Abstract
Histone deacetylase (HDAC) proteins are promising targets for cancer treatment, as shown by the recent FDA approval of the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA, Vorinostat,) for the treatment of cutaneous T-cell lymphoma. To identify additional potent inhibitors and characterize HDAC mutant proteins, there is interest to develop an inexpensive screening method dependent on native substrates. Here, we report the first yeast-based gene reporter screen dependent on the yeast Rpd3, which is a homolog of human class I HDAC proteins. The screen was sensitive to an inactive Rpd3 mutant and various inhibitors in qualitative, agar-based and quantitative, solution-phase formats. Interestingly, inclusion of the lytic enzyme zymolyase enhanced the inhibitor sensitivity of the screen. The gene reporter screen provides a tool to screen Rpd3 mutants and inhibitors of class I HDAC proteins.
Keywords: Histone Deacetylase, HDAC inhibitors, Gene Reporter Screen
1. Introduction
Histone Deacetylase (HDAC) proteins catalyze the removal of acetyl groups from acetylated lysines on histone and other protein substrates. The acetylation state of specific lysine residues in histone proteins can alter the chromatin structure and influence eukaryotic gene transcription.1 Due to their fundamental role in gene expression, HDAC proteins are promising targets for cancer treatment, as shown by the recent FDA approval of the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA, Vorinostat,) for the treatment of cutaneous T-cell lymphoma.2 In addition, HDAC inhibitors are currently in clinical trials to treat various cancers.3–5 Consistent with their clinical effects, inhibitors of HDAC proteins suppress tumor cell proliferation, induce cell differentiation, and regulate the expression of crucial genes associated with cancer.6 HDAC inhibitor drugs represent a promising next generation of anti-cancer therapeutics.
Eighteen human HDAC proteins are known and divided into four classes on the basis on mechanism, phylogenetic analysis, and similarity to yeast HDAC proteins.7 Human HDAC1, HDAC2, HDAC3, and HDAC8 are members of the class I and are homologous to yeast Rpd3 protein.8–12 Human HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10 belong to class II and are homologues of yeast HDA1 protein.13–15 The class III proteins include the NAD-dependent Sirtuin proteins, which are related to Sir2 in yeast. HDAC11 is the only member of class IV in humans and is predicted to have diverged very early in evolution.7,12 Proteins in class I, II, and IV display considerable sequence similarity in their catalytic sites, which suggests that they operate via similar metal-dependent mechanisms.16–18 Most inhibitors discussed in this work nonspecifically target all eleven metal-dependent HDAC isoforms.19,20
Several assays are currently available to screen HDAC inhibitors.21 The most widely used screen is a commercially available fluorimetric assay that is a convenient and high throughput alternative to protocols utilizing radiolabeled substrates or HPLC methods.22 However, recent reports have suggested that the non-native fluorophore on the peptide substrate in the fluorimetric assay can lead to false positive hits during screening. Specifically, the ability for resveratrol and other similar compounds to activate the deacetylase activity of the Sirtuin proteins was dependent on the presence of the fluorophore.23,24 Further, biophysical evidence revealed that the small molecules interacted directly with the fluorophore-labeled peptide substrate.25 As a result of these reports, development of HDAC screening tools utilizing non-radioactive, native substrates is desirable.
In addition to small molecule screening, the fluorimetric assay as been used to monitor HDAC protein activity. Because structural information on HDAC proteins is limited,16,17,26–28 site directed mutagenesis has been the used extensively to identify the catalytic active site of many HDAC isoforms. For example, mutagenesis of HDAC1 has revealed residues critical for catalysis.29,30 The availability of a rapid screening platform to identify active/inactive mutant forms of the HDAC proteins would facilitate their characterization.
Here we report the development of a yeast-based gene reporter system for the screening of HDAC mutant proteins and small molecule inhibitors. Yeast-based screening was used previously to identify small molecule inhibitors of the yeast Sir2 protein.31,32 The Sir2 inhibitors identified in the screens have been used widely to inhibit the human Sirtuin proteins,33,34 which demonstrates the utility of yeast based screens to identify HDAC inhibitors. Based on the success of the Sir2 inhibitor screen, we envisioned developing a similar yeast screen targeting the metal-dependent yeast HDAC proteins. Yeast Rpd3 shares 60% identity to human class I proteins.7 With its high sequence similarity, we rationalize that an Rpd3-dependent yeast assay would provide a facile tool for the screening of HDAC protein mutants or inhibitors. Like with the Sir2 inhibitor assays, the yeast Rpd3 screen can be coupled with the fluorimetric assay to provide a powerful approach to mutant and inhibitor characterization.
2. Materials and methods
2.1 Yeast strains and plasmids
The FT5 Δrpd3::HIS3 yeast strain and pJK1621 reporter plasmid used in this study were generous gifts of Dr. Kevin Struhl.35 YEplac112-Rpd3-LexA-FLAG and YEplac112-Rpd3H150AH151A-LexA-FLAG expression plasmids were generated as follows: Rpd3-LexA-FLAG or Rpd3H150AH151A-LexA-FLAG PCR fragments were generated from YEplac112-Rpd3-LexA or YEplac112-Rpd3H150AH151A-LexA (from Dr. Kevin Struhl)35 using the CGGGGTACCATGGTATATGAAGCAACACCTTTT-GATCCG forward and CTGGTTAATTATAAATTAGCCATGGCCTTTATCATCATCAT-CTTTATAATCCAGCCAGTCGCCGTTGCG reverse primers, which contain the FLAG epitope. Generated PCR fragments were cloned into Nco1 digested YEp112–FLAG (from Dr. Kevin Struhl)35 using homologous recombination. Generation of plasmids was confirmed by DNA sequencing.
2.2 β-galactosidase activity screen
For the agar assay, FT5 Δrpd3::HIS3 yeast cells transformed with the pJK1621 reporter alone, the pJK1621 reporter and YEplac112-Rpd3-LexA-FLAG expression plasmid, or the pJK1621 reporter and YEplac112-Rpd3H150/H151A-LexA-FLAG expression plasmid were plated on selection media (CSM-Ura-His for pJK1621 alone or CSM-Trp-Ura-His for the others) containing 0.5% dextrose with or without small molecule. Cells were grown for 48 h at 30 °C and then overlaid with X-gal solution (0.25 mg/mL 5-Bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Sigma), 6% DMF, 0.1% SDS, 0.5 M KPO4 pH 7.0). Blue color development was monitored visually, with best results observed in less than 4 hours at 30°C.
