Abstract
Inositol auxotrophy (Ino− phenotype) in budding yeast has classically been associated with misregulation of INO1 and other genes involved in lipid metabolism. To identify all non-essential yeast genes that are necessary for growth in the absence of inositol, we carried out a genome-wide phenotypic screening for deletion mutants exhibiting Ino− phenotypes under one or more growth conditions. We report the identification of 419 genes, including 385 genes not previously reported, which exhibit this phenotype when deleted. The identified genes are involved in a wide range of cellular processes, but are particularly enriched in those affecting transcription, protein modification, membrane trafficking, diverse stress responses, and lipid metabolism. Among the Ino− mutants involved in stress response, many exhibited phenotypes that are strengthened at elevated temperature and/or when choline is present in the medium. The role of inositol in regulation of lipid metabolism and stress response signaling is discussed.
Keywords: Yeast, Inositol auxotrophy, Inositol, Lipid metabolism, Stress response
Introduction
Inositol serves as a precursor of inositol-containing lipids and inositol phosphates, which play essential roles in signaling, membrane trafficking, and membrane identity in eukaryotic cells (De Camilli et al. 1996; Lemmon 2003; Jesch and Henry 2005; Majerus and York 2009). Most eukaryotic organisms, including the budding yeast, Saccharomyces cerevisiae, are able to synthesize inositol de novo (Majumder et al. 1997; Michell 2007; Majerus and York 2009), starting with the conversion of glucose-6-phosphate to inositol-3-phosphate (Chen and Charalampous 1964a, b). The first report of isolation of inositol auxotrophs (Ino− mutants) in yeast identified ten independently segregating loci among 52 independently generated mutants (Culbertson and Henry 1975; Culbertson et al. 1976). The majority of these mutants proved to be alleles of INO1, later shown to be the structural gene encoding inositol-3-phosphate synthase (Donahue and Henry 1981; Dean-Johnson and Henry 1989). Two other loci, INO2 and INO4, each represented about 9% of the original Ino− mutants. The INO2 and INO4 loci were later shown to encode basic helix loop helix transcription factors that form a heterodimer that binds to the repeated upstream activation sequence, UASINO, found in the promoter of INO1 and coregulated genes (Carman and Henry 1989; Kodaki et al. 1991; Lopes and Henry 1991; Lopes et al. 1991; Bailis et al. 1992; Nikoloff et al. 1992; Ambroziak and Henry 1994; Nikoloff and Henry 1994; Bachhawat et al. 1995; Schüller et al. 1995; Schwank et al. 1995). Subsequently, mutants with more general defects in RNA polymerase II (RNA-Pol II) transcription were found to have Ino− phenotypes (Henry and Patton-Vogt 1998) due to low levels of INO1 expression (Nonet and Young 1989; Scafe et al. 1990a, b).
INO1 is the most highly regulated of a group of genes containing the inositol-sensitive upstream activating sequence, UASINO, in their promoters (Greenberg and Lopes 1996; Carman and Henry 1999). INO1 and other UASINO-containing genes are maximally repressed when both inositol and choline are present in the growth medium, but inositol alone is sufficient to achieve 30-fold or more repression of INO1 (Hirsch and Henry 1986). The addition of choline when inositol is present results in an additional several-fold repression of INO1 expression (Hirsch and Henry 1986; Lopes et al. 1991; Jesch and Henry 2005; Jesch et al. 2006). Phosphatidic acid (PA), a precursor of all glycerophospholipids in yeast, serves as the metabolic signal for derepression of INO1 and other UASINO-containing genes (Henry and Patton-Vogt 1998; Loewen et al. 2004). Opi1p, a known repressor of INO1 (White et al. 1991), is localized to the endoplasmic reticulum (ER) by direct binding to PA (Loewen et al. 2004) and to the VAP homolog Scs2p (Loewen et al. 2003). Upon addition of inositol to the growth medium of wild-type cells, phosphatidylinositol (PI) synthesis increases dramatically, resulting in consumption of PA. Loss of the ER pool of PA causes Opi1p translocation to the nucleus where it represses the transcription of INO1 and coregulated genes (Loewen et al. 2004). The INO1 gene is also repressed in stationary phase and under conditions of nitrogen or zinc depletion, even when inositol is absent (Carman and Zeimetz 1996; Griac and Henry 1999; Carman and Henry 2007).
Mutants defective in several stress signaling pathways, including the glucose response pathway (Hirschhorn et al. 1992; Ouyang et al. 1999; Shirra and Arndt 1999), the unfolded protein response (UPR) pathway (Nikawa and Yamashita 1992; Cox et al. 1993), and the protein kinase C–cell wall integrity (PKC–CWI) pathway (Nunez et al. 2008) exhibit Ino− phenotypes. Because of their Ino− phenotypes and/or reduced INO1 expression, it was assumed that the signaling pathways defective in these mutants were involved in regulating INO1 transcription. However, the role of each of these pathways in INO1 expression is complex. For example, while PKC–CWI mutants pkc1Δ, bck1Δ, and slt2Δ/mpk1Δ all exhibit Ino− phenotypes, the slt2Δ/mpk1Δ mutant is not defective in expression or regulation of INO1 or other UASINO containing genes, but instead exhibits alterations in PC and neutral lipid homeostasis (Nunez et al. 2008). Thus, while the PKC–CWI pathway is not involved in expression or regulation of INO1, it is essential for survival and lipid homeostasis in cells growing in the absence of inositol.
Interestingly, the Ino− phenotype of PCK-CWI mutants are strengthened at 37°C in the presence of choline (Nunez et al. 2008), growth conditions known to influence PC synthesis and turnover (Dowd et al. 2001). Growing cells in the presence of choline or shifting cells from 30 to 37°C causes increased flux through the CDP-choline pathway (Dowd et al. 2001), one of the two PC biosynthetic pathways present in yeast. Moreover, growth under these conditions results in increased deacylation of CDP-choline-derived PC, which may provide wild-type yeast with the flexibility to adjust their PC acyl chain composition in response to changing environmental conditions (Boumann et al. 2003). However, the membrane stress imposed by inositol limitation may be exacerbated by PC synthesis through the CDP-choline pathway (Fernandez-Murray et al. 2009).
Seeking to develop a comprehensive understanding of the diverse cellular processes that influence INO1 expression or are required for growth when inositol is limiting, we conducted a genome-wide phenotypic screening for mutants exhibiting Ino− phenotypes. Based upon our previous observation that the Ino− phenotype of PKC–CWI mutants is significantly intensified at higher temperatures in the presence of choline (Nunez et al. 2008), we carried out our current screen under these growth conditions. We report the identification of 385 mutations not previously reported to confer an Ino− phenotype. The addition of choline and/or growth at 37°C increased the strength of the Ino− phenotypes of a subset of these mutants. Mutations affecting key members of a number of stress response pathways, including the high-osmolarity glycerol (HOG) pathway, are among the newly identified Ino− mutants. Ino− phenotypes were also detected in mutants defective in transcription, protein modification, membrane trafficking, lipid metabolism, and diverse stress responses.
Materials and methods
Strains and plasmids
The homozygous diploid deletion collection [BY4743 strain background; MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0 (4741/4742)] was purchased originally from Research Genetics (now Invitrogen Corporation). This deletion set consists of a collection of 4,741 strains in which a single non-essential open reading frame (ORF) has been disrupted in each strain. Complete details regarding the collection construction can be found at sequence. http://www.sequence.stanford.edu/group/yeast_deletion_project/deletions3.html.
Other strains were constructed in the BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) strains, which are parent strains of BY4743. BY4741 and BY4742 were originally derived from the S288C strain background (Brachmann et al. 1998). We noted that the ino1Δ strain provided in the original Research Genetics collection has growth deficiencies that are not ascribable to deletion of the INO1 (S. Jesch, unpublished). Therefore, a single gene disruption in the INO1 was separately created in the BY4742 background strain by replacing targeted ORF with the HIS3 marker by PCR-mediated gene replacement using the pFA6a-His3MX6 template (kind gift from M. Longtine) as described previously (Longtine et al. 1998).
Media and growth conditions
YPD liquid medium consisted of 1% yeast extract, 2% bactopeptone, and 2% glucose. Chemically defined synthetic complete media lacking inositol and choline (I−C− media) used in this study was described by (Jesch et al. 2005), except that threonine was omitted. I−C− media contains (per liter): 20 g of glucose, 5 g of ammonium sulfate, 1 g of potassium phosphate, 0.5 of g magnesium sulfate, 0.1 g of sodium chloride, 0.1 g of calcium chloride, 0.5 mg of boric acid, 0.04 mg of cupric sulfate, 0.1 mg of potassium iodide, 0.2 mg of ferric chloride, 0.4 mg of manganese sulfate, 0.2 mg of sodium molybdate, 0.4 mg of zinc sulfate, 2 µg of biotin, 400 µg of calcium pantothenate, 2 µg of folic acid, 400 µg of niacin, 200 µg of p-aminobenzoic acid, 400 µg of pyridoxine hydrochloride, 200 µg of riboflavin, 400 µg of thiamine hydrochloride, 20 mg of adenine sulfate, 20 mg of arginine, 20 mg of histidine, 60 mg of leucine, 230 mg of lysine, 20 mg of methionine, 20 mg of tryptophan, and 40 mg of uracil. Where indicated, I− media was supplemented with 75 µM myo-inositol (I+) and/or 1 mM choline (C+). For example, I+C+ medium contains 75 µM inositol and 1 mM choline, whereas I−C− medium lacks both inositol and choline. Solid media contained 2% agar.
In preliminary experiments leading to the final screening, we observed that the presence of threonine in the I+C− medium as described by Jesch et al. (2005) increased the lag phase of growth of a large number of mutant strains derived from S288C, including those in the BY4743 background, by about 10–12 h at 30°C. When threonine was omitted, this lengthening of the lag phase was not observed. Therefore, in order to compare growth of strains over comparable time intervals with and without inositol, threonine was excluded from all synthetic media used in this study. Similar inhibition of growth in the presence of threonine in synthetic complete media was previously reported by Shirra et al. (2001).
The standard protocol used in the mutant screening was as follows: Each frozen master-plate was thawed completely, and cells were resuspended to homogeneity. A 96-pin microplate replicator (V & P Scientific, Inc) was used to transfer a 2-µl aliquot from each well in a master plate to a corresponding well containing 800 µl of YPD, plus G418 (200 µg/ml) in a deep-well microtiter plate. The deep-well plate was incubated for 2 days at 30°C. A 2-µl aliquots from each well from the YPD + G418 cell culture were then inoculated into a second deep-well plate containing 800 µl of YPD per well and incubated at 30°C for 15 h. A 1:50 dilution of the 15-h culture was carried out by transferring 2-µl aliquots from each well into 100 µl of I−C− media to dilute any carryover of inositol or choline from the YPD culture. Finally, 2 µl from each 100 µl dilution was transferred to Nunc Omni plates containing the following solid media: YPD, I+C+, I+C−, I−C−, I− C+, and 2% agar. Plating was carried out in duplicate to control for variability in inoculation volumes, and plates were incubated for 4 days at 30°C or 37°C. Each plate was photographed on days 2 and 4 using a digital camera to create a permanent record. In all cases, only wells that gave the same result on duplicate plates were scored in the final tally of Ino− phenotypes shown on Tables 1 and 2. Questionable cases and mutants of interest were reassessed in subsequent spotting assays. Growth of each strain on I−C− or I−C+ medium was scored visually relative to growth of the same strain on I+C− or I+C+ medium at the equivalent growth temperature, conducted independently by two different investigators in two separate blind screenings. On Tables 3 and S1, a score of “S” (strong) indicates no visible growth on media lacking inositol. A score “W” (weak) indicates some residual growth on I− medium, but substantially less than on I+ medium, while “VW” (very weak) indicates some growth on I− medium, but still visibly less than on I+ medium. Some strains had strong Ino− phenotypes at 30°C, but failed to grow at all on I+C+ media, I+C− media, and/or YPD at 37°C. Such strains are denoted as “NG” (no growth) at 37°C. This scoring system is described in detail in the legend of Table 3 and Table S1, where the assigned scores of all Ino− mutants identified in this study are listed. Several examples of Ino− phenotypes, as detected in the original screening, are shown in Fig. 1, as are several examples of variable growth on I−C+ medium that were not validated in the duplicates plates and/or in the subsequent spotting assays.
Table 1.
Number of mutations in yeast reported to confer an Ino− phenotype
Table 2.
Sumary of Ino− phenotypes of mutants carrying deletions in non-essential genes described in this study
| I−C− at 30°C |
I−C+ at 30°C |
I−C+ at 37°C |
|
|---|---|---|---|
| Strong | 31 | 45 | 243 |
| Weak | 74 | 106 | 134 |
| Very weak | 57 | 68 | 22 |
| Total | 162 | 219 | 399a |
Of the 419 mutants, 20 which had Ino− phenotypes at 30°C did not grow in either I+C+, I+C− or YPD medium at 37°C
Table 3.
