Abstract
The INK4b-ARF-INK4a locus encodes for two cyclin-dependent kinase inhibitors, p15INK4b and p16INK4a, and a regulator of the p53 pathway, ARF. In addition ANRIL , a non-coding RNA, is also transcribed from the locus. ARF, p15INK4b and p16INK4a are well-established tumor suppressors which function is frequently disabled in human cancers. Recent studies showed that single nucleotide polymorphisms mapping in the vicinity of ANRIL are linked to a wide spectrum of conditions, including cardiovascular disease, ischemic stroke, type 2 diabetes, frailty and Alzheimer disease. The INK4b-ARF-INK4a locus is regulated by Polycomb repressive complexes (PRCs) and its expression can be invoked by activating signals. Other epigenetic modifiers such as the histone demethylases JMJD3 and JHDM1B, the SWI/SNF chromatin remodeling complex and DNA methyltransferases regulate the locus interplaying with PRCs. In view of the intimate involvement of the INK4b-ARF-INK4a locus on disease, to understand its regulation is the first step for manipulate it to therapeutic benefit.
Key words: senescence, p16INK4a, ARF, p15INK4b, ANRIL, polycomb, histone demethylases, DNA methylation
Introduction
The INK4b-ARF-INK4a locus spans around 35 kb on human chromosome 9p21 that contains the INK4b (also termed CDKN2b), ARF and INK4a genes (these two jointly referred as CDKN2a, reviewed in ref. 1 and Fig. 1). They encode for two cyclin dependent kinase inhibitors, p15INK4b and p16INK4a and an unrelated protein, p14ARF. Whereas p15INK4b is translated from its own independent genetic structure, ARF and p16INK4a share their second and third exons. However, ARF splices exon 2 in an alternative reading frame to that of p16INK4a, hence the name ARF.2 ARF and INK4a are transcribed from independent promoters. Both p15INK4b and p16INK4a bind specifically to CDK4 and CDK63 blocking cell proliferation by preventing phosphorylation of RB resulting in a G1 arrest. ARF sequesters MDM2 in the nucleolus.4 This in turn activates p53 resulting in either cell cycle arrest or apoptosis.3 Recently, a new large antisense non-coding RNA termed ANRIL (also known as CDKN2b antisense or CDKN2BAS) has been mapped to the INK4b-ARF-INK4a locus5 (Fig. 1) where it is presumed to play a regulatory role. How ANRIL and other noncoding RNAs regulate the expression of the locus is currently the matter of active investigation.
Figure 1.
Organization of the INK4b-ARF-INK4a locus and disease-associated SNPs. Genetic structure of the human INK4b-ARF-INK4a locus. The coding exons are shown in colors and non-coding exons are shown in light gray for ANRIL and dark gray for the other genes of the locus. The approximate position of single nucleotide polymorphisms (SNPs) associated with disease states is indicated by blue arrows. SNPs associated with type 2 diabetes mellitus (D), vascular heart disease (H) and frailty (F) are indicated. Map is not drawn to scale and positions are approximate.
The INK4b-ARF-INK4a Locus and Disease
The interest on the INK4b-ARF-INK4a locus originated from genetic linkage studies showing the association of mutations or deletions on chromosome 9p21 with familial predisposition to melanoma.6,7 It was subsequently demonstrated that in addition to germ-line mutations, homozygous deletion on 9p21 is one of the most frequent cytogenetic events associated with a wide variety of tumors (reviewed in ref. 8). Loss of the INK4b-ARF-INK4a locus is the most frequent copy number alteration across tumors and cancer cell lines.9,10 Multiple studies have revealed p16INK4a as the main tumor suppressor in the locus while showing that p15INK4b and p14ARF can also act as tumor suppressors. Intragenic mutations that inactivate INK4b or ARF are observed, though rare in comparison to those affecting INK4a. For example, specific inactivation affecting ARF but not INK4a can occur in melanoma,11 while methylation of the INK4b promoter is observed in hematopoietic malignancies.12 Mouse models have confirmed that deficiency for either of the proteins encoded by the INK4b-ARF-INK4a locus, alone or in combination results in tumor-prone animals.8,13 It is worthy to mention that despite mouse models have been clearly useful to dissect the involvement of the INK4b-ARF-INK4a locus in health and disease, significant differences exist in its regulation between mouse and human. Most notably while mouse p19Arf is upregulated during replicative or Ras-induced senescence, human p14ARF is not (reviewed in ref. 1).
