Abstract
C to T mutation hotspots in skin cancers occur primarily at methylated CpG sites that coincide with sites of UV-induced cyclobutane pyrimidine dimer (CPD) formation. These mutations are proposed to arise from the insertion of A by DNA polymerase η opposite the T that results from deamination of the methylC (mC) within the CPD. Although the frequency of CPD formation and repair is modestly modulated by its rotational position within a nucleosome, the effect of position on the rate of mC deamination in a CPD has not been previously studied. We now report that deamination of a TmC CPD whose sugar phosphate backbone is positioned against the histone core surface decreases by a factor of 4.7, whereas that of a TmC CPD positioned away from the surface increases by a factor of 8.9 when compared with unbound DNA. Because the mCs undergoing deamination are in similar steric environments, the difference in rate appears to be a consequence of a difference in the flexibility and compression of the two sites due to DNA bending. Considering that formation of the CPD positioned away from the surface is also enhanced by a factor of two, a TmCG site in this position might be expected to have up to an 84-fold higher probability of resulting in a UV-induced mC to T mutation than one positioned against the surface. These results indicate that rotational position may play an important role in the formation of UV-induced C to T mutation hotspots, as well as in the mutagenic mechanism of other DNA lesions.
Keywords: DNA Damage, DNA Methylation, Histones, Mutagenesis Mechanisms, Nucleic Acid Chemistry, Nucleic Acid Structure, Nucleosome, Deamination, Methylcytosine, Photoproduct
Introduction
Sunlight is a major epidemiological factor for the induction of skin cancer. In basal and squamous cell carcinomas, the p53 tumor suppressor gene exhibits a very high percentage of C to T transition mutations at dipyrimidine sites, including the tandem CC to TT mutation (1–4). The UVB wavelengths in sunlight induce the formation of many types of photoproducts at dipyrimidine sites in DNA, the majority of which are cyclobutane pyrimidine dimers (CPDs)2 (5–8). Methylation of C at 5′-PyCG sites further enhances formation of CPDs 15-fold in sunlight (9, 10), and most C to T mutation hotspots occur at methylated 5′-PyCG sites (10–12). It has been found that 5-methylcytosine is involved in 25–40% of sunlight-induced mutations of the cII and lacI transgenes as well as the p53 gene in skin tumors and that CPDs are responsible for a significant fraction of these mutations (6, 13).
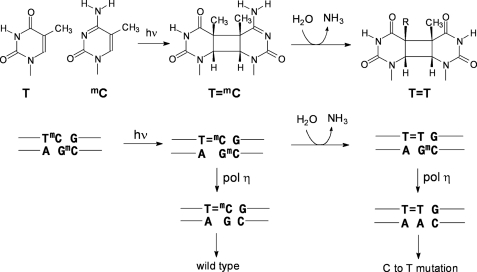
The initially formed cyclobutane pyrimidine dimers are not significantly mutagenic, however, because of the DNA damage bypass polymerase η, which can efficiently bypass T- and C/mC-containing dimers in an essentially error-free manner (14–17). Although the T in a CPD is stable, the C and mC are not, and they deaminate to U and T in a matter of hours or days (18–24) (Fig. 1), unlike their canonical forms, which deaminate with a half-life of about 50,000 years (25, 26). Deamination of Cs or mCs in CPDs is highly mutagenic because polymerase η will faithfully insert A opposite the resulting Us or Ts, thereby producing the observed C to T and CC to TT mutations (the deamination-bypass mechanism) (3, 4, 27–29). The frequency of C to T and CC to TT mutations will depend, however, on the rate of formation of C-containing dimers, their rate of repair, deamination, and bypass by polymerases. All of these processes are expected to be modulated by sequence context, protein interactions, as well as the secondary and tertiary structure of DNA. A detailed understanding of all these processes and interactions may lead to a better understanding of the origin of UV mutation hotspots.
FIGURE 1.
Mutagenic properties of mC-containing cis-syn-cyclobutane dimers and their deamination products. Upper, UVB irradiation of a TmC site leading to a cis-syn-T=mC CPD followed by deamination to give the cis-syn-cyclobutane dimer of T=T. Lower, mutagenic consequences of bypass of a T=mC CPD and its deamination product, T=T by polymerase η (pol η).
Nucleosomes are the primary structural unit of chromatin in eukaryotic cells (30). Nucleosome core particles contain about 150 bp of DNA, which wrap 1.7 times around a histone octamer, made up of two H2A, H2B, H3, and H4 histones (31). UV preferentially induces the formation of CPDs in nucleosomes at sites where the phosphodiester backbone is positioned away from the histone surface and DNA bending is toward the major groove (32, 33). This preference is also seen for bent DNA that is not in contact with a protein (34) and has been attributed to the greater degree of rotational freedom in the phosphate backbone, making it easier to for adjacent pyrimidines to adopt a photoreactive conformation (35, 36). When DNA containing randomly distributed CPDs is assembled into nucleosomes, the CPDs also favor positions away from the surface (37), which is consistent with the 30° bend that they make toward the major grove of DNA (38). Despite the distortion of DNA caused by CPDs, nucleosome core particles containing CPDs in different rotational settings can be readily prepared and isolated (39–41).
