Abstract
Although the first poly(A) polymerase (PAP) was discovered in Escherichia coli in 1962, the study of polyadenylation in bacteria was largely ignored for the next 30 years. However, with the identification of the structural gene for E. coli PAP I in 1992, it became possible to analyze polyadenylation using both biochemical and genetic approaches. Subsequently, it has been shown that polyadenylation plays a multifunctional role in prokaryotic RNA metabolism. While the bulk of our current understanding of prokaryotic polyadenylation comes from studies on E. coli, recent experiments with Cyanobacteria, organelles and Archaea, although limited, have widened our view on the diversity, complexity, and universality of the polyadenylation process.
For example, the identification of polynucleotide phosphorylase (PNPase), a reversible phosphorolytic enzyme that is highly conserved in bacteria, as an additional PAP in E. coli caught everyone by surprise. In fact, PNPase has now been shown to be the source of post-transcriptional RNA modifications in a wide range of cells of prokaryotic origin including those that lack a eubacterial PAP homologue. Accordingly, the past few years have witnessed increased interest in the mechanism and role of post-transcriptional modifications in all species of prokaryotic origin. However, the fact that many of the poly(A) tails are very short and unstable as well as the presence of polynucleotide tails has posed significant technical challenges to the scientific community trying to unravel the mystery of polyadenylation in prokaryotes. This review discusses the current state of knowledge regarding polyadenylation and its functions in bacteria, organelles and Archaea.
Keywords: poly(A) polymerase, polynucleotide phosphorylase, Hfq, RNA degradation
Polyadenylation is a post-transcriptional event that involves the addition of untemplated adenosine residues to the 3’ ends of RNA substrates. The first bacterial poly(A) polymerase (PAP) was identified almost 50 years ago in Escherichia coli [1,2]. A PAP was also identified in eukaryotic cells at about the same time [3,4]. However, polyadenylation in bacteria was virtually ignored for next 30 years, in part because eukaryotic poly(A) tails were relatively long, nearly uniform in length, and were found on almost all mRNAs. Furthermore, even though poly(A) tails were detected in E. coli and Bacillus subtilis [5–9], the overall low abundance of polyadenylated transcripts and the apparent lack of evidence for a physiological role led to the belief that polyadenylation was only important in higher organisms (See definition of polyadenylation in the glossary of Lewin, Genes I through Genes VIII).
PAP I, encoded by the pcnB gene, and polynucleotide phosphorylase (PNPase), a 3’→ 5’ exonuclease encoded by the pnp gene, are responsible for the post-transcriptional addition of 3’ tails to transcripts in exponentially growing E. coli [10,11]. Interestingly, in vivo PAP I synthesized tails exclusively contain A residues, while PNPase synthesized tails are primarily heteropolymeric (the tails contain all four nucleotides but are ~50% A) [10,12]. It has now been shown that polyadenylation in many prokaryotic organisms, Archaea, and organelles of prokaryotic origin lacking a PAP I protein is carried out by PNPase. In fact, PNPase functioning as a polymerase is probably much more prevalent than previously thought [13–16].
Organelles such as mitochondria and chloroplasts are thought to have originated from ancient invasions and successful symbiosis of eukaryotic hosts by eubacteria more than a billion years ago [17,18]. Analysis of the genomic organization of these organelles indicates that while mitochondria evolved from α-proteobacterium, chloroplasts originated from a cyanobacterial ancestor. During the evolutionary development that shaped organelle biogenesis, both chloroplasts and mitochondria lost many of their eubacterial characteristics and acquired many host-derived properties. However, for the most part, their post-transcriptional activities still mimic bacterial systems with only a few significant differences. Recently, post-transcriptional modifications have also been reported in hyperthermophilic and methanogenic Archaea [15,16,19].
In prokaryotes, the exact method of substrate selection by either PAP or PNPase is still not clear. However, it would appear that any transcript that is a substrate for the exonucleolytic activity of PNPase can probably also be modified by the addition of polynucleotide tails [20]. Furthermore, the observation that Hfq, an abundant RNA binding protein, modulates poly(A) levels in E. coli [12,21] has raised the interesting question of whether PAP I acts independently in vivo as suggested by in vitro experiments [22]. The reduced ability of the PAP I protein to add poly(A) tails at the 3’ termini of mRNAs containing Rho-independent transcription terminators in Hfq mutants coupled with the increase in the biosynthetic activity of PNPase has suggested that the regulation of polyadenylation involves a multiprotein complex [12]. The distinct difference in the polyadenylation pattern of transcripts with and without Rho-independent transcription terminators also suggests the presence of a discrete polyadenylation signal in E. coli [20].
While many transcripts decay rapidly following polyadenylation, recent studies indicate that its major role involves quality control for transcriptional or processing errors [23]. In addition, polyadenylation in E. coli has been implicated as a sensing mechanism for adjusting the levels of ribonucleases such as RNase E and PNPase [24]. Although the studies on E. coli transcripts have led the way towards a better understanding of the molecular mechanism and role of prokaryotic polyadenylation, the detection of poly(A) tails in all three domains of life has established their universal presence. In this review, we describe how information gained from experiments over the past decade has expanded our knowledge of the role played by polyadenylation in the post-transcriptional regulation of prokaryotic, archaeal and organellar gene expression. Readers are also encouraged to consult other comprehensive reviews that have been published recently on the subject [25–27].
I. NATURE OF 3’-TAILS: POLY(A), POLY(U), AND POLYNUCLEOTIDE TAILS
The initial discovery of poly(A) tails, comprised of multiple untemplated adenosine residues, at the 3’ ends of RNA substrates dates back to early 1960’s. With the development of new detection techniques and analytical procedures, scientists have identified post-transcriptionally added 3’ tails that contain combinations of all four nucleotides and are present in many different organisms (Table 1). For example, many 3’ tails are A-rich polynucleotide tails (the tails contain all four nucleotides but generally are ~50% A) [10,15,28–32] and some are A/U tails (contain a few U residues besides A) [33]. For sake of clarity, in this article we refer poly(A) tails as homopolymeric adenosine tails and all other kind as polynucleotide tails (Fig. 1). It should also be noted that polyuridylated [poly(U)] tails have recently been reported in both human and Chlamydomonas mitochondrial RNA [34–36].
Table 1.
Nature and source of 3’ tails in bacteria, Archaea and organelles
| Bacteria | Nature of tails | Protein/Gene | Mol. Wt (KDa) | Crystal Structure |
Reference |
|---|---|---|---|---|---|
| E. coli | Poly(A) Polynucleotide |
PAP I/pcnB PNPase/pnp |
53.9 77.1 |
ND Yes |
[12,20,47] [105] |
| Bacillus subtilis | Poly(A) Polynucleotide |
ND PNPase/pnpA, |
- 77.5 |
- ND |
[29] |
| Streptomyces coelicolor | Poly(A) Polynucleotide |
ND ND |
- - |
- - |
[28,56] |
| Pseudomonas aeruginosa | Unknown | ND | - | - | [166] |
| Caulobacter cresentus | Poly(A) | ND | - | - | [167] |
| Rhodospirillum rubrum | Poly(A) | ND | - | - | [168] |
| Mycobacterium | Unknown | ND | - | - | [169] |
| Cyanobacteria | Polynucleotide | PNPase/Sll1043 | - | - | [31] |
| Organelles | |||||
| Spinach Chloroplasts | Polynucleotide | PNPase/ND | ~100 | ND2 | [13,30,57] |
| Algae Chloroplasts | Polynucleotide (>98% A) | ND | [62] | ||
| Arabidopsis Chloroplasts | Unkown | ND | [35,63] | ||
| Plant Mitochondria | Poly(A) Polynucleotide |
ND ND |
[65,67] | ||
| Mammalian Mitochondria | Poly(A) | hmtPAP/hmtPAP | 65–68 | ND | [68,69] |
| Trypanosomes Mitochondria | Poly(A) Polynucleotide (A/U) |
KPAP1/ND | ND | ND | [74] |
| Yeast Mitochondria | Unknown | ND | [76] | ||
| Archaea | |||||
| Hyperthermophiles | Polynucleotide | Exosome* | ~250 | Yes | [19,80] |
| Methanogens (with exosome) | Polynucleotide | Exosome* | ~250 | Yes | [15] [80] |
| Methanogens (no exosome) | None | [15] | |||
| Halophile | None | [19] | |||
The archaeal exosome is a nine subunit complex. Each subunit is composed of two proteins Rrp41 and Rrp42 (homologous to RNase PH) and another protein containing a KH/S1 domain [80].
ND: Not determined
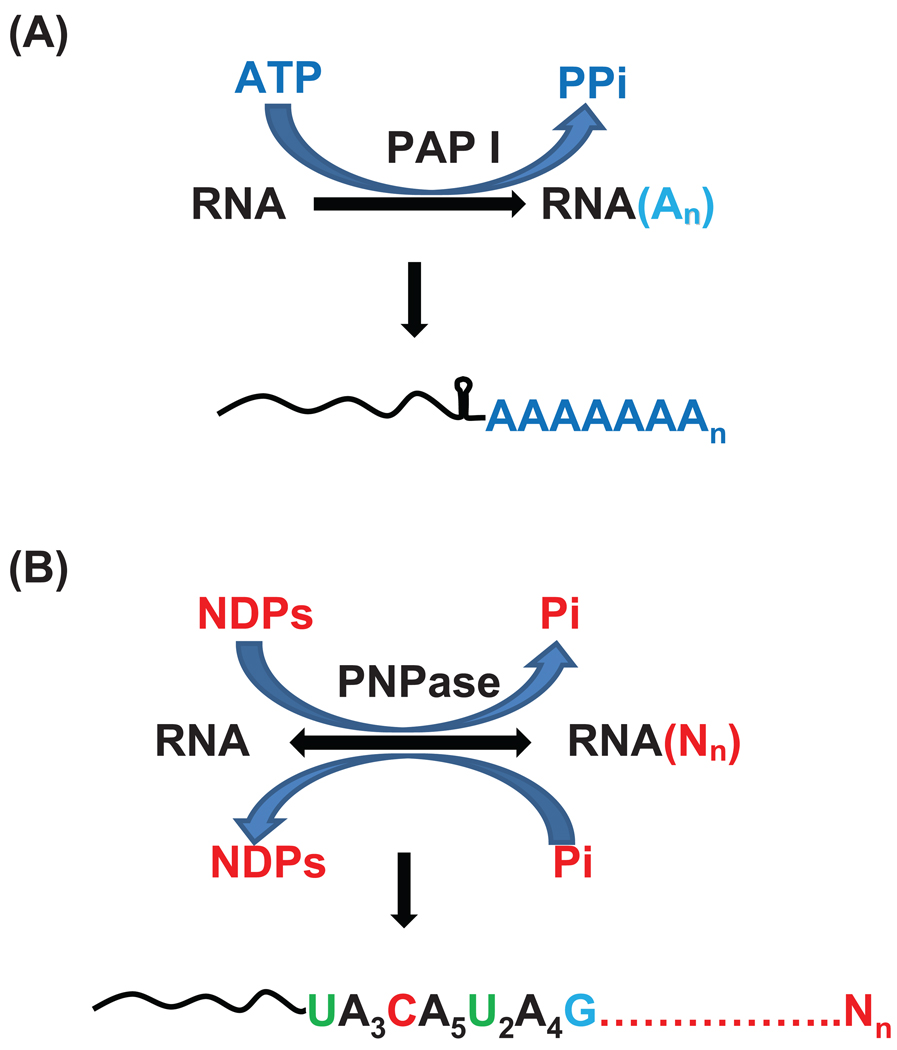
Fig. 1.
Addition of (A) poly(A) tails by PAP I and (B) polynucleotide tails by PNPase in E. coli. Since the equilibrium constant of the PNPase catalyzed reaction is close to one, the enzyme can work either degradatively or biosynthetically depending upon the availability of inorganic phosphate (Pi). High NDP and low Pi concentrations favor the biosynthetic reaction which generates untemplated polynucleotide tails. Low NDP and high Pi concentrations favor the exoribonucleolytic degradation of transcripts. N: any nucleotide.
