Abstract
Through long-term laboratory selection (over 200 generations), we have generated Drosophila melanogaster populations that tolerate severe, normally lethal, levels of hypoxia. Because of initial experiments suspecting genetic mechanisms underlying this adaptation, we compared the genomes of the hypoxia-selected flies with those of controls using deep resequencing. By applying unique computing and analytical methods we identified a number of DNA regions under selection, mostly on the X chromosome. Several of the hypoxia-selected regions contained genes encoding or regulating the Notch pathway. In addition, previous expression profiling revealed an activation of the Notch pathway in the hypoxia-selected flies. We confirmed the contribution of Notch activation to hypoxia tolerance using a specific γ-secretase inhibitor, N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT), which significantly reduced adult survival and life span in the hypoxia-selected flies. We also demonstrated that flies with loss-of-function Notch mutations or RNAi-mediated Notch knockdown had a significant reduction in hypoxia tolerance, but those with a gain-of-function had a dramatic opposite effect. Using the UAS-Gal4 system, we also showed that specific overexpression of the Notch intracellular domain in glial cells was critical for conferring hypoxia tolerance. Unique analytical tools and genetic and bioinformatic strategies allowed us to discover that Notch activation plays a major role in this hypoxia tolerance in Drosophila melanogaster.
Keywords: evolution, next-generation sequencing
Oxygen homoeostasis is essential for development, growth, and integrity of cells, tissues, and organisms. Limited oxygen supply to cells and tissues (hypoxia) has a wide range of physiologic and potentially pathologic consequences, ranging from ischemic/hypoxic heart disease, stroke, and pulmonary hypertension to a number of obstetrical/perinatal complications, to high-altitude illnesses, to organ transplantation, and finally to intratumor hypoxia and cancer progression. Despite the clinical importance and societal disease impact of such a wide range of disorders, the molecular underpinnings of susceptibility or tolerance of cells or tissues to lack of O2 are not well understood. Many studies have investigated the mechanisms that lead to injury when cells are deprived of O2, but to potentially treat or prevent the consequences of hypoxia necessitates also the understanding of the inherent tissue mechanisms that are critical for tolerance and survival. To do so, we use a long-term laboratory selection strategy that unmasks mechanisms that play an important role in hypoxia tolerance in a genetic model, Drosophila melanogaster (1, 2). In this attempt, starting with 27 isofemale D. melanogaster strains, and applying decreasing levels of O2 over >200 generations, we generated Drosophila populations that tolerate severe levels of hypoxia, which are lethal to the original parental lines. These hypoxia-adapted flies (AF) pass the tolerance trait from generation to generation and the trait persists even in the absence of hypoxic stress (i.e., after several generations in a normoxic environment), suggesting that a genetic rather than a physiological mechanism underlies adaptation. We have discovered that a) genetic mechanisms allowed D. melanogaster to adapt and tolerate extremely low O2 environments, b) there were genomic intervals that were selected for, mostly on the X chromosome, that occurred during long-term hypoxia, and c) the Notch pathway played an important role in hypoxia tolerance.
Results
Hypoxia-Selected Regions and Genetic Profiles in the AF Genome.
To determine whether there are DNA signatures of hypoxia selection, we sequenced two control (C1 and C2) and two AF (H1 and H2) populations that had been under hypoxia selection for 180 generations in separate environmental chambers at >60× coverage, using the Illumina GA II sequencer. We aligned between 120 million and 200 million 54-bp paired-end reads per population to the D. melanogaster reference sequence (Table S1). Because individual genotypes and the number of individuals sampled at any given base and standard linkage disequilibrium (LD) information could not be determined from the pooled sequence data, standard tests of selection could not be used (3, 4). Consequently, we used two complementary approaches to determine the hypoxia-selected regions and genetic profiles in the AF genome.
