Abstract
Cytosine DNA methylation is a stable epigenetic mark that is critical for diverse biological processes including gene and transposon silencing, imprinting, and X chromosome inactivation. Recent findings in plants and animals have greatly increased our understanding of the pathways utilized to accurately target, maintain, and modify patterns of DNA methylation and have revealed unanticipated mechanistic similarities between these organisms. Key roles for small RNAs, proteins with methylated DNA binding domains and DNA glycosylases in these processes have emerged. Drawing on insights from both plants and animals should deepen our understanding of the regulation and biological significance of DNA methylation.
The genetic information within a cell is encoded by DNA, which is packaged into chromatin. Epigenetic modifications of DNA and histones, the core components of chromatin, constitute an additional layer of information that influences the expression of the underlying genes. DNA methylation, the addition of a methyl group to a cytosine base, is one such epigenetic modification, and it is evolutionarily ancient and associated with gene silencing in eukaryotes. Attesting to its importance, DNA methylation defects in mammals are embryonic lethal, and in plants they can lead to pleiotropic morphological defects.
In mammals, DNA methylation occurs almost exclusively in the symmetric CG context and is estimated to occur at ~70-80% of CG dinucleotides throughout the genome1. However, a small amount of non-CG methylation is observed in embryonic stem (ES) cells2–4. The remaining unmethylated CG dinucleotides are mostly found near gene promoters in dense clusters, termed CpG islands5,6. In plants, DNA methylation commonly occurs at cytosine bases within all sequence contexts: the symmetric CG and CHG contexts (where H=A, T, or C) and the asymmetric CHH context7. Genome wide, DNA methylation levels of approximately 24%, 6.7% and 1.7% are observed for CG, CHG, and CHH contexts, respectively8. Unlike in mammals, DNA methylation in plants predominantly occurs on transposons and other repetitive DNA elements9.
In mammals, DNA methylation patterns are established by the DNA methyltransferase (Dnmt) 3 family of de novo methyltransferases and maintained by the maintenance methyltransferase, Dnmt110–12 (FIG. 1). In plants, de novo methylation is catalyzed by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), a homolog of the Dnmt3 methyltransferases and maintained by three different pathways: CG methylation by DNA METHYLTRANSFERASE 1 (MET1), the plant homolog of Dnmt1, CHG methylation by CHROMOMETHYLASE (CMT3), a plant specific DNA methyltransferase, and asymmetric CHH methylation through persistent de novo methylation by DRM213 (FIG. 1). However, the pathways controlling the establishment and maintenance of DNA methylation, as well as those involved in the removal of DNA methylation, are less well characterized.
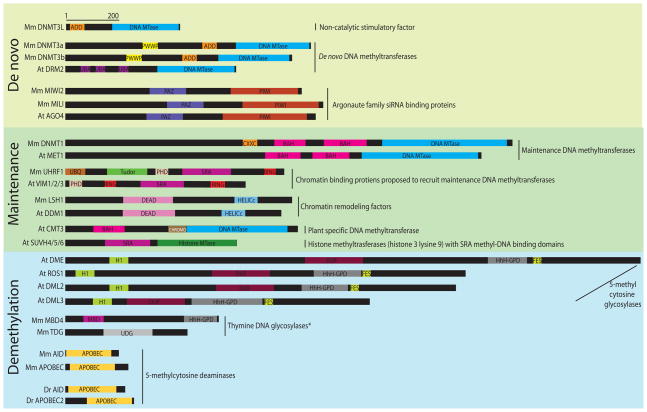
Figure 1. Cartoon representation of select mouse (Mm), Arabidopsis (At), and Zebrafish (Dr) proteins involved in maintenance methylation, de novo methylation, and demethylation.
The MmDnmt3 family and AtDRM2 contain DNA methyltransferase domains (DNA MTase); in AtDRM2 the catalytic motifs are rearranged. The MmDnmt3 proteins also possess a cysteine rich domain that contains a Plant Homeodomain (PHD) zinc finger motif and is referred to as an ATRX-DNMT3-DNMT3L (ADD) domain. MnDnmt3a/b possess a Proline-Tryptophan-Tryptophan-Proline motif (PWWP) that MmDnmt3L lacks. AtDRM2 contains Ubiquitin Associated domains (UBA). MmMIWI2, MmMILI, and AtAGO4 possess a Piwi Argonaute and Zwille domain (PAZ) and a P-element induced wimpy testis domain (PIWI). MmDnmt1 and AtMET1 possess Bromo-Adjacent Homology domains (BAH) and a DNA MTase domain. MmDnmt1 also contains a cysteine rich (CXXC) domain. MmUHRF1 and the AtVIM family contain SET or RING associated (SRA), ring finger E3 ubiquitin protein ligase (RING), and PHD domains. MmUHRF1 also has a Tudor domain and a ubiquitin domain (UBQ). MmLSH1 and AtDDM1 contain DEAD and HELICc helicase domains. AtCMT3 contains a DNA MTase, a Chromatin Organization Modifier (CHROMO) and a BAH domain. AtSUVH4,5, and 6 possess an SRA and a histone methyltransferase domain (Histone MTase). The AtDME/ROS1 family of glycosylases all possesses a helix-hairpin-helix-Gly-Pro-Asp domain (HhH-GDP), a 4Fe-4S cluster (FES), a domain with similarity to histone H1 (H1), and a domain of unknown function (DUF). MmMBD4 contains a HhH-GDP domain and a methyl CpG binding domain (MBD); MmTDG contains a Uracil-DNA glycosylase domain (UDG). Mouse and Zebrafish AID and APOBEC proteins all contain a apolipoprotein B mRNA editing enzyme domain (APOBEC). * indicates activity on additional substrates145
In this review, we focus on recent studies in plants and animals that have greatly expanded our understanding of such pathways. We begin with the establishment of DNA methylation, with a separate section focusing on the dynamics of DNA methylation in reproductive cells and the roles of small RNAs at this stage of development. We then discuss mechanisms governing the maintenance and removal of DNA methylation. In each section, recent advances from both plants and animals will be presented and both similarities and differences will be highlighted. In particular, small RNAs, methyl-binding domain proteins and DNA glycosylases are common components of the pathways that define dynamic DNA methylation patterns in the two taxonomic groups.
DE NOVO DNA METHYLATION
De novo methylation in plants
Throughout plant development, small RNAs target homologous genomic DNA sequences for cytosine methylation in all sequence contexts through a phenomenon initially observed by Wassenegger et al.14 termed RNA-directed DNA methylation (RdDM)7,15. In addition to the canonical RNA interference (RNAi) machinery (that is, members of the Dicer and Argonaute families) and DRM2, RdDM also requires two plant specific RNA polymerases, Pol IV and Pol V, with largely non-redundant functions16,17, two putative chromatin remodeling factors, and several other recently identified proteins15. Through the characterization of these components, an increasingly detailed mechanistic understanding of RdDM is emerging (FIG. 2).
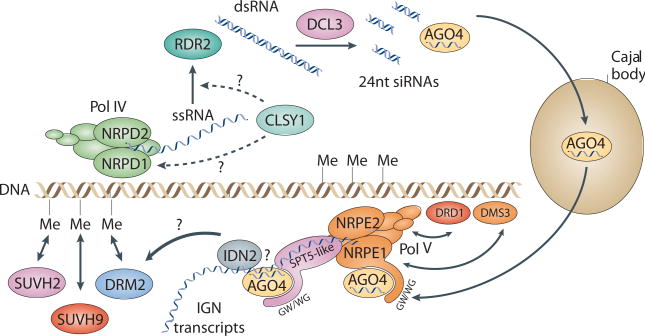
Figure 2. Model for RNA directed DNA methylation (RdDM).
Single-stranded RNA transcripts (ssRNA) corresponding to transposons and repeat elements are hypothesized to be generated by Pol IV. CLSY1, a putative chromatin remodeling factor, likely functions early in RdDM, possibly recruiting Pol IV to chromatin or aiding in ssRNA transcript processing. RDR2, an RNA dependant RNA polymerase, is proposed to generate double-stranded RNA (dsRNA) from the ssRNA transcripts. DCL3, a dicer-like protein, is thought to process the dsRNAs into 24nt siRNAs, which are bound by an argonaute protein, AGO4. AGO4 localizes to Cajal bodies and while the function of this association remains unknown, it appears necessary for wild-type levels of RdDM33. AGO4 also co-localizes with two Pol V subunits, NRPE1 and NRPE2, and DRM2 at a distinct nuclear foci, the AGO4/NRPD1b(NRPE1) body (not depicted), that may represent a site of active RdDM33. Pol V is thought to transcribe intergenic noncoding (IGN) regions throughout the genome. NRPE1 association with chromatin requires another putative chromatin remodeling factor, DRD1, and an SMC domain protein, DMS3. IGN transcripts may serve as a scaffold to recruit AGO4, which interacts with the GW/WG motifs of NRPE1 and SPT5-like, possibly through interactions between AGO4-bound siRNAs and the nascent transcript. An RNA binding protein, IDN2, is proposed to recognize the siRNA/nascent transcript duplex. These associations may aid in targeting DRM2 to genomic loci that produce both 24nt siRNAs and IGN transcripts. Recruitment or retention of DRM2 at such loci may be aided by SUVH9 and SUVH2, two proteins that bind methylated DNA and likely act late in RdDM. Me, DNA methylation.
