Abstract
Nuclear hormone receptors (NHRs) are transcription factors that work in concert with co-activators and co-repressors to regulate gene expression. Some examples of ligands for NHRs include endogenous compounds such as bile acids, retinoids, steroid hormones, thyroid hormone, and vitamin D. This review describes the evolution of liver X receptors α and β (NR1H3 and 1H2, respectively), farnesoid X receptor (NR1H4), vitamin D receptor (NR1I1), pregnane X receptor (NR1I2), and constitutive androstane receptor (NR1I3). These NHRs participate in complex, overlapping transcriptional regulation networks involving cholesterol homeostasis and energy metabolism. Some of these receptors, particularly PXR and CAR, are promiscuous with respect to the structurally wide range of ligands that act as agonists. A combination of functional and computational analyses has shed light on the evolutionary changes of NR1H and NR1I receptors across vertebrates, and how these receptors may have diverged from ancestral receptors that first appeared in invertebrates.
Keywords: Bile acids and salts, Ciona intestinalis, cholesterol, drug modeling, molecular evolution, oxysterols, phylogeny
1. Nuclear hormone receptors
Nuclear hormone receptors (NHRs) are transcription factors that work in concert with co-activators and co-repressors to regulate gene expression. NHRs share a conserved domain structure, which includes, from N-terminus to C-terminus, a modulatory A/B domain, the DNA-binding domain (C domain), the ‘hinge’ D domain, the ligand-binding domain (E domain), and a variable C-terminal F domain that is absent in some NHRs (McEwan, 2009; Steinmetz et al., 2001). Most of the known NHRs are ligand-activated, although some NHRs function in a ligand-independent manner. Examples of family-specific ligands for NHRs include a range of endogenous compounds such as bile acids, retinoids, steroid hormones, thyroid hormone, and vitamin D. A few NHRs, such as the ‘xenobiotic sensors’ pregnane X receptor (PXR, NR1I1; also known as steroid and xenobiotic receptor or SXR) and constitutive androstane receptor (CAR, NR1I3), are activated by structurally diverse exogenous ligands.
The NHR superfamily in mammals is composed of approximately 50 functional genes, with 48 genes in humans, 49 in mice, and 47 in rats (Zhang et al., 2004). Teleost fish have a somewhat larger complement of NHR genes due to gene duplication, exemplified by the 68 and 71 NHR genes, respectively, found in the genomes of the pufferfish (Fugu rubripes) (Maglich et al., 2003) and green-spotted pufferfish (Tetraodon nigriviridis) (Metpally et al., 2007). An expanded role for NHR genes in vertebrates is suggested by the presence of only 18 NHR genes in the fruitfly Drosophila melanogaster (King-Jones and Thummel, 2005) and 17 NHR genes identified so far in Ciona intestinalis (sea squirt), an invertebrate from Urochordata, a subphylum thought to contain the closest extant invertebrate relatives of vertebrates (Delsuc et al., 2006; Yagi et al., 2003).
This review will discuss the evolution of two NHR subfamilies, NR1H and NR1I, that include liver X receptors (LXRs) α and β (NR1H3 and 1H2, respectively), farnesoid X receptor (FXR, NR1H4), vitamin D receptor (VDR, NR1I1), PXR (NR1I2), and CAR (NR1I3). These receptors, particularly PXR and CAR, are promiscuous with respect to the wide range of ligands that act as agonists. This promiscuity may be facilitated by multiple binding sites, a very large binding site, or a binding site with flexibility to alter size and shape depending on the size of the ligand. Selected endogenous and synthetic ligands for NR1H and NR1I receptors are summarized in Fig. 1.
Fig. 1.
Endogenous and synthetic ligands for NR1H and NR1I receptors. A. Most of the known endogenous ligands for LXR, FXR, VDR, PXR, and CAR are products formed from cholesterol, which can be converted to oxysterols, steroid hormones, bile salts, and vitamin D. B. Endogenous ligands for NR1H and NR1I receptors include: 5α-bile alcohols (planar structure, ‘ancestral’ bile salts; FXRs, PXRs), 5β-bile acids (bent structure, evolutionarily ‘recent’ bile salts; FXRs, VDRs, PXRs), calcitriol (VDRs), 5β-pregnan-3,20-dione (PXRs), 5α-androstan-3α-ol (PXRs, CARs), farnesol (FXRs), and 3-aminoethylbenzoate (frog PXRs). The bile alcohol shown is 5α-myxinol disulfate (3β,7α,16α,27-tetrahydroxy-5α-cholestan-3,27-disulfate) from the hagfish. The bile acid shown is taurochenodeoxycholic acid, a common bile acid found in teleost fish, birds, and mammals. C. Synthetic ligands for NR1H and NR1I receptors include: GW4064 (mammalian and zebrafish FXRs), fexaramine (mammalian FXRs), T-0901317 (LXRs, FXRs, PXRs), GW3965 (LXRs), and TCPOBOP (PXRs, CARs).
The only other known member of the NR1H subfamily, the ecdysone receptors (NR1H1), has so far only been found in invertebrates (Riddiford et al., 2000), and will not be discussed in this review. A putative ortholog of NR1I receptors in Drosophila, termed DHR96, has been implicated as a regulator of cholesterol homeostasis and the response to potentially toxic xenobiotics (Bujold et al., 2009; King-Jones et al., 2006). We will focus our review on studies in vertebrates and the invertebrate Ciona intestinalis. In addition, we will describe some of the results from various computational analyses of the NHRs (Ai et al., 2009).
2. Evolution of NR1H and NR1I nuclear hormone receptors
2.1 Liver X receptors
LXRs are key regulators of lipid and cholesterol metabolism (Kalaany and Mangelsdorf, 2006). More recently, LXRs have been shown to regulate uterine contractility (Mouzat et al., 2007) and to negatively regulate the Hedgehog signaling pathway involved in tumorigenesis and embryonic development (Gill et al., 2008; Kim et al., 2009). In all mammals whose genomes have been sequenced so far (including marsupials), two distinct LXR genes are found (Reschly et al., 2008b). LXRα is typically detected at high levels in macrophages, adipose tissues, kidney, lung, and spleen; in contrast, LXRβ is expressed at similar levels in a wide variety of tissues, the basis for an alternative name for this receptor as ‘ubiquitous receptor’ (Song et al., 1994). Based on sequenced genomes, non-mammalian vertebrates appear to generally have only a single LXR gene (Reschly et al., 2008b). The pattern of LXR tissue expression has been determined for the Fugu pufferfish (Maglich et al., 2003). Pufferfish LXR is more closely related to mammalian LXRα genes by sequence similarity, yet the pattern of tissue expression more closely resembles mammalian LXRβ genes in its ubiquity of expression, being found in brain, gill, gut, heart, ovary, and liver tissues. The sequence data suggests that a single LXR gene duplicated concurrent with the evolution of mammals. If this hypothesis is correct, then one of the duplicated genes maintained its ubiquitous tissue expression (LXRβ) while the other (LXRα) assumed new roles in cholesterol and lipid metabolism with a restricted expression in adipose tissue, liver, and macrophages (Maglich et al., 2003; Reschly et al., 2008b).
Of the NHRs in the NR1H and NR1I subfamilies, LXRs are the most conserved across vertebrate species, with sequence identities in the ligand-binding domain (LBD) between human LXRα or LXRβ and non-mammalian LXRs being approximately 75% (Reschly et al., 2008b). Thirty-one amino residues, identified in X-ray crystallographic structures of human LXRα (Svensson et al., 2003), mouse LXRα (Jaye et al., 2005), or LXRβ (Färnegårdh et al., 2003; Hoerer et al., 2003; Williams et al., 2003) as interacting closely with bound ligands (including endogenous oxysterols and synthetic ligands), are entirely conserved across all known vertebrate LXR sequences. Consistent with this high degree of sequence conservation, the ligand specificities of human LXRα, human LXRβ, mouse LXRα, mouse LXRβ, Xenopus laevis LXR, Xenopus tropicalis LXR, and zebrafish LXR are very similar, with all being activated by oxysterols and the synthetic LXR agonists GW3965 and T-0901317 (Collins et al., 2002; Schultz et al., 2000) (see Fig. 1 for chemical structures).
