Abstract
Objective
The microenvironment wherein hematopoietic stem cells (HSC) reside orchestrates HSC self-renewal vs. differentiation decisions. Stromal cells derived from ontogenically divergent hematopoietic microenvironments can support HSC in vitro and have been used to decipher factors that influence HSC fate decisions. Employing stromal cell lines derived from the AGM and embryonic liver, we aim to identify secreted factors that maintain/expand HSC in vitro.
Materials and Methods
We cultured murine lineage antigen negative (Lin−) bone marrow (BM) cells in transwells above the UG26-1B6, urogenitial ridge-, and EL08-1D2, embryonic liver-, derived cell lines. We, also, performed RT-qPCR analysis to identify differentially expressed genes from the Wnt family of proteins in ontogenically different stromal cell lines.
Results
Lin− murine BM cells maintained for 3 weeks in transwells above UG26-1B6 but not EL08-1D2 cells contained competitive repopulating HSC. Addition of as few as 25% UG26-1B6 cells to EL08-1D2 feeders led to maintenance of HSC in non-contact cultures, validating soluble factors are secreted by the UG26-1B6 cells. As we found that Wnt5a was significantly higher expressed in UG26-1B6 than EL08-1D2 cells, we added Wnt5a to EL08-1D2 transwell cultures or an antibody against Wnt5a to UG26-1B6 transwell cultures. Addition of Wnt5a to EL08-1D2 transwell cultures restored maintenance of HSC, whereas addition of an anti-Wnt5a antibody to UG26-1B6 transwell cultures inhibited maintenance of CR-HSC.
Conclusions
We demonstrate that stromal cell lines generated from embryonic microenvironments provide a tool to identify secreted proteins that play a role in the maintenance of HSC, and that at least one of the factors produced by UG26-1B6 cells, responsible for preserving HSC, is Wnt5a.
Introduction
HSC are responsible for maintaining hematopoiesis throughout adult life. HSC fate decisions are regulated by both extrinsic and intrinsic signals. In vivo, extrinsic signals are emanated by the microenvironment where the cell resides. These signals are direct cell-cell interaction based, wherein receptors on HSC bind to ligands on osteoblasts, endothelial cells, and/or marrow stroma cells within their microenvironment, cell-extracellular matrix based (fibronectin, proteoglycans, among others), as well as, due to soluble factors secreted by cells residing in the microenvironment or produced at a more distant location [1–5]. The microenvironment wherein HSC reside differs throughout ontogeny, perhaps because distinct signals are needed for the proper development, expansion and lineage commitment of HSC [6–11].
The nature of signals that govern HSC self-renewal and differentiation is still largely unknown. Although a large number of cytokines and growth factors have been cloned that affect HSC, no combination of such factors applied to HSC ex vivo has lead to significant HSC expansion [12–15]. During ontogeny, hematopoiesis occurs in different locations, including the yolk sac and aorta-gonad-mesonephros region (AGM), where definitive HSC are specified and undergo initial expansion [6, 16–19], the fetal liver, where further HSC expansion as well as differentiation to mature myeloid and lymphoid lineages occurs [10, 20, 21], and the bone marrow (BM), where hematopoiesis is maintained throughout adult life [10]. All studies evaluating stem cell niches, from C.elegans to man, have demonstrated that direct cell-cell interactions are important, as well as secreted factors [1, 4, 5, 22, 23]. Noteworthy, however, is that the soluble signals in these niches are in general not common cytokines or growth factors, but morphogens, including members of the TGF-β, Hedgehog and Wnt family [2, 22, 24–27]. These are known to direct specification and differentiation of stem cells during embryogenesis, but are clearly also still active in regulating stem cells during postnatal life.
To identify signals that are of importance for preventing HSC differentiation while promoting HSC proliferation [28–30], stromal cell lines have been generated from these ontogenically different environments. Here we used cell lines generated from E10.5 urogenital ridge (UG26-1B6) and embryonic liver (EL08-1D2), cells that support mouse and human primitive hematopoietic progenitor and stem cells [28, 31–35]. To elucidate whether some of the factors capable of supporting HSC ex vivo are secreted by feeders, we evaluated the maintenance of competitive repopulating (CR)-HSC from adult mice cultured in transwells above UG26-1B6 and EL08-1D2 cells. We found that the UG26-1B6 but not the EL08-1D2 cells line possesses the ability to maintain CR-HSC for 3 weeks in transwells above the feeder without addition of exogenous cytokines. These studies suggest thus that UG26-1B6 cells may secrete one or more factors that can support murine HSC in vitro. One family of soluble factors that has previously been identified to influence hematopoiesis, is the Wnt family, we therefore performed RT-qPCR to evaluate the transcript levels of all Wnt proteins[2, 36–40]. We identified Wnt5a as one of the cytokines produced by UG26-1B6 cells responsible for HSC maintenance.
Materials and Methods
Mice
8 to 10 week female mice were used as recipient and donor mice. C57BL/6J (CD45.2) and B6.SJL-PTPRCA (CD45.1) mice were purchased from Jackson Laboratories (Bar Harbor, ME). Mice were maintained at the University of Minnesota Research Animal Resources in specific pathogen free conditions or Katholieke Universiteit Leuven. All studies were done with approval of the ethical committees at the University of Minnesota or Katholieke Universiteit Leuven.
Isolation of Lin− BM
Femurs and tibias from mice were removed, BM flushed, and depleted of red blood cells by ammonium chloride (Stem Cell Technologies, Vancouver, BC, Canada). Lin− cells were obtained using the Stem Cell Technologies Lineage Negative Selection Kit (Stem Cell Technologies) per manufacturer’s protocol. Purity was usually between 85-95% Lin− BM.
KLS/Fzd4+ and KLS/Fzd4− cell isolation
Lin− BM cells were stained with 1µg/ml mouse anti-Frizzled 4 (R&D Systems, Minneapolis, MN) or 1µg/ml IgG control antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in 2% FCS (Stem Cell Technologies, Vancouver, BC, Canada) followed by staining with FITC conjugated anti-goat IgG (Santa Cruz Biotechnology). Afterwards cells were stained with the following antibodies: anti-ckit (2B8) allophycocyanin (APC), anti-Sca-1 (E13-161.7) phycoerythrin (PE) and biotinylated antibodies against the lineage markers (Gr-1 (RB6-8C5), Mac-1 (M1/70), B220 (RA3-6B2), CD4 (H129.19), CD8 (53.6.7), Terr-119 (Ly-76)) followed by streptavidin peridinin-chlorophyll-protein complex (PerCp) (BD Pharmingen, San Jose, CA). KLS cells were then sorted on a FacsVantage with Diva update (BD Biosciences, San Jose, CA).
Contact and non-contact cultures
UG26-1B6, and EL08-1D2 cells (a kind gift from Dr. Dzierzak, Erasmus MC) were aintained as previously described [28, 32] For hematopoietic cell cocultures, stromal cells were grown on 0.1% gelatin (Sigma, St. Louis, MO) coated 6-well plates (Corning, Lowell, MA). Once cells reached confluence, they were irradiated at 20–25 Gy. For contact cultures, 1×104 Lin− BM cells were seeded in 3ml of LTC-medium, which contains horse and fetal bovine serum (Stem Cell Technologies) with hydrocortisone (10−6 M, Stem Cell Technologies), and cultured for 3 weeks. Each week 50% of the medium was replaced with fresh medium. After 3 weeks, both adherent and non-adherent cells were collected and assayed for presence of CFC and HSC. For non-contact cultures, 1×104 Lin− BM cells were plated in a 0.4µm collagen coated transwell (Corning) placed above the feeders. Each week, 50% of the medium from beneath the transwell was replaced with fresh medium added to the transwell insert. After 3 weeks, the adherent feeder layer below the transwell and cells in the transwell were collected and assayed for CFC and repopulating HSC. Some cultures were supplemented with 10–100ng/ml recombinant mWnt5a, 1µg/ml anti-Wnt5a, or 1µg/ml anti IgG (R&D Systems, Minneapolis, MN) added once per week.