Biogenesis of the 24nt small interfering RNAs (siRNAs) required to target DNA methylation depends on Pol IV, RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3). Other RdDM components including DRM2, ARGONAUTE 4 (AGO4), and Pol V are needed for siRNA accumulation for a subset of loci; however, these proteins do not appear to be involved in the initial production of siRNAs and are proposed to reinforce siRNA biogenesis by an unknown mechanism7,18. Additional subunits or interacting partners of Pol IV and Pol V have recently been identified19–23. While some subunits are shared with Pol II, others are unique to Pol IV, Pol V, or both20. Although no polymerase activity has been demonstrated for Pol IV, mutations in the largest subunit, NUCLEAR RNA POLYMERASE D1 (NRPD1), including mutations within the conserved metal binding motif, greatly reduce the abundance of siRNAs18,24–30, suggesting Pol IV may be an active polymerase. Pol IV is hypothesized to initiate siRNA biogenesis by producing long single-stranded RNA transcripts. These transcripts are then thought to be acted upon by RDR2, generating double-stranded RNAs that are processed into 24nt siRNAs by DCL3 and loaded into AGO47,15. AGO4 interacts with the Pol V subunit, NUCLEAR RNA POLYMERASE E1 (NRPE1)31,32, and this interaction is required for RdDM31, leading to the hypothesis that this complex functions as a downstream effector of DNA methylation. In vivo, AGO4 co-localizes either with Cajal bodies or with NRPE1, NRPE2 and DRM2 at a separate discrete nuclear body termed the AGO4/NRPDE1 (previously termed NRPD1b) (AB) body32,33. The AB body is located adjacent to 45S ribosomal DNA and may be a site of active RdDM33.
A recent study further clarified the role of Pol V in RdDM by identifying low abundance intergenic noncoding (IGN) transcripts from several loci whose accumulation depends on Pol V34. NRPE1 is present at these transcribed DNA regions and is associated with the RNA transcripts, suggesting Pol V is an active polymerase34. These Pol V-dependent transcripts are required for DNA methylation and silencing of surrounding loci, but their accumulation does not depend on NRPD1, DCL3 or RDR234, suggesting Pol V acts in RdDM via a pathway that is independent of siRNAs. These IGN transcripts are proposed to function as scaffolds for the recruitment of the silencing machinery, possibly facilitated by base pairing interactions between AGO4-bound siRNAs and nascent Pol V transcripts34. A requirement of transcription for silencing is also observed in fission yeast where transcription of heterochromatic DNA by Pol II is required for siRNA mediated heterochromatin formation35.
Current models of RdDM posit that both Pol V-dependent transcripts and siRNAs are required to silence a particular locus. Several studies support the hypothesis that AGO4 and/or SUPPRESSOR OF TY INSERTION 5-LIKE (SPT5-like)/KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR 1 (KTF1) may bridge the siRNA and IGN transcript generating pathways. SPT5-like, a protein with homology to the yeast transcription elongation factor Spt5, was recently identified as a downstream effector of RdDM21,23,36. SPT5-like and NRPE1 are both able to interact with AGO4 through a conserved Glycine/Tryptophan (GW/WG), or Ago hook, motif present in their carboxy-terminal regions23,31,32,36. In vivo, both SPT5-like and AGO4 interact with Pol V-dependent transcripts36,37, prompting speculation that SPT5-like serves as an adaptor protein that binds both AGO4 and nascent Pol V transcripts, aiding in the recruitment of AGO4 to Pol V transcribed loci. This interaction may also be required to recruit the silencing machinery, including DRM2, to establish DNA methylation.
Another factor thought to act as a downstream RdDM effector, INVOLVED IN DE NOVO 2 (IDN2), was recently identified38. IDN2 has homology with SUPPRESSOR OF GENE SILENCING 3 (SGS3), a protein involved in posttranscriptional gene silencing, and like SGS3 it contains an XS domain able to recognize double-stranded RNAs with 5’ overhangs38. A possible RNA substrate for IDN2 is the duplex between AGO4-bound siRNAs and Pol V non-coding transcripts39, which could also be a signal that aids in recruitment of DRM2 to establish DNA methylation.
In addition to Pol V-dependent transcripts, Pol II-dependent noncoding transcripts required for transcriptional gene silencing at some loci have recently been identified in a weak nuclear RNA polymerase B2 (nrpb2) mutant and these transcripts are also proposed to act as scaffolds for the recruitment of RdDM factors including AGO4, and possibly Pol IV and Pol V40. Further supporting a role for Pol II in RdDM, two genetic screens for RdDM factors identified a conserved protein, RNA-DIRECTED DNA METHYLATION 4 (RDM4)/DEFECTIVE IN MERISTEM SILENCING 4 (DMS4), with similarity to the yeast protein, Interacts with Pol II (IWR1)41,42. While the precise relationship between Pol II, Pol V and Pol IV remains elusive, these studies suggest they may be more intimately connected than previously thought.
Although the mechanisms through which Pol IV and Pol V are targeted to specific loci are poorly understood, several recent findings are beginning to shed light on these aspects of RdDM. DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), a putative chromatin remodeling factor43, and DEFECTIVE IN MERISTEM SILENCING 3 (DMS3), an RdDM component with similarity to Structural Maintenance of Chromosome (SMC) proteins38,44, are needed for NRPE1 chromatin association and for accumulation of IGN transcripts34,37, although how these components are targeted is unknown. In addition, the NRPB2 subunit of Pol II aids in the association of both NRPE1 and NRPD1 with chromatin, suggesting Pol II-dependent transcripts, or the act of transcription, may recruit Pol IV and Pol V to specific loci40. Finally, a putative chromatin remodeling factor, CLASSY 1 (CLSY1), may be involved at an early stage of siRNA production, possibly at the level of Pol IV or RDR2 activity45.
De novo methylation in mammals
Unlike in plants, DNA methylation in mammals covers most of the genome, with the main exception being CpG islands. This DNA methylation pattern is largely established during early embryogenesis, around the time of implantation46,47, through the activity of the Dnmt3a and Dnmt3b DNA methyltransferases5,48,49. However, during post-implantation development, further epigenetic reprogramming occurs in primordial germ cells (PGCs). Following a wave of demethylation, which is required to erase DNA methylation imprints established in the previous generation, DNA methylation patterns are reestablished at imprinted loci and transposable elements (TEs) during gametogenesis by Dnmt3a and a non-catalytic paralog, Dnmt3L5,12,48,49. Recent studies suggest DNA methylation may be targeted to TEs and imprinted genes during germ cell development through different mechanisms, with the former involving PIWI-interacting RNAs (piRNAs) (discussed later) and the latter involving an interactions between Dnmt3L and unmethylated histone 3 lysine 4 (H3K4) tails.
Biochemical purification of Dnmt3L lead to the discovery that Dnmt3L interacts with unmethylated H3K4 tails through its cysteine-rich ATRX-Dnmt3-Dnmt3L (ADD) domain50,51. As Dnmt3L also interacts with Dnmt3a12,50, a model in which Dnmt3L binds unmethylated H3K4 tails and recruits the Dnmt3a2 isoform to specific loci, including imprinted loci, was proposed50,52 (FIG. 3A). Indeed, an inverse relationship between H3K4 methylation and allele-specific DNA methylation has been reported at several imprinted loci53–56. Further supporting this model, an oocyte-specific H3K4 demethylase57, lysine demethylase 1B (KDM1B), was recently shown to be required for the establishment of DNA methylation at several differentially methylated regions (DMRs) associated with imprinted genes during oogenesis57, and defects in DNA methylation at such loci resulted in a loss of imprinting in developing embryos57. This model is also consistent with studies showing H3K4 methylation appears anticorrelated with DNA methylation in multiple mammalian cell types58–62 and with findings that H3K4 dimethylation and trimethylation are anticorrelated with DNA methylation in plants63.
Figure 3. Model depicting recruitment of the de novo methylation machinery by unmethylated histone 3 lysine 4 (H3K4) tails.
The amino-terminal domain of Dnmt3L possesses a cysteine rich domain that interacts with unmethylated H3K4 tails and this interaction is proposed to recruit or activate Dnmt3a2. The carboxy-terminal domains of Dnmt3L and Dnmt3a form a tetrameric complex in which two Dnmt3a proteins interact with each other and are flanked by two Dnmt3L proteins. The Dnmt3a active sites (red stars) are thought to be separated by approximately one helical turn and thus could catalyze methylation (filled circles) on opposite DNA strands ~10bps apart. Once recruited to a specific locus, the Dnmt3L/Dnmt3a tetramer might be able to oligomerize, which could result in an ~10bp periodic pattern of DNA methylation along the same DNA strand.
In addition to Dnmt3a and Dnmt3L, there is evidence that transcription across DMRs is also required for imprinting64. Chotalia et al.64 demonstrate that such transcription occurs during oocyte growth (prior to or around the time when de novo methylation occurs) and, at least at the imprinted Gnas locus, is required for the establishment of DNA methylation64. These finding lead to the proposal that the act of transcription, or the transcripts themselves, may alter the chromatin structure of imprinted loci and/or recruit the histone modifying enzymes and DNA methyltransferases required to establish DNA methylation imprints64.