For the solution phase assay, the overnight cultures (described above) were diluted to 0.1 absorbance at OD600 with the appropriate media containing 0.5% dextrose (1 mL total volume) and then incubated without (2% DMSO control) or with small molecule for 6 hours. Small molecule final concentrations are as listed in the Figures. The OD600 was then measured. Cells were harvested by centrifugation from equal culture volumes (typically 500 μL to 800 μL) and then permeabilized by vortexing for 30 seconds in 500 μL of permealization buffer (60mM Na2HPO4, 40mM NaH2PO4, 10mM KCl, 2mM MgSO4, 0.27% β-mercaptoethanol, pH 7.0) containing 25 μL of 0.1% SDS and 20 μl of chloroform. The β-gal substrate 2-Nitrophenyl β-D-galactopyranoside (ONPG, Sigma, 100 μL of 4 mg/mL solution) was added to each solution and the reaction was incubated at 30°C for not more than 1h. Once yellow color developed, the reaction was quenched by adding 100 μL of 1M Na2CO3. The reactions were centrifuged to remove cell debris and absorbance at OD420 was measured of the soluble fraction. Normalized β-galactosidase activity units were calculated using the following equation, Units = (OD420 × 1000)/(OD600 × T × V), where T= ONPG incubation time in min and V= the volume in μL of the soluble fraction used to obtain the OD420.36,37
2.3 Immunoprecipitation and histone deacetylase assay
Whole cell extracts were prepared from FT5 Δrpd3::HIS3 yeast cells transformed with the pJK1621 reporter alone, the pJK1621 reporter and YEplac112-Rpd3-LexA-FLAG expression plasmids, or the pJK1621 reporter and YEplac112-Rpd3H150/H151A-LexA-FLAG expression plasmids. Specifically, the yeast were grown overnight with shaking at 30 °C in 5 mL of appropriate selection media (CSM-Ura-His or CSM-Trp-Ura-His). After centrifugation to collect the cells, glass beads equal to the packed volume of the cell pellet were added and the cell/glass bead mixture was resuspended in 1 mL of yeast lysis buffer (20 mM HEPES pH 7.9, 150 mM, NaCl, 10 mM, 10% glycerol) with 1X protease inhibitor cocktail set V (Calbiochem). Cells were vortexed for 30 seconds and kept on ice for additional 45 seconds. This cycle was repeated 6 to 8 times to complete the lysis. After the last vortex cycle, the sample was incubated on ice for 2 min. The extract was collected and centrifuged to remove cell debris. The soluble fraction was either used immediately or stored at −80 °C.
Expressed wild-type and mutant FLAG-tagged Rpd3 proteins were immunoprecipitated from the whole cell extracts (200 μg of total protein) by incubating with 15 μL of anti-FLAG agarose beads (Sigma) in buffer F (20 mM HEPES at pH 7.6, 1 mM EDTA, 150 mM NaCl, 20% glycerol) at 4 °C for two hours. The deacetylase activity of immunoprecipitated proteins was measured using the Fluor de Lys™ fluorescence activity assay kit (Biomol), as described.38 Briefly, the immunoprecipitates were resuspended in HDAC assay buffer (25 μL; 50 mM Tris/Cl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2) containing 2% DMSO alone or 200 μM TSA in 2% DMSO. After 10 minutes of incubation, the Fluor de Lys™ substrate (100 μM final concentration) was added in a total 50 μL volume and incubated at 37 °C for 45 min with shaking. Developer (50 μL) was added and the reaction was incubated for 5 min with shaking. The fluorescence signal at 465 nm was measured after excitation at 360 nm using a GENios Plus plate-reading fluorimeter (Tecan). The percentage HDAC activity was determined by dividing the fluorescence intensity of the reaction with mutant Rpd3, no protein, or HDAC1 with that of wild type Rpd3. For western blot analysis, immunoprecipitated proteins were separated by 10% SDS-PAGE, transferred to a PVDF membrane (Immobilon PSQ), and probed with LexA antibodies (Santa Cruz).
3. Results
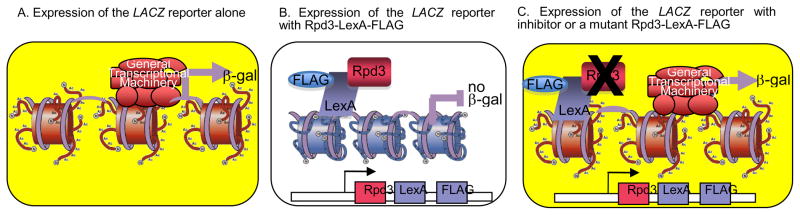
The HDAC-dependent inhibitor screen is based upon a gene reporter assay in yeast reported by the Struhl laboratory.35 As shown in Figure 1, the assay monitors expression of the LACZ gene from a reporter controlled by an intact CYC1 promoter and 4 LexA DNA binding sites. Because CYC1 promotes a basal level of transcription, cells transformed with the reporter alone express the LACZ gene (Figure 1A). The HDAC-dependent screen involves expressing the yeast HDAC protein Rpd3 as a LexA-FLAG fusion protein (Rpd3-LexA-FLAG). In the presence of the reporter, LexA recruits Rpd3 to the LACZ gene via binding the LexA DNA binding sites, which results in deacetylation of the nucleosomal histones and reduction in LACZ gene expression (Figure 1B). In contrast, if the Rpd3 in the LexA-FLAG fusion is catalytically inactive or incubated with a small molecule inhibitor, the nucleosomal histones will remain acetylated and accessible to the transcription machinery, causing expression of LACZ gene (Figure 1C). The LACZ gene encodes the enzyme β-galactosidase (β-gal), which can hydrolyze the substrate 5-Bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal), resulting in a blue color. Therefore, by observing the color of the yeast cell, the screen will monitor inhibition of Rpd3 activity by a small molecule or mutation.
Fig. 1.
Schematic diagram of yeast-based LACZ gene reporter screen. (A) The promoter displays basal LACZ expression and β-gal activity, resulting in a colored cell. (B) The presence of the expression plasmid for the Rpd3-LexA-FLAG fusion (bottom construct) results in the repression of LACZ gene expression due to deacetylation by Rpd3, resulting in minimal β-gal activity and color. (C) Inactivation of the deacetylase activity of the Rpd3-LexA-FLAG fusion due to mutation or inhibition results in expression of the LACZ gene, β-gal activity, and a colored cell.
