SUMMARY
The importance of Methylthioadenosine/S-adenosylhomocysteine (MTA/SAH) nucleosidase in bacteria has started to be appreciated only in the past decade. A comprehensive analysis of its various roles here demonstrates that it is an integral component of the activated methyl cycle, which recycles adenine and methionine through S-adenosylmethionine (SAM)-mediated methylation reactions, and also produces the universal quorum-sensing signal, autoinducer-2 (AI-2). SAM is also essential for synthesis of polyamines, N-acylhomoserine lactone (autoinducer-1), and production of vitamins and other biomolecules formed by SAM radical reactions. MTA, SAH and 5′-deoxyadenosine (5′dADO) are product inhibitors of these reactions, and are substrates of MTA/SAH nucleosidase, underscoring its importance in a wide array of metabolic reactions. Inhibition of this enzyme by certain substrate analogs also limits synthesis of autoinducers, and hence, causes reduction in biofilm formation and may attenuate virulence. Interestingly, the inhibitors of MTA/SAH nucleosidase are very effective against the Lyme disease causing spirochete, Borrelia burgdorferi, which uniquely expresses three homologous functional enzymes. These results indicate that inhibition of this enzyme can affect growth of different bacteria by affecting different mechanisms. Therefore, new inhibitors are currently being explored for development of potential novel broad-spectrum antimicrobials.
Keywords: Methylthioadenosine/S-adenosylhomocysteine nucleosidases, Activated methyl cycle, MTA, SAH (AdoHcy), S-adenosylmethionine, SAM (AdoMet), autoinducer, 5′ deoxyadenosine (5′dADO)
INTRODUCTION
S-adenosylmethionine (SAM) is an important nucleoside that serves as an activated group donor in a broad array of metabolic and biosynthetic reactions, including methylations, propylamine group transfer in polyamine synthesis, and SAM radical-mediated vitamin synthesis. The majority of cellular SAM is used in methylation of macromolecules of prokaryotes and eukaryotes, yielding S-adenosylhomocysteine (SAH) as a product. SAM involvement in synthesis of polyamine and acylhomoserine lactones results in the production of 5′-methylthioadenosine (MTA). In vitro studies have shown that thionucleoside excess inhibits mammalian and bacterial methyltransferase and polyamine synthase activities. Therefore, inhibition of MTA/SAH nucleosidase activity is predicted to cause an accumulation of MTA and SAH within bacterial cells and lead to inhibition of methylase and polyamine synthase activities that are growth inhibitory (Beeston & Surette, 2002). In a recent study, the intracellular concentrations of SAM and SAH in the wild-type E. coli strain MG1655 (OD600=1.62±0.16) were determined to be 0.4mM, and 1.3μM, respectively (Halliday et al., 2010). A deletion of MTA/SAH nucleosidase in this strain caused an intracellular accumulation of SAH to levels that were approximately 50-fold greater than those found in the wild-type bacteria. These levels would be sufficient to inhibit a variety of bacterial methyltransferases in vitro (Reich & Mashhoon, 1991, Reich & Mashhoon, 1990, Simms & Subbaramaiah, 1991). In mammalian cells, buildup of MTA and SAH can affect cAMP metabolism, endothelial expression of adhesin molecules and cytokine secretion (Riscoe et al., 1984, Cerri et al., 1993). Therefore, breakdown of MTA and SAH is critical for the regulation of cellular processes in bacteria as well as in mammals, and these thionucleosides are quickly catabolized so that levels are typically maintained at submicromolar concentrations inside cells. In spite of this, the importance of MTA/SAH nucleosidase (EC 3.2.2.9) remains generally underappreciated. Only recently, it has been reported that the enzyme also catabolizes 5′ deoxyadenosine (5′dADO), the product of SAM radical reactions, suggesting that it is in fact a tri-substrate specific nucleosidase, broadly involved in bacterial metabolism (Choi-Rhee & Cronan, 2005, Challand et al., 2009, Challand et al., 2010). The crucial roles of this enzyme in bacterial metabolism are reviewed here.
Previous studies examined the presence of MTA/SAH nucleosidase and SAH hydrolase (EC 3.3.1.1) among bacterial species with completely sequenced genomes (Sun et al., 2004, Winzer et al., 2002a). Sun and coworkers showed that 51 of 138 bacterial species possess cytoplasmic MTA/SAH nucleosidase, 60 possess SAH hydrolase, while only a few possess both enzymes. Approximately 20% of the examined bacterial species lack both enzymes, but these are primarily symbionts and intracellular pathogens that probably rely on host enzymes for MTA, SAH and 5′dADO catabolism (Sun et al., 2004, Winzer et al., 2002a). Borrelia burgdorferi is the only known species of bacteria to possess three homologs of MTA/SAH nucleosidase, two of which are exported (Fraser et al., 1997, Parveen & Leong, 2000, Parveen et al., 2006). In fact, Bgp, a MTA/SAH nucleosidase that also displays affinity for heparin has been shown to be present on the surface of bacteria (Parveen & Leong, 2000, Parveen et al., 2006). Furthermore, all three homologs are functional MTA/SAH nucleosidases [(Parveen et al., 2006, Cornell et al., 2009), and unpublished data], suggesting that these enzymes play a crucial role in the B. burgdorferi life cycle.
PRODUCTION OF MTA/SAH NUCLEOSIDASE SUBSTRATES
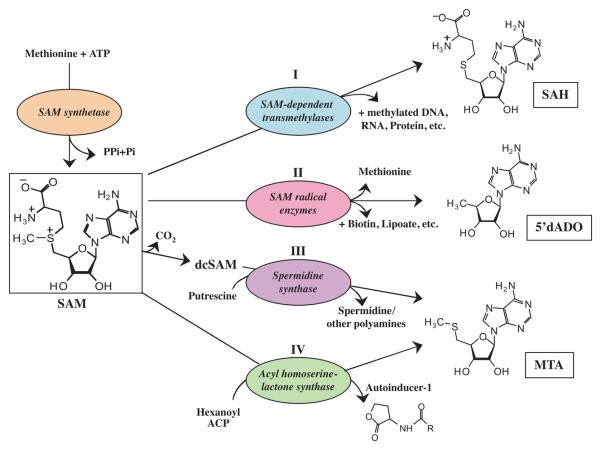
SAH, MTA and 5′dADO are the byproducts of four major pathways (Fig. 1) and they serve as substrates for MTA/SAH nucleosidase in bacteria. These reactions involve SAM, which is the primary methyl donor for methylation of macromolecules including nucleic acids, proteins, carbohydrates and lipids, and small molecules such as sterols and nucleosides. A comprehensive review of the SAM-dependent methyltransferases of both prokaryotes and eukaryotes has been previously published (Cheng & Blumenthal, 1999). A list of bacterial methyltransferases that play roles in the synthesis of secondary metabolites and other unique molecules is provided here (supplementary Table 1). SAM is also a critical activated group donor required for the synthesis of polyamines (predominantly spermidine in bacteria), N-acyl-homoserine lactone, and various vitamins (biotin, thiamine, etc) derived from SAM radical reaction mechanisms. Four pathways producing MTA/SAH nucleosidase substrates are as follows.
Fig. 1. Four metabolic pathways produce substrates of MTA/SAH nucleosidase.
Four pathways involving SAM: (I) methylation of macromolecules, (II) reactions producing different metabolites by SAM radical enzymes, (III) polyamine (including spermidine) synthesis, and (IV) autoinducer-1 (N-acylhomoserine lactone) synthesis. All result in production of nucleosidase substrates: SAH, MTA or 5′dADO (boxed). Methylation of various macromolecules using specific transmethylases produces SAH as a byproduct. SAM radical enzymes produce vitamins, metabolites and antibiotics with methionine and 5′dADO as byproducts. Polyamine synthesis is a two-step process, (i) decarboxylation of SAM followed by (ii) conversion of putrescine and decarboxylated SAM to spermidine and MTA. Hexanoyl Acyl carrier protein (ACP) donates theAcyl group to SAM to form N-acylhomoserine lactone.
I. Methylation of Nucleic acids
SAM-dependent methyltransferases play critical roles in diverse biological reactions including the methylation of DNA. Methylation of cytosine (C-5 and N-4) and adenine (N-6) has regulatory effects on chromosomal structure, DNA mismatch repair and transcription. Methylation plays a key role in regulation of gene expression and the bacterial cell cycle (Cheng & Roberts, 2001, Jeltsch, 2002, Collier, 2009), and is also required for some nuclease recognition, such as the Type I restriction endonuclease EcoK1 (Loenen, 2006). Methylation of DNA at cytosine and adenine is also a common protective mechanism against damage by other restriction endonucleases (Scavetta et al., 2000). In Caulobacter crescentus, cell cycle-regulated DNA methyltransferase (CcrM) is essential for bacterial viability, is transiently expressed at pre-division state, methylates adenine and releases SAH as a byproduct. The newly synthesized hemimethylated DNA is the preferred target of CcrM (Berdis et al., 1998). Post-transcriptional modification of ribosomal RNA (rRNA), especially adenosine residue methylation facilitates bacterial resistance and survival in the presence of ribosome-targeting antibiotics (Vester & Long, 2009, Ero et al., 2008, Yan et al., 2010, Zelinskaya et al., 2010). The X-ray crystal structure of the thiostrepton resistance RNA methyltransferase (Tsr) and in vitro analysis of the enzyme-target interactions show that two structural domains in the 23S rRNA contain methylation of specific nucleotides (Dunstan et al., 2009). Methyltransferases are also involved in the transfer of methyl groups to the 16S rRNA (Gregory et al., 2009) and methylation of tRNA of Gram-negative bacteria (Kealey et al., 1994, Bujnicki et al., 2004, Urbonavicius et al., 2005, Zelinskaya et al., 2010, Ta & Kim, 2010).