Mechanistic insight into how de novo methyltransferases function once targeted to a particular locus was provided by several biophysical studies focused on the interaction between Dnmt3L and Dnmt3a. Co-crystallization of the carboxy-terminal regions of Dnmt3a and Dnmt3L revealed a tetrameric complex that positions two Dnmt3a proteins such that their active sites are adjacent to one another52. Two Dnmt3L proteins are located on either side of the Dnmt3a dimer and residues of Dnmt3L may stabilize the active site loop in Dnmt3a52, which could account for the observed stimulatory effect of Dnmt3L on Dnmt3a/b activity65,66. Superimposition of the Dnmt3a carboxy-terminal structure with that of the M.HhaI methyltransferase complexed with DNA67 provided a model in which the two Dnmt3a active sites are separated by approximately one helical DNA turn, suggesting that each tetrameric complex could simultaneously methylate two cytosine residues at a defined spacing of 8–10 bps52. This tetrameric complex was subsequently shown to oligomerize on DNA substrates, forming a filamentous nucleoprotein complex68 (FIG. 3A).
Consistent with the determined structural parameters, a periodicity for DNA methylation on opposite strands of a DNA duplex as well as along the same strand of DNA was observed in vitro through bisulfite sequencing analyses52,68. In vivo, the spacing of CG dinucleotides at many DMRs are also consistent with an ~8–10 bp periodicity52, as are findings that CG dinucleotides at an 8 bp spacing are overrepresented across the human genome69,70 and to a lesser extent across the mouse genome69. Since Dnmt3a appears to be a non-processive DNA methyltransferase71, formation of an oligomer could help explain the observed periodic pattern of DNA methylation. Although whether oligomerization occurs in vivo remains unknown, it is tempting to hypothesize a model in which interactions between Dnmt3L and unmethylated H3K4 tails, or possibly between Dnmt3a and other histone modifications or histone methyltransferases, might target and set the register for oligomerization of Dnmt3a/Dnmt3L tetramers resulting in an ~8–10 bp periodicity.
In Arabidopsis, nucleotide resolution DNA methylation mapping revealed an element of periodicity for DNA methylation. For CHH methylation (mostly controlled by DRM2) a period of ~10 bps was observed genome wide8, suggesting that the periodicity observed for Dnmt3a may be a common feature of de novo methyltransferases and that it may also occur on a genome wide scale in mammals. For CHG methylation (mostly controlled by CMT3), a period of approximately the size of a nucleosome, 167 nucleotides, was found8, consistent with the chromodomain in CMT3 interacting with methylated H3 tails72.
In addition to interactions with unmethylated H3K4 tails, other mechanisms for targeting DNA methylation to specific loci throughout the genome also shape the overall methylation landscape during mammalian development. These including interactions between Dnmt3a/b and the histone methyltransferases G9a, Enhancer of zeste homolog 2 (EZH2), Suppressor of variegation 3–9 homolog 1 (SUV39H1), and SET domain bifurcated 1 (SETB1)5. More recently, Zhao et al.73 have demonstrated that symmetric methylation of histone 4 arginine 3 (H4R3me2s) by protein arginine methyltransferase 5 (PRMT5) can recruit Dnmt3a to the human β-globin locus, which is required for the DNA methylation and silencing of this gene. In vitro characterization of the interaction between the H4R3me2s modification and Dnmt3a demonstrated that the ADD domain of Dnmt3a is sufficient to mediate this interaction73. While histone arginine methylation has been implicated in gene silencing74, this finding marks the first direct link between arginine methylation and DNA methylation.
DNA METHYLATION IN REPRODUCTIVE CELLS
Transposons and other repetitive DNA elements are highly abundant in both plant and mammal genomes. Due to the high risk TEs pose to genome integrity, their expression must be tightly regulated. Such control is particularly important in cells that transmit genetic information to the subsequent generation. In both plants and mammals, such elements are targeted by the de novo methylation machinery and are maintained in a methylated and silenced state. Recent evidence suggests that in mammals, like in plants, small RNAs play an important role in targeting transposons for methylation.
RdDM in plant reproductive cells
In plants, DNA methylation patterns appear to be maintained in a multigenerational manner leading to the view that DNA methylation in plants is quite static. However, several complimentary studies demonstrate that DNA methylation patterns in plants are dynamic during development by showing that genome-wide losses of DNA methylation occur during both male and female gametogenesis. This observed hypomethylation is similar to the global demethylation observed in PGCs and on the paternal genome during mammalian development48,49. These resent studies suggest that, in Arabidopsis, these changes may reinforce transposon silencing in the sperm and egg cells, respectively75–77 (FIG. 4).
Figure 4. DNA methylation changes during plant development.
Diagram of an Arabidopsis (A) flower, and (B) the male and female gametophytes. Male gametogenesis occurs in stamens (St) and generates tricellular pollen grains that contain a vegetative nucleus (VN) and two sperm cells (SC). Female gametogenesis occurs in ovules (Ov) and produces a multicellular gametophyte with three antipodal cells (AC), two synergid cells (SC), one egg cell (EC) and a diploid (2n) central cell nucleus (CCN)78. (upper) Model showing transposon reactivation and siRNAs production specifically in the VN. These siRNAs may travel to the SCs to reinforce transposon silencing. (lower) Model showing siRNAs in the CCN, which may arise as a consequence of global demethylation. These siRNAs may travel to the EC and reinforce silencing. Reinforced silencing in the sperm and egg cells could account for the observed hypermethylation of the embryo. (C) Fertilization of the EC and CCN generate the embryo and endosperm, respectively. The embryo will give rise to the mature Arabidopsis plant while the endosperm is a terminally differentiated tissue. Imprinting is observed the endosperm, which nourishes the embryo, and is thus analogous to the placenta in mammals, where imprinting also occurs. In plants, maternal imprinting results from demethylation in the CCN by the DME glycosylase, which likely accounts for the observed hypomethylation in this tissue. After fertilization, the unmethylated (open circles) maternal alleles (♀) are expressed in the endosperm, while the paternal allele (♂) is methylated (closed circles) and silent.
During male gametogenesis tricellular pollen grains that contain a vegetative cell nucleus and two sperms cells are produced78 (FIG. 4B). Analysis of transposon expression in different plant tissues revealed that transposons, which are methylated and silenced in most tissues, are expressed and mobile in pollen75. Within the pollen grain, transposon reactivation appears to be restricted to the vegetative nucleus. This is a key distinction, as the sperm cells, but not the vegetative nucleus, provide genetic information to subsequent generations78 and thus their genome integrity must be protected. Consistent with decreased DNA methylation and transposon activation, several RdDM components are down regulated in pollen75,79 and DECREASED DNA METHYLATION 1 (DDM1), a chromatin remodeling factor required for maintenance of CG methylation80, appears to be excluded from the vegetative nucleus75. Sequencing of siRNA populations from pollen and isolated sperm cells showed an increase in 21nt siRNAs in sperm cells75. Since these siRNAs correspond to transposons that do not appear to be expressed in the sperm cells, it was postulated that siRNAs generated in the vegetative nucleus might travel to the sperm cells and reinforce silencing by an unknown mechanism75 (FIG.4C).
The two sperm cells fertilize the central cell and the egg cell of the multicellular female gametophyte in a double fertilization event that generates the embryo and the endosperm, respectively78 (FIG. 4B,C). While previous studies have documented decreased DNA methylation at discrete imprinted loci in endosperm78, two recent studies show endosperm DNA methylation is reduced genome wide, likely originating from demethylation in central cell of the female gametophyte76,77. These findings are in line with observations that chromatin appears less condensed in endosperm nuclei81. Despite this global decrease in DNA methylation, Hsieh et al.76 found increased CHH methylation in both the endosperm and embryo tissues relative to adult shoot tissue and suggest this hypermethylation could result from enhanced RdDM. Consistent with these findings, profiling of Pol IV-dependent siRNA levels in different plant tissues shows maternal-derived siRNAs accumulate to high levels in the endosperm82. Analogous with the model of reinforced silencing in the male gametophyte75, these finding lead to the suggestion that siRNAs potentially generated in the central cell may reinforce silencing in the egg cell and possibly in the developing embryo76 (FIG. 4B,C).
Since potentially deleterious transposition events occurring in the sperm or egg cells would be inherited in subsequent generations, demethylation during gametogenesis may function to reveal TEs within the genome with the potential to be expressed and arm siRNA-based pathways in order to ensure these elements are efficiently silenced. Such a mechanism would be inherently adaptable, as newly integrated transposons would also be expressed, leading to siRNA production and the establishment of silencing. Interestingly, Teixeira et al.83 recently demonstrated that siRNA producing loci, unlike other regions of the Arabidopsis genome, can be re-methylated in all sequence contexts when methylation is lost in previous generations, suggesting a dynamic role for RdDM in correcting DNA methylation defects. However, remethylation to approximately wild-type levels was only observed after multiple generations, as is also the case when newly inserted transgenes become silenced. It is tempting to speculate that decreased methylation and transposon reactivation during gametogenesis might be required to generate siRNA signals and allow the observed reestablishment of silencing.
piRNAs in mammalian germ cells
A small RNA pathway is also required to silence some transposons in mammals during male gametogenesis (FIG. 5). While RdDM in plants utilizes 24nt siRNAs, transposon control in mammals utilizes 25-30nt small RNAs, termed piRNAs, initially identified in Drosophila84. In Drosophila, piRNAs bound by the P-element induced wimpy testis (PIWI) clade of argonautes guide cleavage of transposon transcripts, which results in posttranscriptional gene silencing85. This clade of argonautes is highly conserved in animals86 and initial genetic analysis in mammals and flies suggested that roles for piRNAs in germ cell development and transposon silencing were also conserved85. Yet, early studies of mammalian piRNA populations revealed that, unlike in Drosophila, mammalian piRNAs were not enriched for repetitive regions of the genome87,88 leaving it unclear whether mammalian piRNAs function to silence transposons. However, subsequent analyses of piRNA populations isolated at earlier stages of mouse development revealed an enrichment in repetitive DNA sequences89-91. These piRNA populations possess the characteristic sequence properties of both primary89-91 and secondary89,91 piRNAs, suggesting that they are generated by a mechanism similar to the ping-pong amplification model initially proposed in Drosophila92,93 and that they are indeed involved in the posttranscriptional silencing of transposons (FIG. 5A).
