Abstract
Visualization and monitoring of endogenous mRNA in the cytoplasm of living cells promises a significant comprehension of refined post-transcriptional regulation. Fluorescently labeled linear antisense oligonucleotides can bind to natural mRNA in a sequence-specific way and, therefore, provide a powerful tool in probing endogenous mRNA. Here, we investigated the feasibility of using linear antisense probes to monitor the variable and dynamic expression of endogenous cytoplasmic mRNAs. Two linear antisense 2′-O-methyl RNA probes, which have different interactive fluorophores at the 5′-end of one probe and at the 3′-end of the other, were used to allow fluorescence resonance energy transfer (FRET) upon hybridization to the target mRNA. By characterizing the formation of the probe-mRNA hybrids in living cells, we found that the probe composition and concentration are crucial parameters in the visualization of endogenous mRNA with high specificity. Furthermore, rapid hybridization (within 1 min) of the linear antisense probe enabled us to visualize dynamic processes of endogenous c-fos mRNA, such as fast elevation of levels after gene induction and the localization of c-fos mRNA in stress granules in response to cellular stress. Thus, our approach provides a basis for real time monitoring of endogenous cytoplasmic mRNA in living cells.
INTRODUCTION
In eukaryotic cells, cytoplasmic mRNA plays an important role in gene expression. After moving from the nucleus to the cytoplasm, mRNAs are subject to diverse regulatory processes concerning translational repression, translational activation and degradation. These cytoplasmic processes greatly influence gene activity. Recent studies have revealed that microRNAs (miRNAs) work as modulators of translational processes within cytoplasmic structures, such as the processing body (P-body) or the stress granule (SG) (1–3). Therefore, the cytoplasm is considered to be not only the stage for translation but also the location of translational regulation. Elucidating the characteristics of cytoplasmic mRNA, such as levels and localization inside the cell, will help us to understand the molecular mechanisms of gene expression. In particular, the ability to monitor mRNA in real time would offer significant advantages because the function of mRNA is under fast and dynamic control.
Several techniques have been developed to visualize mRNAs in living cells (4–6). The most popular method is to tag mRNA with green fluorescent protein (GFP). In this approach, a gene encoding a target mRNA is tagged with the binding site for the coat protein of bacteriophage MS2 (7,8). By expressing this gene and another gene-encoding GFP fused with MS2, the target mRNA is bound to GFP via MS2 and becomes fluorescent. This method has been used to probe the localization of mRNAs in the cell; for instance, Shav-Tal et al. (9) observed the localization of β-actin mRNA molecules at single-molecule level. This technique, however, requires the addition of a tag sequence to the target mRNA and hence the mRNA becomes exogenous. Therefore, the endogenous processes of gene expression cannot be fully explored using this approach because the tagged mRNA does not necessarily reflect the characteristics of the endogenous mRNA.
On the other hand, antisense molecules can be used to label endogenous mRNAs because their use does not require the modification of target mRNA. Tyagi and colleagues (10,11) developed such an antisense molecule, which they termed a molecular beacon (MB). An MB is a fluorogenic oligonucleotide that possesses complementary sequences on either end of a probe, enabling the molecule to assume a hairpin configuration in which a fluorophore and quencher are held in close proximity (6). Currently, an MB is a unique reagent that allows us to detect an endogenous cytoplasmic mRNA target in living cells (12–15). MBs are advantageous in imaging mRNAs owing to the simplicity of detection, however, MBs are not suited for monitoring rapidly changing levels of target mRNA because upon hybridization the stem region in the MB structure must open before MB fluoresces and this may reduce the kinetic rate of MB binding (5,16).
Another antisense molecule candidate for labeling RNAs is a linear antisense probe. A linear antisense probe is a fluorescent oligonucleotide that can bind to mRNA in a sequence-specific way. Due to the lack of intramolecular interaction, this probe has superior hybridization kinetics that enables the dynamic fluctuation of endogenous mRNAs to be detected. Molenaar et al. (17) detected nuclear mRNA using linear antisense probes by observing bright foci representing localized mRNAs in small compartments in the nucleus. Santangelo et al. (18) recently developed a bright linear antisense probe and imaged localized mRNAs, such as endogenous β-actin mRNA in the leading edges of cells and viral RNA in SGs. These results indicate the high potency of linear antisense probes in visualizing cytoplasmic mRNAs. To detect mRNAs with high specificity, eliminating the fluorescence of unbound probes from that emitted by probes bound to target mRNA is required. A simple method for this is to use fluorescence resonance energy transfer (FRET); the detection of FRET upon hybridization of a pair of linear antisense probes to adjacent sequences on a target mRNA enables the distinction between bound and unbound probes (19). Using this technique, Tsuji et al. (20) reported the detection of cytoplasmic c-fos mRNA using linear antisense oligodeoxynucleotide (ODN) probes. Despite the relatively simple procedure and the great advantages of this method, linear antisense ODN probes have not been employed to study mRNA in live cells. This implies that the use of linear antisense ODN probes in living cells may be problematic (21).
In this study, to establish a method for monitoring endogenous cytoplasmic mRNA in real time, the drawback of the linear antisense ODN probes was overcome by utilizing 2′-O-methyl ribonucleic acid (2′OMeRNA), an artificial nucleic acid, as the backbone of the linear antisense probe. We characterized the intracellular properties of linear antisense 2′OMeRNA probes in the FRET-based visualization of mRNA, including the effect of probe concentration on FRET efficiency. We achieved reproducible visualization of endogenous c-fos mRNA in the cytoplasm of living COS7 cells. Furthermore, the hybridization kinetics of linear antisense 2′OMeRNA probes to target mRNA was found to be significantly higher than that of MBs. Exploiting the excellent hybridization kinetics, we successfully performed real time monitoring of endogenous c-fos mRNA during cellular events, such as rapid elevation of c-fos mRNA levels after gene induction and the localization of c-fos mRNA in SGs in response to cellular stress.
