Abstract
Adaptive radiation is the rapid origination of multiple species from a single ancestor as the result of concurrent adaptation to disparate environments. This fundamental evolutionary process is considered to be responsible for the genesis of a great portion of the diversity of life. Bacteria have evolved enormous biological diversity by exploiting an exceptional range of environments, yet diversification of bacteria via adaptive radiation has been documented in a few cases only and the underlying molecular mechanisms are largely unknown. Here we show a compelling example of adaptive radiation in pathogenic bacteria and reveal their genetic basis. Our evolutionary genomic analyses of the α-proteobacterial genus Bartonella uncover two parallel adaptive radiations within these host-restricted mammalian pathogens. We identify a horizontally-acquired protein secretion system, which has evolved to target specific bacterial effector proteins into host cells as the evolutionary key innovation triggering these parallel adaptive radiations. We show that the functional versatility and adaptive potential of the VirB type IV secretion system (T4SS), and thereby translocated Bartonella effector proteins (Beps), evolved in parallel in the two lineages prior to their radiations. Independent chromosomal fixation of the virB operon and consecutive rounds of lineage-specific bep gene duplications followed by their functional diversification characterize these parallel evolutionary trajectories. Whereas most Beps maintained their ancestral domain constitution, strikingly, a novel type of effector protein emerged convergently in both lineages. This resulted in similar arrays of host cell-targeted effector proteins in the two lineages of Bartonella as the basis of their independent radiation. The parallel molecular evolution of the VirB/Bep system displays a striking example of a key innovation involved in independent adaptive processes and the emergence of bacterial pathogens. Furthermore, our study highlights the remarkable evolvability of T4SSs and their effector proteins, explaining their broad application in bacterial interactions with the environment.
Author Summary
Adaptive radiation is the rapid origination of an array of species by the divergent colonization of disparate ecological niches. In the case of pathogenic bacteria, radiations can lead to the emergence of novel human pathogens. Being divergently adapted to a range of different mammalian hosts, including humans as reservoir or incidental hosts, the genus Bartonella represents a suitable model to study genomic mechanisms underpinning divergent adaptation of pathogens. Here we show that two distinct lineages of Bartonella have radiated in parallel, resulting in two arrays of evolutionary distinct species adapted to overlapping sets of mammalian hosts. Such parallelisms display excellent models to reveal insights into the genetic mechanisms underlying these independent evolutionary processes. Our genome-wide analysis identifies a striking evolutionary parallelism in a horizontally-acquired protein secretion system in the two lineages. The parallel evolutionary trajectory of this system in the two lineages is characterized by the convergent origination of a wide array of adaptive functions dedicated to the cellular interaction within the mammalian hosts. The parallel evolution of the two radiating lineages on the ecological as well as on the molecular level suggests that the horizontal acquisition and the functional diversification of the secretion system display an evolutionary key innovation underlying adaptive evolution.
Introduction
Adaptation to different ecological niches can lead to rapid diversification of a single ancestor into an array of distinct species or ecotypes. This process, called adaptive radiation, typically occurs after the arrival of a founding population in a novel environment with unoccupied ecological niches (‘ecological opportunity’) and/or by the acquisition of a novel trait (‘evolutionary key innovation’) allowing the exploitation of so far unapproachable niches [1]. Spectacular examples of adaptive radiation come from different metazoan lineages with the cichlid fishes of the East African Great Lakes and the Darwin finches on Galapagos Islands representing the most prominent examples [2], [3]. Although known from a few cases only, bacterial lineages also underwent adaptive radiation - as documented in natural settings as well as in evolution experiments [4]–[6]. It remains a fundamental problem to biology to understand why and how certain lineages diversified; adaptive radiations, and in particular the genetic and genomic basis thereof, provide an ideal set-up to address this question [3], [7].
One of the most fascinating aspects of adaptive radiation is the frequent occurrence of evolutionary parallelism resulting in independent adaptation to same ecological niches [8]–[10]. Such evolutionary parallelisms are excellent examples for the action of similar, yet independent, selective forces and, hence, for the key role of natural selection in evolution [11]. Furthermore, parallel adaptive radiations in a single group indicate the existence of traits conferring a high degree of adaptability allowing the group members to efficiently occupy distinct environments. Therefore, lineages that radiate in parallel are of great value to study the molecular basis of adaptation and their independent evolutionary trajectories [1], [9]. This is of particular interest in case of host-adapted bacteria differentiating into divergent ecological niches and potentially resulting in the emergence of new pathogens. Species of the α-proteobacterial genus Bartonella are specifically adapted to distinct mammalian reservoir hosts where they cause intra-erythrocytic infections [12]. Different animal models revealed that Bartonella upon reservoir host infection colonizes a primary cellular niche from where the bacteria get seeded into the bloodstream adhering to and invading erythrocytes [13]–[15]. In most cases, infections of the reservoir host do not lead to disease symptoms suggesting a highly specific adaptation to the corresponding host niche. The transmission between host individuals is mediated by blood sucking arthropods.
An integrative genome-wide analysis showed that most factors essential for Bartonella to colonize their mammalian reservoir hosts are found within the core genome of this genus [16]. This is not surprising as it reflects the common strategy used by divergently adapted species to colonize their hosts. However, this study also revealed that two type IV secretion systems (T4SS), Trw and VirB, which are essential for host interaction at different stages of the infection cycle represent the few colonization factors exclusively found in the most species rich sub-lineages of bartonellae. It was assumed that the horizontal acquisition of these T4SS substantially refined the infection strategy of Bartonella facilitating concurrent adaptation to a wide range of different hosts [16]. The VirB T4SS translocates a cocktail of evolutionarily related effector proteins into host cells of the primary infection niche where they modulate various cellular processes [17]–[21]. The Trw T4SS is involved in the erythrocyte invasion by binding to the erythrocytic surface with its manifold variants of pilus subunits [22]–[24].
Here, we study the evolutionary relationship of Bartonella species adapted to distinct reservoir hosts and investigate the genetic mechanisms underlying adaptive radiation in different lineages. We uncover two parallel adaptive radiations in the genus Bartonella. Our genome-wide analysis revealed a remarkable evolutionary parallelism in the horizontally acquired VirB T4SS in the two radiating lineages. This parallelism is characterized at the molecular level by the lineage-specific chromosomal integration of the virB loci and the independent origination of versatile sets of effector proteins for the interaction with host cells. Providing an arsenal of host-subverting functions that can be efficiently modulated, the VirB T4SS thus seems to represent an evolutionary key innovation triggering the independent radiations of the two lineages. Our study provides detailed insights into the molecular mechanisms underlying parallel adaptive radiations in a bacterial pathogen. Furthermore, many of the diversified T4SS effector proteins carry a FIC domain recently shown to mediate ‘AMPylation’, a lately recognized post-translational modification [25], [26]. FIC domains are highly conserved in evolution and the diversified variants of the Bartonella effector proteins may display a suitable model to study their activity spectrum in the future.
Results/Discussion
Genome sequencing of ecologically distinct Bartonella strains
To study the adaptive evolution of Bartonella on a genomic level, we aimed for a set of genome sequences from species adapted to distinct mammalian reservoir hosts. To this end, we included in our analysis the published genome sequences of Bartonella bacilliformis (Bb), Bartonella grahamii (Bg), Bartonella henselae (Bh), Bartonella quintana (Bq), and Bartonella tribocorum (Bt) [16], [27], [28]. These five species are adapted to human (Bb and Bq), cat (Bh), mouse (Bg), and rat (Bt). Further, we sequenced the complete genome of Bartonella clarridgeiae (Bc) and generated draft sequences of Bartonella schoenbuchensis (Bs), Bartonella rochalimae (Br), Bartonella sp. AR 15-3 (BAR15), and Bartonella sp. 1-1C (B1-1C). Bs was selected as representative of a solely ruminant-infecting clade [16]. Bc, Br, BAR15, and B1-1C were previously shown to be closely related [29]–[31]. However, they were isolated from different mammalian reservoir hosts and therefore display a suitable set of species to study adaptive processes on the genomic level. BAR15 and B1-1C were recently isolated from American red squirrel and rat, respectively [30], [31], whereas Br was predominantly recovered from canidae like dogs or foxes, and Bc from cats [32]–[34].
Genome sequencing by 454-pyrosequencing resulted in an average sequence coverage of >35x. The single chromosome of the completely assembled genome of Bc was found to be 1,522,743 bp in size and thus belongs to the smaller genomes of Bartonella (Table 1). The draft genomes of Br, BAR15, B1-1C, and Bs consist of 13 to 19 contigs with total genome sizes similar to the one of Bc. On average, 99% of all 454-sequencing reads were assembled into the analyzed 13 to 19 contigs indicating that our draft genomes did not miss essential sequence data for subsequent analysis. Genomic features of the strains used in this study are summarized in Table 1.
Table 1. Genomic features of analyzed Bartonella species and information about their reservoir host and phylogenetic lineage (see also Figure 1).
| Genome features | |||||||||
| Lineage | Host | Size bp | Status | contigs | GC % | CDS | rRNA | tRNA | |
| B. bacilliformisa | 1 | Human | 1,445,021 | complete | 1 | 38.2 | 1,283 | 2×3 | 44 |
| B. schoenbuchensisb | 2 | Deer | 1,667,713 | draft | 18 | 37.6 | 1,584 | 2×3 | 42 |
| B. clarridgeiaeb | 3 | Cat | 1,522,743 | complete | 1 | 35.7 | 1,342 | 2×3 | 41 |
| B. rochalimaeb | 3 | Dog/fox | 1,540,290 | draft | 13 | 35.6 | 1,405 | 3×3 | 44 |
| B. sp. AR 15-3b | 3 | Squirrel | 1,590,248 | draft | 17 | 35.7 | 1,487 | 2×3 | 41 |
| B. sp. 1-1Cb | 3 | Rat | 1,566,739 | draft | 19 | 35.8 | 1,465 | 2×3 | 41 |
| B. grahamiic | 4 | Mouse | 2,341,328 | complete | 1 | 38.1 | 1,737 (31)d | 2×3 | 42 |
| B. henselaea | 4 | Cat | 1,931,047 | complete | 1 | 38.2 | 1,488 | 2×3 | 44 |
| B. quintanaa | 4 | Human | 1,581,384 | complete | 1 | 38.8 | 1,142 | 2×3 | 44 |
| B. tribocoruma | 4 | Rat | 2,619,061 | complete | 1 | 38.8 | 2,136 (18)d | 2×3 | 42 |
A genome-wide phylogeny reveals parallel adaptive radiations in two sister clades of Bartonella
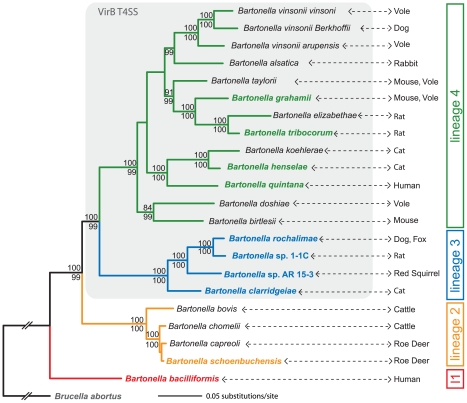
We inferred a robust species tree of the genus Bartonella based on 478 core genome genes of the ten available Bartonella genomes sequenced (Figure S1). To exclude that recombination or horizontal gene transfer within this set of core genome genes was affecting our phylogenetic analysis, we reconstructed single gene trees of the entire data set and performed a recombination analysis using the GARD algorithm [35]. 471 of the 478 genes revealed the same overall topology as our genome-wide phylogeny with the two monophyletic clades of lineage 3 and lineage 4 (see Figure 1). Further, the GARD analysis detected significant recombination breakpoints with a p-value<0.01 in only two out of the 478 core genome genes. Together these analyses show that our genomic data set is suitable for inferring a consistent species tree.
Figure 1. Phylogeny of Bartonella based on a genome-wide dataset.
Maximum likelihood analysis using an alignment of 478 genes (515,751 nt) of ten sequenced Bartonella species (indicated by bold and color type) and Brucella abortus. Based on sequence data from rpoB, gltA, ribC, and groEL genes, additional Bartonella species were included in the analysis. Numbers above the branches represent maximum likelihood bootstraps (100 replicates); numbers below represent Bayesian posterior probabilities. Values ≥80% are shown. Lineages harboring the VirB T4SS [16] are shaded in gray; the primary mammalian hosts are indicated for each species. Phylogenetic trees inferred from either the genomic data set excluding non-sequenced bartonellae or the sequences from only rpoB, gltA, ribC, and groEL genes revealed the same four lineages for the Bartonella ingroup (Figure S1). Estimates of average evolutionary divergence over sequence pairs within lineages and between lineages are presented in Table S1. l1, lineage 1.