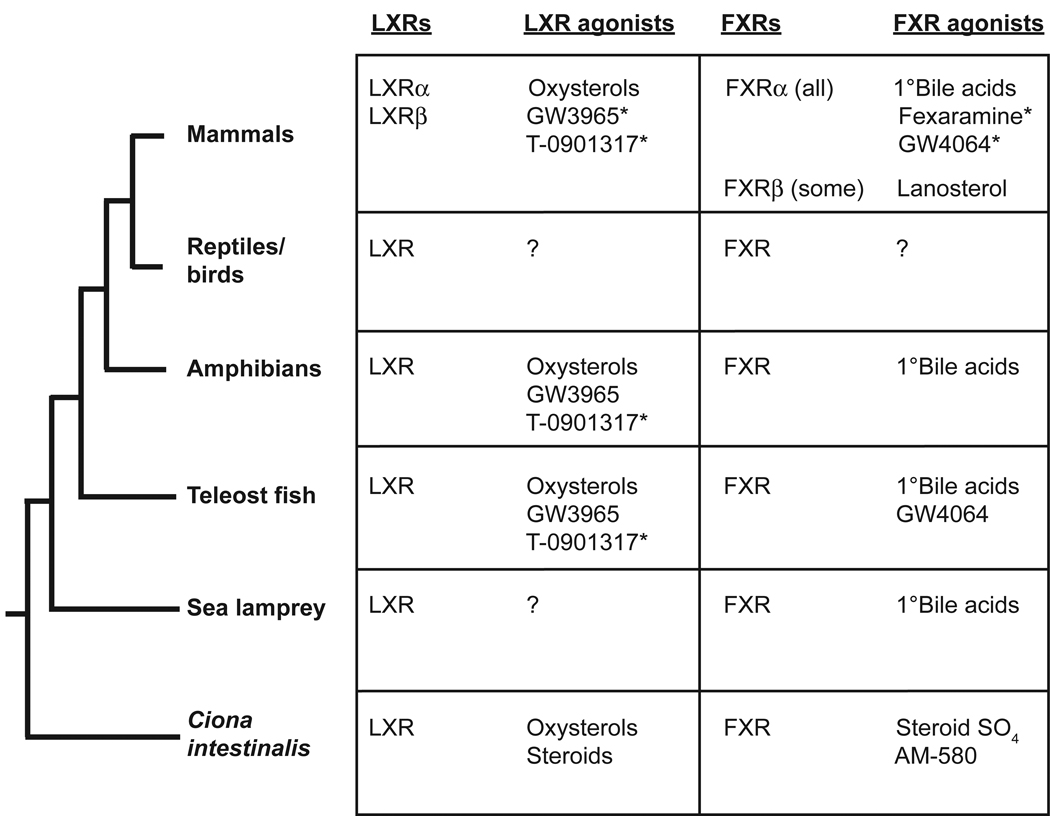
A single putative ortholog to vertebrate LXRs is found in the invertebrate Ciona intestinalis (Reschly et al., 2008b). In contrast to vertebrate LXRs, Ciona LXR is not activated by the agonists T-0901317 or GW3965, but is activated by a limited number of oxysterols, as well as some androstane and pregnane steroids. Homology modeling and docking studies of Ciona LXR predict a receptor with a smaller and more hydrophobic ligand-binding pocket (LBP) compared to human LXRβ (estimated volume of LBP is 1198 Å3 for human LXRβ and 908 Å3 for Ciona LXR). Pharmacophore studies using ligands for each receptor also indicated the Ciona LXR was likely to have a more restrictive LBP compared to human LXRβ. In addition, intrinsic disorder analysis for Ciona LXR showed no predicted disorder in the LBD compared with LXRs from 20 vertebrate species (Krasowski et al., 2008). All of these computational analyses indicated that Ciona LXR would have unique ligand specificity. Fig. 2 summarizes the ligand specificities of LXRs overlaid on a phylogeny of vertebrates and Ciona intestinalis. Ligands that have submicromolar affinities or potencies for activation of FXR or LXR are indicated with an asterisk (*) in Fig. 2. These include the synthetic ligands fexaramine and GW4064 for human FXR and T-0901317 for mammalian LXRs. The endogenous ligands (bile acids, oxysterols) generally have affinities (potencies) in the low micromolar range.
Fig. 2.
Pharmacology of liver X and farnesoid X receptors across species. The tables list the receptors found in the corresponding animal(s) organized according to the standard phylogenetic tree on the left. One liver X receptor (LXR) gene has been detected in non-mammalian species (including the invertebrate Ciona intestinalis) while two LXR genes (termed LXRα and LXRβ) are found in mammals. Most animals have a single farnesoid X receptor (FXR) gene except for a few mammalian species that have an additional functional FXRβ gene. The agonists for LXRs from mammals, amphibians, and teleost fish are very similar, including oxysterols and the synthetic agonists GW3965 and T-0901317. The LXR from Ciona differs in pharmacology from vertebrate LXRs in not being activated by GW3965 and T-0901317. The pharmacology of avian, reptile, and sea lamprey LXRs have not been reported. Vertebrate FXRs studied so far share the common feature of being activated by species-specific primary (1°) bile salts. Outside mammals, the synthetic agonists fexaramine and GW4064 are generally inactive except for GW4064 as an agonist for the zebrafish FXR. The Ciona FXR is activated by sulfated steroids (steroid SO4) and AM-580 but not by bile salts. The synthetic agonists marked by an asterisk (*) have submicromolar potency at the receptor indicated.
2.2 Bile salts, ligands for multiple nuclear hormones receptors
Before proceeding to discussion of FXR, VDR, and PXR, it is useful to first discuss bile salts, which are ligands for all three of these receptors. Bile salts are water-soluble, amphipathic end-metabolites of cholesterol that facilitate intestinal absorption of lipids, exert potent antimicrobial activity in the small intestine, and enhance proteolytic cleavage of dietary proteins (Hofmann and Hagey, 2008). Bile salts are produced by every class of vertebrate animals and show remarkable structural diversity across species (Haslewood, 1967; Moschetta et al., 2005; Une and Hoshita, 1994). Bile salts have not been detected to date in invertebrate animals, although certain species such as Ciona intestinalis synthesize bile salt-like compounds for physiological functions likely unrelated to digestion or cholesterol disposal (Yoshida et al., 2002). Bile salt derivatives are known to be pheromones in the sea lamprey (Petromyzon marinus) (Li et al., 2002). The olfactory systems of a number of teleost fish have been shown to be highly sensitive to the detection of bile salts in water, although the physiologic importance of this is as yet unclear (Hara, 1994).
A broad survey of bile salts in phylogenetically diverse vertebrates, building on the previous efforts of Haslewood and other investigators (Haslewood, 1967; Une and Hoshita, 1994), provides a detailed map of how these small molecules vary across species (Hagey et al., 2010; Hofmann et al., 2010). Two major shifts have happened in bile salt structure across evolution. The first is from bile salts with a 5α (steroid ring A/B trans) configuration of the steroid rings to those with 5β (A/B cis) configuration. This shift of a ring juncture changes the conformation of the steroid rings of the bile salt from flat (planar) to bent. The second major shift in bile salt structure is from bile alcohols with 27 carbon atoms (C27) to bile acids with 24 carbon atoms (C24). The phylogenetically most basal vertebrates are the jawless fish (Agnatha), currently represented by hagfish and lampreys. All species of hagfish that have been analyzed with respect to bile salt composition have essentially the same bile salt profile, specifically a C27 bile alcohol with a 5α configuration (Hagey et al., 2010).
We have hypothesized that the type of bile alcohols found in hagfish represents the ‘ancestral’ bile salt phenotype. If this is true, then the C24 5β-bile acids typical of humans and many other vertebrates are the ‘derived’ or evolutionarily more ‘recent’ phenotype. Fig. 1 shows the structures of the main hagfish bile salt (3β,7α,16α,27-tetrahydroxy-5α-cholestan-3,27-disulfate, also known as 5α-myxinol disulfate) (Haslewood, 1966) and taurochenodeoxycholic acid (common 5β-bile acid), illustrating the differences in steroid ring configuration. Starting with the known bile salt synthetic pathway in mammals, we have hypothesized that animals like hagfish that use C27 5α-bile alcohols have a much simpler, shorter synthetic pathway for bile salts than that found in mammals and many other vertebrates. Other than lampreys and hagfish, teleost fish from the order Cypriniformes (which includes carp and the zebrafish, Danio rerio, a versatile model laboratory fish) also use C27 5α-bile alcohols (Hagey et al., 2010).
2.3 Farnesoid X receptors
FXR serves as one of the major transcriptional regulators of bile salt synthesis in humans, in part by controlling the expression of cytochrome P450 (CYP) 7A1, the ratelimiting enzyme in bile salt synthesis (Kalaany and Mangelsdorf, 2006). Mammalian FXRs are activated by farnesol and its metabolites (Forman et al., 1995) and also by primary bile acids such as chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid), which are likely the more physiologically important endogenous ligands (Makishima et al., 1999; Parks et al., 1999; Wang et al., 1999). FXR is typically expressed at high levels in the liver, adrenal glands, intestine, and kidney. A second functional FXR, termed FXRβ (NR1H5), is found in some mammalian species (e.g., dog, mice, rat, and rabbit) but does not appear to be involved with bile salt binding or regulation; instead it binds the cholesterol precursor lanosterol and some other sterols. In humans and other primates, FXRβ is a non-functional pseudogene (Otte et al., 2003; Zhang et al., 2008b).