In some studies eGFP+ UG26-1B6 and EL08-1D2 cells were mixed at different ratios. To generate eGFP+ UG26-1B6 cells, cells were transduced with the MSCV-eGFP vector [41]. eGFP+ UG26-1B6 cells were sorted using the FacsAria (BD Biosciences) to a purity of 98±1%. To confirm the relative ratios of cells present in mixed feeder, the percent eGFP+ cells was determined by both fluorescent microscopy and flowcytometry (Data Not Shown).
Colony-Forming Cells (CFC) Assay
Fresh or culture progeny were plated in methylcellulose medium (M3234, Stem Cell Technologies) supplemented with 20ng/ml mSCF, 10ng/ml mIL-3, 10ng/ml mIL-6 (all from R&D Systems), and 3U/ml hEpo (Amgen Inc., Thousand Oaks, CA). All cultures were incubated at 37°C and 5%CO2. Colonies were counted between day 10 and 12.
Competitive repopulation
CD45.2 recipient mice were irradiated with 950-1,100 cGy 3 to 12 hours prior to transplantation. 2×105 BM cells from CD45.2 mice were mixed with fresh or 3-week culture progeny of 104 Lin− CD45.1 cells. After 1 to 4 months, PB and/or BM was obtained and stained with FITC- conjugated anti-CD45.1 (A20) and PerCp-conjugated anti-CD45.2 (104). The cells were simultaneously stained with APC-conjugated anti-B220 antibody together with a mixture of PE-conjugated anti-Mac-1 (M1/70) and –Gr-1(RB6-8C5) or anti-CD4 (GK1.5) and –CD8 (53-6.7) antibodies (BD Pharmingen). Fourcolor analysis was performed on a FACSCalibur or FACSCanto (BD Biosciences, San Jose, CA). A recipient mouse was considered multi-lineage repopulated if the percentage of donor cell-derived cells was >1% and donor cells contributed to all three hematopoietic lineages (myeloid, T lymphoid and B lymphoid cells) in PB and/or BM. Competitive repopulation assays were repeated 2 to 4 times with separate isolations of Lin− BM cells.
Animals with >1% donor cells in the BM were used as donors for secondary transplantation. 1×106 total BM cells obtained from primary recipients were injected into 3–5 lethally irradiated CD45.2 secondary recipients, and BM from these animals was analyzed 3–4 months later for CD45.1 derived hematopoietic cells.
Western Blot
Cell pellets from stromal cells were resuspended in ice-cold RIPA buffer (Sigma) with protease inhibitor tablets (Roche, Nutley, NJ). Samples were then centrifuged at 14,000 rpm at 4°C for 10 minutes, and supernatant recovered. An equal volume of 2× sample buffer (Invitrogen, Carlsbad, CA) was added and samples were placed at 90°C for 10 minutes. Samples were separated on 4–12% (SDS-PAGE) gels (Invitrogen). Gels were transferred to Immuno-Blot PVDF membrane (BioRad, Hercules, CA) for 2 hours. Membranes were blocked using 5% nonfat dry milk in TBS-T pH 7.6, 0.1% Tween-20 (Sigma) in TBS (Amresco, Solon, OH) for 1 hour at room temperature and then incubated overnight with specific primary antibodies against Wnt5a (R&D Systems) at 4°C. Blots were then washed 3X for 5 minutes with TBS-T, and incubated with secondary horseradish peroxidase (HRP) conjugated anti-rabbit IgG antibodies (Amersham, GE Healthcare Biosciences Piscataway, NJ) in TBS-T according to the manufacturer's protocol. Bands were visualized using chemiluminescence (Amersham).
Quantitative RT-PCR
Total RNA was harvested from stromal cell lines using the Qiagen RNeasy Kit (Qiagen, Hilden, Germany). Total RNA was Dnase treated using Turbo DNase kit (Ambion, Austin, TX). 2μg of total RNA used for cDNA synthesis using Taqman reverse transcription reagents (Perkin Elmer Applied Biosystems, Boston, MA). Q-RT-PCR was carried out using Taqman SYBR green universal mix PCR reaction buffer (Perkin Elmer Applied Biosystems) using an ABI PRISM 7700 (Applied Biosciences, Norwalk, CT).
Single Cell Immunofluorescent Staining
KLS/CD34− cells were sorted into 30µl Stemspan (Stem Cell Technologies) on Teflon printed 10 well glass slides (Matsunmai Glass Industry, Osaka, Japan). After sorting, slides were placed on ice for 1 hour. Cells were then fixed with 30µl of 10% NBF (Sigma) for 10 minutes, and permeabilized for 10 minutes using 0.02% Triton-X100 (Sigma). Slides were blocked with 10% donkey serum (Abcam, Cambridge, UK), and stained with primary antibodies against Fzd4 overnight. The cells were washed with PBS, and stained with secondary donkey anti-goat Alexa488 conjugated antibodies (Invitrogen) and Hoechst (Sigma) for 30 minutes. The protocol was adapted from Ema, et al.[42].
Statistical analysis
Data for studies determining cell number, cell phenotype, CFC number and transcript levels are shown as the mean of a minimum of three experiments±standard deviation. Statistical significance was determined by student’s two-tail t-test in all experiments. P-value of <0.05 was considered statistically significant.
Results
Long-term repopulating HSC are maintained when cultured in transwells above UG26-1B6 but not EL08-1D2 cells
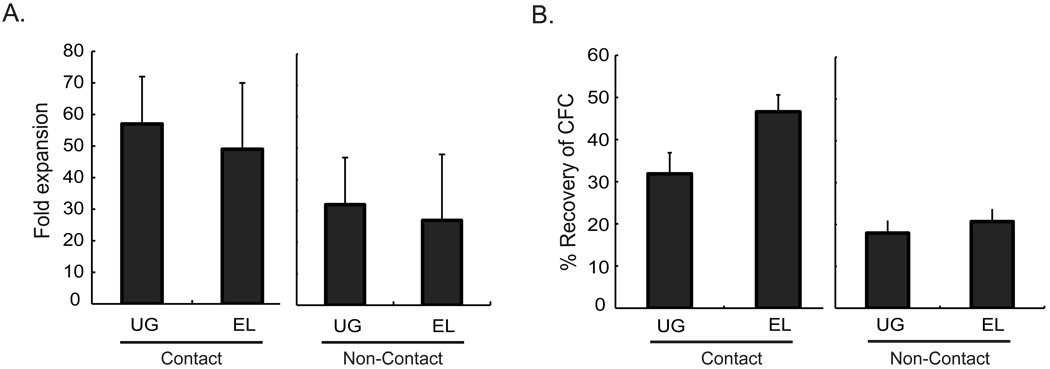
To determine whether contact with hematopoietic supportive embryonic derived stromal cells is required for maintenance of HSC, we plated murine Lin− BM cells in transwells above or in direct contact with the UG26-1B6 and EL08-1D2 feeders in horse and fetal calf serum-containing long-term culture media. Three weeks later, progeny were evaluated for total cell expansion, presence of CFC, and presence of repopulating stem cells. Cell expansion was similar when cells were cultured in contact with the feeders, or in transwells above the feeders (p>0.05) (Fig.1A). FACS analysis of 3 week progeny demonstrated that the majority of cells were Gr-1 and Mac-1 double positive, irrespective of the culture condition (Data not shown). The number of CFC was significantly lower in non-contact cultures compared to contact cultures (UG26-1B6 p=0.002 and EL08-1D2 p<0.001), consistent with what we demonstrated previously for human CFC [43]. When we compared recovery of CFC between cultures supported by UG26-1B6 and EL08-1D2, we found no significant differences between cells cultured in transwells above the feeders (Non-contact cultures: n=3, p=0.59) (Fig. 1B) suggesting that soluble factors from both feeders support committed progenitors.
Figure 1. Total cell and CFC expansion of Lin- BM cells in contact and non-contact stromal cultures.
104 Lin- BM cells were cultured either in direct contact with or in transwells above UG26-1B6 or EL08-1D2 cells that had been irradiated at 25Gy. After 3 weeks of culture, A) total cell number was enumerated to determine cell expansion. Trypan blue was used to distinguish the live cells from the irradiated stromal feeder and B) progeny were plated in CFC assay. CFC were enumerated at day 12. The data represents 4 individual experiments ± standard deviation. % CFC recovery = (#CFC generated by the progeny of 104 Lin- BM cells recovered after 3 weeks of culture / #CFC / 104 fresh Lin- BM cells) × 100. Statistical significance for all experiments was determined by two-tail t-test.