An explanation for the frequent alteration of the locus in cancer is its activation in response to aberrant oncogenic signalling. As such, members of the INK4b-ARF-INK4a locus are key effectors of oncogene-induced senescence (OIS) and are induced in premalignant lesions, limiting tumor progression. Therefore, to progress to a more malignant state, a lesion suffers insurmountable pressure to silence the locus through deletion, mutations or epigenetic regulation. The INK4b-ARF-INK4a locus is also upregulated at replicative senescence and aging.8 In murine tissues, increased expression of p16Ink4a and p19Arf, but not of p15Ink4b, is observed with aging,14,15 making the case for an involvement of the INK4b-ARF-INK4a locus in age-related pathologies. Again, the difference in the locus regulation between mouse and human should be taken into account and although p16INK4a expression increases with aging in humans, there are no reports of a similar increase for p14ARF levels.16 Additional evidence for an extended role of the INK4b-ARF-INK4a locus in disease came from a series of linkage studies in which single nucleotide polymorphisms (SNPs) in a region spanning 120 kb around the INK4b-ARF-INK4a locus were associated with increased susceptibility to frailty,17 coronary artery disease,18,19 myocardial infarction,20 type 2 diabetes21–23 and late onset Alzheimer disease.24 Interestingly different SNPs have been associated with increased disease risk on those studies (Fig. 1), suggesting that not a single polymorphism is responsible for the increased susceptibilities observed.
Regulation of the INK4b-ARF-INK4a Locus by Polycomb Repressive Complexes
Given the extraordinary relevance of the INK4b-ARF-INK4a locus on disease, it is key to maintain it repressed under normal circumstances but without losing the ability to induce its expression when needed. A critical layer to achieve this control is epigenetic regulation through Polycomb (Pc) repressive complexes (PRC1 and 2). The PRC2 complex establishes the repressive H3K27met3 chromatin mark, catalyzed by the histone methyltransferase activity of EZH2.25–27 This epigenetic mark, is recognized by the PRC1 maintenance complex, which in addition mono-ubiquitinates histone H2A.28 The key role of PRCs in regulating the INK4b-ARF-INK4a locus is remarked by the fact that overexpression of different PcG members such as Bmi1, Cbx7 or Cbx8 results in repression of the locus and bypass or delays senescence.29–31 Conversely, cells lacking PRC1 components such as Bmi1 or Ring1b29,32 show aberrant expression of the INK4b-ARF-INK4a locus. Mice knockout for different PcG genes have multiple developmental problems. In particular Bmi1-/- mice have skeletal transformations and severe neurological and hematopoietic defects.33 With the exception of the skeletal alterations that are due to deregulation of the Hox gene cluster, the rest of the defects observed are restored to a great extent by knocking out Ink4a/Arf.34
Non-coding RNAs: ANRIL and Others
A remaining question is how PRCs are targeted to the INK4b-ARF-INK4a locus. The core members of the PRCs do not have a DNA binding motif and the assumption is that they must associate through accessory factors to target DNA. In Drosophila, PREs (polycomb recruiting elements) have been identified, and some transcription factors such as Pho named as responsible of the recruitment of PRCs to those elements. Pho is required for PcG-mediated silencing in Drosophila.35 Interestingly, the homolog of Pho, the transcription factor YY1,36 is also involved in PcG-mediated silencing in mammals.37 However, despite studies showing genome-wide PcG distribution in mammalian chromatin, equivalent PRE and targeting factor(s) has not yet been clearly identified for mammalian systems.38 It has been proposed that a combination of association with transcription factors and long interfering non coding RNAs could be responsible of the recruitment of PRC to their target genes in mammalian cells.38 Recently, a DNA element recognized by YY1 has been shown to target PRC complexes to the HoxD locus.39 However, a similar arrangement in the INK4b-ARF-INK4a locus has not been identified so far. On the other hand, evidence is starting to show that long noncoding RNAs such as ANRIL contribute to the targeting of PRCs to the INK4b-ARF-INK4a locus. LincRNAs can control gene expression through tethering chromatin-modifying complexes to specific genomic loci. Multiple lincRNAs interact with PRC2 and other chromatin modifier complexes.40 In addition, a novel class of short RNAs transcribed from the 5′ end of Polycomb target genes interact with PRC2 and could play a role in PRC association to target genes.41 Chromobox proteins, such as those that are part of the PRC1, cannot only bind methylated histones but also to RNA. For example, treatment with RNAse decreases the association of CBX7 with H3K27me3 and the inactive X chromosome.42 Treatment with RNAse or mutation of residues needed for CBX7 to interact with RNA also results in reduced recruitment of CBX7 to the INK4b-ARF-INK4a locus with the corresponding effects on senescence.43 LincRNAs are currently seen as a platform helping in the recruitment of different chromatin remodeling complexes, such as PRC2 and LSD1/CoREST/REST.40 Although ANRIL seems to control the levels of p15INK4b acting as an antisense transcript,44 ANRIL and probably other non coding RNAs (long and short) can also regulate the INK4b-ARF-INK4a locus by contributing to the recruitment of epigenetic factors. Single nucleotide polymorphisms (SNPs) in a 58 kb-long interval on chromosome 9p21 have previously been associated with an increased susceptibility to coronary heart disease.18,20 ANRIL maps to this same region (Fig. 1).5,45 A relation between disease-associated SNP close to ANRIL and the expression of the members of the INK4b-ARF-INK4a locus has been noted in human samples.46,47 Recently, a deletion of the orthologous 70 kb-long non-coding intervals on mouse chromosome 4 was also found to affect the cardiac expression of the neighboring p15Ink4b and p16Ink4a genes.48 Whether ANRIL or cis-regulatory regions are mediating this effect needs to be investigated.