Methylation of other substrates
SAM is also the major methyl donor for methylation of various proteins, carbohydrates, lipids and other molecules and yields SAH as the byproduct (Fig. 1). For example, the methyltransferase CheR-mediated methylation of the cytoplasmic domain of the methyl-accepting chemotaxis protein (MCP) triggers an adaptive response in E. coli to external stimuli (Antommattei et al., 2004). Recently, proteome analysis of the spirochete pathogen Leptospira interrogans demonstrated the presence of 155 proteins with arginine methylation (Cao et al., 2010). Methylation of mycobacterial heparin-binding hemagglutinin and laminin-binding adhesin results in their resistance to proteolysis (Pethe et al., 2002). The SAM-dependent methyltransferase Hma of Mycobacterium tuberculosis is essential for biosynthesis of both keto- and methoxymycolate components of the cell envelope (Boissier et al., 2006). In Geobacillus stearothermophilus, SAM-dependent methylation of terminal L-rhamnose sugars is required for its incorporation into the S-layer glycan chain (Steiner et al., 2008). Microbes under abiotic stress produce the methylation product betaine (N, N, N-trimethylglycine) as an osmoprotectant (Waditee et al., 2003). The opportunistic pathogen Pseudomonas aeruginosa uses a SAM-dependent methyltransferase PhzM in pyocyanin pigment production (Parsons et al., 2007). These reactions show SAH production by a variety of methylation reactions.
II. SAM-radical dependent enzymes
A superfamily of radical SAM dependent enzymes has been identified that uses SAM as an oxidizing agent to accomplish a diverse array of reactions including anaerobic oxidations, sulfur insertions, isomerizations, ring formation, and unusual methylations (Sofia et al., 2001). The reactions proceed through a radical intermediate that yields methionine and 5′dADO, which is also a substrate for the nucleosidase (Fig. 1). These radical SAM enzymes are involved in biosynthesis of secondary metabolites, vitamins, antibiotics and DNA repair in bacteria (Wang & Frey, 2007). Biotin synthase (EC 2.8.1.6) is a radical SAM- dependent enzyme responsible for sulfur insertion into desthiobiotin to form biotin (Jarrett, 2005, Layer et al., 2004, Marquet et al., 2001). Lipoate synthase (EC 2.8.1.8) shows sequence homology to biotin synthase suggesting that it could also be a radical enzyme. In several bacteria, the enzyme tyrosine lyase (EC 4.3.1.23) uses radical SAM to produce precursors of thiamine (Challand et al., 2010). In addition, the Klebsiella pneumoniae enzyme PqqE is a SAM radical enzyme involved in the initial step in the biosynthesis of Pyrroquinoline quinone (PQQ) (Wecksler et al., 2009). Accumulation of 5′dADO inhibits activities of these radical SAM enzymes (Choi-Rhee & Cronan, 2005), further emphasizing the importance of MTA/SAH nucleosidase in its metabolism (Fig. 2).
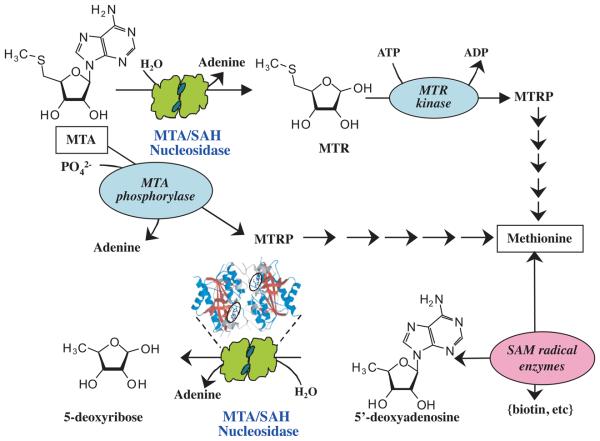
Fig. 2. Recycling of MTA and 5′dADO by MTA/SAH nucleosidase.
In the majority of bacteria, MTA is first converted by MTA/SAH nucleosidase into methylthioribose (MTR). In species with a complete methionine salvage cycle, MTR is subsequently phosphorylated by MTR kinase to 5-methylthioribose-1-phosphate (MTRP). The thiomethyl group from MTRP is recycled in a multistep pathway back to methionine. This critical amino acid is also produced as a byproduct by SAM radical enzyme reactions during synthesis of vitamins and other metabolites. The byproduct, 5′dADO, is also a substrate of MTA/SAH nucleosidase and this enzymatic reaction produces 5′deoxyribose and adenine. In eukaryotes and some environmental bacteria, MTA is catabolized by MTA phosphorylase to MTRP in a single step, which is then converted through the same enzymatic steps to methionine. A ribbon diagram of the MTA/SAH nucleosidase with bound substrate, MTA, (marked by oval) is shown.
III. Polyamine synthesis
In prokaryotic polyamine synthesis, SAM is first decarboxylated and the resulting 5′ propylamine group is then added to putrescine by spermidine synthases (EC 2.5.1.16) to produce spermidine and the corresponding MTA nucleoside (Fig. 1). MTA is recycled to methionine through different paths depending on the species of the organism (Fig. 2). The levels of spermidine and putrescine are tightly controlled in bacteria, and their concentrations within E. coli were reported to be 4.2mM and 28.4mM, respectively (Davis et al., 1992). Polyamines were suggested to exist bound to nucleic acids in the intact cell with negligible amount existing as the free form in the cytoplasm (Kashiwagi et al., 1986). Therefore, the effect of intracellular MTA accumulation on polyamine production in bacteria is difficult to assess. However, mutation in the pfs gene in E. coli resulted in a significant growth defect (Cadieux et al., 2002). Furthermore, MTA accumulation was proposed to inhibit polyamine synthases in bacteria, but the experimental evidence was not provided (Beeston & Surette, 2002).
IV. Production of N-acylhomoserine lactone
MTA is also a byproduct of N-acylhomoserine lactone synthases (EC 2.3.1.184) in Gram-negative bacteria, as previously described in two comprehensive reviews (Winans & Bassler, 2002, Fuqua et al., 2001). N-acylhomoserine lactone (autoinducer 1, AI-1) was identified as the first cell-cell communication or quorum-sensing (QS) signal in bacteria, where it functions in a largely species-specific manner (Nealson & Hastings, 1979).
EFFECT OF ACCUMULATED MTA, SAH, and 5′dADO
Accumulation of MTA, SAH and 5′dADO can have wide-ranging physiological consequences. SAH is a potent feedback inhibitor of SAM-dependent methylation reactions, both in bacteria and mammals (Borchardt, 1986). While data on the in vivo effect of SAH inhibition on methyltransferase activity in bacteria is sparse, the assumption has been made that the bacterial methyltransferases will be affected in a similar fashion to the more extensively studied eukaryotic methyltransferases. This is supported by data from in vitro enzymology demonstrating that a number of bacterial methyltransferases are susceptible to SAH inhibition at low micromolar concentrations (Reich & Mashhoon, 1991, Simms & Subbaramaiah, 1991, Reich & Mashhoon, 1990). This assumption is further validated by several reports of the development of colorimetric assays that use MTA/SAH nucleosidase to alleviate product inhibition of methylation reactions in vitro (Dorgan et al., 2006, Hendricks et al., 2004). More recently, an E. coli MTA/SAH nucleosidase knockout strain has been shown to accumulate SAH to more than 50μM, a concentration that is approximately 50-fold greater than that of the isogenic wild-type. Conversely, SAM, SRH, and methionine levels were decreased in the knockout strain, probably due to the loss of the salvage pathway (Halliday et al., 2010).
MTA is also a potent product inhibitor of polyamine synthase reactions at low micromolar concentrations (Pajula & Raina, 1979, Raina et al., 1982). The subsequent limitation of polyamine availability influences DNA replication, leading to growth arrest. In addition, MTA is a potent feedback inhibitor of AI-1 synthase, inhibiting 50% of the enzyme activity at concentrations as low as 5μM (Hanzelka et al., 1999). Although this latter effect probably has little influence on growth rates in Gram negative bacteria, MTA inhibition of AI-1 synthase could significantly alter quorum sensing-dependent phenotypes such as biofilm formation and virulence.
The buildup of 5′dADO resulting from radical SAM reactions reduces vitamin availability required for numerous metabolic reactions and probably alters flux rates through central glycolytic pathways, resulting in reduced cellular growth (Choi-Rhee & Cronan, 2005). To prevent these growth inhibitory effects, all organisms that produce SAH, MTA, and 5′dADO also possess or exploit mechanisms for removal of these compounds.