We next performed competitive repopulation assays to assess maintenance and/or expansion of HSC cultured in transwells above the two feeders [44–46]. We competed 104 Lin− BM cells from CD45.1+ mice cultured for 3 weeks in contact with or in transwells above UG26-1B6 and EL08-1D2 cells, with 2×105 total BM cells from CD45.2+ mice, in lethally irradiated CD45.2+ mice. Repopulation from CD45.1+ donor cells was defined as presence of >1% CD45.1+ cells in peripheral blood and / or BM, 3–4 months post transplantation, with contribution to the myeloid (Gr-1/Mac-1+), B- (B220+) and T-lymphoid (CD4/CD8+) lineages.
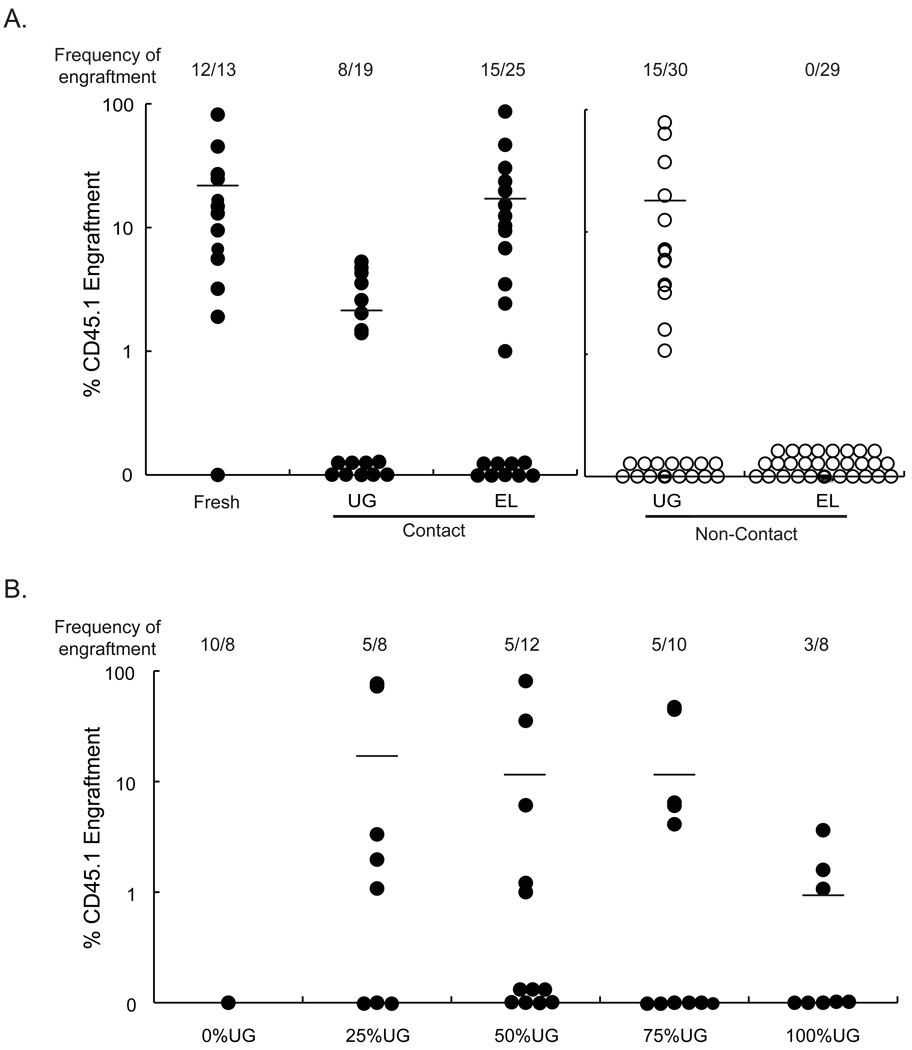
CD45.1+ hematopoiesis was seen in 12/13 mice transplanted with fresh CD45.1+ Lin− cells, (19±22% CD45.1+ chimerism) (Fig.2A). 15/25 (15±21% CD45.1+ chimerism) mice were engrafted with progeny of CD45.1+ Lin− cells harvested from EL08-1D2-contact cultures, whereas 8/19 (2±1% CD45.1+ chimerism) mice were engrafted with progeny of CD45.1+ Lin− cells cultured in contact with UG26-1B6 cells. Although this suggests that chimerism from cells cultured in contact with EL08-1D2 cells was higher compared with UG26-1B6 feeders, multilineage engraftment in animals that received progeny of EL08-1D2-contact cultures was skewed towards T lymphocytes compared to fresh uncultured BM cells and cells cultured in contact with UG26-1B6 cells (p-value=0.05) (Fig.S1). No mice were engrafted with progeny from Lin− cells maintained for 3 weeks in transwells above EL08-1D2 feeders (0/29 mice). In contrast, 15/30 mice transplanted with culture progeny of UG26-1B6 non-contact cultures showed multilineage repopulation with CD45.1+ cells (10±20% CD45.1+ cells). By contrast, engraftment was not statistically different between mice grafted with CD45.1+ Lin− cell progeny cultured in contact or in transwells above with UG26-1B6 cells (p>0.05) (Fig.2A) Oostendorp, et al previously showed that BM derived CD31+/K6 side population cells could also be maintained in transwells above EL08-1D2 cells [47]. The differences between these published studies and the current results may be related to differences in mouse strain used, or HSC enrichment strategy.
Figure 2. UG26-1B6 cells but not EL08-1D2 cells maintain HSC in non-contact cultures.
A) A combination of 2×105 CD45.2+ competitor BM and 104 fresh CD45.1+ Lin- BM cells, or progeny from 104 CD45.1+ Lin− BM cells cultured for 3 weeks in contact with or in transwells above UG26-1B6 or EL08-1D2 cells were transplanted in CD45.2+ recipients. (UG26-1B6 non-contact vs. EL08-1D2 non-contact, p=0.02) B) 2×105 CD45.2+ total BM cells were competed against 104 CD45.1+ Lin− BM cells cultured for 3 weeks in transwells above different mixtures of UG26-1B6 and EL08-1D2 cells in CD45.2+ recipients. Twelve to sixteen weeks after transplantation, PB was collected and analyzed by FACS for presence of CD45.2+ and CD45.1+ cells. Data points represent total CD45.1+ cells in the CD45.2+ recipient animals. The frequency of engraftment represents the number of mice that showed both >1% overall engraftment and evidence of multi-lineage engraftment versus the total number of mice transplanted. (0% UG26-1B6 vs. 25% UG26-1B6, p=0.13)
We performed secondary transplants to ascertain that long-term repopulating (LTR)-HSC had persisted. Supplementary Table 1 summarizes the chimerism of the 4 primary recipients from which the BM was used for secondary transplants. Three and four of four of the secondary recipients of BM cells from animals grafted with EL08-1D2-contact #1 or #2 progeny, respectively, had multilineage CD45.1+ hematopoiesis, but engraftment was skewed towards the B- and T-lymphoid lineage, respectively. Both groups of mice that received cells from primary recipients repopulated with UG26-1B6 non-contact cultured cells repopulated with CD45.1+ cells, which was multilineage in 6/8 secondary recipients.
UG26-1B6 secretes a factor or factors capable of supporting HSC in culture
To determine whether lack of HSC maintenance in EL08-1D2 non-contact cultures is due to secretion of an inhibitor or because EL08-1D2 cells fail to secrete one or more soluble factors capable of maintaining HSC, we mixed UG26-1B6 and EL08-1D2 cells at different ratios. If adding 25% UG26-1B6 cells to 75% EL08-1D2 cells restores HSC maintenance, then UG26-1B6 cells likely secrete a factor that supports HSC. If, by contrast, combining 75% UG26-1B6 cells with 25% EL08-1D2 cells does not lead to HSC maintenance in non-contact culture, a factor produced by EL08-1D2 may inhibit HSC maintenance. To distinguish between the two cell lines, UG26-1B6 cells were transduced with a retroviral vector encoding the eGFP protein, and were then sorted for GFP expression by FACS. The proportion of UG26-1B6 and EL08-1D2 cells present in culture after plating at different ratios was verified by FACS and fluorescence microscopy.