Activation of the INK4b-ARF-INK4a Locus by Oncogenic Signals
To fulfill its function as a stress sensor, the INK4b-ARF-INK4a locus must be kept repressed in basal conditions, but also be quickly activated when needed. A number of transcription factors have been linked with the activation of the locus as a whole or their individual members and this have been reviewed in reference 1, In particular, we have a better knowledge of the transcription factors involved in activating p16INK4a in response to oncogenic stress such as Ras expression, where the relation between Ets2 factors and Id1 is key.49
Parallel to activate or mobilize transcription factors to induce the INK4b-ARF-INK4a locus, stimuli that trigger its induction have to modify the epigenetic status of the locus and wipe their repressive marks. To achieve this during replicative and oncogene-induced senescence there is a change in the expression and recruitment of the key enzymes regulating the methylation of H3K27. Levels of EZH2, the enzyme that methylates H3K27 decrease during replicative senescence and OIS.50–52 In parallel, the H3K27 demethylase JMJD3 is upregulated in response to Ras, recruited to the INK4b-ARF-INK4a locus and regulates the activation of the locus by oncogenic stress. JMJD3 is found upregulated in some preneoplastic lesions as nevi and have credentials to be a tumor suppressor, as it is frequently deleted in different tumor types.51,52 How JMJD3 is regulated by Ras is a matter of active investigation. Another mechanism mediating the activation of the INK4b-ARF-INK4a locus is the direct modification and displacement of polycomb proteins. MAPKAP, which is activated downstream of Ras can phosphorylate Bmi1 prompting its release from chromatin.53 However, whether this phosphorylation-mediated displacement occurs during senescence has not been investigated.
Chromatin Remodeling by the SWI/SNF Complex
Additional chromatin modifiers and epigenetic marks also control the status of the locus, sometimes in direct interplay with PRCs. The upregulation of genes normally repressed by PRCs need of chromatin remodeling complexes, such as the SWI/SNF (or BAF) complex.54 The gene encoding SNF5 (also termed SMARCB1 or BAF47), one of the components of the SWI/SNF complex is frequently deleted in malignant rhabdoid tumors (MRT). By analyzing MRT tumors it was noted that SNF5 is needed to regulate p16INK4a expression through recruitment of the SWI/SNF complex and Polycomb eviction and the deletion of SNF5 results in reduced expression of p16INK4a.55,56 There is a crosstalk between SWI/SNF-mediated chromatin remodeling and the PRC complexes (Fig. 2). An interesting question is whether the SWI/SNF complex plays an active role in activating the INK4b-ARF-INK4a locus in response to oncogenic stress or in specific tissues. Recent evidence seems to suggest so, as Snf5 acts as a prominent mediator of p19Arf expression in murine sarcomas generated upon activation of K-Ras. It has been suggested that this can account for the differences in susceptibility to transformation by Ras of different tissues.57
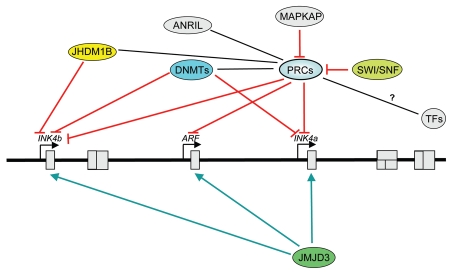
Figure 2.
