Abstract
Glucose-dependent insulinotropic polypeptide (GIP) is a gastrointestinal hormone that exerts insulinotropic and growth and survival effects on pancreatic β-cells. Additionally, there is increasing evidence supporting an important role for GIP in the regulation of adipocyte metabolism. In the current study we examined the molecular mechanisms involved in the regulation of GIP receptor (GIPR) expression in 3T3-L1 cells. GIP acted synergistically with insulin to increase neutral lipid accumulation during progression of 3T3-L1 preadipocytes to the adipocyte phenotype. Both GIPR protein and mRNA expression increased during 3T3-L1 cell differentiation, and this increase was associated with upregulation of nuclear levels of sterol response element binding protein 1c (SREBP-1c) and peroxisome proliferator-activated receptor γ (PPARγ), as well as acetylation of histones H3/H4. The PPARγ receptor agonists LY171883 and rosiglitazone increased GIPR expression in differentiated 3T3-L1 adipocytes, whereas the antagonist GW9662 ablated expression. Additionally, both PPARγ and acetylated histones H3/H4 were shown to bind to a region of the GIPR promoter containing the peroxisome proliferator response element (PPRE). Knockdown of PPARγ in differentiated 3T3-L1 adipocytes, using RNA interference, reduced GIPR expression, supporting a functional regulatory role. Taken together, these studies show that GIP and insulin act in a synergistic manner on 3T3-L1 cell development and that adipocyte GIPR expression is upregulated through a mechanism involving interactions between PPARγ and a GIPR promoter region containing an acetylated histone region.
Keywords: adipocytes, diabetes, gene expression, obesity, peroxisome proliferator-activated receptor
Glucose-dependent insulinotropic polypeptide (GIP) and glucagon-like peptide-1 (GLP-1) are the two major incretin hormones that potentiate glucose-stimulated insulin secretion during a meal and exert additional beneficial effects on β-cell proliferation and survival (1–9). Long-acting incretin mimetics (10–13) and highly selective inhibitors of the incretin-degrading enzyme, dipeptidyl peptidase-IV (DPP-IV) (14–16), have been recently introduced as therapeutic agents for type 2 diabetes. As both incretin hormones exert effects on a number of additional target tissues (2, 4), it is important to understand the functional implications of such actions. There is strong evidence supporting a role for GIP in the regulation of lipogenesis in adipocytes (17–19), a function that is consistent with its anabolic characteristics. Recently, it was demonstrated that expression of the GIP receptor (GIPR) increases during adipogenesis (20, 21), and it was suggested that GIP plays a developmental role, possibly by potentiating the effects of insulin.
Adipose tissue mass is determined by the volume and number of adipocytes, factors that are dependent upon a balance between preadipocyte differentiation and maturation to the adipocyte phenotype and cell loss by apoptosis (22). A complex series of events is involved in preadipocyte-to-adipocyte development, including growth arrest, expression of lipogenic enzymes, lipid accumulation, and development of sensitivity to hormones important for regulation (23). Although insulin-like growth factor-1 (IGF-1) has been considered the major hormone regulator during initiation of differentiation (24), insulin regulation contributes to all subsequent stages of adipogenesis, lipogenesis, and modulation of lipolysis (23, 24). In earlier studies, we demonstrated that insulin antagonizes lipolytic actions of GIP (26), whereas actions of GIP on lipogenesis were found to be insulin-dependent (27–29). The significance of the dual lipolytic/lipogenic actions of GIP is currently unclear. However, lipolytic action may play a role in maintaining circulating FFAs at appropriate levels during fasting, when insulin levels are low, thus priming β-cells for subsequent glucose stimulation. Following nutrient ingestion, GIP stimulates insulin secretion, and the two hormones may then act in combination to stimulate lipogenesis.
In view of the interaction between insulin and GIP in regulating adipocyte lipoprotein lipase (LPL) (27–29), we considered it important to establish whether GIP potentiated the effects of insulin on preadipocyte differentiation. Additionally, expression of the GIPR in preadipocytes is extremely low (20, 21), and mechanisms underlying its induction have not been characterized. An additional objective of the current study, therefore, was to identify factors involved in the regulation of adipocyte GIPR expression.
During progression of 3T3-L1 preadipocytes to the adipocyte phenotype, GIP was found to act synergistically with insulin to increase neutral lipid accumulation. In studies of both differentiating and differentiated 3T3-L1 cells, GIPR expression was shown to be increased by a mechanism involving peroxisome proliferator-activated receptor γ (PPARγ) activation and acetylation of histones H3/H4, and, at the nuclear level, PPARγ bound to a peroxisome proliferator response element (PPRE) in a region of the GIPR promoter that also contained acetylated histones H3/H4. This appears to be the first description of key factors involved in the regulation of adipocyte GIPR expression.
EXPERIMENTAL PROCEDURES
Cell culture and differentiation of 3T3-L1 adipocytes
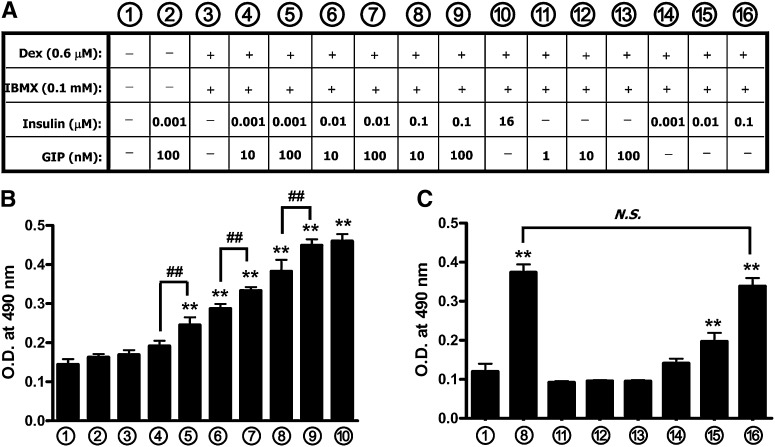
3T3-L1 cells (Zen-Bio Inc.) were cultured in DMEM containing high glucose and supplemented with 5% newborn calf serum plus penicillin/streptomycin (standard medium). Two days after cells were confluent, they were treated for 1, 3, or 7 days under various culture conditions, with medium supplemented with 10% FBS plus penicillin/streptomycin in the absence or presence of dexamethasone (Dex; 0.6 µM) and 3-isobutyl-1-methylxanthine (IBMX; 0.1 mM) and/or GIP (1–100 nM) with or without insulin (0.001–16 µM), as shown in Fig. 1A (culture medium conditions 1–16). Differentiation of cells into the adipocyte phenotype was assessed by Oil Red O or boron-dipyrromethene (BODIPY) 493/503 dye staining. In studies of fully differentiated 3T3-L1 adipocytes, treatment was carried out for 7 days with a specific culture medium condition (Fig. 1A, condition10). After differentiation, adipocytes were treated with a PPARγ agonist (rosiglitazone) or antagonist (GW9662) as indicated in the figure legends.
Fig. 1.
Insulin and GIP synergistically increase lipogenesis in 3T3-L1 cells. A: Conditional treatments of 3T3-L1 cells for differentiation are shown. 3T3-L1 preadipocytes were treated for 7 days with Dex (0.6 µM) and IBMX (0.1 mM) and insulin in the presence or absence of GIP. B, C: Cells were stained with Oil Red O at day 7. 3T3-L1 preadipocytes were treated as described in the legend to panel A for 7 days and then stained with Oil-Red-O, and the level of staining was determined spectrophotometically. All data represent three independent experiments, each carried out in triplicate. Significance was tested using ANOVA with Newman-Keuls post hoc test, where ** represents P <0.05 versus control, conditional culture medium 1 ; ## represents P <0.05 versus the indicated group, conditional culture medium 4 , 6 or 8. N.S., not significant; O.D., optical density.
Oil Red O staining
Cells were fixed and stained for 2 h by complete immersion in a working solution of Oil Red O. The method described by Ramirez-Zacarias et al. (30) was used to determine the level of staining: isopropyl alcohol was added to the stain culture dish, dye was extracted by gentle pipetting, and the absorbance at 490 nm was measured with a spectrophotometer.
Fluorescence image capture
3T3-L1 preadipocytes were treated for 7 days as described in the legend to Fig. 1A and stained with BODIPY 493/503 and Hoechst 33342 dyes (Molecular Probes, Eugene, OR). The fluorophore lipid probe BODIPY 493/503 specifically stains neutral lipids and reveals a broader range of lipids than Oil Red O, including unesterified cholesterol (31). Fluorescent signals were detected by a high-throughput CellomicsTM ArrayScan VTI automated fluorescence imager.