Total cell expansion and recovery of CFC at 3 weeks was similar between cells maintained in transwells above any combination of stromal cell lines (Fig.S2). We transplanted Lin− CD45.1+ BM cell progeny from the different cultures in a competitive repopulation assays with CD45.2+ cells. Mixing as little as 25% UG26-1B6 cells with the EL08-1D2 feeder resulted in the maintenance of HSC in non-contact culture. Fifty percent of transplanted mice demonstrated multi-lineage engraftment with >1% donor derived cells. Similar multilineage engraftment was seen in mice receiving progeny of cultures containing 100%, 75%, 50%, or 25% UG26-1B6 cells (Figure 2B). Three of eight mice transplanted with cells cultured in transwells above 100% UG26-1B6 cells engrafted, whereas 5/10, 5/12, 5/8, and 0/8 mice, respectively, transplanted with cells cultured in transwells above feeders containing 75%, 50%, 0% UG cells, respectively engrafted. Lymphoid skewing was not seen in recipients of cells cultured with mixed stromal feeders (Fig.S3). These studies thus strongly suggest that UG26-1B6 cells secrete a factor or multiple factors required for HSC maintenance in non-contact culture, not produced by EL08-1D2 cells.
Wnt5a is more highly expressed in UG26-1B6 than EL08-1D2 stromal cells
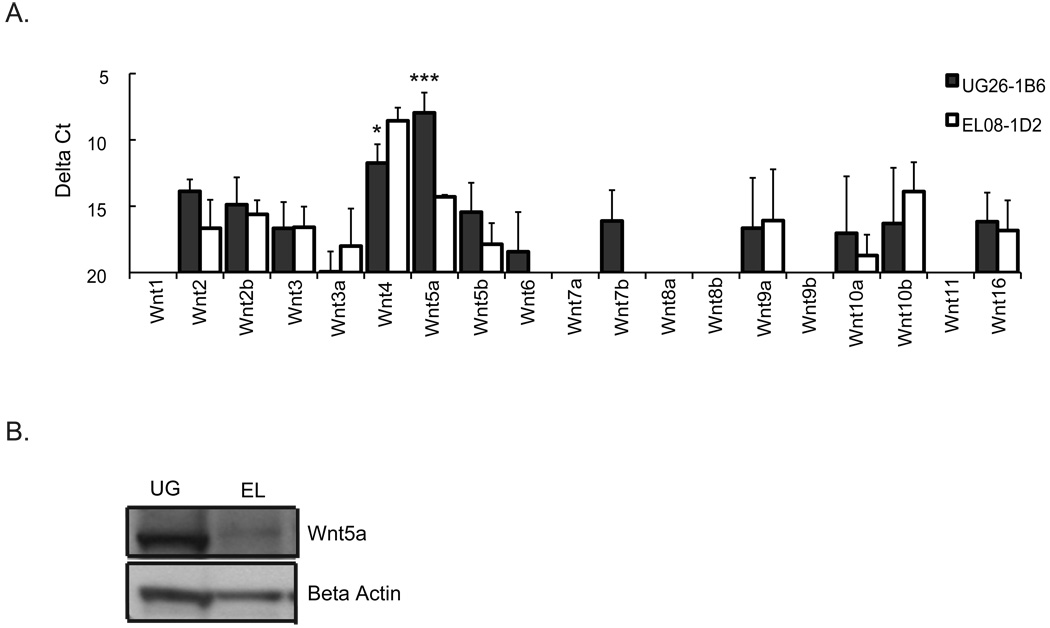
As the mixing studies demonstrated that one or more factors secreted by UG26-1B6 cells maybe responsible for HSC maintenance in non-contact cultures, we evaluated the expression of the Wnt family of proteins in UG26-1B6 and EL08-1D2 cells by RT-qPCR. Wnt proteins are expressed in the developing embryo in sites of hematopoiesis like the fetal liver [36] as well as the in the HSC niche in postnatal BM [2]. There is mounting evidence that Wnts affect the self-renewal of HSC in postnatal life, which led us to investigate the possible role of Wnts in UG26-1B6 cultures. Although many Wnt genes were expressed, the level of expression in general was low, except for Wnt5a that was significantly higher expressed in UG26-1B6 than EL08-1D2 cells (Fig.3A). To confirm the differential expression further, western blot analysis was preformed. The sensitivity of the assay was not sufficient to determine the level of Wnt5a in the medium; however, significantly higher levels of Wnt5a protein were detected in UG26-1B6 than EL08-1D2 total cell lysates (Fig.3B).
Figure 3. Wnt5a is highly expressed in UG26-1B6 compared to EL08-1D2.
UG26-1B6 and EL08-1D2 cells were grown to confluence and irradiated at 25Gy 24 hours prior to RNA or protein extraction. A) RT-qPCR was preformed to assess the expression of different Wnt genes. Transcript levels are represented as the average delta Ct (delta Ct= Ct (gene of interest) −Ct (Gapdh)) of three to five independent experiments ± standard deviation. Gapdh values remained were similar between independent experiments. Statistical significance was determined by a two-tail t-test. (*P<0.05,***P<0.001) B) Expression of Wnt5a was evaluated by western blot on lysates of 106 stromal cells using 1µg/ml anti-Wnt5a antibody. To control for loading, blots were stripped and reprobed with antibodies against β-actin. The western blot is a representative example of three independent experiments.
Wnt5a is at least one of the secreted factors responsible for maintenance of HSC in UG26-1B6 cells
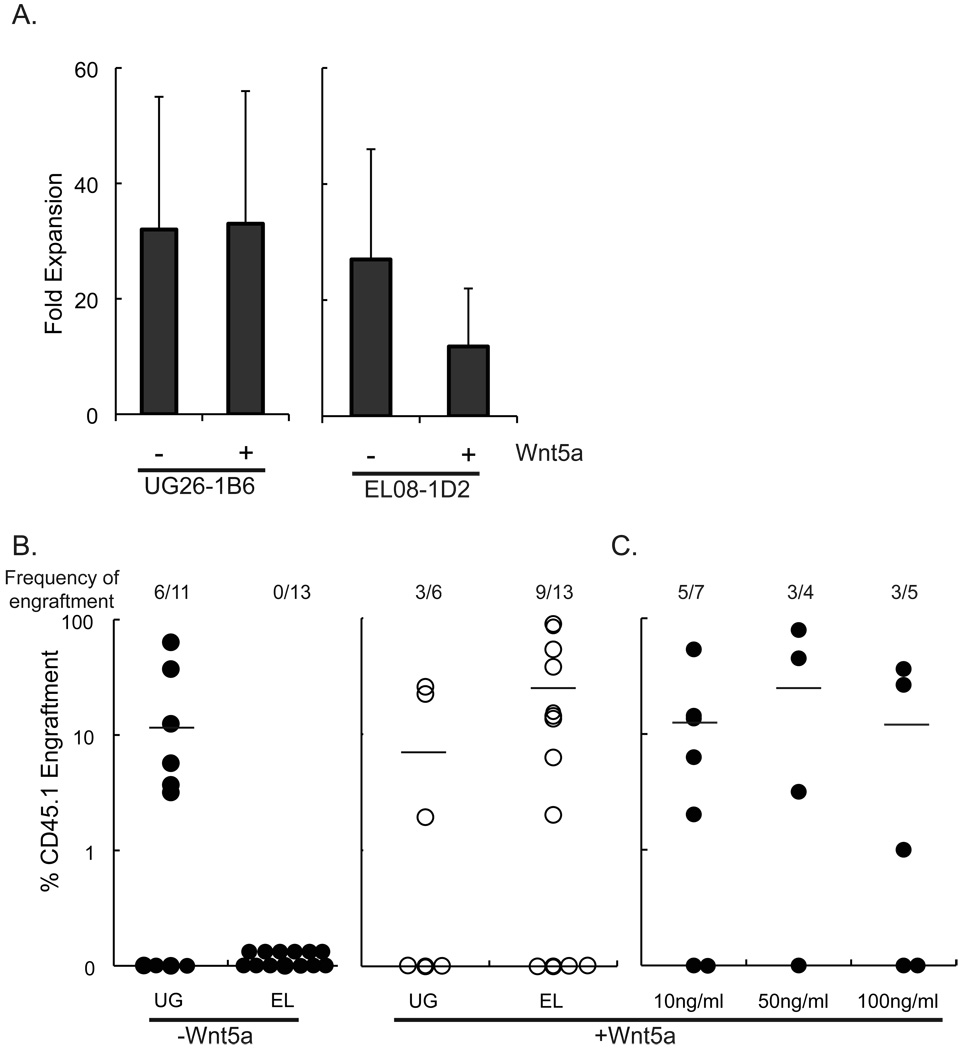
To determine if Wnt5a is responsible for the maintenance of HSC cultured in transwells above UG26-1B6 cells, we cultured CD45.1+ Lin− BM in transwells above EL08-1D2 or UG26-1B6 cells with or without 10ng/ml Wnt5a, added weekly for the 3-week culture period. Addition of Wnt5a to UG26-1B6 non-contact cultures did not affect cell expansion at 3 weeks (Fig.4A). Although not statistically significant cells cultured in EL08-1D2-transwell cultures expanded less in the presence of Wnt5a (12±10 vs. 27±19, p=0.18). Addition of Wnt5a to UG26-1B6-non-contact cultures did not affect the percentage of mice engrafted with