II. PROTEINS INVOLVED IN POLYADENYLATION
Bioinformatic studies have shown that eubacterial PAPs, which belong to a superfamily of nucleotidyl transferases [37,38], are present in only a limited number of bacteria and plant organelles and are absent in Archaea and eukaryotes. In fact, it is believed that eubacterial PAPs have evolved fairly recently such that they do not exist in many bacterial species [39]. In the absence of a PAP I-like activity, post-transcriptional modification of RNA species is carried out by other enzymes such as PNPase and the exosome [13,40]. Recent data suggests that bacterial PAPs have evolved from the CCA-adding enzymes (tRNA nucleotidyltransferases) as evident from the presence of comparable structural elements within these proteins [41,42].
a. Bacteria
PAP I (ATP:polyribonucleotide adenylyl transferase) I is the major polyadenylating enzyme in E. coli. It catalyzes the template independent addition of AMP moieties to 3’-hydroxyl termini of RNA substrates using ATP as a substrate (Fig. 1). Although PAP I can inefficiently add the other three nucleotides in vitro [43,44], in vivo the enzyme only synthesizes homopolymeric poly(A) tails [10]. The structural gene encoding PAP I (pcnB, plasmid copy number) was first identified based on N-terminal sequencing of the purified PAP I protein from E. coli [11]. While deletion of pcnB has only a moderate effect on growth rate [11,45,46], overproduction of PAP I is highly toxic [11,47].
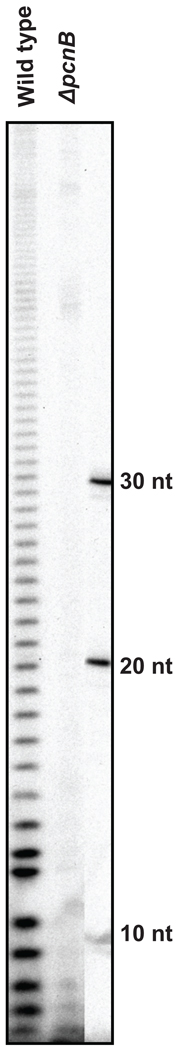
PAP I accounts for ~90% of the poly(A) tails in E. coli, which are on average 15–30 nt long [10,12,20,48]. However, a limited number of the poly(A) tails can be longer than 30 nt, while the bulk of the tails are less than 10 nt in length (Fig. 2). Since deletion of the pcnB gene did not abolish all the polyadenylating activity in cell extracts (Fig. 2), it was speculated that E. coli contained a second PAP (PAP II). The f310 gene, encoding a 36 KDa protein, was initially identified as the putative PAP II [49], even though the protein had no sequence similarity to the nucleotidyl transferase superfamily of proteins [50]. Further investigation of F310, however, failed to support its involvement in polyadenylation [51]. Specifically, a pcnB f310 double mutant was still viable and overexpression of F310 had no significant effect on poly(A) levels or tail length either in vivo or in vitro [51]. Furthermore, cells containing increased levels of F310 did not show a detectable change in the copy number of a ColE1 plasmid [51].
Fig. 2.
Poly(A) tail profiles in wild type and pcnB deletion strains of E. coli. Total RNA was processed for the poly(A) sizing assay as described by Mohanty et al. [55].
Subsequently, it was noted that E. coli contained three additional proteins that belonged to the same nucleotidyl transferase superfamily as PAP I [50]. These are PNPase encoded by pnp, RNase PH encoded by rph, and tRNA nucleotidyl transferase encoded by cca. While PNPase, as well as its homologue RNase PH, are both reversible enzymes that can either degrade RNA by using inorganic phosphate or synthesize RNA by using NDPs as precursors (Fig. 1) [52,53], the high intracellular levels of inorganic phosphate (>10 mM) [54] led to the assumption that this class of enzyme functioned exclusively in vivo as an exoribonuclease. However, Mohanty and Kushner [10] demonstrated that PNPase was, in fact, the second E. coli PAP, which adds A rich polynucleotide tails to the 3’ ends of RNA (Fig. 1). Not only did the identification of PNPase as the second PAP in E. coli significantly alter more than 50 years of thinking about how PNPase works in vivo, but it also provided the much needed basis for identification of post-transcriptional modifications in organisms and organelles lacking an eubacterial PAP (see below).
Unfortunately, current techniques do not allow researchers to accurately measure the length or amount of either poly(A) or polynucleotide tails. Poly(A) sizing assays (Fig. 2) [55], which provide the most accurate status regarding the size of poly(A) tails in the cell, are not helpful for estimating the length of polynucleotide tails since RNase A and RNase T1 cleave after C/U and G residues, respectively. Furthermore, traditionally researchers have used oligo(dT)-dependent detection [55] for identifying polynucleotide tails in all organisms [10,12,19,20,28–30,56,57]. This approach, however, probably does not reflect the accurate length of such tails. With this caveat, the average length of polynucleotide tails have been reported to be in the 100’s of nucleotides compared to poly(A) tails, which are relatively short except for Trypanosomes (Table 2A–B).
Table 2.
| (A) Average length of poly(A) tails reported in bacteria and organelles | ||
|---|---|---|
| Bacteria/Organelles | Length (nt) | References |
| E. coli | 1–50 | [12,47,48] |
| B. subtilis | 1–40 | [29] |
| Plant Chloroplasts | Unknown | [35,63] |
| Plant mitochondria | 5–36 | [65–67] |
| Mammalian Mitochondria | 40–75 | [68–70] |
| Trypanosome Mitochondria | 20–25 120–250 |
[74] |
| Yeast Mitochondria | 1–8 | [76] |
| (B) Actual length of poly(A) tails reported on specific transcripts in wild type prokaryotes and organelles | |||
|---|---|---|---|
| Bacteria/ Organelles |
Transcripts | Length (nt)* | References |
| E. coli | 5S rRNA | 1 | [170] |
| 16S rRNA | 15–18 | [47] | |
| 23S rRNA | 17–18 | [47] | |
| GlmY | 1–8 | [93] | |
| lpp | 16–28 | [12,47] | |
| rmf | 1–5 | [171] | |
| RNA I | 3 | [94] | |
| rpsO | 1–20 | [172]; Mohanty & Kushner, Unpublished results | |
| ompA | 17–31 | [20] | |
| cysT | 1 | [82] | |
| hisR | 1–3 | [82] | |
| leuU | 1–3 | [82] | |
| leuX | 1–5 | [83] | |
| B. subtilis | rnpB | 2–14 | [29] |
| cry1Aa | 2–7 | [29] | |
| 23S rRNA | 2–7 | [29] | |
| tRNACys-LeuU | 1–2 | [29] | |
| Streptomyces | leuA | 17 | [60] |
| 16S rRNA | 12–15 | [28] | |
| 23S rRNA | 9–16 | [28] | |
| Plant Mitochondria | cox2 | 14–36 | [65] |
| atp9 | 5–17 | [67] | |
| 18S rRNA | 4–6 | [66] | |
| Human Mitochondria | |||
| ND3 | 18–58 | [68] | |
| ATP618 | 13–62 | [68] | |
| COX III | 10–56 | [68] | |
| CYTB | 29–36 | [68] | |
| 12S rRNA | 1–2 | [68] | |
The length of poly(A) tails reported were measured by various techniques. In our experience, each technique has specific limits on the types and length of poly(A) tails can be detected.
While post-transcriptional addition of 3’ tails has now been reported in a large number of bacterial species, many of the proteins responsible for these activities have yet to be identified (Table 1). Both poly(A) and polynucleotide tails, with a mean size of ~40 nt, have been reported in the gram positive bacterium B. subtilis [29]. Although the presence of two PAP activities was reported in a pnp mutant of B. subtilis, their identities are still unknown. A protein encoded by papS, with 18.9% identity to E. coli PAP I, was predicted to have PAP activity in B. subtilis [58]. However, careful characterization of the protein in vitro revealed that it had tRNA nucleotidyl transferase activity and the gene was renamed cca [59]. Furthermore, the fact that the absence of PNPase did not change the polyadenylation profile significantly indicated its limited role in the post-transcriptional modifications observed in B. subtilis [29].
Similarly, a putative E. coli PAP I homologue in S. coelicolor with 36% amino acid sequence identity [56] was also identified to be a tRNA nucleotidyl transferase [14]. However, cloning and sequencing data strongly suggest that a PNPase homologue plays an important role in generating the predominately polynucleotide tails in vivo [28,60,61].
b. Organelles
Although no E. coli PAP I homologues have been detected in the chloroplast genome, the identification of PNPase as the second PAP in E. coli [10] led to the demonstration that its homologue was the only enzyme responsible for polyadenylation in the spinach chloroplast [13], producing predominately polynucleotide tails [30,57]. Subsequently, polyadenylation in Cyanobacteria, the ancestor of chloroplasts, was also shown to be carried out by PNPase [31]. Interestingly, the 3’-tails in the chloroplasts of Chlamydomonas are nearly homopolymeric (>98% A) [62], even though recent data suggests that PNPase may be the only polyadenylating enzyme present [35]. The nature of 3’ tails in Arabidopsis chloroplasts is still unknown [35,63]. Moreover, an increase in PNPase levels directly correlated with a decrease in the level of polyadenylation, suggesting a degradative role rather than synthetic role for the enzyme [63]. A more recent study, however, suggests that the poly(A) tails may be synthesized by a nuclear encoded eubacterial PAP homologue targeted to the chloroplast and mitochondria [35].
The nature of post-transcriptional modifications in mitochondria varies significantly among organisms [64] (Table 1). Plant mitochondrial RNA contains mostly poly(A) tails [65–67], although some polynucleotide tails have been reported [65]. However, no PAP-like enzymes have been identified yet. Furthermore, mtPNPase most likely plays a major role as a degradative enzyme [66,67]. Poly(A) tails of 40–60 nt in length have been detected in mammalian mitochondrial mRNAs [68–70], but the occasional incorporation of other nucleotides has also been reported [68,71]. The presence of mostly homopolymeric tails suggests their synthesis by a specific PAP.
While a rat liver mitochondria specific PAP has been reported [72], its identity is still a mystery. In contrast, two groups have independently identified a human mitochondrial poly(A) polymerase (hmtPAP), which directly affects the length of poly(A) tails in mitochondria [68,69]. However, silencing of hmtPAP only reduced poly(A) tail length from ~50 nt to ~8 nt. Recent data suggest that more than one polyadenylating enzyme is responsible for polyadenylation in human mitochondria [36,68]. PNPase is probably not one of them because of the presence of predominantly homopolymeric tails and the localization of PNPase in the intermembrane space whereas RNA polyadenylation occurs in the matrix [68,69,73].
The mitochondria of Trypanosome brucei, one of the earliest branching eukaryotes, contain poly(A) tails that can broadly be divided into two classes, short (~20–25 nt) and long (~120–250 nt) tails [74]. While short tails are adenosine homopolymers, the long tails contain many uridine residues [75]. KPAP1 (kinetoplast poly(A) polymerase 1) was recently identified as the T. brucei mitochondrial PAP responsible for synthesizing both the short and long tails [75]. Interestingly, the enzyme is also essential for parasite viability and mitochondrial function.
Yeast mitochondria lack both PAP and PNPase homologues. However, poly(A) tails up to approximately 8 nt in length have been detected in Saccharomyces cerevisiae mitochondrial RNA [76], although no specific RNAs containing poly(A) tails have been identified. Instead, an A/U-rich dodecamer sequence that is encoded in the yeast mitochondrial genome and is attached to the 3’ ends of mRNAs, providing protection from exonucleolytic degradation has been identified [64,77,78].
c. Archaea
Neither eubacterial PAP or PNPase homologues have been identified in any class of Archaea. However, an archaeal exosome, which bears both structural and functional similarities to prokaryotic PNPases, has been found in both hyperthermophiles and some methanogens, but not in halophiles (Table 1). The archaeal exosome is a nine subunit complex containing three copies of proteins Rrp41 and Rrp423, which are homologuges of RNase PH and another protein containing a KH/S1 RNA binding domain [79]. The crystral structure of the exosome assembly has a ring structure that is very similar to bacterial PNPases [40,80]. Thus, not surprisingly, polynucleotide tails have been detected in hyperthermophiles and methanogens containing exosomal assemblies [15,19].