Preparation of nuclear extracts
Nuclear extracts were prepared from cells as described by Schreiber et al. (32). Briefly, cells were washed with PBS and scraped into 200 µl of ice-cold buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 0.1% Nonidet-P40, and protease inhibitors). Following centrifugation, supernatants (cytoplasmic extracts) were collected, and the resulting pellets were resuspended in 20 µl of buffer B (20 mM HEPES [pH 7.9], 400 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 20% glycerol, and protease inhibitors) and incubated on ice for 10 min. After the mixture was clarified by centrifugation, the supernatants (nuclear extracts) were collected and subjected to Western blot analysis. Histone H3 and β-actin were used as nuclear and nonnuclear markers, respectively, to establish lack of cross-contamination between the fractions.
Western blot analysis
Cytoplasmic and nuclear extracts from 3T3-L1 preadipocytes or adipocytes were separated on a 15% SDS-polyacrylamide gel and transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Mississauga, ON). Probing of the membranes was performed with antibodies against PPARγ, PPARγ2, and SREBP-1 (codes sc-9000, sc-22020, and sc-8984, respectively; Santa Cruz Biotechnology, Santa Cruz, CA); and acetyl-histone H3 (Lys9), acetyl-histone H4 (Lys8), and histone H3 (product no. 9649, no. 2594, and no. 9715, respectively; Cell Signaling Technology, Beverly, MA). GIPR and β-actin antibodies (product nos. NLS1251 and NB600-501, respectively) were from Novus Biologicals (Littleton, CO). Immunoreactive bands were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ) using horseradish peroxidase-conjugated IgG secondary antibodies.
Quantitative real-time reverse transcriptase-PCR
Total RNA was extracted from adipocytes, and cDNA fragments were generated by reverse transcription. cDNA (100 ng) was used in real-time reverse transcriptase (RT)-PCR to measure GIPR expression, whereas 10 ng of cDNA was used in the 18S rRNA control PCR. The primer and probe sequences used for the amplification of GIPR were forward primer 5′-CCG CGC TTT TCG TCA TCC-3′; reverse primer 5′-CCA CCA AAT GGC TTT GAC TT-3′; and probe 5′-FAM-CCC AGC ACT GCG TGT TCT CGT ACA GG-3′-TAMRA (where FAM is 6-carboxyfluorescein and TAMRA is tetramethylrhodamine). The primer and probe sequences used for the amplification of PPARγ2 were forward primer 5′-TTC TCC TGT TGA CCC AGA GCA-3′; reverse primer 5′-CAA CCA TTG GGT CAG CTC TTG-3′; and probe 5′-FAM-GAA TCA GCT CTG TGG ACC TCT-3′-TAMRA. All reactions followed the typical sigmoidal reaction profile, and cycle threshold was used as the measurement of amplicon abundance.
Chromatin immunoprecipitation
Following treatment, cells were fixed to isolate intact chromatin. Acetyl-histone H3, acetyl-histone H4, and PPARγ were immunoprecipitated from intact chromatin, using Dynabead protein A (Invitrogen, Carlsbad, CA) and, respectively, anti-PPARγ (sc-7196; Santa Cruz Biotechnology), anti-acetyl-histone H3 (Lys9), or anti-acetyl-histone H4 (Lys8) (no. 9649 or no. 2594; Cell Signaling) antibodies. Normal rabbit IgG (product code 2729; Cell Signaling) was used as negative control, and 1% Input (a PCR product of 0.01 the total amount of isolated DNA used in the chromatin immunoprecipitation [ChIP] assay) was used as positive control, respectively. Primer sequences used for amplification of the PPRE flanking region were forward primer 5′-ACA CAC ACA CAC ACA CAC ACA CAC ACC-3′ and reverse primer 5′-CCA AGT GAA CCA TTG CTC CAA TCC CTG-3′ (nucleotides −1225 to −956, 270 base pairs [bp]). The primer sequences used for the negative PCR control were forward primer 5′-CCC TAT ATC TGG GGT GAT GGA AGA TCC-3′ and reverse primer 5′-AGG CAG GGG CTC TAC CAC TGA GCC ACA-3′ (nucleotides −2249 to −1959, 291 bp).
Coimmunoprecipitation
Following cell treatment, nuclear extracts were prepared and immunoprecipitated with anti-acetyl-histone H3 (Lys9) or anti-acetyl-histone H4 (Lys8) antibody, using Dynabead protein A (Invitrogen, Carlsbad, CA). The coimmunoprecipitation (CoIP) products were then resolved by SDS-PAGE and probed with PPARγ antibody.
Knockdown of PPARγ by RNA interference
Differentiated 3T3-L1 adipocytes were transfected with a pool of three small interfering RNAs (siRNAs) for PPARγ (sc-29456; Santa Cruz Biotechnology), using Lipofectamine 2000 transfection reagent, and incubated for 72 h. The reduction in PPARγ expression level was determined by Western blot hybridization using antibody against PPARγ2 (sc-22020; Santa Cruz Biotechnology).
Statistical analysis
Data are expressed as means ± standard errors of the mean, and the number of individual experiments is presented in the figure legend. Significance was tested using ANOVA with a Newman-Keuls post hoc test (P < 0.05) or a Student t test, as indicated in figure legends.
RESULTS
Insulin and GIP act synergistically to promote 3T3-L1 cell differentiation
The effects of various concentrations of GIP and insulin on preadipocyte differentiation were determined in 3T3-L1 cells treated for 7 days in the presence or absence of Dex and IBMX, as shown in Fig. 1A. Accumulation of neutral lipid, including triglyceride, during development of the adipocyte phenotype was determined by Oil Red O staining (Fig. 1B, C). Dex and IBMX were essential for differentiation, although insufficient for its initiation. Seven days’ treatment with GIP alone (1–100 nM) did not increase neutral lipid accumulation (Fig. 1C, conditions 11–13), whereas insulin alone (10–100 nM) increased lipid levels (Fig. 1B, C, conditions 10 and 14–16). However, although 1 nM insulin alone had no effect on neutral lipid accumulation (Fig. 1C, treatment 14), the addition of 100 nM GIP significantly promoted accumulation (Fig. 1B, condition 5). Similarly, GIP at concentrations of 10 or 100 nM potentiated insulin-stimulated lipid accumulation in a graded fashion (Fig. 1B). A combination of 100 nM GIP plus 100 nM insulin produced levels of lipid accumulation comparable to those obtained with the high concentration of insulin (16 μM) routinely used for 3T3-L1 cell differentiation (Fig. 1B). Visualization of intracellular lipid by using fluorophore BODIPY 493/503 dye produced qualitative results similar to that using Oil Red O staining, with a graded increase in the presence of increasing concentrations of insulin plus GIP (see supplementary Fig. I). Taken together, these results demonstrated that insulin alone stimulated 3T3-L1 cell differentiation, but GIP acted synergistically to potentiate responses.
Levels of GIPR, SREBP-1, PPARγ2, and histone H3/H4 acetylation increase during 3T3-L1 cell differentiation
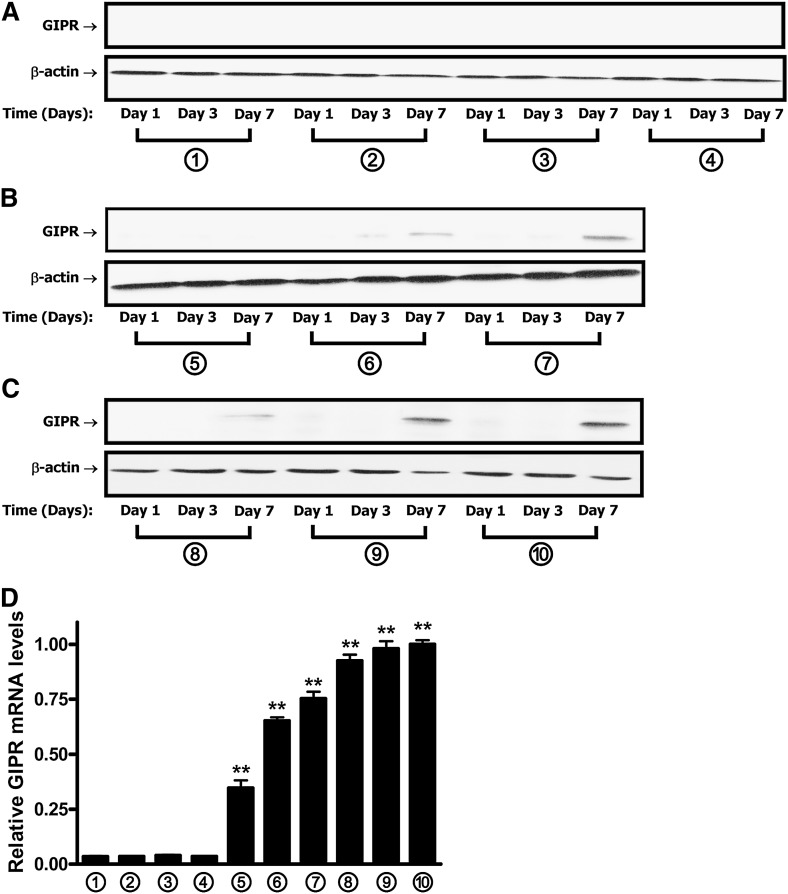
We next examined GIPR expression during the course of adipogenesis in response to the treatment with GIP and insulin. After 7 days’ treatment with 1 nM insulin and 10 nM GIP, GIPR mRNA was undetectable. However, with a 10-fold higher GIP concentration and 1 nM insulin, a large increase in GIPR mRNA levels was detected (see Fig. 2D, treatments 4 and 5). Increasing insulin levels to 10 nM in the presence of 10 or 100 nM GIP produced an approximate doubling of expression levels. GIPR protein expression was detected in medium containing a minimum of only 10 nM insulin (Fig. 2A–C), probably due to the lower sensitivity of protein staining. Overall, GIPR mRNA levels reflected the degree of adipocyte differentiation demonstrated with neutral lipid staining (compare Figs. 2D and 1B).