CATABOLISM OF THIONUCLEOSIDES BY MTA/SAH NUCLEOSIDASE
After examination of 138 bacterial species for which the genomes have been completely sequenced and annotated, the majority of bacterial species (80%) were predicted to use either Pfs (MTA/SAH nucleosidase) or SAH hydrolase to breakdown SAH (Sun et al., 2004, Winzer et al., 2002a) emphasizing the importance of SAH hydrolysis in cellular metabolism. Sun et al (2004) suggested that the remaining 20% of bacterial species, which mainly represent symbionts and intracellular parasites, depend on the host to recycle these toxic intermediates. In 51 bacterial species, including the majority of the pathogens with reductive genome evolution, a MTA/SAH nucleosidase enzyme is responsible for the metabolism of both MTA and SAH (Cornell et al., 1996a, Cornell et al., 1996b, Ferro et al., 1976, Della Ragione et al., 1985, Sun et al., 2004). Bacterial MTA/SAH nucleosidases show comparable efficiency in hydrolyzing the glycosidic bond in these substrates to yield adenine and the corresponding sugar (Lee et al., 2005a, Cornell & Riscoe, 1998, Cornell et al., 1996a, Parveen et al., 2006). In addition, the MTA/SAH nucleosidase substrate 5′dADO is produced as a result of SAM radical enzyme activities (Figs. 1 and 2). In P. aeruginosa, L. interrogans, several Archaea, and a number of other bacterial species that possess relatively large genomes (>6Mb) and exhibit complex metabolism, MTA phosphorylase (EC 2.4.2.28) converts MTA to 5-methylthioribose-1-phosphate (MTRP) in a reversible phosphate dependent reaction (Sekowska et al., 2004, Cacciapuoti et al., 2003) with adenine as a byproduct. In these organisms, SAH is broken down by SAH hydrolase and results in the production of homocysteine and adenosine. Similarly, in various eukaryotes, including mammals, MTA phosphorylase and SAH hydrolase catabolize MTA and SAH, respectively (Sufrin et al., 1995, Riscoe, 1989, Stepkowski et al., 2005). These separate enzymes have narrow substrate specificities in both environmental bacteria and eukaryotic counterparts, but are more efficient in metabolizing MTA and SAH. Efficient recycling of MTA and SAH probably facilitates the survival of these versatile bacteria under varying environmental conditions (Stepkowski et al., 2005).
In a variety of organisms, quick removal of MTA and salvage of the methionyl moiety is facilitated by hydrolysis of MTA into 5-methylthioribose (MTR) and subsequent phosphorylation of MTR to MTRP by MTR kinase (EC 2.7.1.100), as shown in the Fig. 2. Four to five enzymes then convert MTRP into methionine in a sequential manner that has been best elucidated in Klebsiella and Bacillus, but appears to be common to all organisms with a complete salvage pathway (Albers, 2009, Cornell et al., 1996b, Gianotti et al., 1990, Sekowska et al., 2004, Furfine & Abeles, 1988). Other organisms that exist in sulfur rich environments, such as E. coli, often do not possess MTR kinase, and thus do not salvage methionine from MTA, but rather secrete MTR (Hughes, 2006).
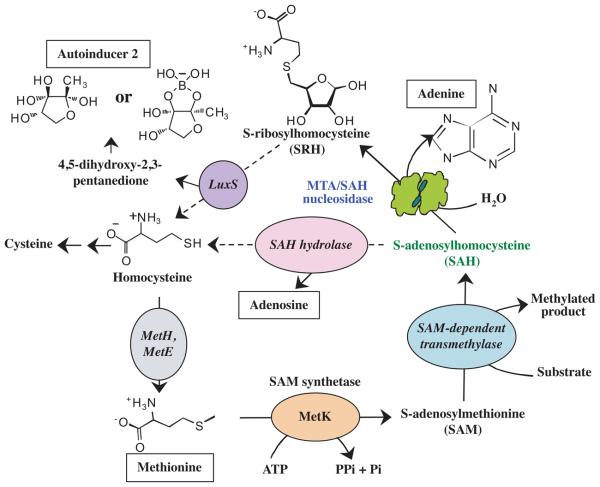
SAH CATABOLISM THROUGH ACTIVATED METHYL CYCLE
The activated methyl cycle (AMC) is responsible for producing SAM required for methylations, and the recycling of the product SAH back to methionine (Fig. 3). Methionine is converted by SAM synthetase (MetK, EC 2.5.1.6) to SAM using ATP as a substrate and energy from the hydrolysis of high-energy phosphate bonds to drive the reaction. In several environmental bacteria, and some plant and animal pathogens/symbionts, such as Caulobacter, Pseudomonas, Xanthomonas, Rhizobium and Brucella spp., SAH hydrolase is used to directly convert SAH to homocysteine (Sun et al., 2004). However, in a majority of eubacteria, detoxification of SAH is carried out by MTA/SAH nucleosidase to produce S-ribosylhomocysteine (SRH) (Sun et al., 2004, Winzer et al., 2002a, Markham & Pajares, 2009). The enzyme LuxS (EC 4.4.1.21) further cleaves SRH to homocysteine and 4,5-dihydroxy-2,3-pentanedione, the precursor of AI-2 (Fig. 3). Homocysteine is then recycled back to methionine using cobalamine-dependent MetH (EC 2.1.1.13) or cobalamine-independent MetE (EC 2.1.1.14) methionine synthases (Kamarthapu et al., 2008). Only strains of H. pylori, Streptococcus pyogenes and Enterococcus faecalis lack MetE and MetH enzymes (Winzer et al., 2002a, Sun et al., 2004). In several bacterial species, AMC is also the sole sulfur source, such that either methionine or homocysteine is converted to cysteine through a series of enzymatic reactions (Hullo et al., 2007, Doherty et al., 2010, Sewald et al., 2007, Markham & Pajares, 2009).
Fig. 3. MTA/SAH nucleosidase is involved in Activated Methyl Cycle.
Methionine is converted into SAM by SAM synthetase (MetK). Donation of the methyl group of SAM to a variety of methyl acceptors results in SAH, which is recycled back to homocysteine in either a one-step or two-step process. In several bacterial species SAH hydrolase directly converts SAH into homocysteine. However, in the majority of eubacteria, MTA/SAH nucleosidase first converts SAH into S-ribosylhomocysteine (SRH), which is then recycled back to homocysteine by LuxS. As a byproduct of this reaction, 4,5 dihydroxy 2,3-pentanedione is produced which spontaneously forms autoinducer-2 (AI-2). In some bacteria, cysteine is produced from homocycteine by multi-step enzymatic reactions since this cycle is the only source of sulfur-containing compounds. Homocycteine is recycled back to methionine in different organisms by the Met E or MetH methionine synthases.
MTA/SAH NUCLEOSIDASE IN QUORUM-SENSING
Quorum-sensing (QS) is a cell density-dependent communication system of bacteria. Some Gram-negative bacteria are known to secrete as many as three signaling molecules or autoinducers (AI): AI-1, AI-2, and AI-3. The accumulation of these autoinducers as a function of culture cell density results in signaling cascades that ultimately change cellular gene expression profiles and lead to population wide adaptive responses (Winzer et al., 2002a, Chen et al., 2002, Sun et al., 2004, Miller et al., 2002, Zhu et al., 2002, Waters & Bassler, 2005, Ng & Bassler, 2009). QS systems regulate pathogen-host cell interactions, bacterial virulence, and the formation of bacterial biofilms. N-Acylhomoserine lactone or AI-1 was first discovered in the marine bioluminescent bacterium Vibrio fischeri (Nealson & Hastings, 1979). AI-1 is produced by acylhomoserine lactone synthase (LuxI) in V. fischeri, or by equivalent enzymes in other bacteria. SAM is a substrate and hexanoyl Acyl carrier protein (ACP) acts as the acyl donor in a reaction that generates AI-1 and MTA as the byproduct (Fig. 1). After reaching a critical threshold, AI-1 interacts with the receptor LuxR and this complex then induces expression of the lux operon encoding luciferase (Fuqua & Greenberg, 2002).
Both Gram-positive and Gram-negative bacteria possess AI-2-mediated QS systems derived from SAH catabolism. Therefore, AI-2 is often known as the “universal” QS-signaling molecule (Winzer et al., 2002a, Sun et al., 2004). In addition to methyltransferases, three enzymes, SAM synthase (MetK), MTA/SAH nucleosidase (Pfs) and LuxS in AMC are involved in the synthesis of 4,5-dihdroxy-2,3-pentanedione from methionine (Fig. 3). SAH hydrolysis by Pfs (MTA/SAH nucleosidase) yields S-ribosylhomocysteine (SRH), a precursor of autoinducer-2 synthesis, a quorum-sensing signal that governs a variety of bacterial phenotypes such as virulence and biofilm formation as reviewed previously (Gospodarek et al., 2009, Federle, 2009, Federle & Bassler, 2003). AI-2 is produced as a result of spontaneous cyclization of 4,5-dihdroxy-2,3-pentanedione to methyltetrahydroxy furan, and in boron rich environments on to the corresponding borate diester (Fig. 3). In the Lyme disease causing spirochete, B. burgdorferi, the three genes of the AI-2 pathway (metK, pfs, luxS) are present in one operon producing all three as cytoplasmic enzymes, while the other two Pfs homologs, Bgp and MtnN, in B. burgdorferi are exported proteins. Genes encoding bgp and mtnN are located elsewhere in the spirochete genome and are not part of any operon (Riley et al., 2007, Fraser et al., 1997, Winzer et al., 2002a). In Porphyromonas gingivalis, the operon contains only pfs and luxS genes, indicating simultaneous production of the encoded enzymes (Riley et al., 2007, Fraser et al., 1997, Winzer et al., 2002a). Indeed, transformation of P. aeruginosa with the pfs-lux operon of P. gingivalis, reconstitution of luxS mutant E. coli strains with luxS gene from other genera, and magnetic nanofactories cell capture modules assembled with Pfs-LuxS fusion chimeras, all showed that these two proteins are sufficient to produce AI-2 from SAH (Winzer et al., 2002a, Fernandes & Bentley, 2009).