III. REGULATION OF POLYADENYLATION
It has been estimated that as much as 15–25% of total RNA is polyadenylated in B. subtilis [8]. In contrast, less than 2% of total RNA in E. coli is estimated to be polyadenylated [5–7], even though recent genome-wide analysis suggests that polyadenylation of E. coli transcripts occurs more frequently than previously envisioned [20]. A comparison of the oligo(dT)-dependent cDNA transcriptome between the wild type and ΔpcnB strains indicated that the majority of transcripts (~90%) undergo some degree of polyadenylation either as full-length transcripts or decay intermediates during exponential growth [20]. However, the most important question, namely what fraction of each full-length transcript gets polyadenylated still remains unanswered.
The limited data available indicate that the level of polyadenylation of specific transcripts actually varies significantly and is independent of both transcript size and overall abundance (Table 3). For example, while both 16S and 23S rRNAs transcripts are highly abundant, only 0.6% of 16S rRNA transcripts are polyadenylated compared to 10% for the 23S rRNA [47]. Similarly, the percentage of polyadenylated transcripts among specific mRNAs varies from as low as 0.4 % to as high as 10% in E. coli (Table 3). Similar observations have been made for plant mitochondrial mRNAs [65].
Table 3.
Percentage of specific transcripts polyadenylated in E. coli
| Transcripts | % polyadenylated | Reference |
|---|---|---|
| 23S rRNAa | 10 ± 2 | [47] |
| 16S rRNAa | 0.6 ± 0.2 | [47] |
| lppa | 0.43–0.74 ± 0.02 | [20] |
| ompAa | 1.3 ± 0.1 | [20] |
| rpsO | 10 | [172] |
Full-length transcripts
The precise reason for the significantly lower levels and limited length of prokaryotic poly(A) tails compared to their eukaryotic counterparts is not clear. It is possible that the tails are added only to a limited number of transcripts in response to specific needs such as for RNA surveillance and/or processing pathways [23,81]. However, there are also multiple levels of regulation both before and after the synthesis of poly(A) tails in E. coli, which are discussed in the following sections.
a. Low intracellular levels of PAP I
It is estimated that there are only 32–50 molecules of PAP I in E. coli [12]. One of the reasons for such a low PAP I level may be related to the toxicity, which is rapid and irreversible, when excess PAP I is synthesized in the cell [47]. Even though the exact reason for the toxicity is not clearly understood at this time, macroarray analysis ruled out the possibility of rapid turnover of one or more mRNAs essential for cell viability during increased polyadenylation (Mohanty & Kushner, unpublished results). Since PAP I uses ATP as substrate, it is possible that rapid ATP depletion and/or excess NDP accumulation may cause the toxicity. However, this hypothesis is unlikely since bacteria growing in a rich medium should be able to quickly replenish ATP levels. Another possibility is the polyadenylation of essential RNAs, which are not usually substrates for PAP I in a wild type cell, such as the mature CCA termini of tRNAs. Addition of even a single A residue will render a mature, uncharged tRNA non-functional, leading to a shutdown of protein synthesis. In fact, the mature 3’ termini of tRNAs in wild type strains do not seem to be polyadenylated [82,83]. Similarly, excessive polyadenylation of small regulatory RNAs (sRNAs) might lead to a change in conformation thereby altering their functionality.
Thus it may not be surprising that PAP I levels in wild-type E. coli appear to be kept very low by a combination of factors. Changes in the in vivo PAP I level as a function of growth rate have been reported [84]. Furthermore, while pcnB transcription does not appear to be autoregulated, the steady-state level of the transcript is low despite having a moderately strong promoter [47,85]. PAP I levels are also downregulated by the presence of a weak non-canonical translation initiation codon [11,85].
Besides transcriptional and translational control, PAP I activity may also be controlled through specific localization or modification(s) of the protein. The recent demonstration that PAP I is either membrane localized or cytosolic based on growth phase is one such example [86,87]. However, it is possible that such localization may be indirect through a loose association with RNase E, which is also membrane localized [88]. Furthermore, a preliminary study indicates that PAP I may be phosphorylated in E. coli [87], a modification that helps regulate human PAP activity [89,90]. While not much is known about the conditions regulating the addition of polynucleotide tails by PNPase, a recent study suggests that the intracellular ATP concentration may play a role in the process [91].
b. Substrate selection
One of the biggest mysteries in prokaryotic polyadenylation is the nature of substrate selection by the various polyadenylating enzymes. In E. coli, the low level of PAP I is most probably one of the major factors in substrate selection, since transient overproduction of PAP I significantly increased the number of polyadenylated transcripts [20,47]. In vitro data suggest that PAP I selects its substrates based on structural features without help from any ancillary protein(s). Thus, it was reported that a substrate with single-stranded segment at either 5’ or 3’ end, along with monophosphorylation at an unpaired 5’ terminus, becomes highly susceptible to 3’ polyadenylation [22]. However, this work has never been reproduced. In fact, PAP I has been reported to have physical interactions both in vivo and in vitro with Hfq, PNPase, RNase E and the DEAD-box RNA helicase encoded by rhlB [12,92]. In fact, the interactions between PAP I, Hfq and PNPase have been proposed to play an important role in deciding between PAP I and PNPase as the polymerizing enzyme (see below).
While it has long been presumed that 3’ polyadenylation is restricted only to mRNAs, another unexpected feature of PAP I and PNPase in prokaryotes appears to be their ability to polyadenylate almost any RNA species, including rRNAs and tRNAs and sRNAs (Table 4). What is also interesting is that the tails found on tRNAs and sRNAs tend to be very short (1–8 nt) (Table 2B) [47,83,93,94], while those on rRNAs resemble the ones found on mRNAs (Table 2B) [12,82,83]. Unfortunately, the abundance of very short poly(A) tails (less than 12 nts) (Fig. 2) has made it technically extremely difficult to accurately assess the true state of polyadenylation in a bacterium such as E. coli.
Table 4.
Identified transcripts with poly(A) or polynucleotide tails in bacteria, organelles and Archaea
| Group | Species | RNA Type | Transcripts | Reference |
|---|---|---|---|---|
| Bacteria | E. coli | mRNA | lpp, rpsO, ompA, secG, rmf, pcnB, trxA | [10,12,20,47,126,171] |
| rRNA | 16S rRNA, 23S rRNA | [47] | ||
| nc RNA | 6S RNA, 4.5S RNA, RNA I, SoK, SraK, SraL, GlmY, SsrA, RnpB |
[94,124,173–175] | ||
| tRNA | cysT, hisR, leuX, trpT, leuU, tyrT, tyrV | [82,83,175] | ||
| B. subtilis | mRNA | rnpB, rpsD, cry1Aa | [29] | |
| rRNA | 23S rRNA | [29] | ||
| tRNA | tRNACys-LeuU | [29] | ||
| Streptomyces | mRNA | redD, actII-orf4, pnp, clpP, leuA | [28,60] | |
| rRNA | 16S rRNA, 23S rRNA | [60] | ||
| Synechocystis | mRNA | rbcL | [31] | |
| rRNA | 23S rRNA | [31] | ||
| tRNA | tRNAFmet | [31] | ||
| Chloroplast | Spinach | mRNA | psbA, petD | [30,176] |
| Algae | mRNA | cox1, atpB, petD | [35,62] | |
| rRNA | 5S rRNA | [62] | ||
| tRNA | tRNAArg, tRNAGlu | [62] | ||
| Plant | mRNA | psbA, rbcL, rps14 | [63] | |
| Mitochondria | Plant | mRNA rRNA |
cox2, atp9 18S |
[65–67] |
| Mammalian | mRNA | co1,co2,co3,atp6, ND3 | [69] | |
| Archaea | M. kandleri | mRNA | Exosome complex exonuclease 2 | [15] |
| rRNA | 16S rRNA | [15] | ||
| S. solfataricus | mRNA | NADH dehydrogenase subunit H | [19] | |
| rRNA | 16S rRNA | [19] | ||
c. Polyadenylation signals
Although it is clear that PAP I adds poly(A) tails and PNPase adds polynucleotide tails to the 3’-ends of a transcripts in E. coli [10], the absence of 100% polyadenylation for any given transcript in the bacterium suggests a regulatory mechanism for substrate selection by both enzymes. In vitro most RNA molecules can be polyadenylated by E. coli PAP I [43,44]. In vivo, however, RNA breakdown products generated by endoribonucleolytic cleavages are considered the most favored substrates for 3’-tailing by either enzyme facilitating in their rapid exonucleolytic degradation in both bacteria [95–97] and organelles [57,65,66].
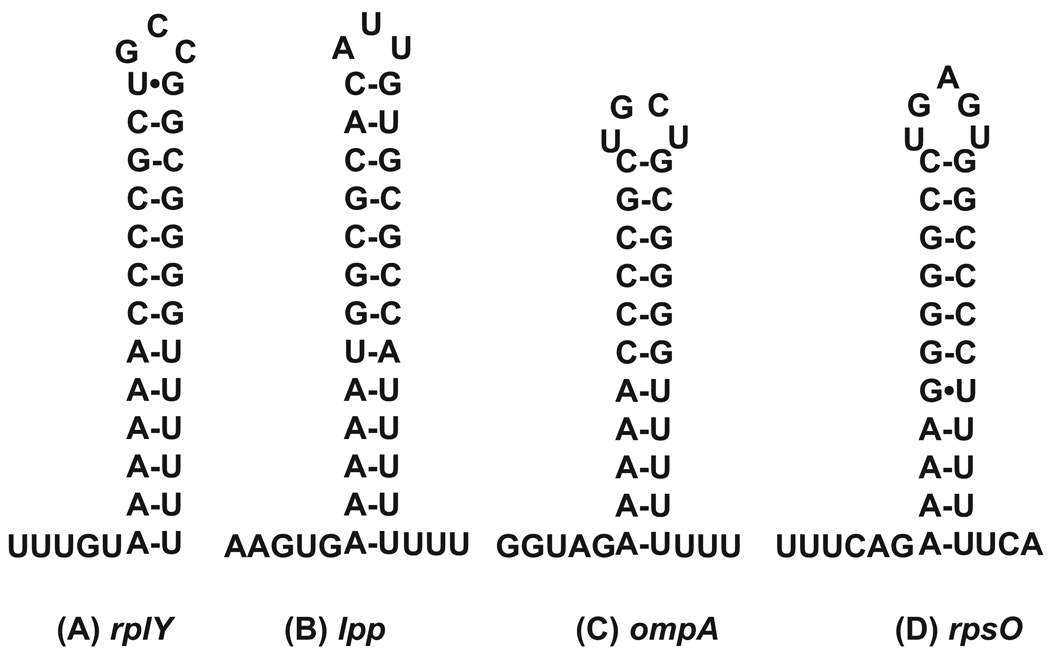
Are there any features of a prokaryotic RNA molecule that could serve as a polyadenylation signal? Clearly, there are no sequence specific motifs such as the AAUAA signal found in eukaryotic pre-mRNAs. However, in bacteria and organelles many transcripts are terminated by Rho-independent transcription terminators, which form stem-loop structures (Fig. 3). Recent data suggest that 3’ ends of the Rho-independent transcription terminators of lpp, rpsO and ompA mRNAs are preferred substrates for polyadenylation by PAP I [10,12,20,98]. Interestingly, all of these terminators have a 3–4 nt 3’ extensions as opposed to no 3’ extension in the rplY Rho-independent transcription terminator where no poly(A) tails were detected [20]. These results are consistent with the in vitro data showing inhibition of PAP I activity by Rho-independent transcription terminators that have no 3’ single-stranded extension [43]. Conversely, a 2–6 nt single-stranded region is sufficient to be recognized as a PAP I substrate [43].
Fig. 3.
Predicted secondary structures of various Rho-independent transcription terminators in E. coli. Sequencing of cDNAs copies of these four transcripts has confirmed the nature of each single-stranded 3’ extension [12,20,47].
Furthermore, a recent genome-wide analysis of the polyadenylated transcripts in E. coli revealed that ~72% of the ORFs showing high levels of polyadenylation were associated with either monocistronic or polycistronic mRNAs containing a Rho-independent transcription terminator [20]. In contrast, transcripts terminated in a Rho-dependent fashion tended to contain only polynucleotide tails, generated by PNPase, which were located throughout the coding sequences [12,20] (Table 5). These observations led to the suggestion that Rho-independent transcription terminators may serve as polyadenylation signals in E. coli [12,20].
