Abstract
Here we present the genomic sequence, with analysis, of a canarypox virus (CNPV). The 365-kbp CNPV genome contains 328 potential genes in a central region and in 6.5-kbp inverted terminal repeats. Comparison with the previously characterized fowlpox virus (FWPV) genome revealed avipoxvirus-specific genomic features, including large genomic rearrangements relative to other chordopoxviruses and novel cellular homologues and gene families. CNPV also contains many genomic differences with FWPV, including over 75 kbp of additional sequence, 39 genes lacking FWPV homologues, and an average of 47% amino acid divergence between homologues. Differences occur primarily in terminal and, notably, localized internal genomic regions and suggest significant genomic diversity among avipoxviruses. Divergent regions contain gene families, which overall comprise over 49% of the CNPV genome and include genes encoding 51 proteins containing ankyrin repeats, 26 N1R/p28-like proteins, and potential immunomodulatory proteins, including those similar to transforming growth factor β and β-nerve growth factor. CNPV genes lacking homologues in FWPV encode proteins similar to ubiquitin, interleukin-10-like proteins, tumor necrosis factor receptor, PIR1 RNA phosphatase, thioredoxin binding protein, MyD116 domain proteins, circovirus Rep proteins, and the nucleotide metabolism proteins thymidylate kinase and ribonucleotide reductase small subunit. These data reveal genomic differences likely affecting differences in avipoxvirus virulence and host range, and they will likely be useful for the design of improved vaccine vectors.
Within the Chordopoxvirinae subfamily (poxviruses of vertebrates) of the Poxviridae, Avipoxvirus is the only characterized genus to infect nonmammalian hosts (57, 60), including more than 60 species of wild birds representing 20 families (60, 97). Variable restriction enzyme profiles suggest significant genomic differences among avipoxviruses (32, 97, 98). Cross-infection studies also suggest genetic differences among avipoxviruses, as specific virus-host combinations may not be cross-protective and result in a wide range of pathogenic effects, from absence of clinical disease to generalized infection and death (8, 97).
Canarypox virus (CNPV) is an avipoxvirus and etiologic agent of canarypox, a disease of birds both in the wild and in commercial aviaries, where significant losses result (15, 16, 33, 45). Canarypox has been described broadly as the poxviral disease of passeriform (song) birds, efficiently causing disease in passerine hosts compared to galliform (domestic fowl) and columbiform (pigeon) hosts (8, 15, 22, 33). However, passerine host preferences exist, and current International Committee on Taxonomy of Viruses classification differentiates between CNPV and several other passerine isolates (60, 97). CNPV produces clinical signs similar to generalized poxviral infections of other birds, including both cutaneous and diptheretic disease forms caused by the prototypical galliform avipoxvirus, fowlpox virus (FWPV), and including proliferative and necrotic changes in epithelial tissues of the dermis, notably around the eyes and commissures of the beak, feet, and respiratory tract (8, 15, 16, 22, 33, 97). Canarypox, however, is generally associated with higher mortality rates than seen in fowlpox, commonly approaching 100%, and may occur without characteristic skin lesions (8, 15, 22, 33). Histologically and ultrastructurally, CNPV undergoes morphogenic stages similar to FWPV and other chordopoxviruses (ChPV), causing type A and B intracytoplasmic inclusions in epithelial and mononuclear cells of permissive hosts; however, CNPV may have a broader tissue tropism than FWPV (12, 15, 33, 77).
CNPV has been successfully used as a host range-restricted mammalian expression vector and is a vaccine vector of increasing importance, with CNPV-based veterinary vaccines commercially available and human vaccines undergoing clinical trials (34, 35, 44, 90). Licensed and experimental CNPV-based vaccines, most of which utilize the highly attenuated ALVAC strain of CNPV (91), encode a range of pathogen and tumor-associated antigens, including those from rabies virus, canine distemper virus, feline leukemia virus, human immunodeficiency virus, human cytomegalovirus, hepatitis B and C viruses, Plasmodium falciparum, melanoma, and colorectal cancer (3, 35, 67-69, 90, 91). Vaccine use takes advantage of the abortive infection that avipoxvirus vectors undergo in mammalian cells while still expressing virally encoded antigens to safely generate cellular, humoral, and protective immune responses (69, 84, 91-94). CNPV-based vaccines have proven effective in prime-boost vaccine strategies and as immunoadjuvants through expression of recombinant cytokines and costimulatory proteins (47, 67-69, 90). Recent evidence suggests that dendritic cell antigen presentation, maturation, and apoptosis are important in CNPV-generated immunity (24, 41, 55, 59). Improved understanding of virus-host interactions should yield improved vaccine vectors, as demonstrated by recent incorporation of vaccinia virus (VACV) host response modification genes to create a third-generation CNPV-based vaccine (28, 44).
Current molecular data indicate that CNPV is related to FWPV, a virus for which the genomic sequence has been determined (2, 5). Features of the 288-kbp FWPV genome included (i) large genomic segments containing replicative genes conserved and syntenic with those of other ChPV, (ii) rearrangement of specific genomic regions relative to other ChPV, (iii) the presence of numerous gene families and novel homologues of cellular genes likely involved in virus-host interaction, viral virulence, and viral host range which account for the large size of the FWPV genome relative to ChPV, and (iv) the location of potential virulence and host range genes both in terminal genomic regions and discrete, variable central genomic regions near the junctions of large genomic rearrangements. While other select avipoxvirus species closely resemble FWPV in restriction fragment patterns and estimated genomic size, the CNPV genome is larger and comparatively divergent (79, 92, 97). In addition, limited sequence data indicate that while CNPV and FWPV share regions of similar gene order and are likely monophyletic within the Poxviridae, amino acid identity between open reading frame (ORF) homologues (55 to 74%) is comparable to that seen between different ChPV genera (5). Thus, there are likely to be substantial differences in the genomic elements responsible for virulence and host range differences of CNPV and FWPV.
The rational design of safer and more effective CNPV vaccines and CNPV-based expression vectors will benefit from a more complete understanding of viral genes functioning in viral virulence, pathogenesis, tissue tropism, and immune evasion. Here we present the genomic sequence of a CNPV, comparing it to FWPV and characterizing novel aspects of the genome likely significant for viral virulence and host range.
MATERIALS AND METHODS
Viral DNA isolation, cloning, sequencing, and sequence analysis.
CNPV strain Wheatley C93 was obtained from the American Type Culture Collection (ATCC VR-111) (8, 39). Viral genomic DNA was extracted from CNPV-infected primary chicken embryo fibroblasts derived from specific-pathogen-free eggs (Charles River SPAFAS, North Franklin, Conn.). Random DNA fragments were obtained by incomplete enzymatic digestion with Tsp509I endonuclease (New England Biolabs, Beverly, Mass.), and DNA fragments larger than 1.0 kbp were cloned, used in dideoxy sequencing reactions as previously described, and analyzed on an ABI Prism 3700 automated DNA sequencer (Applied Biosystems, Foster City, Calif.) (2). Sequence data were assembled with the Phrap and CAP3 software programs (26, 40), and gaps were closed as described previously (1). The final DNA consensus sequence represented on average 7.5-fold redundancy at each base position and a Consed estimated error rate of less than 0.01 per 10 kbp (26, 27, 36).
Sequence analysis was conducted essentially as previously described (1, 2). Briefly, genome DNA composition, structure, repeats, and restriction enzyme patterns were analyzed and ORF maps were created using the GCG version 10 and MacVector version 7.0 software packages, tandem repeats finder, Pipmaker, and EMBOSS (ftp:uk.embnet.org/pub/EMBOSS) programs (9, 20, 80). ORFs longer than 30 codons were evaluated for coding potential and subjected to homology searches as previously described, with additional searches against the Pfam, TIGRFAMs, and SMART databases (1, 2, 7). Based on these criteria, at least 328 ORFs were annotated as potential genes. All CNPV ORFs were compared to FWPV genomic sequences, and vice versa, using the tfasta and tfastx programs (70, 71). Multiple sequences were aligned with ClustalW, and phylogenetic comparisons were done with the PHYLO_WIN software package (30, 95). Database accession numbers are from GenBank, Swissprotein, or PIR databases unless otherwise noted.
Nucleotide sequence accession number.
The CNPV genome sequence has been deposited in GenBank under accession no. AY318871.
RESULTS AND DISCUSSION
Organization of the CNPV genome.
The CNPV genome was assembled into a contiguous sequence of 359,853 bp, which is larger than the 330 kbp estimated for highly attenuated CNPV (60, 92). Because no terminal resolution or hairpin loop sequences were sequenced, the left-most nucleotide of the assembled sequence was arbitrarily designated base number one. The CNPV genome, like FWPV, contains a central genomic region bounded by two identical inverted terminal repeat (ITR) regions. The overall nucleotide composition is 70% A+T, with localized regions of higher or lower A+T composition.
Assembled ITRs of CNPV are 6,491 bp each in length and have a lower-than-average A+T composition (63% A+T). Each ITR contains six ORFs (CNPV001/328, CNPV002/327, CNPV003/326, CNPV004/325, CNPV005/324, and CNPV006/323), and two ORFs (CNPV007 and CNPV322) cross the ITR boundary to share 30 identical amino acids in their carboxyl termini. Notably, CNPV005/324 are likely orthologous to genes in the FWPV ITR, containing a remarkably high level of amino acid identity (86%). The noncoding, most-terminal region of the ITR contains at least 31, 9, and 7 copies of 17-, 41-, and 48-bp tandem repeats, respectively. The CNPV 17-bp tandem repeat (TTACGAGGTAACGAGTG) contains a CGAG(4N)CGAG pattern similar to the 32-bp terminal tandem repeats of FWPV (2), and it contains additional internal sequence to comprise CNPV 41- and 48-bp repeats. Comparison of predicted restriction fragments with those obtained experimentally indicated that additional sequences of less than 500 bp may be present in each ITR (data not shown).
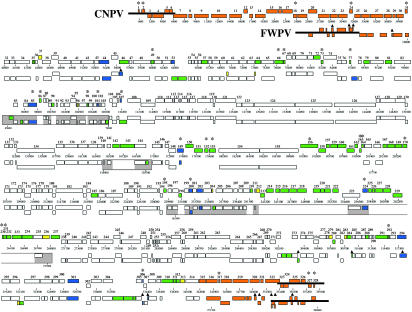
CNPV contains at least 328 ORFs which have been annotated here as putative genes. These genes represent a 91% coding density and encode proteins of 42 to 1,951 amino acids (Table 1). A total of 209 CNPV genes were annotated here as putative orthologues of FWPV genes based on amino acid identity, ORF length, and genomic position (Table 1). Alignment of likely gene orthologues indicated overall genomic synteny between CNPV and FWPV (Fig. 1). Exceptions included terminal genomic regions (CNPV001 to CNPV031 and CNPV315 to CNPV328) which have undergone significant rearrangements relative to FWPV; random individual gene insertions, deletions, and replacements relative to FWPV; and internal genomic regions (CNPV142 to CNPV170 and CNPV195 to CNPV237) that are variable and expanded relative to FWPV (Fig. 1).
TABLE 1.