The variability of bile salt structures across species suggested that FXRs, if involved in bile salt detection throughout vertebrates, might show corresponding cross-species differences in ligand selectivity. Indeed, FXRs from sea lamprey and zebrafish (Danio rerio) are activated by 5α-bile alcohols but not by the evolutionarily more recent 5β-bile acids (Reschly et al., 2008a). The African clawed frog (Xenopus laevis) expresses an unusual FXR (also called FOR, FXR-like orphan receptor) that has a 33 amino acid insert, not found in mammalian FXRs, in helix 7 of the LBD (Seo et al., 2002). Similar to mammalian FXRs, Xenopus FXR is highly expressed in liver and kidney of adults, and also in the liver and kidney of metamorphosing tadpoles. Initial studies of Xenopus FXR showed insensitivity to activation by synthetic human FXR ligands or 5β-bile acids like CDCA; however, the receptor was activated by extracts isolated from frog bile (Seo et al., 2002). Further studies showed activation of Xenopus FXR by purified C27 bile alcohols that are the primary bile salts of Xenopus laevis (Reschly et al., 2008a). Preliminary investigation of the FXR from the green-spotted pufferfish (Tetraodon nigriviridis), a teleost fish whose primary salts are 5β-bile acids such as CDCA (Hagey et al., 2010), shows activation predominantly by 5β-bile acids (Krasowski MD, Hagey LR, unpublished data). Thus, FXRs generally seem to be activated by species-specific primary bile salts.
Homology models of the LBDs of sea lamprey and zebrafish FXRs predict narrow LBPs ideal for binding of planar bile salts such as 5α-bile alcohols, but not for the binding of bent 5β-bile acids (Reschly et al., 2008a). In contrast, the LBPs of human and rat FXRs can accommodate the wider bent shape of 5β-bile acids (Mi et al., 2003; Reschly et al., 2008a). The structural variation of FXRs and their corresponding bile salt ligands across species provides a model system to understand the co-evolution of receptors and ligands. A summary of results indicates a shift in selectivity for FXRs from 5α-bile alcohols (evolutionary early, ‘ancestral’ ligands) to 5β-bile acids (evolutionarily recent ligands).
The differences in ligand specificity for FXRs also extend to the synthetic human FXR agonists fexaramine, GW4064, and T-0901317 (Downes et al., 2003; Houck et al., 2004) (see Fig. 1 for chemical structures). In transactivation assays, these three compounds were generally inactive at Xenopus, zebrafish, and sea lamprey FXRs, with the only activity being T-0901317 activation of zebrafish FXR. The different architectures of the LBPs from the non-mammalian FXRs likely contribute to the ligand selectivity differences (Reschly et al., 2008a).
Analysis of the Ciona intestinalis genome revealed a single putative ortholog to vertebrate FXRs. Ciona FXR was found to be completely insensitive to activation by bile salts, but was activated by sulfated pregnane steroids, suggesting that the endogenous ligands of this receptor may be steroidal in nature. The homology model for Ciona FXR predicted a receptor with a smaller LBP (648 Å3) than that of human FXR (814 Å3). Docking studies predicted that Ciona FXR could bind AM-580, a synthetic ligand that did not activate any of the vertebrate FXRs tested, but that strongly activated Ciona FXR in functional assays (Reschly et al., 2008a).
FXR isolated from the little skate (Leucoraja erinacea, a cartilaginous fish) was found to be insensitive to bile salts, even those from jawless and cartilaginous fish (Cai et al., 2007). Skate FXR, however, showed significant differences in sequence from other vertebrate FXRs, including novel insertions, and there is the possibility that this receptor is actually orthologous to FXRβ. Better resolution of FXR phylogeny requires the study of additional invertebrates and basal vertebrates. Fig. 2 summarizes the ligand specificities of FXRs overlaid on a phylogeny of vertebrates and Ciona intestinalis.
2.4 Vitamin D receptors
VDRs bind 1α,25-(OH)2-vitamin D3 (calcitriol; see Fig. 1) with high affinity and mediate classic calcitriol effects such as regulation of calcium and phosphate homeostasis. Over the last several decades, VDRs have been shown to influence a variety of physiological functions, affecting nearly every organ and tissue (Dusso et al., 2005; Holick, 2003). VDR genes have been detected in mammals, birds, amphibians, reptiles, teleost fish, and even the sea lamprey (Whitfield et al., 2003). All mammalian genomes analyzed to date have a single VDR gene; where expression has been studied, VDR is found in a broad range of tissues that include brain, gut, heart, skeletal muscle, liver, pancreas, and immune tissues (Reschly and Krasowski, 2006). A similarly broad pattern of tissue expression was also seen with African clawed frog (Li et al., 1997) and avian VDRs (Elaroussi et al., 1994). Some teleost fish, including pufferfish and Japanese flounder (Paralichthys olivaceus) have two VDR genes (Maglich et al., 2003; Suzuki et al., 2000). Functional studies of the two VDRs from Japanese medaka (Oryzias latipes) showed differences in ligand transactivation and co-activator recruitment (Howarth et al., 2008).
Until 2002, it was generally thought that the only endogenous ligands for VDR were vitamin D compounds such as calcitriol. Then Makishima and colleagues demonstrated that lithocholic acid (LCA, 3α-hydroxy-5β-cholan-24-oic acid) and its derivatives could activate human and mouse VDRs (Adachi et al., 2005; Adachi et al., 2004; Makishima et al., 2002). LCA is a ‘secondary’ bile acid formed by the action of anaerobic intestinal bacterial on primary bile acids such as CDCA. LCA has limited aqueous solubility and is known to be toxic to humans and some other mammals (Hofmann, 2004; Hofmann and Hagey, 2008).
Unlike human and mouse VDRs, African clawed frog and sea lamprey VDRs are completely insensitive to activation by a wide range of bile salt structure, including LCA and the endogenous bile salts for these species (Krasowski et al., 2005a; Krasowski et al., 2005b; Reschly et al., 2008a). In contrast, we have found that VDRs from chicken (Gallus gallus) and the green-spotted pufferfish, two non-mammalian species that use 5β-bile acids, are weakly activated by LCA (Krasowski MD, Hagey LR, unpublished data). These data suggest that only VDRs from animals that predominantly use 5β-bile acids are activated by bile acids, possibly as an adaptive response to limit the toxicity of secondary bile acids generated in the intestinal tract. The caveat to this hypothesis is that there is little data on the disposition and toxicity of bile salts in the intestine of non-mammalian species, factors that would be influenced by cross-species differences in intestinal anatomy, physiology, and microbial colonization (Reschly et al., 2008a). Structural analysis of non-mammalian VDRs from species such as sea lamprey or Xenopus laevis may provide insight into differences in ligand selectivity. Crystallographic structures of the LBDs of human, rat, and zebrafish VDRs, while showing subtle differences, are quite similar to one another in many aspects including overall volume and shape of the LBPs (Ciesielski et al., 2007; Rochel et al., 2000; Vanhooke et al., 2004).
2.5 Pregnane X receptors
PXRs are activated by a very structurally diverse array of endogenous and exogenous molecules that include antibiotics, bile salts, steroid hormones, fat-soluble vitamins, prescription medications, herbal drugs, and endocrine disruptors (Kliewer and Willson, 2002; Orans et al., 2005; Zhou et al., 2009). PXR regulates the transcription of enzymes and transporters involved in the metabolism and elimination of potentially harmful compounds, including sulfation of toxic bile acids (Sonoda et al., 2002). Transcriptional targets of PXR include genes for the broad specificity enzyme CYP3A4 and the efflux transporter P-glycoprotein (Kliewer and Willson, 2002) to name but a few of clinical significance. Microarray analysis studies have revealed that PXR agonists significantly alter the expression of greater than 200 genes in mouse and rat liver, including genes whose products are important in cell cycle regulation, intracellular metabolism, redox balance, and anion transport, in addition to genes for CYP enzymes and drug efflux transporters (Guzelian et al., 2006; Hartley et al., 2004; Rosenfeld et al., 2003; Slatter et al., 2006). PXR has been implicated in bone homeostasis, apoptosis in cancer cells, and inflammation pathways (Pascussi et al., 2008; Tabb et al., 2003; Verma et al., 2009; Zhang et al., 2008a; Zhou et al., 2006a; Zhou et al., 2006b).