3.1 Qualitative HDAC-dependent Screen
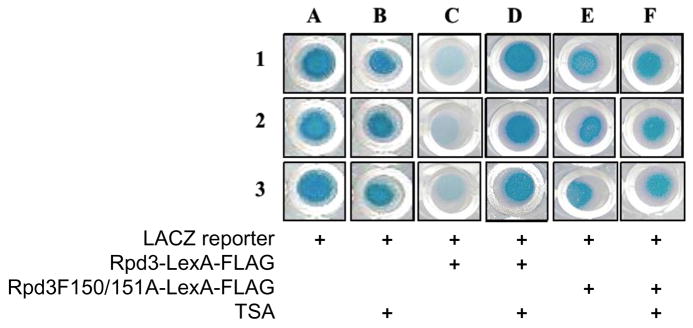
To test the yeast gene reporter assay, the LACZ reporter plasmid, pJK1621,35 was transformed alone or with an expression plasmid encoding Rpd3-LexA-FLAG into the FT5 Δrpd3::HIS3 yeast strain. The RPD3 gene has been deleted in the yeast strain to reduce possible complications from endogenous Rpd3. After growth of the cells on agar, the β-gal activity due to LACZ gene expression was measured qualitatively by overlaying the colony with a solution containing the X-Gal substrate. As shown in Figure 2, yeast transformed with the LACZ reporter plasmid alone showed a high level of β-gal activity and a blue colored cell due to basal LACZ gene expression (Figure 2, column A). As expected, yeast cells transformed with the Rpd3-LexA-FLAG fusion showed a minimum level of β-gal activity and reduced blue color (Figure 2, column C), indicating that recruitment of Rpd3 suppresses the expression of the reporter gene. To assure that the HDAC activity of Rpd3 is responsible for the reduced LACZ expression, cells transformed with the reporter and a catalytically inactive Rpd3H150/151A-LexA-FLAG fusion were also tested. The expectation was that the inactive mutant would not influence LACZ gene expression. As expected, cells containing the catalytically inactive Rpd3H150/151A-LexA-FLAG fusion maintained similar levels of β-gal activity compared to the reporter alone, as observed by the similar blue color (Figure 2, column E versus A). In total, these results suggest that the yeast gene reporter is dependent on Rpd3 activity, which is consistent with previous results from the Struhl lab.35
Fig. 2.
The LACZ reporter plasmid, pJK1621, was transformed with or without wild type Rpd3 or catalytically inactive Rpd3H150/151A mutant as LexA-FLAG fusions into the FT5 Δrpd3::HIS3 yeast strain. Transformed cells were grown with or without TSA (100 μM) for 48 hr in 96-well plates and were overlaid with the X-Gal substrate. β-gal activity was observed qualitatively by blue color formation after 3h of incubation. The components of each experiment are indicated below each column. The experiment was carried out in triplicate (rows 1, 2 and 3).
To establish the sensitivity of the screen to small molecule inhibition, the cells containing the LACZ reporter without or with either Rpd3-LexA-FLAG or Rpd3H150/151A-LexA-FLAG were incubated with the HDAC inhibitor trichostatin A (TSA) prior to X-Gal overlay. The expectation was that the TSA would significantly influence LACZ expression in the cells containing the Rpd3-LexA-FLAG fusion, but not the others. TSA-treated cells expressing the LACZ reporter and wild type Rpd3-LexA-FLAG showed elevated β-gal activity and blue color (Figure 2, column D) compared to the cells without TSA (Figure 2, column C). In addition, the extent of blue color was similar to that observed with the reporter alone (Figure 2, lane A) or with the inactive Rpd3-LexA-FLAG fusion (Figure 2, column E). The data indicate that the expression of LACZ reporter gene is sensitive to TSA. As critical controls, TSA-treatment had no effect on cells expressing LACZ reporter plasmid with or without the inactive Rpd3-LexA-FLAG fusion (Figure 2, columns B and F), indicating that the effect of the endogenous HDAC activity on the expression of the reporter is negligible. In total, the screen is TSA-dependent, suggesting that it is appropriate for small molecule inhibitor screening.
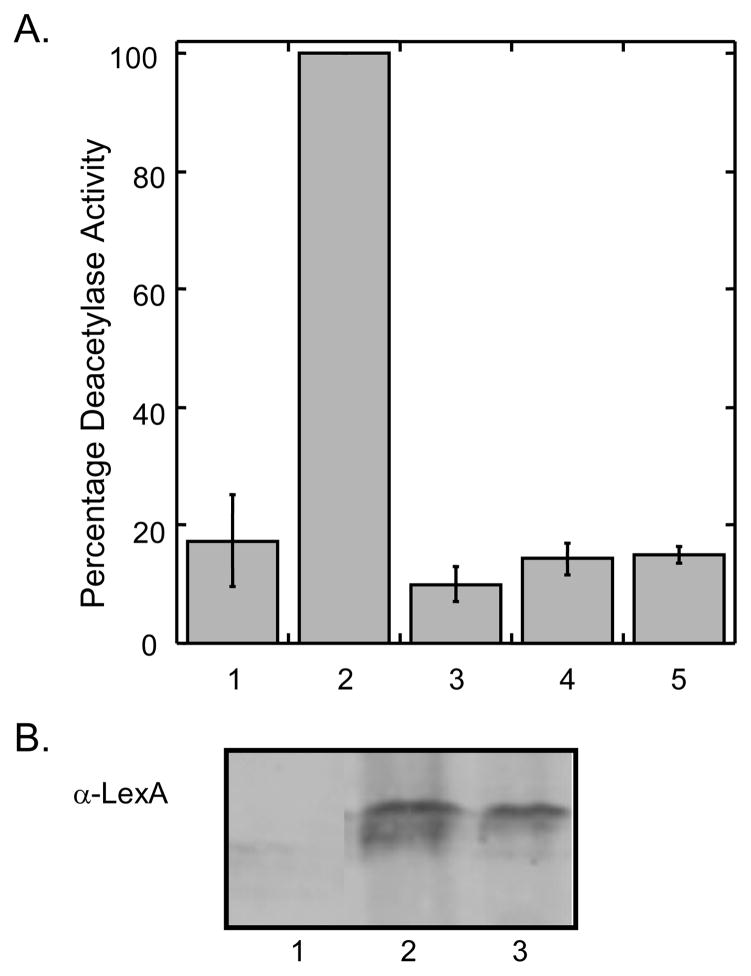
As a critical control, the enzymatic activities of the Rpd3-LexA-FLAG fusion proteins used in the yeast-based screen were assessed. The fusion proteins were immunoprecipitated using the attached C-terminal FLAG tag and enzymatic activities were monitored using the commercially available fluorimetric HDAC assay (Biomol). As expected, the immunoprecipitated Rpd3-LexA-FLAG fusion displayed significant HDAC activity (Figure 3, column 2), while the Rpd3H150/151A-LexA-FLAG fusion displayed activity comparable to the no protein control (Figure 3, column 1 and 4). In addition, the TSA-sensitivity of the immunoprecipitated proteins was assessed. As expected, the immunoprecipitated Rpd3-LexA-FLAG fusion was inactivated by TSA (Figure 3, column 3), while the inactive mutant was unaffected (Figure 3, column 5). The immunoprecipitation experiment confirms that the yeast-based screen is dependent on active Rpd3 and sensitive to inhibitors.