Table 5.
Relationship between transcription terminators and the nature of the added tails in E. coli.
| Transcript | Terminator1 | % of Transcripts with | Reference | |
|---|---|---|---|---|
| Poly(A) tail | Polynucleotide tail | |||
| lpp | RI | >70 | <30 | [10,12,47] |
| rpsO | RI | >73 | <27 | [10] |
| ompA | RI | >77 | <23 | [20] |
| trxA | RD | 0 | 100 | [12] |
| pcnB folK 2 | RD | 0 | 100 | [20] |
Each transcript contains either a Rho-independent (RI) or Rho-dependent (RD) transcription terminator.
In the pcnB folK operon, the pcnB coding sequence overlaps the downstream folK gene. The dicistronic transcript is terminated in a Rho-dependent fashion.
In contrast, decay intermediates of transcripts terminated with Rho-independent terminators (lpp, ompA and rpsO) contained more polynucleotide tails than poly(A) tails [12,20]. Of particular significance is the fact that polynucleotide tails are generally found closer to the 5’ termini of transcripts suggesting that they may arise after PNPase stalls at secondary structures while degrading an RNA molecule and, in the presence of high concentrations of NDPs, biosynthetically adds untemplated nucleotides onto the substrate that it had been degrading [10] (Fig. 4).
Fig. 4.
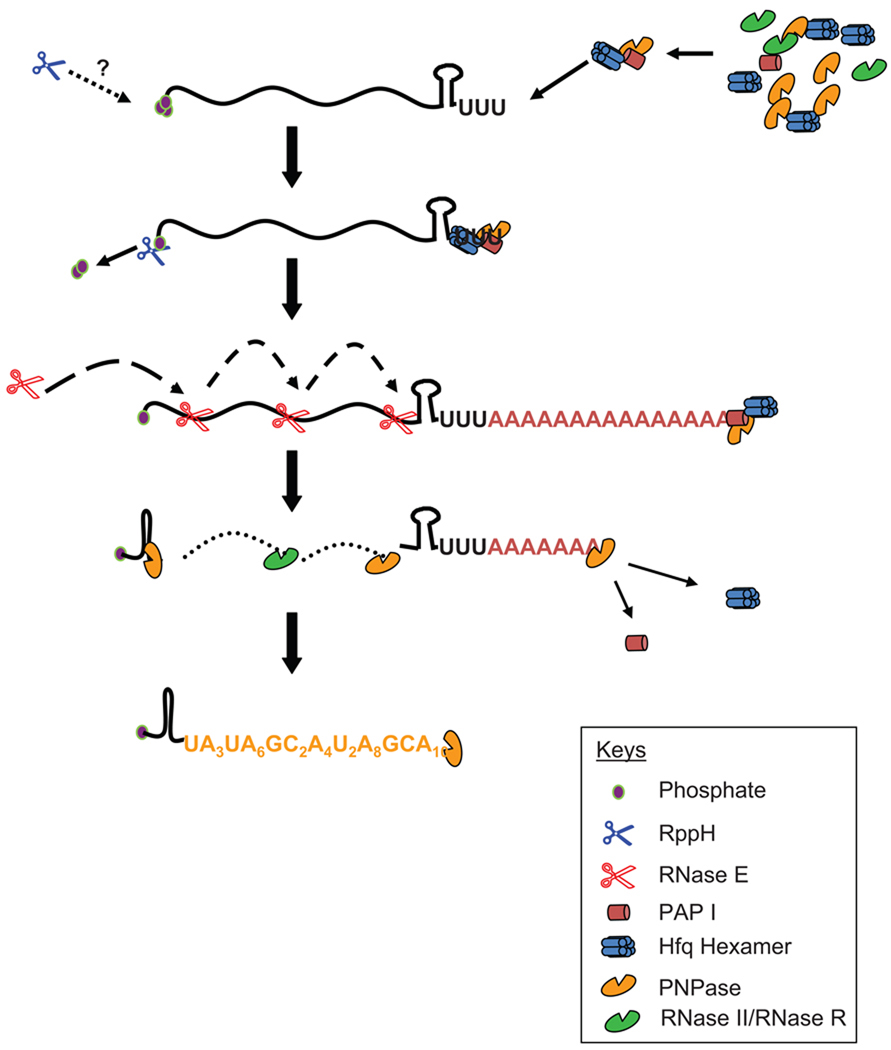
Hfq mediated polyadenylation by PAP I in E. coli. Recent studies suggest that Rho-independent transcription terminators in E. coli transcripts may serve as polyadenylation signals [12,20]. Hfq has been shown to preferentially bind to the base of A/U rich region of the terminator [12]. It has been hypothesized that Hfq in its hexameric form interacts with PAP I and PNPase to form a polyadenylation complex [12], which binds to the base of the stem-loop associated with the Rho-independent transcription terminator. Consequently, some or all of the A/U base pairs of the stem loop melt permitting PAP I to bind to the resulting single-stranded 3’ end and add poly(A) tails processively. The interaction is believed to help PAP I to compete the vast excess of 3’ → 5’ exoribonucleases in order to find its substrate and to also suppress PNPase’s biosynthetic activity [12]. Terminal 5’ triphosphates can be converted to 5’ phosphomonoesters by RppH [161], a requirement for polyadenylation in vitro [22]. However, this requirement has not been demonstrated in vivo. An endoribonuclease such as RNase E may access the transcript from the 5’ end at the same time. The 5’ phosphorylation status, which can affect RNase E activity, probably varies for individual transcripts [162–164]. In addition, it is also possible that RNase E can access the substrate as part of the degradosome by binding to the poly(A) tail (Fig. 5B).
Once PAP I dissociates, along with Hfq, PNPase can degrade the poly(A) tail in the 3’→5’ direction. Endonucleolytically derived decay intermediates are mostly degraded by 3’→5’ exoribonucleases such as PNPase, RNase II, and RNase R. However, some of the decay intermediates contain strong G/C rich secondary structures forcing PNPase to stall and possibly switch to a biosynthetic mode, thereby generating unstructured polynucleotide tails (Fig. 6). These tails either change the conformation of the substrate or provide necessary single-stranded region for either PNPase, RNase II, or RNase R to bind and complete the degradation process.
d. Potential polyadenylation complexes
While in vitro data suggests that PAP I selects its substrate independent of any ancillary proteins, it has been shown both in vivo and in vitro that both the poly(A) tail length and total polyadenylation levels are modulated by the RNA binding protein Hfq [12,21]. It has been suggested that Hfq regulates polyadenylation by changing PAP I from a distributive to a processive enzyme [21,26,99]. Interestingly, increasing Hfq levels in vivo did not change the polyadenylation level [12] similar to what was observed in vitro [21]. In addition, PAP I can easily overcome the absence of Hfq in vivo when overproduced (Mohanty and Kushner, unpublished results). Furthermore, the absence of Hfq did not alter PAP I protein levels but augmented synthesis of polynucleotide tails suggesting increased PNPase activity [12].
Thus, the role of Hfq as a facilitator in RNA polyadenylation by PAP I has been proposed. In this model (Fig. 4), it is hypothesized that Hfq primarily helps PAP I to compete with the more abundant PNPase to find its substrates and to control the biosynthetic activities of PNPase through protein-protein interactions [12]. This process most likely determines which enzyme synthesizes a 3’ tail. Initial support for this model was obtained from the demonstration of RNA-independent protein-protein interactions among PAP I, PNPase and Hfq using immunoprecipitation and co-purification experiments [12,100]. However, the exact nature of the complex(es) is still unknown. It is also possible that more proteins are involved in forming an in vivo polyadenylation complex.
e. Role of ribonucleases
The net rate of poly(A) synthesis in vivo is determined by a combination of the rate of 3’-tail synthesis by polyadenylating enzymes versus degradation by ribonucleases (Fig. 5). Both RNase II and PNPase are considered the major exoribonucleases that degrade poly(A) tails in E. coli [101–105]. Furthermore, recent data also suggest that poly(A) tails stimulate RNA degradation by RNase R [106,107].
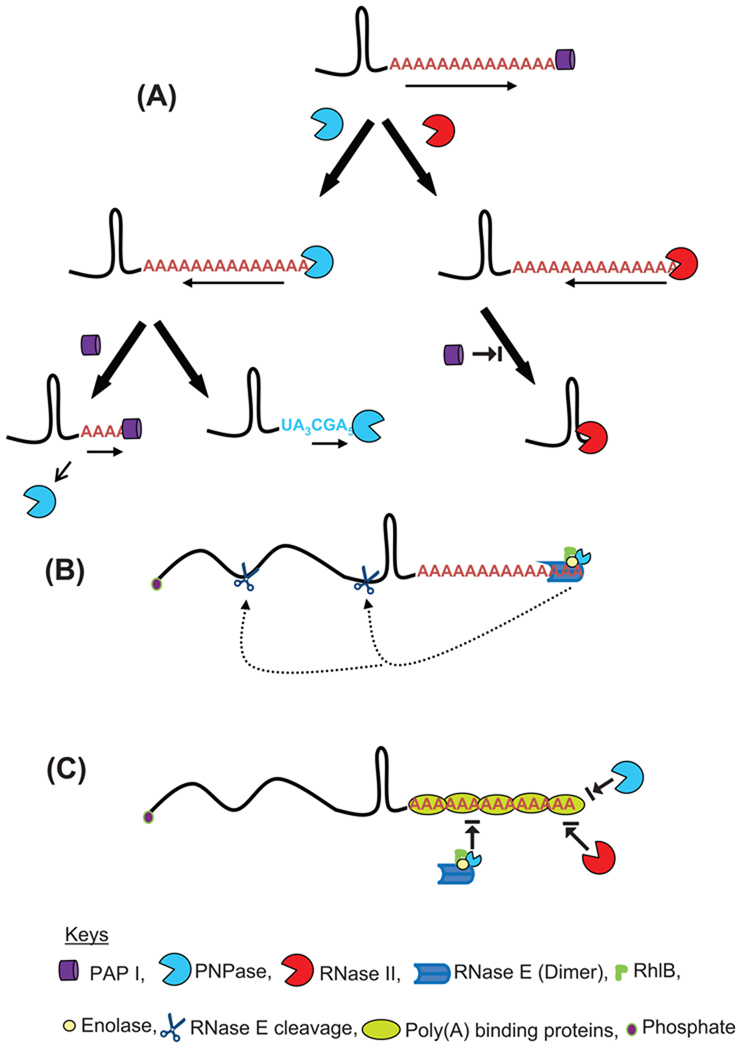
Fig. 5.
Polyadenylation assisted RNA decay in E. coli. (A) Addition of poly(A) tails by PAP I to the 3’ ends of an RNA substrate provides the single stranded binding site for both PNPase and RNase II that initiate the degradation. While PNPase catalyzes both 3’→ 5’ phosphorolytic degradation in presence of in organic phosphate and 5’→ 3’ polymerization in presence of NDPs, RNase II can only degrade RNA hydrolytically in the 3’→ 5’ direction. Both the ribonucleases pause upon encountering a G/C rich secondary structure. PNPase either dissociates relatively quickly or reverses its activity to polymerize polynucleotide tails. Dissociation of PNPase may initiate multiple rounds of polymerization by PAP I. In contrast, RNase II remains bound to the base of the secondary structure thereby effectively blocking the binding of either PAP I or PNPase.
(B) RNase E alone or as part of the multiprotein complex called the degradosome can bind A/U rich poly(A) and polynucleotide tails to initiate degradation of a potential substrate through endonucleolytic cleavage. A full-length polyadenylated RNA substrate may be degraded very fast [114] by direct or internal entry [164] resulting in very few steady-state polyadenylated RNA species. This type of RNase E entry to an RNA substrate has yet to be experimentally demonstrated.
(C) Potential poly(A) binding proteins can block RNA decay in E. coli. Proteins such as CspE, Hfq and ribosomal protein S1 could bind to poly(A) or polynucleotide tails blocking endonucleolytic access by the RNase E-based degradosome through its PNPase moiety or direct exonucleolytic degradation by exoribonucleases such as RNase II, RNase R, and PNPase.