Based on available sequence information for the housekeeping genes rpoB, gltA, ribC, and groEL, we included most other Bartonella species in the analysis resulting in a so-called supertree phylogeny (Figure 1) [36]. Just as the analysis based on the 478 core genome genes of the ten sequences species alone, this supertree revealed four major clades in the monophyletic bartonellae: ancestral lineage 1 represented by the highly virulent human pathogen Bb [37]; lineage 2 comprising of Bs and three other ruminant-infecting species; lineage 3 consisting of the closely related Bc, Br, BAR15, and B1-1C; and the most species-rich lineage 4 with 13 species including Bg, Bh, Bq, and Bt (Figure 1). A phylogeny based on only the four housekeeping genes resulted in the same clustering of these taxa into the four different Bartonella lineages (Figure S1).
In contrast to the ancestral lineage 1, lineages 2, 3, and 4 are ramifying to different degrees comprising species isolated from various hosts. While the species of lineage 2 are limited to infect ruminants and have overlapping host range [38], the diversification of lineage 3 and 4 seems to result from the specific adaptation to distinct mammalian hosts [12]. To substantiate the ecological divergence within these two lineages, we analyzed the genotype-host correlation of Bartonella isolates sampled from diverse mammals. Based on gltA sequences, this analysis revealed clustering of strains isolated from same or similar hosts in lineage 3 and 4 (Figure S2). Further support for the host specific adaptation of different Bartonella species comes from recently published laboratory infections [39], [40] and from our own rat infection experiments with the strains of lineage 3 (Figure S3). It is to mention that some Bartonella species can incidentally be transmitted to other hosts like humans [12]. These so-called zoonotic Bartonella species do not cause intraerythrocytic bacteremia in the accidental human host reflecting the lack of specific adaptation. However, such accidental transmissions might facilitate the emergence of new specificity resulting in host switches and the origination of new species. In particular, several species of lineage 3 and 4 are known to display such zoonotic pathogens, whereas for lineage 1 or 2 to our knowledge no such case has been reported so far [12], [29].
In summary, our genome-wide phylogenetic analysis shows that the sister lineages 3 and 4 have evolved by adaptive radiations into same or similar ecological niches (i.e. hosts). Long internal branches separating the two lineages from each other and preceding the radiations are evidence for their independent occurrence (Figure 1). Due to the lack of calibration time points, the exact timing of these independent radiations cannot be deduced. However, the phylogenetic tree in Figure 1 might suggest that lineage 3 diversified more recently compared to lineage 4. This is supported by the mean p-distances inferred for the sequenced taxa of these two lineages: lineage 3 = 0.07±0.0002, lineage 4 = 0.12±0.0003 (see also Table S1). Two alternative explanations for the observed differences in lineage diversification could be (i) a sampling bias, i.e. the full diversity of lineage 3 was not captured or (ii) smaller population sizes for species of lineage 4 over lineage 3 leading to faster evolution at purifying sites. Significant differences in population size might be rather unlikely as Bartonella species are thought to share a common life style in their respective reservoir host. Whether sampling of Bartonella species in animal populations was exhaustive enough is difficult to assess. However, a newly discovered species would only change the coalescent point of a lineage if it would hold a more ancestral position than the already known species of this lineage. In contrast to these alternative hypothesis, epidemiological studies rather seem to support the scenario of a more ancient onset of the radiation in lineage 4: (i) lineage 4 comprises a much wider range of divergently adapted species and (ii) they represent the most frequently found Bartonella species in natural host populations [30], [41]. In contrast, except for Bc, taxa of lineage 3 were only recently detected and so far only sampled at low prevalence [29]–[32].
The VirB T4SS displays an adaptive trait specific to the radiating lineages of Bartonella
The evolutionary parallelism of the radiating Bartonella lineages provides an ideal setting to study independent evolutionary processes linked to the adaptation to divergent niches. An important driving force for these adaptive radiations might be the presence of ecological opportunities. The niche of Bartonella in the mammalian reservoir host, the bloodstream, displays a privileged environment in which other resource-competing microbes are typically absent. Thus, the adoption of the characteristic intra-erythrocytic infection strategy together with the vector-borne transmission route might have enabled adaptive radiations of Bartonella by the specialization to different hosts. However, not all Bartonella lineages appear to have diversified to the same extent (see Figure 1) suggesting that the availability of such an ecological opportunity alone is not sufficient to explain the pronounced radiation of lineages 3 and 4. Supposedly, key innovations, i.e. lineage-specific traits underlying the adaptation to the mammalian host niches are responsible for the adaptive radiations. Potential adaptive traits would have to be involved in species-environment interactions, such as molecular factors responsible for causing bacteremia. Further, in analogy to the modulation of the adaptive traits in metazoan radiations [2], [3], any molecular factor used to exploit distinct environments in a specific manner should be divergent among niche-specialized species [1]. Molecular evolutionary analyses provide the means to identify divergent adaptive traits as the genes encoding them are expected to show signs of adaptive evolution, i.e. an excess of non-synonymous (dn) over synonymous (ds) substitutions as the result of positive selection. We performed a genome-wide natural selection analysis in the two radiating lineages to detect genes (and therefore traits) with divergent evolution. To this end, we analyzed all orthologous genes from the available genomes of the radiating lineage 3 (Bc, Br, BAR15, and B1-1C) and lineage 4 (Bh, Bg, Bq, and Bt) for signs of adaptive sequence evolution by inferring the natural selection of orthologs by estimation of ω, the ratio of non-synonymous (dn, amino acid change) to synonymous (ds, amino acid conservation) substitution rates (ω = dn/ds). Generally, ω<1, ω = 1, ω>1 represent purifying, neutral, and positive selection (adaptive evolution), respectively [42].
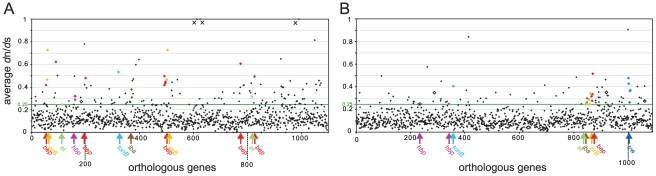
We first calculated “gene-wide” dn/ds for all orthologous genes of the two lineages (lineage 3: 1,097 genes, lineage 4: 1,091 genes). We excluded one gene from this analysis for each of the two lineages, because the GARD analysis detected statistically significant recombination breakpoints. As adaptive evolution is typically affecting only a few sites of a gene rather than the entire gene sequence [42], we first looked for genes exhibiting an elevated value of ω≥0.25 over the entire gene length. This analysis revealed 133 (12%) and 86 (8%) genes in lineage 3 and lineage 4, respectively, under relaxed purifying selection indicating signs of adaptive evolution (Figure 2). To have an additional measurement for adaptive evolution, we subjected our genomic data sets to a maximum likelihood analysis for the detection of site-specific positive selection. To this end, we used the CodeML module implemented in the PAML package. CodeML compares the likelihoods of different evolutionary models for each analyzed gene alignment. When comparing model M2a (PositiveSelection) vs. model M1a (NearlyNeutral), we detected 62 or 34 genes for lineage 3 and 26 or 14 genes for lineage 4 harboring sites under positive selection with a p-value of <0.05 or <0.01, respectively. A large fraction of these genes (29 for lineage 3 and 12 for lineage 4) exhibited also gene-wide dn/ds values ≥0.25 indicating them as good candidates for encoding adaptive traits (Table 2). Comprehensive lists of the genes identified to have dn/ds values ≥0.25 and/or exhibiting site-specific positive selection in the CodeML analysis are provided in Table S2 and Table S3 for lineage 3 and lineage 4, respectively. As non-synonymous mutations accumulate over time, the higher number of genes identified for lineage 3 could be a further indication for its more recent radiation, or alternatively, the effect of larger population sizes compared to lineage 4. Irrespectively, these findings render the dataset derived from lineage 3 to be more sensitive to the detection of adaptive sequence evolution (Figure 2, Table 2).
Figure 2. Gene-wide dn/ds analysis of the core genomes of the two radiating lineages.
dn/ds analysis for lineage 3 (A) and for lineage 4 (B). The mean values plotted on the y-axis were obtained from the dn/ds values of pairs of orthologs. Genes are ordered according to their position in the genome of Bc (A) and Bq (B). Genes with elevated dn/ds values homologous to known colonization factors of Bartonella (based on previously published results [16], [43]–[45]), are depicted by colors. at, putative autotransporter, bep, Bartonella effector protein, hbp, hemin-binding protein, iba, inducible Bartonella autotransporter, virB, VirB T4SS protein, trw, Trw T4SS protein. Genes encoding hypothetical proteins known to be important for colonization are depicted by open black squares (one in lineage 3, four in lineage 4). In lineage 3, three genes exhibit average dn/ds values ≫1 (marked by crosses). This is explained by the fact that between Br and B1-1C, only non-synonymous mutations but no synonymous mutations have been detected in these three genes. As all three genes revealed ω<1 in the other pair-wise comparisons, they were treated as outliers and excluded from further analysis.
Table 2. Number of genes revealing significant positive selection in the CodeML analysis for lineage 3 and lineage 4.
| CodeML results a | |||
| Lineage 3 | Total | M2a vs. M1a (p<0.05) | M2a vs. M1a (p<0.01) |
| All core genes | 1096 | 62 | 34 |
| Genes with dn/ds ≥0.25 | 133 | 29 | 20 |
| Lineage 4 | |||
| All core genes | 1090 | 26 | 14 |
| Genes with dn/ds ≥0.25 | 86 | 12 | 8 |
All genes for which model M2a (PositiveSelection) fitted the data significantly better than model M1a (NearlyNeutral) for two different p-values.
Interestingly, there was a marked number of genes with ω≥0.25 over the entire gene length that overlapped in both lineages (Table 3). Among those were genes encoding autotransporters, hemin-binding proteins, and different components of the VirB T4SS. They all constitute important host colonization factors [43]–[45] and thus are likely to display adaptive traits of Bartonella. For many of these genes, also our analysis of site-specific natural selection detected positive selection (Table 3). Most remarkably, all analyzed Bartonella effector protein (bep) genes of the VirB T4SS were among the genes with ω≥0.25. Particularly in lineage 3, they showed strong signs of adaptive evolution by exhibiting ω>0.4 over the entire gene length. Further, in eight out of nine analyzed bep genes of lineage 3, we detected site-specific positive selection (Table 3). Being exclusively found in the radiating lineages and showing strong signs of adaptive evolution, the VirB system and its effector proteins, thus, fulfill the criteria of an evolutionary key innovation likely contributing to the parallel adaptive radiations of Bartonella. Autotransporters and hemin-binding proteins could represent further adaptive traits important for radiations in Bartonella. They exhibited strong positive selection in our analysis and are known to be important factors for host colonization. Further, their conservation throughout the genus Bartonella indicates an important role for the life style and infection strategy of this pathogen (Table S2, Table S3). However, factors conserved in radiating and non-radiating lineages appear unlikely to represent specific key innovations, unless other factors, such as the absence of ecological opportunities or ecological separation, prevented certain lineages to radiate and to colonize more divergent niches.