Pfs is an integral component of the AI-2 synthesis pathway. Indeed, AI-2 production shows a tighter correlation with Pfs transcription than luxS gene expression in several bacteria including Salmonella enterica serovar typhimurium and Streptococcus suis (Beeston & Surette, 2002, Han & Lu, 2009, Kim et al., 2006). In B. burgdorferi, luxS mutations affect transcription of several genes but this change in the transcriptome did not seem to affect transmission of bacteria through ticks or pathogenesis of the spirochete in the mouse model of infection (Babb et al., 2005, Stevenson & Babb, 2002, Hubner et al., 2003). The luxS mutant of H. pylori also did not show any detectable change in its gene expression pattern relative to the wild-type strain (Joyce et al., 2000, Forsyth & Cover, 2000). However, knock-out mutants of luxS genes in V. cholerae, S. pyogenes, S. pneumoniae, N. meningitidis, and C. perfringens exhibited severe defects in the expression of genes encoding virulence factors (Miller et al., 2002, Zhu et al., 2002, Lyon et al., 2001, Marouni & Sela, 2003, Stroeher et al., 2003, Winzer et al., 2002c, Ohtani et al., 2002). In the meningitis and septicemia causing bacterium Neisseria meningitidis, AI-2 release was completely eliminated in the pfs mutant (Heurlier et al., 2009). These results are further supported by the report that cultures of V. cholerae and enterohemorrhegic E. coli (EHEC) strain O157:H7 treated with MTA/SAH nucleosidase inhibitors did not synthesize AI-2 and showed reduced biofilm formation (Gutierrez et al., 2009). However, the growth defects observed in the N. meningitidis pfs mutant, and probably also of the luxS mutant, were not attributable to loss of AI-2 synthesis but rather due either to the accumulation of toxic SAH and MTA or to metabolic imbalances within the bacteria (Heurlier et al., 2009, Winzer et al., 2002a, Winzer et al., 2002b, Dove et al., 2003, Li et al., 2008).
The majority of bacteria possess either the single step SAH hydrolase that yields adenosine and homocysteine, or the two-step enzymatic system mediated by Pfs and LuxS to recycle the toxic SAH metabolite produced by the AMC. However, a few bacteria appear to possess both systems (Sun et al., 2004). Winzer and coworkers (2002) suggested that the physiological and metabolic state of the organism determines the uptake of different products using ABC transporters or equivalent import systems, after breakdown of SRH by LuxS. In addition, both uptake and degradation of these molecules occurs in a controlled manner (Surette & Bassler, 1999, Surette et al., 1999, Surette & Bassler, 1998, Lyon et al., 2001, Winzer et al., 2002a, Winzer et al., 2002b, Wang et al., 2005, Xavier & Bassler, 2005, Taga et al., 2003, Taga et al., 2001). Therefore, the primary role of LuxS was predicted to be in metabolic pathways, especially in the AMC to produce homocysteine for conversion back to methionine. Several reports have now provided supporting evidence for the metabolic function of Pfs and LuxS in recycling the sulfur group. Although AI-2 production has been evaluated in a wide variety of bacteria by using V. harveyi reporter strains, the proof of a functionally active set of proteins from sensor/receptor to response regulators required for AI-2-signalling cascade has not yet been reported in organisms other than Vibrio spp. (Sun et al., 2004). Therefore, there is no direct evidence that the majority of bacteria actually respond to AI-2 as a QS signal. However, Pfs expression shows a strong correlation with AI-2 synthesis, purine and nutrient availability, and a pfs-mutant of E. coli failed to grow in the absence of exogenous methionine (Cadieux et al., 2002, Kim et al., 2006, Han & Lu, 2009). Furthermore, exogenous supply of AI-2 to cultures failed to rescue the growth defect and competitiveness of N. meningitidis pfs and luxS mutants. These results indicate that intracellular metabolite imbalance rather than the lack of AI-2-mediated QS signaling was responsible for the growth defect of the N. meningitidis mutants (Heurlier et al., 2009). Since SAH was not detectable in the wild-type strain, accumulation of this metabolite, or potentially MTA, was suggested to be responsible for the growth defect of the pfs mutant of N. meningitidis. Furthermore, the luxS mutant of Campylobacter jejuni exhibited metabolic effects and inhibition of methionine cycle gene transcription was reported (Holmes et al., 2009).
MTA/SAH NUCLEOSIDASE AS TARGET OF NOVEL ANTIMICROBIALS
Soon after the first purification of MTA nucleosidase from E. coli, even before SAH was identified as its second substrate, MTA analogs were recognized as effective competitive inhibitors of this enzyme (Ferro et al., 1976). A further analysis of 25 analogs of naturally occurring thioesters showed that several were potent inhibitors of the nucleosidase activity (Della Ragione et al., 1985). MTA/SAH nucleosidase and MTA phosphorylase have been purified and characterized from a variety of bacteria (Appleby et al., 2001, Cacciapuoti et al., 2007, Cacciapuoti et al., 2003, Cacciapuoti et al., 1999, Lee et al., 2001, Lee et al., 2005c). Extensive x-ray crystallographic analysis (Lee et al., 2003, Lee et al., 2001, Lee et al., 2005c, Lee et al., 2005b, Singh et al., 2007, Singh & Schramm, 2007, Singh et al., 2005) unequivocally showed that the MTA/SAH nucleosidase enzyme exists as a dimer. Importantly, the active site of each subunit contains a hydrophobic pocket that appears to play a role in the discrimination of different adenosyl nucleoside substrates and is partially composed of residues from the second subunit. The acidic residues involved in catalysis are highly conserved across species, and the substrate hydrolysis appears to be essentially irreversible (Cornell et al., 1996a, Lee et al., 2005c, Singh et al., 2005). In contrast, MTA phosphorylases appear to be trimeric and display reversible reactions (Cacciapuoti et al., 2003, Cacciapuoti et al., 1999, Cacciapuoti et al., 2007). MTA analog(s) interaction with the purified E. coli enzyme indicated the transition state of the enzyme-substrate complex (Allart et al., 1998). Lee and coworkers solved the crystal structure of the enzyme complexed with adenine and with the substrate analog 5′-methylthiotubercidin (MTT) and transition state analog, formycin A (FMA) (Lee et al., 2003, Lee et al., 2001) to determine the molecular interactions involved in substrate recognition. Additional analysis using a series of early and late transition state analogs and kinetic isotope effects, allowed differentiation of the transition states of N. meningitides and H. pylori nucleosidases that showed an early dissociative transition state and was more similar to bovine purine nucleoside phosphorylases (PNP, EC 2.4.2.1). In contrast, the nucleosidases from E. coli, K. pneumoniae, S. aureus and S. pneumoniae demonstrated a fully dissociative transition state that was shared with human PNP and MTA phosphorylase (Singh et al., 2007, Singh & Schramm, 2007, Singh et al., 2005, Gutierrez et al., 2009, Luo & Schramm, 2008).
Despite similarities in the enzyme transition states, crystallographic evidence points to distinct structural differences in the enzyme active sites between the bacterial nucleosidase and mammalian phosphorylase. A study by Lee et al. (2004) compared the crystallographic structures of the E. coli MTA/SAH nucleosidase to the mammalian MTA phosphorylase (Lee et al., 2004). The active site in the nucleosidase was shown to have a larger 5′ alkylthio binding pocket than MTA phosphorylase, supporting the observation that the bacterial enzyme functions on thionucleosides such as SAH that have extended 5′ structures, whereas the mammalian enzyme has a much narrower substrate specificity. In addition, electrostatic maps showed a negatively charged region in MTA/SAH nucleosidase around the substrate 2′ hydroxyl recognition site, which was positively charged in MTA phosphorylase. These combined differences could be exploited for drug design. More recent work has reported potent late stage transition state analogs with bulky 5′ substitutions (DADMe-immucillin 54; DADMe-immucillin 57) that can indeed discriminate between the E. coli and human enzymes (Longshaw et al., 2010). These analogs show either no activity against MTA phosphorylase and picomolar dissociation constants for MTA/SAH nucleosidase (DADMe-immucillin 54), or nanomolar dissociation constants for the human enzyme that are greater than a thousand-fold higher than for the bacterial enzyme (DADMe-immucillin 57).
Structure-based design was also employed to identify highly potent indazole, purine, and deazapurine-based inhibitors for MTA/SAH nucleosidase by screening virtual compound libraries (Li et al., 2003, Tedder et al., 2004). The most potent of these inhibitors displayed low nanomolar concentration dissociation constants for the enzyme. However, the antimicrobial activity of the inhibitors was only modest. Reported MIC values against N. meningitides, S. pneumoniae, and S. pyogenes were at best 1-2 micromolar, and more frequently were greater than 20-30 micromolar.
Antimicrobial activity reported for immucillin-based transition state analogs also appears modest, with little effect on bacterial planktonic cell growth, possibly due to poor drug transport (Gutierrez et al., 2009). Addition of a million fold concentrations of picomolar transition state inhibitors did not affect the growth of V. cholerae. However, these inhibitors bound tightly to MTA/SAH nucleosidase in V. cholerae and EHEC E. coli O157:H7, and significantly disrupted AI-2 production and biofilm formation at picomolar concentrations (Gutierrez et al., 2009, Schramm et al., 2008). These results demonstrate that sufficient drug influx is occurring to effect metabolic changes. These drugs could be developed into novel antimicrobials based upon their mechanism of action in disrupting AI-2 dependent pathogenicity, rather than direct bactericidal activity. Furthermore, the inhibitors may exert their effects by restricting degradation of 5′ dADO, leading to altered rates of vitamin synthesis that ultimately affect central carbon metabolism. This may explain some of the observations on changes to bacterial growth in response to pharmacologic or genetic blockade of MTA/SAH nucleosidases made by Cadieux et al. (2002), Gutierrez et al. (2009) and Heurlier et al. (2009). Evaluation of the substrate analogs and transition state analogs effective against MTA/SAH nucleosidases of B. burgdorferi led to identification of compounds that either inhibited growth of these spirochetes or showed bactericidal activities (Cornell et al., 2009). Some of these inhibitors showed more potent activities than those exhibited on E. coli, perhaps due to the underlying purine auxotrophy in B. burgdorferi, which makes it more critical for these spirochetes to salvage methionine and adenine through the nucleosidase. In addition, there are three homologous MTA/SAH nucleosidases in B. burgdorferi, two of which are exported and at least one, Bgp, which is present on the surface of the spirochete, presumably to help scavenge purines from the host or the vector (Parveen & Leong, 2000, Parveen et al., 2006). Therefore, in B. burgdorferi drug permeability may not be an issue.