progeny of the CD45.1+ cells, nor the levels of engraftment seen from the CD45.1+ donor cells (3/6 mice, 8±12%CD45.1+ cells) (Fig.4B). By contrast, when Lin− CD45.1+ cells were cultured in EL08-1D2-non-contact cultures supplemented with 10ng/ml Wnt5a, 9/13 mice showed multilineage CD45.1+ cell derived engraftment at 4 months, while no CD45.1+ cells were detected in animals grafted with cells cultured in EL08-1D2-non-contact cultures not supplemented with Wnt5a (25±33% vs. 0±0% CD45.1+ cells, p=0.01) (Fig. 4B). Supplementing EL08-1D2 non-contact cultures with 10ng/ml Wnt5a revealed similar levels of chimerism as when mice were grafted with cells maintained in UG26-1B6 non-contact cultures (p=0.2). When higher concentrations of Wnt5a were added (50 and 100 ng/ml Wnt5a weekly for 3 weeks), we observed no further increase in the frequency of mice engrafted with CD45.1+ progeny (5/7, 3/4, and 3/5 of mice engrafted with cells cultured with for 10, 50, and 100ng/ml Wnt5a, respectively), or the relative contribution of CD45.1+ cells to the lymphoid and myeloid lineages (Fig. 4C). As control, Lin− BM cells were also cultured in direct contact with the feeders with or without Wnt5a.
Figure 4. Addition of Wnt5a to UG26-1B6 and EL08-1D2 non-contact cultures does not affect cell expansion, but enables EL08-1D2 cells to support LTR-HSC in transwells above the feeder.
104 CD45.1+ Lin- BM were cultured in transwells above or in contact with irradiated UG26-1B6 and EL08-1D2 cells. Cultures were supplemented with or without 10ng/ml Wnt5a. After 3 weeks, the total cell number was enumerated to determine cell expansion. Trypan blue was used to distinguish the live cells from the irradiated stromal feeder cells. Data shown is mean ± standard deviation of 4 experiments. Statistical significance was determined by a two-tail t-test. B) Lin- BM were cultured in transwells above irradiated UG26-1B6 and EL08-1D2 cells with or without 10ng/ml Wnt5a. After 3 weeks, progeny of 104 CD45.1+ Lin- BM cells were transplanted together with 105 CD45.2+ fresh BM in lethally irradiated CD45.2+ recipients. After 4 months PB of recipient animals was analyzed for CD45.1+ cells. Data points represent total CD45.1+ cells in the CD45.2+ recipient animals. The frequency of engraftment represents the number of mice that showed both >1% overall engraftment and evidence of multi-lineage engraftment versus the total number of mice transplanted. C) Lin- BM were cultured in transwells above irradiated UG26-1B6 and EL08-1D2 cells with 10ng/ml, 50ng/ml, or 100ng/ml Wnt5a. After 3 weeks, progeny of 104 CD45.1+ Lin- BM cells were transplanted together with 105 CD45.2+ fresh BM in lethally irradiated CD45.2+ recipients. After 4 months PB of recipient animals was analyzed for CD45.1+ cells. Data points represent total CD45.1+ cells in the CD45.2+ recipient animals. The frequency of engraftment represents the number of mice that showed both >1% overall engraftment and evidence of multi-lineage engraftment versus the total number of mice transplanted. (UG26-1B6 vs. EL08-1D2 +Wnt5a, p=0.21)
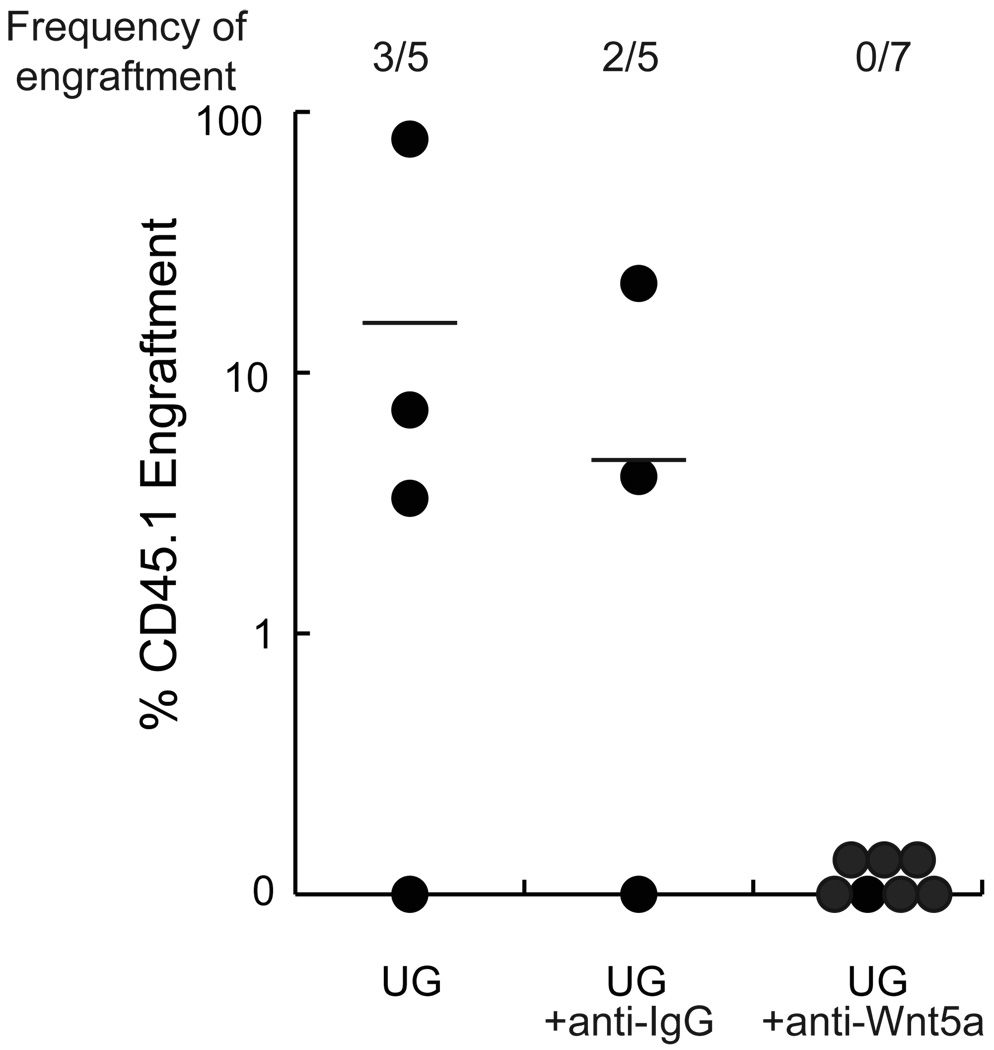
To further substantiate a role for Wnt5a in maintenance of HSC in UG26-1B6 non-contact cultures, we added 1µg/ml-neutralizing antibody against Wnt5a to UG26-1B6- non-contact cultures, weekly for 3 weeks. Anti-Wnt5a inhibited the ability of UG26-1B6 feeders to maintain HSC, as 0 out 7 mice were engrafted with CD45.1+ cells at 4 months (Fig.5), whereas 2/5 animals grafted with progeny of UG26-1B6-noncontact cultures treated with 1µg/ml anti-IgG control antibodies and 3/5 animals grafted with cells maintained in UG26-1B6-non-contact cultures without antibody addition were engrafted with CD45.1+ progeny (5±10% vs. 0±0% CD45.1+ cells, p=0.17). Goat IgG antibody, used to control for non-specific effects on cell engraftment, did not affect engraftment. These studies suggest that Wnt5a may be one of the secreted factors in UG26-1B6 cultures responsible for maintenance of HSC.