CNPV ORFs
| ORF | Position (length [aa])a | Best matchb
|
FWPVe
|
VACVg ORF | Predicted structure and/or function | |||
|---|---|---|---|---|---|---|---|---|
| Accession no.c | Scored | Species | ORF (length [aa]) | % IDf | ||||
| CNPV001 | 1412-1627 (72) | — | ||||||
| CNPV002 | 1900-2412 (171) | — | ||||||
| CNPV003 | 3397-2786 (204) | FPV001 (205) | 27 | C-type lectin-like protein | ||||
| CNPV004 | 5064-3523 (514) | FPV246 (592) | 28 | B4R | Ankyrin repeat protein | |||
| CNPV005 | 5190-5855 (222) | FPV002 (222) | 86 | |||||
| CNPV006 | 5818-6363 (182) | FPV002.5 (47) | 56 | |||||
| CNPV007 | 8422-6401 (674) | FPV242 (358) | 30 | B4R | Ankyrin repeat protein | |||
| CNPV008 | 9117-8611 (169) | AF470404 | 144 | Pongo pygmaeus | FPV239 (163) | 24 | A40R* | C-type lectin-like protein |
| CNPV009 | 11470-9407 (688) | FPV244 (668) | 79 | B4R | Ankyrin repeat protein | |||
| CNPV010 | 14032-11831 (734) | FPV231 (256) | 34 | B4R | Ankyrin repeat protein | |||
| CNPV011 | 16211-14454 (586) | FPV246 (592) | 47 | B4R | Ankyrin repeat protein | |||
| CNPV012 | 16473-17039 (189) | FPV229 (180) | 28 | |||||
| CNPV013 | 17755-17252 (168) | FPV015 (177) | 38 | |||||
| CNPV014 | 19545-18076 (490) | FPV017(245) | 37 | Ig domain protein | ||||
| CNPV015 | 19710-21293 (528) | FPV162(603) | 28 | B4R | Ankyrin repeat protein | |||
| CNPV016 | 21357-21860 (168) | FPV008 (167) | 49 | A40R | C-type lectin-like protein | |||
| CNPV017 | 21941-23398 (486) | FPV034 (415) | 36 | B4R | Ankyrin repeat protein | |||
| CNPV018 | 24067-23495 (191) | AF026277 | 118 | Trichosurus vulpecula | — | IL-10-like protein | ||
| CNPV019 | 25495-24188 (436) | FPV014 (437) | 48 | B4R | Ankyrin repeat protein | |||
| CNPV020 | 25676-26932 (419) | FPV245 (436) | 46 | B4R | Ankyrin repeat protein | |||
| CNPV021 | 28678-27074 (535) | FPV246 (592) | 37 | B4R | Ankyrin repeat protein | |||
| CNPV022 | 29796-28723 (358) | FPV010 (355) | 64 | C12L | Serpin | |||
| CNPV023 | 31161-29881 (427) | FPV006 (418) | 33 | C10L | C4L/C10L-like protein | |||
| CNPV024 | 31439-31972 (178) | — | ||||||
| CNPV025 | 33091-32192 (300) | FPV011 (278) | 54 | α-SNAP | ||||
| CNPV026 | 34330-33185 (382) | FPV246 (592) | 29 | B4R | Ankyrin repeat protein | |||
| CNPV027 | 36279-34402 (626) | FPV242-243 (262-358) | 31-32 | K1L | Ankyrin repeat protein | |||
| CNPV028 | 37502-36417 (362) | FPV240 (410) | 32 | B4R | Ankyrin repeat protein | |||
| CNPV029 | 38040-37615 (142) | FPV235 (143) | 25 | A40R* | C-type lectin-like protein | |||
| CNPV030 | 39147-38128 (340) | FPV012 (331) | 45 | K1L | Ankyrin repeat protein | |||
| CNPV031 | 39373-39729 (119) | — | ||||||
| CNPV032 | 40687-39962 (242) | FPV016 (238) | 32 | Ig domain, IFN-γ binding protein | ||||
| CNPV033 | 41503-40766 (246) | FPV017 (245) | 38 | Ig domain protein | ||||
| CNPV034 | 43578-41602 (659) | FPV018 (700) | 43 | C9L | Ankyrin repeat protein | |||
| CNPV035 | 44369-43968 (134) | FPV003 (123) | 33 | C-type lectin-like protein | ||||
| CNPV036 | 44754-44470 (95) | FPV019 (104) | 35 | |||||
| CNPV037 | 44818-45354 (179) | Fragment | ||||||
| CNPV038 | 46603-45365 (413) | FPV020 (426) | 42 | C10L | C4L/C10L-like protein | |||
| CNPV039 | 46721-47701 (327) | FPV021 (320) | 45 | G protein-coupled receptor | ||||
| CNPV040 | 49497-47725 (591) | FPV022 (578) | 37 | B4R | Ankyrin repeat protein | |||
| CNPV041 | 50862-49573 (430) | FPV023 (434) | 55 | M1L | Ankyrin repeat protein | |||
| CNPV042 | 52728-50914 (605) | FPV024 (596) | 49 | B4R | Ankyrin repeat protein | |||
| CNPV043 | 53439-52837 (201) | FPV025 (203) | 55 | |||||
| CNPV044 | 54923-53484 (480) | FPV024 (596) | 30 | B4R | Ankyrin repeat protein | |||
| CNPV045 | 55197-56192 (332) | FPV027 (336) | 44 | G protein-coupled receptor | ||||
| CNPV046 | 57574-56225 (450) | FPV024 (596) | 29 | Ankyrin repeat protein | ||||
| CNPV047 | 58015-57644 (124) | FPV029 (124) | 52 | Conserved hypothetical protein | ||||
| CNPV048 | 60588-58186 (801) | FPV030 (817) | 57 | Alkaline phosphodiesterase | ||||
| CNPV049 | 61127-60678 (150) | — | ||||||
| CNPV050 | 62239-61184 (352) | FPV031 (341) | 45 | B4R | Ankyrin repeat protein | |||
| CNPV051 | 63516-62314 (401) | FPV032 (232) | 51 | DNase II | ||||
| CNPV052 | 64057-63545 (171) | M88072 | 168 | Gallus gallus | FPV235 (143) | 29 | A34R | C-type lectin-like protein |
| CNPV053 | 64675-64238 (146) | FPV035 (135) | 52 | |||||
| CNPV054 | 65090-64671 (140) | FPV036 (153) | 32 | |||||
| CNPV055 | 65633-65145 (163) | FPV037 (162) | 38 | |||||
| CNPV056 | 66067-65633 (145) | FPV038 (145) | 69 | F2L | dUTPase | |||
| CNPV057 | 67014-66097 (306) | FPV040 (337) | 24 | Serpin | ||||
| CNPV058 | 67586-67062 (175) | FPV039 (175) | 39 | Bcl-2 | ||||
| CNPV059 | 68660-67647 (338) | FPV040 (337) | 50 | C12L* | Serpin | |||
| CNPV060 | 69657-68710 (316) | FPV041 (206) | 29 | |||||
| CNPV061 | 71442-69748 (565) | FPV043 (564) | 70 | A50R | DNA ligase | |||
| CNPV062 | 72533-71484 (350) | FPV044 (358) | 51 | C12L | Serpin | |||
| CNPV063 | 73680-72607 (358) | FPV046 (370) | 60 | A44L | Hydroxysteroid dehydrogenase | |||
| CNPV064 | 74590-73745 (282) | X63427 | 145 | Xenopus laevis | FPV080 (363) | 21 | TGF-β | |
| CNPV065 | 76433-74685 (583) | FPV047 (612) | 48 | A39R | Semaphorin | |||
| CNPV066 | 77758-76544 (405) | — | ||||||
| CNPV067 | 78013-77843 (57) | — | ||||||
| CNPV068 | 78173-78943 (257) | FPV048 (261) | 80 | GNS1/SUR4-like protein | ||||
| CNPV069 | 79038-79502 (155) | FPV049 (154) | 72 | A1L | Late transcription factor VLTF-2 | |||
| CNPV070 | 79522-81174 (551) | FPV050 (552) | 82 | D13L | Rifampin resistance protein, IMV assembly | |||
| CNPV071 | 81209-82075 (289) | FPV051 (289) | 76 | D12L | mRNA capping enzyme small subunit/PICK> | |||
| CNPV072 | 82099-83034 (312) | FPV060 (188) | 30 | CC chemokine-like protein | ||||
| CNPV073 | 83444-83118 (109) | — | ||||||
| CNPV074 | 83515-85419 (635) | FPV052 (637) | 82 | D11L | NPH-1, transcription termination factor | |||
| CNPV075 | 86104-85415 (230) | FPV053 (225) | 67 | D10R | mutT motif, gene expression regulator | |||
| CNPV076 | 86786-86091 (232) | FPV054 (231) | 76 | D9R | mutT motif | |||
| CNPV077 | 87409-87176 (78) | — | ||||||
| CNPV078 | 88879-88400 (160) | FPV056 (161) | 74 | D7R | RNA polymerase subunit RPO18 | |||
| CNPV079 | 90033-89218 (272) | FPV125 (345) | 41 | Ig domain protein | ||||
| CNPV080 | 92060-90162 (633) | FPV057 (633) | 92 | D6R | Early transcription factor small subunit | |||
| CNPV081 | 93244-92246 (333) | FPV125 (345) | 43 | B16R* | Ig domain protein | |||
| CNPV082 | 95922-93541 (794) | FPV058 (791) | 79 | D5R | NTPase, DNA replication | |||
| CNPV083 | 96741-96079 (221) | FPV060 (188) | 30 | CC chemokine-like protein | ||||
| CNPV084 | 97484-96831 (218) | FPV062 (218) | 83 | D4R | Uracil DNA glycosylase | |||
| CNPV085 | 98751-97543 (403) | AF023917 | 464 | Homo sapiens | — | H1L* | PIR1-like RNA phosphatuse | |
| CNPV086 | 99035-99385 (117) | AF156738 | 183 | Salvelinus fontinalis | — | C22L | TNFR-like protein | |
| CNPV087 | 99428-100021 (198) | FPV064 (200) | 74 | Glutathione peroxidase | ||||
| CNPV088 | 100061-100360 (100) | FPV065 (111) | 39 | |||||
| CNPV089 | 100847-100371 (159) | FPV066 (122) | 50 | |||||
| CNPV090 | 101217-100837 (127) | FPV066 (122) | 40 | |||||
| CNPV091 | 101553-101305 (83) | FPV067 (90) | 62 | HT motif protein | ||||
| CNPV092 | 102367-101930 (146) | FPV068 (133) | 40 | |||||
| CNPV093 | 103273-102473 (267) | FPV069 (270) | 51 | D3R | Virion protein | |||
| CNPV094 | 103348-104172 (275) | FPV070 (273) | 54 | T10-like protein | ||||
| CNPV095 | 104320-104186 (45) | FPV070.5 (36) | 64 | |||||
| CNPV096 | 104559-104305 (85) | X63237 | 386 | Homo sapiens | Fragment | Ubiquitin | ||
| CNPV097 | 105686-104670 (339) | FPV071 (289) | 53 | Conserved hypothetical protein | ||||
| CNPV098 | 105951-105712 (80) | — | ||||||
| CNPV099 | 106544-105960 (195) | FPV072 (186) | 60 | β-NGF | ||||
| CNPV100 | 107077-106571 (169) | FPV073 (174) | 52 | Interleukin binding protein | ||||
| CNPV101 | 107389-107135 (85) | — | ||||||
| CNPV102 | 107717-107403 (105) | FPV074 (104) | 31 | |||||
| CNPV103 | 108306-107737 (190) | FPV075 (199) | 47 | N1R/p28-like protein | ||||
| CNPV104 | 108510-108884 (125) | FPV077 (125) | 60 | G4L | Glutaredoxin 2, virion morphogenesis | |||
| CNPV105 | 109528-109833 (102) | FPV078 (103) | 66 | G3L* | ||||
| CNPV106 | 109534-108833 (234) | FPV079 (225) | 62 | G2R | Putative elongation factor | |||
| CNPV107 | 109972-110202 (77) | — | ||||||
| CNPV108 | 110450-112345 (632) | FPV081 (626) | 70 | G1L | Metalloprotease, virion morphogenesis | |||
| CNPV109 | 114377-112335 (681) | D86731 | 1,710 | Canarypox virus | FPV082 (682) | 74 | I8R | NPH-II, RNA helicase |
| CNPV110 | 114412-115677 (422) | D86731 | 2,217 | Canarypox virus | FPV083 (421) | 75 | I7L | Virion core proteinase |
| CNPV111 | 115685-116857 (391) | D86731 | 2,075 | Canarypox virus | FPV084 (390) | 69 | I6L | DNA-binding protein |