Epigenetic regulation of the INK4b-ARF-INK4a locus. Cartoon summarizing different epigenetic mechanisms regulating the locus. Epigenetic silencing of the locus in normal cells is mediated by Polycomb repressive complexes (PRCs). In tumorigenesis, methylation of the INK4a or INK4b promoters is often observed. Chromatin remodeling by the SWI /SNF5 complex result in displacement of PRCs complexes and locus activation. The histone demethylases Jhdm1b regulate the expression of p15INK4b while JMJD3 counteract the effects of H3K27me3 marks and PRC-mediated silencing. MAPKAP phosphorylation of Bmi1 results in PRC1 displacement from chromatin. Recent evidence suggests that ANRIL and maybe other ncRNAs could regulate the locus. We still do not understand how transcription factors interplay with this epigenetic machinery to regulate the locus. Epigenetic modifiers are shown in color. PRCs, Polycomb repressive complexes; DNMTs, DNA methyl transferases; TF s, transcription factors. Black lines show relation; red arrows, activation; green arrows, inhibition. Map is not drawn to scale and positions are approximate.
Regulation of the INK4b-ARF-INK4a Locus by DNA Methylation
In addition to homozygous deletion and inactivating mutations, one of the most common mechanisms of inactivation of INK4a in cancer is through aberrant promoter methylation.58 Similarly, DNA promoter methylation inactivating INK4b is observed in a number of hematological malignancies.12 It is becoming evident that DNA methylation occurs not in isolation but in close relation with other epigenetic modifications such as PRC-mediated silencing. This link is highlighted by the fact that members of the PRC1 and PRC2 complexes, such as EZH2 and CBX7, can interact physically with the DNA methylation machinery, binding to Dnmt3b.59,60 In addition, Polycomb target genes are more often represented among those aberrantly methylated in cancer,61–63 adding functional consequences to these physical interactions. Further highlighting the relation between the different epigenetic remodelers of the INK4b-ARF-INK4a locus, restoration of an active SWI/SNF complex by reintroduction of SNF5 in deficient cells results not only in the eviction of PRC complexes from the INK4b-ARF-INK4a locus but in a loss of DNMT3b localization and DNA methylation in the locus.56
Additional Chromatin Modifiers Controlling the INK4b-ARF-INK4a Locus
Recently another histone demethylase, Jhdm1b/Kdm2b, has also been linked with regulation of INK4b-ARF-INK4a locus. Initial interest on this enzyme arose from insertional mutagenesis studies that showed its association with tumorigenesis, although there was controversy as if a tumor suppressor or as an oncogene.64,65 It was first suggested that Jhdm1b/Kdm2b demethylates H3 lysine 4 (H3K4me3)66 but currently the strongest biochemical evidence in vitro and in vivo suggests that Jhdm1b controls demethylation of H3 lysine 36 (H3K36me3).67 Ectopic expression of Jhdm1b/Kdm2b bypasses replicative senescence.64 Independent studies suggest that regulation of the Ink4b-Arf-Ink4a locus mediates its effects on senescence.68 More specifically it has been proposed that Jhdm1b/Kdm2b controls p15Ink4b expression.67 How modification of H3K36, a mark normally present in recently transcribed genes to prevent reinitiation at intragenic sites can regulate the expression of p15Ink4b is not clear.69 A possible explanation can be found in the observation that Jhdm1b/Kdm2b can form part of complexes containing PcG members,70 suggesting a link between this chromatin remodeling enzyme and PRC (Fig. 2).
Concluding Remarks
Recent GWAS have highlighted that besides being key actors on tumor suppression the members of the INK4b-ARF-INK4a locus may play important roles on other diseases. At the moment multiple laboratories are trying to better understand what the relevant SNPs located around the INK4b-ARF-INK4a locus mean for its expression and how they affect its regulation. In particular the question of whether these SNPs are affecting the levels or function of the non-coding RNA ANRIL or maybe unveil the presence of cis-regulatory elements needs to be answered. The interplay between different chromatin modifiers with PRCs on its centre is complex (Fig. 2). We will need also to better understand how cellular signals such as oncogenic stress are integrated by transcription factors and the epigenetic machinery to regulate the INK4b-ARF-INK4a locus. The INK4b-ARF-INK4a locus has been analyzed on cancer from a diagnostic and prognostic perspective, as very often mutations or deletions make its alterations irreversible. However in a subset of tumors, epigenetic modifications, reversible by nature, contribute to silence the locus. If a functional INK4b-ARF-INK4a locus is also present in other diseases and it is still open to regulation, we could envision mechanisms and drugs that could contribute to restore its normal function. Clever mice models have shown that there is potential to control the locus obtaining beneficial effects (i.e., cancer protection) while averting unwanted side effects (i.e., accelerated aging).71 To fully understand how the INK4b-ARF-INK4a locus is regulated in normal and pathological circumstances is the essential first step for its therapeutic manipulation.