Studies from multiple laboratories have shown substantial cross-species differences in PXR ligand specificity, including selectivity for xenobiotics and bile salts (Iyer et al., 2006; Krasowski et al., 2005a; Milnes et al., 2008; Moore et al., 2002). Most mammalian PXRs studied so far (including human, rhesus macaque, dog, pig, and rabbit) are activated by a broad range of bile salt structures, while chicken and zebrafish PXRs are activated by a narrower range of bile salts (Ekins et al., 2008; Krasowski et al., 2005a; Krasowski et al., 2005b; Moore et al., 2002; Reschly et al., 2008a). We and others have proposed that the evolution of PXRs has been driven by at least two factors: (1) adaptation to changes in bile salt (and perhaps other endogenous molecule) structures and (2) their function as a xenobiotic sensor (Krasowski et al., 2005a; Moore et al., 2002; Reschly et al., 2008a; Schuetz et al., 2001). The size and flexibility of the human PXR LBP make computational prediction of ligand binding difficult (Ekins et al., 2009; Ngan et al., 2009; Yasuda et al., 2008). Prediction of ligand binding in PXRs from non-mammalian species using homology models is even more difficult, although the ligand specificity of each species can be used as a surrogate for understanding the volume of the binding site and its evolution (Ekins et al., 2008; Reschly et al., 2008a).
The PXRs from the African clawed frog deserve special mention, as these receptors have markedly different pharmacology from other PXRs, being activated by a unique class of endogenous benzoate molecules (e.g., 3-aminoethylbenzoate; see Fig. 1) that mediate developmental functions in the frog. Thus, the frog PXRs are also termed benzoate X receptors (BXRs) (Blumberg et al., 1998; Grün et al., 2002). Phylogenetic analysis by maximum likelihood showed evidence for positive evolutionary selection in the LBD of the frog PXRs relative to other PXRs, particularly at amino acid residue positions involved in ligand binding (Krasowski et al., 2005a; Krasowski et al., 2005b). The available evidence suggests that the frog PXRs have lost broad specificity for ligands, gained high efficacy activation by endogenous benzoates (which may be molecules unique to amphibians), and show an altered tissue expression pattern to carry out developmental functions. Using intrinsic disorder prediction we found that whereas the human H1-H3 interhelical domain was disordered, this was not the case for the shorter domain in the frog (Krasowski et al., 2008). The degree of differences in function and ligand specificity of the Xenopus PXRs relative to PXRs from other species is quite unusual and possibly unique in the NHR superfamily in vertebrates, with no other comparable examples yet described (Krasowski et al., 2005b).
Structural studies of the LBD of human PXR reveal an expansive (~1,300 Å3), hydrophobic, roughly spherical pocket with the flexibility to accommodate large molecules such as rifampicin and hyperforin (active component of the herbal antidepressant St. John’s wort) (Chrencik et al., 2005; Watkins et al., 2003; Watkins et al., 2001; Xue et al., 2007). We can also see this by analysis of the co-crystallized ligands that cover a molecular weight range of 273–714 Da (mean 488±147) and a calculated ALogP (measure of hydrophobicity) range of 3.54–10.11 (mean 5.5±2.4) (Ekins et al., 2009). Although X-ray crystallographic structures of PXRs from species other than humans have not yet been reported, homology models of the LBDs of African clawed frog PXRα (~860 Å3), green-spotted pufferfish PXR (~1,230 Å3), and zebrafish PXR (~1,000 Å3) are all predicted to have smaller LBPs than that for human PXR (Reschly et al., 2008a) (Ai N, Krasowski MD, Ekins S, unpublished data). Expansion of the size and topology of the PXR LBP correlates with the general pattern of broadening of PXR ligand specificity across vertebrate species and the shift from planar to non-planar bile acids.
2.6 Constitutive androstane receptors
CARs also have the capacity to bind a structurally broad range of ligands, although not to the extent of PXRs (Honkakoski et al., 2003). There is overlap between CAR and PXR ligands (see Fig. 1), including androstane steroids, clotrimazole, phenobarbital, and 1,4-bis-[2-(3,5-and dichloropyridyloxy)] benzene (TCPOBOP) (Jones et al., 2000; Moore et al., 2002; Moore et al., 2000; Tzameli et al., 2000). Some CARs, particularly from the mouse, show high constitutive activity in cell-based functional assays. Consequently, some CAR ligands function as inverse agonists, i.e., reducing the constitutive activity (Forman et al., 1998). Microarray analysis studies have demonstrated that CAR ligands upregulate or repress genes with diverse functions in mouse liver, including genes involved in regulation of energy metabolism and heme synthesis (Rezen et al., 2009; Ross et al., 2009; Slatter et al., 2006; Ueda et al., 2002). Crystallographic structures of human and mouse CARs have provided insight into both the high level of constitutive activity and the ability of steroidal compounds (e.g., androstenol, 5α-androst-16-en-3α-ol) to act as inverse agonists (Shan et al., 2004; Suino et al., 2004; Xu et al., 2004).
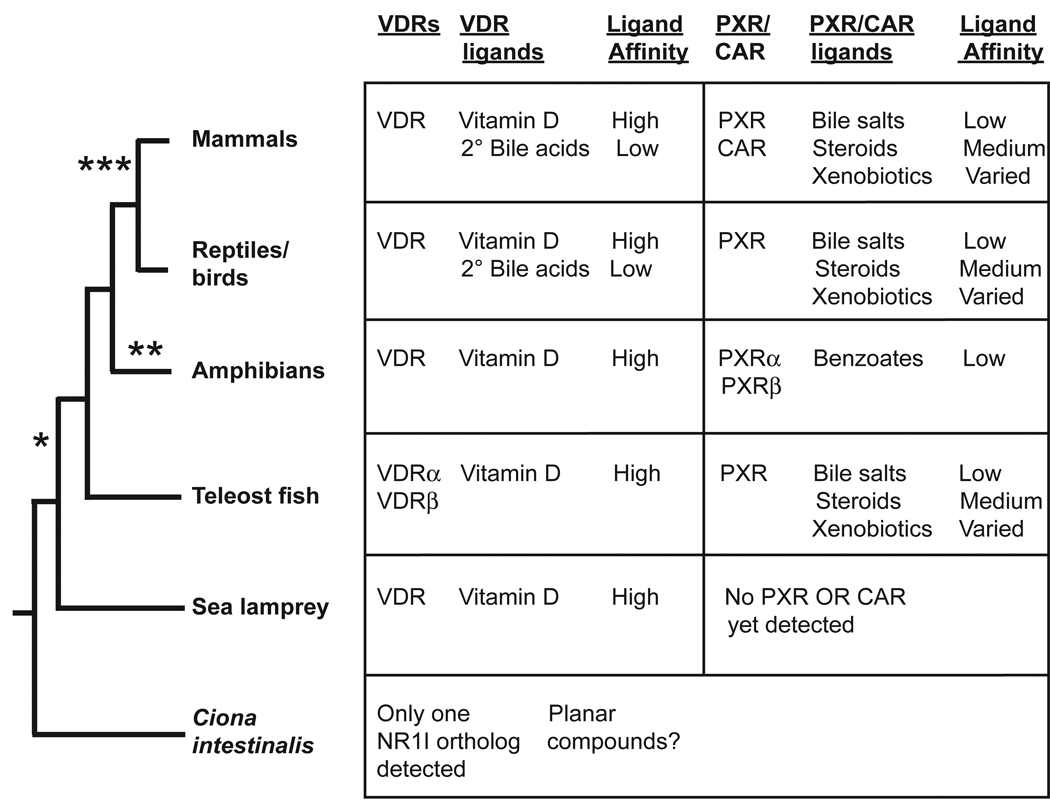
Distinct CAR and PXR genes have only been described in mammals, including marsupials and the monotreme duck-billed platypus (Reschly and Krasowski, 2006). In contrast, non-mammalian vertebrates show a PXR/CAR-like ‘combination’ receptor, and it has been difficult to determine whether either CAR or PXR represents the ‘ancestral’ receptor. For example, the single PXR/CAR-like receptor in the chicken (also termed chicken X receptor) has approximately equal similarities to mammalian PXRs and CARs in terms of sequence identity and ligand specificity (Handschin et al., 2000; Moore et al., 2002). Regardless of which receptor is ancestral, a single PXR/CAR-like ancestral gene likely duplicated concurrent with the evolution of mammals, with subsequent divergence into the separate CAR and PXR genes found in all mammals sequenced so far (Handschin et al., 2004). A summary of VDR, PXR, and CAR phylogeny and ligand specificities is found in Fig. 3. The affinities of the ligands shown in Fig. 3 are summarized as low, medium, and high affinity (see figure legend).
Fig. 3.