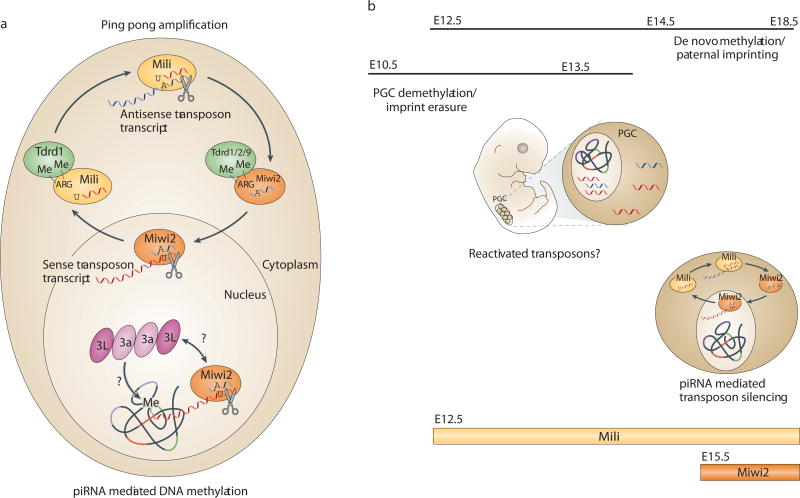
Figure 5. piRNAs and male gametogenesis.
(A) Ping-pong model In mammals, as in flies, piRNAs are proposed to arise via a ping-pong amplification cycle that produces primary piRNAs with a 5’ uridine (U) and secondary piRNAs with an adenine (A) at position 10. In mammals, transposon transcripts, mainly sense oriented, are the presumed substrates for primary piRNA production91. Cleavage of these transcripts produces primary piRNAs proposed to preferentially associate with cytoplasmic Mili91. Mili, bound with sense piRNAs, cleaves antisense transcripts, producing secondary piRNAs that preferentially associate with cytoplasmic and nuclear Miwi291. Nuclear Miwi2, bound with antisense piRNAs, cleaves sense transposon transcripts, producing more primary piRNAs. piRNA complexes are also proposed to guide DNA methylation to homologous genomic loci by potentially interacting with nascent transposon transcripts and directly or indirect recruiting de novo methyltransferases, possibly Dnmt3a/3L complexes. Mili and Miwi2 contain symmetrical dimethylarginines (Me-ARG) and interact with tudor domain-containing (Tdrd) proteins. (B) Model for transposon silencing during male gametogenesis. Genome wide demethylation (E10.5-E12.5) in primordial germ cells (PGC) erases DNA imprints and could briefly reactivate transposons. De novo methylation and paternal imprinting are observed in testes from E14.5 through birth. Consistent with prior transposon expression, piRNAs bound to Mili and Miwi2 (expressed by E12.5 and E15.5, respectively) are enriched for transposon sequences during this time period and are proposed to facilitate targeted reestablishment of DNA methylation at transposons. Although less studied, PIWI argonautes are expressed in female germ cells of flies, mammals85, frogs180 and silkworms181 and piRNAs are present in fly182 and silkworm181,183 ovaries and in frog oocytes184.
In mammals, decreases in DNA methylation and increases in expression were observed at several TEs in two PIWI clade mutants, miliand miwi290,94, suggesting that piRNAs silence transposons at both the transcriptional and posttranscriptional levels. However, these initial methylation studies were carried out at a developmental stage many mitoses after the establishment of DNA methylation, which occurs following a wave of genome wide demethylation in PGCs (FIG. 5B). Thus it could not be determined whether methylation defects were occurring at the level of maintenance or de novo methylation. Two recent studies provide compelling evidence that piRNAs are indeed involved in de novo methylation by demonstrating that DNA methylation defects in mili mutants occur at the stage in development when de novo methylation in male germ cells is observed89 and that piRNA populations from this time period are highly enriched in transposon sequences89,91. Aravin et al.91 further demonstrate that piRNAs are present in Dnmt3L mutants, suggesting the piRNA pathway acts upstream of de novo methylation.
By analogy with models for siRNA and piRNA mediated transposon control in Arabidopsis and Drosophila, respectively, the demethylation in PGCs may reveal TEs with the potential to be expressed when hypomethylated leading to the production of piRNAs and the targeting of DNA methylation to homologous sequences throughout the genome (FIG. 5). It has been hypothesized (FIG. 5A) that PIWI-piRNA complexes could interact with nascent transposon transcripts and directly recruit the de novo methyltransferases. However, preliminary studies failed to show an interaction between PIWI argonautes and Dnmt3 proteins91. Alternatively, this recruitment could be indirect, first involving the recruitment of chromatin modifiers which catalyze modifications that subsequently recruit the DNA methyltransferases95.
Recently, it was found that PIWI family members in mouse96–99, Drosophila, and Xenopus contain symmetrical dimethylarginine (sDMA) modifications97. Methylated arginines can be recognized by tudor domains and purification of Mili, Miwi, or Miwi2 containing complexes demonstrated interactions with various Tudor domain-containing (Tdrd) proteins96,98–101: Tdrd1 was found to interact with Mili96,98–101, while Tdrd1, Tdrd2, Tdrd9 interacted with Miwi298 (FIG. 5A). Like Mili, Tdrd1 is required for DNA methylation and transposon silencing in mouse germ cells98,99. In tdrd1 mutants, the profile of Mili bound piRNAs is altered, containing a higher proportion of non-transposon sequences98,99, as is the profile of Miwi2 bound piRNAs, containing a lower proportion of antisense piRNAs98, which may explain the observed transposon reactivation.
MAINTENANCE OF DNA METHYLATION PATTERNS
Once established, global DNA methylation patterns must be stably maintained in order to ensure that transposons remain in a silenced state and to preserve cell type identity.
Maintenance of CG methylation
In mammals, DNA methylation is maintained by Dnmt1 (FIG. 6A). This methyltransferase is associated with replication foci and functions to restore hemimethylated DNA generated during DNA replication to the fully methylated state10. Early studies illustrated that Dnmt1 is recruited to replication foci via an interaction with the proliferating cell nuclear antigen (PCNA) component of the replication machinery102. However, disruption of this interaction only resulted in a minor reduction in DNA methylation103–105. Recently, it was shown that Dnmt1 also interacts with another chromatin associated protein, Ubiquitin-like PHD and RING finger domain 1 (UHRF1) (FIG. 1), and that UHRF1 is required for the association of Dnmt1 with chromatin106,107. Studies showing that mutations in UHRF1 cause severe decreases in DNA methylation106,107 and that the SRA domain of UHRF1 specifically binds to hemimethylated CG dinucleotides106,108–111 have lead to a model in which UHRF1 recruits Dnmt1 to hemimethylated DNA106,107. In addition, UHRF1 also interacts with Dnmt3a and Dnmt3b112, which may suggest a role for UHRF1 in de novo methylation. Maintenance of DNA methylation also requires the chromatin remodeling factor Lymphoid-specific Helicase (LSH1)113,114, although the mechanism through which LSH1 functions in DNA methylation remains unknown.
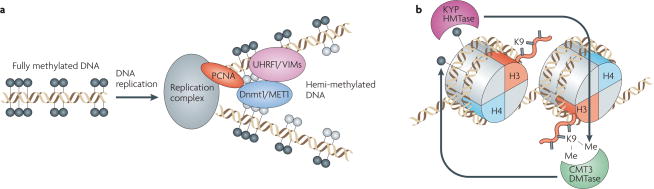
Figure 6. Maintenance of DNA methylation in plants and mammals.
(A) Model depicting maintenance of CG methylation during replication. Dnmt1 is proposed to be recruited to replication foci through interactions with UHRF1, an SRA domain protein that specifically interacts with hemi-methylated DNA, and with PCNA. Once recruited, Dnmt1 functions to maintain methylation patterns by restoring the hemi-methylated DNA to a fully methylated state. In plants, MET1, and the VIM family of SRA domain proteins, homologs of Dnmt1 and UHRF1, respectively, likely function in a similar manner to maintain CG methylation patterns. Black and Gray circles represent methylated and unmethylated cytosines respectively. (B) Model depicting maintenance of CHG methylation in plants. A reinforcing loop of DNA and histone methylation is proposed to maintain CHG methylation in plants. The CMT3 DNA methyltransferase maintains methylation in the CHG context, which is recognized by the SRA domain of the KYP/SUVH4 histone methyltransferase. KYP catalyzes H3K9 dimethylation (H3K9me2), a modification that is required for the maintenance of CHG methylation, and the chromodomain of CMT3 binds methylated histone 3 (H3) tails.