Figure 5. Addition of anti-Wnt5a antibodies to UG26-1B6 non-contact cultures inhibits maintenance of LTR-HSC.
Lin− CD45.1+ BM cells were cultured in transwells above irradiated UG26-1B6 cells without addition of antibodies, with addition 1µg/ml anti-Wnt5a or with 1µg/ml control IgG added weekly. After 3 weeks, progeny of 104 CD45.1+ Lin− BM cells were transplanted together with 105 CD45.2+ fresh BM in lethally irradiated CD45.2+ recipients. After 4 months PB of recipient animals was analyzed for CD45.1+ cells. Data points represent total CD45.1+ cells in the CD45.2+ recipient animals. The frequency of engraftment represents the number of mice that showed both >1% overall engraftment and evidence of multi-lineage engraftment versus the total number of mice transplanted. (UG26-1B6 vs. UG26-1B6 + anti-Wnt5a, p=0.18)
LTR-HSC express the putative Wnt5a Fzd4-receptor
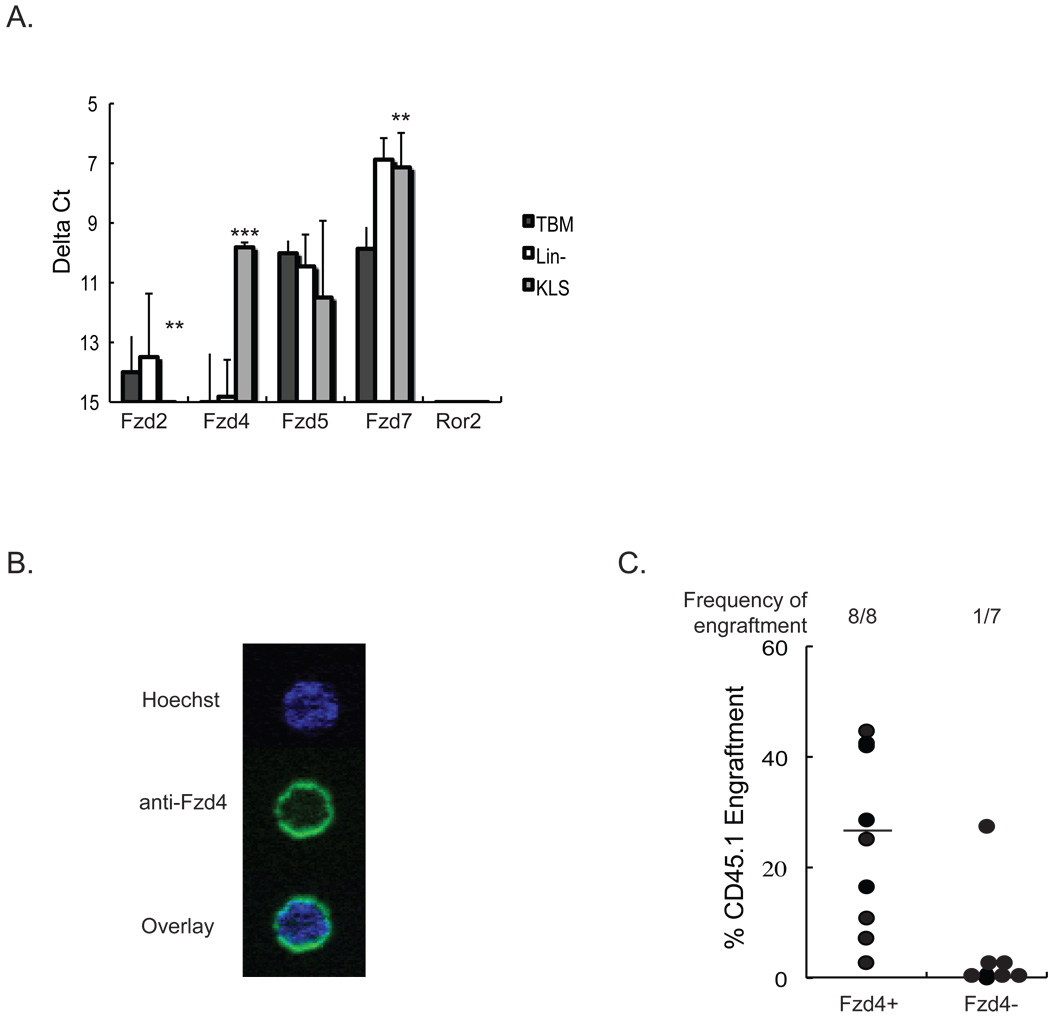
Wnt5a signal via the orphan receptor, Ror2 [48], as well as, Fzd4 and its co-receptor Lrp5[49]. Microarray studies by Zhong, et. al. showed that Fzd4 distinguishes LTR-HSC from hematopoietic progenitor cells (HPC) and STR-HSC[50]. We used RT-qPCR to assess the level of expression of Fzd2, Fzd4, Fzd5, Fzd7, and Ror2 in total BM cells, Lin− BM cells, and HSC-enriched BM-derived KLS cells. Fzd2 was not expressed in either BM, Lin− BM, or KLS cells. By contrast, Fzd5 and Fzd7 were expressed in all three cell populations with higher expression of Fzd7 in Lin− BM cells and KLS cells than total BM cells. Ror2 could not be detected in any of the three cell populations. Fzd4 was solely expressed in the KLS population (Figure 6A).
Figure 6. Frizzled 4 is expressed on nearly all LTR-HSC.
A) RNA was extracted from total BM, Lin− and KLS cells. Levels of Fzd2, Fzd4, Fzd5, Fzd7 and Ror2 mRNA were evaluated by RT-qPCR. Transcript levels are represented as the average delta Ct (delta Ct= Ct (gene of interest) − Ct (Gapdh)) of three independent experiments ± standard deviation. Gapdh values remained were similar between independent experiments. Statistical significance was determined by a two-tail t-test. (*P<0.05, **P<0.01, ***P<0.001). B) Lin− cells were stained with anti- Sca-1-PE, anti-c-kit-APC, and anti-Lineage markers-PerCp, and anti-CD34-FITC antibodies. 50 KLS/CD34− were sorted into a drop of serum-free media and stained with anti-Fzd4 and Hoechst. A representative example of a KLS/CD34− cells stained with anti-Fzd4 antibodies is shown. The percentage of KLS/CD34− cells that was Fzd4 positive was 90±5%. Figure is representative example of two independent experiments. C) 100 Fzd4− or Fzd4+ KLS from CD45.1+ mice were transplanted with 105 CD45.2+ BM cells into lethally irradiated CD45.2+ mice. Four months after transplantation, PB was collected and analyzed by FACS for presence of CD45.1+ cells. Data points represent total CD45.1+ cells in the CD45.2+ recipient animals. Mice that showed both >1% overall engraftment and evidence of multi-lineage CD45.1+ cells were considered engrafted.
