Abstract
The small subunit (SSU) processome is a 2.2 MDa ribonucleoprotein complex involved in the processing, assembly and maturation of the SSU of eukaryotic ribosomes. The identities of many of the factors involved in SSU biogenesis have been elucidated over the past 40 years. However, as our understanding increases, so do the number of questions about the nature of this complicated process. Cataloguing the components is the first step towards understanding the molecular workings of a system. This review will focus on how identifying components of ribosome biogenesis has led to the knowledge of how these factors, protein and RNA alike, associate with one another into sub-complexes, with a concentration on the small ribosomal subunit. We will also explore how this knowledge of sub-complex assembly has informed our understanding of the workings of the ribosome synthesis system as a whole.
Keywords: SSU processome, U3 snoRNA, Utp, ribosomal SSU, RNA processing, RNA chaperone
The process of making a single ribosome is a herculean task, and is one of the most metabolically expensive activities of a cell. In vigorously growing yeast cells, it requires the activity of all three RNA polymerases, accounting for 70% of total transcription, 90% of pre-mRNA splicing and more than 25% of translation.1 In Saccharomyces cerevisiae, nearly 7000 nucleotides of pre-rRNA must be accurately transcribed, cleaved, folded, chemically modified by 71 snoRNPs directing either 2′-O-methylation or pseudouridylation, and assembled with 78 ribosomal proteins (r-proteins) to form one mature ribosome. Despite the immensity of this task, about 2000 new ribosomes are produced each minute in yeast (~7500 subunits per minute in HeLa cells), leading to the presence of ~200 000 ribosomes in each cell (~10 million in each HeLa cell).1, 2
Because of its central importance, defects in ribosome biogenesis can have detrimental effects on cellular metabolism and vitality. Interestingly, a number of diseases have been found to be associated with defects in ribosome synthesis pathways. Several recent reviews contain details on the particular ribosomopathies known to date.3, 4 In addition, ribosome biogenesis is a key component of the cell cycle where it regulates cell size and growth,5–7 and is thus up-regulated in cancer.8 Despite this critical linkage, ribosome biogenesis is understudied and its role in cancer is underappreciated. In addition, new evidence suggests that ribosome biogenesis proteins play critical roles in stem-cell differentiation in Drosophila.9 Furthermore, ribosome synthesis may also be a mechanism through which HIV modulates host response.10
Tremendous efforts over the past 40 years or so have greatly increased our understanding of the molecular mechanisms of ribosome biogenesis. However, there are still many mysteries left to solve. A recent review in a different field outlined a particularly succinct way of describing how researchers can approach learning about a new biological system.11 First, researchers must identify all the parts of the system. Over the past 10 years, great strides have been made in identifying the factors with roles in ribosome synthesis. Especially in the model organism S. cerevisiae, one can say that probably most of these factors have been catalogued. Once the parts list of a system is complete, or nearly complete, researchers can then investigate which parts interact with other parts. In the dynamic process of ribosome biogenesis, this can be a decidedly frustrating task. A number of large- and small-scale studies have identified sub-complexes involved in ribosome biogenesis. However, the scope and coverage of these interactions are limited. Additionally, there are numerous enzymes involved in maturation steps of pre-ribosomes. How these enzymes interact with pre-ribosomal particles and the specific substrates of their reactions is also a challenge for researchers to decipher. Once sub-complexes of the system are defined, a third central question that researchers can ask is how does this system of connected parts work as a whole? Our current understanding of the molecular mechanism by which eukaryotic ribosomes are assembled is largely speculative, owing mainly to the lack of an in vitro reconstitution system. Indeed, our knowledge of the assembly and function of the spliceosome has been greatly aided by the use of in vitro splicing assays (reviewed in12). An advantage of studying ribosome biogenesis in Bacteria is that the bacterial ribosome can be assembled, although inefficiently, in vitro from its protein and rRNA components.13 Unfortunately, no system exists to study eukaryotic ribosome biogenesis in vitro, as coupling of transcription with pre-ribosome processing and maturation seems to be key to the complex journey of ribosome assembly.14, 15
There are a number of excellent reviews on the details of ribosome biogenesis.16–20 This review will focus mainly on biogenesis of the small subunit (SSU) of the ribosome in Eukaryotes, specifically on the current state of our understanding of ribosome biogenesis, and some anticipated future directions in this field. Therefore, we have included a brief overview of the process of SSU biogenesis to serve as a starting point for our commentary.
Overview of ribosome biogenesis in Eukaryotes
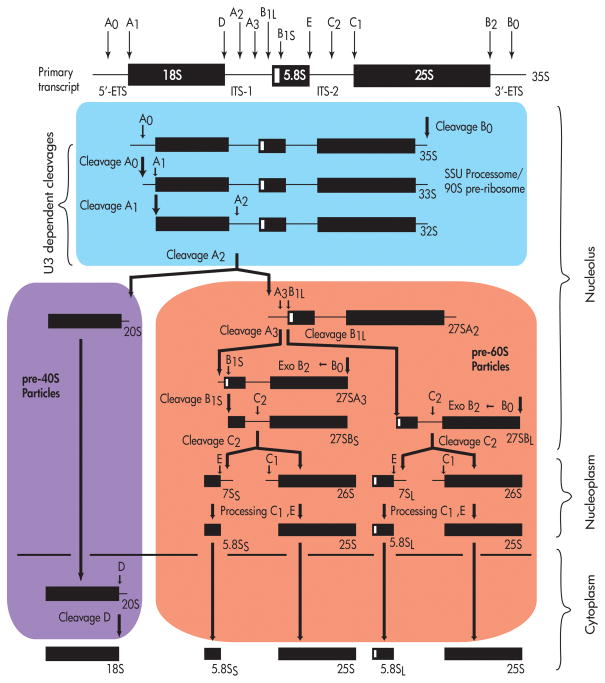
Ribosome biogenesis takes place in the nucleolus, a specialized compartment (or compartments) of the nucleus where the rDNA loci are grouped in several hundred tandem repeats in nucleolar organizing regions (NORs). Although the size, number and structure of nucleoli vary according to cell type and metabolic state, the ultrastructural morphology of the nucleolus is quite conserved in Eukaryotes. Three basic nucleolar regions can be distinguished by EM: the fibrillar center (FC) contains inactive rDNA genes, the dense fibrillar component (DFC) where pre-rRNA synthesis and early processing events occur, and the granular component where late processing events and assembly of ribosomal particles occur (reviewed in21–23). In Eukaryotes, three of the four mature rRNAs are transcribed by RNA polymerase I as a polycistronic precursor (see Figure 1). In yeast, the 35S pre-rRNA contains the sequences for the mature 18S, 5.8S, and 25S rRNAs. The fourth rRNA, the 5S rRNA, is transcribed separately by RNA polymerase III. The 35S precursor must be cleaved at sites A0, A1 and A2 to generate the pre-18S rRNA, a process which requires a large RNP dubbed the SSU processome. A subset of ribosomal and non-ribosomal proteins are assembled onto the pre-rRNA in the nucleolus. The rRNA precursor is also chemically modified by box C/D snoRNPs (43 in yeast), which are responsible for 2′-O-methylation, and box H/ACA snoRNPs (28 in yeast), which are responsible for pseudouridylation (reviewed in24–28). The 90S pre-ribosomal particles are separated into pre-40S and pre-60S particles via cleavage at site A2. Throughout the transcription and folding of the pre-rRNA, r-proteins are assembled into the pre-ribosomes as they migrate through the nucleoplasm. The final maturation steps, which include cleavage at site D, occur after the particles are exported into the cytoplasm. Details of these late steps can be found in several recent reviews.16, 17, 20
Figure 1. S. cerevisiae pre-rRNA processing scheme.
In yeast, the 35S polycistronic precursor rRNA is processed to the mature 18S, 5.8(S+L) and 25S rRNAs. The colored boxes signify pre-ribosomal particles: Blue – SSU processome, Purple – pre-40S particles, Red – pre-60S particles. See text for details on processing steps.
1. WHAT ARE ALL THE PARTS OF THE SYSTEM?
Ribosome biogenesis utilizes snoRNAs for varied functions
U3 was the first small nucleolar (sno) RNA identified and has since become the most extensively studied. It was discovered in 1968 by James L. Hodnett and Harris Busch during their investigations on the small RNAs of animal cells.29 Although it was originally identified with the U1 and U2 small nuclear (sn) RNAs, the U3 snoRNA was found to be enriched in the nucleolus and to co-purify with ribosomes. However, its role in ribosome biogenesis remained enigmatic for several years. In the mid 1970’s, the U3 snoRNA was conclusively implicated in pre-rRNA processing through the identification, and subsequent confirmation in the mid 1990’s by chemical cross-linking and mutational studies, of regions of complementarity between the U3 snoRNA and the pre-rRNA/rRNA transcript.30–34 Knockout of the U3 snoRNA genes was also found to arrest pre-rRNA processing at sites A0, A1 and A2, resulting in the accumulation of unprocessed 35S pre-rRNA transcripts, loss of the mature SSU 18S rRNA and concomitant cell lethality.