Overall, these reports suggest that inhibitors of MTA/SAH nucleosidase could possibly become new anti-infective drugs against various bacterial pathogens. Interestingly, Rrp1, which produces the secondary messenger cyclic diguanylate (c-di-GMP) and the hybrid histidine kinase-response regulator (Hpk1) are involved in the upregulation of expression of Bgp at the cusp of infection, i.e., immediately after tick takes bloodmeal from the mammals (Rogers et al., 2009). Thus, these novel drugs could inhibit infection in the early state in some organisms and restrict QS systems and biofilm formation later in infection in others.
CONCLUDING REMARKS
SAM is an essential nucleoside required for growth of all living organisms due to its involvement in critical biological methylations, polyamine synthesis, and a broad array of other reactions. MTA, SAH, and 5′dADO are toxic byproducts of these reactions that can even be lethal. Most prokaryotes possess either MTA/SAH nucleosidase and/or a combination of SAH hydrolase and MTA phosphorylase to remove these inhibitory nucleosides. In addition, MTA/SAH nucleosidase is an integral component of the AMC responsible for recycling methionine from homocysteine, and is involved in AI-2 synthesis. Indeed, transcription of the MTA/SAH nucleosidase gene, pfs, and not luxS, is tightly correlated with AI-2 production in different bacteria. Therefore, MTA/SAH nucleosidase is being explored as a target for potential novel drugs that could show anti-infective activity due to accumulation of toxic byproducts within the cells or by prevention of AI-2-mediated processes, such as virulence factor synthesis and biofilm formation by the pathogen later in infection.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank John Leong and Purnima Bhanot for helpful comments and the National Research Fund for Tick-borne Diseases and Arthritis Foundation for funding to NP; and the Idaho INBRE Program of the National Center for Research Resources NIH (P20 RR016454) and the Mountain States Tumor and Medical Research Institute for funding to KAC.
BIBLIOGRAPHY
- Aktas M, Narberhaus F. In vitro characterization of the enzyme properties of the phospholipid N-methyltransferase PmtA from Agrobacterium tumefaciens. J Bacteriol. 2009;191:2033–2041. doi: 10.1128/JB.01591-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alahari A, Alibaud L, Trivelli X, Gupta R, Lamichhane G, Reynolds RC, Bishai WR, Guerardel Y, Kremer L. Mycolic acid methyltransferase, MmaA4, is necessary for thiacetazone susceptibility in Mycobacterium tuberculosis. Mol Microbiol. 2009;71:1263–1277. doi: 10.1111/j.1365-2958.2009.06604.x. [DOI] [PubMed] [Google Scholar]
- Albers E. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life. 2009;61:1132–1142. doi: 10.1002/iub.278. [DOI] [PubMed] [Google Scholar]
- Allart B, Gatel M, Guillerm D, Guillerm G. The catalytic mechanism of adenosylhomocysteine/methylthioadenosine nucleosidase from Escherichia coli--chemical evidence for a transition state with a substantial oxocarbenium character. Eur J Biochem. 1998;256:155–162. doi: 10.1046/j.1432-1327.1998.2560155.x. [DOI] [PubMed] [Google Scholar]
- Amachi S, Kamagata Y, Kanagawa T, Muramatsu Y. Bacteria mediate methylation of iodine in marine and terrestrial environments. Appl Environ Microbiol. 2001;67:2718–2722. doi: 10.1128/AEM.67.6.2718-2722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antommattei FM, Munzner JB, Weis RM. Ligand-specific activation of Escherichia coli chemoreceptor transmethylation. J Bacteriol. 2004;186:7556–7563. doi: 10.1128/JB.186.22.7556-7563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby TC, Mathews II, Porcelli M, Cacciapuoti G, Ealick SE. Three-dimensional structure of a hyperthermophilic 5′-deoxy-5′-methylthioadenosine phosphorylase from Sulfolobus solfataricus. J Biol Chem. 2001;276:39232–39242. doi: 10.1074/jbc.M105694200. [DOI] [PubMed] [Google Scholar]
- Babb K, von Lackum K, Wattier RL, Riley SP, Stevenson B. Synthesis of Autoinducer 2 by the Lyme Disease Spirochete, Borrelia burgdorferi. J Bacteriol. 2005;187:3079–3087. doi: 10.1128/JB.187.9.3079-3087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeston AL, Surette MG. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:3450–3456. doi: 10.1128/JB.184.13.3450-3456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdis AJ, Lee I, Coward JK, Stephens C, Wright R, Shapiro L, Benkovic SJ. A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA. Proc Natl Acad Sci U S A. 1998;95:2874–2879. doi: 10.1073/pnas.95.6.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissier F, Bardou F, Guillet V, Uttenweiler-Joseph S, Daffe M, Quemard A, Mourey L. Further insight into S-adenosylmethionine-dependent methyltransferases: structural characterization of Hma, an enzyme essential for the biosynthesis of oxygenated mycolic acids in Mycobacterium tuberculosis. J Biol Chem. 2006;281:4434–4445. doi: 10.1074/jbc.M510250200. [DOI] [PubMed] [Google Scholar]
- Borchardt RT, Creveling CR, Ueland PM. Biological methylation and drug design. Humana Press; Clifton, NJ: 1986. pp. 227–238. [Google Scholar]
- Budin-Verneuil A, Maguin E, Auffray Y, Ehrlich SD, Pichereau V. Transcriptional analysis of the cyclopropane fatty acid synthase gene of Lactococcus lactis MG1363 at low pH. FEMS Microbiol Lett. 2005;250:189–194. doi: 10.1016/j.femsle.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciapuoti G, Bertoldo C, Brio A, Zappia V, Porcelli M. Purification and characterization of 5′-methylthioadenosine phosphorylase from the hyperthermophilic archaeon Pyrococcus furiosus: substrate specificity and primary structure analysis. Extremophiles. 2003;7:159–168. doi: 10.1007/s00792-002-0307-2. [DOI] [PubMed] [Google Scholar]
- Cacciapuoti G, Fusco S, Caiazzo N, Zappia V, Porcelli M. Heterologous expression of 5′-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus: characterization of the recombinant protein and involvement of disulfide bonds in thermophilicity and thermostability. Protein Expr Purif. 1999;16:125–135. doi: 10.1006/prep.1999.1076. [DOI] [PubMed] [Google Scholar]
- Cacciapuoti G, Gorassini S, Mazzeo MF, Siciliano RA, Carbone V, Zappia V, Porcelli M. Biochemical and structural characterization of mammalian-like purine nucleoside phosphorylase from the Archaeon Pyrococcus furiosus. FEBS J. 2007;274:2482–2495. doi: 10.1111/j.1742-4658.2007.05784.x. [DOI] [PubMed] [Google Scholar]
- Cadieux N, Bradbeer C, Reeger-Schneider E, Koster W, Mohanty AK, Wiener MC, Kadner RJ. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J Bacteriol. 2002;184:706–717. doi: 10.1128/JB.184.3.706-717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakici O, Sikorski M, Stepkowski T, Bujacz G, Jaskolski M. Cloning, expression, purification, crystallization and preliminary X-ray analysis of NodS N-methyltransferase from Bradyrhizobium japonicum WM9. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:1149–1152. doi: 10.1107/S174430910803604X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XJ, Dai J, Xu H, Nie S, Chang X, Hu BY, Sheng QH, Wang LS, Ning ZB, Li YX, Guo XK, Zhao GP, Zeng R. High-coverage proteome analysis reveals the first insight of protein modification systems in the pathogenic spirochete Leptospira interrogans. Cell Res. 2010;20:197–210. doi: 10.1038/cr.2009.127. [DOI] [PubMed] [Google Scholar]
- Cerri MA, Beltran-Nunez A, Bernasconi S, Dejana E, Bassi L, Bazzoni G. Inhibition of cytokine production and endothelial expression of adhesion antigens by 5′-methylthioadenosine. Eur J Pharmacol. 1993;232:291–294. doi: 10.1016/0014-2999(93)90787-i. [DOI] [PubMed] [Google Scholar]
- Challand MR, Martins FT, Roach PL. Catalytic activity of the anaerobic tyrosine lyase required for thiamine biosynthesis in Escherichia coli. J Biol Chem. 2010;285:5240–5248. doi: 10.1074/jbc.M109.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challand MR, Ziegert T, Douglas P, Wood RJ, Kriek M, Shaw NM, Roach PL. Product inhibition in the radical S-adenosylmethionine family. FEBS Lett. 2009;583:1358–1362. doi: 10.1016/j.febslet.2009.03.044. [DOI] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. S-Adenosylmethionine-dependent methyltransferases: Structure and function. World Scientific Publishing Co. Pte. Ltd.; Singapore: 1999. [Google Scholar]
- Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi-Rhee E, Cronan JE. A nucleosidase required for in vivo function of the S-adenosyl-L-methionine radical enzyme, biotin synthase. Chem Biol. 2005;12:589–593. doi: 10.1016/j.chembiol.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Collier J. Epigenetic regulation of the bacterial cell cycle. Curr Opin Microbiol. 2009;12:722–729. doi: 10.1016/j.mib.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Cornell KA, Primus S, Martinez JA, Parveen N. Assessment of methylthioadenosine/S-adenosylhomocysteine nucleosidases of Borrelia burgdorferi as targets for novel antimicrobials using a novel high-throughput method. J Antimicrob Chemother. 2009;63:1163–1172. doi: 10.1093/jac/dkp129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell KA, Riscoe MK. Cloning and expression of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase: identification of the pfs gene product. Biochim Biophys Acta. 1998;1396:8–14. doi: 10.1016/s0167-4781(97)00169-3. [DOI] [PubMed] [Google Scholar]