Nucleosomes have also been found to affect the repair of CPDs (42). In vitro studies with human fibroblast extracts found that nucleosomes reduce the rate of nucleotide excision repair of CPDs by 6–9-fold relative to free DNA (43). A similar 10-fold reduction in excision repair has been observed for a site-specific CPD reconstituted with human nucleosome core particles (44). With Xenopus nuclear extracts, the nucleotide excision repair rates of rotationally positioned CPDs were only 2–3 times lower in nucleosomes and no more than 1.5-fold greater when a CPD was positioned away from the histone surface when compared with against the surface (41). In an earlier study with a 5S rRNA gene, no correlation was observed with rotational positioning of the CPD (45).
Although the effects of nucleosome positioning on the rates of CPD formation and repair have been studied, the effects on the deamination rates of C- and mC-containing CPDs have not. The purpose of this study was to determine the extent to which the rotational position of the CPD relative to the histone surface affects the rate of mC to T deamination, and hence, its potential mutagenicity by a deamination bypass mechanism. To address this question, we determined the deamination rate of a TmCG CPD in two different orientations in a nucleosome core particle. We find that a TmC CPD positioned against the surface deaminates 4.7 times slower than the unbound sequence, whereas a TmC CPD facing out deaminates 8.9 times faster than the unbound sequence, corresponding to an overall 42-fold difference in rate.
EXPERIMENTAL PROCEDURES
DNA Substrates
Oligodeoxynucleotides with or without 5′-terminal phosphates were purchased from Integrated DNA Technologies and purified by denaturing gel electrophoresis prior to ligation with T4 DNA ligase and ATP in the presence of complementary 20-mer ligation scaffolds (supplemental Figs. S1 and S2). The 150-mer single strand products were purified by denaturing PAGE. Complementary 150-mers were then annealed to form the 150-mer duplexes and purified by native PAGE (supplemental Fig. S3).
Nucleosome Reconstitution
Nucleosome core particles were isolated and purified from chicken erythrocytes following a detailed procedure provided by the Dr. Michael Smerdon laboratory. Each 150-mer DNA duplex (ds-IN, ds-OUT, and ds-control) was reconstituted with the chicken nucleosome core particles by slow dialysis from high to low salt as described previously (46). Briefly, about 10 nm 150-mer duplex was incubated with an increasing amount of nucleosome core particles (from 100 to 500 nm) in a total volume of 500 μl, containing 2 m NaCl, 10 mm Tris-HCl, 5 mm EDTA at pH 7.5 and room temperature for 2 h, and then dialyzed against 50 mm NaCl, 10 mm Tris-HCl, pH 7.5, at 4 °C, overnight. Finally, the reconstituted particles were recovered from the dialysis tubing and equilibrated at 55 °C for 2 h to fix the nucleosome phasing. The reconstituted particles were assayed by native PAGE (6% acrylamide, 0.2% bisacrylamide in TBE), and the ratio of nucleosome-bound DNA to free DNA was quantified by the Quantity One software (supplemental Fig. S4).
Hydroxyl Radical Footprinting and Dimethylsulfate Mapping
Hydroxyl radical footprinting was performed as described previously (46). Briefly, a 15-μl aliquot of 10 mm sodium ascorbate, a 15-μl aliquot containing 1 mm Fe(NH4)2(SO4)2·6H2O and 2 mm EDTA, and 15 μl of a 0.12% (w/w) H2O2 solution were premixed and added within 5 s to 105 μl of the nucleosome-bound DNA sample. The reaction was incubated for 120 s at room temperature and stopped by the addition of 16 μl of 50% (v/v) glycerol and 4 μl of a 500 mm EDTA solution. The samples were electrophoresed on a native gel (6% acrylamide, 0.2% bisacrylamide in TBE), and the nucleosome bands were electroeluted in TBE. The proteins were extracted with phenol:chloroform:isopropyl alcohol 25:24:1, and the DNA was precipitated with ethanol. The free ds-control was treated in a similar way, except that the reaction was quenched with a solution containing 1 m sodium acetate, 120 mm thiourea, 300 μg/ml salmon sperm DNA, and 60 mm EDTA at pH 6.5 and then ethanol-precipitated. A Maxam-Gilbert G sequencing reaction was also carried out on the free ds-control in 50 mm cacodylate, 50 mm NaCl, 5 mm EDTA in the presence of 10 nm ds-control. For a 50-μl reaction, 0.5 μl of dimethyl sulfate was added to initiate the reaction, and 10-μl aliquots were removed over time and quenched by the addition of 50 μl of 1.5 m sodium acetate, 1 m mercaptoethanol and 50 μg of denatured salmon sperm DNA. The samples were ethanol-precipitated twice, and the resulting pellets were vacuum-dried and then solubilized in 100 μl of 1 m piperidine. The samples were then heated at 90 °C in 1 m piperidine for 30 min and then evaporated to dryness at 60 °C.