Fig. 2.
GIPR expression increases during adipocyte differentiation in 3T3-L1 cells. Protocols for treatment of 3T3-L1 preadipocytes with GIP are as described in the legend to Fig. 1A. A–C: The time course of GIPR protein expression during 3T3-L1 adipocyte differentiation is shown. Western blot analyses were performed using antibodies against GIPR and β-actin. D: GIPR mRNA expression during 3T3-L1 adipocyte differentiation is shown. 3T3-L1 preadipocytes were treated for 7 days, as described in the legend to Fig. 1A, and real-time RT-PCR was performed with extracts to quantify GIPR mRNA levels, shown as fold difference versus control normalized to 18S rRNA expression levels. All data represent three independent experiments, each carried out in duplicate, and Western blots are representative of n = 3 replicates. Significance was tested using ANOVA with Newman-Keuls post hoc test, where ** represents P <0.05 versus control group, treated with conditional culture medium 1 (no IBMX/Dex/insulin/GIP additions) for 7 days.
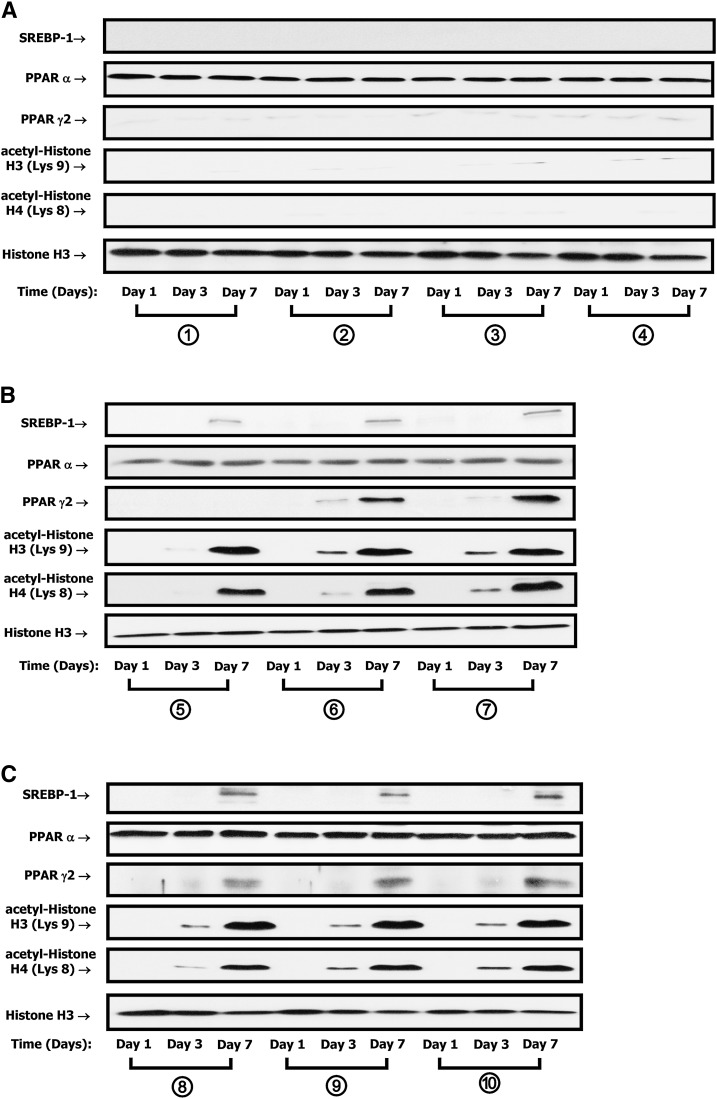
To study potential molecular mechanisms involved in the upregulation of GIPR mRNA during differentiation of 3T3-L1 cells, we focused on transcription factors known to be upregulated during adipogenesis. Among those factors that have been shown to play central roles in the regulation of fat cell development are adipocyte determination differentiation factor 1/sterol response element binding protein 1c (ADD1/SREBP-1c) and the nuclear receptor PPARγ2. A functional PPRE (5′-GGGTACCTCCAGT-3′) was recently identified in the rat GIPR gene (33), and both PPARα (34, 35) and PPARγ (33) have been implicated in the regulation of GIPR mRNA expression in pancreatic β-cells. 3T3-L1 preadipocytes were therefore treated as described in the legend to Fig. 1A, and Western blot analyses of nuclear extracts were performed using antibodies against SREBP-1, PPARα, or PPARγ2. PPARα was detectable in undifferentiated 3T3-L1 cells, but levels did not change further, either during differentiation or with increasing peptide concentrations (Fig. 3A–E). In contrast, nuclear localization of both SREBP-1 and PPARγ2 were strongly upregulated during adipocyte differentiation. However, low concentrations of insulin (1 nM) were incapable of increasing SREBP-1 expression without the synergistic action of GIP (Fig. 3–E).
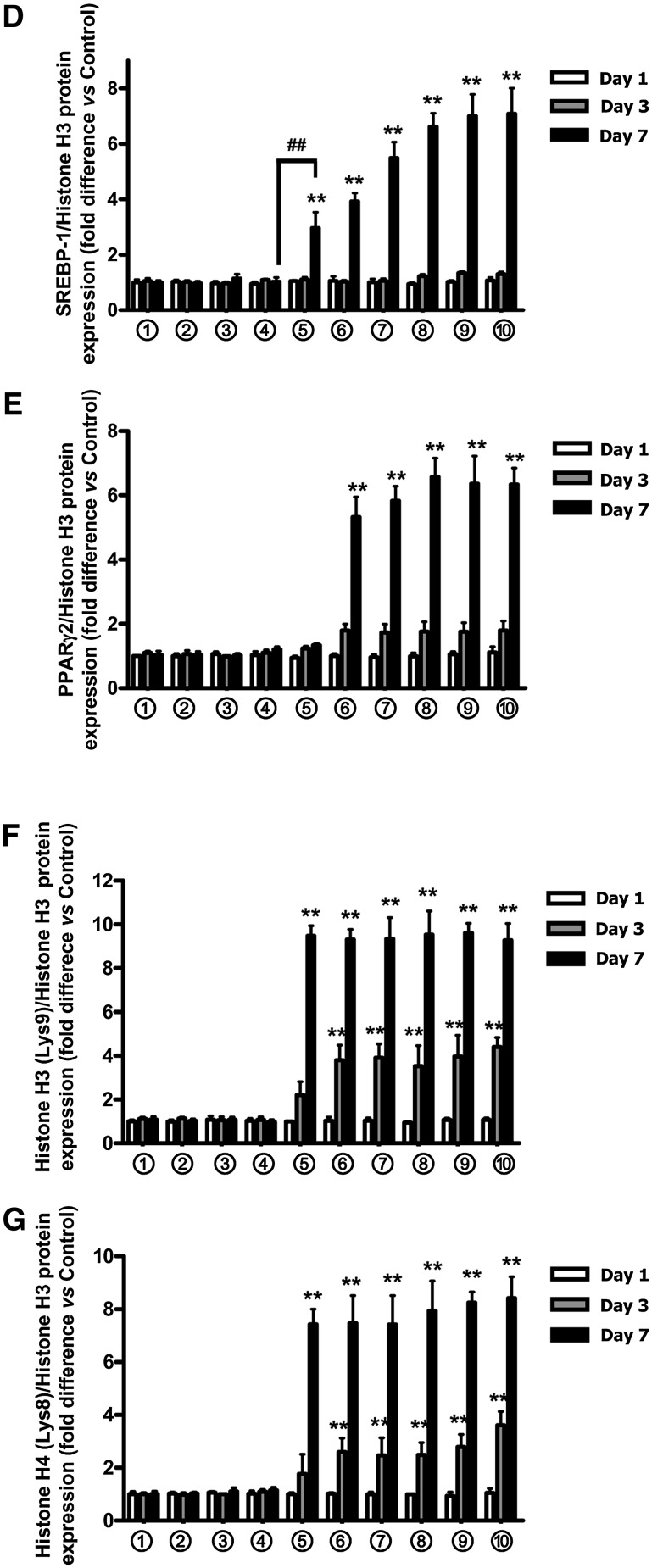
Fig. 3.