Table 3. Common genes of lineage 3 and lineage 4 with average dn/ds values (ω) ≥0.25 and results of the CodeML analysis.
| Lineage 3 | Lineage 4 | |||||||||||
| CodeML resultsa | CodeML resultsa | |||||||||||
| M2a vs. M1a | M2a vs. M1a | |||||||||||
| Gene | aa | ω | LRT | %>1 | dn/ds | Protein product | Geneb | aa | ω | LRT | %>1 | dn/ds |
| 0075 | 103 | 0.47 | 0.43 | VirB T4SS proteins | 10580 | 100 | 0.34 | 1.13 | 4.7 | 3.3 | ||
| 0645 | 103 | 0.47 | 0.43 | (VirB7)b | ||||||||
| 0077 | 150 | 0.73 | 11.02** | 25.0 | 3.4 | VirB T4SS proteins | 10560 | 146 | 0.25 | 1.08 | 1.8 | 7.3 |
| 0647 | 150 | 0.73 | 11.02** | 25.0 | 3.4 | (VirB5)b | ||||||
| 0118 | 223 | 0.62 | 13.84** | 12.2 | 4.8 | VirB T4SS effector proteins | 10630 | 272 | 0.32 | 0 | ||
| 0069 | 547 | 0.42 | 2.46 | (Beps)b | 10650 | 532 | 0.28 | 0 | ||||
| 0245 | 497 | 0.48 | 10.19** | 9.5 | 3.0 | 10660 | 414 | 0.33 | 0 | |||
| 0631 | 538 | 0.42 | 15.09** | 6.7 | 4.5 | 10680 | 555 | 0.58 | 7.26* | 10.6 | 2.8 | |
| 0632 | 528 | 0.50 | 6.51* | 3.2 | 4.6 | |||||||
| 0639 | 625 | 0.43 | 19.73** | 3.1 | 8.8 | |||||||
| 0640 | 539 | 0.45 | 18.16** | 4.0 | 6.8 | |||||||
| 0960 | 430 | 0.61 | 11.42** | 7.1 | 4.5 | |||||||
| 1031 | 547 | 0.49 | 16.29** | 8.1 | 4.5 | |||||||
| 0196 | 277 | 0.32 | 8.84* | 7.4 | 4.4 | Hemin binding proteinsb | 02420 | 267 | 0.25 | 19.27** | 11.1 | 39.4 |
| 0197 | 241 | 0.29 | 35.92** | 9.8 | 8.3 | 04010 | 270 | 0.25 | 17.49** | 7.5 | 6.6 | |
| 0198 | 291 | 0.32 | 9.37** | 15.1 | 2.8 | |||||||
| 0473 | 673 | 0.38 | 108.28** | 8.1 | 25.9 | Inducible autotransportersb | 10380 | 759 | 0.25 | 0 | ||
| 0475 | 823 | 0.31 | 89.02** | 3.1 | 95.7 | 10410 | 792 | 0.3 | 0 | |||
| 1020 | 837 | 0.32 | 0.00 | putative autotransportersb | 10280 | 799 | 0.25 | 0 | 9.6 | 1.0 | ||
| 0145 | 608 | 0.25 | 3.24 | 10290 | 1753 | 0.26 | 202.47** | 16.5 | 48.2 | |||
| 0416 | 244 | 0.53 | 0.37 | TonB protein | 04170 | 248 | 0.41 | 10.25** | 4.5 | 7.0 | ||
| 1236 | 377 | 0.28 | 0.20 | TolA protein | 11830 | 381 | 0.25 | 0 | ||||
| 0171 | 160 | 0.28 | 2.03 | Alanine acetyltransferase | 02150 | 160 | 0.29 | 0 | ||||
| 1171 | 328 | 0.25 | 20.56** | 4.1 | 8.3 | putative phage protein | 11060 | 260 | 0.26 | 0 | ||
| 0847 | 293 | 0.30 | 0.00 | Lysophospholipase l2 | 05220 | 300 | 0.28 | 0 | ||||
| 0125 | 50 | 0.50 | 0.99 | Heme exporter protein CcmD | 01050 | 83 | 0.50 | 0 | ||||
| 0115 | 157 | 0.35 | 2.27 | membrane protein FxsA | 00960 | 163 | 0.26 | 1.63 | ||||
| 1107 | 391 | 0.27 | 0.62 | Filament-A percursor | 01520 | 424 | 0.28 | 0 | ||||
| 1274 | 194 | 0.30 | 2.17 | ATP synthase delta subunit | 12260 | 194 | 0.30 | 0.13 | ||||
| 1270 | 138 | 0.38 | 1.00 | ATP synthase epsilon chain | 12220 | 138 | 0.26 | 0 | ||||
| 1329 | 143 | 0.37 | −9.41 | Antioxidant, AhpC/TSA family | 13100 | 226 | 0.30 | 0.13 | ||||
| 1038 | 144 | 0.34 | 2.25 | twin arginine translocase TatB | 04750 | 134 | 0.84 | 4.26 | ||||
| 0226 | 473 | 0.27 | 0.00 | conserved, unknown function | 02810 | 447 | 0.25 | 0.16 | ||||
| 0279 | 218 | 0.41 | 0.00 | conserved, unknown function | 09660 | 228 | 0.49 | 1.72 | ||||
| 0327 | 84 | 0.30 | 0.20 | conserved, unknown function | 03350 | 85 | 0.27 | 3.11 | ||||
| 0670 | 813 | 0.32 | 4.85 | conserved, unknown function | 07710 | 819 | 0.35 | 11.5** | 1.1 | 11.5 | ||
| 0949 | 181 | 0.27 | 0.00 | conserved, unknown function | 08360 | 184 | 0.28 | 5.59 | ||||
| 0978 | 40 | 0.41 | 0.92 | conserved, unknown function | 08690 | 60 | 0.32 | 5.19 | ||||
| 1013 | 158 | 0.43 | 2.71 | conserved, unknown function | 09010 | 176 | 0.35 | 2.67 | ||||
| 1059 | 141 | 0.42 | 5.04 | conserved, unknown function | 03120 | 135 | 0.57 | 2.13 | ||||
| 1163 | 97 | 0.65 | 2.11 | conserved, unknown function | 11040 | 114 | 0.31 | 0 | ||||
| 1164 | 139 | 0.41 | 3.14 | conserved, unknown function | 11050 | 148 | 0.35 | 0.17 | ||||
| 1199 | 63 | 0.33 | 0.00 | conserved, unknown function | 11430 | 62 | 0.46 | 2.47 | ||||
| 1200 | 126 | 0.33 | 1.08 | conserved, unknown function | 11450 | 129 | 0.26 | 0 | ||||
| 1261 | 95 | 0.30 | 0.00 | conserved, unknown function | 12190 | 93 | 0.31 | 0 | ||||
| 1321 | 85 | 0.82 | 1.36 | conserved, unknown function | 12930 | 110 | 0.40 | 2.55 | ||||
| 1327 | 387 | 0.42 | 5.28 | conserved, unknown function | 13000 | 389 | 0.31 | 1.57 | ||||
| 0768 | 77 | 16.69 | 0.00 | conserved exported protein | 06010 | 103 | 0.35 | 1.27 | ||||
Likelihood ratio test (LRT) statistics for model M2a versus model M1a, percentage of sites under positive selection (%>1) and estimated dn/ds value are shown, if model M2a (PositiveSelection) fitted data significantly better than model M1a (NearlyNeutral) (* p<0.05, ** p<0.01).
no clear ortholog assignment possible between the genes of lineage 3 and lineage 4. Gene indicates locus tag number of the analyzed gene of Bc (BARCL_XXXX) for lineage 3 or Bq (BQXXXX) for lineage 4. aa means analyzed gene length in amino acids. ω represents the average dn/ds value calculated over the entire gene length (see also Figure 2).
Importantly, our analysis revealed some lineage-specific colonization factors to carry signs of adaptive evolution. Among others, surface-exposed pilus-components of the Trw T4SS exclusively present in lineage 4 were found to exhibit elevated dn/ds values and sites under positive selection (Table S3). This is in agreement with previous studies and appears to reflect the adaptation of this putative adhesion factor to the erythrocytic surface of different host species [22], [24]. As the Trw T4SS is only present in lineage 4, it might have specifically contributed to the radiation of this most species-rich clade of Bartonella. Factors known to be important for colonization and exclusively present in lineage 3 were not identified by our analysis.
We cannot exclude that the selective pressure imposed by the immune-system might have contributed to the adaptive evolution detected in our genome-wide analysis. It was previously reported that the arms race between host and pathogen can drive the diversification of secretion system- and effector protein-encoding genes [46], [47]. However, in case of the Trw T4SS, recently published in vitro infections with erythrocytes isolated from different mammals demonstrated that the Trw-dependent binding and invasion of Bartonella is host-specific [22]. Although experimental data is not yet available, our data suggest that the VirB T4SS and its effector proteins evolved by similar mechanisms. Together with the previous finding that the VirB T4SSs belong to the few colonization factors specific to the radiating lineages [16], this analysis reveals these horizontally acquired host interacting systems as potential key innovations facilitating adaptation to new hosts and therefore driving the radiations of Bartonella.
Independent evolutionary history of the VirB T4SS in the two parallel adaptive radiations of Bartonella
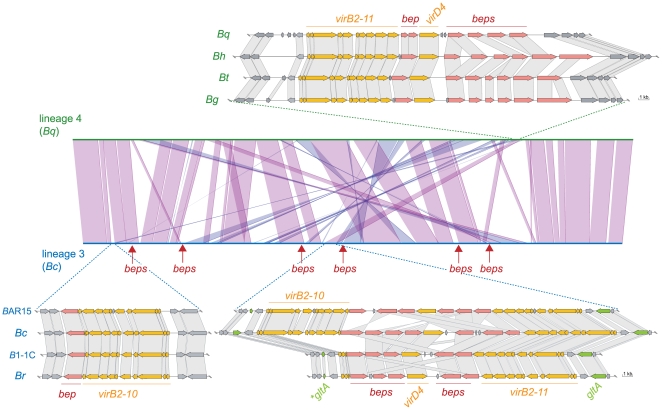
To further assess the role of the VirB T4SS for the independent adaptive radiations of Bartonella, we compared the chromosomal organization of the VirB and effector protein-encoding genes of the two lineages. Remarkably, our analysis uncovered independent evolutionary scenarios for the chromosomal incorporation of this horizontally acquired trait. In the genomes of lineage 4 (Bg, Bh, Bq, and Bt), the virB T4SS genes, virB2-virB11 and the coupling protein gene virD4, are encoded at the same chromosomal location (Figure 3, Figure S4). Also, the bep genes are encoded in this region. In contrast, the genome sequences of lineage 3 (Bc, Br, BAR15, and B1-1C) revealed marked differences in organization, copy number, and chromosomal localization of the genes encoding the VirB T4SS. In the completely assembled genome of Bc, we found three copies of the virB2-virB10 genes encoded at two different chromosomal locations (Figure 3, Figure S4). Two copies are encoded at the same locus and belong to inverted repeats of ∼10kb. They are separated by several bep genes and the gene virD4. A third copy of the virB2-virB10 cluster including an additional bep gene is encoded in another genomic region highly conserved across different Bartonella lineages (Figure 3).
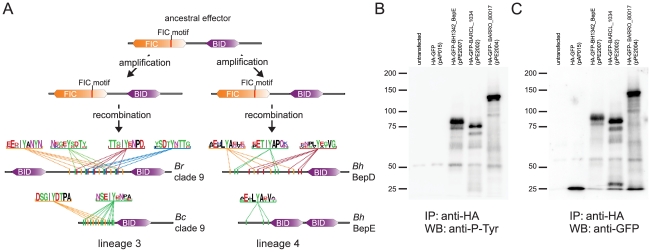
Figure 3. Genomic organization of the virB T4SS and bep gene loci in the two radiating lineages.
Synteny plot of Bc (lineage 3) and Bq (lineage 4) generated in MaGe [58] and genetic organization of the virB T4SS loci. Syntenic relationships comprising at least five genes are indicated by violet and blue lines for genes found on the same or the opposite strand, respectively. The genomic integration sites of virB loci are indicated. Additional bep loci in lineage 3 are marked by arrows. For the depicted genomic loci, orthologous genes are connected via gray boxes. For bep genes, connections are drawn if they belong to the same Bep clade (Figure 4) or if they are top blast hits of one another. The glutamine syntethase I gene (glnA) and its fragments which are flanking the two inverted virB T4SS copies are colored in green. The fragments are indicated by an asterisk.
The same chromosomal integration and amplification of the virB T4SS genes was found in the other three genomes of lineage 3. However, in a common ancestor of Br and B1-1C, one of the three copies must have been partially deleted, as only virB2, virB3, and a remnant of the virB4 gene were found in the corresponding region of these two genomes (Figure 3, Figure S4). Interestingly, the different copies of virB2-virB10 are identical to each other within one species, but divergent across different species indicating the presence of an intra-chromosomal homogenization process. The fact that duplicated components of another T4SS, Trw, also evolved in concert, and the finding of several other identical genes or gene clusters in different Bartonella genomes [24] suggests that sequence homogenization is a common mechanism in Bartonella to conserve paralogous gene copies. The inverted organization of the two virB T4SS gene clusters seems to result from a duplication event subsequent to the integration of a first copy. Evidence comes from a remnant of the glutamine synthetase I gene (glnA) flanking the entire locus at its upstream end. The full-length copy of this vertically inherited housekeeping gene is located directly downstream of the integration site (Figure 3, Figure S4).
In addition to the effector genes adjacently located to the virB genes (as in lineage 4), we found six additional loci encoding bep genes in lineage 3 (Figure S4). These effector genes are not entirely conserved throughout lineage 3, and the existence of gene remnants provides evidence of their deterioration in certain species. Altogether, we identified 12 to 16 bep genes in lineage 3, whereas only five to seven bep genes are present in lineage 4.