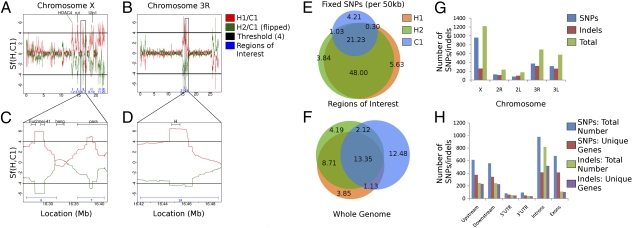
In the first case, we used a coarse-grained approach to compare SNP distributions in both control and AF pools. In this approach, we developed a unique statistic to determine potential regions under selection using the sequence data generated from the pooled populations. Sf(C1, H1) is the log ratio of control and AF scaled mutation rates and provides a comparison of the effective population sizes. We identified Sf(C1, H1) >> 0 as indicative of deviation from neutrality and consistent with a purifying selection in hypoxia (reduction in effective population size). We used Sf(C1, C2) as an empirical control for false discovery rate (FDR) computation. We also investigated the concordance between the AF populations from different chambers by comparing the independent estimates Sf(C1, H1) and Sf(C1, H2). Using a Sf(C1, H1) cutoff of 4, corresponding to an FDR of ∼1%, and overlapping 50-kbp windows, we observed remarkable concordance between regions under selection in H1 and H2. A total of 1,509,436 bp comprising 24 distinct hypoxia-selected regions and containing a total of 188 genes were under selection in both H1 and H2. Twenty of these regions (>80%) were located within a 10-Mb interval on chromosome X and the remaining 4 were located within a 1.2-Mb interval on chromosome 3R (Fig. 1 A–D and Fig. S1). These results demonstrate that two populations of flies, independently selected for hypoxia tolerance (i.e., in different environmental chambers) had the same intervals in the genome undergo a high degree of fixation (hypoxia-selected regions) and suggest that the genes required for adaptation to severe hypoxic conditions are localized rather than distributed across the genome. The latter observation is reinforced by the distribution of C1 vs. H1 or H2 fixed SNPs across the genome: Whereas all three populations have a median value of 28 fixed SNPs (range 0–363) per 50-kb interval across genome, there is a large difference between C1 and H1 and H2 in the hypoxia-selected regions where the AF populations have threefold higher fixed SNPs compared with the control flies (∼74; Poisson P = 2.57 × 10−13). Furthermore, conservation between H1 and H2 of fixed SNPs was also higher in the hypoxia-selected regions compared with those in the non-hypoxia–selected regions (93% vs. 78%; hypergeometric P = 3.12 × 10−43). Consistent with a hypoxic stress-mediated population bottleneck leading to an overall loss of diversity in the AF populations, we observed a much higher genetic similarity between H1 and H2 (66% of the fixed SNPs are common), compared with a concordance of 35–37% between AF and control lines, despite the identical ancestry (Fig. 1 E and F).
Fig. 1.
Genomic regions under selection for hypoxia tolerance. The Sf statistic was computed on overlapping 50-kbp windows, comparing two hypoxia samples (H1 and H2) to a normoxia one (C1). Regions with Sf values >4 are shown, corresponding to 1% FDR (based on comparing two normoxia samples). The results for chromosomes (A) X and (B) 3R show near perfect concordance between H1 vs. C1 (red) and H2 vs. C2 (green, inverted). Regions deemed significant for both hypoxic samples are shown as blue intervals. (C) Regions 6 and 7, including genes Fur2, mei-41, hang, and para, and (D) region 24, containing Hairless, are shown in greater detail. Note that the value of the statistic is plotted at the median of the region and decays as it goes toward the edge of the selected region. (E and F) Venn diagrams, describing the frequency of fixed SNPs (per 50 kbp) in (E) just the regions under selection and (F) the entire genome, were tabulated. Orange represents H1, green represents H2, and blue represents C1. (G and H) Bar graphs indicating the distribution of SNPs and small indels in extended gene regions by chromosome and gene region. Loci falling in the same gene region of more than one gene were counted for each gene.
In the second case, we used a fine-grained approach and focused the analysis on the loci with high-confidence allelic differences (between control and AF) considered likely to represent allele selection or linkage to selected alleles within the AF populations. This analysis was limited to the 45% of the euchromatic genome (including 70–75% of exon loci) with ≥20× coverage and high base-calling quality. This approach identified SNPs and small indels in loci where the two AF populations differed from both the control and reference alleles. This approach confirmed many of the findings described above. The majority of detected high-confidence polymorphisms occurred in chromosome X (959 SNPs, 259 indels), with chromosome 3R (371 SNPs, 318 indels) containing the next largest number. Also detected were a significant number of polymorphisms on chromosome 3L and comparatively few on chromosomes 2L and 2R (Fig. 1G). Polymorphisms were identified across the entire extended gene region, with introns containing the largest number of both SNPs and indels (Fig. 1H).
Candidate Genes Underlying Hypoxia Tolerance in AF.