In plants, genetic analyses have demonstrated that homologs of the above mentioned mammalian proteins (FIG. 1), the MET1 DNA methyltransferase80, the VARIANT IN METHYLATION/ORTHRUS (VIM/ORTH) family of SRA domain proteins115,116, and the DDM1 chromatin remodeling factor80,117 are required to maintain CG methylation, suggesting plants and mammals maintain CG methylation in a similar manner. However, further work is needed to determine mechanistically whether these proteins are indeed functioning in a similar way as observed in mammals. One known difference between plants and mammals, is that mutations in DDM1, but not LSH1, cause a decrease in histone 3 lysine 9 (H3K9) methylation118,119, a modification that is highly correlated with DNA methylation and silencing in plants120 and mammals5.
In Arabidopsis ~1/3 of genes have CG methylation in their coding region, which is maintained by MET18,9,121,122. Unlike methylation at transposons, CG methylation within gene bodies does not appear to cause silencing as these genes tend to be moderately expressed in many tissues9,122. Nonetheless, the expression of some body methylated genes is upregulated in met1 mutants122, and highly or lowly expressed genes tend to lack body methylation, suggesting an interplay between transcription and body methylation. The presence of body CG methylation at some genes has also been reported in other invertebrate organisms, suggesting it may be a common feature of eukaryotic genomes6. Initial studies in Arabidopsis postulated that body methylation might suppress the production of antisense transcripts from cryptic promoters122,123. However, increases in antisense transcripts in met1 mutants were found to be rare and uncorrelated with body methylated genes9. Thus, the function of body methylation remains poorly understood.
Maintenance of non-CG methylation in plants
CHG methylation is thought to be maintained via a reinforcing loop involving histone and DNA methylation124 (FIG. 6B). Genome wide profiling of H3K9 and DNA methylation showed that these marks are highly correlated120. Furthermore, loss of either CMT3, the DNA methyltransferase largely responsible for maintaining CHG methylation125,126, or SU(VAR)3–9 HOMOLOG 4 (SUVH4)/ KRYPTONITE (KYP), the histone methyltransferase largely responsible for H3K9 dimethylation127–129, results in a dramatic decrease in DNA methylation127,128. Two other H3K9 histone methyltransferases, SUVH5 and SUVH6, also contribute to global levels of CHG methylation130,131. The observed interdependence of DNA and histone modifications could arise from the multidomain structure of CMT3 and KYP (FIG. 1). In addition to its histone methyltransferase domain, KYP possesses an SRA domain that specifically binds CHG methylation124, suggesting CHG methylation recruits KYP. In turn, CMT3 possesses a chromodomain that binds methylated histone H3 tails72, suggesting histone methylation by KYP may recruit CMT3. Such cross talk between DNA and histone methylation is also observed in mammals and, in many cases, the connection between these modifications appears to involve protein-protein interactions between the histone and DNA methyltransferases themselves5. Whether direct protein interactions between CMT3 and KYP occur and aid in maintaining CHG methylation in plants is unknown.
Asymmetric DNA methylation is maintained by constant de novo methylation by DRM2 and RdDM. However, at some loci CHH methylation is controlled by CMT3 and DRM2132. Like maintenance of CG and CHG methylation, RdDM also requires proteins with SRA domains. SUVH9 and SUVH2 possess SRA domains that preferentially bind CHH and CG methylation, respectively, and these proteins are thought to act late in the RdDM pathway (FIG. 2), possibly functioning to recruit or retain DRM2 at loci targeted for methylation133.
DNA DEMETHYLATION
Although in most cases DNA methylation is a stable epigenetic mark, reduced levels of methylation are observed during development in both plants and mammals. This net loss of methylation can either occur passively, via replication in the absence of functional maintenance methylation pathways, or actively by removing methylated cytosines.
Active Demethylation in plants
In plants, active demethylation is achieved by DNA glycosylase activity, likely in combination with the base excision repair (BER) pathway134,135 (FIG. 7). DEMETER (DME)136 and REPRESSOR OF SILENCING 1 (ROS1)137 are the founding members of a family of DNA glycosylases in Arabidopsis that also includes DEMETER-LIKE 2 and 3 (DML2 and DML3)138,139. The Arabidopsis glycosylases recognize and remove methylated cytosines from double-stranded DNA oligonucleotides irrespective of sequence context in vitro138–142 and in vivo, mutations in these genes cause increased DNA methylation in all sequence contexts at specific genomic loci121,137–140,143,144. In general, DNA glycosylases involved in BER recognize and remove mutagenic substrates, including oxidized and alkylated bases, as well as thymine/guanine (T:G) mismatches often generated by deamination of methylated cytosines145. The DME/ROS1 glycosylases have homology to the helix-hairpin-helix-Gly-Pro-Asp (HhH-GPD) class of DNA glycosylases and they are bifunctional enzymes able to break both the N-glycosidic bond, removing the base, and the DNA backbone145,146 (FIG. 7). In mammals, the resulting single nucleotide gap is then acted upon by DNA polymerase β and ligase IIIα activities, respectively, in order to repair the DNA through the short patch BER pathway. Clear homologs of these enzymes have not been identified in plants, raising the possibility that plants utilize enzymes traditionally involved in the long patch BER pathway145.
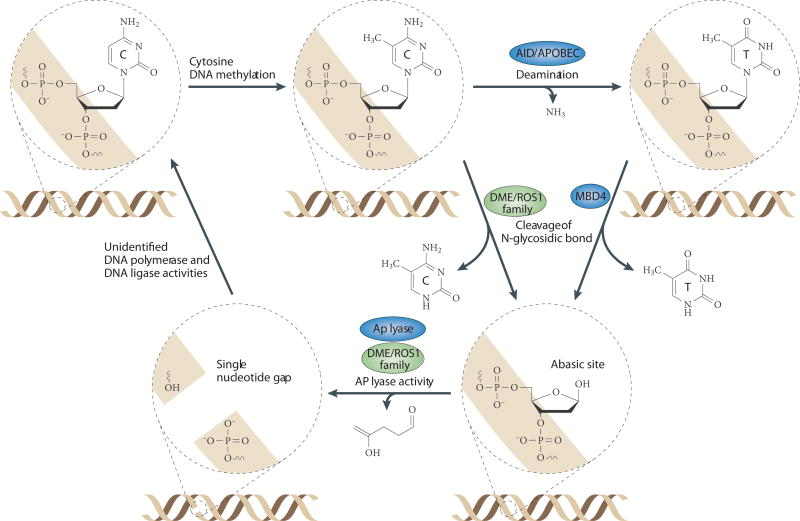
Figure 7. Active DNA Demethylation via DNA glycosylase activity and Base Excision Repair.
In Arabidopsis (green shaded proteins) methylated (CH3) cytosine (C) bases are removed by the DEMETER (DME)/ROS1 family of bifunctional 5-methylcytosine glycosylases. First, the methylated cytosine base is released by cleavage of the N-glycosidic bond, generating an abasic site. Next the phosphodiester linkage is broken both 3’ and 5’ of the abasic site through apyrimidic (AP) lysase activity, generating a single nucleotide gap in the DNA. The DNA is then proposed to be repaired by unknown DNA polymerase and ligase activities, resulting in a net loss of cytosine methylation. In Zebrafish and mammals, no efficient 5-methylcytosine glycoslases have been identified. However, in Zebrafish (blue shaded proteins), it has been proposed that the AID and APOBEC family of deaminases first convert methylated cytosines into thymines (T), generating thymine/guanine (G) (T:G) mismatches. Then, these mismatches could be recognized by the MBD4 glycosylase, resulting in removal of the thymine base and generation of an abasic site. Unlike the DME/ROS1 glycosylases, MBD4 is a monfunctional DNA glycosylase, thus another unidentified protein is likely required to provide the AP lyase activity in order to remove the sugar ring to generate a single nucleotide gap. As in Arabidopsis, this substrate is proposed to be repaired by unidentified DNA polymerase and ligase activities.
Despite similar substrate specificity, the DME/ROS1 glycosylases have distinct biological roles, with DME functioning during gametogenesis to establish imprinting78, and the other family members functioning in vegetative tissues, possibly to counteract robust DNA methylation by the RdDM pathway138,143,144,147. Unlike in mammals, where imprinting is established by the addition of methylation in an allele specific manner and is observed in both the placenta and the developing embryo, in plants, imprinting is restricted to the endosperm, the plant equivalent of the placenta78, and is established by allele specific removal of DNA methylation by DME in the central cell prior to fertilization such that only the maternal allele is expressed in the resulting endosperm78 (FIG. 4C).
Until recently, DME was only known to activate the maternal alleles of three genes: MEDEA (MEA), FLOWERING WAGENINGEN (FWA), and FERTILIZATION INDEPENDENT SEED 2 (FIS2), while in mammals ~80 imprinted genes have been identified (http://www.har.mrc.ac.uk/research/genomic_imprinting). However, findings that the genome wide decrease in CG methylation observed in the endosperm are largely dependent on DME suggest that this glycosylase acts as a global regulator of DNA methylation76,77. By comparing DNA methylation levels in embryo and endosperm tissues Gehring et al.77 were able to identify DMRs and confirm parent of origin expression of five genes, doubling the number of known imprinted genes in Arabidopsis. Approximately 40 other candidates for imprinted genes were identified77 and while imprinting at these loci remains to be experimentally verified, these findings suggest the number of imprinted loci may be more similar in plants and mammals than previously thought.