MATERIALS AND METHODS
Nucleic acid probes
c-fos mRNA, which encodes the transcription factor c-Fos, was chosen as the target endogenous mRNA. Linear antisense probes were designed (Table 1) according to a previous report (20). Cy3 and Cy5 were selected as a pair of fluorophores for the detection of FRET. The donor and acceptor probes were labeled with Cy3 at the 5′-end and Cy5 at the 3′-end, respectively, as illustrated in Figure 1A. Nucleic acid probes having sense sequences of c-fos mRNA (sense probes) were also prepared for negative control experiments. The concentration and the molar ratio of fluorescent dye to oligonucleotide were measured using UV–VIS photospectroscopy (V-570; JASCO, Hachioji, Japan). The molar ratios of dyes to oligonucleotides were in the range from 0.72 to 1.03. 2′OMeRNA MBs for c-fos mRNA were also prepared (Table 1). These MBs consisted of the same antisense sequences as the linear antisense donor probe and contained self-complementary sequences (stem) on either end that were labeled by Cy3 and BHQ2 (22). Pairs of antisense probes bearing a 2′OMeRNA or a DNA backbone and 2′OMeRNA MBs were purchased from Proligo (Kyoto, Japan) and FASMAC (Atsugi, Japan).
Table 1.
Sequences and designs of linear and molecular beacon probes and target region of mRNA
| Construct | Sequencea |
|---|---|
| Linear antisense probes | |
| DNA (donor) | (Cy3-)TCTAGTTGGTCTGTCTCCGC |
| DNA (acceptor) | GCAAAGCAGACTTCTCA(-Cy5) |
| 2′OMe RNA (donor) | (Cy3-)UCUAGUUGGUCUGUCUCCGC |
| 2′OMe RNA (acceptor) | GCAAAGCAGACUUCUCA(-Cy5) |
| Linear sense probes | |
| DNA (donor) | (Cy3-)AGATCAACCAGACAGAGGCG |
| DNA (acceptor) | CGTTTCGTCTGAAGAGT(-Cy5) |
| 2′OMe RNA (donor) | (Cy3-)AGAUCAACCAGACAGAGGCG |
| 2′OMe RNA (acceptor) | CGUUUCGUCUGAAGAGU(-Cy5) |
| Linear antisense probes (biotinylated) | |
| DNA (donor) | (Cy3-)TCTAGTTGGTCTGTCTCCGC(-TEGb-biotin) |
| DNA (acceptor) | (biotin-TEG-)GCAAAGCAGACTTCTCA(-Cy5) |
| 2′OMe RNA (donor) | (Cy3-)UCUAGUUGGUCUGUCUCCGC(-TEG-biotin) |
| 2′OMe RNA (acceptor) | (biotin-TEG-)GCAAAGCAGACUUCUCA(-Cy5) |
| Linear sense probes (biotinylated) | |
| 2′OMe RNA (donor) | (Cy3-)AGAUCAACCAGACAGAGGCG(-TEG-biotin) |
| Molecular beacon | |
| 2′OMe RNA (antisense) | (Cy3-)CGACCUCUAGUUGGUCUGUCUCCGCGGUCG(-BHQ2) |
| Molecular beacon (biotinylated) | |
| 2′OMe RNA (antisense) | (Cy3-)CGACCUCUAGUUGGUCUGUCUCCGCGGUCG(-BHQ2-TEG-biotin) |
| 2′OMe RNA (sense) | (Cy3-)CGACCAGAUCAACCAGACAGAGGCGGGUCG(-BHQ2-TEG-biotin) |
| c-fos mRNA (657–696) | 5′···GCGGAGACAGACCAACUAGAAGAUGAGAAGUCUGCUUUGC···3′ |
aAll sequences are listed from their 5′-end to their 3′-end. The hybridization of antisense probes with the target mRNA is illustrated in Figure 1A.
bTriethyleneglycol (TEG) was used as a spacer between a biotin moiety and an oligonucleotide.
Figure 1.
Hybridization of antisense probes with the target mRNA. (A) Schematic drawing of mRNA detection. When two kinds of fluorescently labeled antisense probes are hybridized to adjacent sequences of the target mRNA, the distance between the two fluorophores becomes short and FRET occurs. (B) The fluorescence spectra of fluorescently labeled probes containing 2′OMeRNA or DNA backbones in the absence (gray line) or presence (red line) of the target c-fos mRNA, prepared by an in vitro transcription system. FRET was observed when mRNA hybridized with antisense probes; in contrast, FRET was not detected when using linear sense probes.
Preparation of c-fos mRNA in vitro
c-fos mRNA was prepared in vitro using the following transcription reaction. pBluescriptII KS(-) containing c-fos cDNA was treated with KpnI and EcoRI to excise the c-fos cDNA. The c-fos fragment was then inserted into pcDNA6/V5 (Invitrogen, Carlsbad, CA, USA). The region containing c-fos and the T7 promoter of pcDNA-cfos was amplified by the polymerase chain reaction (PCR), using DNA polymerase KOD-plus (TOYOBO, Osaka, Japan). One of the primers contained a poly (T)30 sequence, thereby incorporating a poly (A)30 tail into the c-fos DNA template. The transcription reaction was performed using the above PCR product and the T7 Ribomax transcription kit (Promega, Madison, WI, USA). Cap analog (Promega) was incorporated at the 5′-end of the resultant c-fos mRNA to introduce a cap structure.
Fluorescence spectroscopy
Fluorescence spectra were measured using a spectrofluorometer (FP-6500; JASCO). The donor and acceptor probes and the c-fos mRNA were prepared at 2 μM in 1×SSC solution (150 mM NaCl and 15 mM sodium citrate, pH 7.0). The probes and mRNA solutions, at final concentrations of 667 nM, were mixed and incubated at room temperature for 20 min and then diluted with 1×SSC solution to a final concentration of 40 nM. Fluorescence spectra of the mixtures were measured by excitation at 532 nm. Time course studies of linear antisense probes were performed by mixing the three solutions containing the donor, acceptor and c-fos mRNA to a final concentration of 50 nM at 37°C and observing the acceptor fluorescence intensities at 670 nm with an excitation of the donor probe at 532 nm. Likewise, time course studies of MBs were performed by observing Cy3 fluorescence at 560 nm with an excitation at 532 nm.