SREBP-1 and PPARγ expression and acetylation of histones H3/H4 increase during 3T3-L1 adipocyte differentiation. A–C: The time course of expression of SREBP-1, PPARα, and PPARγ and acetylation of histones H3/H4 during 3T3-L1 adipocyte differentiation is shown. 3T3-L1 preadipocytes were treated as described in the legend to Fig. 1A; nuclear extracts were isolated, and Western blot analyses were performed using antibodies against SREBP-1, PPARα, PPARγ, acetyl-histone H3 (Lys9), acetyl-histone H4 (Lys8), and histone H3. Representatives of n = 3 replicates are shown. D–G: Densitometric analyses are shown for SREBP-1 (D), PPARγ (E), acetyl-histone H3 (F), and acetyl-histone H4 (G). Western blots were quantified and normalized with the loading control, histone H3. Shown are fold differences versus control group, treated with medium 1 (no IBMX/Dex/insulin/GIP additions) for 1 day. Significance was tested using ANOVA with Newman-Keuls post hoc test, where ** represents P <0.05 versus control group, treated with conditional culture medium 1 (no IBMX/Dex/insulin/GIP additions) for 1 day; ## represents P <0.05 versus the indicated group.
The identification of chromatin histone proteins that regulate gene transcription by modulating accessibility of transcription factors to target genes and the marked changes in nuclear localization of both SREBP-1 and PPARγ2 observed in the current study prompted us to examine whether chromatin modifications were associated with GIP-and-insulin treatment. 3T3-L1 preadipocytes were therefore treated as described in the legend to Fig. 1A, and Western blot analyses were performed with nuclear extracts, using antibodies against acetyl-histones H3/H4. As shown in Fig. 3A–G, acetylation of histone H3 at lysine (Lys) 9 and histone H4 at Lys8 increased during treatment with GIP (10–100 nM) plus insulin (1–100 nM) or with insulin alone (16 µM). These results suggest that histone modifications were involved in the regulation of adipocyte GIPR gene transcription. In view of the lack of change in PPARα during 3T3-L1 cell adipogenesis and the strong evidence for PPARγ2 involvement in the regulation of β-cell GIPR expression (33), we focused further on its potential involvement in regulating adipocyte GIPR expression.
Adipocyte GIPR expression is regulated via a PPARgamma-mediated pathway
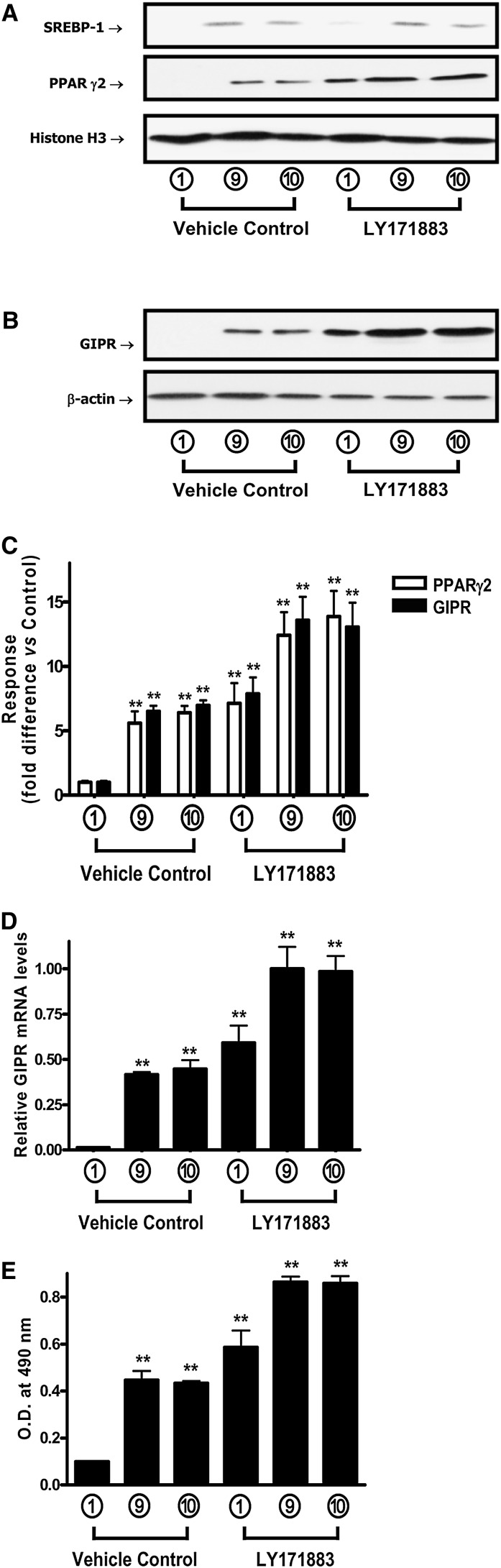
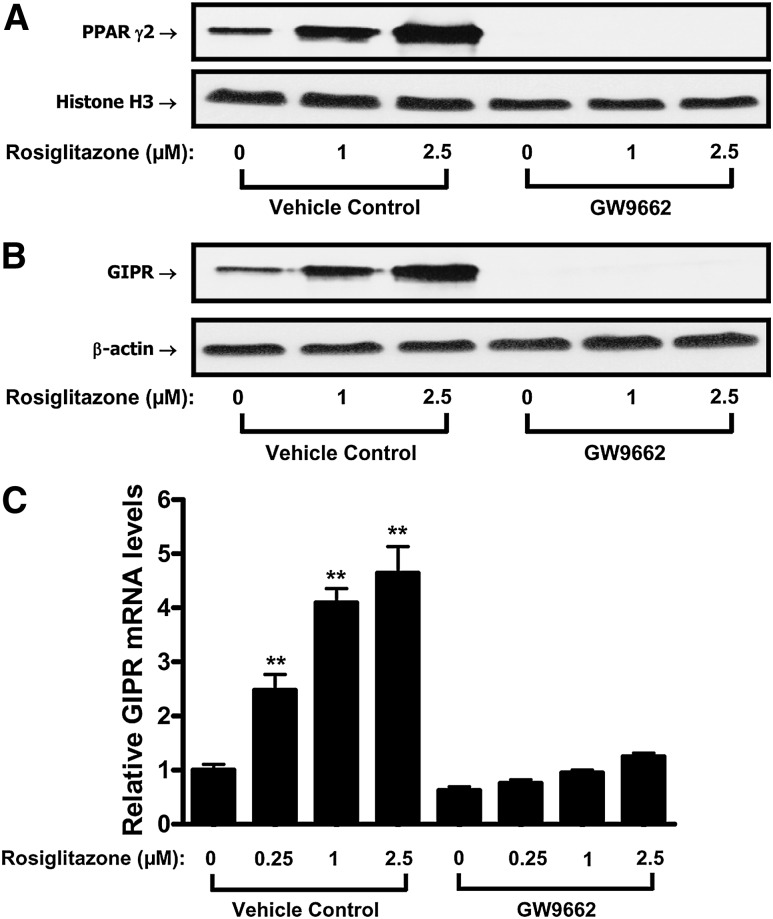
The functional involvement of PPARγ in the regulation of adipocyte GIPR expression was examined using PPARγ agonists and an antagonist. 3T3-L1 preadipocytes first were treated for 7 days in Condition 1 (i.e., no IBMX/Dex/insulin/GIP additions), 9 (100 nM insulin plus 100 nM GIP), or 10 (16 µM insulin) culture medium, as shown in Fig. 1A, in the presence or absence of the PPARγ agonist LY171883 (100 μM). This led to enhanced expression of GIPR protein (Fig. 4) (see Fig. 4B, C) and mRNA (Fig. 4D), as well as accumulation of PPARγ in the nucleus (Fig. 4A, C). LY171883 also potentiated accumulation of lipid (Fig. 4E), but it did not significantly alter nuclear SREBP-1 expression levels (Fig. 4A). In agreement with an earlier study (36), LY171883 caused adipocyte differentiation in the absence of DEX, IBMX, and insulin (Fig. 4E). Fully differentiated 3T3-L1 cells were additionally treated for 20 h with a second PPARγ agonist, rosiglitazone, and this treatment also resulted in marked increases in nuclear PPARγ2 localization (Fig. 5A), as well as GIPR mRNA levels (see Fig. 5C) and protein expression (Fig. 5B). Co-incubation with the PPARγ inhibitor GW9662 led to a complete ablation of detectable nuclear PPARγ2 and GIPR protein and mRNA levels (Fig. 5A–C). In contrast, under identical treatment conditions, PPARγ2 mRNA levels were only reduced by ∼25% compared to those in vehicle control (see supplementary Fig. II). Taken together, these results strongly suggest that adipocyte GIPR expression in 3T3-L1 cells is modulated through a pathway involving increased nuclear localization of PPARγ.