Mutations that confer Ino− phenotypes
| ORF | Gene | I− C− 30°C |
I−C+ 30°C |
I−C+ 37°C |
References |
|---|---|---|---|---|---|
| A. Protein complexes involved in transcription regulation | |||||
| A1. Subunits of the SWI/SNF chromatin remodeling complex | |||||
| YOR290C | SNF2 | + | VW | VW | (Peterson and Herskowitz 1992) |
| YBR289W | SNF5 | S | S | S | (Peterson and Herskowitz 1992) |
| YHL025W | SNF6 | S | S | S | (Peterson and Herskowitz 1992) |
| YPL016W | SWI1a | NS | NS | NS | (Peterson and Herskowitz 1992) |
| YJL176C | SWI3 | W | S | S | (Peterson and Herskowitz 1992) |
| A2. Subunits of the remodel the structure of chromatin (RSC) complex | |||||
| YGR056W | RSC1 | VW | VW | S | This study |
| YLR357W | RSC2 | VW | W | S | This study |
| A3. Subunits and proteins associated with the Ino80 chromatin remodeling complex | |||||
| YNL059C | ARP5a | NG | NG | NG | (Shen et al. 2003a) |
| YOR141C | ARP8 | W | S | NG | (Shen et al. 2003a) |
| YFL013C | IES1 | + | + | S | This study |
| YNL215W | IES2 | W | W | NG | This study |
| YOR189W | IES4 | VW | W | S | This study |
| YER092W | IES5 | + | + | S | This study |
| YEL044W | IES6 | S | S | NG | (Fernandez-Murray et al. 2009) |
| YGL150C | INO80a | NS | NS | NS | (Ebbert et al. 1999) |
| YDL002C | NHP10 | + | + | S | This study |
| A4. Subunits and proteins associated with the ADA and SAGA complexes | |||||
| YDR448W | ADA2 | S | W | W | This study |
| YGR252W | GCN5 | VW | VW | VW | This study |
| YPL254W | HFI1a | NG | NG | NG | (Horiuchi et al. 1997) |
| YDR176W | NGG1 | VW | W | W | This study |
| YGL066W | SGF73 | W | W | S | This study |
| YOL148C | SPT20a | NG | NG | NG | (Roberts and Winston 1996) |
| YDR392W | SPT3 | VW | W | S | This study |
| YBR081C | SPT7a | NS | NS | NS | (Gansheroff et al. 1995) |
| YLR055C | SPT8 | + | + | S | This study |
| A5. Subunit of a histone deacetylase (HAD1) complex | |||||
| YPR179C | HAD3 | + | + | S | This study |
| A6. Subunits of the Set3 deacetylase complex | |||||
| YGL194C | HOS2 | W | W | S | (Cohen et al. 2008) |
| YOL068C | HST1 | VW | VW | VW | This study |
| YKR029C | SET3 | + | W | S | (Cohen et al. 2008) |
| YBR103W | SIF2 | W | W | S | (Cohen et al. 2008) |
| YCR033W | SNT1 | W | S | S | (Cohen et al. 2008) |
| A7. Subunits of the RNA polymerase II SRB/mediator complex | |||||
| YHR041C | SRB2 | S | S | S | (Koleske et al. 1992) |
| YNL236W | SIN4 | S | S | NG | This study |
| YGR104C | SRB5 | VW | VW | NG | (Betz et al. 2002) |
| YGL127C | SOH1 | VW | W | S | This study |
| YGL025C | PGD1 | S | S | S | This study |
| A8. Subunits and proteins associated with the Paf1 complex | |||||
| YLR418C | CDC73 | S | S | S | This study |
| YOL145C | CTR9a | NS | NS | NS | (Betz et al. 2002) |
| YOR123C | LEO1 | W | W | S | This study |
| YBR279W | PAF1 | S | S | NG | (Betz et al. 2002) |
| YGL244W | RTF1 | VW | W | S | (Betz et al. 2002) |
| A9. Subunits and proteins associated with the CCR4-NOT complex | |||||
| YKR036C | CAF4 | + | VW | W | This study |
| YAL021C | CCR4 | + | + | W | (Betz et al. 2002) |
| YGR092W | DBF2 | W | S | S | This study |
| A10. Subunits of the COMPASS (Set1C) complex | |||||
| YLR015W | BRE2 | + | + | VW | This study |
| YDR469W | SDC1 | VW | VW | W | This study |
| YPL138C | SPP1 | + | + | S | This study |
| YAR003W | SWD1 | + | + | S | This study |
| YBR175W | SWD3 | VW | + | S | This study |
| A11. Subunits of the H2B ubiquitination complex | |||||
| YDL074C | BRE1 | W | W | S | This study |
| YPL055C | LGE1 | + | W | S | This study |
| YGL058W | RAD6 | VW | W | S | This study |
| B. Ribosomal biogenesis pathway | |||||
| B1. Components of the small (40S) ribosomal subunit | |||||
| YLR048W | RPS0B | W | W | W | This study |
| YOR096W | RPS7A | VW | VW | W | This study |
| YBR189W | RPS9B | + | W | W | This study |
| YDL083C | RPS16B | + | + | S | This study |
| YOL121C | RPS19A | + | + | S | This study |
| B2. Components of the large (60S) ribosomal subunit | |||||
| YKL006W | RPL14A | VW | W | S | This study |
| YBL027W | RPL19B | + | VW | W | This study |
| YMR242C | RPL20A | + | VW | W | This study |
| YBR191W | RPL21A | + | VW | W | This study |
| YLR061W | RPL22A | W | W | VW | This study |
| YGL031C | RPL24A | + | + | VW | This study |
| YHR010W | RPL27A | W | S | S | This study |
| YDL191W | RPL35A | W | W | S | This study |
| YPR043W | RPL43A | VW | VW | VW | This study |
| B3. Component of small ribosomal subunit (SSU) processosome | |||||
| YOR078W | BUD21 | + | VW | VW | This study |
| B4. Methyltransferase required for rRNA processing and nuclear export of 40S ribosomal subunits | |||||
| YCR047C | BUD23 | + | + | S | This study |
| B5. Components of the ribosomal stalk | |||||
| YDL081C | RPP1A | + | + | S | This study |
| YDL130W | RPP1B | + | + | S | This study |
| B6. Component of eukaryotic eIF3 and required for processing of 20S pre-rRNA | |||||
| YLR192C | HCR1 | + | + | S | This study |
| B7. Elongation factor 2 (EF-2), also encoded by EFT1 | |||||
| YDR385W | EFT2 | + | + | S | This study |
| C. Protein modification pathways | |||||
| C1. Glycosyltransferases of the ER, involved in N-linked protein glycosylation | |||||
| YOR002W | ALG6 | + | S | This study | |
| YOR067C | ALG8 | + | S | This study | |
| YNL219C | ALG9 | + | + | W | This study |
| YGR227W | DIE2 | + | + | S | This study |
| C2. Glycosyltransferases of the Golgi involved in branched glycosylation | |||||
| YJR075W | HOC1 | + | + | W | This study |
| YBR015C | MNN2 | + | + | W | This study |
| C3. Glycosyltransferases of the ER, involved in O-linked protein mannosylation | |||||
| YDL095W | PMT1 | + | VW | S | This study |
| YAL023C | PMT2 | W | W | S | This study |
| C4. GPI-anchored glycoproteins | |||||
| YCR089W | FIG 2 | + | + | W | This study |
| YMR307W | GAS1 | + | VW | S | This study |
| YNL322C | KRE1 | + | + | S | This study |
| YER150W | SPI1 | + | + | W | This study |
| YEL040W | UTR2 | + | + | W | This study |
| C5. Subunits of the N-terminal acetyltransferase NatA (Nat1p, Ard1p, Nat5p) | |||||
| YHR013C | ARD1 | W | S | S | This study |
| YDL040C | NAT1 | VW | W | S | This study |
| C6. Subunit of a palmitoyltransferase complex composed of Shr5p and Erf2p | |||||
| YOL110W | SHR5 | + | + | S | This study |
| D. Membrane trafficking pathways | |||||
| D1. Proteins involved in translocation into the ER (Translocon) | |||||
| YLR292C | SEC72 | + | + | S | This study |
| YKL065C | YET1 | W | S | S | (Wilson and Barlowe 2010) |
| YDL072C | YET3 | S | S | S | (Wilson and Barlowe 2010) |
| YLR242C | ARV1 | W | W | S | This study |
| D2. Proteins involved in COPII vesicle trafficking from the ER to the Golgi | |||||
| YGL054C | ERV14 | W | W | S | This study |
| YLR208W | SEC13a | NS | NS | NS | (Gilstring et al. 1999) |
| YLR268W | SEC22 | W | W | S | This study |
| D3. Proteins involved in Golgi to ER retrival | |||||
| YGL020C | GET1 | + | W | S | This study |
| YER083C | GET2 | W | W | NG | This study |
| D4. Proteins involved in protein ubiquitination/deubiquitination and degradation via proteosome and vacuole | |||||
| YDR069C | DOA4 | S | S | NG | (Henry and Patton-Vogt 1998) |
| YMR276W | DSK2 | + | VW | S | This study |
| YEL037C | RAD23 | + | + | W | This study |
| YBL058W | SHP1 | + | + | W | This study |
| YLL039C | UBI4 | + | + | S | This study |
| YBR058C | UBP14 | + | VW | S | This study |
| YMR304W | UBP15 | + | + | W | This study |
| YFR010W | UBP6 | W | W | S | This study |
| YML013W | UBX2 | VW | VW | S | This study |
| YMR067C | UBX4 | + | + | S | This study |
| YDL190C | UFD2 | + | + | S | This study |
| YDR057W | YOS9 | + | + | W | This study |
| D5. Proteins involved in COPI vesicle intra-Golgi trafficking and the COG complex | |||||
| YNL051W | COG5 | VW | VW | VW | This study |
| YNL041C | COG6 | W | W | W | This study |
| YML071C | COG8 | W | W | W | This study |
| YOR216C | RUD3 | VW | W | W | This study |
| YOL107W | YOL107 W | + | + | S | This study |
| D6. Proteins involved in late secretory trafficking | |||||
| YIL044C | AGE2 | + | + | S | This study |
| YDL192W | ARF1 | + | W | S | This study |
| YCR094W | CDC50 | + | + | W | This study |
| YAL026C | DRS2 | + | + | S | This study |
| YER122C | GLO3 | W | VW | W | This study |
| YGR166W | KRE11 | + | + | S | This study |
| YNL323W | LEM3 | + | VW | W | This study |
| YJL204C | RCY1 | + | VW | S | This study |
| YMR079W | SEC14a | NS | NS | NS | (Kearns et al. 1997) |
| YEL048C | TCA17 | + | + | S | This study |
| D7. Proteins involved in vacuole targeting | |||||
| YOR106W | VAM3 | + | + | S | This study |
| YDL077C | VAM6 | + | + | W | This study |
| YDR080W | VPS41 | + | VW | W | This study |
| YPR139C | VPS66 | + | W | S | This study |
| D8. Proteins involved in retrograde trafficking, from early and late endosomes, to the TGN | |||||
| YLR039C | RIC1 | VW | W | NG | (Kodaki et al. 1995) |
| YOL018C | TLG2 | VW | VW | VW | This study |
| YLR262C | YPT6 | + | + | VW | This study |
| D9. Proteins involved in endocytic trafficking, from plasma membrane to the MVB and vacuole | |||||
| YPL065W | VPS28 | + | + | S | This study |
| YDR486C | VPS60 | + | + | S | This study |
| D10. Subunits of the vacuolar H(+)-ATPase complex and associated proteins with vacuolar acidification | |||||
| YEL027W | CUP5 | S | S | S | This study |
| YMR123W | PKR1 | + | VW | W | This study |
| YHR026W | PPA1 | S | S | S | This study |
| YDL185W | TFP1 | W | S | S | This study |
| YPR036W | VMA13 | S | S | S | This study |
| YBR127C | VMA2 | S | S | S | This study |
| YGR105W | VMA21 | VW | W | S | This study |
| YHR060W | VMA22 | W | W | S | This study |
| YGR020C | VMA7 | VW | W | S | This study |
| YEL051W | VMA8 | S | S | S | This study |
| YOR270C | VPH1 | + | + | W | This study |
| YKL119C | VPH2 | W | S | S | This study |
| E. Stress response pathways | |||||
| E1. Components and associated proteins involved in the PKC–CWI signaling pathway | |||||
| YDL203C | ACK1 | + | + | S | This study |
| YJL095W | BCK1 | S | S | S | (Nunez et al. 2008) |
| YER167W | BCK2 | + | + | W | This study |
| YLR342W | FKS1 | VW | VW | S | This study |
| YLR332W | MID2 | + | VW | W | This study |
| YPL140C | MKK2 | + | + | W | This study |
| YBL105C | PKC1a | NS | NS | NS | (Nunez et al. 2008) |
| YPL089C | RLM1 | + | VW | S | This study |
| YLR371W | ROM2 | VW | W | S | This study |
| YDR389W | SAC7 | W | W | S | This study |
| YOR008C | SLG1 | VW | W | S | This study |
| YHR030C | SLT2 | VW | S | S | (Nunez et al. 2008) |
| YGR229C | SMI1 | + | + | S | This study |
| YLL021W | SPA2 | + | + | S | This study |
| YOL109W | ZEO1 | + | + | S | This study |
| E2. Components and associated proteins involved in the HOG signaling pathway | |||||
| YLR113W | HOG1 | W | W | W | This study |
| YGR014W | MSB2 | + | VW | W | This study |
| YDR162C | NBP2 | + | + | S | This study |
| YDL006W | PTC1 | W | W | NG | This study |
| YER118C | SHO1 | + | + | W | This study |
| YCL032W | STE50 | + | + | S | This study |
| E3. Components and associated proteins involved in the cell cycle progression | |||||
| YBR135W | CKS1a | NS | NS | NS | (Yu and Reed 2004) |
| YAL040C | CLN3 | + | + | W | This study |
| YBR133C | HSL7 | + | + | S | This study |
| YLR079W | SIC1 | + | W | S | This study |
| E4. Components and associated proteins involved in the TOR signaling pathway | |||||
| YMR068W | AVO2 | + | + | S | This study |
| YKR007W | MEH1 | + | + | S | This study |
| YIL105C | SLM1 | + | + | S | This study |
| YBR077C | SLM4 | + | + | W | This study |
| YJR066W | TOR1 | + | + | S | This study |
| YDL077C | VAM6 | + | + | W | This study |
| YMR104C | YPK2 | W | W | W | This study |
| E5. Components and associated proteins involved in the calcineurin signaling pathway | |||||
| YKL190W | CNB1 | VW | VW | W | This study |
| YNL307C | MCK1 | + | VW | W | This study |
| YHR206W | SKN7 | + | + | S | This study |
| E6. Components and associated proteins involved in the cAMP-PKA signaling pathway | |||||
| YER177W | BMH1 | + | VW | S | This study |
| YOR371C | GPB1 | + | + | W | This study |
| YAL056W | GPB2 | + | + | S | This study |
| YMR016C | SOK2 | + | + | S | This study |
| F. Lipid and glycerol metabolic pathways | |||||
| F1. Proteins involved in phospholipid metabolism | |||||
| YJL153C | INO1 | S | S | S | (Culbertson and Henry 1975) |
| YDR123C | INO2 | S | S | S | (Donahue and Henry 1981) |
| YOL108C | INO4 | S | S | S | (Donahue and Henry 1981) |
| YML059C | NTE1 | + | W | S | (Nunez et al. 2008) |
| YPR113W | PIS1a | NS | NS | NS | (Nikawa et al. 1987) |
| YER120W | SCS2 | W | W | S | (Kagiwada et al. 1998) |
| YGL126W | SCS3 | VW | S | S | (Hosaka et al. 1994) |
| F2. Proteins involved in glycerol metabolism | |||||
| YLL043W | FPS1 | + | VW | NG | This study |
| YDL022W | GPD1 | + | + | W | This study |
| YOL059W | GPD2 | W | W | W | This study |
| YHR104W | GRE3 | VW | W | S | This study |
| YBL011W | SCT1 | VW | W | S | This study |
| YDR368W | YPR1 | + | + | W | This study |
| F3. Proteins involved in phosphoinositides and inositol polyphosphates metabolism | |||||
| YDR173C | ARG82 | S | S | NG | This study |
| YFR019W | FAB1 | S | S | NG | This study |
| YDR017C | KCS1 | W | W | S | This study |
| YKL212W | SAC1 | S | S | S | (Whitters et al. 1993) |
| F4. Proteins involved in sphingolipids and ceramides metabolism | |||||
| YBR183W | YPC1 | VW | + | W | This study |
| YDR297W | SUR2 | + | + | W | This study |
| YJL134W | LCB3 | VW | W | S | This study |
| YDR294C | DPL1 | + | + | W | This study |
| YMR272C | SCS7 | VW | W | VW | This study |
| F5. Proteins involved in sterols metabolism | |||||
| YMR202W | ERG2 | W | W | S | This study |
| YLR056W | ERG3 | W | W | S | This study |
| YGL012W | ERG4 | + | W | S | This study |
| YMR015C | ERG5 | + | + | S | This study |
| YML008C | ERG6 | W | W | S | This study |
Table 3 contains only the mutations conferring an Ino− phenotype that were mentioned in the “Introduction”, “Results” or “Discussion” sections, in the approximate order in which they are described in the text. A complete list of all mutants exhibiting inositol auxotrphy detected in this study or previously reported is contained in Table S1. Phenotypes were scored as follows: mutants that grew well on I+C− medium at a given temperature, but whose growth was visibly weaker on either I−C− or I−C+ media, were scored as very weak (VW). Mutants exhibiting very reduced but still detectable growth in I−C− or I−C+ media were scored as weak (W). Mutants exhibiting no visible growth on I−C− or I−C+ media were scored as strong (S) (See Fig. 1). A score of “+” indicates no growth reduction in any inositol lacking media, and compared to growth on I+C− medium at the corresponding temperature. A score of “NS” indicates that the mutant conferring the Ino− phenotype was not present in the homozygous diploid strain collection, and hence was not screened. A score of “NG” (no growth) indicates that the deletion mutant from the homozygous diploid collection failed to grow in either I+C+, I+C− and/or YPD medium at a given temperature. Underlined gene names indicate deletions of essential genes for which conditional alleles have been reported to confer an Ino− phenotype. References are provided for those mutations previously shown to confer an Ino− phenotype
Mutations previously shown to confer an Ino− phenotype that were not confirmed in this study
Fig. 1.