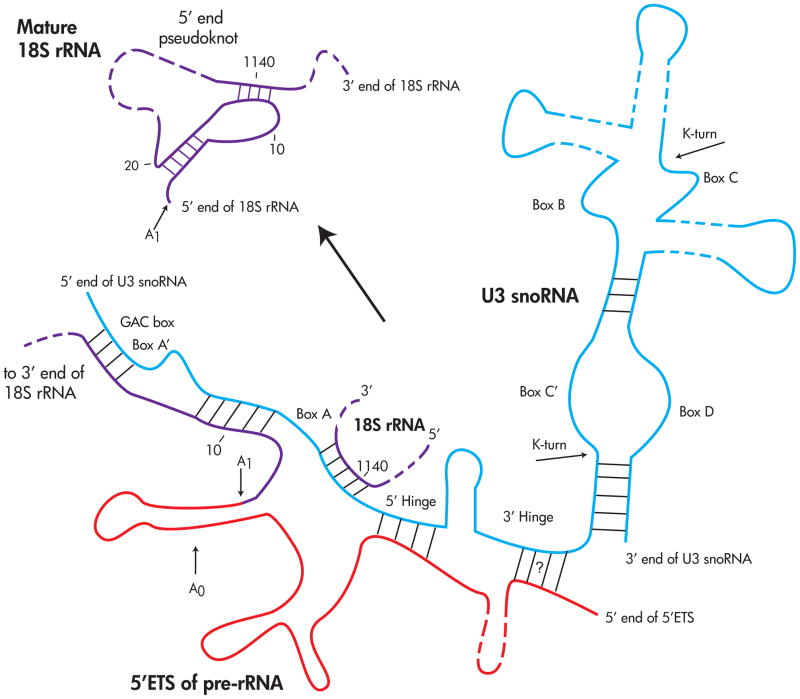
The U3 snoRNA is evolutionarily conserved and has been identified in all Eukaryotes examined thus far, including the phylogenetically diverse protists.35, 36 The secondary structure of U3 snoRNA can be divided into three domains, each with specific sequence elements, protein components, and distinct functions: a 5′ domain involved in base-pairing interactions with the pre-rRNA, a ‘hinge’ region consisting of the 5′ and 3′ hinge sequences, and a 3′ terminal hairpin domain containing the box C/D motifs (see Figure 2).34, 37 The 5′ domain contains the conserved sequence boxes GAC, A′ and A. These sequence elements are complementary to the 5′ end and to an internal region of the 18S rRNA.30–34, 38 Base-paring interactions between these anti-sense guide sequences in the U3 snoRNA and the pre-rRNA have been confirmed by chemical cross-linking,30 mutational analysis,31–33 in vivo structure probing34, 38 and evolutionary conservation.39
Figure 2. The U3 snoRNA base-pairs with the pre-18S rRNA to direct cleavages at A0, A1, and A2.
The U3 snoRNA has been proposed to act as a chaperone for the formation of the conserved 5′ end pseudoknot by base-pairing with the 18S rRNA. Colored lines correspond to different RNAs - Blue – U3 snoRNA, Purple – mature 18S rRNA sequences, Red – 5′ ETS of the pre-rRNA that is cleaved during pre-rRNA processing. See text for details.
The ‘hinge’ region of the U3 snoRNA is typically unstructured40, 41, except for a small centrally located stem-loop structure found in many organisms (including yeast34). The Mpp10 sub-complex of the SSU processome is associated with this region.42–44 The U3 snoRNA 5′ and 3′ hinge sequences have been proposed to provide proper spacing between the pre-rRNA-binding function of the 5′ domain and the protein-complexed 3′ domain of the U3 snoRNA.33, 37 Base-pairing interactions are also known to occur between the ‘hinge’ region of the U3 snoRNA and the 5′ ETS.33, 45–47 In yeast and Xenopus, the 5′ hinge was shown to base-pair with 5′ ETS of the pre-rRNA.30–33 The 3′ hinge in yeast has been predicted to also base-pair with the pre-rRNA,33 but this has not yet been tested.
The 3′ domain of the U3 snoRNA forms an extended stem-loop punctuated by a number of short hairpins forming a cruciform structure in yeast.37 This region is highly variable between different organisms and contains a number of dispensable non-conserved hairpins.35, 37 The 3′ stem-loop contains the box C/D sequence boxes C′, B, C and D, and is important in protein binding, RNA stability and nuclear retention.48–50 This region also contains two kink-turn (K-turn) RNA motifs within the juxtaposed box C′/D and B/C elements.49
There are also other snoRNAs in addition to the U3 snoRNA involved in ribosome maturation. Many of these snoRNAs were identified based on co-immunoprecipitation with fibrillarin (Nop1 in yeast), a component of box C/D snoRNPs.51 The U14 snoRNA was found to be able to base-pair with the 18S rRNA and is essential for cell viability.52 U3 and U14 are both non-canonical box C/D snoRNAs, in that they are required for cleavage of the pre-rRNA.
Subsequent to the discovery of the box C/D snoRNAs, another class of snoRNAs was identified.53, 54 These H/ACA snoRNAs are mainly involved in the chemical modification of uridine to pseudouridine.24, 25 However, two H/ACA snoRNAs also have roles in processing the pre-rRNA. Deletion of snR10, an H/ACA snoRNA, results in a cold-sensitive phenotype – suggestive of defects in RNA folding and ribonucleoprotein assembly55 – and also delays processing of the pre-18S rRNA.56 snR30/U17 is an essential box H/ACA snoRNA, and is involved in processing of the pre-18S rRNA.57, 58 snR30/U17 is unique in that it is the first box H/ACA snoRNA identified to have an evolutionarily conserved role in the cleavage of pre-RNA.59, 60 Although the U14, snR10 and snR30/U17 snoRNAs have been linked to pre-18S rRNA cleavages, it remains unclear whether or not these snoRNAs are incorporated into the SSU processome to perform their function.
Deletion of most modification-guide snoRNAs individually does not have a noticeable effect on ribosome biogenesis or ribosome stability. However, deletion of multiple snoRNAs in tandem, especially those with targets in the decoding region of the ribosome, the peptidyl-transferase center, or helix 69 of the LSU rRNA, results in slower growth rates, slower amino acid incorporation rates, decreased translational fidelity, as well as defects in pre-rRNA processing.28, 61, 62 In addition to their roles as modifying enzymes, it has been postulated that snoRNAs could be instrumental in the correct folding and maturation of the rRNA, although this has yet to be tested directly.63, 64
The ribosome synthesis machinery includes many non-ribosomal factors
The genetically tractable model organism S. cerevisiae has been used to identify many of the protein and RNA players in ribosome biogenesis. A majority of factors involved in ribosome biogenesis are both nucleolar and essential for growth. The characteristic processing intermediates visualized by northern blotting for pre-rRNAs have been useful as an assay to identify defects in ribosome biogenesis. Depletion of ribosome biogenesis factors results in processing defects, visualized by differing patterns of stalled pre-rRNA processing intermediates, and has allowed for the classification of proteins and non-coding RNAs into categories of pre-SSU and pre-LSU biogenesis factors. Sucrose gradients have been useful in identifying how individual proteins associate with pre-ribosomal particles via co-sedimentation experiments.
Biochemical purification of pre-ribosomal particles has largely been achieved through tandem affinity purification – mass spectrometry (TAP-MS) schemes, and has resulted in a large collection of ribosome biogenesis factors. The SSU processome, first purified by our laboratory, identified the so called U three proteins (Utps) that co-purified with the U3 snoRNA and its core proteins65, 66 (discussed below). Not surprisingly, both the U3 snoRNA and nearly all SSU processome proteins are phylogenetically conserved in all Eukaryotes examined thus far,35, 36 although they are not present in either Archaea or Bacteria.67 Concomitant with the discovery of the SSU processome, Grandi et al. identified a 90S pre-ribosomal particle that had many overlapping components with the SSU processome.68 Interestingly, these 90S particles were lacking many of the factors known to be required for LSU biogenesis, implying that pre-SSU and pre-LSU biogenesis are two distinct activities. This finding was in line with previous work showing that the 35S pre-rRNA locus can be physically separated and that stand-alone pre-18S and 5.8/25S rRNA coding units can, when transcribed separately, be processed and assembled into mature, functional ribosomes.69 Studies using TAP-MS or similar methods identified other components of 90S particles, many components of LSU biogenesis, as well as components of mitochondrial ribosomes.70–75
Our current view of the SSU processome is that of a massive 80S/2.2 MDa complex, of similar size to the ribosome, possibly containing as many as 72 protein components (see Table 1). Based on a strict inclusion criteria, ~46 proteins are able to (i) co-immunoprecipitate both U3 snoRNA and (ii) Mpp10, a known protein component of the SSU processome, and (iii) their genetic depletion results in loss of the 18S rRNA. Partial data (i.e., ability to co-IP SSU processome components but pre-18S rRNA processing defects not tested, or vice versa) for an additional 26 proteins warrants their inclusion as probable SSU processome components. With a few exceptions, the exact molecular function of most SSU processome proteins is unknown. However, inspection of the list of protein components reveals a number of proteins with RNA binding and/or protein-protein interaction motifs. Undoubtedly, some of these proteins play structural roles in the SSU processome. In addition, the SSU processome also contains endonucleases, 10 RNA helicases and their co-factors, and ATPases, GTPases, kinases and other regulatory proteins.76–78
Table 1. The known and putative protein components of the yeast SSU processome.