Deamination Rate Assay by Two-dimensional Gel Electrophoresis
The deamination rate was determined by adapting a previously described method (24). The free and nucleosome-bound internally 32P-labeled 150-mer ds-IN and ds-OUT were irradiated with 302 nm UVB light at 4 °C for 1 h and then adjusted to pH 7.2 with Mes buffer (0.5 m) and incubated at 37 °C. Aliquots (10-μl) were removed at various times and quickly frozen on dry ice before storing overnight at −70 °C. The remaining sample was adjusted to pH 6.5 with Mes buffer (0.5 m) and heated at 67 °C overnight to complete the deamination. The aliquots were then warmed to room temperature, extracted with phenol:chloroform:isopropyl alcohol 25:24:1, ethanol-precipitated, redissolved in buffer, and photoreverted with photolyase and 365 nm light for 1 h. After photoreversion, the aliquots were treated with nuclease P1 to degrade the DNA to mononucleotides containing either 32P-mdC or 32P-T, depending on the extent of deamination, and separated by two-dimensional gel electrophoresis. In the first dimension, electrophoresis on a 7 m urea, a TBE gel was used to separate 32P-mdC and 32P-T, which co-migrate, from partially digested material and protein. For the second dimension, the gel surrounding the radioactive band containing the mononucleotides was excised, and a second gel containing 25 mm citric acid, pH 3.5, and 7 m urea was poured around the remaining gel slice. Electrophoresis on this gel separated 32P-mdC from 32P-T with the 32P-T migrating the fastest. The deamination rate constant was obtained from the slope of a linear least squares fit of the log of the fraction of remaining TmC CPD versus deamination time (supplemental Figs. S5 and S6). The fraction of TmC CPD remaining was calculated as 1 − (T/(T + mC))/(T∞/(T∞ + mC)) where T∞/(T∞ + mC∞) is the fraction T/(T + mC) in the fully deaminated sample. The yield of CPD photoproduct was calculated as the T∞/(T∞ + mC∞) − T0/(T0 + mC0), where T0/(T0 + mC0) is the fraction T/(T + mC) at time 0.
RESULTS
Design and Synthesis of the DNA Substrates
The substrate for study was adapted from a previously described 150-mer sequence that was shown by hydroxyl radical footprinting to position a TT CPD near the dyad axis with an inside (IN) or outside (OUT) orientation relative to the histone core surface (41). The CPDs were oriented by flanking the dimer-containing sequence with multiple TG motifs (T/A)3NN(G/C)3 that had been shown to position a glucocorticoid hormone-response element (GRE) with different orientations relative to the histone surface (47, 48). To study the effect of the CPD position on the deamination of cyclobutane pyrimidine dimers of TmCG sites, we simply replaced the ATTA (OUT) and GTTC (IN) CPD sites in the original 150-mer sequence with ATmCG and GTmCG sites respectively (see Fig. 3, top right).3
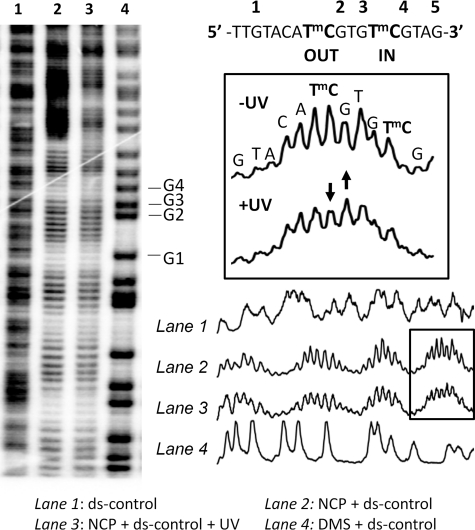
FIGURE 3.
Hydroxyl radical footprinting of the reconstituted nucleosome core particles to determine phasing. The nucleosome core particle reconstituted with 5′-end-labeled 150-mer duplex DNA was subjected to hydroxyl radical footprinting and electrophoresed on a 7 m urea, 10% acrylamide, 0.3% bisacrylamide denaturing gel. Lane 1, footprinting of the free DNA duplex; lane 2, footprinting of the nucleosome-bound DNA duplex; lane 3, footprinting of the nucleosome-bound duplex DNA after 1 h of irradiation at 302 nm at 4 °C; lane 4, Maxam Gilbert G reaction on the free duplex. DMS, dimethyl sulfate.