Incomplete synteny in the corresponding regions may hinder comparison between the two different lineages, however, no gene remnants could be found at the different integration sites across the two lineages. We cannot fully exclude that massive genomic recombination events resulted in the different chromosomal locations and the lineage-specific dissemination of the virB and bep genes. Yet, such a scenario appears unlikely, as the overall genomic backbone is largely conserved (Figure 3) and the flanking regions of the virB T4SS integration sites do not encode vertically-inherited orthologs across the two lineages. Furthermore, the absence of mobile elements adjacent to the virB T4SS genes such as recombinases, transposases, or integrases is not supportive of an intra-chromosomal mobilization of this genomic locus. T4SS are ancestrally related to conjugation machineries [48]. Thus, the virB genes might have been transferred from a conjugative plasmid into the chromosome by independent events after the divergence of lineage 3 and lineage 4. In Bg, a closely related T4SS, the Vbh, is encoded on a plasmid in addition to a chromosomally integrated copy [28]. This indicates that these horizontally acquired elements can be maintained on extra-chromosomal replicons within Bartonella from where they are integrated into the chromosome. Similarly, pathogenic Escherichia coli strains from different phylogenetic clades were shown to have evolved in parallel by the independent incorporation of virulence traits from mobile genetic elements [49].
Bartonella effector proteins of the two radiating lineages evolved by independent gene amplification and diversification processes
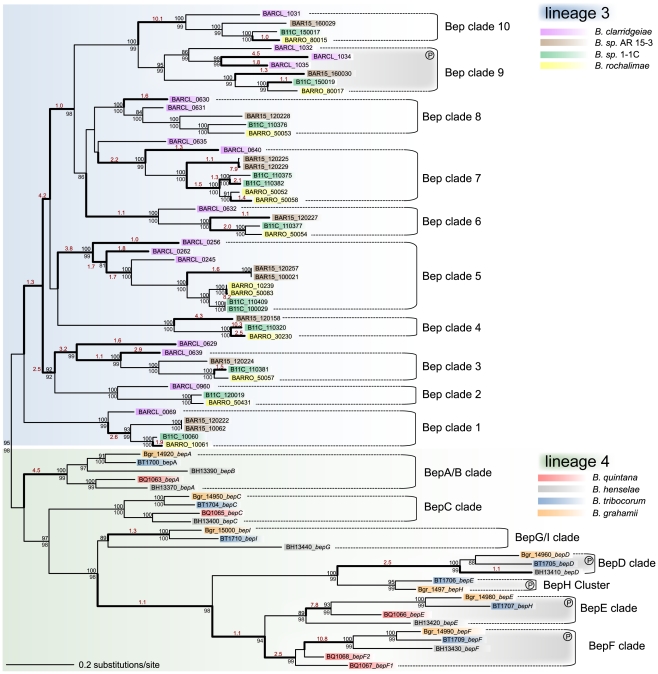
As the chromosomal organization implicates different evolutionary histories of the VirB T4SS in the two radiating lineages, we investigated the relation among the effector proteins translocated by this secretion system. It was previously shown that the bep genes have evolved from a single ancestor by duplication, diversification, and reshuffling of domains resulting in modular gene architectures [17]. The C-terminal BID (Bartonella intracellular delivery) domain is shared by all Beps as it constitutes the secretion signal for the transport via the VirB T4SS. In their N-terminal part, Beps either harbor a FIC (filamentation-induced by cAMP) domain or repeats of additional BID domains (Figure S5). We assessed the evolutionary relationship among the bep genes by inferring phylogenetic trees on the basis of either the BID or the FIC domain, or the entire gene sequence. This revealed that the bep genes of lineage 3 and lineage 4 form two separate clades (Figure 4, Figure S6). Apparently, consecutive rounds of lineage-specific duplications of an ancestral effector gene resulted in the parallel emergence of two distinct arsenals of bep genes. These duplication events preceded the adaptive radiation in both lineages as phylogenetic clusters of effector genes (Bep clades in Figure 4) comprise positional orthologs present in all or a subset of the analyzed genomes of the corresponding lineage (Figure S4). Gene duplications frequently display the primary adaptive response after the acquisition of beneficial factors, because they occur at much higher frequency than other adaptive mutations [50]. This might have been the initial selective pressure for the independent amplification processes. However, in both lineages, the duplicated bep genes subsequently diversified by accumulating mutations as indicated by different branch lengths separating bep genes in Figure 4.
Figure 4. Gene tree of Bartonella effector proteins.
The maximum likelihood tree is based on the BID domain and the C-terminal extension of all bep genes. Numbers above the branches represent maximum likelihood bootstraps (100 replicates); numbers below Bayesian posterior probabilities. Values ≥80% are shown. The independent clustering of the bep genes from lineage 3 and lineage 4 is indicated by blue and green shading, respectively. Different bep genes are named with locus_tag and gene name (if existing) and they are color-coded species-specifically according to the legend. Bold type highlights phylogenetic branches with dn/ds values >1 (the estimated dn/ds value is indicated in red color type). Sub-clades comprising orthologous bep genes are indicated (Bep clades). In lineage 3, BARCL_0629 and BARCL_0635 are not included in any clade since their phylogenetic position is neither supported by bootstraps nor posterior probabilities. In both lineages, clades of bep genes encoding tandem-repeated tyrosine-phosphorylation motifs are indicated by gray shading and an encircled ‘P’ symbol.
To analyze the sequence evolution during the parallel amplification and diversification processes, we used a branch test for positive selection. Since positive selection is not continuously acting during evolution, this analysis allows the detection of episodic adaptive evolution on single phylogenetic branches. We detected positive selection on many of the internal branches suggesting that subsequent to their duplication different Bep clades have undergone adaptive sequence evolution in both lineages (Figure 4, Figure S6). For Bartonella, experimental studies showed that effector proteins exhibit distinct phenotypic properties on host cells indicating that the evolutionary diversification of the duplicated effectors was substantially driven by the acquisition of novel functions [18]–[21]. Not all branches exhibit dn/ds values >1, though, suggesting episodic changes in the selection pressure acting on different effector gene copies. For example, functional redundancy of paralogous effector copies could have resulted in neutral drift, whereas conservation of an advantageous function might have led to purifying selection on certain branches.
The basis for the functional versatility seems to lie in the adaptability of the domains encoded by bep genes. In case of the FIC domain, recently published work showed that this domain mediates a new post-translational modification by transferring an AMP moiety onto a target protein [25]. Proteins ‘AMPylated’ by FIC domains belong to the family of GTPases. The diversity of these targets and their numerous functions in cellular processes might allow the diversified Beps to target and subvert a variety of host cell functions by target-specific ‘AMPylation’. The high degree of conservation of the FIC domain in different kingdoms of life provides further evidence for the remarkable versatility of this domain [51]. Interestingly, also the BID domain, constituting part of the translocation signal of the effector proteins, seems to be capable of adopting various functions in the host cell [17]. In case of BepA, it was shown that the BID domain is sufficient to mediate the anti-apoptotic property of this effector protein [18]. BID domains of other Beps with the same domain constitution as BepA do not exhibit this phenotype indicating specific adaptive modulation of this domain for BepA. This functional adaptability might also explain why certain effector genes carry more than one BID domain.
At last, tandem-repeated tyrosine-phosphorylation motifs found in a subset of effector proteins confer another multifaceted molecular mechanism to modulate cellular processes. Phosphorylated effector proteins are thought to recruit cellular binding partners resulting in the formation of signaling scaffolds that interfere with specific host cell signaling pathways [21]. For several effector proteins of Bh (lineage 4), tyrosine phosphorylation by host cells has been reported and the targeted host interaction partners studied [17], [21]. Beside Bartonella, a number of other pathogens, as E. coli (EPEC), Helicobacter pylori, or Chlamydia trachomatis are using tyrosine-phosphorylation of effector proteins to modulate their hosts in very distinct ways demonstrating the versatility of this type of host subversion [21]. In Bartonella, the tyrosine-phosphorylated effector proteins seem to display an important functionality of the VirB-mediated host modulation as we found effector proteins of this type in both radiating lineages (see below).
Parallel evolution of Bartonella effector proteins harboring tandem-repeated tyrosine-phosphorylation motifs
Our analyses suggest that the domain structure of the ancestral effector gene consisted of an N-terminal FIC and a C-terminal BID domain (FIC-BID). In both lineages, the FIC-BID domain structure displays the most abundant effector protein type. In lineage 3, only effector genes of Bep clade 9 consist of domain architectures different than FIC-BID (Figure S5). The gene tree in Figure 4 shows that bep genes with the shortest evolutionary distance across the two lineages are the ones harboring the FIC-BID structure (BepA clade and Bep clade 1). bep genes with different domain architecture constitute more distantly related clades across the two lineages indicating that they derived by independent recombination from the ancestral domain structure. Furthermore, the distantly related Vbh T4SS of Bg and Bs encodes an effector protein consisting of the FIC-BID domain structure [28].
As mentioned above, it was shown for lineage 4 that some of the derived bep genes become phosphorylated by host cell kinases at conserved tandem-repeated tyrosine-phosphorylation motifs leading to the interference with specific host cell pathways [21]. Strikingly, we found that bep genes with derived domain architecture in lineage 3 also harbour regions with tandem-repeated tyrosine motifs (Figure 5, Figure S5). In silico predictions of tyrosine-phosphorylation sites with three different programs [52]–[54] consistently revealed a high number of potentially phosphorylated motifs within these repetitive regions (Figure 5, Table S4). We ectopically expressed these effector proteins in HEK293T cells and showed that they are indeed phosphorylated within eukaryotic cells by tyrosine kinases implicating their functional importance for host interaction (Figure 5). Interestingly, the motifs found in lineage 3 are clearly different from the ones present in lineage 4 and are also less conserved as depicted by their consensus sequences in Figure 5. This suggests that the motifs in lineage 3 may generally be under weaker purifying selection than in lineage 4, because they target either less conserved or even different pathways in their hosts. Further, the lower degree of conservation and the higher number of motifs per effector found in lineage 3 could also indicate that these proteins and particularly their motifs are under positive selection and evolved more recently than their equivalents in lineage 4.
Figure 5. Parallel evolution of Bartonella effector proteins harboring tandem-repeated tyrosine motifs phosphorylated in host cells.
(A) The lineage-specific amplification of effector proteins harboring the ancestral domain structure was followed by secondary recombination events in both lineages. This resulted in the independent emergence of Beps with a derived domain structure (see also Figure S5). In silico predictions for tyrosine-phosphorylation motifs consistently detected high numbers of tandem-repeated phosphorylation sites in independently derived Beps of both lineages (here shown for two Beps of Bep clade 9 [BARRO_80017 of Br and BARCL_1034 of Bc] from lineage 3 and BepD and BepE of Bh from lineage 4). The depicted motifs were identified with NetPhos2.0 with a score ≥0.8 (for predictions of all motifs in lineage 3 see Table S4). The consensus sequence of tandem-repeated motifs (color-coded) is shown for each of the four Beps by WebLogos (http://weblogo.berkeley.edu/logo.cgi). (B) and (C), Immunoprecipitation/Western blot analysis of Beps with predicted tyrosine-phosphorylation motifs ectopically expressed in HEK293T cells. Immunoprecipitations were performed with HA antibody-coated agarose beads. (B) Western blot analysis was performed with anti-phosphotyrosine antibody. (C) Western blot analysis was performed with anti-GFP antibody. The tested constructs correspond to the Beps of Bep clade 9 (BARCL_1034, 70.8 kDa, and BARRO_80017, 113.1 kDa) and BepE of B. henselae (BH13420, 81.4 kDa) shown in (A). HA-GFP was used as a control (28.7 kDa).
Together with the fact that tandem-repeated phosphorylation motifs are only found in bep genes with derived domain architecture, our findings, thus, suggest parallel evolution of this class of effector proteins within the two radiating lineages. Whether similar pathways are targeted by these effectors in the two lineages remains unknown. Yet, the striking parallelism in the molecular evolution of this class of effector proteins indicates their central role in the VirB T4SS mediated host modulation by Bartonella.
Conclusions
Emerging infectious diseases are frequently caused by zoonotic pathogens which are incidentally transmitted to humans from their reservoir niche (e.g. other animal hosts). Therefore, the understanding of the mechanisms driving diversification of host-adapted bacteria in nature is of relevance for human health. In the present study, we explored the adaptive diversification of host-restricted bartonellae. Our genome-wide phylogeny revealed that two sister clades of this α-proteobacterial pathogen have evolved by parallel adaptive radiations (lineage 3 and lineage 4 in Figure 1). Both lineages comprise species adapted to same or similar reservoir hosts including zoonotic (e.g. Bh and Br) or human specific (Bq) pathogens. The more recent diversification of lineage 3 including the recently recognized incidental human pathogen Br [29] underlines the importance to study the molecular basis of such lineage diversifications.