Pharmacology of vitamin D, pregnane X, and constitutive androstane receptors across species. The tables list the receptors found in the corresponding animal(s) organized as in Fig. 2. We follow the convention of referring to non-mammalian PXR/CAR-like receptors as PXRs, although it is debatable whether PXR or CAR is the ancestral receptor. Vertebrate vitamin D receptors (VDRs) are all activated by vitamin D derivatives. Mammalian VDRs are also activated by secondary (2°) bile acids. The vertebrate PXRs studied so far, with the exception of frog PXRs, are activated by bile salts, steroid hormones, and xenobiotics, although with substantial cross-species differences in ligand specificity. The frog PXRs are selectively activated by a class of benzoate ligands that may be unique to amphibians. Only one putative ortholog to vertebrate NR1I receptors has been cloned and characterized from the invertebrate Ciona intestinalis. This receptor has markedly different pharmacology from vertebrate VDRs, PXRs, and CARs. There are several major evolutionary changes in NR1I receptors indicated on the phylogeny: *, duplication of a single receptor gene to separate VDR and PXR genes; **, divergence of function and ligand specificity for frog PXRs; and ***, duplication of single PXR/CAR gene to separate PXR and CAR genes. The “Ligand affinity” columns classifies the ligands into whether they have EC50 values for activation of the receptor of 10 µM or higher (low affinity), 1–10 µM (medium affinity), or less than 1 µM (high affinity). Xenobiotics at PXRs have a range of affinities, including a small number such as hyperforin than have affinities in the nanomolar range.
2.7 The unusual Ciona intestinalis NR1I receptor
So far, PXR genes have not been identified in jawless or cartilaginous fish, either by cloning efforts or analysis of the partially sequenced genome of the sea lamprey (Reschly et al., 2007). The only NR1I subfamily member identified so far in the sea lamprey is VDR (Whitfield et al., 2003). Similarly, the genome of the invertebrate Ciona intestinalis reveals only a single putative ortholog to vertebrate NR1I receptors. The phylogeny of the Ciona NR1I receptor, as inferred by maximum likelihood analysis, does not clearly group this receptor with VDRs, PXRs, or CARs, although ancestral sequence reconstruction did provide some favor to a closer relationship with vertebrate VDRs (Ekins et al., 2008). The LBD of the Ciona VDR/PXR/CAR has low sequence identity to the LBDs of vertebrate VDRs, PXRs, and CARs (17–27%), in some cases to the extent that reliable sequence alignment is not possible (limiting ancestral reconstruction reliability as well). The DNA-binding domain of the Ciona VDR/PXR/CAR has its highest sequence identity to sea lamprey and zebrafish VDRs (~70%). In functional cell-based assays, the Ciona VDR/PXR/CAR does not respond to vitamin D ligands, bile salts, retinoids, steroid hormones, tocopherols, or typical PXR-activating xenobiotics. The Ciona VDR/PXR/CAR has been shown to be activated only by a small number of planar, synthetic compounds including n-butyl-p-aminobenzoate, carbamazepine, 6-formylindolo-[3,2-b]-carbazole, and 2,3,7,8-tetrachlorodibenzo-p-dioxin (Ekins et al., 2008). Intrinsic disorder analysis showed that the LBD of Ciona VDR/PXR/CAR was most similar to mammalian PXRs, suggesting some ability to adapt to different ligands (Krasowski et al., 2008). This suggests that the natural ligand for Ciona VDR/PXR/CAR may be hard to discern, perhaps a compound in the natural environment of this marine invertebrate.
3. The co-evolution of biochemical pathways and NR1H and NR1I receptors
One common feature of LXR, FXR, VDR, PXR, and CAR is that all receptors are activated (or, in the case of some CARs, repressed) by products of cholesterol: oxysterols (LXR), bile salts (FXR, VDR, PXR), steroid hormones (PXR, CAR, FXRβ), or vitamin D (VDR). These NHRs also participate in complex, overlapping transcriptional regulation networks involving cholesterol synthesis, elimination, and energy metabolism (Handschin and Meyer, 2005; Makishima, 2005). For example, we can examine the overlap of some of the ligands between these NHRs as a network (Ekins, 2006; Ekins et al., 2005) and show bile acids linked to FXR, VDR, and PXR (Supplemental figure 1S). Cholesterol is not unique to vertebrates, being found in some invertebrates; however, vertebrate animals utilize cholesterol to an extent not matched in any invertebrate species studied to date. The increased use of cholesterol by vertebrates compared to invertebrates is thought to have been a major evolutionary shift requiring tightly regulated systems for controlling cholesterol synthesis and elimination from the body (Nes and Nes, 1980). This was achieved by the parallel development of a hepatobiliary tract and synthetic pathways for biosynthesis and conjugation of bile alcohols and bile acids (collectively ‘bile salts’).
Tracing back the early evolution of NR1H and NR1I receptors requires a better understanding of the evolution and basic biological functions of the ligands for these receptors in non-mammalian species. For example, the comparative biology of oxysterols (i.e., derivatives of cholesterol oxidized on the side-chain) in non-mammalian species is not well understood. In mammals, oxysterols inhibit sterol biosynthesis along with other biological functions (Gill et al., 2008). Bile salts and vitamin D have so far been found only in vertebrate animals, although it is possible these compounds are present in invertebrate animals not yet analyzed or that are extinct. Bile salts have been detected in every vertebrate animal analyzed so far, including the phylogenetically basal jawless and cartilaginous fish (Hagey et al., 2010; Hofmann et al., 2010).
The early origins of the vitamin D system are unclear (Holick, 2003). The rise in vertebrate evolution associated with high levels of cholesterol as a nerve insulator also saw a concurrent increase in animal size built on a calcium phosphate base of bone. In terrestrial animals, the need to tightly regulate dietary absorption of calcium and phosphate is clear, especially given a variable dietary intake. However, the biological functions of vitamin D in animals living in salt water (where calcium and phosphate is plentiful) are harder to appreciate. Vitamin D and its cognate receptor are even found in the sea lamprey, a jawless fish lacking a calcified skeleton (Whitfield et al., 2003). This has prompted investigation into the importance of the vitamin D system for other biological functions, including immune regulation and skin development (Kira et al., 2003; Moro et al., 2008).
As discussed above, the model invertebrate Ciona intestinalis has clear orthologs to LXR, FXR, and VDR/PXR/CAR. Pharmacology studies are consistent with these Ciona receptors having different (although possibly structurally similar) ligands to their vertebrate counterparts. In the case of FXR, we have speculated that the ligands for the Ciona receptors are sulfated steroids (Reschly et al., 2008a), compounds that are common in marine invertebrates (Kornprobst et al., 1998). If this is true, there could have been a shift away from sulfated steroids to the growing and ever enlarging pool of cholesterol catabolites (bile salts) as FXR ligands during vertebrate evolution. A similar shift may have happened during the molecular evolution of LXR, e.g., from invertebrate steroidal ligands to vertebrate oxysterols found upstream and downstream of cholesterol biosynthesis (Reschly et al., 2008b).
The properties of the Ciona VDR/PXR/CAR suggest that invertebrate and vertebrate NR1I receptors have diverged markedly in ligand selectivity from an ancestral ‘proto-NR1I receptor’ (Ekins et al., 2008). Given that there are no clear correlates of vitamin D or bile salts yet described in invertebrates, endogenous ligands for the Ciona VDR/PXR/CAR would logically be different from those for vertebrate VDRs and PXRs. Ciona intestinalis is, however, capable of synthesizing steroid hormones and also accumulates cholesterol and other sterols from dietary sources (Delrio et al., 1971; Voogt and van Rheenan, 1975). The endogenous activators of the Ciona VDR/PXR/CAR may be as yet undescribed molecules that have structural similarity to vertebrate vitamins and/or bile salts or they may be structurally unique but sharing a similar three-dimensional pharmacophore to ligands for the vertebrate receptors. Alternatively, this receptor may be activated by exogenous ligands relevant to its marine environment or local habitat. The low sequence identity between the Ciona VDR/PXR/CAR may also be a result of rapid evolution, which has been detected in some gene families (including developmental regulators) in Ciona intestinalis and other tunicates (Dehal et al., 2002; Holland and Gibson-Brown, 2003; Hughes and Friedman, 2005). Intrinsic disorder of Ciona VDR/PXR/CAR may also be an important feature in driving its evolution (Krasowski et al., 2008).
4. Conclusions and perspectives
Studies of further invertebrates and basal vertebrates will be invaluable in better resolving the evolution of the NR1H and 1I receptors. Additional receptor sequences will also facilitate ancestral reconstruction of sequences, as has been elegantly done by Thornton and colleagues for sex and mineralocorticoid receptors, including X-ray crystallography and functional analysis, to understand evolutionary changes in receptor ligand selectivity (Bridgham et al., 2006; Bridgham et al., 2009; Ortlund et al., 2007; Thornton et al., 2003). We have done some ancestral sequence reconstruction for FXR and VDR/PXR but are limited by the high degree of sequence diversity, including insertions and deletions, which makes a parallel approach far more uncertain than for the more highly conserved sex and mineralocorticoid receptors (Ekins et al., 2008; Reschly et al., 2008a). Structural analysis of non-mammalian FXR and PXRs would be particularly helpful in defining how receptors alter ligand specificity across species, and would build on the current homology, pharmacophore, and ligand docking analyses.