PNPase catalyzes both processive 3’→ 5’ phosphorolytic degradation in presence of inorganic phosphate (Pi) and 5’→ 3’ polymerization in presence of NDPs, of an RNA substrate (Fig. 1). In contrast, RNase II and RNase R, which belong to the same RNB exoribonuclease family, degrade an RNA substrate hydrolytically in the 3’→ 5’ direction. With the exception of Mycoplasma, Trypanosomes, yeast and Archaea, all bacteria and organelles contain a PNPase homologue [27]. Some hyperthermophiles and methanogenic Archaea do not have a direct PNPase homologue, but instead contain an multiprotein exosome complex that is structurally similar to PNPase [27]. Homologues of the RNB exoribonuclease family are found in all bacteria, organelles and in some archaeal species with the exception of methanogens that do not contain an exosome [27].
Both PNPase and RNase II stall when they encounter a G/C rich secondary structure (Fig. 5A). While PNPase dissociates relatively rapidly, RNase II does not. Thus one can get multiple rounds of polyadenylation/deadenylation in the case of PNPase but not RNase II (Fig. 5A). In fact, it has been shown that RNase II protects mRNAs from exonucleolytic degradation by either blocking the 3’ ends or generating short single-stranded extensions that are not substrates for either PNPase or RNase R [108–110]. This role of RNase II can also effectively block or significantly slow down polyadenylating enzymes resulting in very short tails [111]. Interestingly, RNase II seems to be involved in modulating the level of poly(A) tails associated with 23S rRNA, the major polyadenylated species in E. coli (Table 3), whereas PNPase is more effective with 16S rRNA and mRNAs [101]. In contrast, if RNase R binds to a substrate containing a poly(A) tail, it can degrade through secondary structures effectively reducing the level of that transcript.
In vitro data suggest that the endoribonuclease RNase E can also act as a poly(A) nuclease to degrade the poly(A) tails [112], although its mode of action is still controversial [113]. However, RNase E does seem to contribute indirectly to increased levels of polyadenylation in vivo by generating new 3’ termini, through endonucleolytic cleavages, which can serve as substrates for the polyadenylating enzymes [101]. In fact, recent in vitro data suggest that polyadenylation of the cspA mRNA by PAP I enhanced its degradation by the RNase E-based degradosome [114]. Thus, it is possible that RNase E can use poly(A) tails or A/U rich polynucleotide tails to bind a potential substrate as part of the multiprotein complex, called the degradosome [115], leading to faster RNA decay (Fig. 5B). Since it has been demonstrated that many mRNAs decay more rapidly in the presence of increased polyadenylation [47], efficient degradation by the RNase E-based degradosome could account for why only a small percentage of E. coli RNA appears to be polyadenylated at any given time.
Although no poly(A) binding proteins have yet been identified in vivo in prokaryotic cells, Hfq, CspE and ribosomal protein S1 have been shown to bind and protect poly(A) tails from ribonucleases in vitro [114,116,117]. While it is possible that these proteins can serve an identical function in vivo (Fig. 5C), similar to eukaryotic poly(A) binding proteins [118], experimental confirmation is still awaited. It has been suggested that binding of Hfq to poly(A) tails protects them from nucleolytic degradation [21,26,116]. However, the fact that there was ~60% reduction in the total poly(A) level along with significant reduction in the poly(A) tail length in an hfq mutant missing all the major ribonucleases compared to the control strain suggests a more complicated role for Hfq [12,99].
IV. ROLE OF POLY(A) AND POLYNUCLEOTIDE TAILS
The exact role(s) of poly(A) and polynucleotide tails in prokaryotic RNA metabolism are still not clear. However, although the specific function of tails in different prokaryotic entities may differ to some extent, the current consensus is that polyadenylation functions in the regulation of RNA stability and quality control [26,27,48,64,119].
It is interesting to note that the majority of the post-transcriptionally added tails in bacteria, organelles, and Archaea are polynucleotide in composition. However, it is currently not clear whether these polynucleotide tails play an identical role to poly(A) tails. Using the RNA Star program [120], we analyzed polynucleotide tails sequenced from bacteria, organelles, and Archaea for predicted secondary structures (Fig. 6). Surprisingly, polynucleotide tails are structurally almost indistinguishable from homopolymeric poly(A) tails. In fact, a 15 nt polynucleotide tail was as effective as a 15 nt poly(A) tail for degrading a transcript by PNPase in vitro [121]. However, if polynucleotide tails affect RNA degradation in vivo their role seems to be limited to breakdown products only, since the half-lives of full-length transcripts in an hfq mutant which contains mostly polynucleotide tails, were identical to a ΔpcnB mutant [12]. This result is consistent with the observation that polynucleotide tails are added mainly to breakdown products, whereas poly(A) tails are added to both breakdown and full-length transcripts.
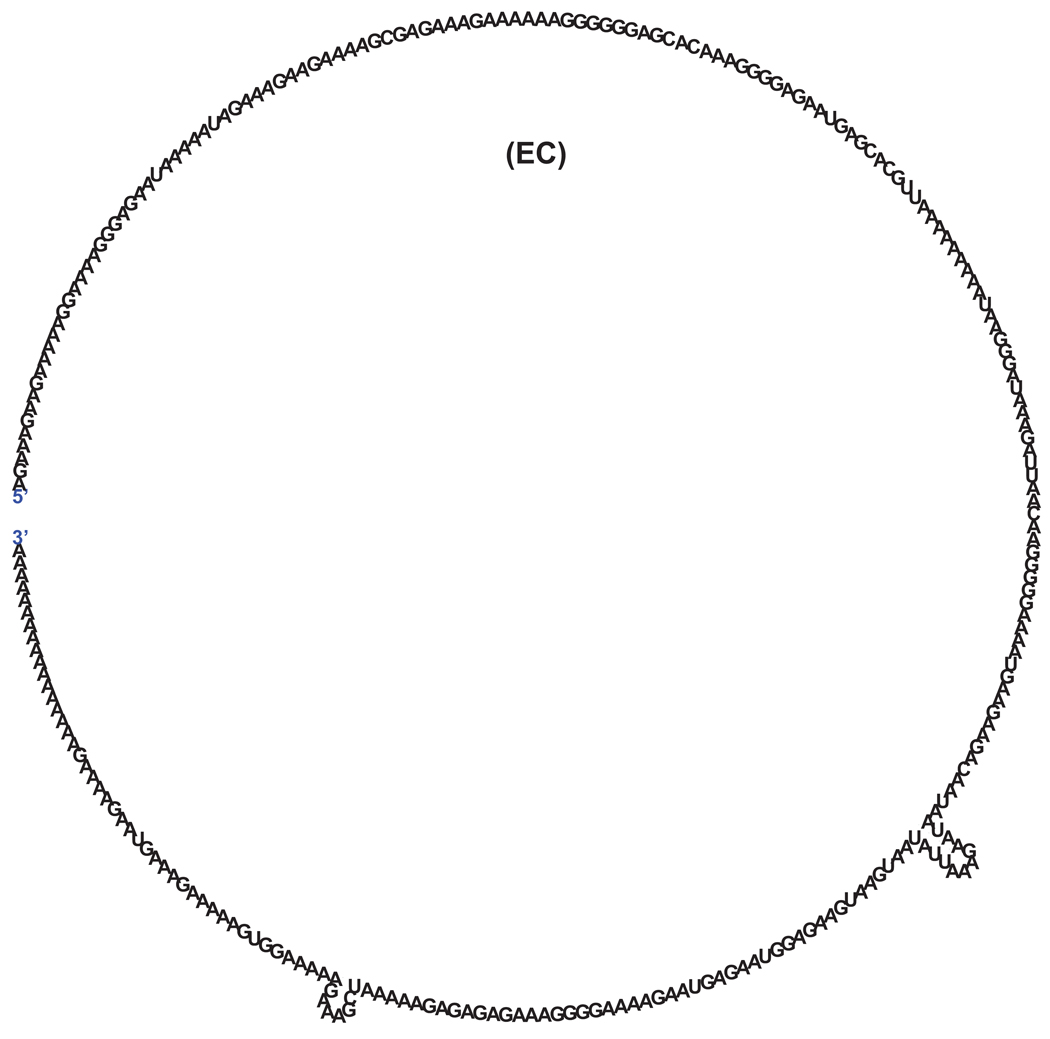
Fig. 6.
Secondary structure of a polynucleotide tail that was cloned and sequenced from a pcnB transcript of E. coli, (257 nt, −3.0 KCal) [20]. Similar analysis of polynucleotide tails derived from a pnp transcript of S. antibioticus, (116 nt, −0.6 KCal) [165], an rpsD transcript of B. subtilis (56 nt, −4.2 KCal) [29], a psbA transcript from spinach chloroplast (177 nt, −2.4 KCal) [30], an rbcL transcript from Synechocystis (172 nt, −1.9 KCal), [31], and an exosome complex exonuclease 2 transcript from M. kandleri (124 nt, −2.5 KCal), [15] yielded identical results (data not shown). The secondary structures and energy level (total energy for all the stems in a structure) were obtained by using RNA STAR program [120].
a. RNA stability
The reduction in plasmid copy number of ColE1 plasmids in a pcnB mutant of E. coli [122] was not fully understood until it was shown that polyadenylation helped regulate plasmid copy number by controlling the stability of an untranslated RNA (RNA I) [94,123]. With the identification of pcnB as the structural gene for PAP I [11], multiple reports demonstrated that 3’ polyadenylation led to decreased stability of variety of mRNAs and sRNAs [48,93,121,124–130]. A direct correlation between increased polyadenylation and decreased mRNA stability was subsequently established [47].
Surprisingly, the effect of 3’ polyadenylation on the stability of specific full-length mRNAs in a pcnB single mutant are minimal when ribonucleases such as RNase E and PNPase are present in the cell [48]. In many cases, such as the rpsO and rpsT mRNAs, the differences in half-lives between wild type and pcnB strains are so subtle that different laboratories have published contradictory reports [47,125,126]. Nevertheless, when one or more of the ribonucleases along with PAP I are missing, many transcripts are significantly stabilized [48]. These results indicate that the presence of poly(A) tails is not required for the initiation of decay but rather that it facilitates the degradation process, possibly identifying a target (Fig. 5). In the absence of poly(A) tail, a ribonuclease such as RNase E still can degrade the transcript, although less efficiently [114].
Interestingly, increased global polyadenylation by transient overexpression of pcnB also led to the stabilization of some transcripts [20,47]. The most notable among these were the pnp (PNPase) and rne (RNase E) mRNAs, transcripts that encode enzymes involved in mRNA decay [47]. The stabilization of these transcripts also resulted in higher protein levels that were directly related to the autoregulation of both transcripts, leading to increase RNA degradative capacity [24,47]. While the mRNAs of other ribonucleases were not affected [24], these findings suggested possible regulatory controls balancing polyadenylation and induced mRNA decay. Thus, the increased polyadenylation of transcripts in E. coli serves as a sensing mechanism to facilitate intracellular adjustments in the levels of both RNase E and PNPase [24].
While the role of poly(A) tails in rRNA and tRNA processing and degradation is still unknown [83], they probably have a destabilizing effect on sRNAs similar to what is observed with mRNAs. In addition to RNA I, the antisense RNAs CopA, which controls the replication frequency of plasmid R1, and Sok, which inhibits translation of hok mRNA of plasmid R1 that mediate plasmid stabilization, are also stabilized by the absence of PAP I [124,129]. The small RNA GlmY has been shown to be polyadenylated by PAP I, which decreases it stability, leading to activation of glmS mRNA translation [93]. Another small RNA, MicA which is required for the accurate expression of outer membrane proteins, is also stabilized in the absence of polyadenylation by PAP I [130].
The destabilizing effects of poly(A) tails in E. coli on various transcripts have led to the assumption that they have similar effects in all bacteria with similar polyadenylation profiles. Detection of poly(A) and polynucleotide tails associated with decay intermediates of B. subtilis and Streptomyces sp. has provided some initial support for a comparable role for polyadenylation in other species [29,60]. However, this hypothesis still awaits direct confirmation.