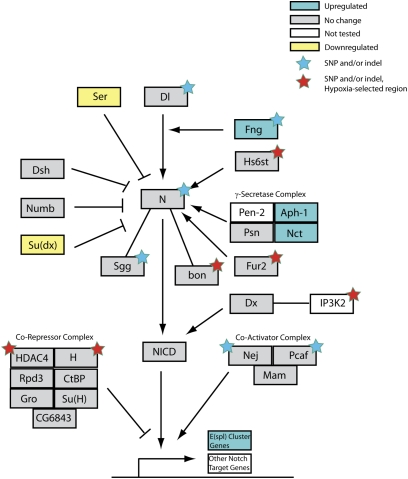
There are 188 genes located in the hypoxia-selected regions (Table S2). To identify the set of potentially causal genes in hypoxia tolerance, we started initially with 28 genes because these were previously implicated in either hypoxia or similar phenotypes (such as oxidative stress and aging). We filtered the remaining 160 genes for evidence of a selective sweep using three complementary tests: i) the McDonald–Kreitman test between H1 and C1 for evidence of adaptive evolution based on correlation between fixed (i.e., >90% frequency) and nonsynonymous mutations in a population (P ≤ 0.05) (5), ii) ≥1.5-fold transcriptomic change under hypoxia (1), and iii) sorting intolerant from tolerant (SIFT) evaluation of the impact of fixed, nonsynonymous mutations on the functions of proteins encoded by the genes in these regions (P ≤ 0.05) (6). A total of 68 genes from the 160 genes were identified and many of these were also observed using the fine-granularity assessment. These include 12 genes that interact with, activate, or are targets of the Notch pathway (Table S2). For example, there are two members of the Notch repressor complex, i.e., Hairless and HDAC4, that are located in the hypoxia-selected regions in AF. HDAC4 contains a SNP fixed in both AF populations with significant impact on function (SIFT P value = 0.05) but virtually absent in both control populations (A1004S in isoform B). The fine-grained genome-wide analysis identified additional polymorphisms in Notch pathway-related genes, including Notch, Delta, fringe, and sgg (Fig. 2) (Table S3 and Table S4).
Fig. 2.
Enrichment of fixed SNPs and Indels in an extended Notch pathway. The Notch pathway was adapted from KEGG (42) by adding Notch interactors from the literature to create an expanded Notch signaling pathway. Genes differentially expressed in larva (expression levels from Zhou et al.) (1) are cyan (up-regulated) or yellow (down-regulated), genes showing no change in expression are gray, and untested genes are white. Genes for which one more SNP and/or indels became fixed are indicated with stars: red for genes located within a hypoxia-selected region and blue for all others.
Because a) there were a number of genes in the Notch pathway or related to Notch signaling that were selected for in AF in contrast to the control flies and b) our previous expression profiling studies demonstrated that the Notch pathway is activated in the AF flies in comparison with control flies (1), we focused our investigation next on the role of the Notch pathway in hypoxia tolerance.
Notch Activation Is Critical for Hypoxia Tolerance in Drosophila melanogaster.
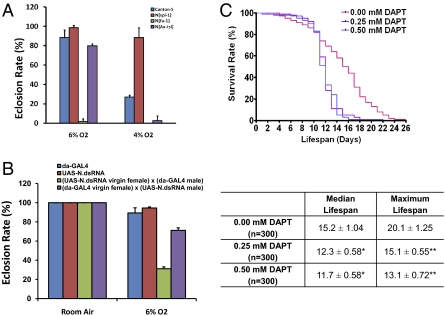
To dissect the contribution of Notch activation to hypoxia tolerance, we used both genetic tools and pharmacologic agents. We first examined the role of Notch in hypoxia tolerance using three homozygous-viable Notch mutants: N[Ax-tsl], N[fa-1], and N[spl-1] (7–9). The N[fa-1] mutation is caused by an insertion of a transposable element (opus) in the second intron of the Notch gene, and N[Ax-tsl] is generated by ethyl methanesulfonate-induced mutagenesis with both N[fa-1] and N[Ax-tsl] having loss-of-function mutations. Unlike N[fa-1] and N[Ax-tsl], N[spl-1] is a Notch gain-of-function allele carrying a point mutation in EGF repeat 14 of the Notch protein that replaces the Isoleucine578 with a Threonine (10). We found that N[fa-1] and N[Ax-tsl] were hypersensitive to hypoxia and had a lowered survival rate, even in much milder hypoxic conditions (i.e., 6% O2) (Fig. 3A). In contrast, N[spl-1] exhibited remarkable hypoxia tolerance and survived 4% O2, much like the AF flies. We next used an RNAi strategy and determined that flies with a knockdown of Notch had a hypoxia-sensitive phenotype (Fig. 3B). These results clearly indicate that Notch function is critical for survival under hypoxia.
Fig. 3.