| CNPV112 | 116861-117103 (81) | D86731 | 386 | Canarypox virus | FPV085 (81) | 72 | I5L* | IMV membrane protein |
| CNPV113 | 117128-117664 (179) | D78347 | 840 | Canarypox virus | FPV086 (183) | 65 | J2R | Thymidine kinase |
| CNPV114 | 117802-118047 (82) | D86731 | 446 | Canarypox virus | FPV087 (91) | 55 | HT motif protein | |
| CNPV115 | 118120-118986 (289) | D86731 | 1,502 | Canarypox virus | FPV088 (290) | 55 | I3L | DNA-binding phosphoprotein |
| CNPV116 | 118990-119196 (69) | D86731 | 334 | Canarypox virus | FPV089 (65) | 63 | I2L* | |
| CNPV117 | 119206-120135 (310) | FPV090 (311) | 84 | I1L | DNA-binding virion protein | |||
| CNPV118 | 120318-122276 (653) | FPV091 (656) | 54 | OIL | ||||
| CNPV119 | 122209-122601 (131) | FPV092 (131) | 63 | E11L | Virion core protein | |||
| CNPV120 | 122882-122604 (93) | FPV093 (94) | 81 | E10R | IMV redox protein, virus assembly | |||
| CNPV121 | 122909-125872 (988) | FPV094 (988) | 78 | E9L | DNA polymerase | |||
| CNPV122 | 126691-125870 (274) | FPV095 (272) | 63 | E8R | ||||
| CNPV123 | 128408-126696 (571) | FPV096 (571) | 76 | E6R | ||||
| CNPV124 | 134226-128473 (1,918) | FPV097 (1,912) | 60 | VARV B22R-like protein | ||||
| CNPV125 | 139596-134296 (1,767) | FPV098 (1,802) | 61 | VARV B22R-like protein | ||||
| CNPV126 | 145731-139879 (1,951) | FPV099 (1,949) | 58 | VARV B22R-like protein | ||||
| CNPV127 | 145821-146366 (182) | FPV100 (182) | 85 | E4L | RNA polymerase subunit RPO30 | |||
| CNPV128 | 146401-148563 (721) | FPV101 (717) | 61 | E2L | ||||
| CNPV129 | 148559-149974 (472) | FPV102 (472) | 75 | E1L | Poly(A) polymerase large subunit PAPL | |||
| CNPV130 | 150330-149974 (119) | FPV103 (114) | 72 | F17R | DNA-binding virion core protein | |||
| CNPV131 | 150406-151026 (207) | FPV104 (210) | 43 | |||||
| CNPV132 | 151123-151566 (148) | FPV105 (148) | 73 | F15L | ||||
| CNPV133 | 151805-152101 (99) | FPV106 (71) | 61 | |||||
| CNPV134 | 157573-152171 (1,801) | FPV107 (1,777) | 59 | VARV B22R-like protein | ||||
| CNPV135 | 157745-158878 (378) | FPV108 (377) | 84 | F13L | Palmitylated EEV envelope lipase | |||
| CNPV136 | 158960-160834 (625) | AX068050 | 3,276 | Canarypox virus | FPV109 (630) | 51 | F12L | EEV maturation protein |
| CNPV137 | 160880-162274 (465) | FPV110 (451) | 43 | F11L | ||||
| CNPV138 | 162368-163699 (444) | FPV111 (444) | 76 | F10L | Ser/Thr protein kinase, virus assembly | |||
| CNPV139 | 163677-164315 (213) | FPV112 (213) | 71 | F9L | ||||
| CNPV140 | 164401-164598 (66) | FPV113 (66) | 69 | F8L* | ||||
| CNPV141 | 164927-165478 (184) | FPV114 (183) | 78 | HAL3 domain protein | ||||
| CNPV142 | 165725-166687 (321) | FPV124 (289) | 44 | N1R/p28-like protein | ||||
| CNPV143 | 166803-168815 (671) | FPV162 (603) | 30 | B4R | Ankyrin repeat protein | |||
| CNPV144 | 168844-170511 (556) | FPV115 (542) | 39 | M1L | Ankyrin repeat protein | |||
| CNPV145 | 170735-172054 (440) | FPV117 (440) | 65 | G5R | ||||
| CNPV146 | 172065-172250 (62) | FPV118 (63) | 75 | G5.5R | RNA polymerase subunit RPO7 | |||
| CNPV147 | 172246-172809 (188) | FPV119 (188) | 68 | G6R | ||||
| CNPV148 | 173823-172780 (348) | FPV120 (343) | 79 | G7L | Virion core protein | |||
| CNPV149 | 174909-173992 (306) | AB037797 | 796 | Homo sapiens | — | Thioredoxin binding protein-like protein | ||
| CNPV150 | 175219-176271 (351) | FPV162 (603) | 31 | B4R | Ankyrin repeat protein | |||
| CNPV151 | 177636-176401 (412) | FPV240 (410) | 35 | B4R | Ankyrin repeat protein | |||
| CNPV152 | 178424-177864 (187) | — | ||||||
| CNPV153 | 179479-178544 (312) | AJ298230 | 328 | Columbid circovirus | — | Rep-like protein | ||
| CNPV154 | 185699-179916 (1,928) | FPV122 (1,870) | 77 | VARV B22R-like protein | ||||
| CNPV155 | 191244-185755 (1,830) | FPV123 (1,766) | 72 | VARV B22R-like protein | ||||
| CNPV156 | 191561-194056 (832) | — | ||||||
| CNPV157 | 195204-195719 (172) | AB088684 | 130 | Hemicentrotus pulcherrimus | FPV080 (363) | 24 | TGF-β | |
| CNPV158 | 195205-194159 (349) | FPV080 (363) | 25 | TGF-β | ||||
| CNPV159 | 195825-196835 (337) | FPV124 (289) | 73 | N1R/p28-like protein | ||||
| CNPV160 | 196882-198069 (396) | FPV124 (289) | 54 | N1R/p28-like protein | ||||
| CNPV161 | 199247-198174 (358) | FPV080 (363) | 24 | TGF-β | ||||
| CNPV162 | 199297-199743 (149) | P15203 | 186 | Sus scrofa | FPV080 (363) | 29 | TGF-β | |
| CNPV163 | 200246-200539 (98) | — | ||||||
| CNPV164 | 200247-199972 (92) | — | ||||||
| CNPV165 | 200678-201715 (346) | FPV124 (289) | 69 | N1R/p28-like protein | ||||
| CNPV166 | 201953-202987 (345) | FPV125 (345) | 59 | B19R* | Ig domain protein | |||
| CNPV167 | 203262-203774 (171) | FPV125 (345) | 42 | A56R* | Ig domain protein | |||
| CNPV168 | 203870-204943 (358) | FPV124 (289) | 59 | N1R/p28-like protein | ||||
| CNPV169 | 204991-205986 (332) | FPV124 (289) | 64 | N1R/p28-like protein | ||||
| CNPV170 | 206069-206704 (212) | PS01331 | 601 | Homo sapiens | — | A48R | Thymidylate kinase | |
| CNPV171 | 206760-207539 (260) | FPV126 (260) | 95 | G8R | Late transcription factor VLTF-1 | |||
| CNPV172 | 207556-208560 (335) | FPV127 (336) | 73 | G9R | Myristylated protein | |||
| CNPV173 | 208564-209292 (243) | FPV128 (243) | 87 | L1R | Myristylated IMV envelope protein | |||
| CNPV174 | 209355-209642 (96) | FPV129 (96) | 32 | L2R* | ||||
| CNPV175 | 210546-209638 (303) | FPV130 (301) | 81 | L3L | ||||
| CNPV176 | 210572-211327 (252) | FPV131 (253) | 70 | L4R | DNA-binding virion core protein | |||
| CNPV177 | 211331-211720 (130) | FPV132 (129) | 77 | L5R | ||||
| CNPV178 | 211677-212120 (148) | FPV133 (148) | 75 | J1R | IMV membrane protein | |||
| CNPV179 | 212157-213062 (302) | FPV134 (308) | 72 | J3R | Poly(A) polymerase small subunit PAPS | |||
| CNPV180 | 213062-213619 (186) | FPV135 (186) | 74 | J4R | RNA polymerase subunit RPO22 | |||
| CNPV181 | 214025-213618 (136) | FPV136 (137) | 75 | J5L | ||||
| CNPV182 | 214068-217931 (1,288) | FPV137 (1,287) | 88 | J6R | RNA polymerase subunit RPO147 | |||
| CNPV183 | 218437-217940 (166) | FPV138 (166) | 79 | H1L | Protein-tyrosine phosphatase, virus assembly | |||
| CNPV184 | 218453-219019 (189) | FPV139 (190) | 88 | H2R | ||||
| CNPV185 | 220084-219101 (328) | FPV232 (482) | 28 | K1L* | Ankyrin repeat protein | |||
| CNPV186 | 221118-220129 (330) | FPV140 (327) | 57 | H3L | IMV envelope protein | |||
| CNPV187 | 223609-221213 (799) | FPV141 (798) | 81 | H4L | RNA polymerase associated protein RAP94 | |||
| CNPV188 | 223778-224287 (170) | FPV142 (174) | 55 | H5R | Late transcription factor VLTF-4 | |||
| CNPV189 | 224291-225238 (316) | FPV143 (316) | 78 | H6R | DNA topoisomerase | |||
| CNPV190 | 225246-225704 (153) | FPV144 (152) | 56 | H7R | ||||
| CNPV191 | 225981-225673 (103) | FPV145 (103) | 51 | |||||
| CNPV192 | 225989-228526 (846) | FPV146 (851) | 73 | D1R | mRNA capping enzyme large subunit | |||
| CNPV193 | 228602-228919 (106) | FPV147 (104) | 54 | HT motif protein | ||||
| CNPV194 | 229341-228922 (140) | FPV148 (139) | 61 | D2L | Virion protein | |||
| CNPV195 | 229395-229826 (144) | — | ||||||
| CNPV196 | 229894-230463 (190) | FPV149 (186) | 52 | |||||
| CNPV197 | 230527-231351 (275) | FPV150 (276) | 44 | N1R/p28-like protein | ||||
| CNPV198 | 231891-231424 (156) | AF320600 | 173 | Mus musculus | FPV008 (167) | 30 | A34R* | C-type lectin-like protein |
| CNPV199 | 232199-232873 (225) | FPV151 (235) | 48 | Deoxycytidine kinase | ||||
| CNPV200 | 232882-233379 (166) | AJ301633 | 165 | Canary circovirus | — | Rep-like protein | ||
| CNPV201 | 233379-233954 (192) | FPV153 (208) | 57 | |||||
| CNPV202 | 234009-234836 (276) | FPV236 (280) | 25 | N1R/p28-like protein | ||||
| CNPV203 | 234912-236057 (382) | FPV155 (408) | 37 | N1R/p28-like protein | ||||
| CNPV204 | 236116-236298 (61) | Fragment | ||||||
| CNPV205 | 236520-237473 (318) | FPV157 (311) | 51 | N1R/p28-like protein | ||||
| CNPV206 | 237537-238952 (472) | FPV158 (464) | 74 | Photolyase | ||||
| CNPV207 | 239086-239634 (183) | FPV159 (241) | 46 | N1R/p28-like protein | ||||
| CNPV208 | 239777-240376 (200) | FPV160 (156) | 40 | |||||
| CNPV209 | 240422-241351 (310) | FPV161 (157) | 46 | N1R/p28-like protein | ||||
| CNPV210 | 241402-241794 (131) | FPV075 (199) | 39 | N1R/p28-like protein | ||||
| CNPV211 | 241852-242013 (54) | FPV161.5 (51) | 48 | |||||
| CNPV212 | 242076-242603 (176) | FPV124 (289) | 40 | N1R/p28-like protein | ||||
| CNPV213 | 243303-242653 (217) | FPV151 (235) | 29 | Deoxycytidine kinase | ||||
| CNPV214 | 243477-244544 (356) | FPV006 (418) | 42 | C10L | C4L/C10L-like protein | |||
| CNPV215 | 244823-245434 (204) | FPV060 (188) | 34 | CC chemokine-like protein | ||||
| CNPV216 | 245527-246738 (404) | FPV063 (400) | 41 | |||||
| CNPV217 | 246839-247828 (330) | FPV124 (289) | 50 | N1R/p28-like protein | ||||
| CNPV218 | 247920-249230 (437) | FPV124 (289) | 45 | N1R/p28-like protein | ||||
| CNPV219 | 250150-251196 (349) | FPV124 (289) | 59 | N1R/p28-like protein | ||||
| CNPV220 | 251243-251776 (178) | FPV124 (289) | 63 | N1R/p28-like protein | ||||