Poly(A)-mediated RNA degradation in plant and algae chloroplasts and mitochondria was found to be very similar to that of bacteria [131], although its effect varied on different types of RNA species [62,132]. The polyadenylated transcripts in plant mitochondria are degraded mainly through 3’→ 5’ exoribonucleolytic activity [57,65,67,133–135]. Endonucleolytically cleaved mRNA decay intermediates containing 3’ polynucleotide tails have been identified in both the spinach and Chlamydomonas chloroplasts [30,57,62], which may be degraded through exoribonucleolytic activity.
No specific role for polyadenylation in yeast mitochondrial RNA metabolism has been identified [64]. The exact function of polyadenylation in mammalian mitochondrial mRNA stability is also still unclear [71]. Long homopolymeric poly(A) tails in human mitochondria provide stability to mRNAs, while deadenylation by PNPase destabilizes the transcripts [69]. Current in vitro data suggest that short poly(A) tails may affect mRNA stability in T. brucei depending on the editing status of individual transcripts [136,137].
b. Quality control
Not long ago it was believed that only ribonucleases were required for RNA degradation. However, E. coli exoribonucleases, such as PNPase, RNase II and RNase R, require 3’ single-stranded regions for initial binding to the RNA substrates. For example, a minimum 10–11 nt single-stranded region is required for PNPase, RNase II, and RNase R to bind to a substrate and initiate degradation [102,104,138]. Thus, in most cases the unstructured nature of poly(A) or polynucleotide tails (Fig. 6) has been hypothesized to provide the required toe-hold for exoribonucleases like RNase II and PNPase, which are particularly inhibited by secondary structures [121,127,139–143]. Accumulation of mRNA breakdown products of the lpp, rpsO, ompA and rpsT mRNAs in a pcnB mutant provides further support for this notion [48,96,97,125,144]. While some full-length mRNAs decay exclusively through the actions of 3’→ 5’ exoribonucleases like PNPase and RNase II [108], the majority of the transcripts are believed to be degraded via initial endonucleolytic cleavages by either RNase E, its homologue RNase G or RNase Z, followed by 3’→ 5’ exonucleolytic decay [145,146].
Mapping of polyadenylation sites by cDNA cloning and sequencing of a variety of transcripts has indicated that poly(A) and polynucleotide tails are frequently attached to RNA breakdown products, which are most probably generated by endonucleolytic cleavages [10,12,19,29,30,57,97,147]. Thus, it is generally believed that prokaryotic polyadenylation is a scavenging mechanism that helps recycle breakdown products, which can form highly structured molecules [81,97,110,148]. RNA degradation by RNase R, which is not inhibited by secondary structures, is also stimulated by the presence of poly(A) tails [106,107], since this enzyme requires single-stranded regions of 10–12 nucleotides to bind [149]. More recently, polyadenylation has been shown to be required in PNPase-mediated degradation of defective tRNAs in E. coli [23].
c. Translation and Editing
Unlike what is observed in eukaryotes, polyadenylation of bacterial RNAs probably has no significant effect on translation [150]. Although absence of polyadenylation leads to increased half-lives of many transcripts in bacteria [12,47,48,126], it is still unclear if the stabilized transcripts contribute to increased protein synthesis. However, a recent report suggests that the glmS mRNA in E. coli, which is highly susceptible to poly(A)-dependent degradation, overproduces glucosamine-6-phosphate synthase in a PAP I deficient strain [151]. Surprisingly, PAP I has been implicated in increasing σs protein levels indirectly by affecting the global regulator RssB, which helps control the levels of σs dependent transcripts [86,152]. A recent study suggests that polyadenylation also has a minor effect on the processing of tRNALeu5 in E. coli [83].
Polyadenylation does play an important role in the translation of both Trypanososme and mammalian mitochondrial mRNAs. Most protein coding transcripts in T. brucei mitochondria undergo massive post-transcriptional editing via the insertion or deletion of U residues [153]. The presence of short or long tails appears to correlate with the editing status of the mRNA. Thus pre-edited forms contain only short tails whereas never-edited and edited forms contain both short and long tails [74,154,155]. It was recently shown that short poly(A) tails are required and sufficient to maintain the steady-state level of partially edited, fully edited and never edited mRNAs, which were extended with long poly(A) tails containing many uridines [75].
In mammalian mitochondria, the polyadenylation of mRNAs is required to create UAA stop codons to be functional that are not encoded in mtDNA. In some cases, polyadenylation is also required for the tRNA maturation by editing of its 3’ terminus [64,156,157].
VI. CONCLUSIONS
Over the past 15 years considerable progress has been made in unraveling the mysteries of polyadenylation in bacteria and organelles. However, it is still unclear what constitutes the polyadenylation complex in E. coli, let alone bacteria such as B. subtilis which does not have a PAP-like protein and still contains poly(A) tails in the absence of PNPase. Another issue that needs further study relates to the nature of substrate selection and how PAP enzymes compete with PNPase for 3’ termini, particularly in E. coli where there is at least 20-fold excess of PNPase. It is also not clear what is the function of the polynucleotide tails that are added by PNPase in E. coli and the large number of other bacterial species.
Perhaps most importantly, the biological significance of polyadenylation in prokaryotes is still not really understood. In contrast, the polyadenylation of eukaryotic mRNAs has long been viewed as a stabilizing element that facilitates localization and improves the translation. However, recent studies showing the polyadenylation induced decay of non-functional RNAs in eukaryotes [158–160] has prompted many to believe that the major role of this ancient trait is quality control where a living cell tags its aberrant and unused transcripts for degradation. While RNA surveillance may only be the common function performed by polyadenylation, it is premature at this time to consider RNA surveillance as its primary function in bacteria, particularly since there is so little PAP I protein found in E. coli. Finally, it should be noted that the study of polyadenylation in prokaryotes is still significantly limited by technical issues relating to the ability to easily identify RNA species with poly(A) tails less than 10 nt in length.
ACKNOWLEDGEMENTS
This work was supported in part by grants from the National Institutes of Health (GM57220 and GM81554) to S.R.K. The authors thank N. Dubose for critically reading the manuscript and providing valuable suggestions.
REFERENCES
- 1.August J, Ortiz PJ, Hurwitz J. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J Biol Chem. 1962;237:3786–3793. [PubMed] [Google Scholar]
- 2.Hardy SJ, Kurland CG. The polynucleotide product of poly(A) polymerase from Escherichia coli. Biochemistry. 1966;5:3668–3676. doi: 10.1021/bi00875a041. [DOI] [PubMed] [Google Scholar]
- 3.Edmonds M, Abrams R. Polynucleotide biosynthesis: Formation of a sequence of adenylate units from adenosine triphosphate by an enzyme from thymus nuclei. J Biol Chem. 1960;235:1142–1148. [PubMed] [Google Scholar]
- 4.Edmonds M, Abrams R. Nature of a polynucleotide required for polyribonucleotide formation from adenosine triphosphate with an enzyme from thymus nuclei. J Biol Chem. 1962;237:2636–2642. [PubMed] [Google Scholar]
- 5.Nakazato H, Venkatesan S, Edmonds M. Polyadenylic acid sequences in E. coli messenger RNA. Nature. 1975;256:144–146. doi: 10.1038/256144a0. [DOI] [PubMed] [Google Scholar]
- 6.Srinivasan PR, Ramanarayanan M, Rabbani E. Presence of polyriboadenylate sequences in pulse-labeled RNA of Escherichia coli. Proc Natl Acad Sci USA. 1975;72:2910–2914. doi: 10.1073/pnas.72.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar N, Langley D, Paulus H. Isolation and characterization of polyadenylate-containing RNA. Biochemistry. 1978;17:3468–3474. doi: 10.1021/bi00610a007. [DOI] [PubMed] [Google Scholar]
- 8.Gopalakrishna Y, Langley D, Sarkar N. Detection of high levels of polyadenylate-containing RNA in bacteria by the use of a single-step RNA isolation procedure. Nucl Acid Res. 1981;9:3545–3554. doi: 10.1093/nar/9.14.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gopalakrishna Y, Sarkar N. The synthesis of DNA complementary to polyadenylate-containing RNA from Bacillus subtilis. J Biol Chem. 1982;257:2747–2750. [PubMed] [Google Scholar]
- 10.Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3' – 5' exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao G-J, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 13.Yehudai-Resheff S, Hirsh M, Schuster G. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Molecular and Cellular Biology. 2001;21:5408–5416. doi: 10.1128/MCB.21.16.5408-5416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohlberg B, Huang J, Cohen SN. The Streptomyces coelicolor polynucleotide phosphorylase homologue, and not the putative poly(A) polymerase, can polyadenylate RNA. J Bacteriol. 2003;185:7273–7278. doi: 10.1128/JB.185.24.7273-7278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Portnoy V, Schuster G. RNA polyadenylation and degradation in different Archaea; roles of the exosome and RNase R. Nucleic Acids Res. 2006;34:5923–5931. doi: 10.1093/nar/gkl763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slomovic S, Portnoy V, Liveanu V, Schuster G. RNA polyadenylation in prokaryotes and organelles: Different tails tell different tales. Critical Reviews in Plant Sciences. 2006;25:65–77. [Google Scholar]
- 17.Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 18.Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 19.Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, Schuster G. RNA polyadenylation in Archaea: not observed in Haloferax while exosome polynucleotidylates RNA. Sulfolobus EMBO Reports. 2005;6:1188–1193. doi: 10.1038/sj.embor.7400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajnsdorf E, Régnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci USA. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y, Cohen SN. Unpaired terminal nucleotides and 5' monophosphorylation govern 3' polyadenylation by Escherichia coli poly(A) polymerase I. Proc Natl Acad Sci USA. 2000;97:6415–6420. doi: 10.1073/pnas.120173797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Reimers S, Pandit S, Deutscher MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohanty BK, Kushner SR. Polyadenylation of Escherichia coli transcripts plays an integral role in regulating intracellular levels of polynucleotide phosphorylase and RNase E. Mol Microbiol. 2002;45:1315–1324. doi: 10.1046/j.1365-2958.2002.03097.x. [DOI] [PubMed] [Google Scholar]
- 25.Edmonds M. A history of poly A sequences: from formation to factors to function A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 26.Regnier P, Hajnsdorf E. Poly(A)-assisted RNA decay and modulators of RNA stability. Prog Mol Biol Transl Sci. 2009;85:137–185. doi: 10.1016/S0079-6603(08)00804-0. [DOI] [PubMed] [Google Scholar]
- 27.Schuster G, Stern D. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol Biol Transl Sci. 2009;85:393–422. doi: 10.1016/S0079-6603(08)00810-6. [DOI] [PubMed] [Google Scholar]
- 28.Bralley P, Jones GH. cDNA cloning confirms the polyadenylation of RNA decay intermediates. Streptomyces coelicolor Microbiol. 2002;148:1421–1425. doi: 10.1099/00221287-148-5-1421. [DOI] [PubMed] [Google Scholar]
- 29.Campos-Guillen J, Bralley P, Jones GH, Bechhofer DH, Olmedo-Alvarez G. Addition of poly(A) and heteropolymeric 3' ends in B. subtilis wild-type and polynucleotide phosphorylase deficient strains. J Bacteriol. 2005;187:4698–4706. doi: 10.1128/JB.187.14.4698-4706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lisitsky I, Klaff P, Schuster G. Addition of destabilizing poly (A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proc Natl Acad Sci U S A. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rott R, Zipor G, Portnoy V, Liveanu V, Schuster G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J Biol Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- 32.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etheridge RD, Clemens DM, Gershon PD, Aphasizhev R. Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei. Mol Biochem Parasitol. 2009;164:66–73. doi: 10.1016/j.molbiopara.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmer SL, Schein A, Zipor G, Stern DB, Schuster G. Polyadenylation in Arabidopsis and Chlamydomonas organelles: the input of nucleotidyltransferases, poly(A) polymerases and polynucleotide phosphorylase. Plant J. 2009;59:88–99. doi: 10.1111/j.1365-313X.2009.03853.x. [DOI] [PubMed] [Google Scholar]