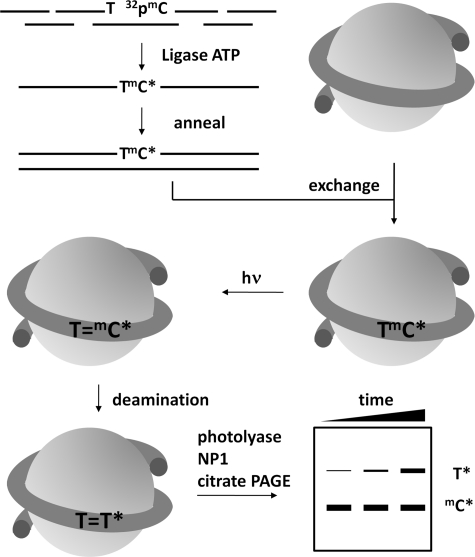
To determine the deamination rates of the TmC CPDs, we used a previously developed method that requires the mC to be 5′-32P-end-labeled (Fig. 2) (24). We therefore prepared the two internally 32P-labeled duplex 150-mers, ds-IN and ds-OUT, along with a 5′-end-labeled control duplex, ds-control. The duplexes were prepared by annealing complementary single strand 150-mers that were prepared by ligating four ODNs together with T4 ligase and ATP in the presence of complementary ligation scaffolds (Fig. 2). For ds-IN and ds-OUT, the second and third ODNs of the top strand were designed so that the mC of interest would be at the 5′-end of the third fragment so that the mC could be 5′-32P-end-labeled prior to ligation (supplemental Figs. S1 and S2). Thus, ds-IN was 32P-labeled at the mC of the GTmCG site, ds-OUT was labeled at the mC of the ATmCG site, and ds-control was labeled at the 5′-end of the 150-mer of the top strand. Each single strand 150-mer was purified by denaturing PAGE, and the final 150-mer duplexes were purified by native PAGE (supplemental Fig. S3).
FIGURE 2.
Strategy for determining the deamination rates of TmCG CPDs in a nucleosome core particle. A 150-mer DNA duplex was constructed by first assembling each strand from four ODNs by ligation in the presence of three complementary 20-mer ligation scaffolds where the ODN containing the mC site of interest was 5′-end-labeled with [γ-32P]ATP and kinase prior to ligation. The internally labeled 150-mer duplex was assembled into isolated nucleosome core particles through an exchange reaction, irradiated with UVB light, and allowed to deaminate. At specific times, the protein was removed by phenol extraction, and the cis-syn-CPDs were photoreverted with photolyase, degraded with nuclease P1, subjected to two-dimensional gel electrophoresis to separate the radiolabeled T from undeaminated radiolabeled mdC, and quantified by radioisotopic imaging analysis.
Nucleosome Core Particle Reconstitution with the 150-mer DNA Duplexes
The 150-mer duplexes, ds-IN, ds-OUT, and ds-control, were assembled into nucleosome core particles according to a previously described procedure that involves exchanging the DNA with that from chicken erythrocyte nucleosome core particles (NCPs) (39). The 150-mer duplexes were titrated with the NCPs and electrophoresed on a native gel to determine the NCP concentration needed to achieve maximal incorporation of the DNA into the NCP (supplemental Fig. S4). We found that about 90% of 10 nm 150-mer could be incorporated into 500 nm nucleosome core particles.
Orientation of the Two TmC Sites on the Nucleosome Core Particle
To verify the IN and OUT positions of the GTmCG and ATmCG sites, the nucleosomes were analyzed by hydroxyl radical footprinting. The hydroxyl radical cleavage intensity on the nucleosome-bound ds-control with and without UVB irradiation exhibited very pronounced 10–11-bp periodicity with the same phasing as described previously for the ATTA and GTTC sites within the same 150-mer sequence (41) (Fig. 3, lanes 2 and 3) when compared with the free ds-control (Fig. 3, lane 1). The cleavage sites were mapped onto the sequence by alignment with the Maxam Gilbert G sequencing reaction bands (Fig. 3, lane 4). Hydroxyl radicals primarily attack the H5′5″ and H4′ hydrogens that are present on the minor groove side of the DNA (49). The mC of the ATmCG CPD site is in the center of a region of maximal cleavage, indicating that its phosphodiester backbone is facing out (Fig. 3, boxed section). The mC of the GTmCG CPD site, which is half a turn from the first site, is at a site of minimal cleavage, indicating that its backbone is facing toward the histone surface.
The similar hydroxyl radical cleavage pattern in the presence and absence of UVB irradiation suggests that the UVB photoproducts do not disrupt the phasing. Furthermore, the decrease of the intensity of the mC band and increase in that of the flanking G at the ATmCG site following UVB radiation is consistent with the formation of a significant amount of CPD photoproduct (Fig. 3, boxed section). It has been previously observed that hydroxyl radical cleavage is suppressed at the 3′-T of a TT CPD and increased at the nucleotide immediately following the 3′-T (41).