The proteins are enumerated by their membership in the known sub-complexes of the SSU processome. Proteins that are confirmed SSU processome components but which have not been ascribed to a specific sub-complex are listed as unclassified. Candidate proteins, for which partial data suggests that they may be SSU processome components, are listed as unknown. The five SSU r-proteins (Rps4, Rps6, Rps7, Rps9 and Rps14) that are confirmed components of the SSU processome66 were not tabulated. Commonly used yeast names are listed, along with their alias when applicable. The name of the human orthologue, along with its GenBank accession number, is also noted.
| Name | Alias | KDa | Human Orthologue | Sub-Complex | Comments | References |
|---|---|---|---|---|---|---|
| Nop1 | Lot3 | 34.5 | Fibrillarin; NP_001427 | Box C/D | 2′-O-methyltransferase; N-terminal glycine/arginine-rich (GAR) domain | 150, 151 |
| Nop56 | Sik1 | 56.9 | NOP56; NP_006383 | Box C/D | KKE/D repeats | 152 |
| Nop58 | Nop5 | 57.0 | NOP58; NP_057018 | Box C/D | KKE/D repeats | 153 |
| Snu13 | 13.6 | 15.5K/NHP2-like protein 1; NP_001003796 | Box C/D | binds to the helix-bulge-helix (K-turn) motifs of U3 snoRNA; also part of the U4/U6-U5 tri-snRNP | 154 | |
| Rrp9 | 65.1 | U3-55K; NP_004695 | U3 snoRNP | binds to the B/C motif of U3 snoRNA; contains WD40 repeats | 155, 156 | |
| Imp3 | 21.9 | IMP3; NP_060755 | Mpp10 | S4 RBD | 79 | |
| Imp4 | 33.5 | IMP4; NP_219484 | Mpp10 | contains a σ70-like motif | 79 | |
| Mpp10 | 67.0 | MPHOSPH10; NP_005782 | Mpp10 | coiled-coils; associated with the hinge region of U3 snoRNA | 157 | |
| t-Utp4 | 87.8 | Cirhin; NP_116219 | UtpA | WD40 repeats | 15, 65, 81 | |
| t-Utp5 | 72.0 | WDR43; NP_055946 | UtpA | WD40 repeats | 15, 65 | |
| t-Utp8 | 80.2 | ? | UtpA | also involved in nuclear tRNA export | 15, 65, 158 | |
| t-Utp9 | 65.3 | ? | UtpA | WD40 repeats | 15, 65, 81 | |
| t-Utp10 | 200.1 | BAP28/HEATR1; NP_060542 | UtpA | HEAT repeats | 15, 65, 81 | |
| t-Utp15 | 57.7 | UTP15; NP_115551 | UtpA | WD40 repeats | 15, 65, 81 | |
| t-Utp17 | Nan1 | 101.2 | WD repeat domain 75; AAH40567 | UtpA | WD40 repeats; also part of RENT complex | 15, 65, 81 |
| Pol5 | 115.9 | Myb-binding protein 1A; AAF33021 | UtpA | required for rRNA synthesis; not required for DNA replication | 15, 81, 159 | |
| Utp1 | Pwp2 | 104.0 | PWP2; NP_005040 | UtpB | WD40 repeats, β transducin family; Pwp2 domain | 65, 81, 82, 89 |
| Utp6 | 52.4 | UTP6; NP_060898 | UtpB | half-a-tetratricopeptide repeat motif [half-a-TPR (HAT)] | 65, 81, 82, 89 | |
| Utp12 | Dip2 | 106.3 | WDR3; NP_006775 | UtpB | WD40 repeats, Dip2 domain | 65, 81, 82, 89 |
| Utp13 | 91.0 | TBL3/transducin β-like 3; NP_006444 | UtpB | WD40 repeats, Utp13 domain | 65, 81, 82, 89 | |
| Utp18 | 66.4 | UTP18/WDR50/CGI-48; NP_057085 | UtpB | WD40 repeats | 66, 81, 82, 89 | |
| Utp21 | 104.8 | WDR36/TA-WDRP; NP_644810 | UtpB | WD40 repeats, coiled-coil domains, Utp21 domain | 66, 81, 82, 89, 160 | |
| Rrp7 | 34.5 | RRP7A; NP_056518 | UtpC | Also part of the CURI complex | 81, 83, 161 | |
| Utp22 | 140.5 | NOL6/NRAP; NP_075068 | UtpC | Also part of the CURI complex | 66, 81, 83, 160, 162 | |
| Cka1 | 44.7 | CSNK2A1; NP_808227 | UtpC | Co-purifies with UtpC; α catalytic subunit of casein kinase 2; also part of the CURI complex; non-essential | 81, 83 | |
| Cka2 | 39.4 | CSNK2A1; NP_808227 | UtpC | Co-purifies with UtpC; α′ catalytic subunit of casein kinase 2; also part of the CURI complex; non-essential | 81, 83 | |
| Ckb1 | 32.3 | CSNK2B; NP_001311 | UtpC | Co-purifies with UtpC; β regulatory subunit of casein kinase 2; also part of the CURI complex; non-essential | 81, 83 | |
| Ckb2 | 29.8 | CSNK2B; NP_001311 | UtpC | Co-purifies with UtpC; β′ regulatory subunit of casein kinase 2; also part of the CURI complex; non-essential | 81, 83 | |
| Rrp36 | 36.1 | C6orf153; NP_149103 | UtpC? | 91 | ||
| Utp2 | Nop14 | 94.3 | Nop14; NP_003694 | unclassified | also involved in SSU nuclear export | 65, 109, 163 |
| Utp3 | Sas10 | 70.3 | UTP3; NP_065101 | unclassified | contains a Utp3 domain; gene silencing | 65 |
| Utp7 | Kre31 | 62.3 | WDR46/BING4; NP_005443 | unclassified | WD40 repeats; adenylate binding site; kinetochore component required for chromosome segregation | 65, 164 |
| Utp11 | 29.7 | UTP11L/CGI-94; NP_057121 | unclassified | coiled-coils | 65 | |
| Utp14 | 103.0 | UTP14A; NP_006640 | unclassified | coiled-coils; ATP/GTP binding site (P-loop) | 65 | |
| Utp16 | Bud21 | 24.4 | yeast-specific | unclassified | coiled-coils; non-essential | 65 |
| Noc4 | Utp19 | 63.6 | NOC4L/MGC3162; NP_076983 | unclassified | Noc domain; also involved in SSU nuclear export | 66, 165 |
| Utp20 | 287.6 | 1A6/DRIM; NP_055318 | unclassified | HEAT repeats, ARM-type fold | 66, 160 | |
| Utp23 | 28.8 | UTP23; NP_115710 | unclassified | PINc nuclease domain not required for function | 118 | |
| Utp24 | Fcf1 | 21.6 | FCF1; NP_057046 | unclassified | PINc nuclease domain required for function | 118, 166 |
| Utp25 | 84.0 | DEF/C1orf107; NP_055203 | unclassified | DEAD-box helicase-like; DUF1253 domain | 167–169 | |
| Utp30 | 31.6 | RSL1D1; NP_056474 | unclassified | non-essential | 160 | |
| Bms1 | 135.6 | BMS1; NP_055568 | unclassified | GTPase, stimulated by Rcl1 | 77, 80 | |
| Dbp8 | 47.9 | DDX49; NP_061943 | unclassified | DEAD-box RNA helicase; stimulated by Esf2 | 119, 128 | |
| Dhr1 | Ecm16 | 145.0 | DHX37; NP_116045 | unclassified | DEAH-box RNA helicase | 119, 170 |
| Dhr2 | 82.7 | DEAH box polypeptide 8; AAH47327 (?) | unclassified | DEAH-box RNA helicase | 119, 170 | |
| Emg1 | Nep1 | 27.9 | EMG1/C2F; NP_006322 | unclassified | member of α/β knot fold methyltransferase (SPOUT) superfamily; displays pseudouridine methyltransferase | 66, 171 |
| Krr1 | 37.2 | KRR1/HRB2; NP_008974 | unclassified | contains a KRR-R motif activity and a KH domain | 66 | |
| Rcl1 | 40.2 | RCL1; NP_005763 | unclassified | RNA terminal phosphate cyclase-like protein; stimulates | 80, 172 | |
| Rok1 | 63.7 | DDX52; NP_008941 | unclassified | DEAD-box RNA helicase Bms1 | 119 | |
| Rrp3 | 56.0 | DDX47; NP_057439 | unclassified | DEAD-box RNA helicase | 119 | |
| Rrp5 | 193.1 | PDCD11/NFBP; NP_055791 | unclassified | 12 S1 RNA-binding motifs; HAT motif; binds single stranded tracts of U′s; participates in A3 cleavage in 5.8S processing | 65, 173 | |
| Sof1 | 56.8 | WDSOF1/DCAF13; NP_056235 | unclassified | WD40 repeats; similar to β subunit of trimeric G-proteins | 174 | |
| Dbp4 | Hca4/Ecm24 | 87.2 | DDX10; NP_004389 | unknown | DEAD-box RNA helicase | 175 |
| Enp1 | Meg1 | 55.1 | bystin/BYSL; NP_004044 | unknown | contains a bystin domain and coiled-coil domains | 66, 176 |
| Esf1 | 72.4 | ESF1; NP_057733 | unknown | 177 | ||
| Esf2 | Abt1 | 36.4 | ABT1; NP_037507 | unknown | binds to RNA and stimulates ATPase activity of Dbp8 | 128, 178 |
| Fal1 | 45.2 | DDX48/EIF4A3; NP_055555 | unknown | member of the eIF4A subfamily of DEAD-box RNA helicases | 179 | |
| Fyv7 | 18.2 | yeast-specific | unknown | contains coiled-coil motifs; non-essential | 162 | |
| Gno1 | Pxr1 | 31.3 | PINX1; NP_060354 | unknown | also involved in snoRNA maturation; contains a G-patch RNA interacting domain and KKE/D repeats | 180 |
| Has1 | 56.7 | DDX18; NP_006764 | unknown | DEAD-box RNA helicase; also required for LSU synthesis | 181 | |
| Kre33 | 119.3 | NAT10; NP_078938 | unknown | contains an N-acetyltransferase and a putative ATPase domain | 68, 169 | |
| Lcp5 | 40.8 | Neuroguidin/NGDN; NP_001036100 | unknown | contains a Utp3 domain | 182 | |
| Ltv1 | Ykl2 | 53.4 | LTV1; NP_116249 | unknown | also required for SSU nuclear export; non-essential | 108 |
| Mrd1 | 101.1 | RBM19; NP_057280 | unknown | contains 5 RNA-binding domains | 183 | |
| Nop9 | 77.7 | C14orf21; NP_777573 | unknown | multiple pumilio-like RNA binding repeats | 184 | |
| Nsr1 | She5 | 44.5 | Nucleolin/NCL; NP_005372 | unknown | contains 2 RNA recognition motifs and a GAR domain | 185, 186 |
| Pfa1 | Sqs1 | 86.9 | GPATCH2; NP_060510 | unknown | stimulates the ATPase and helicase activities of Prp43p; contains a G-patch RNA interacting domain; non-essential | 124 |
| Prp43 | JA1 | 87.6 | DHX15; NP_001349 | unknown | DEAH-box RNA helicase; also involved in mRNA splicing and LSU biosynthesis | 187 |
| Sen1 | Cik3/Nrd2 | 252.5 | Senataxin/SETX/ALS4; NP_055861 | unknown | Upf1-like RNA helicase; also required for LSU synthesis | 188 |
| Sgd1 | 102.9 | NOM1; NP_612409 | unknown | contains armadillo-type fold | 169 | |
| Slx9 | 24.1 | DBF4-type zinc finger-containing protein 2; NP_065974 | unknown | non-essential | 189 | |
| YGR251W | 22.3 | BAF82397 (?) | unknown | 162 |
(?) denotes uncertain human orthologue identification or unclear membership in an SSU processome sub-complex.