Recent characterization of Pol IV-dependent siRNA populations, which are generated from dispersed loci corresponding to >1% of the Arabidopsis genome18,25, suggests they may also be maternally imprinted82. Following reciprocal crosses between two Arabidopsis ecotypes, siRNAs from the resultant silique tissue, which contains the developing embryos, were sequenced and nearly all the Pol IV-dependent siRNAs that could be uniquely distinguished between the two ecotypes where maternal in origin82. What causes these loci to be maternally imprinted, whether this imprinting requires DME, and what function this massive extent of imprinting serves remains unknown. One hypothesis presented is that such maternal imprinting would allow recognition of self from non-self and have a suppressive effect on hybrids82. For example, maternal siRNAs could fail to target and silence a TE present in another Arabidopsis ecotype or they could target and silence a functional gene. Indeed, in Drosophila piRNAs corresponding to TEs other repeat sequences, much like the Pol IV-dependent class of siRNAs in Arabidopsis, are maternally inherited and if female flies lacking piRNAs to a particular TE are crossed to male flies harboring that element the offspring are largely inviable148.
Unlike DME, ROS1, DML2, and DML3 are expressed in vegetative tissues137–139. Comparative analysis of methylation patterns in ros1, dml2, and dml3 single mutants demonstrated that these glycosylases function redundantly, although some locus specificity was observed138. In a ros1 dml2 dml3 triple mutant, 179 loci with increased methylation relative to wild-type controls were identified despite the fact that no global increase in methylation was observed138. These loci are enriched for transposons, repetitive DNA elements, and siRNA generating loci; ~80% are also near or overlap annotated genes and the increase in DNA methylation at genes is primarily located at their 5’ and 3’ ends138,144. Together, these studies suggest ROS1, DML2, and DML3 are acting both at normally silenced loci (i.e. transposons) and at the boundaries between euchromatin and heterochromatin (i.e. genes residing in or near heterochromatic environments). At such boundaries, these glycosylases may function to protect genes that are targeted for methylation through RdDM from silencing by removing DNA methylation. At normally silenced loci, they may be required to maintain a silenced, but readily adaptable state143,144 and perhaps this is important to allow efficient reactivation of transposons during gametogenesis.
The mechanism(s) through which the DME/ROS1 glycosylases are targeted to specific loci to carryout DNA demethylation are unknown. These Arabidopsis glycosylases are quite different from most other glycosylases: they are much larger and contain two conserved domains of unknown function145 (FIG. 1). Whether these domains are required to target demethylation remains unknown. For ROS1, it has been proposed that REPRESSOR OF SILENCING 3 (ROS3), a protein that binds small single-stranded RNAs (21–26nt) in a sequence specific manner and acts in the same demethylation pathway as ROS1, may be involved in targeting ROS1 to certain loci135,149. Recent findings that DME participates in genome wide demethylation, suggests its targeting may be less specific.
Active Demethylation in Zebrafish and mammals
In mammals, genome wide decreases in DNA methylation are observed in PGCs and on the paternal genome of the zygote48,49. While mechanisms for passive demethylation appear to play a role in achieving the observed hypomethylated states, the timing of methylation loss suggests that active mechanisms may also be required150–154. Notably, DNA methylation imprints in the zygote and pre-implantation embryo, but not the PGCs, are resistant to demethylation and several proteins including Stella155, zinc finger protein 57 (Zfp57)156, and methyl-CpG binding 3 (MBD3)157 are proposed to protect specific imprinted loci from demethylation158,159.
Proteins orthologous to the DME/ROS1 family of glycosylases have not been identified in mammals and other enzymes capable of directly removing methylated cytosines have remained largely controversial146,160. However, early work in mammals showing that activation-induced cytosine deaminase (AID) and apolipoprotein B RNA-editing catalytic component 1 (APOBEC1) are expressed in cells thought to undergo active DNA demethylation and catalyze 5-methylcytosine deamination, which results in T:G mismatches, lead to a model for demethylation involving the coupling of 5-methylcytosine deaminase and thymine DNA glycosylase activities161. Such a model is supported by recent findings in the vertebrate, Zebrafish162. Rai et al.162 show that METHYL CPG BINDING DOMAIN 4 (MBD4), a HhH-GPD thymine glycosylase related to the Arabidopsis DME/ROS1 family of glycosylases with active mammalian homologs163,164, is involved in demethylation during Zebrafish development. In addition, they demonstrate that three proteins belonging to the AID/ APOBEC family (FIG. 1), AID and APOBEC2a/2b, are involved in DNA demethylation162.
By overexpressing the AID and APOBEC2a/b deaminases in the absence or presence of overexpressed human MBD4 in Zebrafish embryos, Rai et al.162 find that loss of DNA methylation, as well as deamination of methylated cytosines, appears to be limited by the abundance of the MBD4 glycosylase, suggesting that mechanisms are in place to ensure deamination does not occur unless the resultant T:G mismatch can be efficiently removed. This is an important finding since previous models for DNA demethylation involving 5-methylcytosine deamination have been discounted due to the large mutagenic potential of an uncoupled deamination step. Growth arrest and DNA-damage-inducible protein 45 alpha (Gadd45α) may aid in coupling these processes. Gadd45α interacts with MBD4, AID, and APOBEC in vitro and stimulates demethylation of plasmid DNA transfected into Zebrafish embryos as well as the association of MDB4 and AID with methylated DNA162. In addition, MBD4 possesses a methyl-binding domain (FIG. 1), which may aid in recruitment of the demethylation machinery to methylated DNA. Together, these findings suggest a model (FIG.7) in which tight coupling of 5-methylcytosine deamination by AID and APOBEC to T:G mismatch repair via MBD4 results in DNA demethylation162. Thus, in the case of Zebrafish, (and possibly mammals) there appears to be an addition deamination step in the demethylation pathway as compared to the pathway in plants. However, the downstream events leading to a net loss of cytosine methylation may be similar (FIG. 7).
In mammals, recent data presented by Kim et al.165 suggest that MBD4 may be able to directly remove methylated cytosines at the CYP27B promoter upon hormone induced MBD4 phosphorylation. While previous in vitro analysis of MBD4 glycosylase activity revealed a strong preference for thymine/guanine mismatches over methylated cytosines166, Kim et al.165 find that upon phosphorylation, the activity of MBD4 on methylated cytosines is stimulated. In vivo, the observed decrease in methylation at the CYP27B1 promoter can occur in the absence of DNA replication, suggesting an active mechanism, and is dependent on the presence of a catalytically active MBD4 protein with the serine residues targeted for phosphorylation165. Whether such a mechanism for the direct removal of methylated cytosines could account for DNA demethylation on a larger scale remains unknown.
A role for the 5-hydroxymethylcytosine modification in mammalian DNA demethylation has also been proposed. 5-hydroxymethylcytosine is present in mouse Purkinje neurons, brain tissue, and ES cells167,168 and can be generated from methylated cytosines through hydroxylation of the methyl group. Findings that ten-eleven translocation 1 (TET1) is able to catalyze the conversion of methylated cytosines into 5-hydroxymethylcytosines in vitro and that targeted depletion of TET1 by RNAi in mouse ES cells results in decreased levels of 5-hydroxymethylcytosine167 have lead to the hypothesis that TET1 and possibly other TET family members generate 5-hydroxymethylcytosines. Since proteins known to interact with methylated cytosines have reduced affinity for 5-hydroxymethylcytosine in vitro, namely methyl CpG binding protein 2 (MeCP2) and Dnmt1169,170, potential roles for this modification in the regulation of chromatin structure and in passive DNA demethylation have been proposed167,168. The hypothesis that 5-hydroxymethylcytosine could be an intermediate in an active DNA demethylation pathway involving DNA repair has also been suggested167 given that a 5-hydroxymethylcytosine specific DNA glycosylase activity has been reported in mammalian extracts171.
Passive Demethylation in plants and mammals
In addition to active DNA demethylation by DME in the central cell of the female gametophyte, passive losses of methylation likely contribute to the overall decrease in methylation observed in the endosperm. Using a reporter driven by the MET1 promoter, Jullien et al.172 showed that MET1 expression levels are reduced during female gametogenesis. They further demonstrate that MULTICOPYSUPPRESSOR of IRA1 (MSI1) and RETINOBLASTOMA RELATED 1 (RBR1) are important for the observed repression of MET1172 and conclude that MET1 is transcriptionally repressed during gametogenesis by MSI1, likely through the retinoblastoma pathway and RBR1. Notably, MSI1 and RBR1 are also required for maternal expression of the imprinted FIS2 and FWA genes172, suggesting that passive DNA demethylation resulting from decreased MET1 levels and active demethylation by DME are working together to allow activation of imprinted genes. In mammals, Dnmt1 expression also appears to be regulated through the retinoblastoma pathway, which utilizes Rb and RbAp48, homologs of the plant RBR1 and MSI1 proteins173-176. While a direct role in imprinting has not been established in mammals several observations suggest this role may also be conserved172.
The idea that passive and active demethylation pathways are working together is appealing as it fits well with several other observations: First, DME is more active on hemimethylated DNA, which would be enriched following replication in the absence of MET1, than fully methylated DNA in vitro140,141. Second, enrichment in hemimethylated DNA should decrease the chance of detrimental double strand breaks (DSBs) predicted to arise from the removal of methylated cytosines in symmetric contexts by DME. Indeed, DME is inefficient at removing methylated cytosines across from abasic sites, which should also reduce the production of DSBs140. Finally, in addition to decreasing the workload for DME and the risk of DSBs, down regulation of MET1 might also function to ensure that hemimethylated CG sites generated by DME activity on one strand of the DNA are not efficiently restored to the fully methylated state via active targeting of DNA methyltransferases through interactions with SRA domain containing proteins.