- Cornell KA, Swarts WE, Barry RD, Riscoe MK. Characterization of recombinant Eschericha coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase: analysis of enzymatic activity and substrate specificity. Biochem Biophys Res Commun. 1996a;228:724–732. doi: 10.1006/bbrc.1996.1723. [DOI] [PubMed] [Google Scholar]
- Cornell KA, Winter RW, Tower PA, Riscoe MK. Affinity purification of 5-methylthioribose kinase and 5-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Klebsiella pneumoniae. Biochem J. 1996b;317(Pt 1):285–290. doi: 10.1042/bj3170285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH, Morris DR, Coffino P. Sequestered end products and enzyme regulation: the case of ornithine decarboxylase. Microbiol Rev. 1992;56:280–290. doi: 10.1128/mr.56.2.280-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Ragione F, Porcelli M, Carteni-Farina M, Zappia V, Pegg AE. Escherichia coli S-adenosylhomocysteine/5′-methylthioadenosine nucleosidase. Purification, substrate specificity and mechanism of action. Biochem J. 1985;232:335–341. doi: 10.1042/bj2320335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty NC, Shen F, Halliday NM, Barrett DA, Hardie KR, Winzer K, Atherton JC. In Helicobacter pylori, LuxS is a key enzyme in cysteine provision through a reverse transsulfuration pathway. J Bacteriol. 2010;192:1184–1192. doi: 10.1128/JB.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Dove JE, Yasukawa K, Tinsley CR, Nassif X. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: lack of evidence for a concerted transcriptional response. Microbiology. 2003;149:1859–1869. doi: 10.1099/mic.0.26185-0. [DOI] [PubMed] [Google Scholar]
- Dunstan MS, Hang PC, Zelinskaya NV, Honek JF, Conn GL. Structure of the thiostrepton resistance methyltransferase.S-adenosyl-L-methionine complex and its interaction with ribosomal RNA. J Biol Chem. 2009;284:17013–17020. doi: 10.1074/jbc.M901618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ero R, Peil L, Liiv A, Remme J. Identification of pseudouridine methyltransferase in Escherichia coli. RNA. 2008;14:2223–2233. doi: 10.1261/rna.1186608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib Microbiol. 2009;16:18–32. doi: 10.1159/000219371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, Bassler BL. Interspecies communication in bacteria. J Clin Invest. 2003;112:1291–1299. doi: 10.1172/JCI20195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes R, Bentley WE. AI-2 biosynthesis module in a magnetic nanofactory alters bacterial response via localized synthesis and delivery. Biotechnol Bioeng. 2009;102:390–399. doi: 10.1002/bit.22078. [DOI] [PubMed] [Google Scholar]
- Ferro AJ, Barrett A, Shapiro SK. Kinetic properties and the effect of substrate analogues on 5′-methylthioadenosine nucleosidase from Escherichia coli. Biochim Biophys Acta. 1976;438:487–494. doi: 10.1016/0005-2744(76)90264-3. [DOI] [PubMed] [Google Scholar]
- Forsyth MH, Cover TL. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect Immun. 2000;68:3193–3199. doi: 10.1128/iai.68.6.3193-3199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Venter JC, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Furfine ES, Abeles RH. Intermediates in the conversion of 5′-S-methylthioadenosine to methionine in Klebsiella pneumoniae. J Biol Chem. 1988;263:9598–9606. [PubMed] [Google Scholar]
- Geelen D, Leyman B, Mergaert P, Klarskov K, Van Montagu M, Geremia R, Holsters M. NodS is an S-adenosyl-L-methionine-dependent methyltransferase that methylates chitooligosaccharides deacetylated at the non-reducing end. Mol Microbiol. 1995;17:387–397. doi: 10.1111/j.1365-2958.1995.mmi_17020387.x. [DOI] [PubMed] [Google Scholar]
- Geelen D, Mergaert P, Geremia RA, Goormachtig S, Van Montagu M, Holsters M. Identification of nodSUIJ genes in Nod locus 1 of Azorhizobium caulinodans: evidence that nodS encodes a methyltransferase involved in Nod factor modification. Mol Microbiol. 1993;9:145–154. doi: 10.1111/j.1365-2958.1993.tb01676.x. [DOI] [PubMed] [Google Scholar]
- Gianotti AJ, Tower PA, Sheley JH, Conte PA, Spiro C, Ferro AJ, Fitchen JH, Riscoe MK. Selective killing of Klebsiella pneumoniae by 5-trifluoromethylthioribose. Chemotherapeutic exploitation of the enzyme 5-methylthioribose kinase. J Biol Chem. 1990;265:831–837. [PubMed] [Google Scholar]
- Glickman MS, Cahill SM, Jacobs WR., Jr. The Mycobacterium tuberculosis cmaA2 gene encodes a mycolic acid trans-cyclopropane synthetase. J Biol Chem. 2001;276:2228–2233. doi: 10.1074/jbc.C000652200. [DOI] [PubMed] [Google Scholar]
- Gomez Maqueo Chew A, Frigaard NU, Bryant DA. Bacteriochlorophyllide c C-8(2) and C-12(1) methyltransferases are essential for adaptation to low light in Chlorobaculum tepidum. J Bacteriol. 2007;189:6176–6184. doi: 10.1128/JB.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodarek E, Bogiel T, Zalas-Wiecek P. Communication between microorganisms as a basis for production of virulence factors. Pol J Microbiol. 2009;58:191–198. [PubMed] [Google Scholar]
- Gregory ST, Demirci H, Belardinelli R, Monshupanee T, Gualerzi C, Dahlberg AE, Jogl G. Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG. RNA. 2009;15:1693–1704. doi: 10.1261/rna.1652709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho MC, Almo SC, Schramm VL. Transition state analogs of 5′-methylthioadenosine nucleosidase disrupt quorum sensing. Nat Chem Biol. 2009;5:251–257. doi: 10.1038/nchembio.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday NM, Hardie KR, Williams P, Winzer K, Barrett DA. Quantitative liquid chromatography-tandem mass spectrometry profiling of activated methyl cycle metabolites involved in LuxS-dependent quorum sensing in Escherichia coli. Anal Biochem. 2010;403:20–29. doi: 10.1016/j.ab.2010.04.021. [DOI] [PubMed] [Google Scholar]
- Han XG, Lu CP. Detection of autoinducer-2 and analysis of the profile of luxS and pfs transcription in Streptococcus suis serotype 2. Curr Microbiol. 2009;58:146–152. doi: 10.1007/s00284-008-9291-9. [DOI] [PubMed] [Google Scholar]
- Hanzelka BL, Parsek MR, Val DL, Dunlap PV, Cronan JE, Jr., Greenberg EP. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol. 1999;181:5766–5770. doi: 10.1128/jb.181.18.5766-5770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2004;326:100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Heurlier K, Vendeville A, Halliday N, Green A, Winzer K, Tang CM, Hardie KR. Growth deficiencies of Neisseria meningitidis pfs and luxS mutants are not due to inactivation of quorum sensing. J Bacteriol. 2009;191:1293–1302. doi: 10.1128/JB.01170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K, Tavender TJ, Winzer K, Wells JM, Hardie KR. AI-2 does not function as a quorum sensing molecule in Campylobacter jejuni during exponential growth in vitro. BMC Microbiol. 2009;9:214. doi: 10.1186/1471-2180-9-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Wang Y, Zhou Z, Bao S, Lin Y, Gong W. Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans. J Struct Biol. 2007;159:523–528. doi: 10.1016/j.jsb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang L, Birch RG. Analysis of the genes flanking xabB: a methyltransferase gene is involved in albicidin biosynthesis in Xanthomonas albilineans. Gene. 2000;255:327–333. doi: 10.1016/s0378-1119(00)00320-6. [DOI] [PubMed] [Google Scholar]
- Hubner A, Revel AT, Nolen DM, Hagman KE, Norgard MV. Expression of a luxS gene is not required for Borrelia burgdorferi infection of mice via needle inoculation. Infect Immun. 2003;71:2892–2896. doi: 10.1128/IAI.71.5.2892-2896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JA. In vivo hydrolysis of S-adenosyl-L-methionine in Escherichia coli increases export of 5-methylthioribose. Can J Microbiol. 2006;52:599–602. doi: 10.1139/w06-008. [DOI] [PubMed] [Google Scholar]
- Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, Danchin A, Martin-Verstraete I. Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J Bacteriol. 2007;189:187–197. doi: 10.1128/JB.01273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett JT. The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase. Arch Biochem Biophys. 2005;433:312–321. doi: 10.1016/j.abb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Joyce EA, Bassler BL, Wright A. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J Bacteriol. 2000;182:3638–3643. doi: 10.1128/jb.182.13.3638-3643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamarthapu V, Rao KV, Srinivas PN, Reddy GB, Reddy VD. Structural and kinetic properties of Bacillus subtilis S-adenosylmethionine synthetase expressed in Escherichia coli. Biochim Biophys Acta. 2008;1784:1949–1958. doi: 10.1016/j.bbapap.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K, Kobayashi H, Igarashi K. Apparently unidirectional polyamine transport by proton motive force in polyamine-deficient Escherichia coli. J Bacteriol. 1986;165:972–977. doi: 10.1128/jb.165.3.972-977.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kealey JT, Gu X, Santi DV. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie. 1994;76:1133–1142. doi: 10.1016/0300-9084(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lew CM, Gralla JD. Escherichia coli pfs transcription: regulation and proposed roles in autoinducer-2 synthesis and purine excretion. J Bacteriol. 2006;188:7457–7463. doi: 10.1128/JB.00868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer G, Heinz DW, Jahn D, Schubert WD. Structure and function of radical SAM enzymes. Curr Opin Chem Biol. 2004;8:468–476. doi: 10.1016/j.cbpa.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Lee JE, Cornell KA, Riscoe MK, Howell PL. Structure of E. coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases. Structure (Camb) 2001;9:941–953. doi: 10.1016/s0969-2126(01)00656-6. [DOI] [PubMed] [Google Scholar]
- Lee JE, Cornell KA, Riscoe MK, Howell PL. Structure of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis. J Biol Chem. 2003;278:8761–8770. doi: 10.1074/jbc.M210836200. [DOI] [PubMed] [Google Scholar]
- Lee JE, Luong W, Huang DJ, Cornell KA, Riscoe MK, Howell PL. Mutational analysis of a nucleosidase involved in quorum-sensing autoinducer-2 biosynthesis. Biochemistry. 2005a;44:11049–11057. doi: 10.1021/bi050493q. [DOI] [PubMed] [Google Scholar]
- Lee JE, Settembre EC, Cornell KA, Riscoe MK, Sufrin JR, Ealick SE, Howell PL. Structural comparison of MTA phosphorylase and MTA/AdoHcy nucleosidase explains substrate preferences and identifies regions exploitable for inhibitor design. Biochemistry. 2004;43:5159–5169. doi: 10.1021/bi035492h. [DOI] [PubMed] [Google Scholar]
- Lee JE, Singh V, Evans GB, Tyler PC, Furneaux RH, Cornell KA, Riscoe MK, Schramm VL, Howell PL. Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. J Biol Chem. 2005b;280:18274–18282. doi: 10.1074/jbc.M414471200. [DOI] [PubMed] [Google Scholar]
- Lee JE, Smith GD, Horvatin C, Huang DJ, Cornell KA, Riscoe MK, Howell PL. Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis. J Mol Biol. 2005c;352:559–574. doi: 10.1016/j.jmb.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Li M, Villaruz AE, Vadyvaloo V, Sturdevant DE, Otto M. AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 2008;8:4. doi: 10.1186/1471-2180-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Anzai Y, Kinoshita K, Kato F, Sherman DH. Functional analysis of MycE and MycF, two O-methyltransferases involved in the biosynthesis of mycinamicin macrolide antibiotics. Chembiochem. 2009;10:1297–1301. doi: 10.1002/cbic.200900088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chu S, Feher VA, Khalili M, Nie Z, Margosiak S, Nikulin V, Levin J, Sprankle KG, Tedder ME, Almassy R, Appelt K, Yager KM. Structure-based design, synthesis, and antimicrobial activity of indazole-derived SAH/MTA nucleosidase inhibitors. J Med Chem. 2003;46:5663–5673. doi: 10.1021/jm0302039. [DOI] [PubMed] [Google Scholar]
- Li Y, Muller R. Non-modular polyketide synthases in myxobacteria. Phytochemistry. 2009;70:1850–1857. doi: 10.1016/j.phytochem.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Lobo SA, Brindley A, Warren MJ, Saraiva LM. Functional characterization of the early steps of tetrapyrrole biosynthesis and modification in Desulfovibrio vulgaris Hildenborough. Biochem J. 2009;420:317–325. doi: 10.1042/BJ20090151. [DOI] [PubMed] [Google Scholar]
- Loenen WA. S-adenosylmethionine: jack of all trades and master of everything? Biochem Soc Trans. 2006;34:330–333. doi: 10.1042/BST20060330. [DOI] [PubMed] [Google Scholar]
- Longshaw AI, Adanitsch F, Gutierrez JA, Evans GB, Tyler PC, Schramm VL. Design and synthesis of potent “sulfur-free” transition state analogue inhibitors of 5′-methylthioadenosine nucleosidase and 5′-methylthioadenosine phosphorylase. J Med Chem. 2010;53:6730–6746. doi: 10.1021/jm100898v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Schramm VL. Ribosyl geometry in the transition state of Streptococcus pneumoniae methylthioadenosine nucleosidase from the 3′-(2)H kinetic isotope effect. J Am Chem Soc. 2008;130:11617–11619. doi: 10.1021/ja804578m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Madden JC, Levin JC, Stein JL, Caparon MG. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol Microbiol. 2001;42:145–157. doi: 10.1046/j.1365-2958.2001.02616.x. [DOI] [PubMed] [Google Scholar]
- Markham GD, Pajares MA. Structure-function relationships in methionine adenosyltransferases. Cell Mol Life Sci. 2009;66:636–648. doi: 10.1007/s00018-008-8516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouni MJ, Sela S. The luxS gene of Streptococcus pyogenes regulates expression of genes that affect internalization by epithelial cells. Infect Immun. 2003;71:5633–5639. doi: 10.1128/IAI.71.10.5633-5639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet A, Bui BT, Florentin D. Biosynthesis of biotin and lipoic acid. Vitam Horm. 2001;61:51–101. doi: 10.1016/s0083-6729(01)61002-1. [DOI] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Nakano C, Ozawa H, Akanuma G, Funa N, Horinouchi S. Biosynthesis of aliphatic polyketides by type III polyketide synthase and methyltransferase in Bacillus subtilis. J Bacteriol. 2009;191:4916–4923. doi: 10.1128/JB.00407-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Hastings JW. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979;43:496–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Hayashi H, Shimizu T. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol Microbiol. 2002;44:171–179. doi: 10.1046/j.1365-2958.2002.02863.x. [DOI] [PubMed] [Google Scholar]
- Pajula RL, Raina A. Methylthioadenosine, a potent inhibitor of spermine synthase from bovine brain. FEBS Letters. 1979;99:343–345. doi: 10.1016/0014-5793(79)80988-6. [DOI] [PubMed] [Google Scholar]
- Parsons JF, Greenhagen BT, Shi K, Calabrese K, Robinson H, Ladner JE. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen N, Cornell KA, Bono JL, Chamberland C, Rosa P, Leong JM. Bgp, a secreted GAG-binding protein of B. burgdorferi strain N40, displays nucleosidase activity and is not essential for infection of immunodeficient mice. Infect and Immun. 2006;74:3016–3020. doi: 10.1128/IAI.74.5.3016-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen N, Leong JM. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:1220–1234. doi: 10.1046/j.1365-2958.2000.01792.x. [DOI] [PubMed] [Google Scholar]
- Pethe K, Bifani P, Drobecq H, Sergheraert C, Debrie AS, Locht C, Menozzi FD. Mycobacterial heparin-binding hemagglutinin and laminin-binding protein share antigenic methyllysines that confer resistance to proteolysis. Proc Natl Acad Sci U S A. 2002;99:10759–10764. doi: 10.1073/pnas.162246899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina A, Tuomi K, Pajula RL. Inhibition of the synthesis of polyamines and macromolecules by 5′-methylthioadenosine and 5′-alkylthiotubercidins in BHK21 cells. Biochem J. 1982;204:697–703. doi: 10.1042/bj2040697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V, Fujiwara N, Porcelli SA, Glickman MS. Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J Exp Med. 2005;201:535–543. doi: 10.1084/jem.20041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich NO, Mashhoon N. Inhibition of EcoRI DNA methylase with cofactor analogs. J Biol Chem. 1990;265:8966–8970. [PubMed] [Google Scholar]
- Reich NO, Mashhoon N. Kinetic mechanism of the EcoRI DNA methyltransferase. Biochemistry. 1991;30:2933–2939. doi: 10.1021/bi00225a029. [DOI] [PubMed] [Google Scholar]
- Riley SP, Bykowski T, Babb K, von Lackum K, Stevenson B. Genetic and physiological characterization of the Borrelia burgdorferi ORF BB0374-pfs-metK-luxS operon. Microbiology. 2007;153:2304–2311. doi: 10.1099/mic.0.2006/004424-0. [DOI] [PubMed] [Google Scholar]
- Riscoe MK, Ferro AJ, Fitchen JH. Methionine salvage as a target for antiprotozoal drug development. Parasitology Today. 1989;5:330–333. doi: 10.1016/0169-4758(89)90128-2. [DOI] [PubMed] [Google Scholar]
- Riscoe MK, Tower PA, Ferro AJ. Mechanism of action of 5′-methylthioadenosine in S49 cells. Biochem Pharmacol. 1984;33:3639–3643. doi: 10.1016/0006-2952(84)90150-3. [DOI] [PubMed] [Google Scholar]
- Roessner CA, Warren MJ, Santander PJ, Atshaves BP, Ozaki S, Stolowich NJ, Iida K, Scott AI. Expression of 9 Salmonella typhimurium enzymes for cobinamide synthesis. Identification of the 11-methyl and 20-methyl transferases of corrin biosynthesis. FEBS Lett. 1992;301:73–78. doi: 10.1016/0014-5793(92)80213-z. [DOI] [PubMed] [Google Scholar]