Hypoxia tolerance in Notch mutants and γ-secretase inhibitor-treated flies. (A) Contribution of Notch to hypoxia tolerance was determined by culturing the homozygous-viable Notch mutants in 6% O2 (relatively mild) or 4% O2 (severe) hypoxic conditions. Canton-S was used as a control. N[spl-1], which is a gain-of-function Notch allele, has a dramatically increased survival rate in hypoxic conditions. In contrast, the N[fa-1] and N[Ax-tsl] loss-of-function mutants had little or no eclosion in severe hypoxic condition and showed a reduced survival rate even in mild hypoxic condition (*P < 0.01, compared with Canton-S control). (B) RNAi-mediated Notch knockdown induces increased sensitivity to hypoxia in flies. Flies carrying a UAS-N.dsRNA transgene on the X chromosome were used to determine the function of Notch in hypoxia tolerance. Two crosses were used to generate flies that had N.dsRNA expression either in all progeny [cross A: (UAS-N.dsRNA virgin female) × (da-GAL4 male)] or only in the female progeny [cross B: (da-GAL4) × (UAS-N.dsRNA)]. Cross A progeny showed an increased sensitivity to hypoxia (*P < 0.01 compared with the cross B control). This hypersensitivity was rescued in cross B progeny in which the function of Notch was knocked down only in females. Both male and female progeny were included in the scoring of A and B crosses. (C) γ-Secretase activation plays important role in hypoxia tolerance in hypoxia-selected flies. Five-day-old adult hypoxia-selected flies were collected and treated with 0.25 or 0.50 mM DAPT and their life span was determined under 1.5% O2. Median life span was the time when 50% of death occurred in the sample, and maximum life span was the time when 90% of sample were dead. Compared with the vehicle-treated controls, flies treated with DAPT showed a significant reduction of both median (*P < 0.01) and maximum life span (**P < 0.01). The median and maximum life spans were calculated using GraphPad Prism 4, and the statistical significance was calculated by Student's t test.
Because the up-regulation of genes encoding aph-1 and nct subunits of γ-secretase suggested that activation of Notch signaling in the AF flies might involve γ-secretase, we used a specific γ-secretase inhibitor, N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) (11) and examined their life span in severe hypoxia in the AF population. We found that DAPT treatment indeed reduced significantly both median and maximum life span in the AF flies (Fig. 3C) but had no significant effect on control flies.
Spatial–Temporal Activation of Notch and Its Downstream Genes in Hypoxia Tolerance.
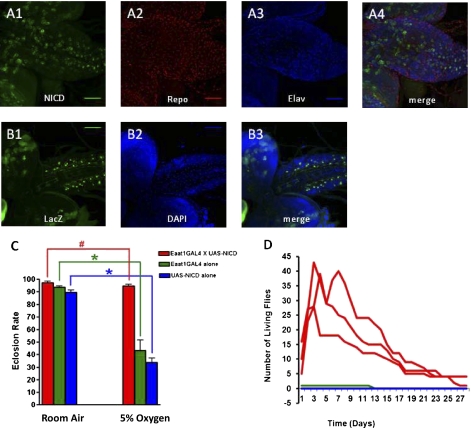
The UAS/GAL4 system was used to determine the critical spatial–temporal activation of the Notch pathway in hypoxia tolerance (12, 13). Several available GAL4 lines were crossed with a UAS–Notch intracellular domain (UAS-NICD) transgenic stock to generate progeny that had specific Notch activation in specific cells/tissues or during specific developmental stages (Table S5). The results showed that specific expression of NICD in the neurons and/or glial cells conferred hypoxia survival in the progeny. For example, the progeny derived from crosses in which NICD was up-regulated in a specific subset of glial cells showed a remarkable increase in hypoxia tolerance and survival. As shown in Fig. 4, Fig. S2, and Table S5, the Eaat-GAL4 and P{GawB}17A-Gal4–driven NICD overexpression in glial cells significantly enhanced both eclosion rate and adult life span (after eclosion) in hypoxia.
Fig. 4.