| CNPV221 | 251832-252674 (281) | FPV124 (289) | 54 | N1R/p28-like protein | ||||
| CNPV222 | 252945-253799 (285) | FPV124 (289) | 51 | N1R/p28-like protein | ||||
| CNPV223 | 256865-254325 (847) | FPV162 (603) | 33 | B4R | Ankyrin repeat protein | |||
| CNPV224 | 257119-257835 (239) | — | ||||||
| CNPV225 | 257913-258389 (159) | FPV124 (289) | 64 | N1R/p28-like protein | ||||
| CNPV226 | 258418-258819 (134) | FPV124 (289) | 61 | N1R/p28-like protein | ||||
| CNPV227 | 258876-259952 (359) | FPV124 (289) | 67 | N1R/p28-like protein | ||||
| CNPV228 | 260009-261121 (371) | FPV124 (289) | 61 | N1R/p28-like protein | ||||
| CNPV229 | 262686-261385 (434) | FPV014 (437) | 42 | B4R | Ankyrin repeat protein | |||
| CNPV230 | 262884-263078 (65) | — | ||||||
| CNPV231 | 263029-263502 (158) | AK027650 | 213 | Homo sapiens | — | MyD116-like domain protein | ||
| CNPV232 | 263535-264146 (204) | FPV060 (188) | 39 | CC chemokine-like protein | ||||
| CNPV233 | 264289-265701 (471) | FPV246 (592) | 29 | B4R | Ankyrin repeat protein | |||
| CNPV234 | 265724-267247 (508) | FPV233 (512) | 28 | B18R | Ankyrin repeat protein | |||
| CNPV235 | 267321-268616 (432) | FPV164 (383) | 33 | |||||
| CNPV236 | 268664-269632 (323) | P31350 | 1,237 | Homo sapiens | Fragment | F4L | Ribonucleotide reductase small subunit | |
| CNPV237 | 269815-271137 (441) | FPV162 (603) | 27 | C9L | Ankyrin repeat protein | |||
| CNPV238 | 271856-271182 (225) | FPV165 (225) | 92 | A2L | Late transcription factor VLTF-3 | |||
| CNPV239 | 272071-271847 (75) | FPV166 (72) | 73 | A2.5L | Virion redox protein | |||
| CNPV240 | 274064-272088 (659) | FPV167 (657) | 83 | A3L | Virion core protein P4b | |||
| CNPV241 | 274798-274154 (215) | FPV168 (288) | 33 | A4L* | Immunodominant virion protein | |||
| CNPV242 | 274837-275343 (169) | FPV169 (167) | 73 | A5R | RNA polymerase subunit RPO19 | |||
| CNPV243 | 276462-275344 (373) | FPV170 (374) | 66 | A6L | ||||
| CNPV244 | 278598-276472 (709) | FPV171 (709) | 87 | A7L | Early transcription factor large subunit | |||
| CNPV245 | 278662-279561 (300) | FPV172 (301) | 79 | A8R | Intermediate transcription factor VITF-3 | |||
| CNPV246 | 279756-279532 (75) | FPV173 (76) | 63 | A9L | IMV membrane protein | |||
| CNPV247 | 282438-279760 (893) | FPV174 (891) | 71 | A10L | Virion core protein P4a | |||
| CNPV248 | 282456-283292 (279) | FPV175 (274) | 64 | A11R | ||||
| CNPV249 | 283798-283295 (168) | FPV176 (171) | 60 | A12L | Virion protein | |||
| CNPV250 | 283813-284109 (99) | FPV177 (68) | 30 | |||||
| CNPV251 | 284310-284104 (69) | FPV178 (71) | 57 | A13L* | IMV membrane protein | |||
| CNPV252 | 284636-284361 (92) | FPV179 (91) | 61 | A14L | IMV membrane protein | |||
| CNPV253 | 284814-284656 (53) | FPV179.5 (53) | 50 | A14.5L | IMV membrane virulence factor | |||
| CNPV254 | 285120-284833 (96) | FPV180 (97) | 60 | A15L* | ||||
| CNPV255 | 286210-285107 (368) | FPV181 (369) | 75 | A16L | Myristylated protein | |||
| CNPV256 | 286804-286229 (192) | FPV182 (198) | 75 | A17L | Phosphorylated IMV membrane protein | |||
| CNPV257 | 286822-288207 (462) | FPV183 (462) | 82 | A18R | DNA helicase, transcriptional elongation | |||
| CNPV258 | 288447-288181 (89) | FPV184 (88) | 67 | A19L | ||||
| CNPV259 | 288792-290093 (434) | FPV185 (433) | 64 | A20R | DNA polymerase processivity factor | |||
| CNPV260 | 288793-288458 (112) | FPV186 (113) | 83 | A21L | ||||
| CNPV261 | 290093-290548 (152) | FPV187 (156) | 68 | A22R | Holliday junction resolvase | |||
| CNPV262 | 290568-291716 (383) | FPV188 (383) | 73 | A23R | Intermediate transcription factor VITF-3 | |||
| CNPV263 | 291745-295215 (1,157) | FPV189 (1,161) | 91 | A24R | RNA polymerase subunit RPO132 | |||
| CNPV264 | 297018-295210 (603) | FPV190 (620) | 69 | A26L | A type inclusion-like protein | |||
| CNPV265 | 298480-297056 (475) | FPV191 (474) | 78 | A26L/A27L | A type inclusion-like/fusion protein | |||
| CNPV266 | 298903-298484 (140) | FPV192 (141) | 84 | A28L | ||||
| CNPV267 | 299825-298911 (305) | FPV193 (302) | 64 | A29L | RNA polymerase subunit RPO35 | |||
| CNPV268 | 300027-299803 (75) | FPV194 (74) | 78 | A30L | ||||
| CNPV269 | 300152-300490 (113) | FPV195 (113) | 63 | A31R | ||||
| CNPV270 | 300502-300861 (120) | FPV196 (120) | 41 | |||||
| CNPV271 | 301707-300856 (284) | FPV197 (301) | 72 | A32L | DNA packaging protein | |||
| CNPV272 | 301822-302364 (181) | FPV198 (173) | 64 | A34R | C-type lectin-like EEV protein | |||
| CNPV273 | 302592-303413 (274) | FPV201 (283) | 45 | |||||
| CNPV274 | 303477-304283 (269) | FPV203 (285) | 51 | Tyrosine protein kinase | ||||
| CNPV275 | 304329-305342 (338) | FPV204 (342) | 56 | C12L | Serpin | |||
| CNPV276 | 306125-305370 (252) | FPV205 (218) | 36 | |||||
| CNPV277 | 306235-307164 (310) | FPV206 (308) | 65 | G protein-coupled receptor | ||||
| CNPV278 | 307178-307465 (96) | FPV207 (100) | 64 | |||||
| CNPV279 | 307534-308040 (169) | Fragment | β-NGF | |||||
| CNPV280 | 308453-308064 (130) | FPV152 (127) | 28 | HT motif gene family | ||||
| CNPV281 | 308557-309198 (214) | FPV208 (67) | 38 | |||||
| CNPV282 | 309574-309215 (120) | FPV209 (130) | 44 | HT motif gene family | ||||
| CNPV283 | 309740-310072 (111) | P28291 | 91 | Bos taurus | Fragment | CC chemokine-like protein | ||
| CNPV284 | 310140-310724 (195) | P01880 | 91 | Homo sapiens | FPV073 (174) | 27 | Interleukin binding protein | |
| CNPV285 | 310837-311214 (126) | FPV211 (125) | 42 | CIIR | EGF-like protein | |||
| CNPV286 | 311219-312133 (305) | FPV212 (303) | 56 | B1R | Ser/Thr protein kinase | |||
| CNPV287 | 312179-312661 (161) | FPV213 (162) | 54 | |||||
| CNPV288 | 312700-313140 (147) | AF009511 | 127 | Rattus norvegicus | FPV239 (163) | 27 | A40R* | C-type lectin-like protein |
| CNPV289 | 313186-313602 (139) | FPV214 (124) | 45 | Interleukin binding protein | ||||
| CNPV290 | 313674-313898 (75) | FPV215 (74) | 79 | |||||
| CNPV291 | 314103-315884 (594) | FPV246 (592) | 31 | B4R | Ankyrin repeat protein | |||
| CNPV292 | 315911-316132 (74) | — | ||||||
| CNPV293 | 316178-317029 (284) | FPV216 (296) | 38 | C18L* | Ankyrin repeat protein | |||
| CNPV294 | 317087-318376 (430) | FPV240 (410) | 29 | B4R | Ankyrin repeat protein | |||
| CNPV295 | 318572-319759 (396) | FPV218 (461) | 37 | B4R | Ankyrin repeat protein | |||
| CNPV296 | 319765-321138 (458) | FPV219 (434) | 45 | B4R | Ankyrin repeat protein | |||
| CNPV297 | 321238-323448 (737) | FPV222 (747) | 39 | B4R | Ankyrin repeat protein | |||
| CNPV298 | 323507-325219 (571) | FPV223-225 (104-146) | 40-55 | B4R | Ankyrin repeat protein | |||
| CNPV299 | 325226-326125 (300) | FPV226 (292) | 57 | B1R | Ser/Thr protein kinase | |||
| CNPV300 | 326201-326932 (244) | FPV227 (361) | 43 | M1L | Ankyrin repeat protein | |||
| CNPV301 | 327525-329105 (527) | FPV233 (512) | 31 | B18R | Ankyrin repeat protein | |||
| CNPV302 | 329704-329126 (193) | FPV229 (180) | 34 | |||||
| CNPV303 | 329772-331271 (500) | FPV230-231 (188-256) | 44-55 | B4R | Ankyrin repeat protein | |||
| CNPV304 | 331490-332887 (466) | FPV232 (482) | 45 | B18R | Ankyrin repeat protein | |||
| CNPV305 | 332961-333746 (262) | FPV236 (280) | 36 | NIR/p28-like protein | ||||
| CNPV306 | 333811-334026 (72) | — | ||||||
| CNPV307 | 334497-334036 (154) | FPV239 (163) | 43 | A34R | C-type lectin-like protein | |||
| CNPV308 | 334673-335743 (357) | FPV240 (410) | 36 | M1L | Ankyrin repeat protein | |||
| CNPV309 | 335897-336484 (196) | FPV241 (106) | 42 | M1L | Ankyrin repeat protein | |||
| CNPV310 | 336592-338202 (537) | FPV246 (592) | 34 | B4R | Ankyrin repeat protein | |||
| CNPV311 | 338239-338610 (124) | FPV247 (124) | 31 | EFc-like protein | ||||
| CNPV312 | 338623-339120 (166) | FPV037 (162) | 22 | |||||
| CNPV313 | 339195-339848 (218) | Fragment | Ig domain protein | |||||
| CNPV314 | 340012-341763 (584) | FPV242-243 (262-358) | 52-58 | B4R | Ankyrin repeat protein | |||
| CNPV315 | 341865-342809 (315) | FPV021 (320) | 38 | G protein-coupled receptor | ||||
| CNPV316 | 342879-344510 (544) | FPV246 (592) | 35 | B4R | Ankyrin repeat protein | |||
| CNPV317 | 344698-344862 (55) | — | ||||||
| CNPV318 | 345044-346585 (514) | FPV233 (512) | 29 | B18R | Ankyrin repeat protein | |||
| CNPV319 | 346733-348949 (739) | FPV246 (592) | 42 | B4R | Ankyrin repeat protein | |||
| CNPV320 | 349143-350549 (469) | FPV017 (245) | 31 | Ig domain protein | ||||
| CNPV321 | 350684-351055 (124) | FPV005 (122) | 63 | EFc-like protein | ||||
| CNPV322 | 351384-353453 (690) | FPV242 (358) | 29 | B4R* | Ankyrin repeat protein | |||
| CNPV323 | 354036-353491 (182) | FPV258.5 (47) | 56 | |||||
| CNPV324 | 354664-353999 (222) | FPV259 (222) | 86 | |||||
| CNPV325 | 354790-356331 (514) | FPV246 (592) | 28 | B4R | Ankyrin repeat protein | |||
| CNPV326 | 356457-357068 (204) | FPV260 (205) | 27 | C-type lectin-like protein | ||||
| CNPV327 | 357954-357442 (171) | — | ||||||
| CNPV328 | 358442-358227 (72) | — | ||||||
aa, amino acids.