Deamination Rates of the Two TmC CPDs
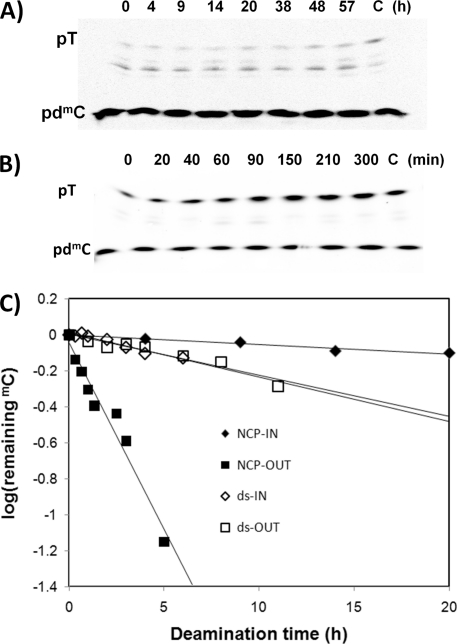
The deamination rates for the IN and OUT TmC CPDs when compared with free DNA were determined by following the conversion of 32P-mdC to 32P-T in the dimer by an enzyme-coupled two-dimensional gel electrophoresis assay (24). In the first step, free or NCP complexed ds-IN and ds-OUT were irradiated with 302 nm light to produce the cis-syn-cyclobutane pyrimidine dimers of the ATmCG and GTmCG sites, along with other photoproducts, at 4 °C to suppress deamination. The samples were then incubated at 37 °C and pH 7.2 to allow for deamination for various times, and one was made to undergo complete deamination by lowering the pH and heating. The aliquots were then incubated with Escherichia coli photolyase and visible light to specifically photorevert the cis-syn-CPDs and then treated with nuclease P1 to degrade all of the undamaged and photoreverted DNA to mononucleotides. Non-photorevertible dipyrimidine photoproducts, such as (6-4) and Dewar photoproducts, would only be digested to trinucleotides. The mononucleotides were then isolated by electrophoresis on a denaturing gel and subjected to a second electrophoresis on a pH 3.5 citrate gel (Fig. 4, A and B) that separates the 32P-T from the 32P-mdC and quantified by radioisotopic imaging analysis.
FIGURE 4.
Deamination of the T=mC CPDs in the free and nucleosome-bound 150-mer DNA duplexes. Free or nucleosome-reconstituted 150-mer DNAs with internal 5′-32P-mdC labeling were irradiated for 1 h with 302 nm light and then allowed to deaminate for the indicated times in hours, after which they were phenol-extracted, photoreverted with E. coli photolyase, and then degraded with nuclease P1. The degradation products were first run on a 7 m urea, 10% acrylamide, 0.3% bisacrylamide gel in TBE (first dimension, downward direction) after which the band containing primarily mononucleotides was run on a 25% acrylamide, 0.8% bisacrylamide gel in 25 mm citric acid, pH 3.5 (second dimension, upward direction). A, representative second dimension gel of nucleosome-bound 150-mer ds-IN. B, representative second dimension gel of nucleosome-bound 150-mer ds-OUT. C, plots of log (fraction of TmC CPD remaining) as a function of different deamination times for the gels shown in A and B and for the free 150-mer ds substrates, after correcting for background 32P-T. Plots for other sets of independent measurements are given in supplemental Figs. S5 and S6. The intermediate bands in the gels shown in A and B are due to non-photorevertible photoproduct-containing trinucleotides and incomplete digestion products and do not significantly affect the rate measurements.
The rate constants for deamination were then determined from the slopes of lines fit to the log of the fraction of remaining TmC CPD (supplemental Figs. S5 and S6) as described under “Experimental Procedures” for multiple experiments and then averaged. The photoproduct yields were determined from the increase in the initial amount of radiolabeled T following complete deamination. The initial amount of T was non-zero in many cases and could be attributed to unintended labeling of the T at the 5′-end of the 150-mer resulting from incomplete heat deactivation of the kinase used to label the mC-containing ODN prior to ligation. Table 1 shows the deamination half-lives and yields of the CPDs in the IN and OUT positions of free and nucleosome-bound DNA. When compared with the free DNA, the nucleosome decreases the rate of deamination of the IN CPD by a factor of 4.7, whereas it increases the rate of deamination of the OUT CPD by a factor of 8.9. We also determined that although the nucleosome did not affect the efficiency of forming the inside CPD, it enhanced the formation of the outside CPD by a factor of two.
TABLE 1.
Yields and deamination half lives of the TmC CPDs in free and nucleosome-bound DNA (37 °C, 50 m NaCl)
| Photoproduct yielda |
Deamination half-livesb |
|||
|---|---|---|---|---|
| ds-IN (%) | ds-OUT (%) | ds-IN t½ | ds-OUT t½ | |
| % | h | |||
| Free | 5.5, 5.5 | 13, 12 | 12.2 ± 0.9 h | 14.0 ± 1.3 h |
| Nucleosome | 6.0 ± 0.5 | 26 ± 4 | 57.8 ± 5.1 h | 1.57 ± 0.13 h |
| -Fold change | 1.1 | 2.1 | 4.7 | 0.11 (1/8.9) |
a Photoproduct yields for free DNA are from two independent experiments (each shown), and for nucleosome DNA, yields are the average of three independent experiments with the standard deviations shown.
b Deamination half-lives were the average of those calculated from linear least squares fits to two (free DNA) or three (nucleosome DNA) independent sets of deamination data (7–9 time points each), with the error shown derived from propagation of the standard deviations for each fit (see supplemental Figs. S5 and S6).