Fig. 4.
PPARγ activation increases GIPR expression during 3T3-L1 adipocyte differentiation. 3T3-L1 preadipocytes were treated for 7 days, as described in the legend to Fig. 1A, in the presence or absence of the PPARγ agonist, LY171883 (100 μM). A: Effects of a PPARγ agonist on nuclear SREBP-1, PPARα, and PPARγ levels are shown. Nuclear extracts were isolated, and Western blot analyses were performed using antibodies against SREBP-1, PPARγ, and histone H3. B: Effects of a PPARγ agonist on GIPR protein expression are shown. 3T3-L1 preadipocytes were treated as described above, and Western blot analyses were performed using antibodies against GIPR and β-actin. C: Densitometric analysis of PPARγ and GIPR levels are shown. Western blots were quantified using densitometric analysis and are representative of n = 3 replicates. D: Effects of a PPARγ agonist on GIPR mRNA expression are shown. 3T3-L1 preadipocytes were treated as described above, and real-time RT-PCR was performed to quantify GIPR mRNA levels, shown as the fold difference versus control normalized to 18S rRNA expression levels. E: Effects of a PPARγ agonist on lipogenesis in 3T3-L1 cells are shown. 3T3-L1 preadipocytes were treated as described above: cells were stained with Oil Red O, and the level of staining was determined spectrophotometically, as described in “Experimental Procedures.” All data represent three independent experiments, each carried out in duplicate, and Western blots are representative of n = 3 replicates. Significance was tested using ANOVA with Newman-Keuls post hoc test, where ** represents P <0.05 versus vehicle control with 0 μM LY17883, treated with conditional culture medium 1 (no IBMX/Dex/insulin/GIP additions) for 7 days.
Fig. 5.
PPARγ activation increases 3T3-L1 GIPR expression in differentiated adipocytes. 3T3-L1 preadipocytes were treated for 7 days with conditional culture medium 10 (see . Following differentiation, adipocytes were treated with the indicated concentrations of rosiglitazone (µM) for 20 h in the presence or absence of the inhibitor GW9662 (20 μM). A: Effects of PPARγ agonist and antagonist on nuclear PPARγ expression are shown. Nuclear extracts were isolated, and Western blot analyses were performed using antibodies against PPARγ and histone H3. B: Effects of PPARγ agonist and antagonist on adipocyte GIPR protein expression are shown. 3T3-L1 adipocytes were treated as described above, and Western blot analyses were performed using antibodies against GIPR and β-actin. C: Effects of PPARγ agonist and antagonist on adipocyte GIPR mRNA expression are shown. 3T3-L1 adipocytes were treated as described above, and real-time RT-PCR was performed to quantify GIPR mRNA levels, shown as the fold difference versus control normalized to 18S rRNA expression levels. All data represent three independent experiments, each carried out in duplicate, and Western blots are representative of n = 3 replicates. Significance was tested using ANOVA with Newman-Keuls post hoc test, where ** represents P <0.05 versus vehicle control with 0 μM rosiglitazone and 0 μM GW9662.
PPARγ and acetylated histones H3/H4 interact with the promoter region of the adipocyte GIPR
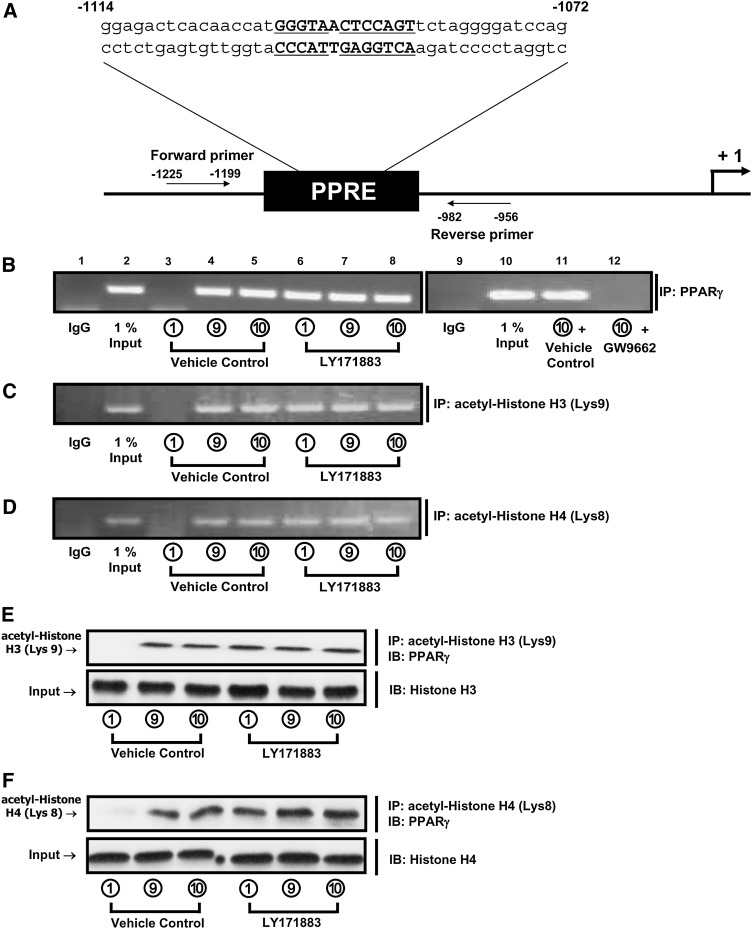
To establish the involvement of PPARγ in transactivation of the adipocyte GIPR, the presence of PPARγ binding to the PPRE in the promoter was next determined by measuring PPRE occupancy, using ChIP assays. Treatment with insulin (100 nM) plus GIP (100 nM) or insulin alone (16 µM) increased the association of both PPARγ (Fig. 6B) and acetylated histones H3/H4 (Fig. 6C, D) with the GIPR PPRE region in the promoter, and these responses were mimicked by treatment with the PPARγ agonist LY171883 (100 μM) (Fig. 6B–D). However, due to the apparent depletion of nuclear PPARγ (Fig. 5A), its interaction with the GIPR PPRE was undetectable following GW9662 treatment (Fig. 6B). Subsequent studies demonstrated close protein interactions between PPARγ and histone H3 acetylated at Lys9 or histone H4 acetylated at Lys8 (Fig. 6E, F). These results therefore strongly support the involvement of PPARγ and acetylated histones H3/H4 in transactivation of the adipocyte GIPR.
Fig. 6.
PPARγ binds to the GIPR promoter PPRE in a region associated with highly acetylated histones H3/H4. A: Identification of a putative PPRE sequence in the promoter of the mouse GIPR gene is shown. The PPRE location and sequence (boldface) and the flanking primers used for ChIP assay are presented. PPRE sequences conserved with the rat GIPR promoter (33) are underlined. B–D: A representative gel of a ChIP assay is shown for the binding of PPARγ (B), acetylated histone H3 (C), and acetylated histone H4 (D) in the GIPR promoter. 3T3-L1 preadipocytes were treated for 7 days as described in the legend to Fig. 1A in the presence or absence of LY171883 (100 μM). In a second set of experiments, 3T3-L1 preadipocytes were treated for 7 days with conditional culture medium 10 (see and, following differentiation, the adipocytes were treated for 20 h with GW9662 (20 μM). PPARγ, acetylated histone H3, or acetylated histone H4 was immunoprecipitated from intact chromatin isolated from 3T3-L1 cells, using, respectively, anti-PPARγ, anti-acetyl histone H3 (Lys9), and anti-acetyl histone H4 (Lys8) antibody. Precipitated DNA fragments were analyzed by PCR using primers flanking the PPRE site in the GIPR promoter. An isotype-matched IgG was used as negative control, and 1% Input (PCR product of 0.01 of the total isolated DNA used in the ChIP assay) was used as positive control. Results from a PPARγ ChIP assay using a negative PCR control amplification of a chromatin region >500 bp away from the PPRE is presented in supplementary Fig. III. E and F: CoIP between acetylated histones H3/H4 and PPARγ is shown. 3T3-L1 preadipocytes were treated for 7 days, as described in the legend to Fig 1A, in the presence or absence of LY171883 (100 μM). Nuclear extracts were isolated and immunoprecipitated with acetyl-histone H3 at Lys9 (E) or acetyl-histone H4 at Lys8 (F), followed by immunoblotting for PPARγ. Input represents 0.10 of the total nuclear extract used in the CoIP assay.