Cell culture and microinjection
In live cell imaging, COS7 cells (African Green Monkey Kidney) were used. COS7 cells were cultured on a 35 mm Glass base dish (ASAHI Techno GLASS, Tokyo, Japan) in Dulbecco’s Modified Eagle’s Medium (D-MEM, Gibco, Carlsbad, CA, USA) supplemented with 5% fetal bovine serum (Gibco). Before live cell imaging, the medium was replaced with D-MEM (containing 25 mM HEPES buffer) (Gibco) supplemented with 5% fetal bovine serum. The temperature of the culture medium was maintained at 37°C using a stage plate heater (TOKAI HIT, Fujinomiya, Japan) and a microscope objective lens heater (TOKAI HIT). A FemtoJet (Eppendorf, Hamburg, Germany), controlled by a micromanipulator (Eppendorf), was used for the microinjection of nucleic acid probes into living cells. Nucleic acid probes were dissolved in a buffer containing 80 mM KCl, 4 mM NaCl, 10 mM KH2PO4–K2HPO4 (pH 7.2) and the solution was then filtered using an Ultrafree-MC (Millipore, Billerica, MA, USA). The filtered solution was microinjected into the cytoplasm of COS7 cells using a Femtotips II glass capillary needle (Eppendorf).
Live cell imaging
Microinjection and live-cell imaging were performed on an inverted microscope (IX70; Olympus, Tokyo, Japan) equipped with an objective lens (UplanApo, 60×, N.A. 1.40; Olympus). To acquire cell images, we used a cooled CCD camera (ORCA-ER; Hamamatsu Photonics, Hamamatsu, Japan). The donor (Cy3) and the acceptor (Cy5) probes were excited with a green solid-state laser (100 mW, 532 nm Compass 315M-100; Coherent, Santa Clara, CA, USA) and a red He–Ne laser (10 mW, 633 nm GLS5360; Showa Optronics, Tokyo, Japan), respectively. The fluorescence image of the donor probe was taken using a dichroic mirror, 565LP (Chroma technology, Rockingham, VT, USA) and an emission filter, 610/75M (Chroma technology). The fluorescence image of the acceptor probe was taken using 660LP and 700/75M (Chroma technology). The FRET image was taken using 565LP and 700/75M (Chroma technology). Fluorescence images were quantitatively analyzed using AQUA-Lite ver. 10 (Hamamatsu Photonics) at various intervals after microinjection. Total fluorescence intensities of donor, FRET and acceptor images inside the cytoplasm of a cell were obtained. The region of interest (ROI), the cytoplasm, was determined according to phase contrast (PC) images of each cell. Background fluorescence intensities were determined by acquiring the signal of the region where cells were absent. In quantitative analysis, the fluorescence intensity of each image was used for calculations after subtracting background intensity. Normalized FRET signals were determined using fluorescence intensities of FRET images (FFRET) divided by those of donor images (Fdonor) acquired from the same cell. To determine the amount of probe in the cytoplasm, acceptor probes were excited with the red laser, and the total fluorescence intensities of acceptor probes in the cytoplasm were measured. Although FRET did not occur when we used a pair of sense probes in solution, all FRET images of cells in which sense probes were introduced contained weak fluorescence signals. This pseudo signal comes from two factors: one is the cross-talk of the filter set occurring between the donor set and the FRET set and another is that the Cy5 fluorophore can be slightly excited by a 532-nm green laser. The fluorescence intensity of this pseudo signal was ∼6% that of the donor image. This signal is far weaker than the signal obtained from actual FRET and thus we could distinguish the FRET signal from the pseudo signal. This pseudo signal was treated as the background noise of the FRET image.
Fluorescence correlation spectroscopy apparatus
An fluorescence correlation spectroscopy (FCS) apparatus was installed on an inverted microscope (IX70, Olympus). The incident excitation from a green laser beam (532 nm, COMPASS 415M, Coherent) was focused with a water immersion objective lens (UplanApo, 60× w, N.A. 1.20, Olympus) onto the diffraction-limited spot in the cytoplasm of COS7 cells. The emitted fluorescence was collected by the objective, passed through an emission filter (Q560LP, Chroma technology) and a pinhole (30 µm in diameter) and detected with an avalanche photo-diode (SPCM-AQA-14PC, PerkinElmer Optoelectronics, Fremont, CA, USA). The fluorescence signal was analyzed in real time with a digital autocorrelator board (ALV-6010/160, ALV-GmbH, Langen, Germany).
FCS measurement and analysis
The fluorescence autocorrelation function (FAF), G (τ) was fitted off-line with Origin software (Origin Lab, Northampton, MA), according to Equation (1).
 |
(1) |
where N is the average number of molecules in the focal volume, Fi and τi are the fraction and diffusion time of component i, respectively (23), and s is a structural parameter, representing the ratio of vertical and axial radius of focus. The average FAF of Cy3-labeled antisense probes were obtained from 10-s measurements. The concentration of probes was calibrated from the pre-determined concentration of Cy3 fluorophore.
Determination of probe concentration in cells
To calibrate the concentration of probes inside cells with respect to total fluorescence intensities, three kinds of fluorescence (i.e. donor, acceptor and FRET fluorescence) were measured from eight cells, in which paired probes were microinjected and imaged. The concentration of probes inside each cell was obtained by averaging five independent FCS measurements. The measured calibration curve was linear (r = 0.988) between total fluorescence intensities and probe concentrations in the range from 107 to 933 nM. The concentration of antisense probes in the cytoplasm was calculated using this calibration curve.
Induction of c-fos mRNA with PMA
Endogenous c-fos mRNA was induced with phorbol 12-myristate 13-acetate (PMA) (24). After the microinjection of a pair of streptavidin-bound linear antisense 2′OMeRNA probes into the cytoplasm, COS7 cells were incubated for 15 min to complete the hybridization reaction of antisense probes to endogenous c-fos mRNA. The cells were then exposed to 100 nM PMA and subjected to time-lapse fluorescence imaging. Normalized FRET fluorescence was determined as fluorescence intensities of FRET images normalized to those of acceptor images acquired from the same cell.
Imaging c-fos mRNA aggregated in SGs
After the microinjection of a pair of streptavidin-bound linear antisense 2′OMeRNA probes into the cytoplasm, COS7 cells were incubated for 15 min to complete the hybridization reaction of antisense probes to endogenous c-fos mRNA. The cells were then exposed to 0.5 mM sodium arsenite. After 30 or 60 min of exposure, endogenous c-fos mRNA and SGs marked by TIA-1-GFP (25) were observed by acquiring FRET fluorescence.