Best-matching, significant non-FWPV protein sequence from Blast2 analysis of nonredundant protein database.
GenBank database accession number.
Blast2 score.
Best-matching ORF from the FWPV challenge strain (accession no. AF198100). Italics indicate ORFs likely nonorthologous based on genome arrangement and/or amino acid identity; bold indicates ORFs that differ in length by greater than 10%; — indicates lack of identifiable homology; fragment indicates presence of potentially orthologous but disrupted ORF sequence in FWPV.
% ID, percent amino acid identity.
Best-matching ORF from the VACV strain Copenhagen genome (accession no. M35027). Asterisks indicate matches with Blast2 scores less than 100; italics indicate ORFs likely nonorthologous.
FIG. 1.
Comparative ORF map of CNPV and FWPV genomes. CNPV ORFs (top) are numbered from left to right based on the position of methionine start codons. ORFs transcribed to the right in each virus are located on top relative to those transcribed to the left; CNPV and selected FWPV genome positions are indicated below each virus. CNPV ORFs (top) are manually aligned with FWPV ORFs (bottom). Colored ORFs indicate differences between CNPV and FWPV: ORFs used to introduce gaps or lacking discernible orthologous sequence in the other virus are marked in green; nonhomologous ORFs in similar genomic positions are marked in blue; ORFs severely disrupted in the opposite virus are marked in yellow; and, due to extensive variability, ORFs in terminal regions marked in orange are unaligned. CNPV ORFs lacking discernible homology to any FWPV ORF are marked above with an asterisk; FWPV ORFs lacking discernible homology to any CNPV ORF are marked above with a triangle. Thick black bars at genomic termini represent ITRs. Boxed regions indicate novel coding regions at junction sites of major genome rearrangements previously identified in FWPV (2), with white indicating gaps between grey sequence.
Specific central genomic regions of CNPV contain homologues of conserved poxviral genes involved in basic replicative mechanisms, including viral transcription and RNA modification, viral DNA replication, and structure and assembly of intracellular mature virions and extracellular enveloped virions (57) (Table 1; Fig. 1). A total of 106 conserved ChPV genes were present in both CNPV and FWPV, sharing on average 70% amino acid identity and all but one differing in length by less than 7% (Table 1) (2). Between CNPV and FWPV, the most conserved protein was CNPV171 (VLTF-1; 95% amino acid identity to FPV126), while the most divergent were CNPV241 (putative VACV A4L-like virion protein; 33% amino acid identity and 25% shorter) and CNPV174 (VACV L2R-like ORF; 32% amino acid identity) (Table 1). Similar to FWPV and molluscum contagiosum virus, CNPV lacks orthologues of the VACV D8L and A27L intracellular mature virion membrane and fusion proteins but contains A27L-like sequence in the carboxyl-terminal region of an A-type inclusion-like protein (CNPV265) (11, 81). Also similar to FWPV, most conserved ChPV genes were present in several large CNPV genomic regions (CNPV069 to CNPV084, CNPV104 to CNPV140, CNPV171 to CNPV194, and CNPV238 to CNPV273) which correspond to the large genomic regions previously noted as rearranged between FWPV and other ChPV (2).
Extensive regions of the CNPV genome contain genes with putative virulence and host range functions similar to those found in FWPV, and they occur predominantly in terminal (CNPV001 to CNPV068 and CNPV274 to CNPV328) and internal (CNPV084 to CNPV103, CNPV141 to CNPV170, and CNPV195 to CNPV237) genomic regions (2). These include gene families and genes likely involved in viral modification or evasion of host cellular, apoptotic, and immune responses or processes (Table 1). These regions and types of genes were more variable between CNPV and FWPV in coding content (average of 43% amino acid identity overall, and half of them differing in length by at least 7%), in gene arrangement, and in overall gene complement.
Nucleotide metabolism.
CNPV contains a unique complement of genes likely involved in nucleotide metabolism. CNPV encodes, similar to FWPV and other ChPVs, thymidine kinase, dUTPase, and glutaredoxin homologues (2, 5). CNPV contains two genes (CNPV199 and CNPV213) similar to both deoxycytidine and deoxyguanosine kinases, thus far avipoxvirus-specific nucleotide metabolism genes. While CNPV199 has a potential orthologue in FWPV, CNPV213 is novel. CNPV170 and CNPV236 are homologues of thymidylate kinase and ribonucleotide reductase small subunit genes, homologues of which are absent or heavily disrupted, respectively, in FWPV. This specific complement of CNPV genes suggests optimization of intracellular nucleotide pools different from that of FWPV, likely affecting differences in cell and tissue tropism.
CNPV gene family proteins.
CNPV contains an extensive array of gene families (138 genes in 14 gene families) comprising over 49% of the CNPV genome (versus 38% in FWPV). CNPV gene families are similar to those in FWPV, including proteins containing ankyrin repeats and those similar to rabbit fibroma virus N1R and ectromelia virus p28 (N1R/p28), variola virus (strain Bangladesh) B22R, and vaccinia virus C4L and C10L (C4/C10-like) ORFs. CNPV and FWPV contain families unique to avipoxviruses (EFc proteins, HT motif proteins, and CNPV012/CNPV302 similar to FPV221 and FPV229) as well as several families with potential roles in immunomodulation, including transforming growth factor-β (TGF-β), β-nerve growth factor (β-NGF), serine proteinase inhibitor (serpin), C-type lectin-like, CC-chemokine, G-protein coupled CC-chemokine receptor (GPCR), immunoglobulin (Ig) domain, and interleukin 18 (IL-18)-binding protein-like proteins (2). However, CNPV and FWPV gene families exhibit significant differences, accounting for much of the variability present between these two avipoxvirus genomes.
Ankyrin repeat and N1R/p28-like genes were the most abundant in the CNPV genome (51 and 26 ORFs, or 21 and 6% of the genome, respectively), containing more copies than FWPV (31 and 10 ORFs, or 14 and 3% of the FWPV genome, respectively) in terminal and internal genomic regions. While CNPV ankyrin repeat genes were predominantly located in the terminal genomic regions (41 ORFs in the most-terminal 110 kbp), 10 ankyrin repeat genes and all but one N1R/p28 gene were notably located in or near the internal variable genomic regions (Table 1). CNPV ankyrin repeat genes ranged in size from 196 to 847 amino acids and had potential FWPV orthologues (syntenic regions) and paralogues (rearranged or expanded regions) that shared from 25 to 79% amino acid identity (average, 37%) and had variable numbers of repeats. Similarly, CNPV N1R/p28-like genes shared 27 to 71% amino acid identity (average, 52%) with FWPV homologues and contained previously described modular domains, including amino-terminal KilA-N (25 genes), bro-C (9 genes), and carboxyl-terminal RING finger domains (2 genes) (2, 43). Notably, 18 CNPV N1R/p28-like genes exhibited characteristics suggestive of CNPV160/CNPV228-like gene duplications, including a high degree of similarity to each other and to FPV124 (58% average amino acid identity) (Table 1). Thus, CNPV contains an extraordinarily large repertoire of ankyrin repeat and N1R/p28-like genes, homologues of which function in virulence and host range in other poxviruses (72, 82).
Variola virus B22R-like genes were a prominent feature in CNPV, as these six large ORFs (average of 1,866 amino acids) comprised 9% of the CNPV genome (Table 1). B22R-like ORFs are relatively well conserved with FWPV orthologues (64% amino acid identity), making the presence of multiple complete copies of these genes in central genomic regions a feature unique to avipoxviruses. As in FWPV, genomic arrangement and phylogenetic analysis suggest that the tandem ORF pairs CNPV124/125 and CNPV154/155 are paralogous. Notably, while all CNPV B22R-like ORFs were present in conserved, ChPV-syntenic genomic regions, four were located in a region unchanged between CNPV and FWPV and two (CNPV154/155) were located in a region further expanded in CNPV (between homologues of VACV G7R and G8R) (Table 1).
TGF-β- and β-NGF-like genes represent cellular homologues that are rare (in the case of TGF-β [C. L. Afonso et al., unpublished data]) or absent (in the case of β-NGF) in other known poxviruses and variable between avipoxviruses. The five CNPV TGF-β-like genes are potential paralogues of the single FWPV TGF-β based on amino acid identity (less than 28%) and genomic location, which along with phylogenetic analysis suggests local gene duplication of CNPV157/158 and CNPV161/162 tandem pairs (data not shown). While two CNPV TGF-β genes (CNPV157 and CNPV162) lack amino-terminal domains, all contain conserved carboxyl-terminal cysteine knot domains present in processed TGF-β-like subunits. Given the homo- or heterodimeric nature of active cellular TGF-β-like proteins, multiple CNPV ORFs potentially provide a combinatorially complex array of CNPV-encoded proteins active in modulating TGF-β-mediated host cell proliferation and differentiation, apoptosis, inflammation, or immune responses (23, 56). Similarly, only one (CNPV099) of the two CNPV β-NGF-like genes had a positional FWPV orthologue, while the second (CNPV279) was heavily disrupted in FWPV (Table 1). In addition to roles in neuronal development and survival, cellular β-NGF affects aspects of inflammation, cytokine production, and apoptosis or cell survival in nonneuronal, immune, and/or virus-infected cells (4, 31, 73, 83, 99). Given the lack of other known viral β-NGF homologues, these genes likely mediate critical virus-host interactions novel for avipoxvirus infections.
Other gene families with potential immunomodulatory functions were variable between CNPV and FWPV. For example, while 78% of CNPV serpin and GPCR genes likely had orthologues in FWPV (averaging 49% amino acid identity), only 32% of CNPV C-type lectin-like, CC-chemokine-like, Ig domain, and IL-18-binding protein-like ORFs likely had orthologues in FWPV (averaging 36% amino acid identity). Four CNPV C-type lectin-like proteins lacking obvious FWPV orthologues were most similar to cellular natural killer (NK) cell receptor-like proteins (Table 1). Four of five CNPV CC-chemokine-like proteins resembled FPV060 in length (average of 235 amino acids) and in CC-chemokine-like cysteine patterns (average of 19.5 cysteines). The remaining ORF (CNPV283) resembled the four smaller FWPV CC-chemokine-like ORFs (average of 123 amino acids and 9 cysteines) and contained a single chemokine-like motif. Amino-terminal domains differentiated CNPV Ig domain proteins into the two groups previously identified in FWPV; however, group 1 ORFs (CNPV014, CNPV032, CNPV033, CNPV313, and CNPV320) were located in more-terminal genomic regions and were less conserved with FWPV homologues than were centrally located group 2 ORFs (36 versus 48% average amino acid identity, respectively). Notably, the CNPV032 Ig domain homologue in FWPV is a novel gamma interferon (IFN-γ)-binding protein with variable activity against avian and mammalian IFN-γ (76). Although three CNPV ORFs (CNPV100, CNPV284, and CNPV289) share cysteine and tryptophan residues present in Ig domains of known cellular and viral IL-18-binding proteins, they are highly divergent from each other and from nonavipoxvirus homologues (20 to 31% amino acid identity), including differences in residues critical for IL-18 binding (63, 101). Given this variability, immunomodulatory gene families may account for major differences in avipoxvirus virulence and host range, possibly binding and interacting with a diverse range of cellular host-specific substrates. Additionally, such viral genes may provide insight for avian immunoregulatory cytokines, which generally are divergent from mammalian homologues and remain poorly characterized (87).