- 36.Slomovic S, Schuster G. Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA. 2008;14:310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aravind L, Koonin EV. DNA polymerase B-like nucleotidyltransferase superfamily: Identification of three new families, classification and evolutionary history. Nucl Acid Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 39.Martin G, Keller W. Sequence motifs that distinguish ATP(CTP):tRNA nucleotidyl transferases from eubacterial poly(A) polymerases. RNA. 2004;10:899–906. doi: 10.1261/rna.5242304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slomovic S, Portnoy V, Yehudai-Resheff S, Bronshtein E, Schuster G. Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases. Biochim Biophys Acta. 2008;1779:247–255. doi: 10.1016/j.bbagrm.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Betat H, Rammelt C, Martin G, Morl M. Exchange of regions between bacterial poly(A) polymerase and the CCA-adding enzyme generates altered specificities. Molec Cell. 2004;15:389–398. doi: 10.1016/j.molcel.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 42.Just A, Butter F, Trenkmann M, Heitkam T, Morl M, Betat H. A comparative analysis of two conserved motifs in bacterial poly(A) polymerase and CCA-adding enzyme. Nucleic Acids Res. 2008;36:5212–5220. doi: 10.1093/nar/gkn494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yehudai-Resheff S, Schuster G. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucl Acid Res. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sippel AE. Purification and characterization of adenosine triphosphate:ribonucleic acid adenyltransferase from Escherichia coli. Eur J Biochem. 1973;37:31–40. doi: 10.1111/j.1432-1033.1973.tb02953.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Parkinson JS. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989;171:1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masters M, Colloms MD, Oliver IR, He L, Macnaughton EJ, Charters Y. The pcnB gene of Escherichia coli, which is required for ColE1 copy number maintenance, is dispensible. J Bacteriol. 1993;175:4405–4413. doi: 10.1128/jb.175.14.4405-4413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohanty BK, Kushner SR. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol Microbiol. 1999;34:1094–1108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 48.O'Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao G-J, Pogliano J, Sarkar N. Identification of the coding region for a second poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:11580–11585. doi: 10.1073/pnas.93.21.11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yue D, Maizels N, Weiner AM. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: Characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA. 1996;2:895–908. [PMC free article] [PubMed] [Google Scholar]
- 51.Mohanty BK, Kushner SR. Residual polyadenylation in poly(A) polymerase I (pcnB) mutants of Escherichia coli does not result from the activity encoded by the f310 gene. Mol Microbiol. 1999;34:1109–1119. doi: 10.1046/j.1365-2958.1999.01674.x. [DOI] [PubMed] [Google Scholar]
- 52.Grunberg-Manago M. Polynucleotide phosphorylase. Progress in Nucleic Acids Research. 1963;1:93–133. [Google Scholar]
- 53.Soreq H, Littauer UZ. Purification and characterization of polynucleotide phosphorylase from Escherichia coli. J Biol Chem. 1977;252:6885–6888. [PubMed] [Google Scholar]
- 54.Shulman RG, Brown TR, Ugurbil K, Ogawa S, Cohen SM, den Hollander JA. Cellular applications of 31P and 13C nuclear magnetic resonance. Science. 1979;205:160–166. doi: 10.1126/science.36664. [DOI] [PubMed] [Google Scholar]
- 55.Mohanty BK, Giladi H, Maples VF, Kushner SR. Analysis of RNA decay, processing, and polyadenylation in Escherichia coli and other prokaryotes. Methods Enzymol. 2008;447:3–29. doi: 10.1016/S0076-6879(08)02201-5. [DOI] [PubMed] [Google Scholar]
- 56.Bralley P, Jones GH. Poly(A) polymerase activity and RNA polyadenylation in Streptomyces coelicolor A3. Molecular Microbiol. 2001;40:1155–1164. doi: 10.1046/j.1365-2958.2001.02457.x. [DOI] [PubMed] [Google Scholar]
- 57.Lisitsky I, Kotler A, Schuster G. The mechanism of preferential degradation of polyadenylated RNA in the chloroplast. The exoribonuclease 100RNP/polynucleotide phosphorylase displays high binding affinity for poly(A) sequence. J Biol Chem. 1997;272:17648–17653. doi: 10.1074/jbc.272.28.17648. [DOI] [PubMed] [Google Scholar]
- 58.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 59.Raynal LC, Krisch HM, Carpousis AJ. The Bacillus subtilis nucleotidyltransferase is a tRNA CCA-adding enzyme. J Bacteriol. 1998;180:6276–6282. doi: 10.1128/jb.180.23.6276-6282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bralley P, Gust B, Chang SA, Chater KF, Jones GH. RNA 3'-tail synthesis in Streptomyces: in vitro and in vivo activities of RNase PH, the SCO3896 gene product and polynucleotide phosphorylase. Microbiol-SGM. 2006;152:627–636. doi: 10.1099/mic.0.28363-0. [DOI] [PubMed] [Google Scholar]
- 61.Bralley P, Jones GH. Organization and expression of the polynucleotide phosphorylase gene (pnp) of Streptomyces: Processing of pnp transcirpts in Streptomyces antibioticus. J Bacteriol. 2004;186:3160–3172. doi: 10.1128/JB.186.10.3160-3172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komine Y, Kwong L, Anguera MC, Schuster G, Stern DB. Polyadenylation of three classes of chloroplast RNA in Chlamydomonas reinhardtii. RNA. 2000;6:598–607. doi: 10.1017/s1355838200992252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walter M, Kilian J, Kudla J. PNPase activity determines the efficiency of mRNA 3'-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J. 2002;21:6905–6914. doi: 10.1093/emboj/cdf686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 65.Lupold DS, Caoile AG, Stern DB. Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell. 1999;11:1565–1578. doi: 10.1105/tpc.11.8.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perrin R, Lange H, Grienenberger JM, Gagliardi D. AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 2004;32:5174–5182. doi: 10.1093/nar/gkh852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perrin R, Meyer EH, Zaepfel M, Kim YJ, Mache R, Grienenberger JM, Gualberto JM, Gagliardi D. Two exoribonucleases act sequentially to process mature 3'-ends of atp9 mRNAs in Arabidopsis mitochondria. J Biol Chem. 2004;279:25440–25446. doi: 10.1074/jbc.M401182200. [DOI] [PubMed] [Google Scholar]
- 68.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 70.Bobrowicz AJ, Lightowlers RN, Chrzanowska-Lightowlers Z. Polyadenylation and degradation of mRNA in mammalian mitochondria: a missing link? Biochem Soc Trans. 2008;36:517–519. doi: 10.1042/BST0360517. [DOI] [PubMed] [Google Scholar]
- 71.Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, Chrzanowska-Lightowlers ZM. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum Mol Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- 72.Jacob ST, Schindler DG. Polyriboadenylate polymerase solubilized from rat liver mitochondria. Biochem Biophys Res Commun. 1972;48:126–134. doi: 10.1016/0006-291x(72)90353-1. [DOI] [PubMed] [Google Scholar]
- 73.Chen HW, Koehler CM, Teitell MA. Human polynucleotide phosphorylase: location matters. Trends Cell Biol. 2007;17:600–608. doi: 10.1016/j.tcb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 74.Bhat GJ, Souza AE, Feagin JE, Stuart K. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 1992;52:231–240. doi: 10.1016/0166-6851(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 75.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3' adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuckenberg PD, Phillips SL. Oligoadenylate is present in the mitochondrial RNA of Saccharomyces cerevisiae. Mol Cell Biol. 1982;2:450–456. doi: 10.1128/mcb.2.4.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 78.Schafer B, Hansen M, Lang BF. Transcription and RNA-processing in fission yeast mitochondria. RNA. 2005;11:785–795. doi: 10.1261/rna.7252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 80.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 81.Dreyfus M, Regnier P. The poly(A) tail of mRNAs:bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;27:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- 82.Mohanty BK, Kushner SR. Rho-independent transcription terminators inhibit RNase P processing of the secG leuU and metT tRNA polycistronic transcripts in Escherichia coli. Nucleic Acids Res. 2008;36:364–375. doi: 10.1093/nar/gkm991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mohanty BK, Kushner SR. Processing of the Escherichia colileuX tRNA transcript, encoding tRNAleu5, requires either the 3'–5' exoribonuclease polynucleotide phosphorylase or RNase P to remove the Rho-independent transcription terminator. Nucleic Acids Res. 2010;38:5306–5318. doi: 10.1093/nar/gkp997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jasiecki J, Wegrzyn G. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Reports. 2003;4:172–177. doi: 10.1038/sj.embor.embor733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Binns N, Masters M. Expression of the Escherichia colipcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Molecular Microbiol. 2002;44:1287–1297. doi: 10.1046/j.1365-2958.2002.02945.x. [DOI] [PubMed] [Google Scholar]
- 86.Carabetta VJ, Mohanty BK, Kushner SR, Silhavy TJ. The response regulator SprE (RssB) modulates polyadenylation and mRNA stability in Escherichia coli. J Bacteriol. 2009;191:6812–6821. doi: 10.1128/JB.00870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jasiecki J, Wegrzyn G. Localization of Escherichia coli poly(A) polymerase I in cellular membrane. Biochem Biophys Res Commun. 2005;329:598–602. doi: 10.1016/j.bbrc.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 88.Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & Develop. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 90.Thuresson AC, Astrom J, Astrom A, Gronvik KO, Virtanen A. Multiple forms of poly(A) polymerases in human cells. Proc Natl Acad Sci U S A. 1994;91:979–983. doi: 10.1073/pnas.91.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Del Favero M, Mazzantini E, Briani F, Zangrossi S, Tortora P, Deho G. Regulation of Escherichia coli polynucleotide phosphorylase by ATP. J Biol Chem. 2008;283:27355–27359. doi: 10.1074/jbc.C800113200. [DOI] [PubMed] [Google Scholar]
- 92.Raynal LC, Carpousis AJ. Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol Microbiol. 1999;32:765–775. doi: 10.1046/j.1365-2958.1999.01394.x. [DOI] [PubMed] [Google Scholar]
- 93.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu F, Lin-Chao S, Cohen SN. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci USA. 1993;90:6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goodrich AF, Steege DA. Roles of polyadenylation and nucleolytic cleavage in the filamentous phage mRNA processing and decay pathways in Escherichia coli. RNA. 1999;5:972–985. doi: 10.1017/s1355838299990398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haugel-Nielsen J, Hajnsdorf E, Régnier P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleotlyic processing and exonucleolytic degradation. EMBO J. 1996;15:3144–3152. [PMC free article] [PubMed] [Google Scholar]
- 97.Hajnsdorf E, Regnier P. E. colirpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J Molecular Biology. 1999;286:1033–1043. doi: 10.1006/jmbi.1999.2547. [DOI] [PubMed] [Google Scholar]
- 98.Folichon M, Marujo PE, Arluison V, Le Derout J, Pellegrini O, Hajnsdorf E, Regnier P. Fate of mRNA extremities generated by intrinsic termination:detailed analysis of reactions catalyzed by ribonuclease II and poly(A) polymerase. Biochimie. 2005;87:819–826. doi: 10.1016/j.biochi.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 99.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Develop. 2005;19:2276–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mohanty BK, Kushner SR. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol Microbiol. 2000;36:982–994. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- 102.Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J Bacteriol. 2000;182:2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 1994;13:3368–3377. doi: 10.1002/j.1460-2075.1994.tb06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 105.Shi Z, Yang WZ, Lin-Chao S, Chak KF, Yuan HS. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA. 2008;14:2361–2371. doi: 10.1261/rna.1244308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheng Z-F, Deutscher MP. An important role for RNase R in mRNA decay. Molec Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 107.Andrade JM, Hajnsdorf E, Regnier P, Arraiano CM. The poly(A)-dependent degradation pathway of rpsO mRNA is primarily mediated by RNase R. RNA. 2009;15:316–326. doi: 10.1261/rna.1197309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol Microbiol. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- 109.Marujo PE, Hajnsdorf E, Le Derout J, Andrade R, Arraiano CM, Regnier P. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA. 2000;6:1185–1193. doi: 10.1017/s135583820000073x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Coburn GA, Mackie GA. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J Mol Biol. 1998;279:1061–1074. doi: 10.1006/jmbi.1998.1842. [DOI] [PubMed] [Google Scholar]