Fig. 3.
A. Deacetylase activity of Rpd3-LexA-FLAG fusions. Rpd3 (column 2 and 3) or Rpd3H150/151A (column 4 and 5) were expressed in FT5 Δrpd3::HIS3 yeast strain as LexA-FLAG fusion proteins and immunoprecipitated using their C-terminal FLAG tag and anti-FLAG-agarose resin (Sigma). A no protein control was included in column 1. The immunoprecipitated proteins were used in fluorescence HDAC assays either in the absence (columns 2 and 4) or presence (columns 3 and 5) of 100 μM trichostatin. The mean percent activities of three independent trials was compared with that of the wild-type protein (100%-column 2) with standard error shown (Table S1). B. The immunoprecipitated proteins were separated by SDS-PAGE and probed with a LexA antibody (Santa Cruz) to ensure protein expression. Lane 1: no protein control; lane 2: Rpd3-LexA-FLAG; lane 3: Rpd3H150/151A-LexA-FLAG.
3.2 Quantitative HDAC-dependent Screen
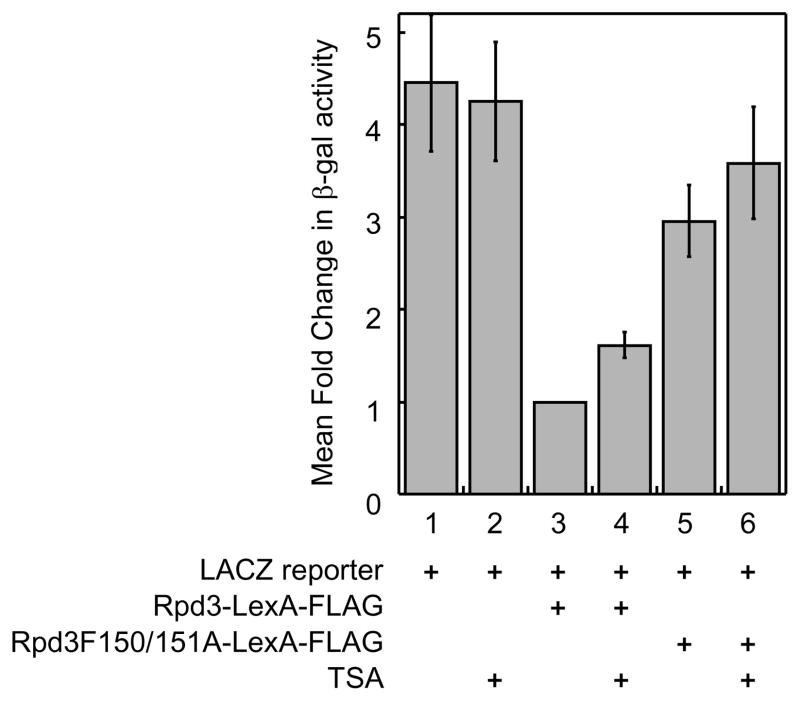
We next tested the possibility of quantitatively monitoring Rpd3-LexA-FLAG activity using the β-gal substrate 2-nitrophenyl β-D-galactopyranoside (ONPG) by detecting the production of yellow color at 420nm. In this case, the assay was performed in liquid format, making it amenable for larger scale screening efforts. Consistent with the agar-based X-Gal format, yeast cells transformed with LACZ reporter plasmid alone showed high levels of β-gal activity (Figure 4, column 1), which was unaffected by TSA-treatment (Figure 4, column 2). The presence of the Rpd3-LexA-FLAG fusion suppressed the expression of LACZ gene, resulting in a 4.4 ± 0.7-fold reduction in β-gal activity (Figure 4, compare columns 1 and 3). In addition, the presence of catalytically inactive Rpd3H150/151A-LexA-FLAG elevated β-gal activity by 3.0 ± 0.4-fold compared to wild type Rpd3-LexA-FLAG (Figure 4, column 5), which was also unaffected by TSA treatment (Figure 4, column 6). We next tested the influence of TSA in the presence of the Rpd3-LexA-FLAG fusion. The β-gal activity was elevated 1.6 ± 0.1-fold after TSA treatment (Figure 4, column 4), indicating that the assay is sensitive to TSA inhibition. Consistent with the X-Gal data, the quantitative analysis confirmed that the yeast gene reporter is Rpd3-dependent and sensitive to inhibitors. We note that the human HDAC1-LexA fusion protein35 produced weak LACZ repression in the ONPG assay (data not shown), which is consistent with the previous reports demonstrating that recombinant human HDAC1 protein from yeast is inactive.39
Fig. 4.
The yeast-based screen is quantitative. The reactions were growth as described in Figure 2 for 6 hr at 30°C and β-gal activity was measured quantitatively by measuring absorbance at 420 nm. Relative fold change in β-gal activity was normalized to the Rpd3-LexA data (set to 1.0) and the mean of three trials is shown with standard error (Table S2).
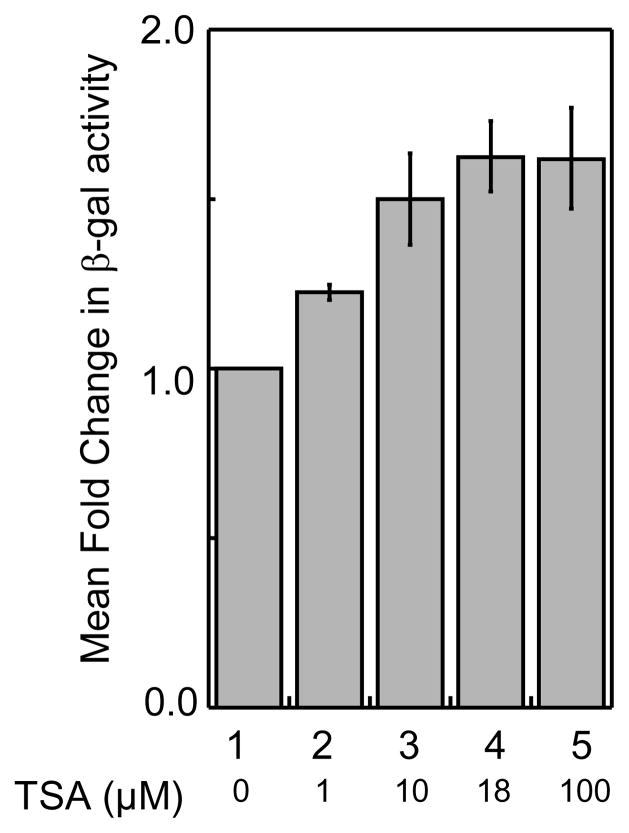
To further explore the screen quantitatively, we assessed the inhibitor dose dependency. Specifically, cells expressing Rpd3-LexA-FLAG were treated with different concentrations of TSA and the β-gal activity was measured using ONPG as the substrate. As shown in Figure 5, TSA showed dose dependent inhibition of HDAC activity. Specifically, while 1μM TSA treatment showed a 1.2 ± 0.03-fold elevated β-gal activity compared to the TSA untreated sample (Figure 5, column 2), 10 μM, 18 μM and 100 μM TSA treatments showed a 1.5 ± 0.1-fold, 1.6 ± 0.1-fold and 1.6 ± 0.1-fold elevated β-gal activity, respectively (Figure 5, columns 3–5). Collectively, data obtained from quantitative analysis suggests that the yeast gene reporter screen is capable of detecting dose-dependent HDAC inhibition.