Acknowledgements
We thank Berenika Gdowska for her expert help designing the figures. Core support from the Medical Research Council and grants from MRC Technology, Cancer Research UK and the Association for International Cancer Research fund the research in J.G.'s laboratory. N.P. is funded by an MRC studentship. J.G. is also supported by the EMBO Young Investigator Programme.
Abbreviations
- ANRIL
antisense noncoding RNA at INK4a/ARF locus
- ARF
alternative reading frame
- GWAS
genome-wide association studies
- INK4a, INK4b
inhibitors of CDK4 a and b
- JMJD3
jumonji-domain containing 3
- LD
linkage disequilibrium
- OIS
oncogene-induced senescence
- PcG
polycomb group
- PRC
polycomb group repressive complexes
- SNP
single nucleotide polymorphism
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/12996
References
- 1.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 2.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 3.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 4.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 5.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 6.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 7.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 8.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, et al. Signatures of mutation and selection in the cancer genome. Nature. 463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedberg DE, Rigas SH, Russak J, Gai W, Kaplow M, Osman I, et al. Frequent p16-independent inactivation of p14ARF in human melanoma. J Natl Cancer Inst. 2008;100:784–795. doi: 10.1093/jnci/djn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boultwood J, Wainscoat JS. Gene silencing by DNA methylation in haematological malignancies. Br J Haematol. 2007;138:3–11. doi: 10.1111/j.1365-2141.2007.06604.x. [DOI] [PubMed] [Google Scholar]
- 13.Krimpenfort P, Ijpenberg A, Song JY, van der Valk M, Nawijn M, Zevenhoven J, et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 14.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 15.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen GP, Stemmer-Rachamimov AO, Shaw J, Roy JE, Koh J, Louis DN. Immunohistochemical survey of p16INK4A expression in normal human adult and infant tissues. Lab Invest. 1999;79:1137–1143. [PubMed] [Google Scholar]
- 17.Melzer D, Frayling TM, Murray A, Hurst AJ, Harries LW, Song H, et al. A common variant of the p16(INK4a) genetic region is associated with physical function in older people. Mech Ageing Dev. 2007;128:370–377. doi: 10.1016/j.mad.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 21.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 23.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuchner S, Gilbert JR, Martin ER, Leon-Guerrero CR, Xu PT, Browning C, et al. Linkage and association study of late-onset Alzheimer disease families linked to 9p21.3. Ann Hum Genet. 2008;72:725–731. doi: 10.1111/j.1469-1809.2008.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 26.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 27.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 30.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 31.Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, et al. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 2007;26:1637–1648. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. P Natl Acad Sci USA. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, et al. Posterior transformation, neurological abnormalities and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 34.Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 37.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 39.Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6:1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 50.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voncken JW, Niessen H, Neufeld B, Rennefahrt U, Dahlmans V, Kubben N, et al. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J Biol Chem. 2005;280:5178–5187. doi: 10.1074/jbc.M407155200. [DOI] [PubMed] [Google Scholar]
- 54.Gebuhr TC, Bultman SJ, Magnuson T. Pc-G/trx-G and the SWI/SNF connection: developmental gene regulation through chromatin remodeling. Genesis. 2000;26:189–197. doi: 10.1002/(sici)1526-968x(200003)26:3<189::aid-gene4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 55.Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn LM, Mohd-Sarip A, et al. P16INK4a is required for hSNF5 chromatin remodelerinduced cellular senescence in malignant rhabdoid tumor cells. J Biol Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 56.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young NP, Jacks T. Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci USA. 2010;107:10184–10189. doi: 10.1073/pnas.1004796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herman JG, Civin CI, Issa JP, Collector MI, Sharkis SJ, Baylin SB. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–841. [PubMed] [Google Scholar]
- 59.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 60.Mohammad HP, Cai Y, McGarvey KM, Easwaran H, Van Neste L, Ohm JE, et al. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 2009;69:6322–6330. doi: 10.1158/0008-5472.CAN-09-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 63.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 64.Pfau R, Tzatsos A, Kampranis SC, Serebrennikova OB, Bear SE, Tsichlis PN. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci USA. 2008;105:1907–1912. doi: 10.1073/pnas.0711865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 2006;25:3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 67.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci USA. 2009;106:2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peters G. An INKlination for epigenetic control of senescence. Nat Struct Mol Biol. 2008;15:1133–1134. doi: 10.1038/nsmb1108-1133. [DOI] [PubMed] [Google Scholar]
- 70.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Serrano M, Blasco MA. Cancer and ageing: convergent and divergent mechanisms. Nat Rev Mol Cell Biol. 2007;8:715–722. doi: 10.1038/nrm2242. [DOI] [PubMed] [Google Scholar]