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- Hille B. Ion channels of excitable membranes. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- Howorka S, Bayley H. Probing distance and electrical potential within a protein pore with tethered DNA. Biophys J. 2002;83:3202–3210. doi: 10.1016/S0006-3495(02)75322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howorka S, Siwy ZS. Nanopore analytics: sensing of single molecules. Chem Soc Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- Howorka S, Movileanu L, Braha O, Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc Natl Acad Sci USA. 2001;98:12996–13001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA-strands using engineered nanopores. Nat Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- Iqbal SM, Akin A, Bashir R. Solid-state nanopore channels with DNA selectivity. Nat Nanotechnol. 2007;2:243–248. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- Kaiser W, Rant U. Conformations of end-tethered DNA molecules on gold surfaces: influences of applied electric potential, electrolyte screening, and temperature. J Am Chem Soc. 2010;132:7935–7945. doi: 10.1021/ja908727d. [DOI] [PubMed] [Google Scholar]
- Karnik R, Duan C, Castelino K, Daiguji H, Majumdar A. Rectification of ionic current in a nanofluidic diode. Nano Lett. 2007;7:547–551. doi: 10.1021/nl062806o. [DOI] [PubMed] [Google Scholar]
- Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathawalla IA, Anderson JL, Lindsey JS. Hindered diffusion of porphyrins and short-chain polystyrene in small pores. Macromolecules. 1989;22:1215–1219. doi: 10.1021/ma00193a037. [DOI] [Google Scholar]
- Keyser UF, Koeleman B, van Dorp S, Krapf D, Smeets RMM, Lemay SG, Dekker NH, Dekker C. Direct force measurements on voltage driven DNA translocation through a nanopore. Nat Phys. 2006;2:473–477. doi: 10.1038/nphys344. [DOI] [Google Scholar]
- Li JL, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA molecules and configurations in a solid-state nanopore microscope. Nat Mater. 2003;2:611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- Mafe S, Ramirez P. Electrochemical characterization of polymer ion-exchange bipolar membranes. Acta Polym. 1997;48:234–250. doi: 10.1002/actp.1997.010480702. [DOI] [Google Scholar]
- Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proc Natl Acad Sci USA. 2008;105:19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marziali A, Akeson M. New DNA sequencing methods. Annu Rev Biomed Eng. 2001;3:195–223. doi: 10.1146/annurev.bioeng.3.1.195. [DOI] [PubMed] [Google Scholar]
- Mathe J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Orientation discrimination of single-stranded DNA inside the alpha-hemolysin membrane channel. Proc Natl Acad Sci USA. 2005;102:12377–12382. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller A, Branton D. Single molecule measurements of DNA transport through a nanopore. Electrophoresis. 2002;23:2583–2591. doi: 10.1002/1522-2683(200208)23:16<2583::AID-ELPS2583>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Meller A, Nivon L, Branton D. Voltage-driven DNA translocation through a nanopore. Phys Rev Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo Z, Johnson ATC, Drndic M. DNA translocation through graphene nanopores. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- Nakane J, Wiggin M, Marziali A. A nanosensor for transmembrane capture and identification of single nucleic acid molecules. Biophys J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen G, Vlassiouk I, Siwy ZS. Comparison of bipolar and unipolar ionic diodes. Nanotechnology. 2010;21:265301. doi: 10.1088/0957-4484/21/26/265301. [DOI] [PubMed] [Google Scholar]
- Odijk T. Polyelectrolytes near the rod limit. J Polym Sci B Polym Phys. 1977;15:477–483. [Google Scholar]
- Powell MR, Vlassiouk I, Martens C, Siwy ZS. Non-equilibrium 1/f noise in rectifying nanopores. Phys Rev Lett. 2009;103:248104. doi: 10.1103/PhysRevLett.103.248104. [DOI] [PubMed] [Google Scholar]