Supplementary Material
Acknowledgements
MDK is supported by National Institutes of Health grant NIH K08-GM074238. SE gratefully acknowledges Ingenuity for providing IPA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi R, Honma Y, Masuno H, Karana K, Shimomura I, Yamada S, Makishima M. Selective activation of vitamin D receptor by lithocholic acid acetate, a bile acid derivative. J. Lipid Res. 2005;46:46–57. doi: 10.1194/jlr.M400294-JLR200. [DOI] [PubMed] [Google Scholar]
- Adachi R, Shulman AI, Yamamoto K, Shimomura I, Yamada S, Mangelsdorf DJ, Makishima M. Structural determinants for vitamin D responses to endocrine and xenobiotic signals. Mol. Endocrinol. 2004;18:43–52. doi: 10.1210/me.2003-0244. [DOI] [PubMed] [Google Scholar]
- Ai N, Krasowski MD, Welsh WJ, Ekins S. Understanding nuclear receptors using computational methods. Drug Discov. Today. 2009;14:486–494. doi: 10.1016/j.drudis.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Kang H, Bolado J, Chen H, Craig AG, Moreno TA, Umesano K, Perlmann T, De Robertis EM, Evans RM. BXR, an embryonic orphan nuclear receptor activated by a novel class of endogenous benzoate metabolites. Genes Dev. 1998;12:1269–1277. doi: 10.1101/gad.12.9.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujold M, Gopalakrishnan A, Nally E, King-Jones K. The nuclear receptor DHR96 acts as a sentinel for low cholesterol concentrations in Drosophila. Mol. Cell. Biol. 2009 doi: 10.1128/MCB.01327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SY, Xiong L, Wray CG, Ballatori N, Boyer JL. The farnesoid X receptor, FXRa/NR1H4, acquired ligand specificity for bile salts late in vertebrate evolution. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;293:R1400–R1409. doi: 10.1152/ajpregu.00781.2006. [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, Wisely GB, Lambert MH, Kliewer SA, Redinbo MR. Structural disorder in the complex of human PXR and the macrolide antibiotic rifampicin. Mol. Endocrinol. 2005;19:1125–1134. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- Ciesielski F, Rochel N, Moras D. Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation. J. Steroid Biochem. Mol. Biol. 2007;103:235–242. doi: 10.1016/j.jsbmb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Collins JL, Fivush AM, Watson MA, Galardi CM, Lewis MC, Moore LB, Parks DJ, Wilson JG, Tippin TK, Binz JG, Plunket KD, Morgan DG, Beaudet EJ, Whitney KD, Kliewer SA, Willson TM. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J. Med. Chem. 2002;45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KEM, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang H-G, Awazu S, Azumi K, Boore J, Branno M, Chin-bow S, DeSantis R, Doyle S, Francina P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Kano S, Kobayashi K, Kobayashi M, Lee B-I, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Galvina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delrio G, D'Istria M, Milone M, Chieffi G. Identification and biosynthesis of steroid hormones in the gonads of Ciona intestinalis. Experientia. 1971;27:1348–1350. doi: 10.1007/BF02136733. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer J-L, Anisfeld AM, Edwards PA, Rosenfeld JM. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell. 2003;11:1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusso AS, Brown AJ, Slatopolsky E. Vitamin. D. Am. J. Physiol. Renal Physiol. 2005;289:F8–F28. doi: 10.1152/ajprenal.00336.2004. [DOI] [PubMed] [Google Scholar]
- Ekins S. Systems-ADME/Tox: resources and network approaches. J. Pharmacol. Toxicol. Methods. 2006;53:38–66. doi: 10.1016/j.vascn.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Ekins S, Kirillov E, Rakhmatulin EA, Nikotskaya T. A novel method for visualizing nuclear hormone receptor networks relevant to drug metabolism. Drug Metab. Dispos. 2005;33:474–481. doi: 10.1124/dmd.104.002717. [DOI] [PubMed] [Google Scholar]
- Ekins S, Kortagere S, Iyer M, Reschly EJ, Lill MA, Redinbo MR, Krasowski MD. Challenges predicting ligand-receptor interactions of promiscuous proteins: the nuclear receptor PXR. PLoS Comput. Biol. 2009;5:e1000594. doi: 10.1371/journal.pcbi.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Reschly EJ, Hagey LR, Krasowski MD. Evolution of pharmacologic specificity in the pregnane X receptor. BMC Evol. Biol. 2008;8:103. doi: 10.1186/1471-2148-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elaroussi MA, Prahl JM, DeLuca HF. The avian vitamin D receptor: primary structures and their origins. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11596–11600. doi: 10.1073/pnas.91.24.11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Färnegårdh M, Bonn T, Sun S, Ljunggren J, Ahola H, Wilhelmsson A, Gustafsson J-A, Carlquist M. The three-dimensional structure of the liver X receptor β reveals a flexible ligand-binding pocket that can accommodate fundamentally different ligands. J. Biol. Chem. 2003;278:38821–38828. doi: 10.1074/jbc.M304842200. [DOI] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi H-S, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-β. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Gill S, Chow R, Brown AJ. Sterol regulators of cholesterol homeostasis and beyond: the oxysterol hypothesis revisited and revised. Prog. Lipid Res. 2008;47:391–404. doi: 10.1016/j.plipres.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Grün F, Venkatesan RN, Tabb MM, Zhou C, Cao J, Hemmati D, Blumberg B. Benzoate X receptors α and β are pharmacologically distinct and do not function as xenobiotic receptors. J. Biol. Chem. 2002;277:43691–43697. doi: 10.1074/jbc.M206553200. [DOI] [PubMed] [Google Scholar]
- Guzelian J, Barwick JL, Hunter L, Phang TL, Quattrochi LC, Guzelian PS. Identification of genes controlled by the pregnane X receptor by microarray analysis of mRNAs from pregnenolone 16α-carbonitrile treated rats. Toxicol. Sci. 2006;94:379–387. doi: 10.1093/toxsci/kfl116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagey LR, Møller PR, Hofmann AF, Krasowski MD. Diversity of bile salts in fish and amphibians: evolution of a complex biochemical pathway. Physiol. Biochem. Zool. 2010;83:308–321. doi: 10.1086/649966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Blaettler S, Roth A, Looser R, Oscarson M, Kaufmann MR, Podvinec M, Gnerre C, Meyer UA. The evolution of drug-activated nuclear receptors: one ancestral gene diverged into two xenosensor genes in mammals. Nucl. Recept. 2004;2:7. doi: 10.1186/1478-1336-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Meyer UA. Regulatory network of lipid-sensing nuclear receptors: roles for CAR, PXR, LXR, and FXR. Arch. Biochem. Biophys. 2005;433:387–396. doi: 10.1016/j.abb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Handschin C, Podvinec M, Meyer UA. CXR, a chicken xenobiotic-sensing orphan nuclear receptor, is related to both mammalian pregnane X receptor (PXR) and constitutive androstane receptor (CAR) Proc. Natl. Acad. Sci. U.S.A. 2000;97:10769–10774. doi: 10.1073/pnas.97.20.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara TJ. Olfaction and gustation in fish: an overview. Acta Physiol. Scand. 1994;152:207–217. doi: 10.1111/j.1748-1716.1994.tb09800.x. [DOI] [PubMed] [Google Scholar]
- Hartley DP, Dai X, He YD, Carlini EJ, Wang B, Huskey SW, Ulrich RG, Rushmore TH, Evers R, Evans DC. Activators of the rat pregnane X receptor differentially modulate hepatic and intestinal gene expression. Mol. Pharmacol. 2004;65:1159–1171. doi: 10.1124/mol.65.5.1159. [DOI] [PubMed] [Google Scholar]
- Haslewood GAD. Comparative studies of bile salts. Myxinol disulphate, the principal bile salt of hagfish (Myxinidae) Biochem. J. 1966;100:233–237. doi: 10.1042/bj1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslewood GAD. Bile salt evolution. J. Lipid Res. 1967;8:535–550. [PubMed] [Google Scholar]
- Hoerer S, Schmid A, Heckel A, Budzinski R-M, Nar H. Crystal structure of the human liver X receptor β ligand-binding domain in complex with a synthetic agoinist. J. Mol. Biol. 2003;334:853–861. doi: 10.1016/j.jmb.2003.10.033. [DOI] [PubMed] [Google Scholar]
- Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab. Rev. 2004;36:703–722. doi: 10.1081/dmr-200033475. [DOI] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionarily significance. J. Lipid Res. 2010;51:226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holick MF. Evolution and function of vitamin D. Recent Results Cancer Res. 2003;164:3–28. doi: 10.1007/978-3-642-55580-0_1. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Gibson-Brown JJ. The Ciona intestinalis genome: when the constraints are off. BioEssays. 2003;25:529–532. doi: 10.1002/bies.10302. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Sueyoshi T, Negishi M. Drug-activated nuclear receptors PXR and CAR. Ann. Med. 2003;35:172–182. doi: 10.1080/07853890310008224. [DOI] [PubMed] [Google Scholar]