Fig. 5.
HDAC inhibition by TSA is dose dependent. Transformed yeast cells (see Figure 2) were grown without or with increasing concentrations of TSA and β-gal activity were measured quantitatively as described. Relative fold change in β-gal activity was calculated with respect to TSA-untreated cells and the mean fold change of four trials is shown with standard error (Table S3).
3.3 Optimization of Reporter Screen to Increase the Inhibitor Sensitivity
The β-gal activity could be elevated up to 1.6-fold by inhibiting Rpd3-LexA-FLAG activity with TSA (Figure 4, column 4). However, the presence of catalytically inactive Rpd3H150/151A mutant elevated β-gal activity up to 3.0-fold compared to wild type Rpd3 (Figure 4, column 5). We speculate that the lower levels of β-gal activity observed with TSA treatment compared to the inactive Rpd3 mutant may be due to the small molecule cell permeability issues in yeast cells, as has been previously reported.40,41
The yeast cell envelope is a protecting capsule that plays a major role in controlling the permeability properties of the cell. The cell wall of a yeast cell envelope is remarkably thick (100 to 200 nm) and its major structural constituents are polysaccharides (80–90%), mainly glucans and mannans.42 Both β-2,6 and β-1,3 linked glucans provide strength to the cell wall, forming a microfibrillar network. Digestion of the yeast cell wall by lytic enzymes, such as zymolyase, is necessary for many experimental procedures, including spheroplasting, immunofluorescence, transformation, and protein purification.43–45 The main enzymatic activities of zymolyase are β-1,3 glucanase and β-1,3 glucan laminaripentao-hydrolase, which hydrolyze glucose polymers at β-1,3 glucan linkages, 46 leading to cell wall digestion.
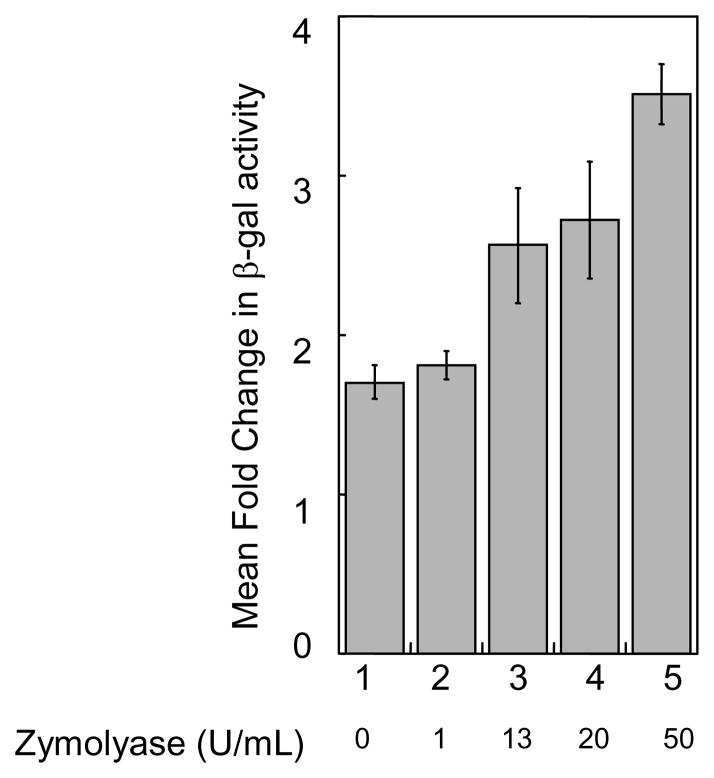
We hypothesized that the weakening of yeast cell wall by treating with zymolyase would facilitate cellular uptake of small molecules, which will augment the HDAC inhibition and β-gal activity. To explore this hypothesis, yeast cells transformed with the LACZ reporter Rpd3-LexA-FLAG fusion were incubated with TSA and increasing amounts of zymolyase prior to measuring β-gal activity. As shown in Figure 6, zymolyase enhanced the β-gal activity of the cells in a dose dependent manner. Specifically, while cells incubated with TSA and 1 U/mL of zymolyase showed the same level of β-gal activity compared to TSA alone (1.8-fold and 1.7-fold, Figure 6, columns 1 and 2), treatment with 13 U/mL or 50 U/mL of zymolyase augmented the β-gal activity to 2.6-fold or 3.5-fold, respectively (Figure 6, columns 3 and 5). Collectively, the increased β-gal activity observed with the presence of zymolyase confirms that the weakening of yeast cell wall facilitates the cellular uptake of small molecules. Importantly, we show that zymolyase treatment increases the sensitivity of the gene reporter screen towards HDAC inhibitors.
Fig. 6.
The influence of zymolyase on TSA dependence. Transformed cells were grown as described in Figure 2 along with the indicated amounts of zymolyase (Zymo Research, Inc.) before β-gal activity was measured as described. An untreated control was performed at each zymolyase concentration to determine the mean fold change, although only the control for the reaction without zymolyase is shown in lane 1. Mean relative fold change of at least three trials is shown with standard error (Table S4).