RESULTS
In this study, we chose c-fos mRNA in COS7 cells as a target to facilitate the comparison of our results with those of a previous report by Tsuji et al. (20), who utilized linear antisense ODN probes to detect the same target. c-fos mRNA, encoding the transcription factor c-fos, is constitutively expressed in many cells (26,27,28), and thus can be an endogenous mRNAs target without requiring the induction of a transfected plasmid.
Linear antisense probes
To detect a fluorescently labeled antisense probe hybridized to the target mRNA in the presence of unbound probe, we designed two linear antisense probes that were complementary to adjacent sequences of c-fos mRNA so that the hybridized probes could be detected by observing a FRET signal under a fluorescence microscope. When a pair of antisense probes was hybridized to the target mRNA, the distance between the two fluorophores became short and FRET occurs, as illustrated in Figure 1A. The target site on c-fos mRNA was selected as the sequence between 657 and 696 because the hybridization of antisense ODN probes within this site was previously confirmed by Tsuji et al. (20) and by our group (29). The number of nucleotides between a pair of antisense probes was determined to be four, as reported previously (20).
As a backbone of the antisense probes, 2′OMeRNAs and DNAs were prepared. 2′OMeRNA is an artificial nucleic acid that has been frequently utilized as an antisense molecule in recent studies because it is resistant to nuclease degradation in living cells and possesses high affinity to nucleic acids (6,30,31,32). Molenaar et al. (17,33) showed that linear antisense 2′OMeRNA probes could hybridize to nuclear RNAs, whereas linear antisense ODN probes could not. In this study, we compared linear 2′OMeRNA and ODN probes for visualizing cytoplasmic mRNAs.
Fluorescence spectra of the fluorescently labeled nucleic acid probes were measured in the presence and absence of c-fos mRNA, which had been prepared by a transcription reaction in vitro (Figure 1B). FRET, which is denoted by a decrease in donor fluorescence and an increase in acceptor fluorescence, was observed in the presence of c-fos mRNA. FRET was observed with both antisense ODN probes and 2′OMeRNA probes. In contrast, FRET was not detected when sense probes, containing the sense sequence of the target mRNA, were mixed with c-fos mRNA. These results indicate that the antisense probes hybridized to a specific target sequence of the mRNA, showing the feasibility of the strategy for the selective detection of target mRNAs.
The stability of the of mRNA and linear antisense probe complexes in living cells
The stability of the linear antisense 2′OMeRNA and ODN probes hybridized to c-fos mRNA in living cells was examined to evaluate the feasibility of these antisense molecules as probes for the visualization of mRNA in living cells. Two linear antisense probes were hybridized to c-fos mRNA in test tubes and then the solutions were microinjected into the cytoplasm of living COS7 cells. The distributions of antisense probes were observed under an epi-fluorescence microscope. Three sources of fluorescence, Cy3 (donor), Cy5 (acceptor) and FRET fluorescence were imaged after microinjection. By acquiring the FRET image, in which the fluorescence of Cy5 is observed with the excitation of Cy3, the distribution of mRNA could be explored because FRET occurred only in the presence of the target mRNA.
Cells in which the mRNA-antisense ODN probe complexes were introduced showed FRET fluorescence in the cytoplasm immediately after microinjection (see the middle fluorescence image of the upper panel of Figure 2A), but the FRET signal soon disappeared. Ten minutes after injection, no FRET signal was detected in the cytoplasm; a weak FRET fluorescence observed in the nucleus was at the background noise level. Donor and acceptor images in the lower panel of Figure 2A show that both probes moved into the nucleus. Free oligonucleotides accumulate in the nucleus when they are introduced into the cytoplasm (12), therefore, ODN probes might exist alone and redistribute as free oligonucleotides 10 min after microinjection. This suggests that ODN probes, even though once hybridized to c-fos mRNA in vitro, were dissociated from the mRNA in living cells and that free ODN probes moved into the nucleus. Since the mRNA-ODN complex is unstable in living cells, linear ODN probes were inappropriate for the visualization of mRNAs in living cells.
Figure 2.
Antisense 2′OMeRNA probes can form a stable complex with mRNA in living cells whereas antisense ODN probes cannot. (A–C) PC images and fluorescence images of the mRNA complex that was hybridized with probes in vitro and then microinjected into the cytoplasm of living COS7 cells; Antisense ODN probes (A), antisense 2′OMeRNA probes (B) and sense 2′OMeRNA probes (C). D, F and A in each image indicate donor image, FRET image and acceptor image, respectively. Scale bars, 10 µm.
Cells in which mRNA-antisense 2′OMeRNA probe complexes were introduced showed strong FRET fluorescence in the cytoplasm, even 10 min after the microinjection (see the middle fluorescence image of the lower panel of Figure 2B). It also should be noted that donor and the acceptor fluorescence was detected both in the cytoplasm and in the nucleus, while FRET was detected only in the cytoplasm (Figure 2B, lower panel). This result suggests that antisense 2′OMeRNA probes that were hybridized to c-fos mRNA were distributed in the cytoplasm, whereas those that had dissociated from the complex accumulated in the nucleus. As we expected, FRET signal, reflecting the distribution of mRNAs, could be detected only when a pair of probes were hybridized to the target mRNA, highlighting the specificity of this method. In contrast, mRNA-sense 2′OMeRNA probe complexes resulted in no FRET signal and nearly all of both sense probes were detected in the nucleus (Figure 2C). These results demonstrate that antisense 2′OMeRNA probes can form stable complexes with mRNAs in living cells and, therefore, are well suited for the visualization of mRNAs.