- Houck KA, Borchert KM, Hepler CD, Thomas JS, Bramlett KS, Michael LF, Burris TP. T0901317 is a dual LXR/FXR agonist. Mol. Genet. Metab. 2004;83:184–187. doi: 10.1016/j.ymgme.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Howarth DL, Law SH, Barnes B, Hall JM, Hinton DE, Moore L, Maglich JM, Moore JT, Kullman SW. Paralogous vitamin D receptors in teleosts: transition of nuclear receptor function. Endocrinology. 2008;149:2411–2422. doi: 10.1210/en.2007-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol. Dev. 2005;7:196–200. doi: 10.1111/j.1525-142X.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- Iyer M, Reschly EJ, Krasowski MD. Functional evolution of the pregnane X receptor. Expert Opin. Drug Metab. Toxicol. 2006;2:381–387. doi: 10.1517/17425255.2.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaye MC, Krawiec JA, Campobasso N, Smallwood A, Qiu C, Lu Q, Kerrigan JJ, Alvaro MDF, Laffitte B, Liu W-S, Marino JP, Meyer CR, Nichols JA, Parks DJ, Perez P, Sarov-Blat L, Seepersaud SD, Steplewski KM, Thompson SK, Wang P, Watson MA, Webb CL, Haigh D, Caravella JA, Macphee CH, Willson TM, Collins JL. Discovery of substituted maleimides as liver X receptor agonists and determination of a ligand-bound crystal structure. J. Med. Chem. 2005;48:5419–5422. doi: 10.1021/jm050532w. [DOI] [PubMed] [Google Scholar]
- Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, McKee DD, Tomkinson NCO, LeCluyse EL, Lambert MH, Willson TM, Kliewer SA, Moore JT. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol. Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. LXRs and FXR: the Yin and Yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- Kim WK, Meliton V, Park KW, Hong C, Tontonoz P, Niewiadomski P, Waschek JA, Tetradis S, Parhami F. Negative regulation of Hedgehog signaling by liver X receptors. Mol. Endocrinol. 2009;23:1532–1543. doi: 10.1210/me.2008-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors - a perspective from Drosophila. Nat. Rev. Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Kira M, Kobayashi T, Yoshikawa K. Vitamin D and the skin. J. Dermatol. 2003;30:429–437. doi: 10.1111/j.1346-8138.2003.tb00412.x. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Willson TM. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J. Lipid Res. 2002;43:359–364. [PubMed] [Google Scholar]
- Kornprobst J-M, Sallenave C, Barnathan G. Sulfated compounds from marine organisms. Comp. Biochem. Physiol. B. 1998;119:1–51. doi: 10.1016/s0305-0491(97)00168-5. [DOI] [PubMed] [Google Scholar]
- Krasowski MD, Reschly EJ, Ekins S. Intrinsic disorder in nuclear hormone receptors. J. Proteome Res. 2008;7:4359–4372. doi: 10.1021/pr8003024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolution of the pregnane X receptor: adaptation to cross-species differences in biliary bile salts. Mol. Endocrinol. 2005a;19:1720–1739. doi: 10.1210/me.2004-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolutionarily selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors) Nucl. Recept. 2005b;3:2. doi: 10.1186/1478-1336-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Scott AP, Siefkes MJ, Yan H, Liu Q, Yun SS, Gage DA. Bile acid secreted by male sea lamprey that acts as a sex pheromone. Science. 2002;296:138–141. doi: 10.1126/science.1067797. [DOI] [PubMed] [Google Scholar]
- Li YC, Bergwitz C, Juppner H, Demay MB. Cloning and characterization of the vitamin D receptor from Xenopus laevis. Endocrinology. 1997;138:2347–2353. doi: 10.1210/endo.138.6.5210. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Caravella JA, Lambert MH, Willson TM, Moore JT, Ramamurthy L. The first completed genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 2003;31:4051–4058. doi: 10.1093/nar/gkg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M. Nuclear receptors as targets for drug development: regulation of cholesterol and bile acid metabolism by nuclear receptors. J. Pharmacol. Sci. 2005;97:177–183. doi: 10.1254/jphs.fmj04008x4. [DOI] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- Makishima M, Okamato AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- McEwan IJ. Nuclear receptors: one big family. Methods Mol. Biol. 2009;505:3–18. doi: 10.1007/978-1-60327-575-0_1. [DOI] [PubMed] [Google Scholar]
- Metpally RP, Vigneshwar R, Sowdhamini R. Genome inventory and analysis of nuclear hormone receptors in Tetraodon nigroviridis. J. Biosci. 2007;32:43–50. doi: 10.1007/s12038-007-0005-4. [DOI] [PubMed] [Google Scholar]
- Mi L-Z, Devarakonda S, Harp JM, Han Q, Pellicciari R, Willson TM, Khorasanizadeh S, Rastinejad F. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol. Cell. 2003;11:1093–1100. doi: 10.1016/s1097-2765(03)00112-6. [DOI] [PubMed] [Google Scholar]
- Milnes MR, Garcia A, Grossman E, Grun F, Shiotsugu J, Tabb MM, Kawashima Y, Katsu Y, Watanabe H, Iguchi T, Blumberg B. Activation of steroid and xenobiotic receptor (SXR, NR1I2) and its orthologs in laboratory, toxicologic, and genome model species. Environ. Health Perspect. 2008;116:880–885. doi: 10.1289/ehp.10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, Lambert MH, Moore JT. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol. 2002;16:977–986. doi: 10.1210/mend.16.5.0828. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- Moro JR, Iwata M, von Andriano UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat. Rev. Immunol. 2008;8:685–698. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschetta A, Xu F, Hagey LR, van Berge Henegouwen GP, van Erpecum KJ, Brouwers JF, Cohen JC, Bierman M, Hobbs HH, Steinbach JH, Hofmann AF. A phylogenetic survey of biliary lipids in vertebrates. J. Lipid Res. 2005;46:2221–2232. doi: 10.1194/jlr.M500178-JLR200. [DOI] [PubMed] [Google Scholar]
- Mouzat K, Prod'homme M, Volle DH, Sion B, Dechelotte P, Gauthier K, Vanacker JM, Lobaccaro JM. Oxysterol nuclear receptor LXRβ regulates cholesterol homeostasis and contractile functions in mouse uterus. J. Biol. Chem. 2007;282:4693–4701. doi: 10.1074/jbc.M606718200. [DOI] [PubMed] [Google Scholar]
- Nes WR, Nes WD. Lipids in evolution. New York: Plenum Press; 1980. [Google Scholar]
- Ngan CH, Beglov D, Rudnitskaya AN, Kozakov D, Waxman DJ, Vajda S. The structural basis of pregnane X receptor binding promiscuity. Biochemistry. 2009;48:11572–11581. doi: 10.1021/bi901578n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orans J, Teotico DG, Redinbo MR. The nuclear xenobiotic receptor PXR: recent insights and new challenges. Mol. Endocrinol. 2005;19:2891–2900. doi: 10.1210/me.2005-0156. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte K, Kranz H, Kober I, Thompson P, Hoefer M, Haubold B, Remmel B, Voss H, Kaiser C, Albers M, Cheruvallath Z, Jackson D, Casari G, Koegl M, Pääbo S, Mous J, Kremoser C, Deuschle U. Identification of farnesoid X receptor β as a novel mammalian nuclear receptor sensing lanosterol. Mol. Cell. Biol. 2003;23:864–872. doi: 10.1128/MCB.23.3.864-872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Duret C, Daujat-Chavanieu M, Vilarem MJ, Maurel P. The tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequences. Annu. Rev. Pharmacol. Toxicol. 2008;48:1–32. doi: 10.1146/annurev.pharmtox.47.120505.105349. [DOI] [PubMed] [Google Scholar]
- Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hofmann AF, Krasowski MD. Evolution of the bile salt nuclear receptor FXR in vertebrates. J. Lipid Res. 2008a;49:1577–1587. doi: 10.1194/jlr.M800138-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Ai N, Welsh WJ, Ekins S, Hagey LR, Krasowski MD. Ligand specificity and evolution of liver X receptors. J. Steroid Biochem. Mol. Biol. 2008b;110:83–94. doi: 10.1016/j.jsbmb.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Bainy ACD, Mattos JJ, Hagey LR, Bahary N, Mada SR, Ou J, Venkataramanan R, Krasowski MD. Functional evolution of the vitamin D and pregnane X receptors. BMC Evol. Biol. 2007;7:222. doi: 10.1186/1471-2148-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Krasowski MD. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr. Drug Metab. 2006;7:349–365. doi: 10.2174/138920006776873526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezen T, Tamasi V, Lovgren-Sandblom A, Bjorkhem I, Meyer UA, Rozman D. Effect of CAR activation on selected metabolic pathways in normal and hyperlipidemic mouse livers. BMC Genomics. 2009;10:384. doi: 10.1186/1471-2164-10-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam. Horm. 2000;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell. 2000;5:173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JM, Vargas R, Xie W, Evans RM. Genetic profiling defines the xenobiotic gene network controlled by the nuclear receptor pregnane X receptor. Mol. Endocrinol. 2003;17:1268–1282. doi: 10.1210/me.2002-0421. [DOI] [PubMed] [Google Scholar]