Currently, it would seem that the component list of the SSU processome is essentially complete. However, a caveat to the methods utilized thus far is that they may be excluding low abundance and/or low affinity proteins, regulatory components and transiently-associated factors, such as enzymes. Experiments that can cross-link proteins and/or RNA in vivo may still yield unexpected additions to the long (and expanding?) list of ribosome biogenesis players.
2. WHICH PARTS INTERACT WITH OTHER PARTS?
The SSU processome enters the Systems Biology era
With a relatively complete, and extensive, parts list for the SSU processome, the next question then becomes: how do these many proteins assemble and interact together during SSU biogenesis? As a box C/D snoRNA, U3 was known, de facto, to associate with the four box C/D snoRNP proteins fibrillarin/Nop1, Nop56, Nop58 and Snu13/15.5K and with the U3 snoRNA-specific Rrp9/U3-55K thereby forming the so called U3 snoRNP. A yeast two-hybrid screen for proteins that physically interact with Mpp10 identified the second sub-complex of the SSU processome. Dubbed the Mpp10 sub-complex, it contains the Mpp10 protein along with Imp3 and Imp4.79 Similarly, a yeast two-hybrid screen of the cyclase-like protein Rcl1 identified the regulatory GTPase Bms1 as a second member of this (as of yet) two component sub-complex.80
The breakthrough in defining the sub-complexes of the SSU processome came through a TAP-MS study of the RNA processing complexes in yeast.81 This study identified three large sub-complexes of the SSU processome, namely the transcriptional Utps (t-Utps) or UtpA (t-Utp4, 5, 8, 9, 10, 15, 17 and Pol5), UtpB (Utp1, 6, 12, 13, 18 and 21) and UtpC (Utp22, Rrp7 and the four subunits of casein kinase II: Cka1, Cka2, Ckb1 and Ckb2).81 While these sub-complexes were initially described by Krogan et al., not all sub-complexes were present in their complete form.15, 82, 83 Indeed, work is still needed to both verify existing sub-complex information and to identify other putative sub-complexes that were not observed in these previous studies.
The t-Utps/UtpA is believed to form one of the earliest sub-complexes in the assembly of the SSU processome.15, 84, 85 This sub-complex, formed independently of the U3 snoRNA,15, 81 is required both for the transcription of the pre-rRNA and its processing, whereas the remainder of the SSU processome components are only known to play a role in pre-rRNA processing.65, 66 A protein-protein interaction (PPI) map of the t-Utp/UtpA sub-complex, determined by yeast two-hybrid assay, has recently been published,86 and generally overlaps with a partial PPI map determined by protein-fragment complementation assay.87 While the t-Utp/UtpA components are generally conserved in Eukaryotes, it is noteworthy that t-Utp8 and 9 are yeast-specific and have no known human homologues.65, 88 This raises the intriguing possibility that currently unidentified components of the human t-Utp/UtpA sub-complex are the functional homologues of the yeast t-Utp8 and 9.88
Little is known of the function of the UtpB sub-complex. Our laboratory previously determined the architecture of the UtpB sub-complex by yeast two-hybrid methodology augmented by the validation of a key PPI by surface plasmon resonance.89 From this PPI map, we obtained some insight into the assembly of the sub-complex from its individual components.
The UtpC sub-complex is the least studied of the sub-complexes. The presence of the casein kinase II complex within the UtpC sub-complex is unexpected and raises a number of intriguing possibilities. In a different yeast species, casein kinase II is known to phosphorylate SSU r-proteins,90 suggesting the possibility that casein kinase II complex phosphorylates components of the SSU processome, and thereby temporally and/or spatially regulates their activities. It is noteworthy that the casein kinase II complex has also been identified as part of two additional complexes and is therefore not unique to the UtpC sub-component of the SSU processome.81 Finally, a recently identified SSU processome component, Rrp36, may also be a member of the UtpC sub-complex based on TAP-MS studies,91 though this has not been firmly established.
In all, six sub-complexes of the SSU processome have been described: (i) the U3 snoRNP “monoparticle,” (ii) the Mpp10 sub-complex, (iii) the Bms1/Rcl1 sub-complex and the (iv) t-Utp/UtpA, (v) UtpB and (vi) UtpC sub-complexes (see Table 1). However, the protein components of these various sub-complexes account for only 31 of the 72 (43%) protein components of the SSU processome; thus, more than half of the SSU processome proteins have yet to be ascribed to a sub-complex. A certain number of these proteins may individually associate with the SSU processome, as one would expect for regulatory proteins. The remainder of these proteins may form currently undescribed sub-complexes, be present in low-abundance, or engage in transient associations with components of the currently known sub-complexes. It is unknown whether the components of the SSU processome sub-complexes remain tethered to one another in some fashion during the ribosome assembly process. It is plausible that many of these sub-complexes exist as discrete functional units at different points of ribosome biogenesis. An example of one such complex is the t-Utp/UtpA subcomplex, which putatively assembles first onto the rRNA during its transcription.15 The t-Utps/UtpA could then recruit other subcomplexes, resulting in the formation of the SSU processome as a whole. While the SSU processome, by definition,65 contains at least the U3 snoRNA, Nop5/58, and Mpp10, the function of this large RNP may require that sub-complex association be somewhat dynamic. However, we know that components of the SSU processome, as originally purified, co-migrate on sucrose gradients in a large (80S) RNP.65 Exploring the interaction profiles of these components will yield much information about the assembly and architecture of the SSU processome, along with the multiple dynamic interactions necessary for assembly of pre-ribosomes.
Most of our knowledge of the protein interactions within the sub-complexes of the SSU processome has been obtained by small-scale studies. Indeed, an examination of high-throughput genetic and binary PPI data has revealed a lack of information on the SSU processome protein components.87, 92–94 In genetic interaction studies, either by epistatic mini-array profile (E-MAP) or by synthetic genetic array (SGA), the function of each gene in the genome is perturbed, often by deletion, and the phenotypic consequence is measured. The SSU processome is particularly recalcitrant to such approaches, most likely due to the fact that the overwhelming majority of its components (62/72) are essential proteins. Thus, alternative strategies for genetic perturbation, such as the decreased abundance by mRNA perturbation (DAmP)95 or the creation of conditional or hypomorphic alleles, must be used. As a result, recent large scale E-MAP96 and SGA97 have included few SSU processome components in their studies. The need for such data is critical, and in its absence, important questions, such as the genetic and physical PPIs linking the various sub-complexes remain unknown.
From PPI maps to structural studies of the sub-complexes
Biochemical studies to characterize sub-complexes of the SSU processome and other pre-40S particles have provided structural biologists with numerous problems to solve. Protein-protein and protein-RNA interaction data support the hypothesis that pre-ribosomes are assembled sequentially from multiple independently formed “modules.”15, 81, 85 While limited information is available for how these proteins assemble in low resolution maps, even less is known about the atomic or near atomic resolution of these “modules.” Indeed, most of the high-resolution structures available for ribosome maturation factors is limited to snoRNPs. Even so, the majority of data that exists for the snoRNPs is from crystal structures of archaeal homologues (reviewed in27).
Although limited to lower resolution than can be achieved via X-ray diffraction, electron microscopy (EM) remains a valuable tool to use for structural studies. Recently, our laboratory solved the structure of an archaeal box C/D sRNP using single particle negative stain EM.98 Importantly, it challenged the conventional view of box C/D sRNPs as monomeric moieties containing one sRNA and two copies of each of the three core proteins: L7Ae, Nop5, and fibrillarin. Instead, the structure showed that the archaeal box C/D sRNP is a di-sRNP with two sRNAs and four copies of each core protein. Formation of this dimeric structure correlates with enzymatic activity. Around the same time, Ye et al.99 solved an atomic resolution structure of an archaeal box C/D sRNP. Their model conflicts with our di-sRNP reconstruction. This discrepancy could be a result of their sRNA construct, as they did not use a full-length sRNA in their structure, increasing the likelihood of artificial crystal packing interactions influencing their proposed model. Previously, results were limited by the available crystal structures of solely the individual core proteins, co-crystal structures containing the Nop5-fibrillarin dimer without RNA, or L7Ae bound to a kink-turn RNA.100–103 In order to obtain pseudo-atomic resolution, it is possible to “fit” crystal structures into EM volumes (see Figure 3). This fitting allows more detailed information to be gleaned from lower-resolution structures, and is an alternative to using antibody or gold nanoparticle labeling to interpret individual protein placement in large protein complexes.104, 105
Figure 3. X-ray crystal structures can be docked into EM reconstruction volumes to give pseudo-atomic resolution of large complexes.