Imaging endogenous c-fos mRNA in the cytoplasm of COS7 cells
Next, we targeted cytoplasmic c-fos mRNA, which is endogenously expressed in COS7 cells, for visualization. The 2′OMeRNA probe, shown to be stable when hybridized to mRNAs in living cells, was microinjected into the cytoplasm of COS7 cells and images of the cells were obtained. However, within 1 min, 2′OMeRNA probes were sequestered in the nucleus and fluorescence was detected only in the nucleus (Supplementary Figure S1, upper panel). This accumulation was so rapid that the probe could not hybridize to the cytoplasmic c-fos mRNA, even though they were capable of hybridization. To prevent the accumulation of antisense probes in the nucleus, the probes were modified with biotin and streptavidin so that they were too large to pass through the nuclear pores (12,20). 2′OMeRNA probes containing a biotin moiety at the end opposite the fluorescent dye were prepared, as listed in Table 1, and then reacted with streptavidin. As we expected, the streptavidin-bound probe distributed only in the cytoplasm of COS7 cells (Supplementary Figure S1, lower panel), suggesting that the sequestration of linear antisense probe into the nucleus could be abolished.
A pair of streptavidin bound 2′OMeRNA probes were microinjected into the cytoplasm of the cell and three fluorescence images (i.e. donor, acceptor and FRET fluorescence) of each cell at each time point were obtained. When the probe at 1–3 µM was prepared and injected, the probe concentration in the cytoplasm was 0.1–0.5 µM. Under these conditions no significant FRET fluorescence was detected. We assumed that this result was due to the low concentration of antisense probes compared to the endogenously expressed c-fos mRNAs. If the concentration of antisense probes was much lower than that of the target mRNA, the probability of a pair of antisense probes hybridizing to the same mRNA would be low and consequently the occurrence of FRET would be rare. When the probe was prepared at a concentration ranging from 5 to 15 μM the injected probe concentration in the cytoplasm was 0.5–3 µM. Under these conditions strong FRET fluorescence was observed in the cytoplasm of cells (Figure 3A). The FRET fluorescence was distributed throughout the cytoplasm, reflecting the distribution of endogenous c-fos mRNA in COS7 cells. To examine the effect of probe concentration on the efficiency of FRET occurrence, we introduced various concentrations of paired antisense probes into cells and fluorescence intensities of FRET images were analyzed. Here, FRET occurrence was evaluated using a FRET signal, defined as the ratio of fluorescence intensity of a FRET image (FFRET) to that of a donor image (Fdonor). When FRET occurs, donor fluorescence decreases and FRET fluorescence increases, and thus the ratio (FFRET/Fdonor) increases. The quantitative analysis of fluorescence images of cells showed that a variety of FRET signals were obtained. Probe concentrations were determined from the total fluorescence intensities inside cells using a calibration curve; this calibration curve was obtained by comparing fluorescence intensities with probe concentration measured by FCS. Figure 3B shows the relationship between FRET signals of each COS7 cell and the concentration of paired antisense probes inside cells. FRET signals were dependent on probe concentration and, interestingly, these two values were generally proportional. This indicates that a higher concentration of linear antisense probes resulted in a higher probability of forming a hybrid with endogenous mRNA. When the cytoplasmic concentration of the antisense probe was >0.5 µM, strong FRET signals were obtained. Therefore, this relatively high concentration of antisense probes was used in the following experiments to obtain high-specificity FRET signals. Interestingly, the FRET signals were dispersed to some extent (Figure 3B), which might reflect the variation of c-fos mRNA concentrations expressed in COS7 cells.
Figure 3.
The detection of endogenous c-fos mRNA in the cytoplasm of living cells using streptavidin-bound antisense 2′OMeRNA probes. (A) Fluorescence images of streptavidin-bound 2′OMeRNA probes microinjected into the cytoplasm of COS7 cells. When antisense probes were introduced, strong fluorescence due to FRET was observed, while no fluorescence was detected when sense probes or ODN probes were introduced. D, F and A in each image indicate donor image, FRET image and acceptor image, respectively. Scale bars, 10 µm. (B) Relationship between FRET signal and probe concentration. FRET signal, defined as the ratio of the fluorescence intensity of the FRET image (FFRET) to that of the donor probe (Fdonor) in each cell, was plotted against the probe concentration inside the cytoplasm. The probe concentration was determined from the total fluorescence intensities inside cells using the calibration curve of fluorescence intensities and probe concentration measured by FCS. Results obtained for paired antisense 2′OMeRNA probes, control 2′OMeRNA probes (sense donor probe and antisense acceptor probe), paired antisense ODN probes and the hybrid paired probes (antisense ODN donor probe and antisense 2′OMeRNA acceptor probe) were compared. Closed red circles indicate antisense 2′OMeRNA probe, open red circles indicate control sense 2′OMeRNA probe, open blue circles indicate antisense ODN probe and open black circles indicate the hybrid paired probes.
As a control experiment, sense 2′OMeRNA donor probes were used instead of the 2′OMeRNA donor antisense probes. In contrast to the paired 2′OMeRNA antisense probes, FRET was not observed, even though these probes were distributed in the cytoplasm (see middle panels of Figure 3A). Introduction of various concentrations of paired probes (2′OMeRNA donor sense probe and 2′OMeRNA acceptor antisense probe) into cells resulted in no significant FRET signal regardless of the probe concentration (Figure 3B). A pair of antisense ODN probes was also microinjected into cells and imaged. Antisense ODN probes introduced into the cells resulted in a low-FRET signal, similar to that from the sense 2′OMeRNA probe (Figure 3A and B). When the combination of donor antisense ODN and acceptor antisense 2′OMeRNA probe was used, FRET was also not detected. These results strongly demonstrate that linear antisense ODN probes cannot hybridize stably to target mRNAs in living cells and that using 2′OMeRNA is crucial for the visualization of target mRNAs in living cells.
We also visualized endogenous c-fos mRNA in COS7 cells using a different method to support our mRNA images. The single-molecule FISH method, recently developed by Raj and Tyagi is one of the most sensitive approaches to image mRNA in a fixed cell (34, www.singlemoleculefish.com). The use of 48 single-labeled fluorescent oligonucleotide probes to hybridize with mRNA renders it quite bright enough to allow detection of the individual mRNA molecule in fixed cells. The results for endogenous c-fos mRNA using 48 antisense probes, each labeled with Cy3 showed that many bright spots corresponding individual c-fos mRNA were dispersed throughout the cytoplasm of fixed COS7 cells (Supplementary Figure S2 and Supplementary Experiments), demonstrating that our mRNA images are validated by a more sensitive method.