Increased hypoxia tolerance in flies with NICD overexpression in specific glial cells. (A) Overexpression of NICD driven by the Eaat1-GAL4 in third instar larval brain. The pattern of Eaat1-GAL4–driven NICD (green) overexpression was determined in the progeny of the (Eaat1-GAL4) × (UAS-NICD) cross. Counterstaining with Repo (red) and Elav (blue) reveals that NICD overexpression is mostly in glia (repo+) cells. Z-projection of 27 slices was obtained by confocal microscopy. (B) NICD overexpression-induced activation of Notch in glial cells was determined by crossing (Eaat1-GAL4/Eaat1-GAL4; UAS-NICD/UAS-NICD) to Su(H)-LacZ reporter. The LacZ protein (green) was detected in the cells in the same locations as in A, demonstrating that the Eaat1-GAL4–driven NICD overexpression is functionally active. Z-projection of 9 slices was obtained by confocal microscopy. (C) Glial-specific overexpression of NICD enhances the eclosion rate in hypoxia. #P value = 0.205; *P value <0.001 (unpaired Student's t test). Bars represent the mean ± SEM for six vials (n = 6). (D) Glial-specific overexpression of NICD enhances adult survival in hypoxia. Adult survival during hypoxia was evaluated by counting the number of newly eclosed experimental and control flies and transferring them to a new individual vial. The following day, the number of dead flies in the adult-only vial was subtracted from the previous day's total and the newly eclosed flies from the original vial were counted and added to the adult-only vial to avoid a cumbersome number of vials for all tests done. Hence, the number of flies in an adult-only vial could go down (due to adult death), but later increase (due to addition of newly eclosed adults from the original vial to the adult-only vial).
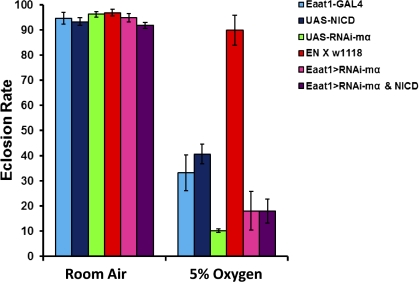
To further explore the mechanisms underlying Notch-mediated hypoxia tolerance, we tested the role of a Notch downstream gene, E(spl)mα, that is located at the E(spl) genomic region and that is significantly up-regulated in the hypoxia-selected flies (1). We generated first a homozygous fly to use in a subsequent cross to provide a Notch overexpression background [i.e., (Eaat1-GAL4/Eaat1-GAL4; UAS-NICD/UAS-NICD) stock, EN line]. This EN line was then crossed to homozygous flies that carry UAS-E(spl)mαRNAi. The progeny of this cross carried Eaat1-GAL4, UAS-NICD, and UAS-E(spl)mαRNAi that allowed us to knock down the target E(spl)mα gene on the Notch overexpression background and determine if Notch activation requires E(spl)mα to regulate hypoxia tolerance. As shown in Fig. 5, Notch activation, which conferred hypoxia tolerance without the RNAi for E(spl)mα, was totally abolished with E(spl)mα knockdown, demonstrating a critical role of E(spl)mα in regulating Notch-induced hypoxia tolerance.
Fig. 5.
Notch activation-conferred hypoxia tolerance requires E(spl)mα. The EN-line flies, which are homozygous for both the Eaat1-GAL4 driver and UAS-NICD, were crossed to homozygous UAS-E(spl)mα. RNAi stock to knock down E(spl)mα on the background of Notch overactivation. Hypoxia tolerance of the progeny was determined by the eclosion rate at a 5% O2 hypoxic condition. Compared with the Notch overactivation control (EN/w1118), knocking down E(spl)mα on the Notch overactivation background abolished hypoxia tolerance (P < 0.01; t test). Bars represent the mean ± SEM (n = 3) for each group/treatment.
Discussion
D. melanogaster has been used as a powerful genetic model for about a century. Because many genes and pathways are evolutionarily conserved between D. melanogaster and humans, Drosophila has become one of the most effective tools for dissecting the genetic mechanisms of human diseases, including developmental and neurological disorders, cancer, cardiovascular disease, and metabolic and storage diseases (for selected reviews see refs. 14–17). The current study has taken advantage of this Drosophila model and used population genetics, deep-sequencing, and molecular strategies to identify potential causative gene(s) underlying hypoxia tolerance. The use of experimental selection methods followed by deep sequencing of pooled individuals is a unique experimental technique in its own right. In a recent publication (18), a similar evolutionary experiment was performed, but with a different phenotype. Their analysis (different from ours) does not point to any specific regions with a genome-wide signature of selection. Rather, they suggest that the adaptation is due to “incomplete sweep models.” By contrast, our experiments and analysis provide a more definitive evidence of a selective sweep. Specifically, we have identified 24 distinct hypoxia-selected regions containing a total of 188 genes in the hypoxia-selected population compared with the naive ones. Interestingly, 20 of these regions (>80%) were located on chromosome X. Because Drosophila males are hemizygous on the X chromosome with no possibility for recombination, recessive X-linked alleles are readily available for selection in males. A consequence may be a more rapid accumulation of favorable mutations as well as stronger purifying selection of deleterious recessive alleles on the X chromosome relative to autosomes (19–21).