RNA interference-mediated knockdown of PPARγ reduces expression of the adipocyte GIPR
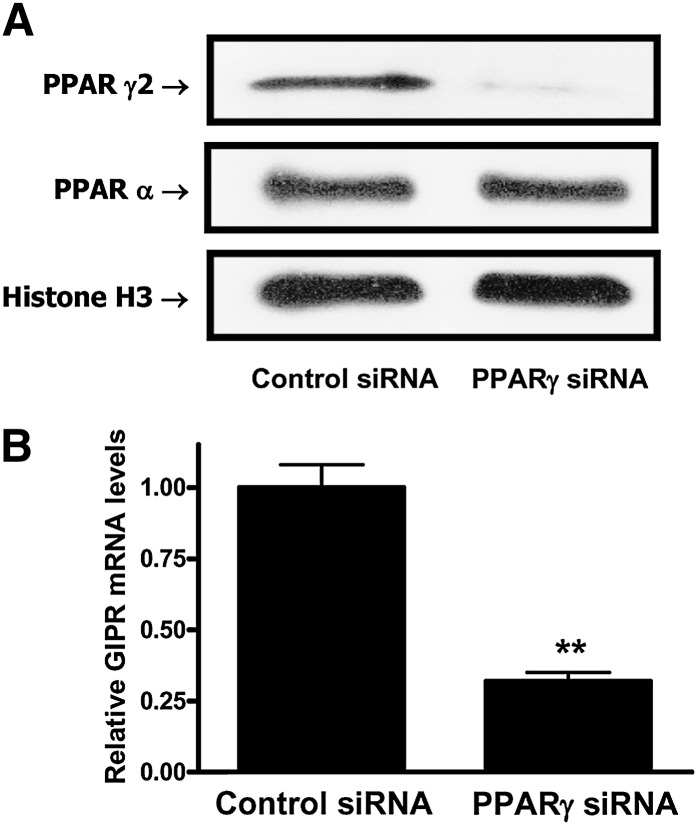
To establish the functional involvement of PPARγ in transactivation of the adipocyte GIPR, RNA interference (RNAi) was used. As shown in Fig. 7A, B, RNAi-mediated knockdown resulted in specific reductions in PPARγ2 protein expression, associated with greatly reduced GIPR mRNA levels. PPARγ is therefore likely to be a key regulator of adipocyte GIPR expression.
Fig. 7.
RNAi-mediated suppression of PPARγ reduces adipocyte GIPR expression. A: Effects of PPARγ siRNAs on PPARγ expression are shown. Differentiated 3T3-L1 adipocytes were transfected with a pool of three siRNAs for PPARγ or control scrambled siRNAs (100 nM) and incubated for 72 h. Nuclear extracts were isolated, and Western blot analyses were performed as described in “Experimental Procedures,” using antibodies against PPARγ2, PPARγ, and histone H3. B: Effects of PPARγ siRNAs on GIPR expression are shown. 3T3-L1 adipocytes were treated as described in the legend to panel A, and real-time RT-PCR was performed with extracts to quantify GIPR mRNA levels, shown as the fold difference versus control normalized to 18S rRNA expression levels. All data represent three independent experiments, each carried out in duplicate, and Western blots are representative of n = 3 replicates. Significance was tested using a Student t test, where ** represents P <0.05 versus control siRNA.
DISCUSSION
Although GIP plays an established role in the regulation of β-cell function (4), its role in regulating the adipocyte has only recently been highlighted. In the current study, we focused on two main questions: does GIP potentiate insulin-stimulated differentiation of 3T3-L1 preadipocytes, a well-characterized mouse cell line model of adipogenesis, and what factors are involved in the regulation of adipocyte GIPR expression?
Adipogenesis is a complex process with multiple steps, including growth arrest, clonal expansion, withdrawal from the cell cycle, and terminal differentiation. IGF-1 has been considered to play a dominant role in the initiation of preadipocyte differentiation (37, 38), but recent studies have emphasized the critical contribution made by insulin (39–41). Both IGF-I and insulin receptor are expressed early in the differentiation of human preadipocytes (42). Insulin receptor knockout mice display underdeveloped adipose tissue (41), and mouse embryonic fibroblasts derived from these mutants fail to differentiate into adipocytes (40). The crucial importance of insulin/IGF-I signaling in the adipocyte developmental program is highlighted by its failure in mice bearing a double-knockout of insulin receptor substrate-1 (IRS-1) and IRS-2 (41). In the current study, as expected, insulin strongly stimulated 3T3-L1 cell differentiation in a concentration-dependent manner. However, low concentrations of insulin (1 nM) were incapable of increasing neutral lipid accumulation without the synergistic action of GIP (Fig. 1). In agreement with earlier studies of human (21) and 3T3-L1 (20) preadipocytes, GIPR expression was found to be extremely low in the preadipocytes, and this probably accounts for the relatively high concentration (10–100 nM) required for potentiation of insulin effects. However, both GIPR mRNA and protein levels increased greatly during insulin-stimulated differentiation (Fig. 2), suggesting that insulin played a key role in regulating GIPR expression. Since GIP potentiated insulin-stimulated differentiation, it is difficult to establish whether its synergistic effects on GIPR mRNA levels were direct effects of GIP on gene transcription or occurred secondarily to the progression of differentiation. Additionally, as both insulin and IGF-1 receptors, as well as insulin/IGF-I hybrid receptors, are expressed in 3T3-L1cells during differentiation (43), multiple signaling pathways could be involved in regulating GIPR expression during development of the adipocyte phenotype.
A number of key transcription factors for adipocyte differentiation have been identified, among which PPARγ and members of the CCAAT/enhancer binding protein (C/EBP) and ADD1/ SREBP-1 families play particularly important roles. Activation of these key regulators during adipogenesis results in increased expression of adipocyte-specific target genes, including fatty-acid-binding protein aP2, whose expression is increased by GIP (20). We chose to examine the possible involvement of PPARγ and PPARα in insulin-stimulated GIPR expression because there was evidence that these transcription factors played such a role in β-cells (33–35, 44). Additionally, SREBP-1 was studied because insulin is an established regulator of SREBP-1c, increasing its adipocyte expression through a PI3K-mediated pathway (45) and modulating its processing (46). In turn, SREBP-1c has been shown to induce the production of endogenous ligands for PPARγ and regulate its activity (47). PPARα was detectable in undifferentiated 3T3-L1 cells, but levels did not correlate with the progression of differentiation or changes in GIPR expression. In contrast, both SREBP-1 and PPARγ nuclear protein levels were increased by insulin-plus-GIP treatment (Fig. 3) in a concentration-dependent manner by day 7 of 3T3-L1 cell differentiation, and these changes were associated with increased GIPR expression. Findings from studies that used selective pharmacological activators of PPARγ (Figs. 4 and 5) supported a functional involvement of PPARγ in the regulation of the GIPR gene expression. The PPARγ inhibitor GW9662 completely ablated rosiglitazone-induced increases in GIPR protein and nuclear PPARγ and profoundly reduced GIPR mRNA levels (Fig. 5A, B, and C). This was surprising but supported a central role for the transcription factor in the regulation of GIPR expression. Interaction between PPARγ and the GIPR PPRE (Fig. 6B) was also undetectable following GW9662 treatment (Fig. 6B), presumably due to the loss of protein. Blockade of PPARγ expression may partially explain its loss, as identical GW9662 treatment resulted in a reduction of ∼25% in PPARγ2 mRNA expression levels (see supplementary Fig. II). However, because GW9662 is an irreversible inhibitor of PPARγ (48), it is also possible that PPARγ/inhibitor binding results in proteasomal degradation. Alternatively, blockade of nuclear/cytoplasmic shuttling could be involved.