- Ross PK, Woods CG, Bradford BU, Kosyk O, Gatti DM, Cunningham ML, Rusyn I. Time-course comparison of xenobiotic activators of CAR and PPARα in mouse liver. Toxicol. Appl. Pharmacol. 2009;235:199–207. doi: 10.1016/j.taap.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ, Venkataramanan R, Cai H, Sinal CJ, Gonzalez FJ, Schuetz JD. Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome P450. J. Biol. Chem. 2001;276:39411–39418. doi: 10.1074/jbc.M106340200. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y-W, Sanyal S, Kim H-J, Won DH, An J-Y, Amano T, Zavacki AM, Kwon H-B, Shi Y-B, Kim W-S, Kang H, Moore DD, Choi H-S. FOR, a novel orphan nuclear receptor related to farnesoid X receptor. J. Biol. Chem. 2002;277:17836–17844. doi: 10.1074/jbc.M111795200. [DOI] [PubMed] [Google Scholar]
- Shan L, Vincent J, Brunzelle JS, Dussault I, Lin M, Ianculescu I, Sherman MA, Forman BM, Fernandez EJ. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol. Cell. 2004;16:907–917. doi: 10.1016/j.molcel.2004.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatter JG, Cheng O, Cornwell PD, de Souza A, Rockett J, Rushmore T, Hartley D, Evers R, He Y, Dai X, Hu R, Caguyong M, Roberts CJ, Castle J, Ulrich RG. Microarray-based compendium of hepatic gene expression profiles for prototypical ADME gene-inducing compounds in rats and mice in vivo. Xenobiotica. 2006;36:902–937. doi: 10.1080/00498250600861694. [DOI] [PubMed] [Google Scholar]
- Song C, Kokontis JM, Hiipakka RA, Liao S. Ubiquitous receptor: a receptor that modulates gene activation by retinoic acid and thyroid hormone receptors. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10809–10813. doi: 10.1073/pnas.91.23.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. Regulation of a xenobiotics sulfonation cascade by nuclear pregnane X receptor (PXR) Proc. Natl. Acad. Sci. U.S.A. 2002;99:13801–13806. doi: 10.1073/pnas.212494599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz ACU, Renaud J-P, Moras D. Binding of ligands and activation of transcription by nuclear receptors. Annu. Rev. Biophys. Biomol. Struct. 2001;30:329–359. doi: 10.1146/annurev.biophys.30.1.329. [DOI] [PubMed] [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha J-Y, Repa JJ, Kliewer SA, Xu HE. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol. Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Suzuki N, Srivastava AS, Kurokawa T. Identification of cDNAs encoding two subtypes of vitamin D receptor in flounder, Paralichthys olivaceus. Biochem. Biophys. Res. Commun. 2000;270:40–45. doi: 10.1006/bbrc.2000.2378. [DOI] [PubMed] [Google Scholar]
- Svensson S, Östberg T, Jacobsson M, Norström C, Stefansson K, Hallén D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L. Crystal structure of the heterodimeric complex of LXRα and RXRβ ligand-binding domains in a fully agonistic conformation. EMBO J. 2003;22:4625–4633. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb MM, Sun A, Zhou C, Grun F, Errandi J, Romero K, Pham H, Inoue S, Mallick S, Lin M, Forman BM, Blumberg B. Vitamin K2 regulation of bone homeostasis is mediated by the steroid and xenobiotic receptor SXR. J. Biol. Chem. 2003;278:43919–43927. doi: 10.1074/jbc.M303136200. [DOI] [PubMed] [Google Scholar]
- Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-Bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol. Cell. Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol. Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Une M, Hoshita T. Natural occurrence and chemical synthesis of bile alcohols, higher bile acids, and short side chain bile acids. Hiroshima J. Med. Sci. 1994;43:37–67. [PubMed] [Google Scholar]
- Vanhooke JL, Benning MM, Bauer CB, Pike JW, DeLuca HF. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. 2004;43:4101–4110. doi: 10.1021/bi036056y. [DOI] [PubMed] [Google Scholar]
- Verma S, Tabb MM, Blumberg B. Activation of the steroid and xenobiotic receptor, SXR, induces apoptosis in breast cancer cells. BMC Cancer. 2009;9:3. doi: 10.1186/1471-2407-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voogt PA, van Rheenan JWA. On the sterols of some ascidians. Arch. Int. Physiol. Biochim. 1975;83:563–572. doi: 10.3109/13813457509071400. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Maglich JM, Moore LB, Wisely GB, Noble SM, Davis-Searles PR, Lambert MH, Kliewer SA, Redinbo MR. 2.1 Å crystal structure of human PXR in complex with the St. John's wort compound hyperforin. Biochemistry. 2003;42:1430–1438. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- Whitfield GK, Dang HTL, Schluter SF, Bernstein RM, Bunag T, Manzon LA, Hsieh G, Dominguez CE, Youson JH, Haussler MR, Marchalonis JJ. Cloning of a functional vitamin D receptor from the lamprey (Petromyzon marinus), an ancient vertebrate lacking a calcified skeleton and teeth. Endocrinology. 2003;144:2704–2716. doi: 10.1210/en.2002-221101. [DOI] [PubMed] [Google Scholar]
- Williams S, Bledsoe RK, Collins JL, Boggs S, Lambert MH, Miller AB, Moore J, McKee DD, Moore L, Nichols J, Parks D, Watson M, Wisely B, Willson TM. X-ray crystal structure of the liver X β ligand binding domain: regulation by a histidine-tryptophan switch Biol. J. Chem. 2003;278:27138–27143. doi: 10.1074/jbc.M302260200. [DOI] [PubMed] [Google Scholar]
- Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, Moore JT. A structural basis for constitutive activity in the human CAR/RXRα heterodimer. Mol. Cell. 2004;16:919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Xue Y, Moore LB, Orans J, Peng L, Bencharit S, Kliewer SA, Redinbo MR. Crystal structure of PXR-estradiol complex provides insights into endobiotic recognition. Mol. Endocrinol. 2007;21:1028–1038. doi: 10.1210/me.2006-0323. [DOI] [PubMed] [Google Scholar]
- Yagi K, Satou Y, Mazet F, Shimeld SM, Degnan B, Rokhsar D, Levine M, Kohara Y, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. III. Genes for Fox, ETS, nuclear receptors and NFkB. Dev. Genes Evol. 2003;213:235–244. doi: 10.1007/s00427-003-0322-z. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Ranade A, Venkataramanan R, Strom S, Chupka J, Ekins S, Schuetz E, Bachmann K. A comprehensive in vitro and in silico analysis of antibiotics that activate pregnane X receptor and induce CYP3A4 in liver and intestine. Drug Metab. Dispos. 2008;36:1689–1697. doi: 10.1124/dmd.108.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Murata M, Inaba K, Morisawa M. A chemoattractant for ascidian spermatozoa is a sulfated steroid. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14831–14836. doi: 10.1073/pnas.242470599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Xie W, Krasowski MD. PXR: a xenobiotic receptor of diverse function implicated in pharmacogenetics. Pharmacogenomics. 2008a;9:1695–1709. doi: 10.2217/14622416.9.11.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J, Gibbs RA, Weinstock G, Wheeler DA. Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004;14:580–590. doi: 10.1101/gr.2160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZD, Cayting P, Weinstock G, Gerstein M. Analysis of nuclear receptor pseudogenes in vertebrates: how the silent tell their stories. Mol. Biol. Evol. 2008b;25:131–143. doi: 10.1093/molbev/msm251. [DOI] [PubMed] [Google Scholar]
- Zhou C, Assem M, Tay JC, Watkins PB, Blumberg B, Schuetz EG, Thummel KE. Steroid and xenobiotic receptor and vitamin D receptor crosstalk mediates CYP24 expression and drug-induced osteomalacia. J. Clin. Invest. 2006a;116:1703–1712. doi: 10.1172/JCI27793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Tabb MM, Nelson EL, Grun F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE, Blumberg B. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J. Clin. Invest. 2006b;116:2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Verma S, Blumberg B. The steroid and xenobiotic receptor (SXR), beyond xenobiotic metabolism. Nucl. Recept. Signal. 2009;7:e001. doi: 10.1621/nrs.07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.