Cellular homologues shared between CNPV and FWPV.
CNPV encodes numerous cellular homologues for which orthologues are present in FWPV. These include homologues of genes also present in nonavian poxviruses, including hydroxysteroid dehydrogenase, glutathione peroxidase, and a CPD-photolyase whose FWPV homologue mediates photoreactive DNA repair and affects viral infectivity (86). Homologues of Bcl-2-like apoptosis regulators are present in other viruses, but within the poxviridae they are found only in CNPV (CNPV058) and FWPV. Notably, CNPV and FWPV contain the only known viral homologues of cellular proteins, including α-SNAP, PC-1-like alkaline phosphodiesterase, Arabidopsis thaliana Hal3 (AtHal3), GNS1/SUR4, mouse T-10, and two conserved hypothetical proteins (CNPV047 and CNPV097) (2, 51) (Table 1). CNPV141 and FPV114 contain potential histidine active-site residues and substrate recognition and binding clamp motifs similar to AtHal3-like flavoproteins, suggesting similar roles in redox-mediated decarboxylation reactions (10, 37). CNPV068 and FPV048 are similar to cellular SUR4/ELO3 and GNS1/ELO2/FEN1 proteins, indicating potential roles in long-chain fatty acid elongation (64). These cellular homologues present only in CNPV and FWPV suggest involvement in novel avipoxvirus-host interactions (2).
CNPV genes absent in FWPV.
CNPV encodes 39 genes for which any homologue is absent from or extensively fragmented in FWPV. In addition to two additional nucleotide metabolism genes, CNPV contains eight ORFs similar to other cellular or viral proteins and 29 unique hypothetical proteins. These CNPV-specific genes likely affect novel aspects of the CNPV-host interaction.
CNPV086 is similar to cellular and poxviral tumor necrosis factor receptor (TNFR) homologues, containing an N-terminal signal sequence and two TNFR- or NGF receptor-like cysteine-rich domains (Prosite signature PS00652). CNPV086 is truncated compared to cellular membrane-bound receptors, lacking carboxyl-terminal transmembrane and cytoplasmic signaling domains. Compared to soluble viral homologues, CNPV086 lacks additional TNFR- or NGF receptor-like domains and/or unique carboxyl-terminal domains. Avian TNFR-like molecules serve as receptors for avian leukosis virus and as cell death receptors (13, 14). Although homologues of TNF and the majority of TNFR-mediated signaling pathways remain to be identified in birds, CNPV086 may function as a secreted TNFR to interfere with TNF-like activities such as inflammation, apoptosis, and induction of antiviral immunity.
CNPV018 has limited similarity to cellular IL-10, including conserved cysteine residues and residues likely involved in protein structure (29). CNPV018 may represent a homologue of a currently unidentified avian IL-10, and similar to parapoxvirus and gammaherpesvirus homologues of mammalian IL-10 it may be immunomodulatory (42, 85). Conversely, CNPV018, like yatapoxvirus and betaherpesvirus IL-10-like proteins, is quite divergent from mammalian IL-10 and diverse members of the IL-10 family and may have a novel biological function.
CNPV096 represents an intact and highly conserved homologue of cellular ubiquitin that is heavily disrupted in FWPV (Table 1) (2). Cellular ubiquitin affects widely diverse functions, including cell cycle, apoptosis, transcription, DNA repair, inflammation, and surface signaling, by marking proteins for specific intracellular trafficking and degradation (74, 75). Virally encoded ubiquitin is present in entomopox, pesti, and baculoviruses, and other viruses utilize ubiquitination for affecting viral gene transcription, virion assembly, and immune evasion (18, 25, 88). CNPV096 may affect one or many diverse ubiquitination-mediated cellular functions to impact CNPV virulence or host range.
CNPV085 is similar to human PIR1, a member of the protein tyrosine phosphatase (PTP) superfamily. PIR1 binds RNA with high affinity, has 5′ RNA di- and triphosphatase activity, shares similarity to other cellular mRNA capping enzymes, and is thought to have a role in RNA processing apart from capping or splicing (19, 102). CNPV085 similarity to PIR1 includes the PTP catalytic domain present in the amino terminus. CNPV085 is distinct from conserved poxviral VACV H1L-like dual-specificity PTPs (CNPV183) and capping enzyme triphosphatases (CNPV192), but it has similarity to a PTP-like ORF (AMV246) in Amsacta moorei entomopoxvirus. PIR1-like 5′ RNA phosphatases are also present along with functional capping enzymes in certain baculoviruses, where they play nonessential but potentially cell-type-specific roles in viral replication (52, 89). Similarly, CNPV085 may play a novel role in virus-avian host interaction.
CNPV149 is similar to cellular proteins, including thioredoxin-binding protein 2/vitamin D3 upregulated protein 1 (TBP-2/VDUP-1). CNPV149 is most similar (53% amino acid identity [Table 1]) to a human protein of unknown function and is less similar to known, mammalian TBP-2/VDUP-1 (34% amino acid identity) and divergent metazoan homologues, lacking the carboxyl quarter of cellular homologues. TBP-2/VDUP-1 binds to and inhibits the reducing activity of thioredoxin, is strongly upregulated during in vitro models of stress, and may play an important role in stress-induced, thioredoxin-mediated cellular responses, including proliferation and apoptosis (46, 61, 62, 78). Conceivably, CNPV149 could have a similar function.
CNPV231 contains a carboxyl-terminal domain similar to those of several cellular and viral proteins involved in regulation of apoptosis and in viral virulence and host range, including cellular myeloid differentiation primary response and growth arrest DNA damage proteins MyD116/Gadd34, African swine fever virus 23NL protein, and herpes simplex virus ICP34.5 protein (53). In other systems, these conserved and functionally interchangeable carboxyl domains mediate binding to cellular proteins, including protein phosphatase 1, to affect phosphorylation of the α subunit of eukaryotic initiation factor 2, cellular translation, and apoptotic and virulence functions (17, 53). These features enable ICP34.5 to antagonize dsRNA-dependent protein kinase activity and overcome IFN-induced translational inhibition (58). Other than CNPV231, the only poxviral gene with this domain is AMV193 of A. moorei entomopoxvirus (6). CNPV231 may provide CNPV an additional means of overcoming host cell antiviral responses.
CNPV153 and CNPV200 are similar to replication-associated (Rep) proteins of avian and mammalian circoviruses and less so to Rep-like proteins of other single-stranded DNA viruses (nanoviruses, geminiviruses, and parvoviruses) (Table 1). CNPV153 contains amino-terminal motifs similar to those of Rep proteins and other proteins, including bacterial, involved in initiation of rolling circle replicative mechanisms (50). These include regions important for binding, cleavage, and ligation within specific double-stranded DNA hairpin loop origin sequences by geminivirus Rep protein during initiation and termination of viral plus-strand replication (65). The shorter CNPV200 lacks these domains but shares putative NTP-binding P-loop and carboxyl-terminal domains conserved with CNPV153 and circovirus Rep proteins (data not shown). Geminivirus Rep proteins affect viral DNA replication and transcriptional repression, induce accumulation of proliferating cell nuclear antigen and bind cellular replication factor C, and bind the cell cycle-regulatory protein retinoblastoma to affect tissue specificity during infection (49, 54, 66). Human herpesvirus 6 and rat cytomegalovirus contain homologues of parvovirus Rep proteins, endonucleases or helicases which are also multifunctional in affecting viral replication, viral and cellular transcription, site-specific viral integration into host genomes, and interference of helper-virus replication (21, 96, 100). To our knowledge, CNPV Rep-like proteins are the first to be identified in a poxvirus and could conceivably affect similar functions in CNPV-infected cells.
CNPV066 and CNPV156 are large, novel ORFs of unknown function located in variable or virulence and host range-related genomic regions (Fig. 1; Table 1). Each ORF is largely comprised of perfect and imperfect tandem repeat motifs, covering the central 80 and 72% of CNPV066 and CNPV156, respectively. Repeats are unique to each ORF, having core nucleotide repeats of 15 to 75 bp and imparting compositional bias to CNPV066 (52% Ser/Asn/Gly) and CNPV156 (47% Glu/Ile/Gln/Met). CNPV066 contains a putative amino-terminal signal sequence, suggesting its secretion. Notably, CNPV156 contains a 21-amino-acid tandem repeat present at the amino-terminal region of CNPV156 tandem repetition (amino acids 106 to 147) that is perfectly or partially conserved in CNPV097 (conserved hypothetical protein), CNPV208, and CNPV218 (N1R/p28-like protein) but is absent in FWPV orthologues. These large, repetitive ORFs may impart novel functions to CNPV.
CNPV lacks genes present in FWPV.
CNPV lacks homologues of 15 genes present in terminal genomic regions of FWPV (Fig. 1). Thirteen of these, six of which are located in the FWPV ITRs and overlap larger genes, encode hypothetical proteins of small size (average, 86 amino acids) which may or may not be expressed. Notably, CNPV also lacks homologues of FPV217 and FPV250, ORFs similar to those from insect baculoviruses and from avian herpesviruses and adenoviruses, respectively, and possibly affecting virulence and host range functions in FWPV (2). CNPV lacks sequences similar to the reticuloendotheliosis virus long terminal repeat, confirming previous PCR analysis of CNPV which suggested a lack of the reticuloendotheliosis virus integration found in various strains of FWPV (38, 48).
CNPV relationship to avipoxviruses.
The CNPV C93 strain is very similar to limited available sequences of other CNPV strains. This includes 98% nucleotide and 91 to 100% amino acid identity with strain Tokyo CG-2 in the TK region (5) and up to 99% local nucleotide identity with patented CNPV sequences, suggesting a relatively conserved genotype for isolates causing canarypox and supporting the species designation for CNPV.
As described above, CNPV is most closely related to FWPV, having overall syntenic genomic arrangement and similar gene complements. These genomic data and the phylogenetic analysis of individual ORFs support a monophyletic origin of the two avipoxviruses relative to other ChPV (reference 5 and data not shown). CNPV and FWPV exhibit significant differences in variable genomic regions and between orthologous ORFs, suggesting divergence comparable to that seen between other ChPV genera. This level of divergence also suggests that considerable genomic diversity may exist within the avipoxvirus genera, consistent with both the breadth of potential host species within the class Aves and differences in host range, cross-protection, and restriction patterns among avipoxvirus isolates (8, 33, 79, 97, 98). Genomic sequencing of additional avipoxvirus isolates should clarify this issue.
Notably, internal genomic regions exhibit considerable variability between CNPV and FWPV, contrasting the overall synteny and conservation observed among central genomic regions of other ChPV. These internal variable regions correspond to regions identified in FWPV as located near junctions of major genome rearrangement relative to other ChPV and containing genes likely involved in virus-host interaction (Fig. 1) (2). The high level of variability between CNPV and FWPV near these internal junction regions suggests that changes continued to accrue there after the genomic rearrangement of avipoxviruses relative to other ChPV. Given the conservation between central genomic regions of other ChPV, the extent of this avipoxvirus central region variability is intriguing and conceivably reflects either increased selective pressures on the potential virulence and host range genes in these regions or unique aspects of avipoxvirus genome replication.