Genome-wide screen for the Ino− phenotype. Representative images of 1 out of a total of 54 96-well microtiter plates from the screen. a A set of mutant strains from the primary screening that includes the doa4Δ, ino2Δ, vps41Δ, ydr049wΔ, mutants on I+C+, and I−C+ media at 30°C. b Same set of mutant strains grown on I+C+ and I−C+ media at 37°C. The detailed screening protocol is described in “Materials and methods” and complete description of the phenotypes of these mutants is presented in Table S1. The position of each mutant colony, which confers an Ino− phenotype is surrounded by a square. Colony positions surrounded by a circle indicate lack of growth in I+C+ medium, scored as no growth (NG) at 37°C in Tables 3, S1. Positions surrounded by a diamond indicates a growth defect on I−C+ medium that were not validated on a duplicate plate or in a subsequent spotting assay
Spotting assay
Inositol auxotrophy was confirmed for a subset of 73 strains detected in the original screen as having Ino− phenotypes under one or more growth conditions using a standard spotting assay. Overnight cultures were grown in I+C− medium at 30°C. The cultures were diluted back to OD600 = 0.15 in 10 ml of the same medium and allowed to grow to mid-logarithmic phase at 30°C. Cells were harvested at OD600 = 0.5 and washed in I−C− medium at 30°C. A series of tenfold dilutions were made in a microtiter plate and a 5-µl aliquot of each dilution was spotted onto I+C−, I+C+, I−C−, and I−C+ media with 2% agar. The plates were incubated at 30°C and at 37°C. Strains presenting a growth defect in I−C− or I−C+ medium in comparison with their growth in I+C− or I+C+ medium were considered inositol auxotrophs.
Mutant identification and functional classification
Identification of the gene deleted in each strain that exhibited an Ino− phenotype was accomplished by comparison with a reference spreadsheet of the homozygous diploid deletion collection supplied by Invitrogen Corporation. Description and gene name information about particular ORFs was obtained from the Saccharomyces Genome Database (SGD) (Cherry et al. 1998; Issel-Tarver et al. 2002; Hirschman et al. 2006) (http://www.yeastgenome.org/). Genes that confer an Ino− phenotype when deleted were functionally classified (clustered) by biological process using the Gene Ontology (GO) Slim mapper tool in SGD (Ashburner et al. 2000) (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl). The P value was calculated by performing a hypergeometric test followed by Benjamini and Hochberg false discovery rate correction (Benjamini and Yekutieli 2001; Maere et al. 2005). The P value is defined as the probability of seeing at least x number of genes out of the total n genes in the list annotated to a particular GO term, with respect to the proportion of genes in the whole genome that are annotated for that GO Term.
Results
419 mutants from the genome-wide collection of deletions of non-essential genes confer Ino− phenotypes
We screened the entire set of 4,741 strains from the Saccharomyces cerevisiae homozygous diploid knockout collection for inositol auxotrophy. Each yeast strain that was screened carried a unique deletion of a single, non-essential gene. The goal of our screen was to identify all mutants present in the knockout collection that exhibit inositol auxotrophy under one or more of the following three conditions: I−C− and I−C+ media at 30°C, and I−C+ medium at 37°C. These growth conditions were chosen based on our previous finding that certain mutants exhibit Ino− phenotypes that are progressively strengthened by growth at higher temperatures and/or in the presence of choline (Nunez et al. 2008). For example, the lcb3Δ mutant, which lacks the sphingolipid long-chain base-1-phosphate phosphatase, grew well on I+C− medium at 30°C but grew visibly weaker on I−C− medium (scored as very weak, VW). In medium containing choline (I−C+), even greater growth reduction was observed (scored as weak, W) and on I−C+ medium at 37°C, no visible growth was observed (scored as strong, S).
The total number of mutants displaying an Ino− phenotype increased when grown in the presence of choline and at 37°C. In I−C− medium at 30°C, we identified 162 Ino− mutants, including 27 already reported in the literature. This number increased to 219 Ino− mutants in I−C+ medium at 30°C and 399 Ino− mutants in I−C+ medium at 37°C (Table 2). Virtually all mutants that failed to grow on I−C− medium at 30°C also failed to grow at 37°C on I−C+ medium. A small number of mutants exhibiting Ino− phenotypes at 30°C failed to grow on any media at 37°C, including I+C+ and YPD. The full list of all the mutants identified in the screen is listed in Table S1, and all mutants described in the “Results” or “Discussion” are listed in Table 3.
Figure 1 illustrates a typical set of plates from the screen at 30°C and 37°C, comparing growth on I−C+ medium to growth on I+C+ medium. Panel A shows the Ino− phenotypes at 30°C of the ino2Δ and doa4Δ deletion mutants, both of which were previously reported (Donahue and Henry 1981; Henry and Patton-Vogt 1998). Panel B, shows phenotypes at 37°C of vps41Δ and ydr049wΔ mutants reported in this study to exhibit Ino− phenotypes. Full descriptions of the phenotypes of these mutants, including phenotype scoring under different growth conditions, can be found in Table S1.
In all, a total of 419 strains carrying deletions of non-essential genes were observed to exhibit growth impairment of varying degrees in the absence of inositol (Ino− phenotype). Among the 419 Ino− mutants identified, 385 had not previously been reported to have this phenotype (Table 1). The screening also confirmed 34 Ino− mutants previously reported to exhibit an Ino− phenotype, providing validation of the methods used in the current study (Tables 1, S1). In addition, rescreening of 73 randomly selected Ino− mutants identified in the current study confirmed the inositol auxotrophy phenotype of all 73 strains. This indicates that the false-positive rate among the Ino− mutants identified in our screen is below 2%. Another nine mutants previously reported to have an Ino− phenotype could not be confirmed in our screening. One such mutant did not exhibit an Ino− phenotype under any of the conditions tested, while the remaining eight mutants either failed to grow in YPD medium or were absent from the collection. These nine mutants are listed in Table S1, as are all of the previously reported mutants with Ino− phenotypes confirmed in this study.
Gene ontology classification of non-essential gene deletions conferring Ino− phenotypes
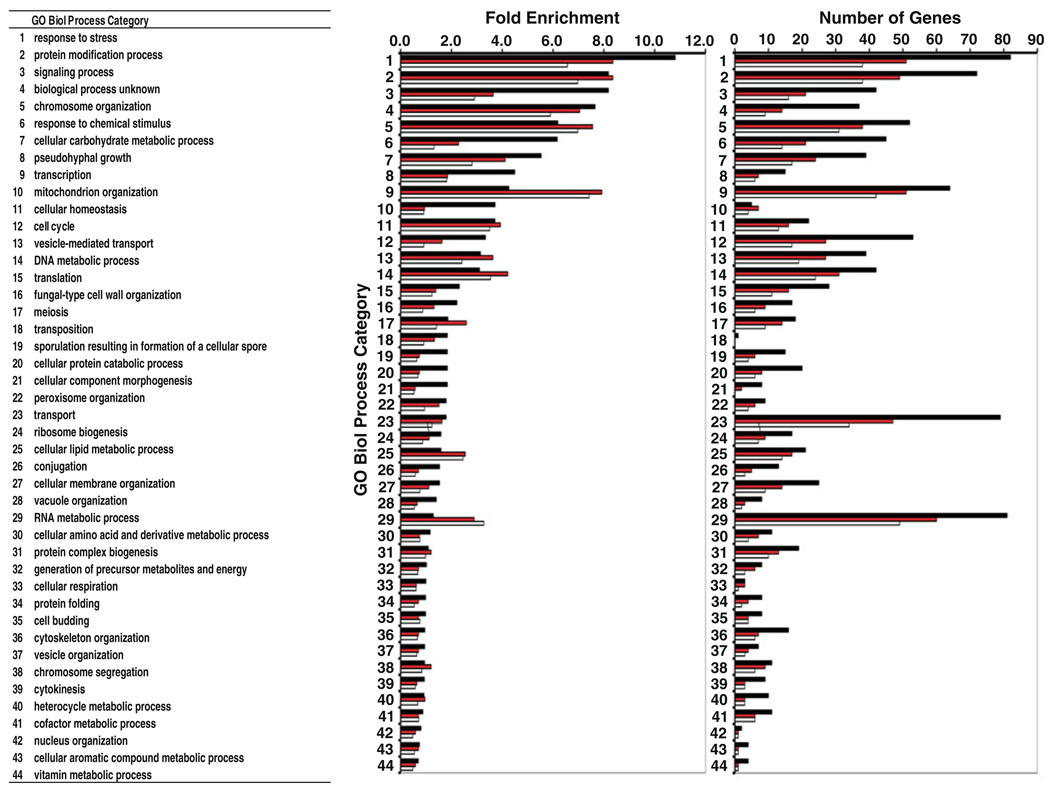
Each non-essential gene mutation that exhibited an Ino− phenotype was grouped according to the gene ontology (GO) Slim classification (Ashburner et al. 2000) into 44 GO categories and ranked according to GO category enrichment for each growth condition (Fig. 2; Table S2). The current screening revealed that many categories of mutants showed progressive strengthening of their Ino− phenotypes in response to the exposure to exogenous choline and elevation of growth temperature to 37°C. For example, genes in the GO category “response to stress” were enriched relative to other categories following addition of choline and shift of the growth temperature to 37°C, suggesting that stress response pathways are critical for survival in cells grown without inositol and at higher temperatures. Likewise, genes present in the GO category “transcription” were highly enriched only at 30°C. Overall, the greatest enrichment of Ino− mutants was observed in the following GO categories: response to stress, protein modification process, and signaling process (Fig. 2; Table S2). Phenotypes of mutants specifically discussed in the remainder of the “Results” are documented in Table 3 in the approximate order of the groupings described below.
Fig. 2.
Clustering of non-essential genes that confer Ino− phenotypes by enrichment in GO biological process categories. Ino− mutants were classified according to the GO Slim classification as described in the “Materials and methods”. In each category, color bars indicate the fold enrichment or number of mutant strains conferring Ino− phenotypes that were detected under each growth condition: I−C− medium at 30°C (white bars), I−C+ medium at 30°C (red bars) and I−C+ medium at 37°C (black bars). Fold enrichment is defined as −Log10 (adjusted P value). The P value was calculated by performing a hypergeometric test followed by Benjamini and Hochberg false discovery rate correction (Benjamini and Yekutieli 2001; Maere et al. 2005) as described in “Materials and methods”. A complete list of non-essential genes in each GO category is provided in Table S2
Ino− phenotypes are associated with mutations affecting general RNA-pol II mediated transcription
Ino− phenotypes associated with 21 mutants defective in subunits of RNA-Pol II (Nonet and Young 1989; Scafe et al. 1990a, b), the TATA binding protein (Arndt et al. 1995), and subunits of various transcriptional coactivation, and nucleosome remodeling complexes were previously reported. We report Ino− phenotypes associated with 30 additional mutants affecting these complexes and several other related transcriptional complexes. The cause of the Ino− phenotype in these strains is most likely due to reduced transcription of INO1, whose expression is necessary in cells growing in the absence of inositol. Many of these mutants are defective in genes clustering in the GO category “transcription” as well as “RNA metabolic process,” “Response to stress,” “Protein modification,” “Chromosome organization,” and “DNA metabolic process” (Fig. 2; Table S2).