- 111.Folichon M, Allemand F, Regnier P, Hajnsdorf E. Stimulation of poly(A) synthesis by Escherichia coli poly(A) polymerase I is correlated with Hfq binding to poly(A) tails. FEBS Journal. 2005;272:454–463. doi: 10.1111/j.1742-4658.2004.04485.x. [DOI] [PubMed] [Google Scholar]
- 112.Huang H, Liao J, Cohen SN. Poly(A)- and poly(U)-specific RNA 3' tail shortening by E. coli ribonuclease E. Nature. 1998;391:99–102. doi: 10.1038/34219. [DOI] [PubMed] [Google Scholar]
- 113.Walsh AP, Tock MR, Mallen MH, Kaberdin VR, von Gabain A, McDowall KJ. Cleavage of poly(A) tails on the 3'-end of RNA by ribonuclease E of Escherichia coli. Nucleic Acids Res. 2001;29:1864–1871. doi: 10.1093/nar/29.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hankins JS, Denroche H, Mackie GA. Interactions of the RNA-binding protein Hfq with cspA mRNA, encoding the major cold shock protein. J Bacteriol. 2010;192:2482–2490. doi: 10.1128/JB.01619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 116.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucl Acid Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Feng Y, Huang H, Kiao J, Cohen SN. Escherichia coli poly(A) binding proteins that interact with components of degradosomes or impede RNA decay mediated by polynucleotide phosphorylase and RNase E. J Biol Chem. 2001;276:31651–31656. doi: 10.1074/jbc.M102855200. [DOI] [PubMed] [Google Scholar]
- 118.Wahle E, Keller W. The biochemistry of polyadenylation. Trends Biochem Sci. 1996;21:247–250. [PubMed] [Google Scholar]
- 119.Anderson JT. RNA turnover: unexpected consequences of being tailed. Curr Biol. 2005;15:R635–R638. doi: 10.1016/j.cub.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 120.Gultyaev AP, van Batenburg FHD, Pleij CWA. The computer simulation of RNA folding pathways using a genetic algorithm. J Molecular Biology. 1995;250:37–51. doi: 10.1006/jmbi.1995.0356. [DOI] [PubMed] [Google Scholar]
- 121.Blum E, Carpousis AJ, Higgins CF. Polyadenylation promotes degradation of 3'-structured RNA by the Escherichia coli mRNA degradosome in vitro. J Biol Chem. 1999;274:4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- 122.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Molecular and General Genetics. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 123.He L, Soderbom F, Wagner EG, Binnie U, Binns N, Masters M. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol Microbiol. 1993;9:1131–1142. doi: 10.1111/j.1365-2958.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 124.Mikkelsen ND, Gerdes K. Sok antisense RNA from plasmid R1 is functionally inactivated by RNase E and polyadenylated by poly(A) polymerase I. Mol Microbiol. 1997;26:311–320. doi: 10.1046/j.1365-2958.1997.5751936.x. [DOI] [PubMed] [Google Scholar]
- 125.Coburn GA, Mackie GA. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3'-exonucleases is dependent on oligoadenylation and RNA secondary structure. J Biol Chem. 1996;271:15776–15781. doi: 10.1074/jbc.271.26.15776. [DOI] [PubMed] [Google Scholar]
- 126.Hajnsdorf E, Braun F, Haugel-Nielsen J, Régnier P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu F, Cohen SN. RNA degradation in Escherichia coli regulated by 3' adenylation and 5' phosphorylation. Nature. 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- 128.Wrobel B, Herman-Antosiewicz A, Szalewska-Palasz A, Wegrzyn G. Polyadenylation of oop RNA in the regulation of biacteriophage lambda development. Gene. 1998;212:57–65. doi: 10.1016/s0378-1119(98)00127-9. [DOI] [PubMed] [Google Scholar]
- 129.Soderbom F, Binnie U, Masters M, Wagner EGH. Regulation of plasmid R1 replication: PcnB and RNase E expedite the decay of the antisense RNA, CopA. Mol Microbiol. 1997;26:493–504. doi: 10.1046/j.1365-2958.1997.5871953.x. [DOI] [PubMed] [Google Scholar]
- 130.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hayes R, Kudla J, Gruissem W. Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem Sci. 1999;24:199–202. doi: 10.1016/s0968-0004(99)01388-2. [DOI] [PubMed] [Google Scholar]
- 132.Komine Y, Elise K, Schuster G, Stern D. Evidence for in vivo modulation of chloroplast RNA stability by 3'-UTR homopolymeric tails in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:4085–4090. doi: 10.1073/pnas.052327599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gagliardi D, Leaver CJ. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 1999;18:3757–3766. doi: 10.1093/emboj/18.13.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gagliardi D, Perrin R, Marechal-Drouard L, Grienenberger JM, Leaver CJ. Plant mitochondrial polyadenylated mRNAs are degraded by a 3'- to 5'-exoribonuclease activity, which proceeds unimpeded by stable secondary structures. J Biol Chem. 2001;276:43541–43547. doi: 10.1074/jbc.M106601200. [DOI] [PubMed] [Google Scholar]
- 135.Kuhn J, Tengler U, Binder S. Transcript lifetime is balanced between stabilizing stem-loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol Cell Biol. 2001;21:731–742. doi: 10.1128/MCB.21.3.731-742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ryan CM, Militello KT, K RL. Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J Biol Chem. 2003;278:32753–32762. doi: 10.1074/jbc.M303552200. [DOI] [PubMed] [Google Scholar]
- 137.Kao C-Y, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol Cell Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–21649. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 139.Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3' exonuclease and a DEAD-box RNA helicase. Genes & Develop. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Plamann MD, Stauffer GV. E. coli glyA mRNA decay: The role of 3' secondary structure and the effects of the pnp and rnb mutations. Molecular and General Genetics. 1990;220:301–306. doi: 10.1007/BF00260498. [DOI] [PubMed] [Google Scholar]
- 141.Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol. 2004;51:777–790. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- 142.McLaren RS, Newbury SF, Dance GSC, Causton H, Higgins CF. mRNA degradation by processive 3'-5' exonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J Mol Biol. 1991;221:81–95. [PubMed] [Google Scholar]
- 143.Mackie GA, Genereaux JL. The role of RNA structure in determining RNase E-dependent cleavage sites in the mRNA for ribosomal protein S20 in vitro. J Mol Biol. 1993;234:998–1012. doi: 10.1006/jmbi.1993.1654. [DOI] [PubMed] [Google Scholar]
- 144.Mackie GA. Stabilization of the 3' one-third of Escherichia coli ribosomal protein S20 mRNA in mutants lacking polynucleotide phosphorylase. J Bacteriol. 1989;171:4112–4120. doi: 10.1128/jb.171.8.4112-4120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol Microbiol. 2006;60:723–737. doi: 10.1111/j.1365-2958.2006.05124.x. [DOI] [PubMed] [Google Scholar]
- 146.Kushner SR. In: Escherichia coli and Salmonella: cellular and molecular biology. Böck A, Curtis R III, Gross CA, Kaper JB, Neidhardt FC, Nyström T, Rudd KE, L SC, editors. Washington, DC: American Society for Microbiology Press; 2007. http://www.ecosal.org. [Google Scholar]
- 147.Cao GJ, Sarkar N. Poly(A) RNA in Bacillus subtilis: identification of the polyadenylylation site of flagellin mRNA. FEMS Microbiol Lett. 1993;108:281–285. doi: 10.1111/j.1574-6968.1993.tb06116.x. [DOI] [PubMed] [Google Scholar]
- 148.Hajnsdorf E, Braun F, Haugel-Nielsen J, Le Derout J, Régnier P. Multiple degradation pathways of the rpsO mRNA of Escherichia coli. RNase E interacts with the 5' and 3' extremities of the primary transcript. Biochimie. 1996;78:416–424. doi: 10.1016/0300-9084(96)84748-1. [DOI] [PubMed] [Google Scholar]
- 149.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 150.Lee K, Cohen SN. Effects of 3' terminus modifications on mRNA functional decay during in vitro protein synthesis. J Biol Chem. 2001;276:23268–23274. doi: 10.1074/jbc.M102408200. [DOI] [PubMed] [Google Scholar]
- 151.Joanny G, Le Derout J, Brechemier-Baey D, Labas V, Vinh J, Regnier P, Hajnsdorf E. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucl Acid Res. 2007;35:2494–2502. doi: 10.1093/nar/gkm120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Santos JM, Freire P, Mesquita FS, Mika F, Hengge R, Arraiano CM. Poly(A)-polymerase I links transcription with mRNA degradation via sigmaS proteolysis. Mol Microbiol. 2006;60:177–188. doi: 10.1111/j.1365-2958.2006.05078.x. [DOI] [PubMed] [Google Scholar]
- 153.Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in Trypanosome mitochondria: a complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Militello KT, Read LK. Coordination of kRNA editing and polyadenylation in Trypanosoma brucei mitochondria: complete editing is not required for long poly(A) tract addition. Nucleic Acids Res. 1999;27:1377–1385. doi: 10.1093/nar/27.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Read LK, Stankey KA, Fish WR, Muthiani AM, Stuart K. Developmental regulation of RNA editing and polyadenylation in four life cycle stages of Trypanosoma congolense. Mol Biochem Parasitol. 1994;68:297–306. doi: 10.1016/0166-6851(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 156.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 157.Montoya J, Ojala D, Attardi G. Distinctive features of the 5'-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 158.Chanfreau GF. CUTting genetic noise by polyadenylation-induced RNA degradation. Trends in Cellular Biology. 2005;15:635–637. doi: 10.1016/j.tcb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 159.Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 160.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 161.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 162.Mackie GA. Stabilization of circular rpsT mRNA demonstrates the 5'-end dependence of RNase E action in vivo. J Biol Chem. 2000;275:25069–25072. doi: 10.1074/jbc.C000363200. [DOI] [PubMed] [Google Scholar]
- 163.Jiang X, Belasco JG. Catalytic activation of multimeric RNase E and RNase G by 5'-monophosphorylated RNA. Proc Natl Acad Sci USA. 2004;101:9211–9216. doi: 10.1073/pnas.0401382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kime L, Jourdan SS, Stead JA, Hidalgo-Sastre A, McDowall KJ. Rapid cleavage of RNA by RNase E in the absence of 5'-monophosphate stimulation. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bralley P, Jones GH. Overexpression of the polynucleotide phosphorylase (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiol. 2003;149:2173–2182. doi: 10.1099/mic.0.26334-0. [DOI] [PubMed] [Google Scholar]
- 166.Saravanamuthu SS, von Gotz F, Salunkhe P, Chozhavendan R, Geffers R, Buer J, Tummler B, Steinmetz EJ. Evidence for polyadenylated mRNA in Pseudomonas aeruginosa. J Bacteriol. 2004;186:7015–7018. doi: 10.1128/JB.186.20.7015-7018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ohta N, Sanders M, Newton A. Characterization of unstable poly (A)-RNA in Caulobacter crescentus. Biochim Biophys Acta. 1978;517:65–75. doi: 10.1016/0005-2787(78)90034-5. [DOI] [PubMed] [Google Scholar]
- 168.Majumdar PK, McFadden BA. Polyadenylated mRNA from the photosynthetic procaryote Rhodospirillum rubrum. J Bacteriol. 1984;157:795–801. doi: 10.1128/jb.157.3.795-801.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Adilakshmi T, Ayling PD, Ratledge C. Polyadenylation in mycobacteria: evidence for oligo(dT)-primed cDNA synthesis. Microbiol-UK. 2000;146:633–638. doi: 10.1099/00221287-146-3-633. [DOI] [PubMed] [Google Scholar]
- 170.Li Z, Pandit S, Deutscher MP. 3' Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Aiso T, Yoshida H, Wada A, Ohki R. Modulation of mRNA stability participates in stationary-phase-specific expression of ribosome modulation factor. J Bacteriol. 2005;187:1951–1958. doi: 10.1128/JB.187.6.1951-1958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Le Derout J, Folichon M, Briani F, Deho G, Regnier P, Hajnsdorf E. Hfq affects the length and the frequency of short oligo(A) tails at the 3' end of Escherichia coli rpsO mRNAs. Nucl Acid Res. 2003;31:4017–4023. doi: 10.1093/nar/gkg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 174.Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Gorke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008;36:2570–2580. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li Z, Pandit S, Deutscher MP. Polyadenylation of stable RNA precursors in vivo. Proc Natl Acad Sci USA. 1998;95:12158–12162. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kudla J, Hayes R, Gruissem W. Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 1996;15:7137–7146. [PMC free article] [PubMed] [Google Scholar]