In mammals, in addition to the reported active demethylation of the paternal genome of the zygote152–154, passive demethylation is proposed to occur during pre-implantation development of the embryo48,177. This passive decrease in methylation is likely due to exclusion of the oocyte specific form of Dnmt1, Dnmt1o, from nuclei until just prior to blastocyst formation178,179. This is reminiscent of the observed localization pattern of DDM1 in pollen, where DDM1 is observed in the sperm cells, but not in the vegetative nucleus75. Thus, plants and mammals appear to employ similar mechanisms for passive DNA demethylation, including transcriptional repression of DNA methyltransferases and exclusion of the methylation machinery from the nucleus.
CONCLUSIONS
Plants and animals employ similar mechanistic strategies for controlling DNA methylation. Both utilize small RNA based pathways to target DNA methylation to transposons, both require methyl-DNA binding proteins to maintain DNA methylation patterns, and both display intimate connections between histone and DNA methylation marks. Furthermore, a growing body of evidence suggests that active demethylation may occur in animals through the use of DNA glycosylases and the BER pathway, as has been documented in Arabidopsis.
Several pathways unique to plants or mammals have also been elucidated and likely contribute to the observed differences in global methylation patterns between plants and mammals. For example, in mammals where methylation is not restricted to repeat elements, the DNA methylation machinery is recruited to specific genomic loci through interactions with chromatin marks as well as with the chromatin modifying enzymes themselves. In addition, structural studies of the mammalian de novo methyltransferases suggests a mechanism in which a Dnmt3a/Dnmt3L tetramer may oligomerize on DNA, potentially leading to the nearly global methylation status of the mammalian genome. In plants, where DNA methylation occurs in all sequence contexts, a plant specific methyltransferase, CMT3, is required to maintain CHG methylation and maintenance of CHH methylation is achieved through constant de novo methylation by DRM2.
Despite the significant advances in our understanding of DNA methylation pathways, several key questions remain, especially surrounding the issue of targeting. How DNA methyltransferases are targeted by siRNAs and piRNAs in plants and mammals, respectively, remains elusive. In terms of DNA demethylation, whether DME is specifically targeted to many sites throughout the genome during gametogenesis or whether it non-selectively removes methylation remains unclear. Similarly, whether demethylation by the other DME/ROS1 family members is specifically directed to certain loci or whether the observed methylation pattern simply reflects a balance between the RdDM and demethylation pathways requires further investigation. For mammals, determination of whether mammals accomplish DNA demethylation in a manner akin to that observed for Zebrafish will be key, and surely the question of mammalian demethylation targeting will follow.
- Epigenetic modifications
Chemical additions to DNA and histones that are associated with changes in gene expression and are heritable but do not alter the primary DNA sequence
- Histone
The main protein component of chromatin. The four core histones, H2A, H2B, H3 and H4, form a globular octameric complex called a nucleosome upon which DNA is wrapped. The N-terminal regions of histone proteins are largely unstructured and are subject to various chemical modifications including methylation
- CpG island
A sequence of at least 200 bp with a greater number of CpG sites than expected for its GC content. These regions are often GC rich, typically undermethylated, and are found upstream of many mammalian genes
- RNA-directed DNA Methylation (RdDM)
Pathway through which small RNAs (24nt) target the de novo methyltransferase, DRM2, to homologous genomic loci to establish DNA methylation, leading to transcriptional gene silencing
- RNA interference
A process ofpost-transcriptional gene silencing in which small RNAs, often generated by the activity of an RNA-dependent RNA (RDR) polymerase and a Dicer (DCR) endoribonuclease, are bound by argonaute (AGO) proteins and target cleavage of homologous mRNA transcripts
- Dicer
An RNase III family endonuclease that processes dsRNA into small interfering RNAs
- Argonautes
Effector proteins of small RNA-directed silencing. Small RNAs guide Argonautes to their RNA targets. Argonaute proteins are characterized by two domains — Piwi (a ribonuclease domain) and PAZ (a ssRNA-binding module)
- Chromatin remodeling factors
Proteins that have the capacity to remodel chromatin, often using the energy of ATP, so that gene transcription can be activated or silenced
- Small interfering RNAs
(20–25 nucleotides long) RNAs that are generated from double stranded RNAs and serve as guides for the cleavage of homologous mRNAs in RNA interference or for the addition of chromatin modifications including histone and DNA methylation at homologous genomic sequences in transcriptional gene silencing
- Cajal bodies
Nuclear bodies associated with the maturation of ribonucleoprotein complexes
- Heterochromatin
Densely packaged form of chromatin associated with repressive histone modifications, DNA methylation, and gene silencing
- Primordial germ cells
Population of embryonic cells from which germ cells are formed
- Imprinted genes
Genes in which one allele is expressed in a parent of origin specific manner
- Bisulfite sequencing
Technique where treatment of DNA with bisulfite, which converts cytosines into uracils but does not modify methylated cytosines, is used to determine the DNA methylation pattern
- Vegetative cell nucleus
Nucleus of the terminally differentiated vegetative cell which does not contribute genetic information tosubsequent generations
- Gametophyte
A multicellular structure that is generated from a haploid spore through mitotic cell divisions and contains themale or female gamete
- Endosperm
Product of the fertilization of the central cell of the female gametophyte, whichis present in the seeds of most flowering plants and provides nutrition to the developing embryo
- Primary piRNAs
The product of piRNA precursor transcript processing. These piRNAs have a preference for a 5′ U
- Secondary piRNAs
The product of a ping-pong amplification cycle. These piRNAs are antisence to primary piRNAs and have a preference for an A at position 10
- Tudor domain
conserved protein motifthat is able to recognize symmetrically dimethylatedarginines
- Base excision repair
A cellular mechanism that repairs damaged DNA and is initiated by the activity of DNA glycosylases
- Ecotype
A genetically distinct population within a widely spread species
- Silique
An elongated seed capsule thatis formed after fertilization
- Hybrids
Offspring produced by crossing two different populations within a single species
- Zygote
Single diploid cell formed by the union of two haploid germ cells
- Blastocyst
an embryonic stage characterized by the first definitive lineages
References
- 1.Ehrlich M, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–42. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009 doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 7.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–24. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 8.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. This study, along with ref. 119, provide single nucleotide resolution, genome wide mapping of DNA methylation patterns in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci. 2009;66:596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 12.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–50. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–60. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 14.Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–76. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 15.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–76. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Huettel B, et al. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769:358–74. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008;13:390–7. doi: 10.1016/j.tplants.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:3145–50. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lahmy S, et al. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci U S A. 2009;106:941–6. doi: 10.1073/pnas.0810310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ream TS, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L, et al. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16:91–3. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 22.He XJ, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–30. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bies-Etheve N, et al. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009 doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–20. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A. 2007;104:4536–41. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haag JR, Pontes O, Pikaard CS. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS ONE. 2009;4:e4110. doi: 10.1371/journal.pone.0004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–22. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Pontier D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–40. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. Embo J. 2006;25:2828–36. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–44. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li CF, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 33.Li CF, et al. Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana. PLoS Genet. 2008;4:e27. doi: 10.1371/journal.pgen.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–48. doi: 10.1016/j.cell.2008.09.035. This study identifies RNA transcripts that are dependent on the Pol V polymerase and DRD1 and demonstrates they are required for DNA methylation and silencing but not for siRNA production. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He XJ, et al. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4-and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–4. doi: 10.1038/ng.365. This study identifies IDN2 as a novel component of the RdDM pathway that encodes an RNA binding protein able to interact with double stranded RNA duplexes containing 5′ overhangs in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausin I, Mockler TC, Chory J, Jacobsen S. IDN1 and IDN2: two proteins required for de novo DNA methylation in Arabidopsis thaliana. Nature Structural and Molecular Biology. doi: 10.1038/nsmb.1690. (Accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng B, et al. Intergenic transcription by RNA Polymerase IIcoordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009 doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanno T, et al. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep. 11:65–71. doi: 10.1038/embor.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He XJ, et al. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23:2717–22. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanno T, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–5. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 44.Kanno T, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–5. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 45.Smith LM, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–21. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kafri T, et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–14. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 47.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–82. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 48.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. Biochemical purification of Dnmt3L revealing and it co-purifies with the de novo DNA methyltransferases as well as with the four core histone proteins. The interaction between Dnmt3L and histones what further characterized and shown to be specific for unmethylated histone 3 lysine 9 tails. [DOI] [PubMed] [Google Scholar]
- 49.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–40. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 50.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. Crystal structure showing the C-terminal regions of Dnmt3a and Dnmt3L form a tetrameric complex. Superimposition of a DNA fragment from another crystal structure onto this structure suggests the Dnmt3a active sites are positioned approximately 10 base pairs apart. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otani J, et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–41. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–51. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fournier C, et al. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. Embo J. 2002;21:6560–70. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delaval K, et al. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. Embo J. 2007;26:720–9. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vu TH, Li T, Hoffman AR. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum Mol Genet. 2004;13:2233–45. doi: 10.1093/hmg/ddh244. [DOI] [PubMed] [Google Scholar]
- 56.Yamasaki Y, et al. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum Mol Genet. 2005;14:2511–20. doi: 10.1093/hmg/ddi255. [DOI] [PubMed] [Google Scholar]
- 57.Ciccone DN, et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–8. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 58.Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–57. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 60.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–66. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 62.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di-and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chotalia M, et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–17. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–23. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 66.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002;99:16916–21. doi: 10.1073/pnas.262443999. Biophysical characterization of a the tetrameric complex described in ref.55 showing formation of nucleoprotein filaments and demonstrating this complex generates periodic DNA methylation patterns in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–69. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 68.Jurkowska RZ, et al. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 2008;36:6656–63. doi: 10.1093/nar/gkn747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glass JL, Fazzari MJ, Ferguson-Smith AC, Greally JM. CG dinucleotide periodicities recognized by the Dnmt3a-Dnmt3L complex are distinctive at retroelements and imprinted domains. Mamm Genome. 2009 doi: 10.1007/s00335-009-9232-3. [DOI] [PubMed] [Google Scholar]