We stained KLS cells with an anti-Fzd4 antibody and found that Fzd4 was expressed by a small subpopulation of KLS cells (12.9±12.0%) (data not shown). Although the KLS population is enriched for HSC it still contains a mixture of HPC, STR-HSC and LTR-HSC [51]. To determine if Fzd4 is expressed on the LTR-HSC subpopulation of KLS cells we sorted CD34−KLS cells, which are more highly enriched in LTR-HSC [51] into a drop of serum-free media and performed single-cell immunostaining for Fzd4. 90±5% of CD34−KLS cells stained positive for Fzd4 (Figure 6.B). We also subfractionated KLS cells using the anti-Fzd4 antibody and preformed competitive repopulation assays. 100 Fzd4+ KLS or 100 Fzd4− KLS cells from CD45.1+ mice were co-transplanted with 105 BM cells from CD45.2+ mice in lethally irradiated CD45.2+ mice. Only 1/7 mice transplanted with Fzd4− KLS showed CD45.1+ cell engraftment whereas 8/8 mice transplanted with Fzd4+ KLS cells were repopulated with CD45.1+ cells (35±12% vs. 0.6±0.7% CD45.1+ cells, p=0.05) (Figure 6.C). This data demonstrates that CR-LTRHSC express Fzd4.
Discussion
Throughout development, HSC reside in multiple different sites [10, 52]. It is therefore thought that characterization of the microenvironment of these developmentally diverse sites may yield important insights in factors that regulate HSC behavior. Stromal cell lines have been generated from the different hematopoietic supportive organs, including yolk sac, AGM, embryonic liver, fetal liver, fetal BM and adult BM with differing abilities to support murine or human HSC [28, 30, 32, 53].
Here we demonstrate that UG26-1B6 cells generated from E10.5-AGM but not EL08-1D2 cells generated from E10.5 embryonic liver, support LTR-HSC in the absence of contact between the feeder and HSC. As mixed feeders comprising as little as 25% UG26-1B6 cells also supported HSC in non-contact cultures, we concluded that UG26-1B6 must secrete one or more soluble factors can support HSC in vitro not produced by EL08-1D2 cells.
Because HSC are influenced by Wnts, shown in numerous publications [36, 38, 40, 54–56], we assessed the expression of Wnts in UG26-1B6 and EL08-1D2 cells, and found that Wnt5a is significantly higher expressed in UG26-1B6 than EL08-1D2 cells. We further show that addition of Wnt5a restores the ability EL08-1D2 cells to support LTRHSC in non-contact cultures. We further show that although KLS-CD34- cells do not express Ror2, one of the receptors known to be responsible for signaling from Wnt5a, all LTR-HSC express Fzd4, recently shown to be involved in signaling via Wnt5a.
The role of Wnt5a in hematopoiesis is unclear. Wnt5a−/− mice are embryonic lethal, precluding evaluating the effect of Wnt5a loss on adult hematopoiesis [57]. Wnt5a is expressed at the time of hematopoietic emergence in vivo, and may mediate hematopoietic development from ESC in vitro [58–60]. Two groups [36, 37] demonstrated that Wnt5a increases proliferation in vitro of primitive murine and human hematopoietic progenitors. Murdoch, et al demonstrated that Wnt5a added during in vitro culture does not significantly affect the proliferative and differentiation capacity of human primitive hematopoietic progenitors, but that administration of Wnt5a containing conditioned medium to mice transplanted with human umbilical cord blood CD34+CD38− Lin− cells significantly increased the repopulation ability of these cells [56]. Although this suggests that Wnt5a may affect the most primitive compartment of human hematopoietic cells, the study did not address whether this effect is by direct interaction of Wnt5a and HSC or indirect, via for instance Wnt5a-mediated stimulation of non-hematopoietic cells that then release other factors that increase HSC engraftment. Nemeth, et al demonstrated that highly enriched murine HSC cultured with SCF, Flt3L and Wnt5a engraft significantly better at 4 weeks and 4 months than cells cultured with SCF and Flt3L alone or a combination of SCF, Flt3L and Wnt3a. They suggest that this may be due to increasing the maintenance of a quiescent state of the ex vivo cultured HSC, mediated by inhibiting β-catenin activation [61].
In conclusion, we demonstrate here that use of stromal feeders from different origins can aid in identifying soluble growth factors / signals that play a role in maintenance of HSC. The studies were not developed to assess whether Wnt5a expands HSC in vitro. However, the levels of chimerism seen with cells cultured for 3 weeks in either UG26-1B6-non-contact cultures or Wnt5a-supplemented EL08-1D2-non-contact cultures are not higher than what would be expected for freshly isolated Lin- cell competitive repopulation assays. Comparing genes from the Wnt family of an HSC supportive and a non-supportive feeder identified Wnt5a, as a candidate factor responsible for HSC maintenance. We here demonstrate that Wnt5a increases maintenance of true long-term repopulating HSC in vitro. Also, further investigation of proteins encoded by genes differentially expressed in HSC supportive and non-supportive stromal cell lines may yield more gene candidates involved in HSC cell fate decisions including self-renewal, and differentiation.
Supplementary Material
Acknowledgements
This work was supported by PO1-CA-65493-06 (CMV), FWO G.0451.06 (CMV), KUL CoE (CMV), FWO-Odysseus Fund (CMV), FP6-STREP CHRYSTAL (CMV), NIH RO1 DK0504077 (ED), and DFG-008/2 (RO). The authors also wish to thank Vick Van Duppen for help with FACS sorting, Sarah Schouteden for technical assistance, and Dr. Rik Snoeckx for his critical review of all data for accuracy and the manuscript prior to submission.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosure: The authors declare no competing financial interests.
References
- 1.Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 2.Fleming HE, Janzen V, Lo Celso C, et al. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell stem cell. 2008;2:274–283. doi: 10.1016/j.stem.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackney JA, Charbord P, Brunk BP, Stoeckert CJ, Lemischka IR, Moore KA. A molecular profile of a hematopoietic stem cell niche. Proceedings of the National Academy of Sciences of the United States of America. 2009;99:13061–13066. doi: 10.1073/pnas.192124499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nature reviews. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 6.Baron MH. Embryonic origins of mammalian hematopoiesis. Experimental hematology. 2003;31:1160–1169. doi: 10.1016/j.exphem.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Baron MH, Fraser ST. The specification of early hematopoiesis in the mammal. Current opinion in hematology. 2005;12:217–221. doi: 10.1097/01.moh.0000163217.14462.58. [DOI] [PubMed] [Google Scholar]
- 8.Dzierzak E. The emergence of definitive hematopoietic stem cells in the mammal. Current opinion in hematology. 2005;12:197–202. doi: 10.1097/01.moh.0000160736.44726.0e. [DOI] [PubMed] [Google Scholar]
- 9.Dzierzak E. Embryonic beginnings of definitive hematopoietic stem cells. Annals of the New York Academy of Sciences. 1999;872:256–262. doi: 10.1111/j.1749-6632.1999.tb08470.x. discussion 262-254. [DOI] [PubMed] [Google Scholar]
- 10.Dzierzak E, Medvinsky A. Mouse embryonic hematopoiesis. Trends Genet. 1995;11:359–366. doi: 10.1016/s0168-9525(00)89107-6. [DOI] [PubMed] [Google Scholar]
- 11.Mikkola HK, Gekas C, Orkin SH, Dieterlen-Lievre F. Placenta as a site for hematopoietic stem cell development. Experimental hematology. 2005;33:1048–1054. doi: 10.1016/j.exphem.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Miller CL, Eaves CJ. Expansion in vitro of adult murine hematopoietic stem cells with transplantable lympho-myeloid reconstituting ability. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13648–13653. doi: 10.1073/pnas.94.25.13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang CC, Kaba M, Ge G, et al. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nature medicine. 2006;12:240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang CC, Lodish HF. Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells. Blood. 2004;103:2513–2521. doi: 10.1182/blood-2003-08-2955. [DOI] [PubMed] [Google Scholar]
- 15.Ueda T, Tsuji K, Yoshino H, et al. Expansion of human NOD/SCID-repopulating cells by stem cell factor, Flk2/Flt3 ligand, thrombopoietin, IL-6, and soluble IL-6 receptor. The Journal of clinical investigation. 2000;105:1013–1021. doi: 10.1172/JCI8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palis J, Yoder MC. Yolk-sac hematopoiesis: the first blood cells of mouse and man. Experimental hematology. 2001;29:927–936. doi: 10.1016/s0301-472x(01)00669-5. [DOI] [PubMed] [Google Scholar]