DISCUSSION
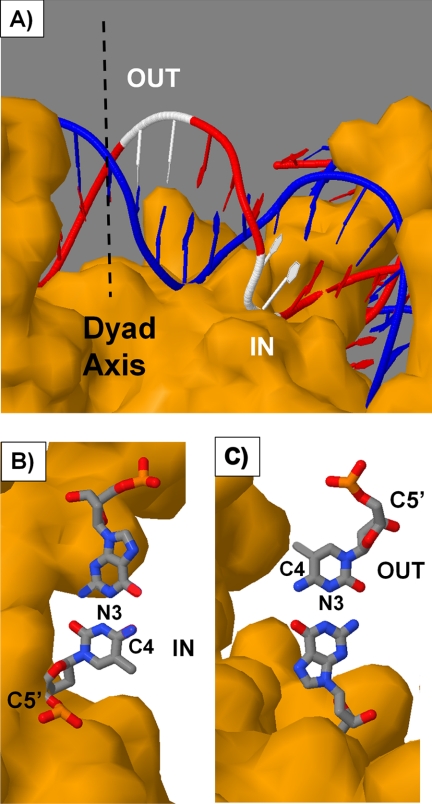
The aim of this study was to determine whether or not the rotational position of a cis-syn-cyclobutane dimer of a TmCG site on a nucleosome would affect its deamination rate and thereby contribute to its relative mutagenicity via a deamination bypass mechanism. Two positions were studied, one with the phosphodiester backbone of the CPD against the histone core surface (IN) and one with the backbone away from the surface (OUT). These positions can be mapped onto a crystal structure of the nucleosome core particle as shown in Fig. 5A. Because the outside CPD site precedes the inside CPD site when going in the 5′- to 3′-direction, the two sites must lie to one side of the nucleosome pseudo dyad axis.
FIGURE 5.
Positions of the TmCG sites on the nucleosome. A, nucleosome core particle structure highlighting in white the positions of the inside and outside TmCG CPD sites that are consistent with the hydroxyl radical footprinting data. B and C, orientation of the mC·G base pair for the inside (B) and outside (C) CPD sites. The structure was rendered from 1KX5.pdb (50) with Jmol using T1C2 and C6T7 of chain J to represent the outside and inside CPD sites with editing to produce the mCG base pair. The pseudo dyad axis is shown as a dashed line. It is also possible that the sites are displaced translationally by 10 bp to the other side of the dyad axis, but if so, they would only be present in minor amounts.
The photoreactivity of the two sites in free DNA (Table 1) was consistent with an earlier study of ours showing that an ATmCG site is about 2-fold more photoreactive than a GTmCG site (5.4 versus 12% yield) due to the quenching effect of flanking Gs (24). Complexing the DNA to the histone core particle did not affect the photoreactivity of the inside GTmCG site but enhanced the photoreactivity of the outside ATmCG site 2-fold to give a 26% yield of the CPD when compared with 5.5% for the inside site. The enhanced photoreactivity of the outside position is in accord with the higher photoreactivity previously noted for dipyrimidine sites positioned away from the histone surface (32, 33).
In contrast to their photoreactivity, the deamination rates of the two CPD sites in the nucleosome were dramatically different from those in the free DNA, with the CPD positioned inside deaminating 4.7 times slower and the CPD positioned outside deaminating 8.9 times faster (Table 1). The relative reactivity of the two CPDs toward deamination parallels the relative reactivity of the two CPD sites to hydroxyl radical cleavage, but we believe for a different reason. Although hydroxyl radical cleavage of DNA results mainly from initial abstraction of the hydrogens on the sugar backbone of DNA (49), deamination of an mC in a cis-syn-dimer involves attack of water on the C4 carbon (20), which lies in the major groove of the DNA.
Hydroxyl radical cleavage mainly involves attack at the C5′ position and 2–5-fold less at the other sugar sites in the order C4′ > C3′ > C2′ > C1′ (49). Maximum inhibition occurs when these hydrogens face toward the histone core surface, which sterically blocks the approach of the hydroxyl radical. Conversely, maximum cleavage is observed when the sugar hydrogens face out toward the solvent. When hydroxyl radical cleavage at a CPD is minimal (inside position), the sugar phosphate backbone faces the histone surface, but the C4 carbon of the mC faces in a direction parallel with the surface (Fig. 5B). On the other hand, when hydroxyl radical cleavage at the CPD is maximal (outside position), the sugar phosphate backbone faces away from the histone surface, but the C4 position of the mC faces in a direction parallel with the surface but in an opposite direction when compared with the CPD in the inside position (Fig. 5C).