The hypoxia-selected regions in the hypoxia-selected flies contain several genes that belong to the Notch signaling pathway and/or regulate the activity of Notch. For example, both Hairless and HDAC4 are important in Notch signaling because downstream genes in the Notch pathway are activated through binding of the NICD to the DNA-binding complex CSL (22). NICD competes for CSL binding with a corepressor complex, which consists of Hairless, CtBP, and Groucho, and additionally recruits histone deacetylases (HDAC) (23). Such enrichment of polymorphic differences in Notch pathway-related genes and the evidence of Notch activation revealed by expression profiling suggested a potentially important role of Notch in hypoxia tolerance. This role of Notch was further investigated using Notch mutants, RNAi-mediated knockdown, and pharmacological inhibitory reagents. These experiments demonstrated that Notch activation confers hypoxia survival. In addition, a remarkable hypoxia survival was observed in flies with NICD overexpression in specific neuronal and/or glial cells, demonstrating the importance of maintaining the integrity of neuronal function in hypoxia tolerance for the whole organism. Although the precise underlying mechanisms regarding the mode of action of Notch remain elusive, we show in this work that the specific NICD activation in particular cells in the central nervous system confers survival and protects from hypoxia-induced death. Indeed, previous studies have demonstrated an anti-apoptotic function of Notch (24–31) and the current study provides evidence demonstrating that the activation of Notch in highly differentiated cells can maintain its survival-promoting property. Furthermore, a recent whole-exome sequencing study of 50 Tibetan subjects revealed that the frequency of a noncoding intronic SNP located at the EPAS1/HIF2α locus is significantly higher in the high altitude-adapted Tibetan subjects than in the lowlander controls, indicating possible involvement of HIF2α in hypoxia tolerance during evolution over many generations (32). It is interesting to note that several studies have demonstrated the interaction between Notch and HIF under hypoxic condition (33–36). In the current study, in addition to proving that Notch is critical for hypoxia tolerance, we have also demonstrated with our newly generated lines overexpressing NICD that the role of Notch in hypoxia tolerance requires its downstream gene E(spl)mα. A previous study has shown that E(spl)mα is activated by Notch and functions in a negative feedback loop to accurately adjust Notch signaling (37), which may be important in distinguishing a cell survival property of Notch from its function of inhibiting cell differentiation. Accumulating evidence demonstrates that hypoxia induces Notch activation in mammals, including humans. For example, chronic constant hypoxia up-regulates Notch1 expression in mouse heart (38) and Notch1 regulates melanoma development by protecting cells from hypoxia-induced cell death (39). Furthermore, Notch3 plays a major role in the development of hypoxic pulmonary hypertension (40) in both humans and rodents. The role of Notch during hypoxia discovered in this study is bound to be important not only in Drosophila but also in humans, raising the distinct possibility of Notch signaling as a potential target for translational and therapeutic strategies.
Materials and Methods
Drosophila Stocks and Culture.
The UAS-NICD and 4XSu(H)-lacZ stocks were provided by J. Posakony. All GAL4 driver lines and N[Ax-tsl], N[fa-1], and N[spl-1] mutants were obtained from Bloomington Drosophila Stock Center at Indiana University. Drosophila stocks were cultured on standard cornmeal/yeast media. To assay for cell type specificity of NICD overexpression homozygous UAS-NICD flies were crossed to Eaat1-GAL4 flies. To test for NICD transcriptional up-regulation, double-homozygous (E/E; N/N) males were crossed to 4xSu(H)-lacZ virgin females. The UAS-RNAi-mα stock was obtained from the Vienna Stock Collection.
Whole-Genome Resequencing.
Genomic DNA was isolated from a pool of 100 male and 100 female adult flies collected from hypoxia-selected populations or generation-matched control populations by standard phenol:chloroform extraction followed by treatment with DNase-free RNase. DNA quality was assessed by spectrophotometry (260/280 and 260/230) and gel electrophoresis. A total of 3 μg was sheared DNA (Covaris) and was used to construct a library for paired-end sequencing. The DNA fragments were subjected to end repair using the End-IT DNA End-Repair Kit (Epicentre) and then ligated to Illumina PE adapters. The adapter-ligated products were purified on Qiaquick spin columns (Qiagen) and PCR amplified with high-fidelity DNA Polymerase in 12 cycles using Illumina’s PE primer set. Cluster generation was performed using the Illumina cluster station and cluster generation kit v2. The 54 + 54 paired-end sequencing was performed using genome analyzer II (Illumina) and sequencing kit v3. The fluorescent images were processed to sequences using the Illumina base-calling pipeline (GA Pipeline-1.4.0). The D. melanogaster reference genome, together with the annotation of genes and repeats, was downloaded from the University of California (Santa Cruz, CA) (UCSC) database (http://genome.ucsc.edu/).