Features of the CNPV genome suggest both genomic expansion in CNPV and reduction in FWPV. CNPV contains many genes that are shorter, disrupted, or fragmented in FWPV. This includes in CNPV 19 genes longer (>10%) than potential FWPV orthologues and 7 genes extensively fragmented in FWPV, 5 of which are nonoverlapping ORFs with putative function (Table 1). In comparison, eight FWPV genes are longer than potential CNPV orthologues and four are fragmented in CNPV; however, these four are largely overlapping and likely represent minor ORFs annotated in FWPV. This discrepancy suggests that FWPV reflects genomic reduction from a more CNPV-like ancestor, assuming that obvious gene degradation indicates a more derived virus. Conversely, CNPV contains relative to FWPV additional, potentially duplicated family genes (N1R/p28 and TGF-β genes between CNPV157 and CNPV169 and between CNPV217 and CNPV228) and genes present in regions syntenic between FWPV and other ChPV (CNPV072, CNPV073, CNPV079, CNPV081, and CNPV185), features suggestive of genomic expansion relative to FWPV and potential ancestors. Alternatively, expanded gene family regions may be ancestral and lost in FWPV. These genomic tendencies could conceivably reflect adaptation of CNPV and FWPV to broader and more-restricted host range potentials, respectively.
Conclusions.
The CNPV genome provides new insight as to avipoxviral genomics and diversity. Analysis revealed the coding capacity of the largest poxviral genome currently sequenced, with much novel sequence and genes comprised of extensive gene families and homologues of cellular genes. Such genes, especially those conserved phylogenetically, are potentially significant in the interaction of CNPV with avian or nonavian hosts and may prove suitable targets for modification in engineering improved CNPV-based vaccines. Major genomic differences between CNPV and FWPV indicate a relatively distant relationship and suggest that, although monophyletic, avipoxviruses may exhibit diversity commensurate with a broad range of avian hosts.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, E. Oma, G. F. Kutish, and D. L. Rock. 1999. The genome of Melanoplus sanguinipes entomopoxvirus. J. Virol. 73:533-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguiar, J. C., R. C. Hedstrom, W. O. Rogers, Y. Charoenvit, J. B. Sacci, Jr., D. E. Lanar, V. F. Majam, R. R. Stout, and S. L. Hoffman. 2001. Enhancement of the immune response in rabbits to a malaria DNA vaccine by immunization with a needle-free jet device. Vaccine 20:275-280. [DOI] [PubMed] [Google Scholar]
- 4.Aloe, L., M. D. Simone, and F. Properzi. 1999. Nerve growth factor: a neurotrophin with activity on cells of the immune system. Microsc. Res. Tech. 45:285-291. [DOI] [PubMed] [Google Scholar]
- 5.Amano, H., S. Morikawa, H. Shimizu, I. Shoji, D. Kurosawa, Y. Matsuura, T. Miyamura, and Y. Ueda. 1999. Identification of the canarypox virus thymidine kinase gene and insertion of foreign genes. Virology 256:280-290. [DOI] [PubMed] [Google Scholar]
- 6.Bawden, A. L., K. J. Glassberg, J. Diggans, R. Shaw, W. Farmerie, R. W. Moyer, D. A. Liebermann, and B. Hoffman. 2000. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology 274:120-139. [DOI] [PubMed] [Google Scholar]
- 7.Baxevanis, A. D., F. Puehler, H. Schwarz, B. Waidner, J. Kalinowski, B. Kaspers, S. Bereswill, and P. Staeheli. 2003. The molecular biology database collection: 2003 update. Nucleic Acids Res. 31:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaudette, F. R. 1953. The identity of canary pox and “Schnappkrankheit” with notes on vaccination and modification of the virus. Proc. U.S. Livestock Sanitary Assoc. 57:249-272. [Google Scholar]
- 9.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaesse, M., T. Kupke, R. Huber, and S. Steinbacher. 2000. Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate. EMBO J. 19:6299-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulanger, D., P. Green, B. Jones, G. Henriquet, L. G. Hunt, S. M. Laidlaw, P. Monaghan, and M. A. Skinner. 2002. Identification and characterization of three immunodominant structural proteins of fowlpox virus. J. Virol. 76:9844-9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boulanger, D., T. Smith, and M. A. Skinner. 2000. Morphogenesis and release of fowlpox virus. J. Gen. Virol. 81:675-687. [DOI] [PubMed] [Google Scholar]
- 13.Brojatsch, J., J. Naughton, H. B. Adkins, and J. A. Young. 2000. TVB receptors for cytopathic and noncytopathic subgroups of avian leukosis viruses are functional death receptors. J. Virol. 74:11490-11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845-855. [DOI] [PubMed] [Google Scholar]
- 15.Burnet, F. M., and J. E. Bernard. 1933. A virus disease of the canary of the fowl-pox group. J. Pathol. Bacteriol. 37:107-122. [Google Scholar]
- 16.Cavill, J. P. 1964. Canary pox: report of an outbreak in roller canaries (Serinus canarius). Vet. Rec. 76:463-465. [Google Scholar]
- 17.Connor, J. H., D. C. Weiser, S. Li, J. M. Hallenbeck, and S. Shenolikar. 2001. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol. Cell. Biol. 21:6841-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshpande, T., T. Takagi, L. Hao, S. Buratowski, and H. Charbonneau. 1999. Human PIR1 of the protein-tyrosine phosphatase superfamily has RNA 5′-triphosphatase and diphosphatase activities. J. Biol. Chem. 274:16590-16594. [DOI] [PubMed] [Google Scholar]
- 20.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Pasquale, G., and J. A. Chiorini. 2003. PKA/PrKX activity is a modulator of AAV/adenovirus interaction. EMBO J. 22:1716-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durant, A. J., and H. C. McDougle. 1938. Investigation of pox in canaries. Proc. U.S. Livestock Sanitary Assoc. 42:181-188. [Google Scholar]
- 23.Durum, S. K., and J. J. Oppenheim. 1993. Proinflammatory cytokines and immunity, p. 801-835. In W. E. Paul (ed.), Fundamental immunology, 3rd ed. Raven Press, New York, N.Y.
- 24.Engelmayer, J., M. Larsson, A. Lee, M. Lee, W. I. Cox, R. M. Steinman, and N. Bhardwaj. 2001. Mature dendritic cells infected with canarypox virus elicit strong anti-human immunodeficiency virus CD8+ and CD4+ T-cell responses from chronically infected individuals. J. Virol. 75:2142-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 27.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 28.Fang, Z. Y., K. Limbach, J. Tartaglia, J. Hammonds, X. Chen, and P. Spearman. 2001. Expression of vaccinia E3L and K3L genes by a novel recombinant canarypox HIV vaccine vector enhances HIV-1 pseudovirion production and inhibits apoptosis in human cells. Virology 291:272-284. [DOI] [PubMed] [Google Scholar]
- 29.Fickenscher, H., S. Hor, H. Kupers, A. Knappe, S. Wittmann, H. Sticht, H. Huang, A. Saxena, and J. Xiang. 2002. The interleukin-10 family of cytokines. Trends Immunol. 23:89-96. [DOI] [PubMed] [Google Scholar]
- 30.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biol. Sci. 12:543-554. [DOI] [PubMed] [Google Scholar]
- 31.Garaci, E., M. C. Caroleo, L. Aloe, S. Aquaro, M. Piacentini, N. Costa, A. Amendola, A. Micera, R. Calio, C. F. Perno, and R. Levi-Montalcini. 1999. Nerve growth factor is an autocrine factor essential for the survival of macrophages infected with HIV. Proc. Natl. Acad. Sci. USA 96:14013-14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghildyal, N., W. M. Schnitzlein, and D. N. Tripathy. 1989. Genetic and antigenic differences between fowlpox and quailpox viruses. Arch. Virol. 106:85-92. [DOI] [PubMed] [Google Scholar]
- 33.Giddens, W. E., Jr., L. J. Swango, J. D. Henderson, Jr., R. A. Lewis, D. S. Farner, A. Carlos, and W. C. Dolowy. 1971. Canary pox in sparrows and canaries (Fringillidae) and in weavers (Ploceidae). Pathology and host specificity of the virus. Vet. Pathol. 8:260-280. [DOI] [PubMed] [Google Scholar]
- 34.Gilbert, P. B., Y. L. Chiu, M. Allen, D. N. Lawrence, C. Chapdu, H. Israel, D. Holman, M. C. Keefer, M. Wolff, and S. E. Frey. 2003. Long-term safety analysis of preventive HIV-1 vaccines evaluated in AIDS vaccine evaluation group NIAID-sponsored phase I and II clinical trials. Vaccine 21:2933-2947. [DOI] [PubMed] [Google Scholar]
- 35.Gonczol, E., and S. Plotkin. 2001. Development of a cytomegalovirus vaccine: lessons from recent clinical trials. Expert Opin. Biol. Ther. 1:401-412. [DOI] [PubMed] [Google Scholar]
- 36.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:192-202. [DOI] [PubMed] [Google Scholar]
- 37.Hernandez-Acosta, P., D. G. Schmid, G. Jung, F. A. Culianez-Macia, and T. Kupke. 2002. Molecular characterization of the Arabidopsis thaliana flavoprotein AtHAL3a reveals the general reaction mechanism of 4′-phosphopantothenoylcysteine decarboxylases. J. Biol. Chem. 277:20490-20498. [DOI] [PubMed] [Google Scholar]
- 38.Hertig, C., B. E. Coupar, A. R. Gould, and D. B. Boyle. 1997. Field and vaccine strains of fowlpox virus carry integrated sequences from the avian retrovirus, reticuloendotheliosis virus. Virology 235:367-376. [DOI] [PubMed] [Google Scholar]
- 39.Hitchner, S. B. 1981. Canary pox vaccination with live embryo-attenuated virus. Avian Dis. 25:874-881. [PubMed] [Google Scholar]
- 40.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ignatius, R., M. Marovich, E. Mehlhop, L. Villamide, K. Mahnke, W. I. Cox, F. Isdell, S. S. Frankel, J. R. Mascola, R. M. Steinman, and M. Pope. 2000. Canarypox virus-induced maturation of dendritic cells is mediated by apoptotic cell death and tumor necrosis factor alpha secretion. J. Virol. 74:11329-11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imlach, W., C. A. McCaughan, A. A. Mercer, D. Haig, and S. B. Fleming. 2002. Orf virus-encoded interleukin-10 stimulates the proliferation of murine mast cells and inhibits cytokine synthesis in murine peritoneal macrophages. J. Gen. Virol. 83:1049-1058. [DOI] [PubMed] [Google Scholar]
- 43.Iyer, L. M., E. V. Koonin, and L. Aravind. 2002. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol. 3:0012. [Online.] http://genomebiology.com/2002/3/3/research/0012. [DOI] [PMC free article] [PubMed]
- 44.Jin, X., M. Ramanathan, Jr., S. Barsoum, G. R. Deschenes, L. Ba, J. Binley, D. Schiller, D. E. Bauer, D. C. Chen, A. Hurley, L. Gebuhrer, R. El Habib, P. Caudrelier, M. Klein, L. Zhang, D. D. Ho, and M. Markowitz. 2002. Safety and immunogenicity of ALVAC vCP1452 and recombinant gp160 in newly human immunodeficiency virus type 1-infected patients treated with prolonged highly active antiretroviral therapy. J. Virol. 76:2206-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson, B. J., and A. E. Castro. 1986. Canary pox causing high mortality in an aviary. J. Am. Vet. Med. Assoc. 189:1345-1347. [PubMed] [Google Scholar]
- 46.Junn, E., S. H. Han, J. Y. Im, Y. Yang, E. W. Cho, H. D. Um, D. K. Kim, K. W. Lee, P. L. Han, S. G. Rhee, and I. Choi. 2000. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J. Immunol. 164:6287-6295. [DOI] [PubMed] [Google Scholar]
- 47.Kawakita, M., G. S. Rao, J. K. Ritchey, D. K. Ornstein, M. A. Hudson, J. Tartaglia, E. Paoletti, P. A. Humphrey, T. J. Harmon, and T. L. Ratliff. 1997. Effect of canarypox virus (ALVAC)-mediated cytokine expression on murine prostate tumor growth. J. Natl. Cancer Inst. 89:428-436. [DOI] [PubMed] [Google Scholar]
- 48.Kim, T. J., and D. N. Tripathy. 2001. Reticuloendotheliosis virus integration in the fowl poxvirus genome: not a recent event. Avian Dis. 45:663-669. [PubMed] [Google Scholar]
- 49.Kong, L. J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koonin, E. V., and T. V. Ilyina. 1993. Computer-assisted dissection of rolling circle DNA replication. Biosystems 30:241-268. [DOI] [PubMed] [Google Scholar]
- 51.Laidlaw, S. M., M. A. Anwar, W. Thomas, P. Green, K. Shaw, and M. A. Skinner. 1998. Fowlpox virus encodes nonessential homologs of cellular α-SNAP, PC-1, and an orphan human homolog of a secreted nematode protein. J. Virol. 72:6742-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, Y., and L. K. Miller. 1995. Properties of a baculovirus mutant defective in the protein phosphatase gene. J. Virol. 69:4533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liebermann, D. A., and B. Hoffman. 2002. Myeloid differentiation (MyD)/growth arrest DNA damage (GADD) genes in tumor suppression, immunity and inflammation. Leukemia 16:527-541. [DOI] [PubMed] [Google Scholar]
- 54.Luque, A., A. P. Sanz-Burgos, E. Ramirez-Parra, M. M. Castellano, and C. Gutierrez. 2002. Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology 302:83-94. [DOI] [PubMed] [Google Scholar]
- 55.Marovich, M. A., J. R. Mascola, M. A. Eller, M. K. Louder, P. A. Caudrelier, R. El-Habib, S. Ratto-Kim, J. H. Cox, J. R. Currier, B. L. Levine, C. H. June, W. B. Bernstein, M. L. Robb, B. Schuler-Thurner, R. M. Steinman, D. L. Birx, and S. Schlesinger-Frankel. 2002. Preparation of clinical-grade recombinant canarypox-human immunodeficiency virus vaccine-loaded human dendritic cells. J. Infect. Dis. 186:1242-1252. [DOI] [PubMed] [Google Scholar]
- 56.Massague, J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 57.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 58.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Motta, I., F. Andre, A. Lim, J. Tartaglia, W. I. Cox, L. Zitvogel, E. Angevin, and P. Kourilsky. 2001. Cross-presentation by dendritic cells of tumor antigen expressed in apoptotic recombinant canarypox virus-infected dendritic cells. J. Immunol. 167:1795-1802. [DOI] [PubMed] [Google Scholar]
- 60.Moyer, R. W., B. Arif, D. N. Black, D. B. Boyle, R. M. Buller, K. R. Dumbell, J. J. Esposito, G. McFadden, B. Moss, A. A. Mercer, S. Ropp, D. N. Tripathy, and C. Upton. 2000. Family Poxviridae. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Academic Press, New York, N.Y.