In line with the ‘ecological’ parallelism of their radiations, our comparative genomic analyses between lineage 3 and lineage 4 uncover striking evolutionary parallelisms at the molecular level of a likely key innovation - the VirB T4SS – essentially involved in the infection of the mammalian hosts. Chromosomal fixation of this horizontally transferred trait occurred by independent evolutionary events. In both lineages, the arsenal of effector proteins translocated via the VirB T4SS was shaped independently by gene duplications and positive selection of diversified gene copies. This amplification process mostly occurred before the onset of the radiations. Strikingly, beside the diversification of effector proteins encoding the evolutionary conserved ‘AMPylase’ domain (FIC), both lineages have convergently evolved a novel effector class with derived domain structure and tandem-repeated tyrosine-phosphorylation motifs. By these evolutionary processes, large reservoirs of distinct biological functions were invented from a single ancestral effector gene. This functional versatility provides the framework for the adaptive potential of the VirB T4SS. Apparently, the plasticity of the underlying genomic loci seems to have favored the parallel occurrence of these adaptive processes in two distinct lineages, thereby essentially contributing to the parallel radiations of Bartonella.
Materials and Methods
Ethics statement
Animals were handled in strict accordance with good animal practice as defined by the relevant European (European standards of welfare for animals in research), national (Information and guidelines for animal experiments and alternative methods, Federal Veterinary Office of Switzerland) and/or local animal welfare bodies. Animal work was approved by the Veterinary Office of the Canton Basel City on June 2003 (licence no. 1741).
Bacterial strains and growth conditions
Bc strain 73 [34], B1-1C [31], and Br strain ATCC BAA-1498 [29] were grown routinely for 3–5 days on tryptic soy agar containing 5% defibrinated sheep-blood in a water-saturated atmosphere with 5% CO2 at 35°C. BAR15 [30] and Bs strain R1 [55] were grown under the same conditions on heart infusion agar and Colombia base agar, respectively.
Genome sequencing, assembly, and annotation
Using the QIAGEN Genomic DNA Isolation kit (Qiagen), DNA was isolated from bacteria grown from single colonies. For 454-sequencing, the DNA was prepared with an appropriate kit supplied by Roche Applied Science and sequenced on a Roche GS-FLX [56]. To assemble the reads, Newbler standard running parameters with ace file output were used. Newbler assemblies were considerably improved by linking overlapping contigs on the basis of the “_to” and “_from” information appended to the read name in the ace files. For the assemblies of Bc, BAR15, B1-1C, Br, and Bs, we obtained a 454-sequence coverage of 35x, 37x, 39x, 39x, and 29x, respectively (for details on 454-sequencing see Table S5). Repeats were identified by analyzing the coverage of each Newbler contig. If the link between two contigs was ambiguous, PCR and long-range PCR were used to confirm contig joins. For the complete assembly of the Bc genome, a library of 35 kb inserts was generated using the CopyControl Fosmid Kit (Epicentre). By end-sequencing of library clones with Sanger technology, 983 high-quality reads were obtained and mapped onto the 454-sequencing-based assembly. Remaining sequence gaps were closed by PCR. The final singular contig was fully covered by staggered fosmid clones indicating a correct assembly of the circular chromosome of Bc. Gene predictions of the genome of Bc and the draft genomes of Bs, Br, BAR15, and B1-1 were performed using AMIGene software [57]. Automated functional gene annotation was conducted with the genome annotation system MaGe [58]. For orthologous genes, the annotation was adopted from the manually annotated genome of Bt [16]. Manual validation of the annotation was performed for the virB and bep genes. By using the “FusionFission” tool of MaGe [58] fragmented genes were identified and the corresponding sequences subsequently examined for 454-sequencing errors. After correcting these errors, the updated sequences were re-annotated as described above. The sequence data of the genome of Bc and the contigs of the draft genomes of Br, BAR15, B1-1C, and Bs is stored on the web-based interface MaGe (Bartonella2Scope, https://www.genoscope.cns.fr/agc/mage/bartonella2Scope) and has been deposited in the EMBL Nucleotide Sequence Database under accession numbers FN645454–FN645524.
Phylogenetic analyses
Phylogenetic trees were based on nucleotide sequence data. Alignments were generated on protein sequences with ClustalW [59] and back-translated into aligned DNA sequences using MEGA4 [60]. Tree topologies were calculated with maximum likelihood and Bayesian inference methods as implemented in the programs PAUP* [61] and MrBayes [62], respectively. The genome-wide phylogeny of Bartonella was calculated on the basis of 478 orthologous genes of the ten sequenced Bartonella genomes and the genome of Brucella abortus (bv. 1 str. 9-941). Orthologs were determined by using the “PhyloProfile Synteny” tool of MaGe [58] with a threshold of 60% protein identity over at least 80% of the length of proteins being directional best hits of each other. The alignments of the 478 identified genes were concatenated resulting in a total of 515,751 aligned nucleotide sites. Tree topology and branch lengths were obtained by maximum likelihood analysis using the HKY85 model. Bootstrap support values were calculated for 100 replicates. For Bayesian inference, the program MrBayes [62] was run for one million iterations with standard parameters (two runs with four heated Monte-Carlo Markov chains in parallel; number of substitutions = 6; burnin = 25%). For the Bartonella ingroup, single gene trees were calculated with maximum likelihood and tree topology congruency assessed with PAUP*. 471 of the 478 single gene trees revealed the same monophyletic clustering of the eight taxa into lineage 3 and lineage 4 as the genome-wide phylogeny. Further, we performed a recombination analysis for each of the 478 single gene alignments using the GARD algorithm as implemented in the HYPHY package [35]. The GARD analysis was run with the GTR model using a general discrete distribution with three rate classes. To identify statistical significant recombination breakpoints in our alignments, we used the Kishino-Hasegawa test as implemented in the GARDProcess.bf algorithm of the HYPHY package. To include non-sequenced Bartonella species in the genome-wide phylogeny, we used available sequence data from the gltA, groEL, ribC, and rpoB genes (7731 aligned sites). Trees were obtained as described above. MrBayes [62] was run for five million iterations. Branch lengths for tip branches of non-sequenced taxa are calculated on the basis of the four housekeeping genes. Branch lengths for tip branches of sequenced taxa and internal branches separating sequenced and non-sequenced taxa are based on the genomic data set. The maximum likelihood tree only based on the gltA, groEL, ribC, and rpoB genes was inferred as described for the genome-wide phylogeny. Bep gene trees were inferred from nucleotide alignments of either the most C-terminal BID domain including the C-terminus (948 sites), the FIC domain including the N-terminal extension (1,305 sites), or the entire bep sequence of genes harboring FIC domains (3,972 sites). To select an appropriate substitution model, the Akaike information criterion of Modeltest 3.7 [63] and MrModeltest 2.0 [64] was used for the maximum likelihood and Bayesian inference analysis, respectively. For the alignments based on the BID domain or the entire bep gene sequence, we obtained the GTR+G+I model with both programs. For the alignments based on the FIC domain, the TVM+I+G model (Modeltest 3.7) and GTR+G+I model (MrModeltest 2.0) were selected. Trees were inferred with the parameters provided by these models as described above. MrBayes [62] was run for one million iterations. The Neighbor-joining phylogeny of different Bartonella isolates in Figure S2 was inferred from a 242 nt segment of the gltA gene with the program MEGA4 [60]. Bootstrap values were calculated for 1,000 replicates.
Natural selection analyses
Based on the four available genomes, orthologous genes for each of the two lineages 3 and 4 were determined by using the “PhyloProfile Synteny” tool of MaGe [58]. The threshold was set to 30% protein identity over at least 60% of the length of proteins being directional best hits of each other. The same tool was used to detect genes without orthologs. By comparing these automatically identified orthologs and non-orthologs, genes present in neither of the two lists were detected and manually assigned to one of the two lists. Alignments were generated and a GARD recombination analysis conducted as described above. To obtain the average dn/ds value (ω) of each ortholog, the arithmetic mean of pair-wise dn/ds values (calculated by the method of Yang and Nielsen implemented in PAML 4.1 [65]) was used. Site tests of positive selection were performed with PAML 4.1 using the CodeML module [65]. To detect positive selection model M1a (NearlyNeutral) vs. model M2a (PositiveSelection) and model M7 (beta) vs. model M8 (beta+ω) were analyzed. PAUP* [61] was used to infer maximum likelihood trees for each set of orthologs. For the CodeML control file, standard parameters were used. The relative significance of model M2a (PositiveSelection) vs. model M1a (NearlyNeutral) and model M8 (beta+ω) vs. model M7 (beta) was assessed using likelihood-ratio-tests (two degrees of freedom). Genes for which significant positive selection was detected were inspected for alignment errors potentially affecting the results of this analysis. If necessary, the alignments were manually modified and the CodeML analysis repeated. Phylogenetic branches were tested for positive selection by using the TestBranchDNDS.bf module implemented as standard analysis tool in HyPhy [66].
Rat infections
Ten weeks old female WISTAR rats obtained from RCC-Füllinsdorf were housed in an BSL2-animal facility for two weeks prior to infection allowing acclimatization. For inoculation, bacterial strains were grown as described above, harvested in phosphate-buffered saline (PBS), and diluted to OD595 = 1. Rats were anesthetized with a 2–3% Isuflurane/O2 mixture and infected with 10 µl of the bacterial suspension in the dermis of the right ear. Blood samples were taken at the tail vein and immediately mixed with PBS containing 3.8% sodium-citrate to avoid coagulation. After freezing to −70°C and subsequent thawing, undiluted and diluted blood samples were plated on tryptic soy agar and heart infusion agar containing 5% defibrinated sheep-blood. CFUs were counted after 8–12 days of growth.
Nucleotide distances
Nucleotide distances were calculated with the program MEGA4 [60] for the alignments based on the genome-wide dataset and the four housekeeping genes. The numbers of base substitutions per site from averaging over all sequence pairs within and between groups were calculated. Codon positions included were 1st, 2nd, and 3rd. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option).
Cloning of plasmids for expression of HA-GFP-Bep fusion proteins
To construct the plasmids pPE2002 and pPE2004, bep genes BARCL_1034 (Bc) and BARRO_80017 (Br) were amplified from genomic DNA with primer pairs containing flanking BamHI/NotI sites: prPE453 (ATAAGAATGCGGCCGCGATGAAAAC-CCATAACACTCCTG)/prPE454 (CGGG-ATCCTTAATGTGTTATAACCATCGTTC) and prPE455 (ATAAGAATGCGGCCGCG-ATGAATTTTGGAGAAAAGAAAAAAATG)/prPE456 (CGGGATCCTTAAATAGC-TACAGCTAACGATTTTTTC), respectively. PCR products were digested with the enzymes BamHI and NotI and ligated into the BamHI/NotI sites of the backbone of plasmid pAP013 (kindly provided by Arto Pulliainen). The resulting constructs pPE2001 (BARCL_1034) and pPE2003 (BARRO_80017) were cut with NotI and ligated with a GFP fragment obtained from NotI digested pAP013. The plasmid pPE2007 was constructed by cutting bepE of B. henselae from plasmid pRO1100 (kindly provided by Rusudan Okujava) with NotI and BamHI and ligating it into pAP013. All plasmid DNA isolations and PCR purifications were performed with Macherey-Nagel and Promega columns according to manufacturer's instructions.
Immunoprecipitation and Western blot analysis of HA-GFP-Bep fusion proteins
The protocol for growth and transfection of HEK293T was performed as described previously [18]. 36 h after transfection, cells were incubated for 10 minutes with 10 ml Pervanadate medium (5 ml PBS containing 100 mM orthovanadate and 200 mM H2O2, incubated for 10 min with 500 µl Catalase [2 mg/ml in PBS] before 45 ml M199 medium were added). After washing three times with 7 ml of PBS at room temperature, cells were scraped off and resuspended in 1 ml of ice-cold PBS containing 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM orthovanadate, 1 mM leupeptin, and 1 mM pepstatin and collected by centrifugation (3,000g at 4°C for 60 sec). The resulting pellet was lysed in 300 µl of ice cold modified RIPA buffer (50 mM Tris-HCl [pH 7.4], 75 mM NaCl, 1 mM EDTA, 1 mM orthovanadate, 1 mM leupeptin, 1 mM pepstatin) for 1 hour at 4°C. The lysate was centrifuged (16,000g at 4°C for 15 min) and 12 µl of anti-HA-agarose (Sigma) added to the supernatant. After 150 min of incubation at 4°C on a slowly turning rotation shaker, the agarose was washed three times with 300 µl of modified RIPA buffer (3,000g for 10 sec). The affinity-gel pellet was then resuspended in 20 µl of modified RIPA buffer, 20 µl of SDS-sample buffer (2×) were added, and the sample was heated for 5 min at 95°C. Proteins were separated on a 10% SDS-polyacrylamide gel, blotted on a nitrocellulose membrane (Hybond-C Extra, Amersham Pharmacia), and examined for tyrosine phosphorylation by using monoclonal antibody 4G10 (Millipore) and anti-mouse IgG-horseradish peroxidase (HRP) afterwards. The HRP-conjugated antibody was visualized by enhanced chemiluminescence (PerkinElmer). For visualization of the signal from GFP-fusion proteins, the membrane was subsequently incubated in 4% PBS-Tween containing 0.02% NaN3 and anti-GFP antibody (Invitrogen), followed by incubation with anti-mouse IgG-HRP and visualization by enhanced chemiluminescence.