Data Analysis.
The next-generation sequencing data for each of the pools were derived from 200 flies descended from 27 parental strains. Neither individual genotypes nor the number of individuals sampled at a region could be determined, precluding use of standard analysis tools to identify differences between control and hypoxia-tolerant populations. We therefore used two complementary analysis methods. One focused on identifying individual loci with high-confidence allelic differences between the control and hypoxia tolerant populations. The other identified genomic regions characterized by allelic frequencies that differed between control and hypoxia-tolerant flies, representing regions of potential selection. Both analyses used Maq v.0.7.1 (41) under its default parameters to map reads from the four populations (H1, H2, C1, and C2) to the D. melanogaster reference genome downloaded from FlyBase (http://www.flybase.org). As in experimental science, the concordance in results obtained by the two methods provides validation not only for the results obtained but also for the methods used.
DAPT Treatment.
Adult flies were collected from each control and hypoxia-selection chamber (10 vials per chamber, 10 flies per vial, 5 vials of male, and 5 vials of female) (n = 100 for each chamber). DAPT was dissolved with ethanol and diluted with 5% sucrose solution to reach a final experimental concentration of 0.25 or 0.50 mM (final ethanol concentration <1%). Five percent sucrose with 1% ethanol was used as a control. The DAPT or control solution was applied in 150 μL on a filter paper in each vial every other day. The dead flies were counted every 24 h to determine life span. The median and maximum life spans were calculated using GraphPad Prism 4 (GraphPad Software), and the statistical significance was calculated by Student's t test.
Hypoxia Tolerance and Vulnerability Tests.
The survival rate of Notch mutants in hypoxia was determined by culturing them in a computer-controlled environmental chamber. After 3 wk in culture, the numbers of eclosed and total pupae were counted. The ratio between elcosed pupae and the total number of pupae was calculated and presented as eclosion rate.
The UAS-NICD stock was crossed with specific GAL4 transgenic flies to determine the effect of specific spatiotemporal NICD overexpression on hypoxia tolerance. Each cross contained 10 virgin female homozygous UAS-NICD flies and 5 male homozygous GAL4 transgenic flies and allowed them to lay eggs for 48 h in normoxia. The flies were then moved to a control vial (for another 48 h before being discarded) and the vial with the eggs was moved to a 5% oxygen chamber with a 12-h dark and 12-h light cycle with a temperature of 22 ± 1 (°C. In parallel, the parental line without the cross wasere tested in normoxia as a control. After 4 wk, both sets of flies were assayed for the number of pupal cases that were empty or full. Six vials of each condition were completed in two2 different experiments for a minimum of 500 pupal cases scored to calculate the eclosion rate for each condition. Adult survival during hypoxia was evaluated by counting the number of newly eclosed experimental and control flies and transferring them to a new individual vial. The following day, the number of adult dead flies in the adult-only vial was subtracted from the previous day's total and the newly eclosed flies from the original vial were counted and added to the adult-only vial to avoid a cumbersome number of vials for all tests done. Hence, the number of flies in the adult-only vial could go down (due to adult death), but later increase (due to the addition of newly eclosed adults from the original vial to the adult-only vial).
The statistical significance of eclosion rate between mutants, NICD-overexpressed flies, and controls was calculated by unpaired t test.
See SI Materials and Methods for full methods.
Supplementary Material
Acknowledgments
We thank O. Gavrialov, M. Y. Hsiao, Y. Lu-Bo, J. Wang, and N. Morgan for technical assistance. Confocal microscopy was performed in the University of California at San Diego Neuroscience Microscopy Shared Facility (P30 NS047101). This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (5P01HD032573); the National Institute of Neurological Disorders and Stroke (5R01NS037756); the National Heart, Lung, and Blood Institute (5R33HL087375); the National Human Genome Research Institute (5R01HG004962); the National Science Foundation (DBI-0641037 and NSF-III-#0810905); and an American Heart Association Award (0835188N).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010643108/-/DCSupplemental.