- 70.Ferguson-Smith AC, Greally JM. Epigenetics: perceptive enzymes. Nature. 2007;449:148–9. doi: 10.1038/449148a. [DOI] [PubMed] [Google Scholar]
- 71.Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J Mol Biol. 2001;309:1201–8. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 72.Lindroth AM, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. Embo J. 2004;23:4286–96. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao Q, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–11. doi: 10.1038/nsmb.1568. This study investigates the expression pattern of transposons during plant development and demonstrates they are expressed and active in pollen but that this reactivation likely only occurs only in the vegetative nucleus and not the sperm cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fabbrizio E, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–5. doi: 10.1093/embo-reports/kvf136. This study, along with ref. 75, compares the methylation status of the endosperm and embryo tissues and demonstrates that the endosperm is globally hypomethylated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–72. doi: 10.1016/j.cell.2008.12.038. In addition to demonstrating a global decrease in methylation in the endosperm(see ref. 74), this study identifies several novel imprinted genes in the Arabidopsis genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–4. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–51. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huh JH, Bauer MJ, Hsieh TF, Fischer RL. Cellular programming of plant gene imprinting. Cell. 2008;132:735–44. doi: 10.1016/j.cell.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 79.Pina C, Pinto F, Feijo JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005;138:744–56. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–8. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 81.Baroux C, Pecinka A, Fuchs J, Schubert I, Grossniklaus U. The triploid endosperm genome of Arabidopsis adopts a peculiar, parental-dosage-dependent chromatin organization. Plant Cell. 2007;19:1782–94. doi: 10.1105/tpc.106.046235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mosher RA, et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009 doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 83.Teixeira FK, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–4. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 84.Aravin AA, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–27. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 85.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 86.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 87.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. This study demonstrates that methylation defects in a PIWI clade mutant occur at the time of de novo methylation in male germ cells, suggesting piRNAs are required for de novo DNA methylation at transposons rather than for the maintenance of pre-existing DNA methylation. [DOI] [PubMed]
- 88.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 89.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–17. doi: 10.1101/gad.1640708. This study, along with ref. 87,isolate piRNAs from fetal germ cellsandshow they are highly enrich for transposon sequences and possess the characteristics of primary and secondary piRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–7. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 91.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 93.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 94.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 95.Aravin AA, Bourc'his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–5. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen C, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci U S A. 2009;106:20336–41. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kirino Y, et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–8. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–62. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reuter M, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 100.Kojima K, et al. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells. 2009;14:1155–65. doi: 10.1111/j.1365-2443.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- 101.Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19:640–4. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chuang LS, et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 103.Schermelleh L, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–12. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Egger G, et al. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci U S A. 2006;103:14080–5. doi: 10.1073/pnas.0604602103. This study,along with ref. 105, demonstrates that UHRF1 is required to maintain DNA methylation in mammals. This study further demonstrates that the SRA domain of UHRF1 specifically interacts with hemimethylated CG sites and is required for the association of Dnmt1 with chromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Spada F, et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–71. doi: 10.1083/jcb.200610062. See ref.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 107.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 108.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–21. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 109.Hashimoto H, et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–9. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qian C, et al. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J Biol Chem. 2008;283:34490–4. doi: 10.1074/jbc.C800169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Avvakumov GV, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–5. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 112.Meilinger D, et al. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep. 2009;10:1259–64. doi: 10.1038/embor.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–4. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang J, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32:5019–28. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Woo HR, Dittmer TA, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 2008;4:e1000156. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21:267–77. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hirochika H, Okamoto H, Kakutani T. Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell. 2000;12:357–69. doi: 10.1105/tpc.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–3. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- 119.Yan Q, Huang J, Fan T, Zhu H, Muegge K. Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. Embo J. 2003;22:5154–62. doi: 10.1093/emboj/cdg493. See ref. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–9. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 123.Tran RK, et al. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol. 2005;15:154–9. doi: 10.1016/j.cub.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 124.Johnson LM, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–84. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lindroth AM, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–80. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 126.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–8. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–60. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 128.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. Embo J. 2002;21:6842–52. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jackson JP, et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–15. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 130.Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol. 2005;25:10507–15. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–76. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cao X, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–7. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 133.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4:e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ikeda Y, Kinoshita T. DNA demethylation: a lesson from the garden. Chromosoma. 2009;118:37–41. doi: 10.1007/s00412-008-0183-3. [DOI] [PubMed] [Google Scholar]
- 135.Zhu JK. Active DNA Demethylation Mediated by DNA Glycosylases. Annu Rev Genet. 2009 doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 137.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–14. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 138.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci U S A. 2007;104:6752–7. doi: 10.1073/pnas.0701861104. This study, along with ref. 139, demonstrates that DME is an active 5-methylcytosine DNA glycosylase in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol. 2008;67:671–81. doi: 10.1007/s11103-008-9346-0. See ref. 138 and 140. [DOI] [PubMed] [Google Scholar]
- 140.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. This study, along with ref. 139, demonstrates that ROS1 is an active 5-methylcytosine DNA glycosylase in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Morales-Ruiz T, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci U S A. 2006;103:6853–8. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci U S A. 2006;103:11796–801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–9. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 144.Penterman J, Uzawa R, Fischer RL. Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 2007;145:1549–57. doi: 10.1104/pp.107.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Baute J, Depicker A. Base excision repair and its role in maintaining genome stability. Crit Rev Biochem Mol Biol. 2008;43:239–76. doi: 10.1080/10409230802309905. [DOI] [PubMed] [Google Scholar]
- 146.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 147.Kapoor A, Agius F, Zhu JK. Preventing transcriptional gene silencing by active DNA demethylation. FEBS Lett. 2005;579:5889–98. doi: 10.1016/j.febslet.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 148.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zheng X, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–62. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 151.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–81. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 153.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–8. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 154.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 155.Nakamura T, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 156.Li X, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–57. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Reese KJ, Lin S, Verona RI, Schultz RM, Bartolomei MS. Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genet. 2007;3:e137. doi: 10.1371/journal.pgen.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ciccone DN, Chen T. Histone lysine methylation in genomic imprinting. Epigenetics. 2009;4:216–20. doi: 10.4161/epi.8974. [DOI] [PubMed] [Google Scholar]
- 159.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–9. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 160.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–8. doi: 10.1016/j.cell.2008.06.009. This study uses Zebrafish embryos as a model to study DNA demethylation and provide evidence that cytosine methylation can be removed through the coordinated activities 5-methylcytosine deaminaseas and thymine mismatch DNA glycosylases. [DOI] [PubMed] [Google Scholar]
- 161.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–60. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 162.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–12. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–4. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 164.Petronzelli F, et al. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J Cell Physiol. 2000;185:473–80. doi: 10.1002/1097-4652(200012)185:3<473::AID-JCP19>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 165.Kim MS, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–12. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 166.Zhu B, et al. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000;28:4157–65. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Valinluck V, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–8. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–50. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 171.Cannon SV, Cummings A, Teebor GW. 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem Biophys Res Commun. 1988;151:1173–9. doi: 10.1016/s0006-291x(88)80489-3. [DOI] [PubMed] [Google Scholar]
- 172.Jullien PE, et al. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008;6:e194. doi: 10.1371/journal.pbio.0060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.McCabe MT, Davis JN, Day ML. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 2005;65:3624–32. doi: 10.1158/0008-5472.CAN-04-2158. [DOI] [PubMed] [Google Scholar]
- 174.Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 2003;31:3101–13. doi: 10.1093/nar/gkg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McCabe MT, Low JA, Imperiale MJ, Day ML. Human polyomavirus BKV transcriptionally activatesDNA methyltransferase 1 through the pRb/E2F pathway. Oncogene. 2006;25:2727–35. doi: 10.1038/sj.onc.1209266. [DOI] [PubMed] [Google Scholar]
- 176.Nicolas E, Ait-Si-Ali S, Trouche D. The histone deacetylase HDAC3 targets RbAp48 to the retinoblastoma protein. Nucleic Acids Res. 2001;29:3131–6. doi: 10.1093/nar/29.15.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 178.Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6:2536–41. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- 179.Mertineit C, et al. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–97. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- 180.Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. Rna. 2009;15:337–45. doi: 10.1261/rna.1422509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kawaoka S, et al. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. Rna. 2009;15:1258–64. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lau NC, et al. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009 doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kawaoka S, et al. Bombyx small RNAs: genomic defense system against transposons in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38:1058–65. doi: 10.1016/j.ibmb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 184.Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. Embo J. 2009 doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]