Supporting Information
Phylogenetic trees inferred from (A) the nucleotide alignment of 478 genes of ten sequenced Bartonella species and (B) the nucleotide alignment of the four housekeeping genes gltA, rpoB, ribC, and groEL for sequenced and non-sequenced Bartonella species. Brucella abortus (bv. 1 str. 9-941) was used as outgroup. The tree topology and branch lengths were obtained by maximum likelihood analysis (as implemented in PAUP* [61]). Numbers above the branches represent maximum likelihood bootstrap values (100 replicates); numbers below the branches represent posterior probabilities obtained with MrBayes (one million iterations) [62]. Values ≥80% are shown. The two monophyletic clades harboring the VirB T4SS are marked by the shaded area. The four different lineages of Bartonella are indicated by red, orange, blue, and green color (l1, lineage 1, l2, lineage 2).
(0.49 MB PDF)
Genotype-host correlation of Bartonella isolates. Neighbor-joining tree of 220 Bartonella isolates based on a 242 nt sequence of the gltA gene. Bootstrap values ≥50% are shown (1000 replicates). Clusters of strains isolated from the same mammalian family, subfamily, or genus are indicated by coloring. For each strain the name, accession number, and host (the strain was isolated from) are given. The typing strains and their described reservoir hosts are depicted by arrows. For Bartonella alsatica and Bq, only one and two isolates, respectively, are included in the tree, as sequence data for additional isolates is not available for the analyzed fragment of the gltA gene. However, for both species, a number of different isolates originating from their corresponding reservoir hosts (Bq, human and B. alsatica, rabbit) were described [68], [69]. For B1-1C and BAR15, only one strain was described so far [30], [31]. All strains included in the analysis have been isolated and cultured from the bloodstream of their corresponding host [55], [68]–[105]. Exceptions display some typing strains which were isolated from incidentally infected humans. Strains sampled from other tissue than blood or only detected by PCR were not included in the analysis.
(2.62 MB PDF)
Experimental infections of rats with Bartonella species of lineage 3. Groups of rats (n = 5) were inoculated intradermally with one of the four different species Bc, Br, BAR15, or B1-1C. At the time points indicated in the graphs, blood was drawn, diluted, and plated for counting of colony-forming units (CFUs). Plotted graphs represent single animals from which bacteria could be recovered at a given time point post-infection. In all five animals infected with the rat-specific strain B1-1C, bacteria were detected between day 8 and 28 post-infection (green graphs). In only three of five animals infected with the red squirrel-specific strain BAR15, bacteria could be detected (red graphs). In these three animals, bacteria came up much later and did not reach the same blood titer as B1-1C. From none of the animals infected with either Br or Bc bacteria could be recovered (threshold of detection ≤102 CFUs/ml). Although Br is closer related to B1-1C than BAR15 (Figure 1), the adaptation to Canoidae hosts seems to have abolished the potential to colonize rodents. In contrast, the closer relationship between the two rodent host species, rat and red squirrel, might explain the limited potential of BAR15 to infect rats. Additionally, a recent study showed that Br is reliably infecting dogs, its natural reservoir host, but neither cats nor guinea pigs [39]. These independent studies confirm the host specificity of Bartonella species.
(0.31 MB PDF)
Comparison of virB T4SS- and bep effector-encoding loci. (A) Comparison of the two virB T4SS loci of Bc, Br, BAR15, and B1-1C (lineage 3). (B) Comparison of the genomic regions of Bc, Br, BAR15, and B1-1C encoding additional bep genes (lineage 3). (C) Comparison of the virB T4SS loci of Bg, Bh, Bq, and Bt (lineage 4). virB T4SS genes are colored in yellow, bep genes in red (locus_tag numbers are given), and flanking regions and putative coding sequences in gray. The monophyletic clades of different bep genes (see Figure 4) are indicated and show that most genes belonging to the same Bep clade constitute positional orthologs of each other. Bep clade 11 does not appear in Figure 4, since these bep genes consist of only the FIC domain. Genes are connected by gray boxes if they are orthologs of each other. For bep genes, connections are drawn if genes belong to the same Bep clade (see Figure 4), or if they are top blast hits of one another. The bep gene tagged by an asterisk (BAR15_120222) belongs to Bep clade 1. In (A), the glutamine synthetase I (glnA) gene and its fragments flanking the two inverted virB T4SS loci are colored in green. In (B), contig breaks are indicated by the interruption of the sequence line.
(0.45 MB PDF)
Domain structure of Bartonella effector proteins. Domain structure for (A) lineage 3 and (B) lineage 4. FIC and BID domains are shown in orange and violet, respectively. The FIC motif (HPFxxGNG) is depicted. Motifs differing from this consensus sequence are indicated. Beps are grouped according to the monophyletic clades indicated in Figure 4 (Bep clades). BARCL_0629 and BARCL_0635 are excluded from groups, because their phylogenetic positions are not well supported in the inferred gene trees (Figure 4 and Figure S6). Tandem-repeated tyrosine-phosphorylation motifs depicted for Bep clade 9, BepD, BepE, BepF, and BepH clade (green bars) were identified with the program NetPhos2.0 [52] using a threshold of 0.8. For other bep genes, no tandem-repeated motifs were identified. bep genes from different species of lineage 3 are only indicated by their locus_tags (BARCL_, Bc, BARRO_, Br, B11C_, B1-1C, BAR15_, BAR15).
(0.37 MB PDF)
Bartonella effector protein gene trees based on (A) the FIC domain containing N-terminus or (B) the entire sequence of effector genes harboring an N-terminal FIC and C-terminal BID domain. The topology and branch lengths are according to maximum likelihood analysis. Numbers above the branches represent maximum likelihood bootstraps obtained with PAUP* [61]; numbers below the branches represent posterior probabilities obtained with MrBayes [62]. Values ≥80% are shown. Locus_tags and gene names (if existing) are given. beps are color-coded according to the species. The clustering of bep genes from lineage 3 and 4 is indicated by blue and green shading, respectively. Bold type highlights phylogenetic branches with dn/ds values >1 (the estimated dn/ds value is indicated in red color type).
(1.65 MB PDF)
Estimates of average evolutionary divergence over sequence pairs within lineages (shaded in gray) and between lineages.
(0.09 MB PDF)
Genes of lineage 3 with average dn/ds values (ω) ≥0.25 and/or site-specific positive selection detected by CodeML (model M2a versus model M1a). Genes are ordered according to ω. Genes with ω<0.25, but with sites under positive selection are indicated by gray color. virB T4SS and bep genes are indicated by orange color. Orthologous genes present in genomes of other lineages are shown. Likelihood-ratio-test (LRT) results of the CodeML selection analysis are given for comparison of model M2a versus M1a and model M8 versus M7.
(0.33 MB PDF)
Genes of lineage 4 with average dn/ds values (ω) ≥0.25 and/or site-specific positive selection detected by CodeML (model M2a versus model M1a). Genes are ordered according to ω. Genes with ω<0.25, but with sites under positive selection are indicated by gray color. virB T4SS, bep, and trw T4SS genes are indicated by orange color. Orthologous genes present in genomes of other lineages are shown. Likelihood-ratio-test (LRT) results of the CodeML selection analysis are given for comparison of model M2a versus M1a and model M8 versus M7.
(0.20 MB PDF)
Prediction of tyrosine-phosphorylation sites for Beps of Bep clade 9 using NetPhos2.0 [52], ScanSite [53], and KinasePhos [54]. Except for BARCL_1035, tandem-repeated motifs were detected in all genes of Bep clade 9.
(0.14 MB PDF)
454-sequencing and newbler-assemby data for genomes sequenced in this study.
(0.09 MB PDF)
Acknowledgments
We thank Rusudan Okujava and Arto Pulliainen for providing plasmids pRO1100 and pAP015, respectively.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grant 31003A-132979 from the Swiss National Science Foundation (to CD), grant 55005501 from the Howard Hughes Medical Institute (to CD), and grant 51RT 0_126008 (InfectX) in the frame of the SystemsX.ch Swiss Initiative for Systems Biology (to CD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press; 2000. p. viii, 288. [Google Scholar]
- 2.Grant PR, Grant BR. How and why species multiply : the radiation of Darwin's finches. Princeton: Princeton University Press; 2008. p. xix, 218. [Google Scholar]
- 3.Salzburger W. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol Ecol. 2009;18:169–185. doi: 10.1111/j.1365-294X.2008.03981.x. [DOI] [PubMed] [Google Scholar]
- 4.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 5.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 6.Sikorski J, Nevo E. Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at “Evolution Canyons” I and II, Israel. Proc Natl Acad Sci U S A. 2005;102:15924–15929. doi: 10.1073/pnas.0507944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brakefield PM, Roskam JC. Exploring evolutionary constraints is a task for an integrative evolutionary biology. Am Nat. 2006;168(Suppl 6):S4–13. doi: 10.1086/509049. [DOI] [PubMed] [Google Scholar]
- 8.Duponchelle F, Paradis E, Ribbink AJ, Turner GF. Parallel life history evolution in mouthbrooding cichlids from the African Great Lakes. Proc Natl Acad Sci U S A. 2008;105:15475–15480. doi: 10.1073/pnas.0802343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kronforst MR, Kapan DD, Gilbert LE. Parallel genetic architecture of parallel adaptive radiations in mimetic Heliconius butterflies. Genetics. 2006;174:535–539. doi: 10.1534/genetics.106.059527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457:830–836. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- 11.Harvey P.H. PMD. The comparative method in evolutionary biology. London/New York/Oxford: Oxford University Press; 1991. [Google Scholar]
- 12.Dehio C. Bartonella-host-cell interactions and vascular tumour formation. Nat Rev Microbiol. 2005;3:621–631. doi: 10.1038/nrmicro1209. [DOI] [PubMed] [Google Scholar]
- 13.Marignac G, Barrat F, Chomel B, Vayssier-Taussat M, Gandoin C, et al. Murine model for Bartonella birtlesii infection: New aspects. Comp Immunol Microbiol Infect Dis. 2010;33:95–107. doi: 10.1016/j.cimid.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Schulein R, Seubert A, Gille C, Lanz C, Hansmann Y, et al. Invasion and persistent intracellular colonization of erythrocytes. A unique parasitic strategy of the emerging pathogen Bartonella. J Exp Med. 2001;193:1077–1086. doi: 10.1084/jem.193.9.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, Chomel BB, Schau MK, Goo JS, Droz S, et al. A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc Natl Acad Sci U S A. 2004;101:13630–13635. doi: 10.1073/pnas.0405284101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, et al. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat Genet. 2007;39:1469–1476. doi: 10.1038/ng.2007.38. [DOI] [PubMed] [Google Scholar]
- 17.Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, et al. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci U S A. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid MC, Scheidegger F, Dehio M, Balmelle-Devaux N, Schulein R, et al. A translocated bacterial protein protects vascular endothelial cells from apoptosis. PLoS Pathog. 2006;2:e115. doi: 10.1371/journal.ppat.0020115. doi: 10.1371/journal.ppat.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhomberg TA, Truttmann MC, Guye P, Ellner Y, Dehio C. A translocated protein of Bartonella henselae interferes with endocytic uptake of individual bacteria and triggers uptake of large bacterial aggregates via the invasome. Cell Microbiol. 2009;11:927–945. doi: 10.1111/j.1462-5822.2009.01302.x. [DOI] [PubMed] [Google Scholar]