Chromatin core histone proteins can exert a regulatory effect on gene transcription by modulating accessibility of transcription factors to target genes. Histone H3 has been shown to be acetylated at Lys residues 9, 14, 18, and 23, whereas histone H4 is acetylated at Lys residues 5, 8, 12, and 16. In previous studies, histone acetylation was shown to be increased during differentiation of 3T3-L1 cells, and inhibition of histone deacetylase activity stimulated adipocyte differentiation (49–51). Additionally, both incretin hormones GIP and GLP-1 regulated β-cell chromatin structure by modulating histone acetylation (52). In the present study, acetylation of histone H3 at Lys9 and histone H4 at Lys8 increased during differentiation of 3T3-L1 cells with the treatment of GIP (10–00 nM) plus insulin (1–100 nM) or insulin alone (16 µM) (Fig. 3A–G), and PPARγ was shown to interact with the PPRE of the GIPR promoter along with acetylated histones H3/H4 (Fig. 6). This complex interplay between PPARγ binding to the PPRE of the GIPR promoter and core histone proteins appears to play an important role in the regulation of adipocyte GIPR expression. A functional link between PPARγ binding and adipocyte GIPR expression was further supported by the greatly reduced GIPR expression observed following RNAi-mediated knockdown of PPARγ (Fig. 7).
In an earlier study of islet β-cells, it was shown that palmitate and the PPARα agonist WY14643 stimulated GIPR gene transcription, and gene expression was reduced with a dominant negative form of PPARα (34). More recently, PPARα was shown to be essential for the upregulating effects of metformin on β-cell incretin receptors (53). In contrast, Gupta and co-workers (33) provided compelling evidence for PPARγ playing an important role in the regulation of β-cell GIPR expression. Both of the members of the PPAR family promote expression through binding to PPREs and forming heterodimers with members of the retinoid X receptor family of steroid receptors (54). From the 3T3-L1 cell studies, evidence was obtained for PPARγ but not PPARα involvement in regulation of the adipocyte GIPR gene expression, and further studies are clearly needed to establish whether both transcription factors are capable of regulating expression in the β-cell.
In conclusion, GIP and insulin act in a synergistic manner on 3T3-L1 cell differentiation, and during this process, adipocyte GIPR expression is upregulated through interactions between PPARγ and the PPRE in the GIPR promoter containing highly acetylated histones. In view of the widespread application of long-acting incretins and DPP-IV inhibitors for type 2 diabetes therapy and the demonstration of tissue-selective actions of GIP analogs (9), it is important to establish whether expression of the GIPR demonstrates tissue-selective regulation and whether changes occur in humans with type 2 diabetes.
Supplementary Material
Footnotes
Abbreviations:
- ADD1/SREBP-1c
- adipocyte determination differentiation factor 1/sterol response element binding protein 1c
- BODIPY
- boron-dipyrromethene
- ChIP
- chromatin immunoprecipitation
- CoIP
- coimmunoprecipitation
- Dex
- dexamethasone
- DPP-IV
- dipeptidyl peptidase-IV
- GIP
- glucose-dependent insulinotropic polypeptide
- GIPR
- GIP receptor
- PPARγ
- peroxisome proliferator-activated receptor γ
- GLP-1
- glucagon-like peptide-1
- IBMX
- 3-isobutyl-1-methylxanthine
- IGF-1
- insulin-like growth factor-1
- PPRE
- peroxisome proliferator response element
- siRNA
- small interfering RNA
These studies were generously supported by funding from the Canadian Diabetes Association and the Canadian Institutes for Health Research (to C.H.S.M.).
References
- 1.Brubaker P. L., Drucker D. J. 2004. Minireview: glucagon-like peptides regulate cell proliferation and apoptosis in the pancreas, gut, and central nervous system. Endocrinology. 145: 2653–2659. [DOI] [PubMed] [Google Scholar]
- 2.Drucker D. J. 2007. The role of gut hormones in glucose homeostasis. J. Clin. Invest. 117: 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yusta B., Baggio L., Estall J. L., Koehler J. A., Holland D. P., Li H., Pipeleers D., Ling Z., Drucker D. J. 2006. GLP-1 receptor activation improves beta cell function and survival following induction of endoplasmic reticulum stress. Cell Metab. 4: 391–406. [DOI] [PubMed] [Google Scholar]
- 4.McIntosh C. H. S., Widenmaier S., Kim S. J. 2009. Glucose-dependent insulinotropic polypeptide (gastric inhibitory polypeptide; GIP). Vitam. Horm. 80: 409–471. [DOI] [PubMed] [Google Scholar]
- 5.Ehses J. A., Casilla V. R., Doty T., Pospisilik J. A., Winter K. D., Demuth H. U., Pederson R. A., McIntosh C. H. S. 2003. Glucose-dependent insulinotropic polypeptide promotes beta-(INS-1) cell survival via cyclic adenosine monophosphate-mediated caspase-3 inhibition and regulation of p38 mitogen-activated protein kinase. Endocrinology. 144: 4433–4445. [DOI] [PubMed] [Google Scholar]
- 6.Kim S. J., Winter K., Nian C., Tsuneoka M., Koda Y., McIntosh C. H. S. 2005. Glucose-dependent insulinotropic polypeptide (GIP) stimulation of pancreatic beta-cell survival is dependent upon phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB) signaling, inactivation of the forkhead transcription factor Foxo1, and downregulation of bax expression. J. Biol. Chem. 280: 22297–22307. [DOI] [PubMed] [Google Scholar]
- 7.Kim S. J., Nian C., Widenmaier S., McIntosh C. H. S. 2008. Glucose-dependent Insulinotropic Polypeptide (GIP) mediated upregulation of β-cell anti-apoptotic Bcl-2 gene expression is coordinated by cAMP-response element binding protein (CREB) and cAMP-responsive CREB coactivator 2 (TORC2). Mol. Cell. Biol. 28: 1644–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widenmaier S. B., Ao Z., Kim S. J., Warnock G., McIntosh C. H. S. 2009. Suppression of p38 MAPK and JNK via Akt-mediated inhibition of Apoptosis Signal regulating Kinase 1 constitutes a core component of the beta-cell pro-survival effects of glucose-dependent insulinotropic polypeptide. J. Biol. Chem. 284: 30372–30382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Widenmaier S. B., Kim S. J., Yang G. K., De Los Reyes T., Nian C., Asadi A., Seino Y., Kieffer T. J., Kwok Y. N., McIntosh C. H. S. 2010. A GIP receptor agonist exhibits β-cell anti-apoptotic actions in rat models of diabetes resulting in improved β-cell function and glycemic control. PLoS ONE. 5: e9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drucker D. J., Nauck M. A. 2006. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 368: 1696–1705. [DOI] [PubMed] [Google Scholar]
- 11.Davidson M. B., Bate G., Kirkpatrick P. 2005. Exenatide. Nat. Rev. Drug Discov. 4: 713–714. [DOI] [PubMed] [Google Scholar]
- 12.Harder H., Nielsen L., Tu D. T. T., Astrup A. 2004. The effect of liraglutide, a long-acting glucagon-like peptide 1 derivative, on glycemic control, body composition, and 24-h energy expenditure in patients with type 2 diabetes. Diabetes Care. 27: 1915–1921. [DOI] [PubMed] [Google Scholar]
- 13.DeFronzo R. A., Ratner R. E., Han J., Kim D. D., Fineman M. S., Baron A. D. 2005. Effects of exenatide (exendin-4) on glycemic control and weight over 30 weeks in metformin-treated patients with type 2 diabetes. Diabetes Care. 28: 1092–1100. [DOI] [PubMed] [Google Scholar]
- 14.Deacon C. F. 2007. Dipeptidyl peptidase 4 inhibition with sitagliptin: a new therapy for type 2 diabetes. Expert Opin. Investig. Drugs. 16: 533–545. [DOI] [PubMed] [Google Scholar]
- 15.Rosenstock J., Baron M. A., Dejager S., Mills D., Schweizer A. 2007. Comparison of vildagliptin and rosiglitazone monotherapy in patients with type 2 diabetes: a 24-week, double-blind, randomized trial. Diabetes Care. 30: 217–223. [DOI] [PubMed] [Google Scholar]
- 16.McIntosh C. H. 2008. Dipeptidyl peptidase IV inhibitors and diabetes therapy. Front. Biosci. 13: 1634–1645. [DOI] [PubMed] [Google Scholar]
- 17.Miyawaki K., Yamada Y., Ban N., Ihara Y., Tsukiyama K., Zhou H., Fujimoto S., Oku A., Tsuda K., Toyokuni S., et al. 2002. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat. Med. 8: 738–742. [DOI] [PubMed] [Google Scholar]
- 18.Kieffer T. J. 2003. GIP or not GIP? That is the question. Trends Pharmacol. Sci. 24: 110–112. [DOI] [PubMed] [Google Scholar]