Ino− phenotypes have been described for mutants defective in nucleosome remodeling, including SWI/SNF (Peterson and Herskowitz 1992) and INO80 chromatin remodeling complexes (Ebbert et al. 1999; Shen et al. 2003a; Fernandez-Murray et al. 2009) (Tables 3, S1). We report Ino− phenotypes in rsc1Δ and rsc2Δ mutants, affecting the Remodel the Structure of Chromatin (RSC) complex (Chai et al. 2002; Bungard et al. 2004), which is related to SWI/SNF complex (Tables 3, S1) and ies1Δ, ies2Δ, ies4Δ, ies5Δ, and nhp10Δ mutants, which affect INO80 complex (Tables 3, S1).
Mutants defective in histone acetylation were previously reported to exhibit Ino− phenotype, including mutants in the SAGA (Spt-Ada-Gcn5 acetyltransferase) complex (Gansheroff et al. 1995; Roberts and Winston 1996; Horiuchi et al. 1997). In the current screening, we report that six additional SAGA mutants, ada2Δ, ngg1Δ, gcn5Δ, sgf73Δ, spt3Δ, and spt8Δ, have Ino− phenotypes (Tables 3, S1). Mutations affecting the SET complex, which is involved in histone deacetylation, RNA-Pol II transcription and repression of sporulation genes (Pijnappel et al. 2001) and response to secretory stress (Cohen et al. 2008) were reported to have an Ino− phenotype. We confirmed that set1Δ, hos2Δ, snt1Δ, and sif2 exhibit a weak Ino− phenotype at 30°C and report a strong Ino− phenotype for these mutants at in I−C+ at 37°C. We also observed an Ino− phenotype for hda3Δ, defective in a subunit of HDA1, a histone deacetylation complex (Carman and Zeimetz 1996; Wu et al. 2001) (Tables 3, S1).
In addition, we report Ino− phenotypes for mutants in complexes affecting ubiquitination and methylation of histones. These mutants include swd3Δ, bre2Δ, sdc1Δ, spp1Δ, and swd1Δ, defective in subunits of the COMPASS complex involved in histone H3 methylation (Miller et al. 2001; Krogan et al. 2002) (Tables 3, S1). The Swd3p subunit is shared with the related histone H2B ubiquitination complex (Weake and Workman 2008), and mutations in three additional histone H2B ubiquitination subunits, Bre1p, Lge1p, and Rad6p, also confer Ino− phenotypes (Tables 3, S1).
Mutations in several genes that interact with the carboxy-terminal domain of RNA-Pol II were previously reported to have Ino− phenotypes (Koleske et al. 1992; Betz et al. 2002). We report that mutations sin4Δ/med16Δ, srb5Δ/med18Δ, soh1Δ/med31Δ, and pgd1Δ/med3Δ, affecting the SRB/MED transactivation complex, cdc73Δ and leo1Δ mutations affecting the Paf1 complex, and dbf2Δ and caf4Δ mutants affecting CCR4-NOT subunits (Tables 3, S1) exhibit Ino− phenotypes (Tables 3, S1).
Mutations affecting protein modification pathways confer Ino− phenotypes
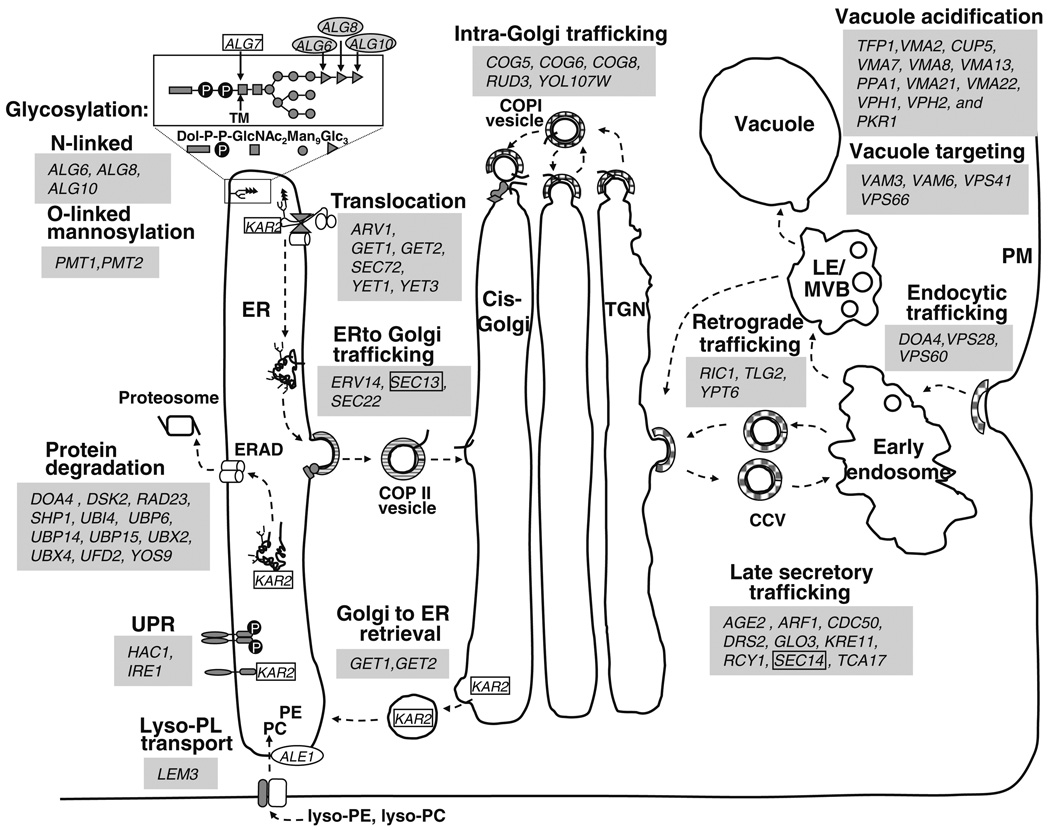
ALG7, which encodes UDP-N-acetylglucosamine-1P transferase (Hanson and Lester 1980; Hanson and Lester 1982), is essential for viability and is necessary for the synthesis of branched oligosaccharides during N-linked glycoprotein biosynthesis (Lehle et al. 2006) (Fig. 3). The reaction catalyzed by Alg7p is inhibited by tunicamycin (Takatsuki and Tamura 1971), a drug well known for its effect in triggering UPR signaling in the ER (Cox et al. 1993). Moreover, wild-type cells treated with sub-lethal doses of tunicamycin exhibit inositol auxotrophy (Fernandez-Murray et al. 2009). Deletion mutations affecting several subsequent steps in the N-linked glycosylation pathway confer Ino− phenotypes, including alg6Δ, alg8Δ and die2Δ/alg10Δ, and alg9Δ (Fig. 3; Tables 3, S1) as well as hoc1Δ (Neiman et al. 1997) and mnn2Δ (Yip et al. 1994) mutations, affecting protein glycosylation in the Golgi (Tables 3, S1). In addition, we found that the pmt1Δ and pmt2Δ mutations, which affect protein O-mannosylation, also confer Ino− phenotypes (Tables 3, S1). Pmt2p is specifically required for O-mannosylation of Mid2p and Slg1p/Wsc1p (Philip and Levin 2001), both of which are sensors in the PKC–CWI pathway (Fig. 4a), to be described in a subsequent section.
Fig. 3.
Mutations in non-essential genes involved in membrane trafficking result in Ino− phenotypes. Mutations in genes enclosed in gray boxes confer Ino− phenotypes. Essential genes are enclosed by a rectangle, for example, KAR2. Conditional alleles that confer Ino− phenotypes have been reported for essential genes in gray boxes. ER endoplasmic reticulum, UPR unfolded protein response, ERAD ER-associated degradation, Lyso-PL lyso-phospholipids, TGN trans golgi network, PM plasma membrane, TM tunicamycin, LE/MVB late endosome/multivesicular bodies
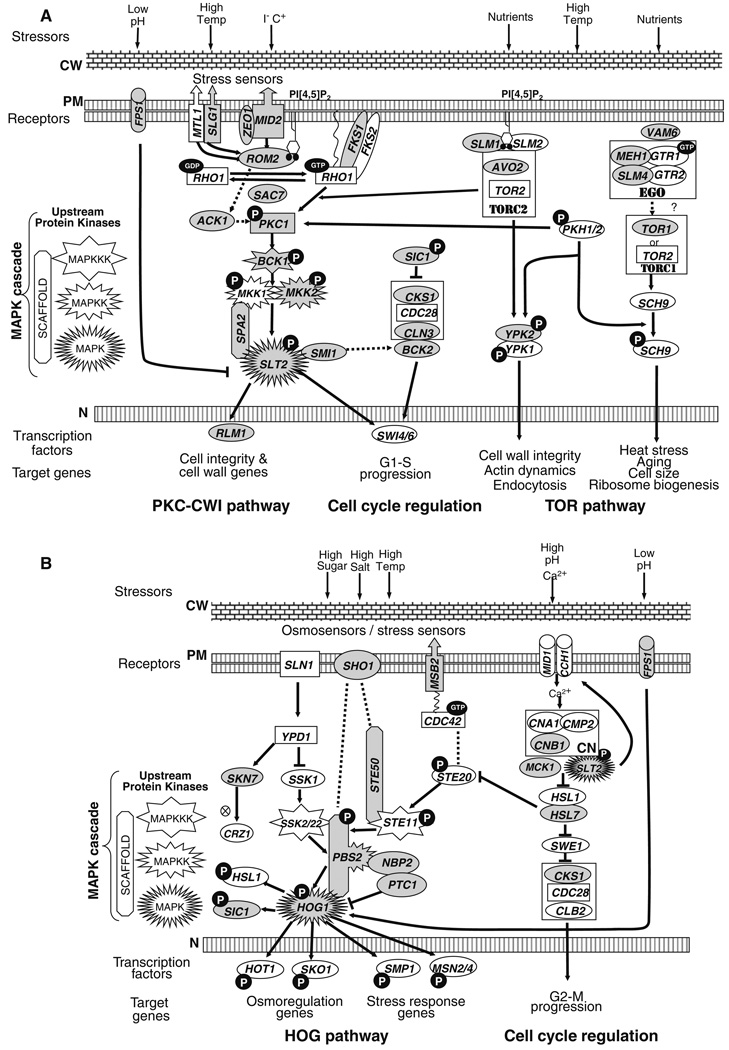
Fig. 4.
Mutations in non-essential genes affecting stress response pathways confer Ino− phenotypes. a Protein kinase C–Cell Wall integrity (PKC–CWI) pathway, signaling required for G1-S progression and Target of Rapamycin (TOR) pathway. b High osmolarity glycerol (HOG) pathway and signaling required for G2-M progression. Mutations in genes shaded in gray confer an Ino− phenotype. Genes enclosed in a rectangle are essential. Genes enclosed in a cylinder encode pore-forming proteins. Genes enclosed in a box with an upward arrow encode glycosylated transmembrane sensors. A black circle with a white letter “P” represents a phosphorylated target. A black oval with white letters “GTP” indicates a GTP-binding protein. Genes encoding mitogen activated protein kinase (MAPK) cascade components (kinases and scaffold proteins) are indicated with star symbols and elongated octagons according to the key on the left side of the figure. CW cell wall, PM plasma membrane, N nucleus
Biosynthesis and addition of the glycosylphosphatidylinositol (GPI) anchor to target proteins in the ER involves modification of PI by step-wise addition of sugars and ethanolamine phosphate (Orlean and Menon 2007; Pittet and Conzelmann 2007). Although mutations in any of the non-essential genes of this pathway did not result in Ino− phenotypes, deletion of the ARV1 gene, required for the efficient delivery of an early GPI intermediate to the first mannosyltransferase involved in GPI synthesis (Kajiwara et al. 2008), results in an Ino− phenotype (Tables 3, S1). In addition, deletion of several structural genes encoding specific GPI-anchored proteins result in an Ino− phenotype, including gas1Δ, kre1Δ, utr2Δ, fig2Δ, and spi1Δ (Tables 3, S1).
Ino− phenotypes were also detected in mutants defective in Nα-terminal protein acetylation (Driessen et al. 1985; Polevoda and Sherman 2002), a modification that affects about half of the abundant proteins in yeast (Polevoda and Sherman 2003). Mutations in two subunits of NatA N-terminal acetyltransferase complex, Ard1p and Nat1p, confer Ino− phenotypes (Tables 3, S1). Significantly, Ino1p is partially acetylated and NatA is responsible for this activity (Perrot et al. 2008). Finally, the shr5Δ mutant, defective in protein palmitoylation (Nadolski and Linder 2007) has an Ino− phenotype (Tables 3, S1). Shr5p is a subunit of a palmitoyltransferase, that together with Erf2p, palmitoylates Ras2p (Nadolski and Linder 2007), a protein that plays a significant role in several important signaling pathways (Fig. S1).
Mutations resulting in defects in membrane trafficking pathways confer Ino− phenotypes
Several temperature sensitive mutants, defective in essential genes involved in the secretory pathway, have previously been reported to have Ino− phenotypes at temperatures that are semipermissive for growth (Tables 3, S1). These include sec13-1 (Gilstring et al. 1999; Chang et al. 2004), defective in a COPII vesicle component required for budding from the ER (Barlowe 2002) and sec14-1 (Kearns et al. 1997; Chang et al. 2004), defective in a PI/PC transporter (Skinner et al. 1993) required for exit from the Golgi (Novick et al. 1980). We report that deletions of 51 additional non-essential genes encoding products involved in membrane trafficking and organelle homeostasis that exhibit Ino− phenotypes (Fig. 3). In addition, we also confirmed four non-essential genes involved in membrane trafficking previously reported to have an Ino− phenotype (Table 3; Fig. 3). These genes are largely grouped within the GO category “transport” but are also found in the categories “cell cycle” and “vesicle mediated transport.” (Fig. 2; Table S2).
We found mutations affecting multiple steps spanning early, middle, and late steps in the secretory pathway pathway that exhibited Ino− phenotypes, including two mutants in translocation of nascent proteins into the ER, two mutants in ER-to-Golgi trafficking, two mutants in Golgi-to-ER retrieval, five mutants intra-Golgi trafficking, and nine mutants in trafficking out of the Golgi, and four mutants involved in protein trafficking to the vacuole (Fig. 3; Table 3). We also found four mutants involved in endocytic trafficking that exhibit Ino− phenotypes (Fig. 3; Table 3). Mutations in organelle homeostasis also exhibited Ino− phenotypes, including 11 mutants in ER-associated protein degradation (ERAD) pathway and 12 mutants in vacuolar acidification (Fig. 3; Table 3). While this paper was under review, a similar study reported Ino− phenotypes associated with mutations in genes involved in vacuole acidification (Young et al. 2010).