- 17.Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 19.Yoder MC, Hiatt K, Mukherjee P. In vivo repopulating hematopoietic stem cells are present in the murine yolk sac at day 9.0 postcoitus. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6776–6780. doi: 10.1073/pnas.94.13.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medvinsky AL, Dzierzak EA. Development of the definitive hematopoietic hierarchy in the mouse. Developmental and comparative immunology. 1998;22:289–301. doi: 10.1016/s0145-305x(98)00007-x. [DOI] [PubMed] [Google Scholar]
- 22.Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annual review of cell and developmental biology. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yen TH, Wright NA. The gastrointestinal tract stem cell niche. Stem cell reviews. 2006;2:203–212. doi: 10.1007/s12015-006-0048-1. [DOI] [PubMed] [Google Scholar]
- 24.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science (New York, NY. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 25.Song X, Call GB, Kirilly D, Xie T. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development (Cambridge, England) 2007;134:1071–1080. doi: 10.1242/dev.003392. [DOI] [PubMed] [Google Scholar]
- 26.Song X, Wong MD, Kawase E, et al. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development (Cambridge, England) 2004;131:1353–1364. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- 27.Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science (New York, NY. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- 28.Oostendorp RA, Harvey KN, Kusadasi N, et al. Stromal cell lines from mouse aorta-gonads-mesonephros subregions are potent supporters of hematopoietic stem cell activity. Blood. 2002;99:1183–1189. doi: 10.1182/blood.v99.4.1183. [DOI] [PubMed] [Google Scholar]
- 29.Dexter TM, Allen TD, Lajtha LG. Conditions controlling the proliferation of haemopoietic stem cells in vitro. Journal of cellular physiology. 1977;91:335–344. doi: 10.1002/jcp.1040910303. [DOI] [PubMed] [Google Scholar]
- 30.Moore KA, Ema H, Lemischka IR. In vitro maintenance of highly purified, transplantable hematopoietic stem cells. Blood. 1997;89:4337–4347. [PubMed] [Google Scholar]
- 31.Harvey K, Dzierzak E. Cell-Cell Contact and Anatomical Compatibility in Stromal Cell-Mediated HSC Support During Development. Stem Cells. 2004;22:253–258. doi: 10.1634/stemcells.22-3-253. [DOI] [PubMed] [Google Scholar]
- 32.Oostendorp RA, Medvinsky AJ, Kusadasi N, et al. Embryonal subregion-derived stromal cell lines from novel temperature-sensitive SV40 T antigen transgenic mice support hematopoiesis. Journal of cell science. 2002;115:2099–2108. doi: 10.1242/jcs.115.10.2099. [DOI] [PubMed] [Google Scholar]
- 33.Kusadasi N, Oostendorp RA, Koevoet WJ, Dzierzak EA, Ploemacher RE. Stromal cells from murine embryonic aorta-gonad-mesonephros region, liver and gut mesentery expand human umbilical cord blood-derived CAFC(week6) in extended long-term cultures. Leukemia. 2002;16:1782–1790. doi: 10.1038/sj.leu.2402615. [DOI] [PubMed] [Google Scholar]
- 34.Durand C, Robin C, Bollerot K, Baron MH, Ottersbach K, Dzierzak E. Embryonic stromal clones reveal developmental regulators of definitive hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20838–20843. doi: 10.1073/pnas.0706923105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durand C, Robin C, Dzierzak E. Mesenchymal lineage potentials of aorta-gonad-mesonephros stromal clones. Haematologica. 2006;91:1172–1179. [PubMed] [Google Scholar]
- 36.Austin TW, Solar GP, Ziegler FC, Liem L, Matthews W. A role for the Wnt gene family in hematopoiesis: expansion of multilineage progenitor cells. Blood. 1997;89:3624–3635. [PubMed] [Google Scholar]
- 37.Van Den Berg DJ, Sharma AK, Bruno E, Hoffman R. Role of members of the Wnt gene family in human hematopoiesis. Blood. 1998;92:3189–3202. [PubMed] [Google Scholar]
- 38.Brandon C, Eisenberg LM, Eisenberg CA. WNT signaling modulates the diversification of hematopoietic cells. Blood. 2000;96:4132–4141. [PubMed] [Google Scholar]
- 39.van de Wetering M, de Lau W, Clevers H. WNT signaling and lymphocyte development. Cell. 2002;109 Suppl:S13–S19. doi: 10.1016/s0092-8674(02)00709-2. [DOI] [PubMed] [Google Scholar]
- 40.Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 41.Zhao RC, Jiang Y, Verfaillie CM. A model of human p210(bcr/ABL)-mediated chronic myelogenous leukemia by transduction of primary normal human CD34(+) cells with a BCR/ABL-containing retroviral vector. Blood. 2001;97:2406–2412. doi: 10.1182/blood.v97.8.2406. [DOI] [PubMed] [Google Scholar]
- 42.Ema H, Morita Y, Yamazaki S, et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat Protoc. 2006;1:2979–2987. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- 43.Verfaillie CM. Direct contact between human primitive hematopoietic progenitors and bone marrow stroma is not required for long-term in vitro hematopoiesis. Blood. 1992;79:2821–2826. [PubMed] [Google Scholar]
- 44.Harrison DE. Competitive repopulation: a new assay for long-term stem cell functional capacity. Blood. 1980;55:77–81. [PubMed] [Google Scholar]
- 45.Harrison DE, Jordan CT, Zhong RK, Astle CM. Primitive hemopoietic stem cells: direct assay of most productive populations by competitive repopulation with simple binomial, correlation and covariance calculations. Exp Hematol. 1993;21:206–219. [PubMed] [Google Scholar]
- 46.Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci U S A. 1990;87:8736–8740. doi: 10.1073/pnas.87.22.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oostendorp RA, Robin C, Steinhoff C, et al. Long-term maintenance of hematopoietic stem cells does not require contact with embryo-derived stromal cells in cocultures. Stem cells (Dayton, Ohio) 2005;23:842–851. doi: 10.1634/stemcells.2004-0120. [DOI] [PubMed] [Google Scholar]
- 48.Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. The Journal of biological chemistry. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 49.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS biology. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong JF, Zhao Y, Sutton S, et al. Gene expression profile of murine long-term reconstituting vs. short-term reconstituting hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2448–2453. doi: 10.1073/pnas.0409459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science (New York, NY. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 52.Muller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 53.Thiemann FT, Moore KA, Smogorzewska EM, Lemischka IR, Crooks GM. The murine stromal cell line AFT024 acts specifically on human CD34+CD38- progenitors to maintain primitive function and immunophenotype in vitro. Experimental hematology. 1998;26:612–619. [PubMed] [Google Scholar]
- 54.Khan NI, Bendall LJ. Role of WNT signaling in normal and malignant hematopoiesis. Histol Histopathol. 2006;21:761–774. doi: 10.14670/HH-21.761. [DOI] [PubMed] [Google Scholar]
- 55.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nature immunology. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 56.Murdoch B, Chadwick K, Martin M, et al. Wnt-5A augments repopulating capacity and primitive hematopoietic development of human blood stem cells in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3422–3427. doi: 10.1073/pnas.0130233100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development (Cambridge, England) 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y, Haviernik P, Bunting KD, Yang YC. Cited2 is required for normal hematopoiesis in the murine fetal liver. Blood. 2007;110:2889–2898. doi: 10.1182/blood-2007-01-066316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen D, Wang P, Lewis RL, et al. A microarray analysis of the emergence of embryonic definitive hematopoiesis. Experimental hematology. 2007;35:1344–1357. doi: 10.1016/j.exphem.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Schuringa JJ, Wu K, Morrone G, Moore MA. Enforced activation of STAT5A facilitates the generation of embryonic stem-derived hematopoietic stem cells that contribute to hematopoiesis in vivo. Stem cells (Dayton, Ohio) 2004;22:1191–1204. doi: 10.1634/stemcells.2004-0033. [DOI] [PubMed] [Google Scholar]
- 61.Nemeth MJ, Topol L, Anderson SM, Yang Y, Bodine DM. Wnt5a inhibits canonical Wnt signaling in hematopoietic stem cells and enhances repopulation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15436–15441. doi: 10.1073/pnas.0704747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.