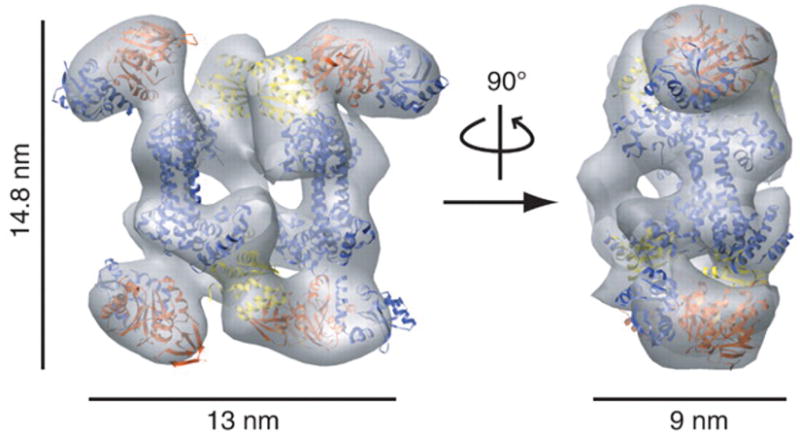
Shown here is a docking of the crystal structures of Pyrococcus furiosus Nop5-fibrillarin [PDB 2nnw]100 and Methanococcus jannaschii L7Ae [1xbi]101 in the isodensity map of the M. jannaschii di-sRNP [EMBD accession code EMD-1636].98
The other class of rRNA modifying enzymes, the box H/ACA s(no)RNPs, has also been a target of structural studies. In 2006, an X-ray crystal structure of a holo-archaeal box H/ACA sRNP was solved.106 This work provided a glimpse into how its architecture affects the enzymatic activity of the sRNP, specifically how the guide sRNA is positioned by the core proteins that bind to it to position the guide sRNA to base-pair with the substrate. Recently, the structure of an archaeal box H/ACA sRNP, albeit lacking one of the four core proteins, Gar1, bound to a substrate was solved to atomic resolution.107 The architecture of this structure confirmed that the sRNA and associated proteins serve to position both guide and substrate RNA for correct enzymatic activity.
Neither high- nor low-resolution structures have been solved for protein complexes involved in folding of the eukaryotic pre-rRNA, nor for the enzymes responsible for the cleavages of the pre-rRNA, although some low-resolution PPI mapping has been done for the UtpA and UtpB sub-complexes.86, 87, 89 In order to gain a better understanding of how different “modules” of the pre-ribosomes are assembled, both structural and biochemical studies need to be correlated to appreciate the concerted and coordinated efforts that are required for the assembly of mature ribosomes.
3. HOW DOES THIS SYSTEM OF CONNECTED PARTS WORK AS A WHOLE?
How is ribosome production regulated by cellular activity?
As emphasized above, ribosome synthesis is an extraordinarily metabolically expensive endeavor for a cell.1, 5–7 Thus, when conditions for cell growth become favorable, mechanisms for up-regulating ribosome biogenesis become extremely important. Ribosome production is therefore tightly coordinated with environmental conditions and nutrient availability.108, 109 In yeast, a cell will only divide once it reaches a critical size, called the “setpoint,” which is dependent on the cell’s protein synthesis capacity - otherwise stated as having a sufficient number of mature ribosomes. In addition, results from our laboratory suggest that yeast cells can, in addition to sensing mature ribosome levels, also sense ribosome biogenesis.110 Genetic depletion of SSU processome components results in G1 arrest, implying that if ribosomes cannot be assembled at G1, progression through the cell cycle is arrested.111 Thus, ribosome biogenesis directly promotes passage through the START phase of the cell cycle.5, 110
A number of signal transduction pathways, such as the target of rapamycin (TOR), have been implicated in linking nutrient availability to ribosome production.112 Recent work is also starting to implicate a number of additional signaling pathways to have effects on ribosome synthesis, many of which have well established links to cancer8, such as the p53 pathway113, 114 and the c-Myc pathway.115, 116 Although in yeast the direct link of metabolism to ribosome biogenesis is well known, a number of disease states in humans are attributed to defects in ribosome synthesis pathways. It is unlikely that the whole story is clear surrounding this apparent linkage (for detailed reviews of ribosomopathies see 3, 4). In all, ribosome biogenesis is directly linked to the proliferative capacity of the cell, including cancer cells. Yet, this link remains understudied and underappreciated.
Similarly, regulation of the SSU processome’s activity is poorly understood. A number of SSU processome proteins have ATPase, GTPase or kinase activities, yet their role in regulating the activity of the SSU processome is just beginning to emerge.76, 77 It is noteworthy that the UtpC sub-complex contains the casein kinase II components that are presumably regulating aspects of the SSU processome’s activity.
Pre-rRNA transcription is spearheaded by UtpA
The tUtps/UtpA form a complex that is interesting in that the proteins are required both for the transcription of the pre-rRNA and for its processing, whereas the other SSU processome components are only known to play a role in pre-rRNA processing. They were first discovered during the initial characterization of the SSU processome components through the observation that genetic depletion of a subset of Utps resulted in the loss of the mature 18S and an unexpected decrease in 25S rRNA levels,65 suggesting a decrease in the levels of the 35S pre-rRNA that is a precursor to both. Subsequent work found that the t-Utps/UtpA are required for optimal transcription of the pre-rRNA in both yeast15 and humans88, as assayed by both transcription run-on assays and by the quantification of the number of rRNA transcripts in Miller chromatin spreads. Furthermore, the t-Utps are associated with rDNA and with the pre-18S rRNAs independent of the presence of the U3 snoRNA.15, 88 Therefore, it was suggested that the t-Utps/UtpA form one of the earliest sub-complexes in the assembly of the SSU processome and thus link pre-rRNA transcription and processing.15, 84 This finding was unexpected because it had been previously assumed that pre-rRNA transcription and processing were two separate processes. Unfortunately, the mechanism by which the t-Utp/UtpA sub-complex links pre-rRNA transcription by RNA polymerase I to processing of the pre-rRNA by the SSU processome remains unknown.
The SSU processome as a pre-rRNA chaperone
The pre-rRNA, containing roughly 7000 nucleotides, must be correctly cleaved and assembled into a functional ribosome. With such a large molecule, one can imagine a nearly infinite number of thermodynamic traps into which the pre-rRNA can misfold. Both the protein and RNA components of the SSU processome have been hypothesized to be involved in ribosome chaperone activities.39, 43, 44, 117 Indeed, the SSU processome is mainly regarded as a chaperone for the RNA folding that must occur prior to the multiple sequence-specific pre-rRNA cleavage events that eventually liberate the mature 5′ end of the 18S rRNA. A recent study by the Correll laboratory used in vitro FRET-based assays to demonstrate the assembly of the Imp3 and Imp4 ‘chaperone complex.’ This complex was shown to possess RNA chaperone activities that stabilize the U3 snoRNA:pre-rRNA duplex.44 In addition to the Imp3 and Imp4 proteins playing a role in promoting the correct U3:pre-rRNA interactions, the U3 snoRNA has been hypothesized to be a chaperone for the folding of the pre-RNA.16, 39 The role of the U3 snoRNA as an RNA chaperone stems from the observation that sequences within the conserved box A of U3 snoRNA are complementary to, and base-pair with, highly conserved regions of the mature SSU rRNA which fold into the universally conserved 5′ end pseudoknot (see Figure 2). Base-pairing interactions between the U3 snoRNA and the 5′ end pseudoknot of the 18S rRNA form a secondary structure that is incompatible with that of the mature 18S rRNA.39, 117 This interaction may serve to chaperone the folding of the SSU rRNA by sequestering the 5′ domain of the 18S rRNA, thereby preventing it from folding prematurely. A chaperone role has also been proposed for the U14 snoRNA.63
In addition to avoiding incorrect folding around the central pseudoknot, the interactions between the U3 snoRNA and the pre-18S rRNA also guide, by a complex and poorly understood mechanism, the multiple sequence-specific pre-rRNA cleavage events at sites A0, A1 and A2.31, 32, 34, 39, 117 The chaperone activity may serve to fold the pre-rRNA into a structure that can be recognized as a substrate by the putative endonucleases.118 Directly testing the chaperone roles of the SSU processome and its components, in particular the U3 snoRNA, is important for understanding their roles in ribosome biogenesis.
Is the SSU processome an RNA “untanglease”?
A remarkably large number of RNA helicases participate in ribosome biogenesis, with 10 such helicases participating in SSU assembly.78, 119 Most of these are members of the DEAH/D box families of RNA helicases, aptly named for their conserved D-E-A-H/D sequence. Their exact role in SSU biogenesis remains a matter of speculation, however, one can presume that RNA helicases mediate the many highly dynamic pre-rRNA folding and conformational rearrangements, rRNA duplex annealing and unwinding, and rRNA/protein remodeling and displacement events (such as the removal of non-ribosomal proteins from the pre-rRNA) that occur during ribosome biogenesis. Intriguingly, it has also been proposed that helicases may play a role in the unwinding of snoRNAs from their pre-rRNA/rRNA target. This is seen by the retention of snoRNAs on the pre-rRNA in the absence of the helicases Dbp4, Has1, Rok1 or Prp43, the latter in the pre-LSU rRNA,120–123 though this activity has not been directly demonstrated. This concept has recently been bolstered by the finding that Prp43 interacts with both snoRNAs and their corresponding substrate regions in the pre-LSU rRNA, as seen by the cross-linking and analysis of cDNA (CRAC) approach.123 Lastly, it has also been suggested that RNA helicases may act as molecular clamps, thereby holding their RNA substrate until its release by ATP hydrolysis.124 This is in line with the finding that RNA helicases can display both duplex unwinding and annealing activities.125
RNA helicase biochemical activities have been fairly well characterized.126, 127 However, the mechanism by which they temporally and spatially reach their target substrate(s) during ribosome biogenesis remains generally unclear and represents one of the most pressing questions in RNA helicase biology. It is generally believed that protein co-factors recruit, through PPIs, the helicase to its site of activity,128 as a few reported examples suggest. The SSU processome helicase Dbp8 is known to interact with Esf2, another SSU processome component. Esf2 can bind to pre-rRNAs, presumably providing substrate specificity, and can stimulate the activity of Dbp8, possibly providing temporal specificity.128 Similarly, the helicase Prp43 associates with the glycine-rich motif (G-patch) containing proteins Pfa1 and Gno1, both components of the pre-SSU particle.70, 124, 129, 130 A G-patch motif is a short conserved domain, consisting of seven highly conserved glycines, found in a number of RNA-binding proteins.131 The binding of Pfa1 to Prp43 was found to stimulate its helicase activity in both yeast124 and human cells,130 thus possibly providing temporal and spatial specificity. These examples involve only 2 of the 10 helicases involved in SSU biogenesis. It remains unknown which co-factors provide temporal and spatial specificity to the 8 remaining helicases. Also intriguing is whether these co-factors all play a stimulatory role, or do some also play an inhibitory role? Furthermore, it will be interesting to investigate the target specificity of these co-factors.