References
- 1.Zhou D, et al. Mechanisms underlying hypoxia tolerance in Drosophila melanogaster: Hairy as a metabolic switch. PLoS Genet. 2008;4:e1000221. doi: 10.1371/journal.pgen.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou D, et al. Experimental selection for Drosophila survival in extremely low O(2) environment. PLoS ONE. 2007;2:e490. doi: 10.1371/journal.pone.0000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 6.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artavanis-Tsakonas S, et al. The Notch locus of Drosophila melanogaster: A molecular analysis. Dev Genet. 1984;4:233–254. [Google Scholar]
- 8.Kidd S, Young MW. Transposon-dependent mutant phenotypes at the Notch locus of Drosophila. Nature. 1986;323:89–91. doi: 10.1038/323089a0. [DOI] [PubMed] [Google Scholar]
- 9.Shellenbarger DL, Mohler JD. Temperature-sensitive mutations of the notch locus in Drosophila melanogaster. Genetics. 1975;81:143–162. doi: 10.1093/genetics/81.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartley DA, Xu TA, Artavanis-Tsakonas S. The embryonic expression of the Notch locus of Drosophila melanogaster and the implications of point mutations in the extracellular EGF-like domain of the predicted protein. EMBO J. 1987;6:3407–3417. doi: 10.1002/j.1460-2075.1987.tb02664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dovey HF, et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 12.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 13.Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- 14.Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- 16.Lu B, Vogel H. Drosophila models of neurodegenerative diseases. Annu Rev Pathol. 2009;4:315–342. doi: 10.1146/annurev.pathol.3.121806.151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ocorr K, et al. Genetic control of heart function and aging in Drosophila. Trends Cardiovasc Med. 2007;17:177–182. doi: 10.1016/j.tcm.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke MK, et al. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature. 2010;467:587–590. doi: 10.1038/nature09352. [DOI] [PubMed] [Google Scholar]
- 19.Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- 20.Singh ND, Davis JC, Petrov DA. Codon bias and noncoding GC content correlate negatively with recombination rate on the Drosophila X chromosome. J Mol Evol. 2005;61:315–324. doi: 10.1007/s00239-004-0287-1. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi KH, Tanaka K, Itoh M, Takano-Shimizu T. Reduced X-linked rare polymorphism in males in comparison to females of Drosophila melanogaster. J Hered. 2009;100:97–105. doi: 10.1093/jhered/esn078. [DOI] [PubMed] [Google Scholar]
- 22.Bray SJ. Notch signalling: A simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 23.Nagel AC, et al. Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol Cell Biol. 2005;25:10433–10441. doi: 10.1128/MCB.25.23.10433-10441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Androutsellis-Theotokis A, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 25.Ciofani M, Zúñiga-Pflücker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 26.Eliasz S, et al. Notch-1 stimulates survival of lung adenocarcinoma cells during hypoxia by activating the IGF-1R pathway. Oncogene. 2010;29:2488–2498. doi: 10.1038/onc.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jögi A, et al. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc Natl Acad Sci USA. 2002;99:7021–7026. doi: 10.1073/pnas.102660199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne BA, Minter LM. Notch signalling during peripheral T-cell activation and differentiation. Nat Rev Immunol. 2007;7:64–75. doi: 10.1038/nri1998. [DOI] [PubMed] [Google Scholar]
- 29.Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: The complex cross talk between Notch and NF-kappaB. Lab Invest. 2008;88:11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- 30.Perumalsamy LR, Nagala M, Sarin A. Notch-activated signaling cascade interacts with mitochondrial remodeling proteins to regulate cell survival. Proc Natl Acad Sci USA. 2010;107:6882–6887. doi: 10.1073/pnas.0910060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci USA. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gustafsson MV, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Pistollato F, et al. Interaction of HIF1alpha and Notch signaling regulates medulloblastoma precursor proliferation and fate. Stem Cells. 2010;28:1918–1929. doi: 10.1002/stem.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, et al. Transcriptional regulation of APH-1A and increased gamma-secretase cleavage of APP and Notch by HIF-1 and hypoxia. FASEB J. 2006;20:1275–1277. doi: 10.1096/fj.06-5839fje. [DOI] [PubMed] [Google Scholar]
- 36.Zheng X, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci USA. 2008;105:3368–3373. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Apidianakis Y, Nagel AC, Chalkiadaki A, Preiss A, Delidakis C. Overexpression of the m4 and malpha genes of the E(spl)-complex antagonizes notch mediated lateral inhibition. Mech Dev. 1999;86:39–50. doi: 10.1016/s0925-4773(99)00099-4. [DOI] [PubMed] [Google Scholar]
- 38.Fan C, et al. Gene expression and phenotypic characterization of mouse heart after chronic constant or intermittent hypoxia. Physiol Genomics. 2005;22:292–307. doi: 10.1152/physiolgenomics.00217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bedogni B, Warneke JA, Nickoloff BJ, Giaccia AJ, Powell MB. Notch1 is an effector of Akt and hypoxia in melanoma development. J Clin Invest. 2008;118:3660–3670. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, et al. Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat Med. 2009;15:1289–1297. doi: 10.1038/nm.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.