- Rogers EA, Terekhova D, Zhang HM, Hovis KM, Schwartz I, Marconi RT. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol. 2009;71:1551–1573. doi: 10.1111/j.1365-2958.2009.06621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavetta RD, Thomas CB, Walsh MA, Szegedi S, Joachimiak A, Gumport RI, Churchill ME. Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases. Nucleic Acids Res. 2000;28:3950–3961. doi: 10.1093/nar/28.20.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm VL, Gutierrez JA, Cordovano G, Basu I, Guha C, Belbin TJ, Evans GB, Tyler PC, Furneaux RH. Transition state analogues in quorum sensing and SAM recycling. Nucleic Acids Symp Ser (Oxf) 2008;52:75–76. doi: 10.1093/nass/nrn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AI, Roessner CA, Stolowich NJ, Spencer JB, Min C, Ozaki SI. Biosynthesis of vitamin B12. Discovery of the enzymes for oxidative ring contraction and insertion of the fourth methyl group. FEBS Lett. 1993;331:105–108. doi: 10.1016/0014-5793(93)80306-f. [DOI] [PubMed] [Google Scholar]
- Sekowska A, Denervaud V, Ashida H, Michoud K, Haas D, Yokota A, Danchin A. Bacterial variations on the methionine salvage pathway. BMC Microbiol. 2004;4:9. doi: 10.1186/1471-2180-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewald X, Saum SH, Palm P, Pfeiffer F, Oesterhelt D, Muller V. Autoinducer-2-producing protein LuxS, a novel salt- and chloride-induced protein in the moderately halophilic bacterium Halobacillus halophilus. Appl Environ Microbiol. 2007;73:371–379. doi: 10.1128/AEM.01625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms SA, Subbaramaiah K. The kinetic mechanism of S-adenosyl-L-methionine: glutamylmethyltransferase from Salmonella typhimurium. J Biol Chem. 1991;266:12741–12746. [PubMed] [Google Scholar]
- Singh V, Lee JE, Nunez S, Howell PL, Schramm VL. Transition state structure of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli and its similarity to transition state analogues. Biochemistry. 2005;44:11647–11659. doi: 10.1021/bi050863a. [DOI] [PubMed] [Google Scholar]
- Singh V, Luo M, Brown RL, Norris GE, Schramm VL. Transition-state structure of neisseria meningitides 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. J Am Chem Soc. 2007;129:13831–13833. doi: 10.1021/ja0754204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Schramm VL. Transition-state analysis of S. pneumoniae 5′-methylthioadenosine nucleosidase. J Am Chem Soc. 2007;129:2783–2795. doi: 10.1021/ja065082r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Novotny R, Werz DB, Zarschler K, Seeberger PH, Hofinger A, Kosma P, Schaffer C, Messner P. Molecular basis of S-layer glycoprotein glycan biosynthesis in Geobacillus stearothermophilus. J Biol Chem. 2008;283:21120–21133. doi: 10.1074/jbc.M801833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepkowski T, Brzezinski K, Legocki AB, Jaskolski M, Bena G. Bayesian phylogenetic analysis reveals two-domain topology of S-adenosylhomocysteine hydrolase protein sequences. Mol Phylogenet Evol. 2005;34:15–28. doi: 10.1016/j.ympev.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Babb K. LuxS-mediated quorum sensing in Borrelia burgdorferi, the lyme disease spirochete. Infect Immun. 2002;70:4099–4105. doi: 10.1128/IAI.70.8.4099-4105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storbeck S, Walther J, Muller J, Parmar V, Schiebel HM, Kemken D, Dulcks T, Warren MJ, Layer G. The Pseudomonas aeruginosa nirE gene encodes the S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase required for heme d(1) biosynthesis. FEBS J. 2009;276:5973–5982. doi: 10.1111/j.1742-4658.2009.07306.x. [DOI] [PubMed] [Google Scholar]
- Stroeher UH, Paton AW, Ogunniyi AD, Paton JC. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect Immun. 2003;71:3206–3212. doi: 10.1128/IAI.71.6.3206-3212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sufrin JR, Meshnick SR, Spiess AJ, Garofalo-Hannan J, Pan XQ, Bacchi CJ. Methionine recycling pathways and antimalarial drug design. Antimicrob Agents Chemother. 1995;39:2511–2515. doi: 10.1128/aac.39.11.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Daniel R, Wagner-Dobler I, Zeng AP. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol Biol. 2004;4:36. doi: 10.1186/1471-2148-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Bassler BL. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci U S A. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Bassler BL. Regulation of autoinducer production in Salmonella typhimurium. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta HM, Kim KK. Crystal structure of Streptococcus pneumoniae Sp1610, a putative tRNA methyltransferase, in complex with S-adenosyl-L-methionine. Protein Sci. 2010;19:617–624. doi: 10.1002/pro.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003;50:1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- Taga ME, Semmelhack JL, Bassler BL. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol Microbiol. 2001;42:777–793. doi: 10.1046/j.1365-2958.2001.02669.x. [DOI] [PubMed] [Google Scholar]
- Tedder ME, Nie Z, Margosiak S, Chu S, Feher VA, Almassy R, Appelt K, Yager KM. Structure-based design, synthesis, and antimicrobial activity of purine derived SAH/MTA nucleosidase inhibitors. Bioorg Med Chem Lett. 2004;14:3165–3168. doi: 10.1016/j.bmcl.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria--evolutionary implications. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B, Long KS. Antibiotic Resistance in Bacteria Caused by Modified Nucleosides in 23S Ribosomal RNA. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Landes Bioscience; Austin, Texas: 2009. p. 682. [Google Scholar]
- Wada K, Yamaguchi H, Harada J, Niimi K, Osumi S, Saga Y, Oh-Oka H, Tamiaki H, Fukuyama K. Crystal structures of BchU, a methyltransferase involved in bacteriochlorophyll c biosynthesis, and its complex with S-adenosylhomocysteine: implications for reaction mechanism. J Mol Biol. 2006;360:839–849. doi: 10.1016/j.jmb.2006.05.057. [DOI] [PubMed] [Google Scholar]
- Waditee R, Tanaka Y, Aoki K, Hibino T, Jikuya H, Takano J, Takabe T. Isolation and functional characterization of N-methyltransferases that catalyze betaine synthesis from glycine in a halotolerant photosynthetic organism Aphanothece halophytica. J Biol Chem. 2003;278:4932–4942. doi: 10.1074/jbc.M210970200. [DOI] [PubMed] [Google Scholar]
- Wang L, Li J, March JC, Valdes JJ, Bentley WE. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J Bacteriol. 2005;187:8350–8360. doi: 10.1128/JB.187.24.8350-8360.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Warren MJ, Bolt E, Woodcock SC. 5-Aminolaevulinic acid synthase and uroporphyrinogen methylase: two key control enzymes of tetrapyrrole biosynthesis and modification. Ciba Found Symp. 1994;180:26–40. doi: 10.1002/9780470514535.ch3. discussion 40-29. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Wecksler SR, Stoll S, Tran H, Magnusson OT, Wu SP, King D, Britt RD, Klinman JP. Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry. 2009;48:10151–10161. doi: 10.1021/bi900918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans SC, Bassler BL. Mob psychology. J Bacteriol. 2002;184:873–883. doi: 10.1128/jb.184.4.873-883.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Burgess N, Doherty N, Kirke D, Holden MT, Linforth R, Cornell KA, Taylor AJ, Hill PJ, Williams P. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology. 2002a;148:909–922. doi: 10.1099/00221287-148-4-909. [DOI] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can't talk now - gone to lunch! Curr Opin Microbiol. 2002b;5:216–222. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- Winzer K, Sun YH, Green A, Delory M, Blackley D, Hardie KR, Baldwin TJ, Tang CM. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect Immun. 2002c;70:2245–2248. doi: 10.1128/IAI.70.4.2245-2248.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelinskaya N, Rankin CR, Macmaster R, Savic M, Conn GL. Expression, purification and crystallization of adenosine 1408 aminoglycoside-resistance rRNA methyltransferases for structural studies. Protein Expr Purif. 2010 doi: 10.1016/j.pep.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J. Biosynthesis of bacterial aromatic polyketides. Curr Top Med Chem. 2009;9:1958–1610. doi: 10.2174/156802609789941906. [DOI] [PubMed] [Google Scholar]
- Zhang W, Tang Y. In vitro analysis of type II polyketide synthase. Methods Enzymol. 2009;459:367–393. doi: 10.1016/S0076-6879(09)04616-3. [DOI] [PubMed] [Google Scholar]
- Zhu H, Thuruthyil SJ, Willcox MD. Determination of quorum-sensing signal molecules and virulence factors of Pseudomonas aeruginosa isolates from contact lens-induced microbial keratitis. J Med Microbiol. 2002;51:1063–1070. doi: 10.1099/0022-1317-51-12-1063. [DOI] [PubMed] [Google Scholar]
- Zimmermann K, Engeser M, Blunt JW, Munro MH, Piel J. Pederin-type pathways of uncultivated bacterial symbionts: analysis of o-methyltransferases and generation of a biosynthetic hybrid. J Am Chem Soc. 2009;131:2780–2781. doi: 10.1021/ja808889k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.