Analysis of the crystal structure of a nucleosome core particle shows that the C4 position of what would be the mC of both the IN and the OUT CPDs is unobstructed by protein within a radius of 8.5 Å (1KX5.pdb) (50). The same would be expected to hold true for the CPDs as the 3′-pyrimidine of a CPD has been found to adopt roughly the same position as it does in the undamaged DNA (38). Thus, the C4 position of the mC of both the inside and the outside CPDs would appear to be in a similar, unobstructed environment, suggesting that factors other than steric interference by the histone proteins must play a role in inhibiting or enhancing deamination of the CPDs.
Previous studies of the deamination of C have found that deamination can be both inhibited and enhanced by protein binding. In one case, the α/β-type small, acid-soluble proteins of Bacillus subtilis spores have been found to suppress deamination by as much as 10-fold (51). It was suggested that the protein might be inhibiting deamination by a variety of means, such as restricting the “breathing” of the DNA, excluding water from the DNA, or enforcing an A conformation on the DNA. In a subsequent study, a restriction enzyme was found to both suppress and enhance deamination of Cs within its binding site by 7- and 15-fold, respectively (52). The large enhancement was proposed to arise from enzyme-mediated flipping of one of the Cs out of the helix. In addition, we recently showed that the methyl CpG binding domain of MeCP2 drastically inhibited deamination of a TmCG CPD, most probably by restricting attack of water on the CPD (53).
In the case of the nucleosome, it may be that the same factors that increase CPD formation at outside positions also increase the deamination rate. It has been shown that CPD dimers form preferentially in outside positions in protein-free DNA loops, demonstrating that DNA curvature rather than protein-DNA contacts is controlling reactivity (34). It was suggested that the inside positions of curved DNA are more compressed and less mobile, whereas the outside positions are less compressed and more mobile. Thus, although the C4 positions of the mC in the inside and outside CPDs may be in similar steric environments, the increased flexibility and/or more open conformation of the outside position may facilitate protonation of N3 and/or attack by water at C4. Conversely, protonation of N3 or attack of water on C4 might be inhibited by restricted movements and the compressed nature of an inside CPD site. It is interesting to note that the observed rate of deamination of the outside CPD is very similar to if not faster than what we observed for a CPD with the same flanking bases in single strand DNA (t½ = 1.57 versus 3.5 h). This could indicate that CPD might be able to flip out of the helix or is being held in a favorable conformation for deamination. A more detailed study of CPDs in different rotational and translational positions will be required to sort out these effects.
The 8.9-fold increase in the rate of deamination of an outside TmCG dimer when compared with a 4.7-fold decrease for the inside dimer corresponds to an overall 42-fold difference in rate. When one couples the difference in deamination rate with a 2-fold enhancement of outside CPD formation, there would appear to be an 84-fold higher propensity for a UVB-induced mC to T mutation at an outside TmCG site when compared with an inside site. Given that little difference has been observed in excision repair rates between inside and outside TT CPDs (41), the large difference in deamination rates could explain, at least in part, the origin of UV mutation hotspots and coldspots in phased nucleosomes that would arise following polymerase η bypass. On the other hand, deamination of an mC-containing CPD will result in a T·G mismatch, which has been shown to destabilize the DNA duplex by 0.7 kcal/mol (54). This duplex destabilization may further destabilize the nucleosome, which has already been shown to be destabilized by 0.14–0.24 kcal/mol by a TT CPD (41), and facilitate recognition by histone modification and/or excision repair systems. The extent of nucleosome destabilization, and hence recognition, may depend, however, on the rotational position of the mismatch. Deaminated C-containing CPDs have been shown to be much more readily detected and repaired than TT CPDs (55, 56).
CONCLUSION
We have found that the rotational position of a TmCG CPD in a nucleosome greatly affects its deamination rate, and this may explain at least in part the origin of UV mutation hotspots and coldspots in phased nucleosomes. Deamination of CPDs may also be accelerated in unphased nucleosomes if the CPDs are in dynamic exchange with outside positions. It remains to been seen how the nucleosome affects the deamination rate of CmCG CPDs, which are much more slowly deaminated than TmCG CPDs in free DNA (24), as well as the deamination of (6-4) and Dewar photoproducts, which also form at this site (22). Rotational position is also expected to affect the spontaneous deamination of C and mC and their more readily deaminated oxidized products (57), as well as the chemistry of many other bases and adducts.
Supplementary Material
Acknowledgments
We thank Aziz Sancar for a gift of photolyase, Stephen Lloyd for a gift of T4 endonuclease V, and Michael Smerdon for a detailed protocol for preparation of the nucleosome core particles. We also thank John Hinz and Zeljko Svedruzic for helpful discussions on the design of the substrates.
This work was supported, in whole or in part, by National Institutes of Health Grant CA40463 (to J.-S. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
The IN and OUT substrates were mislabeled in the materials and methods section of the original publication (41), as described in a recent erratum statement.
- CPD
- cyclobutane pyrimidine dimer
- mC
- methylC
- dC
- deoxycytidine
- ODN
- oligodeoxynucleotide
- NCP
- nucleosome core particle
- TBE
- Tris-borate-EDTA.