- 20.Scheidegger F, Ellner Y, Guye P, Rhomberg TA, Weber H, et al. Distinct activities of Bartonella henselae type IV secretion effector proteins modulate capillary-like sprout formation. Cell Microbiol. 2009;11:1088–1101. doi: 10.1111/j.1462-5822.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- 21.Selbach M, Paul FE, Brandt S, Guye P, Daumke O, et al. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe. 2009;5:397–403. doi: 10.1016/j.chom.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Vayssier-Taussat M, Le Rhun D, Deng HK, Biville F, Cescau S, et al. The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 2010;6:e1000946. doi: 10.1371/journal.ppat.1000946. doi: 10.1371/journal.ppat.1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seubert A, Hiestand R, de la Cruz F, Dehio C. A bacterial conjugation machinery recruited for pathogenesis. Mol Microbiol. 2003;49:1253–1266. doi: 10.1046/j.1365-2958.2003.03650.x. [DOI] [PubMed] [Google Scholar]
- 24.Nystedt B, Frank AC, Thollesson M, Andersson SG. Diversifying selection and concerted evolution of a type IV secretion system in Bartonella. Mol Biol Evol. 2008;25:287–300. doi: 10.1093/molbev/msm252. [DOI] [PubMed] [Google Scholar]
- 25.Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, et al. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- 26.Yarbrough ML, Orth K. AMPylation is a new post-translational modiFICation. Nat Chem Biol. 2009;5:378–379. doi: 10.1038/nchembio0609-378. [DOI] [PubMed] [Google Scholar]
- 27.Alsmark CM, Frank AC, Karlberg EO, Legault BA, Ardell DH, et al. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc Natl Acad Sci U S A. 2004;101:9716–9721. doi: 10.1073/pnas.0305659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berglund EC, Frank AC, Calteau A, Vinnere Pettersson O, Granberg F, et al. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet. 2009;5:e1000546. doi: 10.1371/journal.pgen.1000546. doi: 10.1371/journal.pgen.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, et al. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;356:2381–2387. doi: 10.1056/NEJMoa065987. [DOI] [PubMed] [Google Scholar]
- 30.Inoue K, Maruyama S, Kabeya H, Hagiya K, Izumi Y, et al. Exotic small mammals as potential reservoirs of zoonotic Bartonella spp. Emerg Infect Dis. 2009;15:526–532. doi: 10.3201/eid1504.081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin JW, Chen CY, Chen WC, Chomel BB, Chang CC. Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J Med Microbiol. 2008;57:1496–1501. doi: 10.1099/jmm.0.2008/004671-0. [DOI] [PubMed] [Google Scholar]
- 32.Henn JB, Gabriel MW, Kasten RW, Brown RN, Koehler JE, et al. Infective endocarditis in a dog and the phylogenetic relationship of the associated “Bartonella rochalimae” strain with isolates from dogs, gray foxes, and a human. J Clin Microbiol. 2009;47:787–790. doi: 10.1128/JCM.01351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kordick DL, Hilyard EJ, Hadfield TL, Wilson KH, Steigerwalt AG, et al. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease). J Clin Microbiol. 1997;35:1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heller R, Artois M, Xemar V, De Briel D, Gehin H, et al. Prevalence of Bartonella henselae and Bartonella clarridgeiae in stray cats. J Clin Microbiol. 1997;35:1327–1331. doi: 10.1128/jcm.35.6.1327-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 36.Bininda-Emonds OR. Supertree construction in the genomic age. Methods Enzymol. 2005;395:745–757. doi: 10.1016/S0076-6879(05)95038-6. [DOI] [PubMed] [Google Scholar]
- 37.Ihler GM. Bartonella bacilliformis: dangerous pathogen slowly emerging from deep background. FEMS Microbiol Lett. 1996;144:1–11. doi: 10.1111/j.1574-6968.1996.tb08501.x. [DOI] [PubMed] [Google Scholar]
- 38.Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, et al. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet Res. 2009;40:29. doi: 10.1051/vetres/2009011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chomel BB, Henn JB, Kasten RW, Nieto NC, Foley J, et al. Dogs are more permissive than cats or guinea pigs to experimental infection with a human isolate of Bartonella rochalimae. Vet Res. 2009;40:27. doi: 10.1051/vetres/2009010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosoy MY, Saito EK, Green D, Marston EL, Jones DC, et al. Experimental evidence of host specificity of Bartonella infection in rodents. Comp Immunol Microbiol Infect Dis. 2000;23:221–238. doi: 10.1016/s0147-9571(99)00075-2. [DOI] [PubMed] [Google Scholar]
- 41.Berglund EC, Ellegaard K, Granberg F, Xie Z, Maruyama S, et al. Rapid diversification by recombination in Bartonella grahamii from wild rodents in Asia contrasts with low levels of genomic divergence in Northern Europe and America. Mol Ecol. 2010;19:2241–2255. doi: 10.1111/j.1365-294X.2010.04646.x. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 43.Carroll JA, Coleman SA, Smitherman LS, Minnick MF. Hemin-binding surface protein from Bartonella quintana. Infect Immun. 2000;68:6750–6757. doi: 10.1128/iai.68.12.6750-6757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dehio C. Molecular and cellular basis of Bartonella pathogenesis. Annu Rev Microbiol. 2004;58:365–390. doi: 10.1146/annurev.micro.58.030603.123700. [DOI] [PubMed] [Google Scholar]
- 45.Minnick MF, Sappington KN, Smitherman LS, Andersson SG, Karlberg O, et al. Five-member gene family of Bartonella quintana. Infect Immun. 2003;71:814–821. doi: 10.1128/IAI.71.2.814-821.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCann HC, Guttman DS. Evolution of the type III secretion system and its effectors in plant-microbe interactions. New Phytol. 2008;177:33–47. doi: 10.1111/j.1469-8137.2007.02293.x. [DOI] [PubMed] [Google Scholar]
- 47.Ma W, Dong FF, Stavrinides J, Guttman DS. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2006;2:e209. doi: 10.1371/journal.pgen.0020209. doi: 10.1371/journal.pgen.0020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frank AC, Alsmark CM, Thollesson M, Andersson SG. Functional divergence and horizontal transfer of type IV secretion systems. Mol Biol Evol. 2005;22:1325–1336. doi: 10.1093/molbev/msi124. [DOI] [PubMed] [Google Scholar]
- 49.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandegren L, Andersson DI. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:578–588. doi: 10.1038/nrmicro2174. [DOI] [PubMed] [Google Scholar]
- 51.Kinch LN, Yarbrough ML, Orth K, Grishin NV. Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS ONE. 2009;4:e5818. doi: 10.1371/journal.pone.0005818. doi: 10.1371/journal.pone.0005818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 53.Huang HD, Lee TY, Tzeng SW, Horng JT. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005;33:W226–229. doi: 10.1093/nar/gki471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dehio C, Lanz C, Pohl R, Behrens P, Bermond D, et al. Bartonella schoenbuchii sp. nov., isolated from the blood of wild roe deer. Int J Syst Evol Microbiol. 2001;51:1557–1565. doi: 10.1099/00207713-51-4-1557. [DOI] [PubMed] [Google Scholar]
- 56.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bocs S, Cruveiller S, Vallenet D, Nuel G, Medigue C. AMIGene: Annotation of MIcrobial Genes. Nucleic Acids Res. 2003;31:3723–3726. doi: 10.1093/nar/gkg590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 61.Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics Chapter. 2003;6:Unit 6.4.1–6.4.28. doi: 10.1002/0471250953.bi0604s00. [DOI] [PubMed] [Google Scholar]
- 62.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 63.Posada D. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr Protoc Bioinformatics Chapter. 2003;6:Unit 6.5.1–6.5.14. doi: 10.1002/0471250953.bi0605s00. [DOI] [PubMed] [Google Scholar]
- 64.Nylander JAA. MrModeltest v2. Program distributed by the author; 2004. [Google Scholar]
- 65.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 66.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 67.Engel P, Dehio C. Genomics of Host-Restricted Pathogens of the Genus Bartonella. Genome Dyn. 2009;6:158–169. doi: 10.1159/000235769. [DOI] [PubMed] [Google Scholar]
- 68.Arvand M, Raoult D, Feil EJ. Multi-locus sequence typing of a geographically and temporally diverse sample of the highly clonal human pathogen Bartonella quintana. PLoS ONE. 2010;5:e9765. doi: 10.1371/journal.pone.0009765. doi: 10.1371/journal.pone.0009765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bai Y, Kosoy MY, Cully JF, Bala T, Ray C, et al. Acquisition of nonspecific Bartonella strains by the northern grasshopper mouse (Onychomys leucogaster). FEMS Microbiol Ecol. 2007;61:438–448. doi: 10.1111/j.1574-6941.2007.00364.x. [DOI] [PubMed] [Google Scholar]
- 70.Bai Y, Kosoy MY, Lerdthusnee K, Peruski LF, Richardson JH. Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am J Trop Med Hyg. 2009;81:811–816. doi: 10.4269/ajtmh.2009.09-0294. [DOI] [PubMed] [Google Scholar]
- 71.Bemis DA, Kania SA. Isolation of Bartonella sp. from sheep blood. Emerg Infect Dis. 2007;13:1565–1567. doi: 10.3201/eid1310.070570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berglund EC, Ehrenborg C, Vinnere Pettersson O, Granberg F, Naslund K, et al. Genome dynamics of Bartonella grahamii in micro-populations of woodland rodents. BMC Genomics. 2010;11:152. doi: 10.1186/1471-2164-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bermond D, Boulouis HJ, Heller R, Van Laere G, Monteil H, et al. Bartonella bovis Bermond et al. sp. nov. and Bartonella capreoli sp. nov., isolated from European ruminants. Int J Syst Evol Microbiol. 2002;52:383–390. doi: 10.1099/00207713-52-2-383. [DOI] [PubMed] [Google Scholar]
- 74.Bermond D, Heller R, Barrat F, Delacour G, Dehio C, et al. Bartonella birtlesii sp. nov., isolated from small mammals (Apodemus spp.). Int J Syst Evol Microbiol. 2000;50:1973–1979. doi: 10.1099/00207713-50-6-1973. [DOI] [PubMed] [Google Scholar]
- 75.Birtles RJ, Harrison TG, Saunders NA, Molyneux DH. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 76.Birtles RJ, Hazel SM, Bennett M, Bown K, Raoult D, et al. Longitudinal monitoring of the dynamics of infections due to Bartonella species in UK woodland rodents. Epidemiol Infect. 2001;126:323–329. doi: 10.1017/s095026880100526x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Birtles RJ, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol. 1996;46:891–897. doi: 10.1099/00207713-46-4-891. [DOI] [PubMed] [Google Scholar]
- 78.Celebi B, Carhan A, Kilic S, Babur C. Detection and Genetic Diversity of Bartonella vinsonii Subsp. berkhoffii Strains Isolated from Dogs in Ankara, Turkey. J Vet Med Sci. 2010;72:969–73. doi: 10.1292/jvms.09-0466. [DOI] [PubMed] [Google Scholar]
- 79.Chamberlin J, Laughlin LW, Romero S, Solorzano N, Gordon S, et al. Epidemiology of endemic Bartonella bacilliformis: a prospective cohort study in a Peruvian mountain valley community. J Infect Dis. 2002;186:983–990. doi: 10.1086/344054. [DOI] [PubMed] [Google Scholar]
- 80.Chang CC, Chomel BB, Kasten RW, Heller RM, Ueno H, et al. Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg Infect Dis. 2000;6:306–311. doi: 10.3201/eid0603.000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clarridge JE, 3rd, Raich TJ, Pirwani D, Simon B, Tsai L, et al. Strategy to detect and identify Bartonella species in routine clinical laboratory yields Bartonella henselae from human immunodeficiency virus-positive patient and unique Bartonella strain from his cat. J Clin Microbiol. 1995;33:2107–2113. doi: 10.1128/jcm.33.8.2107-2113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dillon B, Valenzuela J, Don R, Blanckenberg D, Wigney DI, et al. Limited diversity among human isolates of Bartonella henselae. J Clin Microbiol. 2002;40:4691–4699. doi: 10.1128/JCM.40.12.4691-4699.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ellis BA, Regnery RL, Beati L, Bacellar F, Rood M, et al. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–224. doi: 10.1086/314824. [DOI] [PubMed] [Google Scholar]
- 84.Gundi VA, Davoust B, Khamis A, Boni M, Raoult D, et al. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol. 2004;42:3816–3818. doi: 10.1128/JCM.42.8.3816-3818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gurfield AN, Boulouis HJ, Chomel BB, Kasten RW, Heller R, et al. Epidemiology of Bartonella infection in domestic cats in France. Vet Microbiol. 2001;80:185–198. doi: 10.1016/s0378-1135(01)00304-2. [DOI] [PubMed] [Google Scholar]
- 86.Harrus S, Bar-Gal GK, Golan A, Elazari-Volcani R, Kosoy MY, et al. Isolation and genetic characterization of a Bartonella strain closely related to Bartonella tribocorum and Bartonella elizabethae in Israeli commensal rats. Am J Trop Med Hyg. 2009;81:55–58. [PubMed] [Google Scholar]