- 61.Nishinaka, Y., H. Masutani, H. Nakamura, and J. Yodoi. 2001. Regulatory roles of thioredoxin in oxidative stress-induced cellular responses. Redox Rep. 6:289-295. [DOI] [PubMed] [Google Scholar]
- 62.Nishiyama, A., M. Matsui, S. Iwata, K. Hirota, H. Masutani, H. Nakamura, Y. Takagi, H. Sono, Y. Gon, and J. Yodoi. 1999. Identification of thioredoxin-binding protein-2/vitamin D3 up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J. Biol. Chem. 274:21645-21650. [DOI] [PubMed] [Google Scholar]
- 63.Novick, D., S. H. Kim, G. Fantuzzi, L. L. Reznikov, C. A. Dinarello, and M. Rubinstein. 1999. Interleukin-18 binding protein: a novel modulator of the Th1 cytokine response. Immunity 10:127-136. [DOI] [PubMed] [Google Scholar]
- 64.Oh, C. S., D. A. Toke, S. Mandala, and C. E. Martin. 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272:17376-17384. [DOI] [PubMed] [Google Scholar]
- 65.Orozco, B. M., and L. Hanley-Bowdoin. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 273:24448-24456. [DOI] [PubMed] [Google Scholar]
- 66.Orozco, B. M., L. J. Kong, L. A. Batts, S. Elledge, and L. Hanley-Bowdoin. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275:6114-6122. [DOI] [PubMed] [Google Scholar]
- 67.Pancholi, P., D. H. Lee, Q. Liu, C. Tackney, P. Taylor, M. Perkus, L. Andrus, B. Brotman, and A. M. Prince. 2001. DNA prime/canarypox boost-based immunotherapy of chronic hepatitis B virus infection in a chimpanzee. Hepatology 33:448-454. [DOI] [PubMed] [Google Scholar]
- 68.Pancholi, P., Q. Liu, N. Tricoche, P. Zhang, M. E. Perkus, and A. M. Prince. 2000. DNA prime-canarypox boost with polycistronic hepatitis C virus (HCV) genes generates potent immune responses to HCV structural and nonstructural proteins. J. Infect. Dis. 182:18-27. [DOI] [PubMed] [Google Scholar]
- 69.Paoletti, E. 1996. Applications of pox virus vectors to vaccination: an update. Proc. Natl. Acad. Sci. USA 93:11349-11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pearson, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 72.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, K. Limbach, E. K. Norton, and E. Paoletti. 1990. Vaccinia virus host range genes. Virology 179:276-286. [DOI] [PubMed] [Google Scholar]
- 73.Pica, F., A. Volpi, A. Serafino, M. Fraschetti, O. Franzese, and E. Garaci. 2000. Autocrine nerve growth factor is essential for cell survival and viral maturation in HHV-8-infected primary effusion lymphoma cells. Blood 95:2905-2912. [PubMed] [Google Scholar]
- 74.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 75.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8:499-504. [DOI] [PubMed] [Google Scholar]
- 76.Puehler, F., H. Schwarz, B. Waidner, J. Kalinowski, B. Kaspers, S. Bereswill, and P. Staeheli. 2003. An interferon-gamma-binding protein of novel structure encoded by the fowlpox virus. J. Biol. Chem. 278:6905-6911. [DOI] [PubMed] [Google Scholar]
- 77.Sadasiv, E. C., P. W. Chang, and G. Gulka. 1985. Morphogenesis of canary poxvirus and its entrance into inclusion bodies. Am. J. Vet. Res. 46:529-535. [PubMed] [Google Scholar]
- 78.Saitoh, T., S. Tanaka, and T. Koike. 2001. Rapid induction and Ca2+ influx-mediated suppression of vitamin D3 up-regulated protein 1 (VDUP1) mRNA in cerebellar granule neurons undergoing apoptosis. J. Neurochem. 78:1267-1276. [DOI] [PubMed] [Google Scholar]
- 79.Schnitzlein, W. M., N. Ghildyal, and D. N. Tripathy. 1988. Genomic and antigenic characterization of avipoxviruses. Virus Res. 10:65-75. [DOI] [PubMed] [Google Scholar]
- 80.Schwartz, S., Z. Zhang, K. A. Frazer, A. Smit, C. Riemer, J. Bouck, R. Gibbs, R. Hardison, and W. Miller. 2000. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 10:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Senkevich, T. G., E. V. Koonin, J. J. Bugert, G. Darai, and B. Moss. 1997. The genome of Molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 233:19-42. [DOI] [PubMed] [Google Scholar]
- 82.Senkevich, T. G., E. V. Koonin, and R. M. L. Buller. 1994. A poxvirus protein with a RING zinc finger motif is of crucial importance for virulence. Virology 198:118-128. [DOI] [PubMed] [Google Scholar]
- 83.Sofroniew, M. V., C. L. Howe, and W. C. Mobley. 2001. Nerve growth factor signaling, neuroprotection, and neural repair. Annu. Rev. Neurosci. 24:1217-1281. [DOI] [PubMed] [Google Scholar]
- 84.Somogyi, P., J. Frazier, and M. A. Skinner. 1993. Fowlpox virus host range restriction: gene expression, DNA replication, and morphogenesis in nonpermissive mammalian cells. Virology 197:439-444. [DOI] [PubMed] [Google Scholar]
- 85.Spencer, J. V., K. M. Lockridge, P. A. Barry, G. Lin, M. Tsang, M. E. Penfold, and T. J. Schall. 2002. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 76:1285-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Srinivasan, V., W. M. Schnitzlein, and D. N. Tripathy. 2001. Fowlpox virus encodes a novel DNA repair enzyme, CPD-photolyase, that restores infectivity of UV light-damaged virus. J. Virol. 75:1681-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Staeheli, P., F. Puehler, K. Schneider, T. W. Gobel, and B. Kaspers. 2001. Cytokines of birds: conserved functions—a largely different look. J. Interferon Cytokine Res. 21:993-1010. [DOI] [PubMed] [Google Scholar]
- 88.Strack, B., A. Calistri, M. A. Accola, G. Palu, H. G. Gottlinger, R. E. Means, S. Ishido, X. Alvarez, and J. U. Jung. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takagi, T., G. S. Taylor, T. Kusakabe, H. Charbonneau, and S. Buratowski. 1998. A protein tyrosine phosphatase-like protein from baculovirus has RNA 5′-triphosphatase and diphosphatase activities. Proc. Natl. Acad. Sci. USA 95:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tartaglia, J., M. C. Bonnet, N. Berinstein, B. Barber, M. Klein, and P. Moingeon. 2001. Therapeutic vaccines against melanoma and colorectal cancer. Vaccine 19:2571-2575. [DOI] [PubMed] [Google Scholar]
- 91.Tartaglia, J., M. E. Perkus, J. Taylor, E. K. Norton, J. C. Audonnet, W. I. Cox, S. W. Davis, J. van der Hoeven, B. Meignier, M. Riviere, B. Languet, and E. Paoletti. 1992. NYVAC: a highly attenuated strain of vaccinia virus. Virology 188:217-232. [DOI] [PubMed] [Google Scholar]
- 92.Taylor, J., B. Meignier, J. Tartaglia, B. Languet, J. VanderHoeven, G. Franchini, C. Trimarchi, and E. Paoletti. 1995. Biological and immunogenic properties of a canarypox-rabies recombinant, ALVAC-RG (vCP65) in non-avian species. Vaccine 13:539-549. [DOI] [PubMed] [Google Scholar]
- 93.Taylor, J., and E. Paoletti. 1988. Fowlpox virus as a vector in non-avian species. Vaccine 6:466-468. [DOI] [PubMed] [Google Scholar]
- 94.Taylor, J., C. Trimarchi, R. Weinberg, B. Languet, F. Guillemin, P. Desmettre, and E. Paoletti. 1991. Efficacy studies on a canarypox-rabies recombinant virus. Vaccine 9:190-193. [DOI] [PubMed] [Google Scholar]
- 95.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thomson, B. J., S. Efstathiou, and R. W. Honess. 1991. Acquisition of the human adeno-associated virus type-2 rep gene by human herpesvirus type-6. Nature 351:78-80. [DOI] [PubMed] [Google Scholar]
- 97.Tripathy, D. N., and W. M. Reed. 1997. Pox, p. 643-659. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames, Iowa.
- 98.Tripathy, D. N., W. M. Schnitzlein, P. J. Morris, D. L. Janssen, J. K. Zuba, G. Massey, and C. T. Atkinson.1 2000. Characterization of poxviruses from forest birds in Hawaii. J. Wildl. Dis. 36:225-230. [DOI] [PubMed] [Google Scholar]
- 99.Villoslada, P., S. L. Hauser, I. Bartke, J. Unger, N. Heald, D. Rosenberg, S. W. Cheung, W. C. Mobley, S. Fisher, and C. P. Genain. 2000. Human nerve growth factor protects common marmosets against autoimmune encephalomyelitis by switching the balance of T helper cell type 1 and 2 cytokines within the central nervous system. J. Exp. Med. 191:1799-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xiang, Y., and B. Moss. 2001. Correspondence of the functional epitopes of poxvirus and human interleukin-18-binding proteins. J. Virol. 75:9947-9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yuan, Y., D. M. Li, and H. Sun. 1998. PIR1, a novel phosphatase that exhibits high affinity to RNA · ribonucleoprotein complexes. J. Biol. Chem. 273:20347-20353. [DOI] [PubMed] [Google Scholar]