Hybridization kinetics of antisense probes
A highly advantageous characteristic of linear antisense probes for real time monitoring of endogenous mRNA in living cells would be a fast hybridization reaction with target mRNA. However, the intracellular kinetics of linear antisense probe hybridization has been unclear. Here, hybridization time course studies were performed in vitro and in living cells by measuring the intensities of FRET fluorescence and resultant time constants were calculated. These were compared with those obtained with an MB that has an identical antisense sequence for the target (Supplementary Figure S3). Results obtained in 1×SSC solution are shown in Figure 4A and B. The time constants for hybridization of the linear antisense 2′OMeRNA probes and the MB were 3.97 min and 4.72 h, respectively. Linear antisense probes were, therefore, shown to hybridize more quickly than the MB, which had a time constant 71.3 times larger than that of the linear antisense probes. The hybridization kinetics of MB strongly depends on nucleotide composition and stem length (16). Therefore, MBs that possess different composition (ODN) and shorter stem (4 nt) were also tested (Supplementary Table S1 and Figure S4). Time constants for hybridization of ODN MB and 2′OMeRNA MB with short stem in 1×SSC solution were 2.35 and 2.57 h, respectively (Supplementary Figure S5), suggesting that these alterations of probe chemistry result in a better MB kinetics.
Figure 4.
Hybridization time course studies of antisense probes with mRNA. Cases using linear antisense 2′OMeRNA probes in a 1×SSC solution (A), MB in a 1×SSC solution (B), streptavidin-bound linear antisense 2′OMeRNA probes in living COS7 cells (C) and streptavidin-bound MB in living COS7 cells (D) are presented. A c-fos mRNA prepared by in vitro transcription and the endogenously expressed c-fos mRNA in the cytoplasm of living COS7 cells was targeted in (A and B) and (C and D), respectively. The concentration of mRNA and antisense probes was 50 nM in a 1×SSC solution (A and B). In living cell studies, 3 µM of probes (in a microinjection needle) were introduced into the cell (C and D). The estimated concentration of probes inside the cell is ∼0.5 µM. The time course plots were fitted to a single-exponential function of the time constant (t) for the hybridization reaction in each case (red fit line for linear antisense probe and blue fit line for MB). Error bas in (C and D) represent SD.
Next, the hybridization time course of both antisense probes with endogenous c-fos mRNA in living cells was examined by monitoring the signal fluorescence (i.e. FRET fluorescence for the linear antisense probes and Cy3 fluorescence for the MB) in COS7 cells. As shown in Figure 4C and D, hybridization of the linear antisense 2′OMeRNA probes reached a plateau in almost 1 min, while this took >2 h for the MB. Time constants obtained for the linear antisense probes and the MB in living COS7 cells were 0.664 min (39.8 s) and 52.1 min, respectively. This indicates that the linear antisense probes can hybridize to target mRNA in living cells 78.5 times more quickly than the MB and thus they have a great advantage over MBs in terms of hybridization kinetics. This rapid kinetics for linear antisense hybridization suggests a high temporal resolution for the monitoring of mRNA with linear antisense probes. This feature is advantageous in exploring mRNA dynamics and fast fluctuations of mRNA expression.
Real time monitoring of the induction of endogenous c-fos mRNA
Linear antisense 2′OMeRNA probe was applied to the real time monitoring of fast-response gene expression. c-fos is an early response gene that is rapidly induced by the stimulation of serum responsive elements (SREs) (24,26,27). Expression of c-Fos protein is upregulated within 1 h upon PMA stimulation, which activates the SRE driven c-Fos promoter (24). In this study, expression of endogenous c-fos mRNA in COS7 cells was monitored by observing FRET fluorescence of linear antisense probes after PMA stimulation. As shown in Figure 5, FRET fluorescence from the cytoplasm of PMA-treated cells began to rise 10 min after the PMA stimulus and a gradual increase continued over time. More than a 6% increase of FRET fluorescence was observed. Nevertheless, we cannot convert this value into the actual increased quantity of induced target mRNA because there is no linear relationship between the FRET intensity and the quantity of the target mRNA. FRET efficiency strongly depends on the concentration ratio of antisense probe to mRNA. The addition of trace diluents (0.0001% DMSO) resulted in a slight decrease of the signal. When linear sense probes were used, no FRET signal higher (defined by FFRET divided by Fdonor) than background noise was observed at any time point. These results indicated that rapid changes in endogenous mRNA expression can be monitored in real time using linear antisense probes by virtue of their fast hybridization kinetics.
Figure 5.
Real time monitoring of endogenous c-fos mRNA induction. Normalized FRET fluorescence from streptavidin-bound linear antisense 2′OMeRNA probes was recorded over time. Red line indicates the result from PMA-treated COS7 cells and gray line indicates the result from COS7 cells treated with diluents (0.0001% DMSO). The probe concentration inside the cell was 12 ± 4.8 µM (five cells). Error bars represent SD.
Real time imaging of endogenous c-fos mRNA in SGs
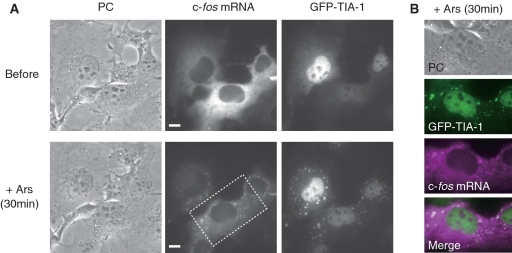
To demonstrate the feasibility of using linear antisense 2′OMeRNA probes in the observation of rapid and dynamic changes in the intracellular distribution of mRNA, the accumulation of endogenous mRNA in SGs was imaged. Real time imaging of RNA-binding proteins that reside in SGs and of RNAs, such as poly(A)+ mRNA and viral RNA, revealed that mRNA accumulated in SGs within ∼30 min after the induction of stress with arsenite (18,35). Here, the accumulation of endogenous c-fos mRNA in COS7 cells after sodium arsenite treatment, a conventional stress inducer, was monitored using linear antisense probes. As shown in Figure 6A, FRET fluorescence from stressed cells showed that a fraction of endogenous c-fos mRNA localized in granular domains in the cytoplasm. These granules were confirmed to be SGs by merging the images of c-fos mRNA with those of GFP-TIA-1, an SG marker (Figure 6B). When linear sense probes were used, no significant FRET fluorescence was detected at any time point, demonstrating the mRNA specificity of the linear antisense probe. It is also noteworthy that a fraction of endogenous c-fos mRNA was also detected diffusely throughout the cytoplasm, in contrast to the distribution of TIA-1, which was mostly detected only in SGs and in the nucleus (Figure 6A and B). Even 60 min after stress induction, when the accumulation of mRNA in SGs would have reached saturation, endogenous c-fos mRNA was still detected both in SGs and in the cytoplasm (Supplementary Figure S6). This distribution of a specific endogenous mRNA agrees with a former investigation which showed that only a portion of poly(A)+ mRNAs accumulate in SGs, while a further portion remains in the cytoplasm (35).