- 87.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, et al. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48 Pt 4:1333–1339. doi: 10.1099/00207713-48-4-1333. [DOI] [PubMed] [Google Scholar]
- 88.Henn JB, Chomel BB, Boulouis HJ, Kasten RW, Murray WJ, et al. Bartonella rochalimae in raccoons, coyotes, and red foxes. Emerg Infect Dis. 2009;15:1984–1987. doi: 10.3201/eid1512.081692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hofmeister EK, Kolbert CP, Abdulkarim AS, Magera JM, Hopkins MK, et al. Cosegregation of a novel Bartonella species with Borrelia burgdorferi and Babesia microti in Peromyscus leucopus. J Infect Dis. 1998;177:409–416. doi: 10.1086/514201. [DOI] [PubMed] [Google Scholar]
- 90.Holmberg M, Mills JN, McGill S, Benjamin G, Ellis BA. Bartonella infection in sylvatic small mammals of central Sweden. Epidemiol Infect. 2003;130:149–157. doi: 10.1017/s0950268802008075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inoue K, Kabeya H, Kosoy MY, Bai Y, Smirnov G, et al. Evolutional and geographical relationships of Bartonella grahamii isolates from wild rodents by multi-locus sequencing analysis. Microb Ecol. 2009;57:534–541. doi: 10.1007/s00248-009-9488-x. [DOI] [PubMed] [Google Scholar]
- 92.Inoue K, Maruyama S, Kabeya H, Yamada N, Ohashi N, et al. Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl Environ Microbiol. 2008;74:5086–5092. doi: 10.1128/AEM.00071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jardine C, Appleyard G, Kosoy MY, McColl D, Chirino-Trejo M, et al. Rodent-associated Bartonella in Saskatchewan, Canada. Vector Borne Zoonotic Dis. 2005;5:402–409. doi: 10.1089/vbz.2005.5.402. [DOI] [PubMed] [Google Scholar]
- 94.Kosoy M, Murray M, Gilmore RD, Jr, Bai Y, Gage KL. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol. 2003;41:645–650. doi: 10.1128/JCM.41.2.645-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kosoy MY, Regnery RL, Tzianabos T, Marston EL, Jones DC, et al. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am J Trop Med Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- 96.Li DM, Meng FX, Song XP, Qin ZJ, Yang XR, et al. [Study on Bartonella vinsonii berkhoffii isolated from blood of native dogs in China]. Zhonghua Liu Xing Bing Xue Za Zhi. 2006;27:333–338. [PubMed] [Google Scholar]
- 97.Lydy SL, Eremeeva ME, Asnis D, Paddock CD, Nicholson WL, et al. Isolation and characterization of Bartonella bacilliformis from an expatriate Ecuadorian. J Clin Microbiol. 2008;46:627–637. doi: 10.1128/JCM.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maillard R, Riegel P, Barrat F, Bouillin C, Thibault D, et al. Bartonella chomelii sp. nov., isolated from French domestic cattle (Bos taurus). Int J Syst Evol Microbiol. 2004;54:215–220. doi: 10.1099/ijs.0.02770-0. [DOI] [PubMed] [Google Scholar]
- 99.Marquez FJ. Molecular Detection of Bartonella alsatica in European Wild Rabbits (Oryctolagus cuniculus) in Andalusia (Spain). Vector Borne Zoonotic Dis. 2010;10:731–4. doi: 10.1089/vbz.2009.0135. [DOI] [PubMed] [Google Scholar]
- 100.Marston EL, Finkel B, Regnery RL, Winoto IL, Graham RR, et al. Prevalence of Bartonella henselae and Bartonella clarridgeiae in an urban Indonesian cat population. Clin Diagn Lab Immunol. 1999;6:41–44. doi: 10.1128/cdli.6.1.41-44.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pretorius AM, Beati L, Birtles RJ. Diversity of bartonellae associated with small mammals inhabiting Free State province, South Africa. Int J Syst Evol Microbiol. 2004;54:1959–1967. doi: 10.1099/ijs.0.03033-0. [DOI] [PubMed] [Google Scholar]
- 102.Rolain JM, Fournier PE, Raoult D, Bonerandi JJ. First isolation and detection by immunofluorescence assay of Bartonella koehlerae in erythrocytes from a French cat. J Clin Microbiol. 2003;41:4001–4002. doi: 10.1128/JCM.41.8.4001-4002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tea A, Alexiou-Daniel S, Papoutsi A, Papa A, Antoniadis A. Bartonella species isolated from rodents, Greece. Emerg Infect Dis. 2004;10:963–964. doi: 10.3201/eid1005.030430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am J Trop Med Hyg. 2002;66:622–627. doi: 10.4269/ajtmh.2002.66.622. [DOI] [PubMed] [Google Scholar]
- 105.Zeaiter Z, Fournier PE, Ogata H, Raoult D. Phylogenetic classification of Bartonella species by comparing groEL sequences. Int J Syst Evol Microbiol. 2002;52:165–171. doi: 10.1099/00207713-52-1-165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic trees inferred from (A) the nucleotide alignment of 478 genes of ten sequenced Bartonella species and (B) the nucleotide alignment of the four housekeeping genes gltA, rpoB, ribC, and groEL for sequenced and non-sequenced Bartonella species. Brucella abortus (bv. 1 str. 9-941) was used as outgroup. The tree topology and branch lengths were obtained by maximum likelihood analysis (as implemented in PAUP* [61]). Numbers above the branches represent maximum likelihood bootstrap values (100 replicates); numbers below the branches represent posterior probabilities obtained with MrBayes (one million iterations) [62]. Values ≥80% are shown. The two monophyletic clades harboring the VirB T4SS are marked by the shaded area. The four different lineages of Bartonella are indicated by red, orange, blue, and green color (l1, lineage 1, l2, lineage 2).
(0.49 MB PDF)
Genotype-host correlation of Bartonella isolates. Neighbor-joining tree of 220 Bartonella isolates based on a 242 nt sequence of the gltA gene. Bootstrap values ≥50% are shown (1000 replicates). Clusters of strains isolated from the same mammalian family, subfamily, or genus are indicated by coloring. For each strain the name, accession number, and host (the strain was isolated from) are given. The typing strains and their described reservoir hosts are depicted by arrows. For Bartonella alsatica and Bq, only one and two isolates, respectively, are included in the tree, as sequence data for additional isolates is not available for the analyzed fragment of the gltA gene. However, for both species, a number of different isolates originating from their corresponding reservoir hosts (Bq, human and B. alsatica, rabbit) were described [68], [69]. For B1-1C and BAR15, only one strain was described so far [30], [31]. All strains included in the analysis have been isolated and cultured from the bloodstream of their corresponding host [55], [68]–[105]. Exceptions display some typing strains which were isolated from incidentally infected humans. Strains sampled from other tissue than blood or only detected by PCR were not included in the analysis.
(2.62 MB PDF)
Experimental infections of rats with Bartonella species of lineage 3. Groups of rats (n = 5) were inoculated intradermally with one of the four different species Bc, Br, BAR15, or B1-1C. At the time points indicated in the graphs, blood was drawn, diluted, and plated for counting of colony-forming units (CFUs). Plotted graphs represent single animals from which bacteria could be recovered at a given time point post-infection. In all five animals infected with the rat-specific strain B1-1C, bacteria were detected between day 8 and 28 post-infection (green graphs). In only three of five animals infected with the red squirrel-specific strain BAR15, bacteria could be detected (red graphs). In these three animals, bacteria came up much later and did not reach the same blood titer as B1-1C. From none of the animals infected with either Br or Bc bacteria could be recovered (threshold of detection ≤102 CFUs/ml). Although Br is closer related to B1-1C than BAR15 (Figure 1), the adaptation to Canoidae hosts seems to have abolished the potential to colonize rodents. In contrast, the closer relationship between the two rodent host species, rat and red squirrel, might explain the limited potential of BAR15 to infect rats. Additionally, a recent study showed that Br is reliably infecting dogs, its natural reservoir host, but neither cats nor guinea pigs [39]. These independent studies confirm the host specificity of Bartonella species.
(0.31 MB PDF)
Comparison of virB T4SS- and bep effector-encoding loci. (A) Comparison of the two virB T4SS loci of Bc, Br, BAR15, and B1-1C (lineage 3). (B) Comparison of the genomic regions of Bc, Br, BAR15, and B1-1C encoding additional bep genes (lineage 3). (C) Comparison of the virB T4SS loci of Bg, Bh, Bq, and Bt (lineage 4). virB T4SS genes are colored in yellow, bep genes in red (locus_tag numbers are given), and flanking regions and putative coding sequences in gray. The monophyletic clades of different bep genes (see Figure 4) are indicated and show that most genes belonging to the same Bep clade constitute positional orthologs of each other. Bep clade 11 does not appear in Figure 4, since these bep genes consist of only the FIC domain. Genes are connected by gray boxes if they are orthologs of each other. For bep genes, connections are drawn if genes belong to the same Bep clade (see Figure 4), or if they are top blast hits of one another. The bep gene tagged by an asterisk (BAR15_120222) belongs to Bep clade 1. In (A), the glutamine synthetase I (glnA) gene and its fragments flanking the two inverted virB T4SS loci are colored in green. In (B), contig breaks are indicated by the interruption of the sequence line.
(0.45 MB PDF)
Domain structure of Bartonella effector proteins. Domain structure for (A) lineage 3 and (B) lineage 4. FIC and BID domains are shown in orange and violet, respectively. The FIC motif (HPFxxGNG) is depicted. Motifs differing from this consensus sequence are indicated. Beps are grouped according to the monophyletic clades indicated in Figure 4 (Bep clades). BARCL_0629 and BARCL_0635 are excluded from groups, because their phylogenetic positions are not well supported in the inferred gene trees (Figure 4 and Figure S6). Tandem-repeated tyrosine-phosphorylation motifs depicted for Bep clade 9, BepD, BepE, BepF, and BepH clade (green bars) were identified with the program NetPhos2.0 [52] using a threshold of 0.8. For other bep genes, no tandem-repeated motifs were identified. bep genes from different species of lineage 3 are only indicated by their locus_tags (BARCL_, Bc, BARRO_, Br, B11C_, B1-1C, BAR15_, BAR15).
(0.37 MB PDF)
Bartonella effector protein gene trees based on (A) the FIC domain containing N-terminus or (B) the entire sequence of effector genes harboring an N-terminal FIC and C-terminal BID domain. The topology and branch lengths are according to maximum likelihood analysis. Numbers above the branches represent maximum likelihood bootstraps obtained with PAUP* [61]; numbers below the branches represent posterior probabilities obtained with MrBayes [62]. Values ≥80% are shown. Locus_tags and gene names (if existing) are given. beps are color-coded according to the species. The clustering of bep genes from lineage 3 and 4 is indicated by blue and green shading, respectively. Bold type highlights phylogenetic branches with dn/ds values >1 (the estimated dn/ds value is indicated in red color type).
(1.65 MB PDF)
Estimates of average evolutionary divergence over sequence pairs within lineages (shaded in gray) and between lineages.
(0.09 MB PDF)
Genes of lineage 3 with average dn/ds values (ω) ≥0.25 and/or site-specific positive selection detected by CodeML (model M2a versus model M1a). Genes are ordered according to ω. Genes with ω<0.25, but with sites under positive selection are indicated by gray color. virB T4SS and bep genes are indicated by orange color. Orthologous genes present in genomes of other lineages are shown. Likelihood-ratio-test (LRT) results of the CodeML selection analysis are given for comparison of model M2a versus M1a and model M8 versus M7.
(0.33 MB PDF)
Genes of lineage 4 with average dn/ds values (ω) ≥0.25 and/or site-specific positive selection detected by CodeML (model M2a versus model M1a). Genes are ordered according to ω. Genes with ω<0.25, but with sites under positive selection are indicated by gray color. virB T4SS, bep, and trw T4SS genes are indicated by orange color. Orthologous genes present in genomes of other lineages are shown. Likelihood-ratio-test (LRT) results of the CodeML selection analysis are given for comparison of model M2a versus M1a and model M8 versus M7.
(0.20 MB PDF)
Prediction of tyrosine-phosphorylation sites for Beps of Bep clade 9 using NetPhos2.0 [52], ScanSite [53], and KinasePhos [54]. Except for BARCL_1035, tandem-repeated motifs were detected in all genes of Bep clade 9.
(0.14 MB PDF)
454-sequencing and newbler-assemby data for genomes sequenced in this study.
(0.09 MB PDF)