Mutants with defects in a number of major stress response pathways exhibit Ino− phenotypes
Prior to this screening, Ino− phenotypes had been reported for mutants in several signaling pathways, including the UPR (Nikawa and Yamashita 1992) the PKC–CWI pathway (Nunez et al. 2008), and the Glucose Response Pathway (Hirschhorn et al. 1992; Shirra et al. 2001). The current study expands the number of mutants shown to have Ino− phenotypes in certain stress response pathways, including the PKC–CWI pathway (Fig. 4a). Mutants exhibiting Ino− phenotypes are also reported here for the first time in the HOG pathway (Fig. 4b): the target of rapamycin (TOR) pathway (Fig. 4a), the cAMP-Protein Kinase A (PKA) and the calcineurin, and filamentous growth pathways (Fig. S1; Tables 3, S1). These pathways collectively respond to stress caused by extreme growth conditions and also respond in an interconnected fashion to stress via pathway cross talk (Fuchs and Mylonakis 2009). The majority of mutations in these pathways conferring Ino− phenotypes are associated with the GO categories, “response to stress,” “RNA metabolism,” “protein modification,” “transcription,” and “response to chemical stimulus” (Fig. 2; Table S2).
The HOG pathway, which contains a mitogen-activated protein kinase (MAPK) cascade (Fig. 4b), responds to multiple stress conditions, including high temperature (Winkler et al. 2002), high osmolarity (Hohmann et al. 2007), oxidative stress (Staleva et al. 2004), and high salt (Posas et al. 2000). We found that deletion of the HOG1 gene, which encodes a MAP kinase, confers an Ino− phenotype, as do the HOG pathway mutations, msb2Δ, sho1Δ, ste50Δ, ptc1Δ, and nbp2Δ (Fig. 4b; Tables 3, S1). Deletion of the SIC1 gene, a Hog1p target that is a negative regulator of the Cdc28p-Cln3p cyclin-dependent kinase complex (Fig. 4a, b) results in an Ino− phenotype (Tables 3, S1). In total mutations in four genes that promote cross-talk between cell cycle regulation and the HOG pathway exhibit Ino− phenotypes.
Previously we reported the Ino− phenotypes of mutants in the well-characterized PKC–CWI pathway (Levin 2005), including pkc1Δ, slt2Δ/mpk1Δ, bck1Δ, and rlm1Δ (Fig. 4a) (Nunez et al. 2008; Fernandez-Murray et al. 2009). However, in the course of this screening, many additional mutants related to PKC–CWI signaling were found to have Ino− phenotypes. These mutants include mkk2Δ defective in one of two redundant MAPK kinases, as well as mutants affecting upstream sensors and regulatory components, slg1Δ/wsc1Δ, mid2Δ, zeo1Δ, rom2Δ, sac7Δ, ack1Δ, and spa2Δ, depicted in Fig. 4a and documented in Tables 3 and S1. Other Ino− mutants related to PKC–CWI signaling detected in this screen include fks1Δ, smi1Δ/knr4Δ, bck2Δ, drs2Δ, ypk2Δ, and cln3Δ (Fig. 4a).
The TOR pathway (Fig. 4a) regulates metabolism and cellular growth in response to nutrient availability, as well as environmental stress, via two distinct multiprotein complexes, TORC1 and TORC2 (Torres et al. 2002; Wullschleger et al. 2006). Deletion of the TOR1 gene confers an Ino− phenotype, while TOR2 is essential (Giaever et al. 2002). However, slm1Δ and avo1Δ mutations, which affect the activity of the TORC2 complex, also have Ino− phenotypes (Fig. 4a; Table 3, S1). The TORC1 Complex has been implicated in response to heat stress, aging, cell size, and ribosome biogenesis (Morano and Thiele 1999; Kaeberlein et al. 2005; Wei et al. 2009) as well as cell wall integrity (Heinisch et al. 1999; Torres et al. 2002; Ho et al. 2005) (Fig. 4a). We also found that mutations in calcineurin, which is involved in calcium stress response (Zhang and Rao 2008) results in Ino− phenotypes (Figs. 4b, S1; Tables 3, S1). The interconnected cyclic PKA and filamentous growth pathways (Fig. S1) also play major roles in stress resistance to high temperature and nutrient availability. The gpb2Δ/krh1Δ, gpb1Δ/krh2Δ, and sok2Δ mutants affecting these pathways display Ino− phenotypes (Fig. S1).
Mutations in genes involved in the metabolism of diverse lipid classes, including sphingolipids, ceramides, phospholipids, fatty acids, and sterols confer Ino− phenotypes
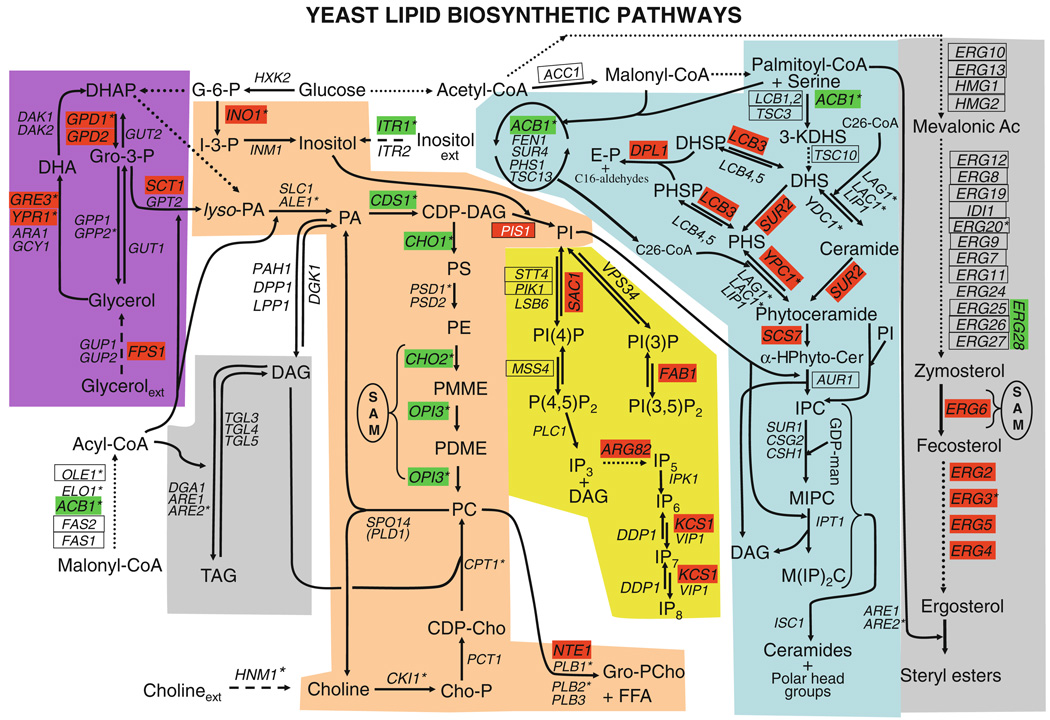
Our screening methodology was, in part, validated by detection of mutations in genes affecting lipid metabolism already reported to confer the Ino− phenotype. These mutations include ino1Δ, ino2Δ, ino4Δ (Culbertson and Henry 1975; Donahue and Henry 1981), sac1Δ (Fig. 5) (Whitters et al. 1993; Rivas et al. 1999), nte1Δ (Fig. 5) (Nunez et al. 2008; Fernandez-Murray et al. 2009), scs2Δ (Kagiwada et al. 1998; Loewen et al. 2003, 2004; Brickner and Walter 2004), and scs3Δ (Hosaka et al. 1994) (Tables 3, S1). However, the current study has significantly extended the list of mutations affecting lipid metabolism that confer an Ino− phenotype, revealing Ino− phenotypes for the first time in mutants defective in structural genes encoding enzymes of sphingolipid and sterol metabolism (Fig. 5). Mutations in structural genes encoding enzymes of lipid metabolism that result in Ino− phenotypes occur in the five interconnected pathways, including glycerophospholipid, glycerol, phosphoinositide (PIP), sphingolipid, and the sterol metabolism (Fig. 5; Tables 3). While the genes defective in these mutants cluster primarily in the GO category lipid metabolism (Fig. 2; Table S2), many are also found in the GO categories “RNA metabolic process,” “transcription,” “cellular homeostasis” and “cellular carbohydrate metabolism.”
Fig. 5.
Major pathways of lipid metabolism in yeast. The pathways are color coded: (tan) glycerophospholipid pathway (violet) glycerol pathway, (yellow) phosphoinositides and inositol phosphates pathway, (blue) ceramide and sphingolipid pathway and (gray) sterol and TAG pathways. Solid arrows represent routes of metabolic conversion. Dotted lines represent multistep metabolic conversions. The names of structural genes (italicized) encoding enzymes catalyzing specific metabolic conversions are shown adjacent to arrows. An asterisk next to the gene name indicates genes regulated by inositol. Mutations in genes colored in green confer an overproduction of inositol (Opi−) phenotype due to elevated PA levels (Henry and Patton-Vogt 1998). Mutations in genes colored in red confer an Ino− phenotype. An allele of the essential PIS1 gene (white letters on red background) has an Ino− phenotype. Genes enclosed in a rectangle are essential. Metabolites are indicated in plain text: DHAP dihydroxyacetone phosphate, DHA dihydroxyacetone, Gro-3-P glycerol 3-phosphate, G-6-P glucose 6-phosphate, I-3-P inositol 3-phosphate, lyso-PA lyso-phosphatidic acid, PA phosphatidic acid, DAG diacylglycerol, TAG triacylglycerol, CDP-DAG cytidine-diphosphate diacylglycerol, PS phosphatidylserine, PE phosphatidylethanolamine, SAM S-adenosyl methionine, PMME phosphatidyl-N-monomethylethanolamine, PDME phosphatidyl-N,N-dimethylethanolamine, PC phosphatidylcholine, Gro-PCho glycerophosphocholine, FFA free fatty acids, CDP-Cho cytidine-diphosphate-choline, Cho-P choline-phosphate, PI phosphatidylinositol, PI3P phosphatidylinositol 3-phosphate, PI4P phosphatidylinositol 4-phosphate, PI(3,5)P2, phosphatidylinositol 3,5-bisphosphate, PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate, IP3 inositol 1,4,5-triphosphate, IP5 inositol 1,3,4,5,6-pentakisphosphate, IP6 inositol 1,2,3,4,5,6-hexakisphosphate, IP7 diphosphoinositol pentakisphosphate, IP8 bis-diphosphoinositol tetrakisphosphate, 3-KDHS 3-ketodihydrosphingosine, DHS dihydrosphingosine, PHS phytosphingosine, DHSP dihydrosphingosine 1-phosphate, PHSP phytosphingosine 1-phosphate, E-P ethanolamine-phosphate, α-HPhyto-Cer alpha-hydroxyphytoceramide, IPC inositol-phosphoceramide, MIPC mannose-inositol-phosphoceramide, M(IP)2C mannosyl-diinositol-phosphorylceramide
We report that the glycerol pathway mutations gpd1Δ, gpd2Δ, sct1Δ, and fps1Δ confer Ino− phenotypes (Tables 3, S1). Gpd1p and Gpd2p are NAD-dependent glycerol-3-phosphate dehydrogenases that produce glycerol-3-phosphate (Gro-3-P), which serves as a precursor for synthesis of PA, which serves as the signal for expression for INO1 (Loewen et al. 2004). The SCT1/GAT1 and the GPT2/GAT1 genes encode Gro-3-P acyltransferases that catalyze the reaction of acyl CoA and Gro-3-P to produce lyso-phosphatidic acid (Lyso-PA), the immediate precursor of PA (Zheng and Zou 2001). Among these mutants potentially affecting PA production, only the sct1Δ mutant has an Ino− phenotype. However, lyso PA can also be made from dihydroxyacetone phosphate (DHAP) (Fig. 5). In yeast, four gene products Gre3p, Ypr1p, Gcy1p, and Ara1p are associated with NADP+ dependant glycerol dehydrogenase (GDH) activity responsible for conversion of glycerol into dihydroxyacetone (DHA), the precursor of DHAP. Triple mutant strains carrying deletions of three of the four genes responsible for GHD activity in yeast are Ino− (Chang and Petrash 2008). We report that the gre3Δ and ypr1Δ single deletions confer Ino− phenotypes when grown at 37°C in I−C+ medium, and the gre3Δ mutant also displays an Ino− phenotype in I−C+ medium even at 30°C (Tables 3, S1; Fig. 5).
The inositol containing lipids of yeast include, in addition to PI, PIPs, and inositol containing sphingolipids, all of which are derived from PI (Fig. 5). The PIS1 gene encoding synthase is essential, but a pis1 mutant, which expresses a partially active PI synthase, is able to synthesize sufficient levels of PI to enable it to grow only when supplied with high levels of exogenous inositol (i.e., it behaves as an Ino− mutant) (Nikawa et al. 1987). The sac1Δ mutant, defective in PI4P phosphatase, also exhibits an Ino− phenotype (Whitters et al. 1993) but reportedly still expresses the INO1 gene (Rivas et al. 1999). PI4P is the precursor of PI(4,5)P2, which, as previously discussed, plays an essential role in both PKC–CWI and TORC2 signaling (Figs. 4a, 5) (Audhya and Emr 2002; Tabuchi et al. 2006). Fab1p, located in the vacuolar membrane, is a PI3P 5-kinase that generates PI(3,5)P2, the levels of which rise dramatically upon osmotic stress (Bonangelino et al. 2002). We report here for the first time that the fab1Δ mutant displays a strong Ino− phenotype at 30°C (Fig. 5; Tables 3, S1).
Turnover of PI(4,5)P2 by phospholipase C, Plc1p, yields diacylglycerol (DAG) and inositol 1,4,5-triphosphate (IP3) (Fig. 5). IP3 is further phospholylated in a series of sequential reactions to yield inositol polyphosphates and diphosphate inositol polyphosphates which, in turn, mediate signaling regulating diverse cellular functions, including vacuole morphology and the response to salt stress (Dubois et al. 2002; Alcazar-Roman and Wente 2008; Demczuk et al. 2008). Two mutations affecting inositol polyphosphate metabolism, arg82Δ, and kcs1Δ, confer Ino− phenotypes. The arg82Δ/ipk2Δ mutant was previously reported to have reduced INO1 expression (Shen et al. 2003b). The kcs1Δ strain exhibits a strong Ino− phenotype at 37°C, but arg82Δ, which is Ino− at 30°C, fails to grow entirely, even in YPD medium at 37°C (Tables 3, S1).