- 19.Gault V. A., O'Harte F. P., Flatt P. R. 2003. Glucose-dependent insulinotropic polypeptide (GIP): antidiabetic and anti-obesity potential? Neuropeptides. 37: 253–263. [DOI] [PubMed] [Google Scholar]
- 20.Song D. H., Getty-Kaushik L., Tseng E., Simon J., Corkey B. E., Wolfe M. M. 2007. Glucose-dependent insulinotropic polypeptide enhances adipocyte development and glucose uptake in part through Akt activation. Gastroenterology. 133: 1796–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weaver R. E., Donnelly D., Wabitsch M., Grant P. J., Balmforth A. J. 2008. Functional expression of glucose-dependent insulinotropic polypeptide receptors is coupled to differentiation in a human adipocyte model. Int. J. Obes. (Lond). 32: 1705–1711. [DOI] [PubMed] [Google Scholar]
- 22.Prins J. B., O'Rahilly S. 1997. Regulation of adipose cell number in man. Clin. Sci. 92: 3–11. [DOI] [PubMed] [Google Scholar]
- 23.Rosen E. D., Spiegelman B. M. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16: 145–171. [DOI] [PubMed] [Google Scholar]
- 24.Gregoire F. M., Smas C. M., Sul H. S. 1998. Understanding adipocyte differentiation. Physiol. Rev. 78: 783–809. [DOI] [PubMed] [Google Scholar]
- 25.Sethi J. K., Vidal-Puig A. J. 2007. Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J. Lipid Res. 48: 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McIntosh C. H. S., Bremsak I., Lynn F. C., Gill R., Hinke S. A., Gelling R., Nian C., McKnight G., Jaspers S., Pederson R. A. 1999. Glucose-dependent insulinotropic polypeptide stimulation of lipolysis in differentiated 3T3–L1 cells: wortmannin-sensitive inhibition by insulin. Endocrinology. 140: 398–404. [DOI] [PubMed] [Google Scholar]
- 27.Kim S. J., Nian C., McIntosh C. H. S. 2007. Activation of lipoprotein lipase by glucose-dependent insulinotropic polypeptide in adipocytes. A role for a protein kinase B, LKB1, and AMP-activated protein kinase cascade. J. Biol. Chem. 282: 8557–8567. [DOI] [PubMed] [Google Scholar]
- 28.Kim S. J., Nian C., McIntosh C. H. S. 2007. Resistin is a key mediator of glucose-dependent insulinotropic polypeptide (GIP) stimulation of lipoprotein lipase (LPL) activity in adipocytes. J. Biol. Chem. 282: 34139–34147. [DOI] [PubMed] [Google Scholar]
- 29.Kim S. J., Nian C., McIntosh C. H. 2010. GIP increases human adipocyte LPL expression through CREB and TORC2-mediated trans-activation of the LPL gene. J. Lipid Res. 51: 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez-Zacarias J. L., Castro-Mufiozledo F., Kuri-Harcuch W. 1992. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry. 97: 493–497. [DOI] [PubMed] [Google Scholar]
- 31.Rudolf M., Curcio C. A. 2009. Esterified cholesterol is highly localized to Bruch's membrane, as revealed by lipid histochemistry in wholemounts of human choroid. J. Histochem. Cytochem. 57: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber E., Matthias P., Muller M. M., Schaffner W. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’ prepared from a small number of cells. Nucleic Acids Res. 17: 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta D., Peshavaria M., Monga N., Jetton T. L., Leahy J. L. 2010. Physiologic and pharmacologic modulation of GIP receptor expression in beta-cells by PPARγ signaling: possible mechanism for the GIP resistance in Type 2 diabetes. Diabetes. 59: 1445–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynn F. C., Thompson S. A., Pospisilik J. A., Ehses J. A., Hinke S. A., Pamir N., McIntosh C. H. S., Pederson R. A. 2003. A novel pathway for regulation of glucose-dependent insulinotropic polypeptide (GIP) receptor expression in beta cells. FASEB J. 17: 91–93.. [DOI] [PubMed] [Google Scholar]
- 35.Lynn F. C., Pamir N., Ng E. H., McIntosh C. H. S., Kieffer T. J., Pederson R. A. 2001. Defective glucose-dependent insulinotropic polypeptide receptor expression in diabetic fatty Zucker rats. Diabetes. 50: 1004–1011. [DOI] [PubMed] [Google Scholar]
- 36.Chawla A., Lazar M. A. 1994. Peroxisome proliferator and retinoid signaling pathways co-regulate preadipocyte phenotype and survival. Proc. Natl. Acad. Sci. U S A. 91: 1786–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith P. J., Wise L. S., Berkowitz R., Wan C., Rubin C. S. 1988. Insulin-like growth factor-I is an essential regulator of the differentiation of 3T3–L1 adipocytes. J. Biol. Chem. 263: 9402–9408. [PubMed] [Google Scholar]
- 38.Benito M., Valverde A., Lorenzo M. 1996. IGF-I: a mitogen also involved in differentiation processes in mammalian cells. Int. J. Biochem. Cell Biol. 28: 499–510. [DOI] [PubMed] [Google Scholar]
- 39.Cinti S., Eberbach S., Castellucci M., Accili D. 1998. Lack of insulin receptors affects the formation of white adipose tissue in mice. A morphometric and ultrastructural analysis. Diabetologia. 41: 171–177. [DOI] [PubMed] [Google Scholar]
- 40.Nakae J., Kitamura T., Kitamura Y., Biggs W. H., Arden K., Accili D. 2003. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 4: 119–129. [DOI] [PubMed] [Google Scholar]
- 41.Miki H., Yamauchi T., Suzuki R., Komeda K., Tsuchida A., Kubota N., Terauchi Y., Kamon J., Kaburagi Y., Matsui J., et al. 2001. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol. Cell. Biol. 21: 2521–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bäck K., Arnqvist H. J., Bäck K., Arnqvist H. J. 2009. Changes in insulin and IGF-I receptor expression during differentiation of human preadipocytes. Growth Horm. IGF Res. 19: 101–111. [DOI] [PubMed] [Google Scholar]
- 43.Modan-Moses D., Janicot M., McLenithan J. C., Lane M. D., Casella S. J. 1998. Expression and function of insulin/insulin-like growth factor I hybrid receptors during differentiation of 3T3–L1 preadipocytes. Biochem. J. 333: 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu G., Kaneto H., Laybutt D. R., Duvivier-Kali V. F., Trivedi N., Suzuma K., King G. L., Weir G. C., Bonner-Weir S. 2007. Downregulation of GLP-1 and GIP receptor expression by hyperglycemia: possible contribution to impaired incretin effects in diabetes. Diabetes. 56: 1551–1558. [DOI] [PubMed] [Google Scholar]
- 45.Nadeau K. J., Leitner J. W., Gurerich I., Draznin B. 2004. Insulin regulation of sterol regulatory element-binding protein-1 expression in L-6 muscle cells and 3T3 L1 adipocytes. J. Biol. Chem. 279: 34380–34387. [DOI] [PubMed] [Google Scholar]
- 46.Yellaturu C. R., Deng X., Park E. A., Raghow R., Elam M. B. 2009. Insulin enhances the biogenesis of nuclear sterol regulatory element-binding protein (SREBP)-1c by posttranscriptional downregulation of Insig-2A and its dissociation from SREBP cleavage-activating protein (SCAP).SREBP-1c complex. J. Biol. Chem. 284: 31726–31734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J. B., Wright H. M., Wright M., Spiegelman B. M. 1998. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. U S A. 95: 4333–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leesnitzer L. M., Parks D. J., Bledsoe R. K., Cobb J. E., Collins J. L., Consler T. G., Davis R. G., Hull-Ryde E. A., Lenhard J. M., Patel L., et al. 2002. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry. 41: 6640–6650. [DOI] [PubMed] [Google Scholar]
- 49.Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 324: 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo E. J., Chung J. J., Choe S. S., Kim K. H., Kim J. B. 2006. Downregulation of histone deacetylases stimulates adipocyte differentiation. J. Biol. Chem. 281: 6608–6615. [DOI] [PubMed] [Google Scholar]
- 51.Steger D. J., Grant G. R., Schupp M., Tomaru T., Lefterova M. I., Schug J., Manduchi E., Stoeckert C. J., Jr., Lazar M. A. 2010. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 24: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S. J., Nian C., McIntosh C. H. S. 2009. Glucose-dependent insulinotropic polypeptide and glucagon-like peptide-1 modulate beta-cell chromatin structure. J. Biol. Chem. 284: 12896–12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maida A., Lamont B. J., Cao X., Drucker D. J. 2011. Metformin regulates the incretin receptor axis via a pathway dependent on peroxisome proliferator-activated receptor-α in mice. Diabetologia. 54: 339–349. [DOI] [PubMed] [Google Scholar]
- 54.Kota B. P., Huang T. H., Roufogalis B. D. 2005. An overview on biological mechanisms of PPARs. Pharmacol. Res. 51: 85–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.