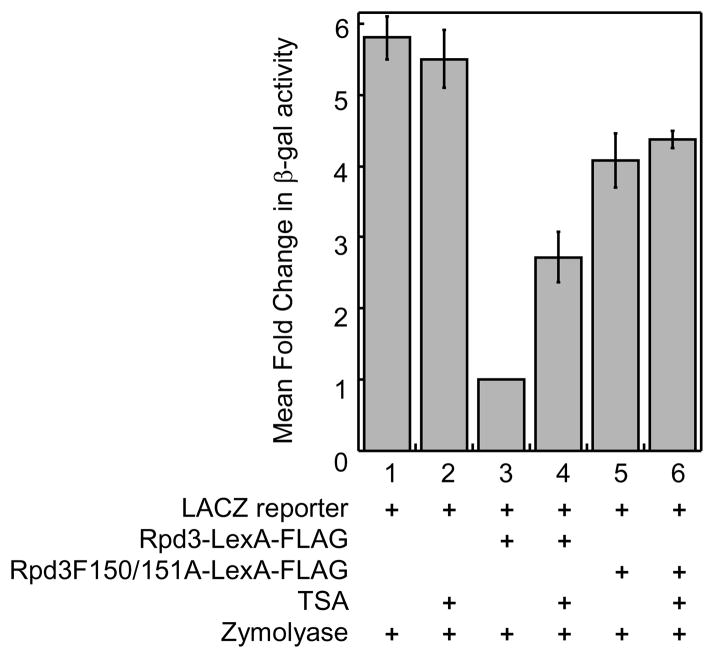
To further evaluate the optimized yeast gene reporter screen, the screen with proper controls was repeated in the presence of zymolyase (20 U/mL). Specifically, the LACZ reporter plasmid, pJK1621, was transformed with or without the LexA-FLAG fused to wild type Rpd3 or catalytically inactive Rpd3 mutant into FT5 Δrpd3::HIS3 yeast strain and the β-gal activity was measured quantitatively. As shown in Figure 7, cells transformed with LACZ reporter plasmid alone showed the same high levels of β-gal activity in the presence or absence of TSA (Figure 7, columns 1 and 2), consistent with the data obtained without zymolyase (Figure 4). While recruitment of Rpd3-LexA-FLAG reduced β-gal activity (Figure 7, column 3), inhibition by TSA restored that activity by 2.7-fold (Figure 7, column 4). It is important to note that only 1.6-fold elevation of β-gal activity was observed with the treatment of 100 μM TSA alone (Figure 4), indicating that zymolyase treatment results in greater sensitivity to TSA. Interestingly, the presence of catalytically inactive Rpd3 mutant-LexA-FLAG elevated β-gal activity to 4.1-fold (Figure 7, column 5), which is greater than the 3.0-fold increase observed without zymolyase (Figure 4). The enhanced β-gal activity observed in all reactions with zymolyase (compare Figures 4 and 7) suggests that the zymolyase facilitates the permeability of yeast cell and release of β-galactosidase.
Fig. 7.
Zymolyase enhances the small molecule sensitivity of the screen. Transformed cell (see Figure 2) were grown with the presence of zymolyase (20 U/mL) and with or without TSA (50 μM) before β-gal activity was measured quantitatively as described. Mean relative fold change of at least three trials is shown with standard error (Table S5).
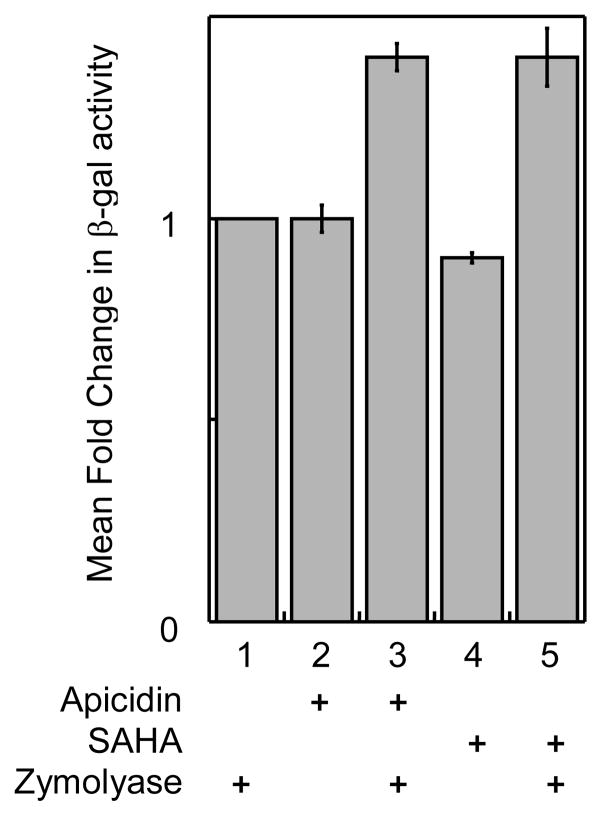
To establish the yeast gene reporter screen as a general tool, we tested two other known HDAC inhibitors, apicidin and SAHA. Unlike TSA, apicidin (100 μM, Figure 8, column 2) and SAHA (2 mM, Figure 8, column 4) did not show the expected elevated levels of β-gal activity compared to untreated cells (Figure 8, column 1). We wondered if issues of cell permeability explained the lack of β-gal activity,47,48 because apicidin and SAHA are potent nM inhibitors of class I HDAC proteins.19 To test the issue of cell permeability, zymolyase was included during inhibitor incubation. In the presence of zymolyase, elevated levels of β-gal activity were observed with apicidin (1.4 ± 0.03-fold, Figure 8, column 3) and SAHA (1.4 ± 0.07-fold, Figure 8, column 5). These results confirm that zymolyase treatment enhances the sensitivity of the yeast gene reporter screen to small molecule inhibitors. Collectively, the data indicate that the yeast gene reporter screen is well suited for screening HDAC inhibitors.
Fig. 8.
Inhibition of Rpd3-LexA-FLAG activity by Apicidin and SAHA. Transformed cells (see Figure 2) were grown with or without Apicidin (100 μM) or SAHA (2 mM) in the presence or absence of zymolyase (100 U/mL) before β-gal activity was measured as described. Mean relative fold change of at least three trials is shown with standard error (Table S6).
4. Discussion
Yeast assays have been used successfully as initial screening tools for the discovery of small molecule modulators.49–51 Most notably, yeast screens were used to identify small molecule inhibitors of the NAD-dependent Sirtuin HDAC proteins.31,32,52–54 Despite the utility, no yeast based assay against metal-dependent HDAC proteins has been reported. We report here development of a yeast-based gene reporter screen centered on the class I yeast homolog Rpd3. The screen is dependent on Rpd3 deacetylase activity and sensitive to the small molecule inhibitors TSA, apicidin, and SAHA. The screen was validated in both a qualitative, agar-based format and a quantitative, solution-based format, making it practical for many applications. In total, the yeast-based gene reporter screen is a tool for testing small molecule inhibitors and mutant HDAC proteins for activity.