On a road to understanding function via structure
A monumental breakthrough occurred nearly a decade ago when atomic resolution structures of the bacterial and archaeal ribosomes were solved.132, 133 An atomic model of eukaryotic ribosomes, however, has been more elusive. Most recently, a pseudo-atomic model of the eukaryotic ribosome has been achieved through docking and modeling yeast rRNA and r-proteins into a cryo-EM volume of the Thermomyces lanuginosus ribosome.134 Understanding the movement of ribosomes during the process of translation has been immensely aided by deciphering the structures of both bacterial and eukaryotic ribosomes with different accessory factors.135–141
If one considers the problem of understanding the structure of a mature ribosome a difficult puzzle to solve, an equally or perhaps more daunting problem is delineating the stages of assembly and deconstructing the associated molecular machines involved in producing this massive molecular motor in vivo. For these steps, practically all of the current structural knowledge relies on negatively stained and cryo EM reconstructions. Work from the Hurt laboratory on the yeast late pre-40S subunit identified a maturation step involving the r-protein Rps3 and the protein kinase Hrr25.142 Based on their EM models, as well as biochemical experiments testing the stability of Rps3 association with the 40S in Hrr25 depleted cells, they determined that Hrr25-dependent phosphorylation and subsequent de-phosphorylation was required for beak formation in the mature 40S subunit.
Electron microscopy has also been effectively used to observe LSU biogenesis. Nissan et al.105 took advantage of the TAP method to investigate pre-60S subunits that contain the Rix1 complex. They were able to use antibody cross-linking in combination with electron microscopy to localize the AAA-type ATPase Rea1 to the “tail” region of the “tadpole-like” pre-60S structure. Interestingly, antibodies against ribosomal proteins Rpl3 or Rpl10 yielded predominantly “head-to-head” cross-links, suggesting that the Rea1 protein is distal to ribosomal proteins incorporated into the pre-60S particles. A more recent study on this particle shows that the positioning of Rea1 may enable the protein to use its metal ion-dependent adhesion site (MIDAS) as a mechanochemical lever to release non-ribosomal factors from the pre-60S particle.143 The combination of EM imaging with the biochemical and genetic experiments in this study provided strong and compelling evidence for their pre-60S maturation model.
Low resolution structural data remains informative
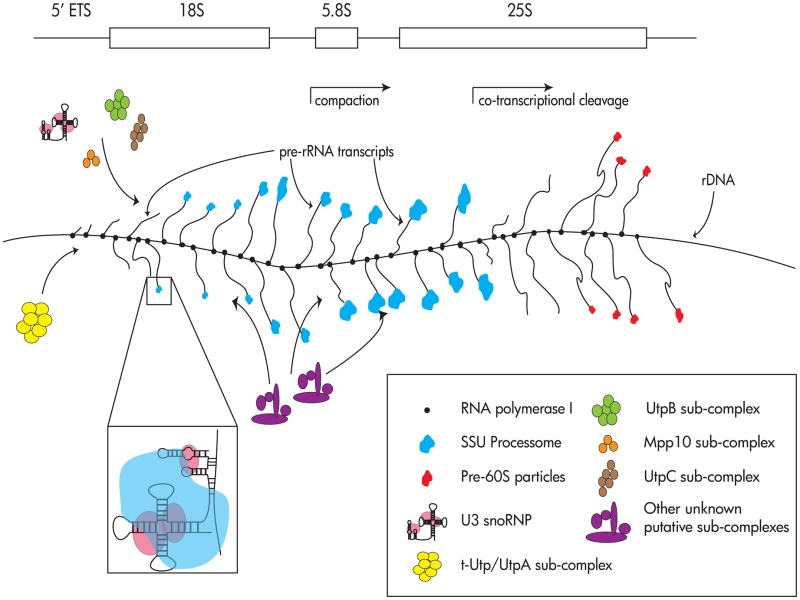
Despite the relative dearth of atomic or lower-resolution structural information available for the early pre-SSU, techniques used since the 1970s have yielded useful information about when, temporally, events in processing and assembly occur. Chromatin spreads, first described by Miller and colleagues,144 have long been used to study ribosome biogenesis in many organisms, including the yeast S. cerevisiae.145 The rDNA is arranged on chromosomes in tandem repeats, and is one of the most actively transcribed loci in the genome. Miller chromosome spreads have been dubbed “Christmas trees,” because the same repeat is transcribed at the same time by multiple molecules of RNA Pol I (see Figure 4). The pre-rRNA transcripts (the “branches” of the trees) have different lengths depending on the location of the polymerase, and, under the Miller spreading conditions, are splayed out from the “trunk” of the tree, corresponding to the rDNA. These pre-rRNA transcripts were also observed to have knobs of high electron density, which are involved in rRNA processing.146 Our laboratory, in collaboration with the Beyer laboratory, showed that depletion of any essential SSU processome component resulted in loss of these terminal knobs. Indeed, Miller spreads showed that depletion of any of the components of the UtpA sub-complex of the SSU processome resulted in reduced transcription at the rDNA loci.15 Interestingly, pre-rRNA processing can also be observed using Miller chromatin spreads. The Beyer laboratory also used EM chromatin spreads to visualize release of terminal knobs, as well as compaction of the pre-rRNA during SSU biogenesis.147, 148 Recently, the Tollervey laboratory has confirmed these co-transcriptional events using new techniques for harvesting and analyzing metabolically labeled pre-rRNA.14
Figure 4. Miller chromatin spreads, so-called “Christmas trees,” can be used to visualize pre-rRNA transcription and steps of pre-rRNA processing.
This diagram represents a Miller spread, and indicates how the SSU processome is assembled via sub-complexes onto the pre-rRNA transcript. See text for details. One repeat of the rDNA tandem chromosomal repeats is shown above the Miller chromatin spread diagram to indicate the relative positions of the 35S pre-rRNA.
FUTURE DIRECTIONS
A great deal of thought and experimental effort has been devoted to understanding the process of ribosome biogenesis, and ongoing experiments hope to address the remaining questions in the field. In S. cerevisiae, probably most of the factors involved in ribosome synthesis have been identified, addressing our first concern of the parts of the system. However, a great deal of information remains missing about the connectivity of the system. Despite a plethora of data generated by high-throughput studies,87, 92–94, 96, 97 most genetic and PPI linkages are still missing when describing which parts of the system interact with other parts. More effort needs to be applied to understand the interactome of the yeast nucleolus, as many proteins in the SSU processome are absent from these studies.149 These putative interactions could give insight into how the SSU processome is assembled on a larger scale, yielding structural information that may aid in understanding how the sub-complexes chaperone and regulate ribosome assembly. Recent work from the Tollervey laboratory has given the field an interesting look into where RNA helicases and other ribosome biogenesis factors bind to RNA.50, 123 However, it remains to be seen and validated how these helicases are directed to their site(s) of action and how their activities are regulated. Finally, we would like to know how the system of connected parts works as a whole. Gaining structural insight into both large and small functional units of the pre-ribosome could both answer and inspire new questions of how ribosome biogenesis is regulated through its many interacting factors. Although much is known about the SSU processome’s components, and some is known about the function of this massive molecular machine, there are assumptions in the field that have yet to be tested and proven. The SSU processome is predicted to mediate the correct folding of the pre-18S rRNA, however this attractive hypothesis has not yet been experimentally validated. An in vitro reconstitution system to study eukaryotic ribosome biogenesis could lead to an understanding of how the U3 snoRNA directs the cleavages of the 35S pre-rRNA. Availability of in vitro assays for ribosome biogenesis could also facilitate single-molecule experiments that may answer questions about the base-pairing interactions of the U3 snoRNA to the pre-rRNA. Are the multiple U3:pre-rRNA interactions simultaneous or sequential? Also, can these interactions occur within a single molecule of U3 snoRNA or from multiple molecules, as might be predicted if one imagines evolutionary conservation of the dimeric architecture of archaeal box C/D snoRNPs.98 While yeast is an incredibly versatile model organism, more focus should be applied to studying ribosome biogenesis in vertebrates in order to flesh out the differences and highlight the similarities between humans and yeast. Above all, despite the predictions that current results allow, it is important to experimentally validate hypotheses, because conclusions are strongest when supported by data.
Acknowledgments
The authors wish to thank members of the Baserga laboratory for the helpful reading of and comments on this manuscript. This material is based upon work supported by NIH GM 52581 (to SJB), NRSA NIH/NIGMS T32 GM007223 Training Grant and a National Science Foundation Graduate Research Fellowship (to KRP), and NIH T32 CA009259 Post-doctoral NIH-Ruth L. Kirschstein National Research Service Award, Institutional Research Training Grant: Radiation Therapy, Biology, Physics (to JMC).