- Randall GC, Schultz KM, Doyle PS. Methods to electrophoretically stretch DNA: microcontractions, gels, and hybrid gel-microcontraction devices. Lab Chip. 2006;6:516–525. doi: 10.1039/b515326c. [DOI] [PubMed] [Google Scholar]
- Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LM, Dekker C. DNA translocation through graphene nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- Sigalov G, Comer J, Timp G, Aksimentiev A. Detection of DNA sequences using an alternating electric field in a nanopore capacitor. Nano Lett. 2008;8:56–63. doi: 10.1021/nl071890k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwy Z, Fulinski A. Fabrication of a synthetic nanopore ion-pump. Phys Rev Lett. 2002;89:198103. doi: 10.1103/PhysRevLett.89.198103. [DOI] [PubMed] [Google Scholar]
- Skolnick J, Fixman M. Electrostatic persistence length of a wormlike polyelectrolyte. Macromolecules. 1977;10:944–948. doi: 10.1021/ma60059a011. [DOI] [Google Scholar]
- Smeets RM, Keyser UF, Krapf D, Wu MY, Dekker NH, Dekker C. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 2006;6:89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- Spohr R (1983) Methods and device to generate a predetermined number of ion tracks. German Patent DE2951376 C2, 15 Sept 1983; US Patent 4369370
- Steinbock LJ, Otto O, Chimerel C, Gornall J, Keyser UF. Detecting DNA folding with nanocapillaries. Nano Lett. 2010;10:2493–2497. doi: 10.1021/nl100997s. [DOI] [PubMed] [Google Scholar]
- Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys Rev E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- Tinland B, Pluen A, Sturm J, Weill G. Persistence length of single-stranded DNA. Macromolecules. 1997;30:5763–5765. doi: 10.1021/ma970381+. [DOI] [Google Scholar]
- Vercoutere W, Akeson M. Biosensors for DNA sequence detection. Curr Opin Chem Biol. 2002;6:816–822. doi: 10.1016/S1367-5931(02)00395-2. [DOI] [PubMed] [Google Scholar]
- Vlassiouk I, Siwy Z. Nanofluidic diode. Nano Lett. 2007;7:552–556. doi: 10.1021/nl062924b. [DOI] [PubMed] [Google Scholar]
- Vlassiouk I, Smirnov S, Siwy ZS. Nanofluidic ionic diodes. Comparison of analytical and numerical solutions. ACS Nano. 2008;2:1589–1602. doi: 10.1021/nn800306u. [DOI] [PubMed] [Google Scholar]
- Vlassiouk I, Smirnov S, Siwy ZS. Ionic selectivity of single nanochannels. Nano Lett. 2008;8:1978–1985. doi: 10.1021/nl800949k. [DOI] [PubMed] [Google Scholar]
- Wang H, Dunning JE, Huang AP, Nyamwanda JA, Branton D. DNA heterogeneity and phosphorylation unveiled by single-molecule electrophoresis. Proc Natl Acad Sci USA. 2004;101:13472–13477. doi: 10.1073/pnas.0405568101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M, Chakrabarti B, Mathe J, Nelson DR, Meller A. Orientation-dependent interactions of DNA with an alpha-hemolysin channel. Phys Rev E. 2008;77:031904. doi: 10.1103/PhysRevE.77.031904. [DOI] [PubMed] [Google Scholar]
- Wanunu M, Soni GV, Meller A. Single-molecule studies on nucleic acid interactions using nanopores. In: Hinterdorfer P, Van Oijen AM, editors. Handbook of single-molecule biophysics. New York: Springer; 2010. pp. 265–291. [Google Scholar]
- Wharton JE, Jin P, Sexton LT, Horne LP, Sherrill SA, Mino WK, Martin CR. A method for reproducibly preparing synthetic nanopores for resistive-pulse biosensors. Small. 2007;3:1424–1430. doi: 10.1002/smll.200700106. [DOI] [PubMed] [Google Scholar]
- White HS, Bund A. Ion current rectification at nanopores in glass membranes. Langmuir. 2008;24:2212–2218. doi: 10.1021/la702955k. [DOI] [PubMed] [Google Scholar]
- Wiggin M, Tropini C, Tabard-Cossa V, Jetha NN, Marziali A. Nonexponential kinetics of DNA escape from alpha-hemolysin nanopores. Biophys J. 2008;95:5317–5323. doi: 10.1529/biophysj.108.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Reber A (2002) Aufbau eines Rasterionenleitwertmikroskops. Stromfluktuationen in Nanoporen. PhD dissertation, ISBN 3-89825-490-9