The yeast assay adds to the available tools for screening HDAC small molecule inhibitors. While the fluorimetric assay widely used in the field is easy to use and high throughput, it suffers from the use of non-native substrates, which can result is false positives, and high cost (from $1 to $4 per reaction).38 As a result, there is interest to develop alternative HDAC inhibitor screening methods that use HDAC activity from inexpensive lysates or cells and involve native substrates. The reported yeast screen employs native histone substrates in the cell-based assay, which will avoid artifacts from non-natural peptide substrates. In addition, by relying on commercially available and relatively inexpensive reagents, we estimate the assay to cost several cents per reaction. With the yeast assay available for initial hit identification, the fluorimetric assay can be used to confirm hits and determine IC50 values towards discovery of new HDAC inhibitors from small molecule libraries.
One limitation of the yeast-based screen is the difficulty for small molecules to penetrate the yeast cell wall. In fact, the problem of cell permeability likely explains the relative high concentrations of inhibitors required to influence β-gal activity. For example, while TSA inhibits Rpd3 and human class I HDAC proteins at nM concentrations,19,55 μM concentrations of TSA were required to influence β-gal activity in the screen (see Figure 4). Despite the issue of cell permeability, yeast-based assays have been used previously to screen libraries of small molecules.31,32,49,53,56 In each of these cases, μM concentrations of small molecules were used successfully. However, methods to augment the cell permeability of yeast to small molecules would be useful. To increase the sensitivity of yeast gene reporter screen to small molecules, we tested zymolyase. Because zymolyase is routinely used for partial digestion of yeast cell wall, we wondered if including zymolyase in the screen would augment screening sensitivity. We found that cells treated with TSA and zymolyase showed an increased level of β-gal activity compared to that of TSA alone (2.7-fold vs. 1.6-fold, respectively). In addition, we found that zymolyase enhanced detection of poorly cell permeable inhibitors, such as apicidin and SAHA. To our knowledge, this is the first report of a lytic enzyme enhancing the sensitivity of a yeast-based reporter screen to small molecule inhibitors. We intend to explore further the general use of zymolyase to overcome the cell permeability concern of yeast-based assays.
The yeast reporter screen is also appropriate for screening Rpd3 mutants for catalytic activity. Identification of inactive mutants has been helpful to characterize the residues involved in activity for yeast and human HDAC proteins.30,35,57,58 The availability of a gene reporter screening platform will facilitate the identification of additional mutants. For example, catalytically active mutants of kinases have been exploited as tools in cell biology experiments and inhibitor identification studies.59,60 In addition, inactive mutants have been used to create dominant negative conditions.61 A screening platform for HDAC protein mutants can be used to create similar cell biology tools and inhibitor design paradigms.
In summary, we developed a yeast gene reporter screen dependent on histone deacetylase Rpd3 activity that is sensitive to inhibitors. To our knowledge, the screen is the first yeast-based method sensitive to class I HDAC proteins. The yeast gene reporter screen relies on native substrates in a cell based assay and can be coupled with the commercially available fluorimetric assay to rigorously characterize identified inhibitors with IC50 values. In addition, the screen can be used to test Rpd3 mutants for enzymatic activity. The availability of multiple complimentary tools for HDAC activity screening will facilitate development of new genetic and chemical tools targeting HDAC proteins.
Supplementary Material
Acknowledgments
We thank NIH (GM067657) for funding, Dr. Kevin Struhl for yeast and plasmid reagents, Dr. David Kadosh for helpful technical advise, Fabiola Bittencourt for experimental assistance, and Emily Aubie, Sun Choi, and Geetha Padige for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grunstein M. Nature. 1997;389:349. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Grant S, Easley C, Kirkpatrick P. Nat Rev Drug Discov. 2007;6:21. doi: 10.1038/nrd2227. [DOI] [PubMed] [Google Scholar]
- 3.Dokmanovic M, Clarke C, Marks PA. Mol Cancer Res. 2007;5:981. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 4.Ryan QC, Headlee D, Acharya M, Sparreboom A, Trepel JB, Ye J, Figg WD, Hwang K, Chung EJ, Murgo A, Melillo G, Elsayed Y, Monga M, Kalnitskiy M, Zwiebel J, Sausville EA. J Clin Oncol. 2005;23:3912. doi: 10.1200/JCO.2005.02.188. [DOI] [PubMed] [Google Scholar]
- 5.Kelly WK, O’Connor OA, Krug LM, Chiao JH, Heaney M, Curley T, MacGregore-Cortelli B, Tong W, Secrist JP, Schwartz L, Richardson S, Chu E, Olgac S, Marks PA, Scher H, Richon VM. J Clin Oncol. 2005;23:3923. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mai A, Masse S, Rotili D, Cerbara I, Valente S, Pezzi R, Simeoni S, Ragno R. Medicinal Research Reviews. 2005;25:261. doi: 10.1002/med.20024. [DOI] [PubMed] [Google Scholar]
- 7.Gregoretti I, Lee YM, Goodson HV. J Mol Biol. 2004;338:17. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Taunton J, Hassig CA, Schreiber SL. Science. 1996;272:408. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 9.Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Proc Natl Acad Sci USA. 1996;93:12845. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang WM, Yao YL, Sun JM, Davie JR, Seto E. J Biol Chem. 1997;272:28001. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 11.Van den Wyngaert I, de Vries W, Kremer A, Neefs J, Verhasselt P, Luyten WHML, Kass SU. FEBS Lett. 2000;478:77. doi: 10.1016/s0014-5793(00)01813-5. [DOI] [PubMed] [Google Scholar]
- 12.Gao L, Cueto MA, Asselbergs F, Atadja P. J Biol Chem. 2002;277:25748. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 13.Grozinger CM, Hassig CA, Schreiber SL. Proc Natl Acad Sci USA. 1999;96:4868. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao HY, Downes M, Ordentlich P, Evans RM. Genes Dev. 2000;14:55. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Marks PA, Rifkind RA, Richon VM. Proc Natl Acad Sci USA. 2001;98:10572. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, Francesco RD, Gallinari P, Steinkuhler C, Marco SD. Proc Natl Acad Sci U S A. 2004;101:15064. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang BC, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structure. 2004;12:1324. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Pavletich NP. Nature. 1999;401:188. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 19.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, Qian X, Mills E, Berghs SC, Carey N, Finn PW, Collins LS, Tumber A, Ritchie JW, Jensen PB, Lichenstein HS, Sehested M. Biochem J. 2008;409:581. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 20.Bieliauskas AV, Pflum MKH. Chemical Society Reviews. 2008;37:1402. doi: 10.1039/b703830p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegener D, Hildmann C, Schwienhorst A. Mol Genet Metab. 2003:139. doi: 10.1016/j.ymgme.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Wegener D, Hildmann C, Riester D, Schwienhorst A. Analytical Biochemistry. 2003;321:202. doi: 10.1016/s0003-2697(03)00426-3. [DOI] [PubMed] [Google Scholar]