The yeast complex sphingolipids inositol-phosphorylceramide (IPC), mannosyl-inositol-phosphorylceramide (MIPC), and mannosyl-diinositol-phosphorylceramide (M[IP]2C) contain inositol. PI serves as the donor for the inositol phosphate moiety in these lipids in a reaction that also generates DAG (Fig. 5). De novo sphingolipid biosynthesis is required for heat stress response (Cowart et al. 2003), and sphingolipid metabolism may be regulated by TORC2 signaling (Fig. 4a) (Tabuchi et al. 2006), suggesting that sphingolipid levels may contribute to stress response signaling during inositol deprivation. Several mutations affecting ceramide metabolism, ypc1Δ, sur2Δ, lcb3Δ and dpl1Δ, and scs7Δ, confer Ino− phenotypes (Fig. 5; Tables 3, S1).
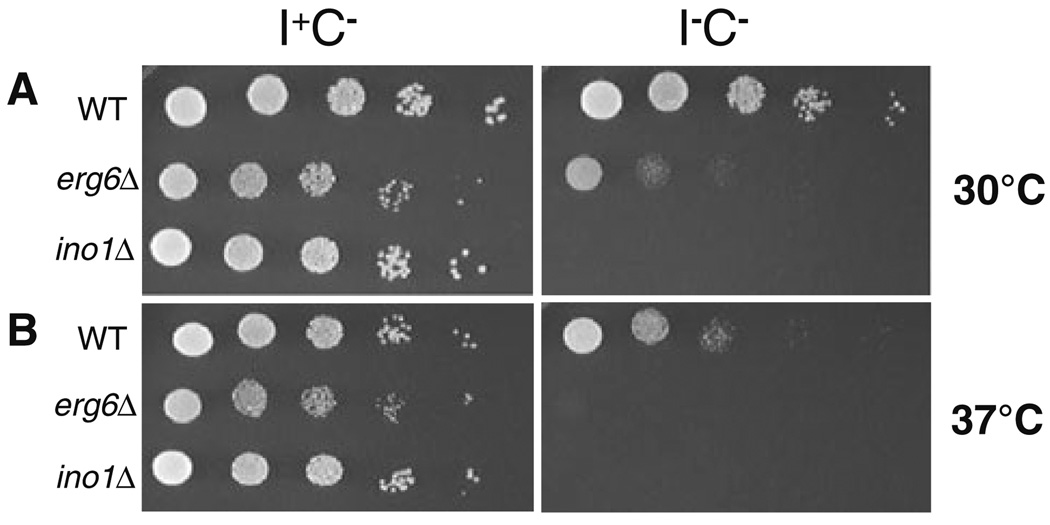
On the other hand, ergosterol synthesis (Fig. 5) does not contribute to the production of PA, use PA or PI as a precursor, or consume inositol. The pathways for sterol and glycerolipid synthesis, however, do share the common precursor acetyl-CoA (Fig. 5), which is also used in protein acetylation. The ERG2, ERG3, ERG4, ERG5, and ERG6 genes are not essential for growth under normal conditions, but mutants carrying deletions of these genes all exhibit abnormal sterol compositions and also display alterations in sphingolipid composition (Guan et al. 2009). The erg2Δ, erg3Δ, erg4Δ, erg5Δ, and erg6Δ mutants all exhibit strong Ino− phenotypes at 37°C, and all but erg5Δ have weak Ino− phenotypes at 30°C (Tables 3, S1). The Ino− phenotype of erg6Δ is shown in Fig. 6 in a spotting assay. At 30°C in I−C− medium, erg6Δ exhibits some residual growth (Fig. 6a), but at 37°C in I−C− medium it has an Ino− phenotype as strong as that of the ino1Δ mutant (Fig. 6b).
Fig. 6.
Inositol auxotrophy phenotype of erg6Δ. Analysis of the inositol auxotrophy (Ino− phenotype). Spotting assays were performed as described in the “Materials and methods”. 5-µl aliquots of a series of tenfold dilutions were spotted onto I+C− and I−C− media containing 2% agar and incubated at 30°C (a) and at 37°C (b). The ino1Δ strain is provided as an example of a strong Ino− mutant
Discussion
This study considerably expands the number of genes in yeast known to be necessary for growth in the absence of the phospholipid precursor, inositol. While these mutations affect a wide range of biological processes, they are heavily enriched in a discrete number of functional categories, including transcription, membrane trafficking, lipid metabolism, and stress response pathways. These results suggest that the Ino− phenotype, which is classically associated with misregulation of lipid metabolism, is not limited to functions directly involved in regulating the INO1 gene. Rather, defects in numerous stress response pathways processes conferred Ino− phenotypes, suggesting that future work should focus on the identification of relevant lipid signals that mediate between inositol-dependent lipid metabolism and stress response signaling. We will discuss several mechanisms that may underlie the Ino− phenotype, thus providing at least a partial explanation as to the number and range of functions that are deficient in these mutants.
Significant deficiency in Ino1p activity or INO1 transcription results in inositol auxotrophy
The most direct cause of an Ino− phenotype is lack of inositol-3-phosphate synthase (Ino1p) activity, as in the ino1Δ mutant. The ino1Δ mutant, which sets the benchmark for a strong Ino− phenotype, shows no residual growth in the absence of inositol in plate assays at any temperature. The ino2Δ and ino4Δ mutants defective in the specific transcription factors required for expression of INO1 and other UASINO containing genes (Ambroziak and Henry 1994) exhibit equally strong Ino− phenotypes (Loewy and Henry 1984). Defects in RNA-Pol II mediated transcription, chromatin remodeling, and histone modification can also result in Ino− phenotypes because of the sensitivity of INO1 transcription to defects in general transcription (Scafe et al. 1990a). However, only a very small proportion of the mutants in this category have Ino− phenotypes as strong as ino1Δ, ino2Δ or ino4Δ (Table 3). Variability in the strength of Ino− phenotypes among mutants defective in RNA-Pol II transcription is presumably due to different levels of residual INO1 transcription. However, direct assessment of INO1 transcription has been carried out only in a subset of such mutants, and it remains an open question as to why INO1 transcription is especially sensitive to sublethal mutations affecting RNA-Pol II transcription. Importantly, expression of several other highly regulated genes, such as GAL10, are similarly affected by defects in RNA-Pol II transcription (Scafe et al. 1990a), suggesting that this phenomenon is not unique to INO1, but may be a more general characteristic of highly regulated genes.
Mutations affecting diverse aspects of lipid metabolism confer Ino− phenotypes
The signal for derepression of INO1 and other UASINO containing genes involves sensing PA levels in the ER by the Opi1p transcriptional repressor. When PA levels are high, Opi1p is tethered to the ER by binding to both PA and Scs2p in the ER (Loewen et al. 2004). Consistent with this model of INO1 regulation by Opi1p, the scs2Δ mutant exhibits an Ino− phenotype (Hosaka et al. 1992) (Tables 3, S1), presumably due to continuous repression of INO1 by Opi1p. Defects in lipid metabolism have the potential to influence INO1 expression by raising or lowering PA production or its utilization, thereby influencing the degree of binding of the Opi1p repressor to the ER membrane and, as a consequence, the level of expression of INO1 and other UASINO-containing genes (Henry and Patton-Vogt 1998; Loewen et al. 2004; Carman and Henry 2007). Mutations that lead to reduced PA levels should have the effect of lowering INO1 expression, possibly to the extent of creating an Ino− phenotype (Carman and Henry 2007). Likely candidates for this mechanism among the Ino− mutant strains depicted in Fig. 5 include the gpd1Δ and gpd2Δ mutants, defective in homologous genes encoding glycerol- 3-phosphate dehydrogenases (Larsson et al. 1993; Eriksson et al. 1995), which catalyze the major route for the synthesis of Gro-3-P, precursor of lysoPA, the immediate precursor of PA. The sct1Δ mutant (Zheng and Zou 2001) defective in one of two acyl transferases responsible for the conversion of Gro-3-P to lysoPA (Fig. 5) should also have the effect of lowering PA levels. However, the correlation of INO1 expression to PA levels has not yet actually been documented in any of these mutants.
Defects in other lipid metabolic steps also produce Ino− phenotypes, including numerous mutants in PIP, sphingolipid, and sterol metabolism depicted in Fig. 5. However, unlike mutations affecting PA metabolism, the cause of the inositol auxotrophy in these mutants may not be due to a deficiency in INO1 expression. For example, the sac1Δ mutant, defective in PI4P phosphatase, exhibits a strong Ino− phenotype, but expresses INO1 at wild-type levels (Rivas et al. 1999). Likewise, PI4P and PI(4,5)P2 levels, which are affected in sac1 mutants (Audhya and Emr 2002), have been implicated in PKC–CWI and TOR signaling (Fig. 4a). However, while the slt2Δ/mpk1Δ mutant in the PKC–CWI pathway exhibits a strong Ino− phenotype, it regulates INO1 normally (Nunez et al. 2008). Interestingly, sac1Δ exhibits other defects in lipid metabolism including reduced levels of PI and inositol-containing sphingolipids (Brice et al. 2009). Sphingolipids, like PIPs, are known to play complex roles in activating stress responses (Dickson 2008), and may functionally interact with sterols to generate membrane domains that function as signaling platforms (Guan et al. 2009). Thus, the root cause(s) of the Ino− phenotypes observed in the mutants defective in PIP, sphingolipid, and sterol metabolism could be interrelated and may be due to their complex roles in regulating stress response pathways.
Mutations affecting membrane trafficking confer Ino− phenotypes
We found that deletion of many non-essential genes affecting diverse steps in the secretory pathway confer Ino− phenotypes. Significantly, Sec− mutants, raised to their semi-restrictive or restrictive temperatures, and wild-type cells, deprived of inositol, exhibit UPR activation (Cox et al. 1997; Chang 2001; Chang et al. 2004; Gaspar et al. 2008). Moreover, the ER stress caused by inositol deprivation appears to be additive with the stress caused by the effect of the sec13-1 secretory defect (Chang et al. 2004). The most extreme form of inositol deprivation occurs in ino1 mutants, which are unable to synthesize any inositol and rapidly lose vitality after transfer to inositol free media, a phenomenon known as “inositol-less death” or “unbalanced growth” (Becker and Lester 1977; Henry et al. 1977; Keith et al. 1977). Prior to dying in the absence of inositol, ino1Δ cells exhibit levels of UPR induction that substantially exceed those seen in wild-type cells growing in inositol-free medium (S. Jesch, unpublished). In the absence of inositol, ino1 mutants also stop dividing and cease expansion of the plasma membrane, while continuing active metabolism including protein synthesis, for a period equivalent to about two doubling times before they lose viability (Atkinson et al. 1977; Henry et al. 1977). A similar phenomenon of “unbalanced growth” occurs in temperature-sensitive Sec− mutants undergoing secretory stress following a shift to their restrictive temperatures. Under these circumstances, Sec− mutants continue protein metabolism for a time after plasma membrane expansion ceases (Ramirez et al. 1983) and become dense, a property that was used as an enrichment procedure in their isolation (Novick and Schekman 1979; Novick et al. 1980). The similarity of events in Sec− mutants shifted to their restrictive temperatures and inositol starved ino1 cells suggests that cells undergoing both types of stress experience similar uncoupling of metabolism from membrane expansion.
Growth in the absence of inositol elicits profound changes in lipid metabolism and activates numerous stress responses
The fully derepressed level of INO1 expression in wild-type cells supports only limited synthesis of inositol which is sufficient to permit cells to survive, but not to attain the level of PI synthesis observed in cells growing in the presence of exogenous inositol (Gaspar et al. 2006). As a consequence, wild-type yeast cells growing in the absence of inositol have PI levels that are 4–5 times lower than cells supplemented with exogenous inositol and exhibit other changes in lipid metabolism (Kelley et al. 1988; Loewen et al. 2004; Gaspar et al. 2006), including changes in the levels of ceramide, precursor to the complex inositol containing sphingolipids (Fig. 5) (Alvarez-Vasquez et al. 2005). Moreover, changes in the expression of literally hundreds of genes accompany the changes in lipid composition that occur in wild-type cells in response inositol (Santiago and Mamoun 2003; Jesch et al. 2005, 2006). Many of these genes are known targets of stress response pathways (Jesch et al. 2005, 2006). While the UPR is the best studied example of a stress response that is activated in the absence of inositol (Cox et al. 1997; Chang 2001), many other stress responses are also activated (Jesch et al. 2006; Nunez et al. 2008).
In the genome-wide screening reported here, Ino− phenotypes are reported for the first time for many mutants defective in the HOG, TOR, cAMP-PKA, filamentous growth, and calcineurin stress response pathways (Figs. 4a, b, S1; Tables 3, S1). Mutants in the glucose response pathway (Hirschhorn et al. 1992; Shirra and Arndt 1999; Shirra et al. 2001), UPR (Nikawa and Yamashita 1992; Sidrauski et al. 1996; Nikawa et al. 1997) and PKC–CWI pathways (Nunez et al. 2008) were previously reported to have Ino− phenotypes. Given their diversity, it is unlikely that all of these signaling pathways mentioned above are involved directly in regulating INO1 expression. Rather, we propose that growth in the absence of inositol is a stress-activating condition, similar to growth at elevated temperature, high or low osmolarity and/or exposure to agents such as tunicamycin, caffeine, or calcoflour white. The fact that mutations in these pathways confer an Ino− phenotype indicates that the signaling through these pathways is essential for survival in the absence of inositol.