References
- 1.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 2.Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 3.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst. 2010;6:481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Jorgensen P, Rupeš I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 9.Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant JA, Bellaiche Y, Huynh JR. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol. 2009;11:685–693. doi: 10.1038/ncb1874. [DOI] [PubMed] [Google Scholar]
- 10.Ponti D, Troiano M, Bellenchi GC, Battaglia PA, Gigliani F. The HIV Tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol. 2008;9:32. doi: 10.1186/1471-2121-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Connolly K, Culver G. Deconstructing ribosome construction. Trends Biochem Sci. 2009;34:256–263. doi: 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 18.Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Hage AE, Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. [PubMed] [Google Scholar]
- 20.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 21.Raška I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Int Rev Cytol. 2006;255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- 22.Prieto JL, McStay B. Nucleolar biogenesis: the first small steps. Biochem Soc Trans. 2005;33:1441–1443. doi: 10.1042/BST0331441. [DOI] [PubMed] [Google Scholar]
- 23.Thiry M, Lafontaine DL. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 2005;15:194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Kiss T, Fayet-Lebaron E, Jády BE. Box H/ACA small ribonucleoproteins. Mol Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 25.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 26.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichow SL, Hamma T, Ferré-D’Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodnett JL, Busch H. Isolation and characterization of uridylic acid-rich 7 S ribonucleic acid of rat liver nuclei. J Biol Chem. 1968;243:6334–6342. [PubMed] [Google Scholar]
- 30.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA GAC-Box A′ and Box A sequences play distinct functional roles in rRNA processing. Mol Cell Biol. 2001;21:6210–6221. doi: 10.1128/MCB.21.18.6210-6221.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borovjagin AV, Gerbi SA. The spacing between functional cis-elements of U3 snoRNA is critical for rRNA processing. J Mol Biol. 2000;300:57–74. doi: 10.1006/jmbi.2000.3798. [DOI] [PubMed] [Google Scholar]
- 34.Méreau A, Fournier R, Grégoire A, Mougin A, Fabrizio P, Lührmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 35.Marz M, Stadler PF. Comparative analysis of eukaryotic U3 snoRNA. RNA Biol. 2009;6:503–507. doi: 10.4161/rna.6.5.9607. [DOI] [PubMed] [Google Scholar]
- 36.Charette JM, Gray MW. U3 snoRNA genes are multi-copy and frequently linked to U5 snRNA genes in Euglena gracilis. BMC Genomics. 2009;10:528. doi: 10.1186/1471-2164-10-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samarsky DA, Fournier MJ. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antal M, Mougin A, Kis M, Boros E, Steger G, Jakab G, Solymosy F, Branlant C. Molecular characterization at the RNA and gene levels of U3 snoRNA from a unicellular green alga, Chlamydomonas reinhardtii. Nucleic Acids Res. 2000;28:2959–2968. doi: 10.1093/nar/28.15.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes JM. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 40.Jeppesen C, Stebbins-Boaz B, Gerbi SA. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988;16:2127–2148. doi: 10.1093/nar/16.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker KA, Steitz JA. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987;7:2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wormsley S, Samarsky DA, Fournier MJ, Baserga SJ. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA. 2001;7:904–919. doi: 10.1017/s1355838201010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gérczei T, Correll CC. Imp3p and Imp4p mediate formation of essential U3-precursor rRNA (pre-rRNA) duplexes, possibly to recruit the small subunit processome to the pre-rRNA. Proc Natl Acad Sci U S A. 2004;101:15301–15306. doi: 10.1073/pnas.0406819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gérczei T, Shah BN, Manzo AJ, Walter NG, Correll CC. RNA chaperones stimulate formation and yield of the U3 snoRNA-Pre-rRNA duplexes needed for eukaryotic ribosome biogenesis. J Mol Biol. 2009;390:991–1006. doi: 10.1016/j.jmb.2009.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA. 2004;10:942–953. doi: 10.1261/rna.5256704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borovjagin AV, Gerbi SA. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005;33:4995–5005. doi: 10.1093/nar/gki815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenwood SJ, Schnare MN, Cook JR, Gray MW. Analysis of intergenic spacer transcripts suggests ‘read-around’ transcription of the extrachromosomal circular rDNA in Euglena gracilis. Nucleic Acids Res. 2001;29:2191–2198. doi: 10.1093/nar/29.10.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cléry A, Senty-Ségault V, Leclerc F, Raué HA, Branlant C. Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair. Mol Cell Biol. 2007;27:1191–1206. doi: 10.1128/MCB.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marmier-Gourrier N, Cléry A, Senty-Ségault V, Charpentier B, Schlotter F, Leclerc F, Fournier R, Branlant C. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA. 2003;9:821–838. doi: 10.1261/rna.2130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci U S A. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schimmang T, Tollervey D, Kern H, Frank R, Hurt EC. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li HD, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 54.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 55.Guthrie C, Nashimoto H, Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci U S A. 1969;63:384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J. 2009;28:1260–1270. doi: 10.1038/emboj.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol Cell Biol. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enright CA, Maxwell ES, Eliceiri GL, Sollner-Webb B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA. 1996;2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 61.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 62.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 63.Liang WQ, Fournier MJ. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 1995;9:2433–2443. doi: 10.1101/gad.9.19.2433. [DOI] [PubMed] [Google Scholar]
- 64.Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 65.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryot Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Russell AG, de Sa MM, Dennis PP. A U3-like small nucleolar RNA in Archaea. Science. 1997;277:1189. doi: 10.1126/science.277.5330.1185d. [DOI] [PubMed] [Google Scholar]
- 68.Grandi P, Rybin V, Baßler J, Petfalski E, Strauß D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 69.Liang WQ, Fournier MJ. Synthesis of functional eukaryotic ribosomal RNAs in trans: development of a novel in vivo rDNA system for dissecting ribosome biogenesis. Proc Natl Acad Sci U S A. 1997;94:2864–2868. doi: 10.1073/pnas.94.7.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 72.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 73.Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fatica A, Cronshaw AD, Dlakic M, Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 75.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karbstein K, Jonas S, Doudna JA. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 78.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 79.Lee SJ, Baserga SJ. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wegierski T, Billy E, Nasr F, Filipowicz W. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA. 2001;7:1254–1267. doi: 10.1017/s1355838201012079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 82.Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J Biol Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 83.Rudra D, Mallick J, Zhao Y, Warner JR. Potential interface between ribosomal protein production and pre-rRNA processing. Mol Cell Biol. 2007;27:4815–4824. doi: 10.1128/MCB.02062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Granneman S, Baserga SJ. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol. 2005;17:281–286. doi: 10.1016/j.ceb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 85.Pérez-Fernández J, Román A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Freed EF, Baserga SJ. The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tarassov K, Messier V, Landry CR, Radinovic S, Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 88.Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007;21:2041–2054. doi: 10.1101/gad.436707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Champion EA, Lane BH, Jackrel ME, Regan L, Baserga SJ. A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol Cell Biol. 2008;28:6547–6556. doi: 10.1128/MCB.00906-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wojda I, Cytryñska M, Frajnt M, Jakubowicz T. Protein kinases CKI and CKII are implicated in modification of ribosomal proteins of the yeast Trichosporon cutaneum. Acta Biochim Pol. 2002;49:947–957. [PubMed] [Google Scholar]
- 91.Gérus M, Bonnart C, Caizergues-Ferrer M, Henry Y, Henras AK. Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit. Mol Cell Biol. 2010;30:1130–1144. doi: 10.1128/MCB.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 94.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 96.Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bleichert F, Gagnon KT, Brown BA, 2nd, Maxwell ES, Leschziner AE, Unger VM, Baserga SJ. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science. 2009;325:1384–1387. doi: 10.1126/science.1176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ye K, Jia R, Lin J, Ju M, Peng J, Xu A, Zhang L. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc Natl Acad Sci U S A. 2009;106:13808–13813. doi: 10.1073/pnas.0905128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oruganti S, Zhang Y, Li H, Robinson H, Terns MP, Terns RM, Yang W. Alternative conformations of the archaeal Nop56/58-fibrillarin complex imply flexibility in box C/D RNPs. J Mol Biol. 2007;371:1141–1150. doi: 10.1016/j.jmb.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 101.Suryadi J, Tran EJ, Maxwell ES, Brown BA., 2nd The crystal structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry. 2005;44:9657–9672. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- 102.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Luhrmann R, Carlomagno T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 103.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 104.Hainfeld JF. A small gold-conjugated antibody label: improved resolution for electron microscopy. Science. 1987;236:450–453. doi: 10.1126/science.3563522. [DOI] [PubMed] [Google Scholar]
- 105.Nissan TA, Galani K, Maco B, Tollervey D, Aebi U, Hurt E. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell. 2004;15:295–301. doi: 10.1016/j.molcel.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 106.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 107.Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol. 2009;16:740–746. doi: 10.1038/nsmb.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Loar JW, Seiser RM, Sundberg AE, Sagerson HJ, Ilias N, Zobel-Thropp P, Craig EA, Lycan DE. Genetic and biochemical interactions among Yar1, Ltv1 and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics. 2004;168:1877–1889. doi: 10.1534/genetics.104.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu PC, Thiele DJ. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol Biol Cell. 2001;12:3644–3657. doi: 10.1091/mbc.12.11.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol Biol Cell. 2007;18:953–964. doi: 10.1091/mbc.E06-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bernstein KA, Baserga SJ. The small subunit processome is required for cell cycle progression at G1. Mol Biol Cell. 2004;15:5038–5046. doi: 10.1091/mbc.E04-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 113.Gilkes DM, Chen J. Distinct roles of MDMX in the regulation of p53 response to ribosomal stress. Cell Cycle. 2007;6:151–155. doi: 10.4161/cc.6.2.3719. [DOI] [PubMed] [Google Scholar]