Figure 6.
The real time imaging of endogenous c-fos mRNA in SGs. (A) PC and fluorescence images of endogenous c-fos mRNA and GFP-TIA1 in COS7 cells before and 30 min after arsenite treatment. Endogenous c-fos mRNA was visualized by acquiring FRET fluorescence from streptavidin-bound linear antisense 2′OMeRNA probes. The probe concentration inside the cell was 0.76 ± 0.24 µM. (B) Co-localization of TIA-1 (green) and endogenous c-fos mRNA (magenta) in SGs. Images of cells are enlarged from boxed area in (A). Scale bars, 10 µm.
DISCUSSION
To monitor the variable and dynamic expression of a specific endogenous mRNA in the cytoplasm of a cell, we investigated the advantages and drawbacks of using linear antisense 2′OMeRNA probes. We used a pair of streptavidin-bound linear antisense 2′OMeRNA probes bearing different fluorophores, which allowed us to observe the target mRNA in the presence of unbound probes by imaging FRET fluorescence from the probe pair. We performed detailed studies on the hybridization of linear antisense 2′OMeRNA probes with endogenous mRNA, such as the effect of probe backbone composition, probe concentrations and time constants in vitro and in living cells. These studies provided us with some critical parameters for imaging endogenous mRNA, enabling us to monitor with high specificity the fast and dynamic behavior of endogenous mRNA in the cytoplasm.
Adopting 2′OMeRNA as the backbone of linear antisense probes is crucial in detecting mRNAs based on hybridization in living cells. In particular, the stable hybridization of linear antisense 2′OMeRNA probes with mRNA is essential for real time monitoring since the procedure requires repeated or sequential imaging. ODN probes are generally utilized as antisense probes in fixed cells for fluorescence in situ hybridization (FISH) or in experiments such as micro arrays that are performed in solutions. However, we failed to visualize mRNA with ODN probes in living cells. The introduction of a pair of linear antisense ODN probes into the cytoplasm of the cell resulted in no significant FRET signal; the signal was similar to that from 2′OMeRNA sense probes (negative control). This result together with the observation that the FRET fluorescence emitted from the ODN probe-mRNA complex quickly disappeared, showed that linear antisense ODN probes could not form a complex with mRNA in living cells. The necessity to utilize linear 2′OMeRNA probes, not linear ODN probes, was also confirmed in previous reports in which the differences in the localization (17) or in the mobility (33) of these two probes were compared for the detection of nuclear mRNAs. Tsuji et al. (20) detected c-fos mRNA in living COS7 cells using ODN linear antisense probes, while in our hands we failed to detect c-fos mRNA using ODN probes. A possible reason for this discrepancy is that Tsuji et al. targeted c-fos mRNA over-expressed from a plasmid, while we visualized endogenously expressed c-fos mRNA in COS7 cells. It is also noteworthy that we showed FRET occurrence to be dependent on probe concentration for linear antisense 2′OMeRNA probes, whereas increased concentration of linear antisense ODN probes and chimeric combinations containing antisense ODN probe did not result in an increase of FRET signal. This suggests that linear antisense ODN probe and mRNA hybrids are unlikely to form in living cells, even with high-ODN-probe concentrations. This implies that the incapability of linear antisense ODN probes to bind to mRNA in living cells is not due to the low affinity for mRNA, but to a certain active interference mechanism of ODN–mRNA hybrid formation. To overcome this problem, we also used an artificial nucleic acid, peptide nucleic acid (PNA). Although PNA formed a stable complex with mRNA in living cells, the hybridization kinetics were so slow that it required a longer time to hybridize to mRNAs (Supplementary Figure S7) than 2′OMeRNA probe.
The concentration of antisense probe was also an important factor in the FRET-based detection of mRNAs. Our results showed that the concentration of the antisense probes had to be higher than that of target mRNA to obtain a strong FRET fluorescence. This is because the probability of the hybridization of pair of probes with the same mRNA molecule would be low when the probe concentration was less than that of the mRNA. The cytoplasmic concentration of paired antisense probes for c-fos mRNA that is required for the detection of FRET with high specificity was >1–2 µM. Furthermore, the FRET signals are strongly dependent on the probe concentration inside the cell and a higher concentration resulted in stronger FRET signals. These results indicate that the quantity of antisense probe is a major element in the detection of FRET in the cytoplasm of the cell.
The most representative feature of linear antisense probes is the excellent mRNA-binding kinetics, demonstrated by monitoring the hybridization reaction with target mRNA in vitro and in living cells. We showed for the first time that the time constant for a linear antisense 2′OMeRNA probe in living cells was as fast as 40 s. This excellent kinetics allowed us to assay the fast and dynamic behavior of endogenous mRNA, such as early response of gene induction and localization of endogenous mRNA in SGs. These results indicate that the technique can be applied to any investigation of quantitative and spatial variation of endogenous mRNA with a temporal resolution of 1 min. Indeed, we could monitor the fast elevation of endogenous c-fos mRNA levels after gene induction using a pair of linear antisense probes. This advantage of linear antisense molecules can be exploited by any hybridization probes for the visualization of mRNA.
Recently, sequence-specific degradation of mRNA and translational repression (and also activation) have elicited considerable attention because they are key processes in post-transcriptional regulation (1–3). Monitoring endogenous cytoplasmic mRNA, using linear antisense probes, will be an asset to this field of research. Furthermore, antisense-mediated silencing of pathogenic miRNAs is a new way of utilizing antisense molecules for the medical treatment of some intractable diseases, such as cancer (36,37). This class of antisense molecules, called antimiR or antagomiR, can antagonize aberrantly expressed endogenous miRNAs that are pathogenic, for example, by repressing tumor suppressor genes. Our results showing the stability of 2′OMeRNA–mRNA hybrids, the fast binding of 2′OMeRNA to target RNA, including newly transcribed mRNA and the localization of 2′OMeRNA–mRNA hybrids in a cytoplasmic structure indicate linear antisense 2′OMeRNA oligonucleotides to be a promising candidate for antisense therapy.