Elevated growth temperature and the inclusion of choline in the growth medium add to the stress produced by growth in the absence of inositol
The presence of choline has the effect of enhancing the stringency of the Ino− phenotypes of a number of mutants, a phenomenon first reported by Hosaka et al. (1992) for a dominant choline sensitive mutation (CSE1), the gene for which was never isolated. High copy suppressors of the choline sensitivity of the CSE1 dominant mutation include the SCS3 and SCS2 genes (Hosaka et al. 1994), both of which when deleted confer Ino− phenotypes that are strengthened by growth at 37°C and the presence of choline (Tables 3, S1). Interestingly, the scs2Δ mutant also exhibits increased PC synthesis via the CDP choline pathway (Kagiwada et al. 1998). PC turnover in wild-type cells grown at 30°C in the absence of choline occurs via phospholipase D mediated mechanism, which generates PA and free choline (Patton-Vogt et al. 1997; Sreenivas et al. 1998). In contrast, when choline is present in the growth medium of wild-type cells, or when cells are grown at 37°C, turnover of PC shifts to a phospholipase B mediated mechanism (Dowd et al. 2001), catalyzed by Nte1p (Zaccheo et al. 2004), which generates free fatty acids and glycerol-P-choline (GroPCho) (Fig. 5). Similar to the PKC–CWI mutants (Fig. 4a), the nte1Δ mutant exhibits an Ino− phenotype, which is strongest at 37°C in the presence of choline (Tables 3, S1) (Nunez et al. 2008; Fernandez-Murray et al. 2009). When the CDP-choline pathway is blocked (Fig. 5), turnover of PC continues via a phospholipase D mediated route regardless of temperature or the presence of choline (Dowd et al. 2001). Significantly, mutations that block the incorporation of exogenous choline via the CDP-choline pathway (Fig. 5), not only suppress the choline sensitivity of the scs2Δ mutant, but also suppress its Ino− phenotype (Kagiwada and Zen 2003).
In wild-type cells, the presence of choline causes increased flow through the CDP-choline pathway, increasing utilization of DAG, which is derived from PA (Fig. 5) (Carman and Henry 2007). Thus, the presence of choline inherently affects PA metabolism, which in turn has the potential to affect INO1 expression. The effect of choline on PA levels and INO1 expression potentially explains both the effect of choline in strengthening of Ino− phenotypes and the ability of mutations in the CDP-choline pathway to suppress such phenotypes in certain mutants, such as scs2Δ (Carman and Henry 2007). The lowering of PA levels in the presence of choline may also result in further lowering of INO1 expression in mutants in which INO1 transcription is already impaired (Tables 3, S1).
However, the effect of choline on INO1 expression does not explain the choline sensitivity of the Ino− phenotypes of mutants in the PCK-CWI pathway (Nunez et al. 2008) (Tables 3, S1). The slt2Δ/mpk1Δ mutant (Fig. 4a), exhibits no defect in INO1 transcription, even in the presence of choline at 37°C despite its strong Ino− phenotype under these conditions (Nunez et al. 2008) (Tables 3, S1). However, in comparison with wild-type cells, the slt2Δ/mpk1Δ mutant exhibits higher levels of PC synthesis and turnover via a phospholipase B mediated route, when grown in I−C+ medium at 37° (Nunez et al. 2008). The Ino− phenotype of slt2Δ/mpk1Δ is also suppressed by overexpression of the NTE1 gene, suggesting that altered PC metabolism plays a significant role in its phenotype (Nunez et al. 2008; Fernandez-Murray et al. 2009). Thus, while the PKC–CWI pathway does not regulate INO1 expression in response to inositol supplementation, it does appear to be essential for maintaining lipid homeostasis in the absence of exogenous inositol, especially at high temperature in the presence of choline. We propose that the strengthening of the Ino− phenotype of slt2Δ/mpk1Δ and other PKC–CWI mutants is due to the additive effects of high temperature, lack of inositol, and the presence of choline on the underlying stress responsible for activating PKC–CWI signaling. Significantly, all of these environmental factors have been shown individually to perturb lipid metabolism in wild-type cells (Gaspar et al. 2006, 2008) and may contribute to the Ino− phenotypes observed in mutants in other stress response pathways as discussed earlier.
Many of the mutants identified here as showing Ino− phenotypes that are strengthened by growth at 37°C in the presence of choline are defective in a stress response, or other cellular function, such as lipid metabolism or membrane trafficking, that are predicted to contribute to cellular stress when partially impaired. However, some mutants with defects related to partial impairment of INO1 expression due to defects in RNA-Pol II transcription, also show strengthening of their Ino− phenotypes in response to choline and temperature. In such mutants, INO1 expression maybe at a level that is barely sufficient for survival in the absence of inositol. Since higher growth temperature increases the demand for PI synthesis (Gaspar et al. 2008), while the presence of choline reduces in INO1 expression (Hirsch and Henry 1986), growth of strains with marginal INO1 expression may not be possible under these conditions. Overall, the results of our genome-wide screen for Ino− phenotypes strongly suggest a role for lipid metabolism in stress response signaling. Future work will focus on identifying relevant lipid signals that mediate between inositol-dependent lipid metabolism and stress response signaling.
Supplementary Material
Ackowledgments
This work was supported by National Institutes of Health Grant GM019629 (to SAH.)
Abbreviations
- Ino−
Inositol auxotrophy
- RNA-Pol II
RNA polymerase II
- PI
Phosphatidylinositol
- PA
Phosphatidic acid
- ER
Endoplasmic reticulum
- DAG
Diacylglycerol
- PC
Phosphatidylcholine
- PKC–CWI
Protein kinase C–cell wall integrity
- UPR
Unfolded protein response
- AMPK
AMP-dependant kinase
- HOG
High osmolarity glycerol
- ERAD
ER-associated protein degradation
- TOR
Target of rapamycin
- PKA
cAMP-protein kinase A
- MAPK
Mitogen activated protein kinase
- ORF
Open reading frame
- GO
Gene Ontology
- SGD
Saccharomyces genome database
- YPD
1% yeast extract, 2% bactopeptone, 2% glucose
- I
Inositol
- C
Choline
- GroPCho
Glycerol-phospho-choline
- GPI
Glycosylphosphatidylinositol
- Gro-3-P
Glycerol-3-phosphate
- lyso-PA
Lyso-phosphatidic acid
- DHA
Dihydroxyacetone
- DHAP
Dihydroxyacetone phosphate
- PIP
Phosphoinositides
- IP3
Inositol 1,4,5-triphosphate
- IPC
Inositol-phosphorylceramide
- MIPC
Mannosyl-inositol-phosphorylceramide
- M(IP)2C
Mannosyl-diinositol-phosphorylceramide
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00438-010-0592-x) contains supplementary material, which is available to authorized users.
References
- Alcazar-Roman AR, Wente SR. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- Alvarez-Vasquez F, Sims KJ, Cowart LA, Okamoto Y, Voit EO, Hannun YA. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 2005;433:425–430. doi: 10.1038/nature03232. [DOI] [PubMed] [Google Scholar]
- Ambroziak J, Henry SA. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- Arndt KM, Ricupero-Hovasse S, Winston F. TBP mutants defective in activated transcription in vivo. EMBOJ. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson KD, Kolat AI, Henry SA. Osmotic imbalance in inositol-starved spheroplasts of Saccharomyces cerevisiae. J Bacteriol. 1977;132:806–817. doi: 10.1128/jb.132.3.806-817.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Bachhawat N, Ouyang Q, Henry SA. Functional characterization of an inositol-sensitive upstream activation sequence in yeast: a cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J Biol Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- Bailis AM, Lopes JM, Kohlwein SD, Henry SA. cis and trans regulatory elements required for regulation of the CHO1 gene of Saccharomyces cerevisiae. Nucl Acids Res. 1992;20:1411–1418. doi: 10.1093/nar/20.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. COPII-dependent transport from the endoplasmic reticulum. Curr Opin Cell Biol. 2002;14:417–422. doi: 10.1016/s0955-0674(02)00348-4. [DOI] [PubMed] [Google Scholar]
- Becker GW, Lester RL. Changes in phospholipids of Saccharomyces cerevisiae associated with inositol-less death. J Biol Chem. 1977;252:8684–8691. [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol Genet Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS. Osmotic stress-induced increase of phosphatidylinositol 3, 5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol. 2002;156:1015–1028. doi: 10.1083/jcb.200201002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumann HA, Damen MJ, Versluis C, Heck AJ, de Kruijff B, de Kroon AI. The two biosynthetic routes leading to phosphatidylcholine in yeast produce different sets of molecular species. Evidence for lipid remodeling. Biochemistry. 2003;42:3054–3059. doi: 10.1021/bi026801r. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brice SE, Alford CW, Cowart LA. Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem. 2009;284:7588–7596. doi: 10.1074/jbc.M808325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard D, Reed M, Winter E. RSC1 and RSC2 are required for expression of mid-late sporulation-specific genes in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:910–918. doi: 10.1128/EC.3.4.910-918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Henry SA. Phospholipid biosynthesis in yeast. Ann Rev Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- Carman GM, Henry SA. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- Carman GM, Henry SA. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 2007;282:37293–37297. doi: 10.1074/jbc.R700038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Zeimetz GM. Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. J Biol Chem. 1996;271:13293–13296. doi: 10.1074/jbc.271.23.13293. [DOI] [PubMed] [Google Scholar]
- Chai B, Hsu JM, Du J, Laurent BC. Yeast RSC function is required for organization of the cellular cytoskeleton via an alternative PKC1 pathway. Genetics. 2002;161:575–584. doi: 10.1093/genetics/161.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HJ. Ph.D. thesis. Pittsburgh: Carnegie Mellon University; 2001. Role of the unfolded protein response pathway in phospholipid biosynthesis and membrane trafficking in Saccharomyces cerevisiae. [Google Scholar]
- Chang Q, Petrash JM. Disruption of aldo-keto reductase genes leads to elevated markers of oxidative stress and inositol auxotrophy in Saccharomyces cerevisiae. Biochim Biophys Acta. 2008;1783:237–245. doi: 10.1016/j.bbamcr.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HJ, Jesch SA, Gaspar ML, Henry SA. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics. 2004;168:1899–1913. doi: 10.1534/genetics.104.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IW, Charalampous FC. Biochemical studies on inositol. VII. Biosynthesis of inositol by a soluble enzyme system. J Biol Chem. 1964;239:1905–1910. [PubMed] [Google Scholar]
- Chen IW, Charalompous FC. Mode of conversion of glucose-6- P to inositol and the role of DPN and NH4+ ions. Biochem Biophys Res Commun. 1964;17:521–526. [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces genome database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Mallory MJ, Strich R, Yao TP. Hos2p/Set3p deacetylase complex signals secretory stress through the Mpk1p cell integrity pathway. Eukaryot Cell. 2008;7:1191–1199. doi: 10.1128/EC.00059-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart LA, Okamoto Y, Pinto FR, Gandy JL, Almeida JS, Hannun YA. Roles for sphingolipid biosynthesis in mediation of specific programs of the heat stress response determined through gene expression profiling. J Biol Chem. 2003;278:30328–30338. doi: 10.1074/jbc.M300656200. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, Henry SA. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics. 1975;80:23–40. doi: 10.1093/genetics/80.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, Donahue TF, Henry SA. Control of inositol biosynthesis in Saccharomyces cerevisiae: inositol-phosphate synthetase mutants. J Bacteriol. 1976;126:243–250. doi: 10.1128/jb.126.1.243-250.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Emr SD, McPherson PS, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- Dean-Johnson M, Henry SA. Biosynthesis of inositol in yeast: Primary structure of myo-inositol 1-phosphate synthase locus and functional characterization of its structural gene, the INO1 locus. J Biol Chem. 1989;264:1274–1283. [PubMed] [Google Scholar]
- Demczuk A, Guha N, Nguyen PH, Desai P, Chang J, Guzinska K, Rollins J, Ghosh CC, Goodwin L, Vancura A. Saccharomyces cerevisiae phospholipase C regulates transcription of Msn2p-dependent stress-responsive genes. Eukaryot Cell. 2008;7:967–979. doi: 10.1128/EC.00438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res. 2008;49:909–921. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue TF, Henry SA. Inositol mutants of Saccharomyces cerevisiae: mapping the ino1 locus and characterizing alleles of the ino1, ino2 and ino4 loci. Genetics. 1981;98:491–503. doi: 10.1093/genetics/98.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SR, Bier ME, Patton-Vogt JL. Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J Biol Chem. 2001;276:3756–3763. doi: 10.1074/jbc.M003694200. [DOI] [PubMed] [Google Scholar]
- Driessen HP, de Jong WW, Tesser GI, Bloemendal H. The mechanism of N-terminal acetylation of proteins. CRC Crit Rev Biochem. 1985;18:281–325. doi: 10.3109/10409238509086784. [DOI] [PubMed] [Google Scholar]
- Dubois E, Scherens B, Vierendeels F, Ho MM, Messenguy F, Shears SB. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J Biol Chem. 2002;277:23755–23763. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- Ebbert R, Birkmann A, Schuller HJ. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol Microbiol. 1999;32:741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- Eriksson P, Andre L, Ansell R, Blomberg A, Adler L. Cloning and characterization of GPD2, a second gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol Microbiol. 1995;17:95–107. doi: 10.1111/j.1365-2958.1995.mmi_17010095.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Murray JP, Gaspard GJ, Jesch SA, McMaster CR. NTE1-encoded phosphatidylcholine phospholipase b regulates transcription of phospholipid biosynthetic genes. J Biol Chem. 2009;284:36034–36046. doi: 10.1074/jbc.M109.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BB, Mylonakis E. Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot Cell. 2009;8:1616–1625. doi: 10.1128/EC.00193-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansheroff LJ, Dollard C, Tan P, Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar ML, Aregullin MA, Jesch SA, Henry SA. Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J Biol Chem. 2006;281:22773–22785. doi: 10.1074/jbc.M603548200. [DOI] [PubMed] [Google Scholar]