- 23.Borra MT, Smith BC, Denu JM. J Biol Chem. 2005;280:17187. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 24.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Journal of Biological Chemistry. 2005;280:17038. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 25.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. Journal of Biological Chemistry. 2010;285:8340. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bressi JC, Jennings AJ, Skene R, Wu Y, Melkus R, Jong RD, O’Connell S, Grimshaw CE, Navre M, Gangloff AR. Bioorganic & Medicinal Chemistry Letters. 2010;20:3142. doi: 10.1016/j.bmcl.2010.03.091. [DOI] [PubMed] [Google Scholar]
- 27.Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Ferrigno F, Jones P, Neddermann P, De Francesco R, Steinkuhler C, Gallinari P, Carfi A. Journal of Biological Chemistry. 2008;283:26694. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, McLean TH, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Journal of Biological Chemistry. 2008;283:11355. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weerasinghe SVW, Estiu G, Wiest O, Pflum MKH. J Med Chem. 2008;51:5542. doi: 10.1021/jm800081j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pflum MKH, Tong JK, Lane WS, Schreiber SL. J Biol Chem. 2001;276:47733. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- 31.Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. J Biol Chem. 2001;276:38837. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 32.Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Proc Natl Acad Sci U S A. 2001;98:15113. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SR, Lee KS, Park SJ, Min KH, Choe YH, Moon H, Yoo WH, Chae HJ, Han MK, Lee YC. Journal of Allergy and Clinical Immunology. 2010;125:449. doi: 10.1016/j.jaci.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Yao Y, Li H, Gu Y, Davidson NE, Zhou Q. Carcinogenesis. 2010;31:382. doi: 10.1093/carcin/bgp308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadosh D, Struhl K. Genes Dev. 1998;12:797. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JH. Experiments in Molecular Genetics. New York: Cold Springs Harbor; 1972. [Google Scholar]
- 37.Zhang X, Bremer H. Journal of Biological Chemistry. 1995;270:11181. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- 38.Bieliauskas A, Weerasinghe S, Pflum MH. Bioorg Med Chem Lett. 2007;17:2216. doi: 10.1016/j.bmcl.2007.01.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Staver MJ, Curtin ML, Holms JH, Frey RR, Edalji R, Smith R, Michaelides MR, Davidsen SK, Glaser KB. Life Sciences. 2004;74:2693. doi: 10.1016/j.lfs.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 40.Clark DD, Peterson BR. Chem Bio Chem. 2003;4:101. doi: 10.1002/cbic.200390001. [DOI] [PubMed] [Google Scholar]
- 41.Khazak V, Golemis EA, Weber L. Methods Mol Biol. 2005;310:253. doi: 10.1007/978-1-59259-948-6_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipke PN, Ovalle R. J Bacteriol. 1998;180:3735. doi: 10.1128/jb.180.15.3735-3740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phalip V, Hatsch D, Jeltsch JM. Biotechnol Lett. 2004;26:409. doi: 10.1023/b:bile.0000018260.26171.6f. [DOI] [PubMed] [Google Scholar]
- 44.Codlin S, Haines RL, Burden JJE, Mole SE. J Cell Sci. 2008;121:2860. doi: 10.1242/jcs.030122. [DOI] [PubMed] [Google Scholar]
- 45.Ovalle R, Spencer M, Thiwanont M, Lipke PN. Appl Environ Microb. 1999;65:3325. doi: 10.1128/aem.65.8.3325-3327.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitamura K. Agr Biol Chem Tokyo. 1982;46:2093. [Google Scholar]
- 47.Mai A, Rotili D, Massa S, Brosch G, Simonetti G, Passariello C, Palamara AT. Bioorganic & Medicinal Chemistry Letters. 2007;17:1221. doi: 10.1016/j.bmcl.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 48.Simonetti G, Passariello C, Rotili D, Mai A, Garaci E, Palamara AT. FEMS Yeast Research. 2007;7:1371. doi: 10.1111/j.1567-1364.2007.00276.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Smith DL, Meriin AB, Engemann S, Russel DE, Roark M, Washington SL, Maxwell MM, Marsh JL, Thompson LM, Wanker EE, Young AB, Housman DE, Bates GP, Sherman MY, Kazantsev AG. Proc Natl Acad Sci U S A. 2005;102:892. doi: 10.1073/pnas.0408936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Nebane NM, Wennerberg† K, Li† Y, Neubauer† V, Hobrath JV, McKellip S, Rasmussen L, Shindo N, Sosa M, Maddry JA, Ananthan S, Piazza GA, White EL, Harsay E. Chem Bio Chem. 2010:9999. doi: 10.1002/cbic.200900681. NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colomer J, Schmitt AA, Toone EJ, Means AR. Biochemistry. 2010;49:4244. doi: 10.1021/bi1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Posakony J, Hirao M, Bedalov A. Comb Chem High Throughput Screen. 2004;7:661. doi: 10.2174/1386207043328346. [DOI] [PubMed] [Google Scholar]
- 53.Posakony J, Hirao M, Stevens S, Simon JA, Bedalov A. J Med Chem. 2004;47:2635. doi: 10.1021/jm030473r. [DOI] [PubMed] [Google Scholar]
- 54.Chung KS, Ahn J, Choi CH, Yim NH, Kang CM, Kim CH, Lee K, Park HM, Song KB, Won M. Molecules and Cells. 2008;26:93. [PubMed] [Google Scholar]
- 55.Carmen AA, Griffin PR, Calaycay JR, Rundlett SE, Suka Y, Grunstein M. Proceedings of the National Academy of Sciences. 1999;96:12356. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin H, Abida WM, Sauer RT, Cornish VW. J Am Chem Soc. 2000;122:4247. [Google Scholar]
- 57.Grozinger CM, Schreiber SL. Proc Natl Acad Sci U S A. 2000;97:7835. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, Schreiber SL. Proc Natl Acad Sci U S A. 1998;95:3519. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shah K, Liu Y, Deirmengian C, Shokat KM. Proc Natl Acad Sci USA. 1997;94:3565. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zunder ER, Knight ZA, Houseman BT, Apsel B, Shokat KM. Cancer Cell. 2008;14:180. doi: 10.1016/j.ccr.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weatherill DB, Dyer J, Sossin WS. Journal of Biological Chemistry. 2010;285:12255. doi: 10.1074/jbc.M109.071142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.