In conclusion, we show here significant characteristics and advantages of linear antisense probes in monitoring an endogenous mRNA to yield novel findings. First of all, utilizing an artificial nucleic acid (2′OMeRNA) as the backbone of the linear antisense probe is mandatory in selectively detecting mRNA by means of FRET; ODN based linear antisense probes yielded no signal. Second, linear antisense probes can make a hybrid with endogenous mRNA in only 1 min. Third, the expression of endogenous c-fos mRNA in the cytoplasm starts within 10 min after an extracellular stimulus. Finally, a fraction of endogenous c-fos mRNA is accumulated in SGs in response to stress, while the remaining fraction remains diffused in the cytoplasm.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.O., in part); Japan Society for the Promotion of Science (JSPS) through its ‘Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program)’ (to T.F.). Funding for open access charge: Grant-in-Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Paul Anderson (Brigham and Women’s Hospital, Harvard Medical School) for providing the GFP-TIA-1 expression plasmid.
REFERENCES
- 1.Anderson P, Kedarsha N. RNA granules: post transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2009;23:75–105. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Chekulaeva M, Filipowicz W. Mechanism of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Dirks RW, Tanke HJ. Advances in fluorescent tracking of nucleic acids in living cells. Biotechniques. 2006;40:489–496. doi: 10.2144/000112121. [DOI] [PubMed] [Google Scholar]
- 5.Bao G, Rhee WJ, Tsourkas A. Fluorescence probes for live-cell RNA detection. Annu. Rev. Biomed. Eng. 2009;11:25–47. doi: 10.1146/annurev-bioeng-061008-124920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 8.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living Mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 11.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi S, Alsmadi O. Imaging native beta-actin mRNA in motile fibroblasts. Biophys. J. 2004;87:4153–4162. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perlette J, Tan W. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal. Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 15.Sokol DL, Zhang X, Lu P, Gewirtz AM. Real time detection of DNA.RNA hybridization in living cells. Proc. Natl Acad. Sci. USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santangelo P, Nitin N, Bao G. Nanostructured probes for RNA detection in living cells. Ann. Biomed. Eng. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 17.Molenaar C, Marras SA, Slats JC, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29:e89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santangelo PJ, Lifland AW, Curt P, Sasaki Y, Bassell GJ, Lindquist ME, Crowe JE., Jr Single molecule-sensitive probes for imaging RNA in live cells. Nat. Methods. 2009;6:347–349. doi: 10.1038/nmeth.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardullo RA, Agrawal S, Flores C, Zamecnik PC, Wolf DE. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 1998;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuji A, Koshimoto H, Sato Y, Hirano M, Sei Iida Y, Kondo S, Ishibashi K. Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys. J. 2000;78:3260–3274. doi: 10.1016/S0006-3495(00)76862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zamaratski E, Pradeepkumar PI, Chattopadhyaya J. A critical survey of the structure-function of the antisense oligo/RNA heteroduplex as substrate for RNase H. J. Biochem. Biophys. Methods. 2001;48:189–208. doi: 10.1016/s0165-022x(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 22.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brock R, Vamosi G, Vereb G, Jovin TM. Rapid characterization of green fluorescent protein fusion proteins on the molecular and cellular level by fluorescence correlation microscopy. Proc. Natl Acad. Sci. USA. 1999;96:10123–10128. doi: 10.1073/pnas.96.18.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh J, Rhee HJ, Kim SW, Kim SB, You HJ, Kim JH, Na DS. Annexin-I inhibits PMA-induced c-fos SRE activation by suppressing cytosolic phospholipase A2 signal. FEBS Lett. 2000;477:244–248. doi: 10.1016/s0014-5793(00)01812-3. [DOI] [PubMed] [Google Scholar]
- 25.Kedarsha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curran T, Morgan JI. Fos: an immediate-early transcription factor in neurons. J. Neurobiol. 1995;26:403–412. doi: 10.1002/neu.480260312. [DOI] [PubMed] [Google Scholar]
- 27.Maturana A, Van Haasteren G, Piuz I, Castelbou C, Demaurex N, Schlegel W. Spontaneous calcium oscillations control c-fos transcription via the serum response element in neuroendocrine cells. J. Biol. Chem. 2002;277:39713–39721. doi: 10.1074/jbc.M200464200. [DOI] [PubMed] [Google Scholar]
- 28.Schiller M, Bohm M, Dennler S, Ehrchen JM, Mauviel A. Mitogen- and stress-activated protein kinase 1 is critical for interleukin-1-induced, CREB-mediated, c-Fos gene expression in keratinocytes. Oncogene. 2006;25:4449–4457. doi: 10.1038/sj.onc.1209479. [DOI] [PubMed] [Google Scholar]
- 29.Yanagihara N, Tadakuma H, Ishihama Y, Okabe K, Funatsu T. Determination of potent antisense oligonucleotides in vitro by semiempirical rules. J. Biosci. Bioeng. 2007;103:270–277. doi: 10.1263/jbb.103.270. [DOI] [PubMed] [Google Scholar]
- 30.Pitts AE, Corey DR. Inhibition of human telomerase by 2′-O-methyl-RNA. Proc. Natl Acad. Sci. USA. 1998;95:11549–11554. doi: 10.1073/pnas.95.20.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majlessi M, Nelson NC, Becker MM. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998;26:2224–2229. doi: 10.1093/nar/26.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsourkas A, Behlke MA, Bao G. Hybridization of 2′-O-methyl and 2′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002;30:5168–5174. [PMC free article] [PubMed] [Google Scholar]
- 33.Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J. Cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raj A, van den Bogaard P, Rifkin S, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 2009;122:3619–3626. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- 36.Naldini L, Brown BD. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat. Rev. Genet. 2009;10:587–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 37.Garzon R, Caalin GA, Croce CM. MicroRNAs in Cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.