- Gaspar ML, Jesch SA, Viswanatha R, Antosh AL, Brown WJ, Kohlwein SD, Henry SA. A block in endoplasmic reticulum-to-Golgi trafficking inhibits phospholipid synthesis and induces neutral lipid accumulation. J Biol Chem. 2008;283:25735–25751. doi: 10.1074/jbc.M802685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gilstring CF, Melin-Larsson M, Ljungdahl PO. Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles. Mol Biol Cell. 1999;10:3549–3565. doi: 10.1091/mbc.10.11.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ML, Lopes JM. Genetic regulation of phospholipid biosynthesis in yeast. Microbiol Rev. 1996;60:1–20. doi: 10.1128/mr.60.1.1-20.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griac P, Henry SA. The yeast inositol-sensitive upstream activating sequence, UASINO, responds to nitrogen availability. Nucleic Acids Res. 1999;27:2043–2050. doi: 10.1093/nar/27.9.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XL, Souza CM, Pichler H, Dewhurst G, Schaad O, Kajiwara K, Wakabayashi H, Ivanova T, Castillon GA, Piccolis M, et al. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell. 2009;20:2083–2095. doi: 10.1091/mbc.E08-11-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson BA, Lester RL. Effects of inositol starvation on phospholipid and glycan syntheses in Saccharomyces cerevisiae. J Bacteriol. 1980;142:79–89. doi: 10.1128/jb.142.1.79-89.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson BA, Lester RL. Effect of inositol starvation on the in vitro syntheses of mannan and N-acetylglucosaminylpyrophosphoryldolichol in Saccharomyces cerevisiae. J Bacteriol. 1982;151:334–342. doi: 10.1128/jb.151.1.334-342.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinisch JJ, Lorberg A, Schmitz HP, Jacoby JJ. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol Microbiol. 1999;32:671–680. doi: 10.1046/j.1365-2958.1999.01375.x. [DOI] [PubMed] [Google Scholar]
- Henry SA, Patton-Vogt JL. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog Nucleic Acid Res Mol Biol. 1998;61:133–179. doi: 10.1016/s0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- Henry SA, Atkinson KD, Kolat AI, Culbertson MR. Growth and metabolism of inositol-starved Saccharomyces cerevisiae. J Bacteriol. 1977;130:472–484. doi: 10.1128/jb.130.1.472-484.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JP, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Hirschman JE, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hong EL, Livstone MS, Nash R, et al. Genome Snapshot: a new resource at the Saccharomyces Genome Database (SGD) presenting an overview of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2006;34:D442–D445. doi: 10.1093/nar/gkj117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HL, Shiau YS, Chen MY. Saccharomyces cerevisiaeTSC11/ AVO3 participates in regulating cell integrity and functionally interacts with components of the Tor2 complex. Curr Genet. 2005;47:273–288. doi: 10.1007/s00294-005-0570-8. [DOI] [PubMed] [Google Scholar]
- Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Methods Enzymol. 2007;428:29–45. doi: 10.1016/S0076-6879(07)28002-4. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Pina B, Marcus GA, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka K, Nikawa J, Kodaki T, Yamashita S. A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae. J Biochem. 1992;111:352–358. doi: 10.1093/oxfordjournals.jbchem.a123761. [DOI] [PubMed] [Google Scholar]
- Hosaka K, Nikawa J, Kodaki T, Ishizu H, Yamashita S. Cloning and sequence of the SCS3 gene which is required for inositol prototrophy in Saccharomyces cerevisiae. J Biochem. 1994;116:1317–1321. doi: 10.1093/oxfordjournals.jbchem.a124681. [DOI] [PubMed] [Google Scholar]
- Issel-Tarver L, Christie KR, Dolinski K, Andrada R, Balakrishnan R, Ball CA, Binkley G, Dong S, Dwight SS, Fisk DG, et al. Saccharomyces genome database. Methods Enzymol. 2002;350:329–346. doi: 10.1016/s0076-6879(02)50972-1. [DOI] [PubMed] [Google Scholar]
- Jesch SA, Henry SA. Yeast inositol phospholipids: synthesis, regulation, and involvement in membrane trafficking and lipid signaling. In: Daum G, editor. Cell biology and dynamics of yeast lipids. Kerala: Research Signpost; 2005. pp. 105–131. [Google Scholar]
- Jesch SA, Zhao X, Wells MT, Henry SA. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J Biol Chem. 2005;280:9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch SA, Liu P, Zhao X, Wells MT, Henry SA. Multiple endoplasmic reticulum-to-nucleus signaling pathways coordinate phospholipid metabolism with gene expression by distinct mechanisms. J Biol Chem. 2006;281:24070–24083. doi: 10.1074/jbc.M604541200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kagiwada S, Zen R. Role of the yeast VAP homolog, Scs2p, in INO1 expression and phospholipid metabolism. J Biochem. 2003;133:515–522. doi: 10.1093/jb/mvg068. [DOI] [PubMed] [Google Scholar]
- Kagiwada S, Hosaka K, Murata M, Nikawa J, Takatsuki A. The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J Bacteriol. 1998;180:1700–1708. doi: 10.1128/jb.180.7.1700-1708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K, Watanabe R, Pichler H, Ihara K, Murakami S, Riezman H, Funato K. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol Biol Cell. 2008;19:2069–2082. doi: 10.1091/mbc.E07-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith AD, Pollard EC, Snipes W, Henry SA, Culbertson MR. Inositol-less death in yeast results in a simultaneous increase in intracellular viscosity. Biophys J. 1977;17:205–212. doi: 10.1016/S0006-3495(77)85650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MJ, Bailis AM, Henry SA, Carman GM. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by inositol: inositol is an inhibitor of phosphatidylserine synthase activity. J Biol Chem. 1988;263:18078–18084. [PubMed] [Google Scholar]
- Kodaki T, Nikawa J, Hosaka K, Yamashita S. Functional analysis of the regulatory region of the yeast phosphatidylserine synthase gene, PSS. J Biochem. 1991;173:7992–7995. doi: 10.1128/jb.173.24.7992-7995.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T, Hosaka K, Nikawa J, Yamashita S. The SNF2/SWI2/GAM1/TYE3/RIC1 gene is involved in the coordinate regulation of phospholipid synthesis in Saccharomyces cerevisiae. J Biochem. 1995;117:362–368. doi: 10.1093/jb/117.2.362. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Buratowski S, Nonet M, Young RA. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992;69:883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- Larsson K, Ansell R, Eriksson P, Adler L. A gene encoding snglycerol 3-phosphate dehydrogenase (NAD+) complements an osmosensitive mutant of Saccharomyces cerevisiae. Mol Microbiol. 1993;10:1101–1111. doi: 10.1111/j.1365-2958.1993.tb00980.x. [DOI] [PubMed] [Google Scholar]
- Lehle L, Strahl S, Tanner W. Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew Chem Int Ed Engl. 2006;45:6802–6818. doi: 10.1002/anie.200601645. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJR, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- Loewy BS, Henry SA. The INO2 and INO4 loci of Saccharomyces cerevisiae are pleiotropic regulatory genes. Mol Cell Biol. 1984;4:2479–2485. doi: 10.1128/mcb.4.11.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopes JM, Henry SA. Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucl Acids Res. 1991;19:3987–3994. doi: 10.1093/nar/19.14.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JM, Hirsch JP, Chorgo PA, Schulze KL, Henry SA. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucl Acids Res. 1991;19:1687–1693. doi: 10.1093/nar/19.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Majerus PW, York JD. Phosphoinositide phosphatases and disease. J Lipid Res. 2009;50 Suppl:S249–S254. doi: 10.1194/jlr.R800072-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder AL, Johnson MD, Henry SA. 1L-myo-inositol 1-phosphate synthase. Biochim Biophys Acta. 1997;1348:245–256. doi: 10.1016/s0005-2760(97)00122-7. [DOI] [PubMed] [Google Scholar]
- Michell RH. Evolution of the diverse biological roles of inositols. Biochem Soc Symp. 2007:223–246. doi: 10.1042/BSS0740223. [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Thiele DJ. The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. EMBO J. 1999;18:5953–5962. doi: 10.1093/emboj/18.21.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadolski MJ, Linder ME. Protein lipidation. FEBS J. 2007;274:5202–5210. doi: 10.1111/j.1742-4658.2007.06056.x. [DOI] [PubMed] [Google Scholar]
- Neiman AM, Mhaiskar V, Manus V, Galibert F, Dean N. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics. 1997;145:637–645. doi: 10.1093/genetics/145.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J-I, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol prototrophy in Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Kodaki T, Yamashita S. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J Biol Chem. 1987;262:4876–4881. [PubMed] [Google Scholar]
- Nikawa J, Sugiyama M, Hayashi K, Nakashima A. Suppression of the Saccharomyces cerevisiae hac1/ire15 mutation by yeast genes and human cDNAs. Gene. 1997;201:5–10. doi: 10.1016/s0378-1119(97)00418-6. [DOI] [PubMed] [Google Scholar]
- Nikoloff DM, Henry SA. Functional characterization of the INO2 gene of Saccharomyces cerevisiae. J Biol Chem. 1994;269:7402–7411. [PubMed] [Google Scholar]
- Nikoloff DM, McGraw P, Henry SA. The INO2 gene of Saccharomyces cerevisiae encodes a helix-loop-helix protein that is required for activation of phospholipid synthesis. Nucl Acids Res. 1992;20:3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Nunez LR, Jesch SA, Gaspar ML, Almaguer C, Villa-Garcia M, Ruiz-Noriega M, Patton-Vogt J, Henry SA. Cell wall integrity MAPK pathway is essential for lipid homeostasis. J Biol Chem. 2008;283:34204–34217. doi: 10.1074/jbc.M806391200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- Ouyang Q, Ruiz-Noriega M, Henry SA. The REG1 gene product is required for repression of INO1 and other inositol-sensitive upstream activating sequence-containing genes of yeast. Genetics. 1999;152:89–100. doi: 10.1093/genetics/152.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton-Vogt JL, Griac P, Sreenivas A, Bruno V, Dowd S, Swede MJ, Henry SA. Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation. J Biol Chem. 1997;272:20873–20883. doi: 10.1074/jbc.272.33.20873. [DOI] [PubMed] [Google Scholar]
- Perrot M, Massoni A, Boucherie H. Sequence requirements for Nalpha-terminal acetylation of yeast proteins by NatA. Yeast. 2008;25:513–527. doi: 10.1002/yea.1602. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Philip B, Levin DE. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol Cell Biol. 2001;21:271–280. doi: 10.1128/MCB.21.1.271-280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet M, Conzelmann A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. The diversity of acetylated proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-reviews0006. reviews 0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Arino J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- Ramirez RM, Ishida-Schick T, Krilowicz BL, Leish BA, Atkinson KD. Plasma membrane expansion terminates in Saccharomyces cerevisiae secretion-defective mutants while phospholipid synthesis continues. J Bacteriol. 1983;154:1276–1283. doi: 10.1128/jb.154.3.1276-1283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p#x0201D; and inositol auxotrophy. Mol Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SM, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago TC, Mamoun CB. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J Biol Chem. 2003;278:38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- Scafe C, Chao D, Lopes J, Hirsch JP, Henry S, Young RA. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990a;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- Scafe C, Martin C, Nonet M, Podos S, Okamura S, Young RA. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol Cell Biol. 1990b;10:1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller H-J, Richter K, Hoffmann B, Ebbert R, Schweizer E. DNA binding site of the yeast heteromeric Ino2p/Ino4p basic helix-loop-helix transcription factor: Structural requirements as defined by saturation mutagenesis. FEBS Lett. 1995;370:149–152. doi: 10.1016/0014-5793(95)00818-t. [DOI] [PubMed] [Google Scholar]
- Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schuller H-J. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucl Acids Res. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003a;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003b;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Shirra MK, Arndt KM. Evidence for the involvement of the Glc7-Reg1 phosphatase and the Snf1-Snf4 kinase in the regulation of INO1 transcription in Saccharomyces cerevisiae. Genetics. 1999;152:73–87. doi: 10.1093/genetics/152.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirra MK, Patton-Vogt J, Ulrich A, Liuta-Tehlivets O, Kohlwein SD, Henry SA, Arndt KM. Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5710–5722. doi: 10.1128/MCB.21.17.5710-5722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Skinner HB, Alb JG, Jr, Whitters EA, Helmkamp GM, Jr, Bankaitis VA. Phospholipid transfer activity is relevant to but not sufficient for the essential function of the yeast SEC14 gene product. EMBO J. 1993;12:4775–4784. doi: 10.1002/j.1460-2075.1993.tb06166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J Biol Chem. 1998;273:16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
- Staleva L, Hall A, Orlow SJ. Oxidative stress activates FUS1 and RLM1 transcription in the yeast Saccharomyces cerevisiae in an oxidant-dependent Manner. Mol Biol Cell. 2004;15:5574–5582. doi: 10.1091/mbc.E04-02-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. The phosphatidylinositol 4, 5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuki A, Tamura G. Effect of tunicamycin on the synthesis of macromolecules in cultures of chick embryo fibroblasts infected with Newcastle disease virus. J Antibiot (Tokyo) 1971;24:785–794. doi: 10.7164/antibiotics.24.785. [DOI] [PubMed] [Google Scholar]
- Torres J, Di Como CJ, Herrero E, De La Torre-Ruiz MA. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J Biol Chem. 2002;277:43495–43504. doi: 10.1074/jbc.M205408200. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Madia F, Hu J, Ge H, Li LM, Longo VD. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, Hirsch JP, Henry SA. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J Biol Chem. 1991;266:863–872. [PubMed] [Google Scholar]
- Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JD, Barlowe C. Yet1p and Yet3p, the yeast homologs of BAP29 and BAP31, interact with the ER translocation apparatus and are required for inositol prototrophy. J Biol Chem. 2010 Apr 8; doi: 10.1074/jbc.M109.080382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler A, Arkind C, Mattison CP, Burkholder A, Knoche K, Ota I. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot Cell. 2002;1:163–173. doi: 10.1128/EC.1.2.163-173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Carmen AA, Kobayashi R, Suka N, Grunstein M. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc Natl Acad Sci USA. 2001;98:4391–4396. doi: 10.1073/pnas.081560698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yip CL, Welch SK, Klebl F, Gilbert T, Seidel P, Grant FJ, O’Hara PJ, MacKay VL. Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc Natl Acad Sci USA. 1994;91:2723–2727. doi: 10.1073/pnas.91.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BP, Shin JJ, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- Yu VP, Reed SI. Cks1 is dispensable for survival in Saccharomyces cerevisiae. Cell Cycle. 2004;3:1402–1404. doi: 10.4161/cc.3.11.1208. [DOI] [PubMed] [Google Scholar]
- Zaccheo O, Dinsdale D, Meacock PA, Glynn P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J Biol Chem. 2004;279:24024–24033. doi: 10.1074/jbc.M400830200. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Rao R. A spoke in the wheel: calcium spikes disrupt yeast cell cycle. Cell Cycle. 2008;7:870–873. doi: 10.4161/cc.7.7.5616. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Zou J. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J Biol Chem. 2001;276:41710–41716. doi: 10.1074/jbc.M104749200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.