- 114.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010 doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 115.Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105:670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 117.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci USA. 2006;103:9464–9469. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Granneman S, Bernstein KA, Bleichert F, Baserga SJ. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol Cell Biol. 2006;26:1183–1194. doi: 10.1128/MCB.26.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kos M, Tollervey D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 121.Liang XH, Fournier MJ. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol Cell Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bohnsack MT, Kos M, Tollervey D. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008;9:1230–1236. doi: 10.1038/embor.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lebaron S, Papin C, Capeyrou R, Chen YL, Froment C, Monsarrat B, Caizergues-Ferrer M, Grigoriev M, Henry Y. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009;28:3808–3819. doi: 10.1038/emboj.2009.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 126.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 127.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 128.Granneman S, Lin C, Champion EA, Nandineni MR, Zorca C, Baserga SJ. The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res. 2006;34:3189–3199. doi: 10.1093/nar/gkl419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pertschy B, Schneider C, Gnädig M, Schäfer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284:35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lin ML, Fukukawa C, Park JH, Naito K, Kijima K, Shimo A, Ajiro M, Nishidate T, Nakamura Y, Katagiri T. Involvement of G-patch domain containing 2 overexpression in breast carcinogenesis. Cancer Sci. 2009;100:1443–1450. doi: 10.1111/j.1349-7006.2009.01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aravind L, Koonin EV. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem Sci. 1999;24:342–344. doi: 10.1016/s0968-0004(99)01437-1. [DOI] [PubMed] [Google Scholar]
- 132.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 133.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 136.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 138.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 139.Gilbert RJ, Gordiyenko Y, von der Haar T, Sonnen AF, Hofmann G, Nardelli M, Stuart DI, McCarthy JE. Reconfiguration of yeast 40S ribosomal subunit domains by the translation initiation multifactor complex. Proc Natl Acad Sci U S A. 2007;104:5788–5793. doi: 10.1073/pnas.0606880104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wild K, Halic M, Sinning I, Beckmann R. SRP meets the ribosome. Nat Struct Mol Biol. 2004;11:1049–1053. doi: 10.1038/nsmb853. [DOI] [PubMed] [Google Scholar]
- 141.Nilsson J, Sengupta J, Gursky R, Nissen P, Frank J. Comparison of fungal 80 S ribosomes by cryo-EM reveals diversity in structure and conformation of rRNA expansion segments. J Mol Biol. 2007;369:429–438. doi: 10.1016/j.jmb.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schafer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi U, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 143.Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Bottcher B, Hurt E. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 2009;138:911–922. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 144.Miller OL, Jr, Beatty BR. Visualization of nucleolar genes. Science. 1969;164:955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- 145.Saffer LD, Miller OL., Jr Electron microscopic study of Saccharomyces cerevisiae rDNA chromatin replication. Mol Cell Biol. 1986;6:1148–1157. doi: 10.1128/mcb.6.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mougey EB, O’Reilly M, Osheim Y, Miller OL, Jr, Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- 147.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 148.Segerstolpe A, Lundkvist P, Osheim YN, Beyer AL, Wieslander L. Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes. Nucleic Acids Res. 2008;36:4364–4380. doi: 10.1093/nar/gkn384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lim YH, Charette JM, Baserga SJ. Assembling a protein-protein interaction map of the SSU processome from existing datasets. doi: 10.1371/journal.pone.0017701. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt EC. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lischwe MA, Ochs RL, Reddy R, Cook RG, Yeoman LC, Tan EM, Reichlin M, Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG,NG-dimethylarginine. J Biol Chem. 1985;260:14304–14310. [PubMed] [Google Scholar]
- 152.Lafontaine DL, Tollervey D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol Cell Biol. 2000;20:2650–2659. doi: 10.1128/mcb.20.8.2650-2659.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wu P, Brockenbrough JS, Metcalfe AC, Chen S, Aris JP. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 155.Venema J, Vos HR, Faber AW, van Venrooij WJ, Raue HA. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA. 2000;6:1660–1671. doi: 10.1017/s1355838200001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lukowiak AA, Granneman S, Mattox SA, Speckmann WA, Jones K, Pluk H, Venrooij WJ, Terns RM, Terns MP. Interaction of the U3-55k protein with U3 snoRNA is mediated by the box B/C motif of U3 and the WD repeats of U3-55k. Nucleic Acids Res. 2000;28:3462–3471. doi: 10.1093/nar/28.18.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dunbar DA, Wormsley S, Agentis TM, Baserga SJ. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Strub BR, Eswara MB, Pierce JB, Mangroo D. Utp8p is a nucleolar tRNA-binding protein that forms a complex with components of the nuclear tRNA export machinery in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:3845–3859. doi: 10.1091/mbc.E06-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shimizu K, Kawasaki Y, Hiraga S, Tawaramoto M, Nakashima N, Sugino A. The fifth essential DNA polymerase phi in Saccharomyces cerevisiae is localized to the nucleolus and plays an important role in synthesis of rRNA. Proc Natl Acad Sci U S A. 2002;99:9133–9138. doi: 10.1073/pnas.142277999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Samanta MP, Liang S. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc Natl Acad Sci U S A. 2003;100:12579–12583. doi: 10.1073/pnas.2132527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Baudin-Baillieu A, Tollervey D, Cullin C, Lacroute F. Functional analysis of Rrp7p, an essential yeast protein involved in pre-rRNA processing and ribosome assembly. Mol Cell Biol. 1997;17:5023–5032. doi: 10.1128/mcb.17.9.5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- 163.Milkereit P, Kuhn H, Gas N, Tschochner H. The pre-ribosomal network. Nucleic Acids Res. 2003;31:799–804. doi: 10.1093/nar/gkg165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jwa M, Kim JH, Chan CS. Regulation of Sli15/INCENP, kinetochore, and Cdc14 phosphatase functions by the ribosome biogenesis protein Utp7. J Cell Biol. 2008;182:1099–1111. doi: 10.1083/jcb.200802085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Milkereit P, Strauss D, Bassler J, Gadal O, Kuhn H, Schutz S, Gas N, Lechner J, Hurt E, Tschochner H. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J Biol Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- 166.Rempola B, Karkusiewicz I, Piekarska I, Rytka J. Fcf1p and Fcf2p are novel nucleolar Saccharomyces cerevisiae proteins involved in pre-rRNA processing. Biochem Biophys Res Commun. 2006;346:546–554. doi: 10.1016/j.bbrc.2006.05.140. [DOI] [PubMed] [Google Scholar]
- 167.Goldfeder MB, Oliveira CC. Utp25p, a nucleolar Saccharomyces cerevisiae protein, interacts with U3 snoRNP subunits and affects processing of the 35S pre-rRNA. FEBS J. 2010;277:2838–2852. doi: 10.1111/j.1742-4658.2010.07701.x. [DOI] [PubMed] [Google Scholar]
- 168.Charette JM, Baserga SJ. The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA. 2010 doi: 10.1261/rna.2359810. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Colley A, Beggs JD, Tollervey D, Lafontaine DL. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol Cell Biol. 2000;20:7238–7246. doi: 10.1128/mcb.20.19.7238-7246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wurm JP, Meyer B, Bahr U, Held M, Frolow O, Kotter P, Engels JW, Heckel A, Karas M, Entian KD, et al. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 2010;38:2387–2398. doi: 10.1093/nar/gkp1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Billy E, Wegierski T, Nasr F, Filipowicz W. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 2000;19:2115–2126. doi: 10.1093/emboj/19.9.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 174.Jansen R, Tollervey D, Hurt EC. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liang WQ, Clark JA, Fournier MJ. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol Cell Biol. 1997;17:4124–4132. doi: 10.1128/mcb.17.7.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen W, Bucaria J, Band DA, Sutton A, Sternglanz R. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 2003;31:690–699. doi: 10.1093/nar/gkg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Peng WT, Krogan NJ, Richards DP, Greenblatt JF, Hughes TR. ESF1 is required for 18S rRNA synthesis in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:1993–1999. doi: 10.1093/nar/gkh518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hoang T, Peng WT, Vanrobays E, Krogan N, Hiley S, Beyer AL, Osheim YN, Greenblatt J, Hughes TR, Lafontaine DL. Esf2p, a U3-associated factor required for small-subunit processome assembly and compaction. Mol Cell Biol. 2005;25:5523–5534. doi: 10.1128/MCB.25.13.5523-5534.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Guglielmi B, Werner M. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J Biol Chem. 2002;277:35712–35719. doi: 10.1074/jbc.M205526200. [DOI] [PubMed] [Google Scholar]
- 181.Emery B, de la Cruz J, Rocak S, Deloche O, Linder P. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol Microbiol. 2004;52:141–158. doi: 10.1111/j.1365-2958.2003.03973.x. [DOI] [PubMed] [Google Scholar]
- 182.Wiederkehr T, Prétôt RF, Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jin SB, Zhao J, Bjork P, Schmekel K, Ljungdahl PO, Wieslander L. Mrd1p is required for processing of pre-rRNA and for maintenance of steady-state levels of 40 S ribosomal subunits in yeast. J Biol Chem. 2002;277:18431–18439. doi: 10.1074/jbc.M112395200. [DOI] [PubMed] [Google Scholar]
- 184.Thomson E, Rappsilber J, Tollervey D. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA. 2007;13:2165–2174. doi: 10.1261/rna.747607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kondo K, Inouye M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem. 1992;267:16252–16258. [PubMed] [Google Scholar]
- 186.Lee WC, Zabetakis D, Mélèse T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Combs DJ, Nagel RJ, Ares M, Jr, Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bax R, Raué HA, Vos JC. Slx9p facilitates efficient ITS1 processing of pre-rRNA in Saccharomyces cerevisiae. RNA. 2006;12:2005–2013. doi: 10.1261/rna.159406. [DOI] [PMC free article] [PubMed] [Google Scholar]