REFERENCES
- 1. Brash D. E., Rudolph J. A., Simon J. A., Lin A., McKenna G. J., Baden H. P., Halperin A. J., Pontén J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10124–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ziegler A., Leffell D. J., Kunala S., Sharma H. W., Gailani M., Simon J. A., Halperin A. J., Baden H. P., Shapiro P. E., Bale A. E., Brash D. E. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4216–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giglia-Mari G., Sarasin A. (2003) Hum. Mutat. 21, 217–228 [DOI] [PubMed] [Google Scholar]
- 4. Pfeifer G. P., You Y. H., Besaratinia A. (2005) Mutat. Res. 571, 19–31 [DOI] [PubMed] [Google Scholar]
- 5. Yoon J. H., Lee C. S., O'Connor T. R., Yasui A., Pfeifer G. P. (2000) J. Mol. Biol. 299, 681–693 [DOI] [PubMed] [Google Scholar]
- 6. You Y. H., Pfeifer G. P. (2001) J. Mol. Biol. 305, 389–399 [DOI] [PubMed] [Google Scholar]
- 7. Cadet J., Sage E., Douki T. (2005) Mutat. Res. 571, 3–17 [DOI] [PubMed] [Google Scholar]
- 8. Douki T., Cadet J. (2001) Biochemistry 40, 2495–2501 [DOI] [PubMed] [Google Scholar]
- 9. Tommasi S., Denissenko M. F., Pfeifer G. P. (1997) Cancer Res. 57, 4727–4730 [PubMed] [Google Scholar]
- 10. Drouin R., Therrien J. P. (1997) Photochem. Photobiol. 66, 719–726 [DOI] [PubMed] [Google Scholar]
- 11. You Y. H., Li C., Pfeifer G. P. (1999) J. Mol. Biol. 293, 493–503 [DOI] [PubMed] [Google Scholar]
- 12. You Y. H., Szabó P. E., Pfeifer G. P. (2000) Carcinogenesis 21, 2113–2117 [DOI] [PubMed] [Google Scholar]
- 13. You Y. H., Lee D. H., Yoon J. H., Nakajima S., Yasui A., Pfeifer G. P. (2001) J. Biol. Chem. 276, 44688–44694 [DOI] [PubMed] [Google Scholar]
- 14. Washington M. T., Johnson R. E., Prakash S., Prakash L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3094–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu S. L., Johnson R. E., Prakash S., Prakash L. (2001) Mol. Cell. Biol. 21, 185–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCulloch S. D., Kokoska R. J., Masutani C., Iwai S., Hanaoka F., Kunkel T. A. (2004) Nature 428, 97–100 [DOI] [PubMed] [Google Scholar]
- 17. Vu B., Cannistraro V. J., Sun L., Taylor J. S. (2006) Biochemistry 45, 9327–9335 [DOI] [PubMed] [Google Scholar]
- 18. Setlow R. B. (1966) Science 153, 379–386 [DOI] [PubMed] [Google Scholar]
- 19. Fix D., Bockrath R. (1981) Mol. Gen. Genet. 182, 7–11 [DOI] [PubMed] [Google Scholar]
- 20. Lemaire D. G., Ruzsicska B. P. (1993) Biochemistry 32, 2525–2533 [DOI] [PubMed] [Google Scholar]
- 21. Tu Y., Dammann R., Pfeifer G. P. (1998) J. Mol. Biol. 284, 297–311 [DOI] [PubMed] [Google Scholar]
- 22. Douki T., Cadet J. (1994) Biochemistry 33, 11942–11950 [DOI] [PubMed] [Google Scholar]
- 23. Celewicz L., Mayer M., Shetlar M. D. (2005) Photochem. Photobiol. 81, 404–418 [DOI] [PubMed] [Google Scholar]
- 24. Cannistraro V. J., Taylor J. S. (2009) J. Mol. Biol. 392, 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frederico L. A., Kunkel T. A., Shaw B. R. (1990) Biochemistry 29, 2532–2537 [DOI] [PubMed] [Google Scholar]
- 26. Shen J. C., Rideout W. M., 3rd, Jones P. A. (1994) Nucleic. Acids. Res. 22, 972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taylor J. S., O'Day C. L. (1990) Biochemistry 29, 1624–1632 [DOI] [PubMed] [Google Scholar]
- 28. Jiang N., Taylor J. S. (1993) Biochemistry 32, 472–481 [DOI] [PubMed] [Google Scholar]
- 29. Lee D. H., Pfeifer G. P. (2003) J. Biol. Chem. 278, 10314–10321 [DOI] [PubMed] [Google Scholar]
- 30. Luger K. (2006) Chromosome Res. 14, 5–16 [DOI] [PubMed] [Google Scholar]
- 31. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 32. Gale J. M., Nissen K. A., Smerdon M. J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6644–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pehrson J. R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 9149–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pehrson J. R., Cohen L. H. (1992) Nucleic. Acids Res. 20, 1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Becker M. M., Wang Z. (1989) J. Mol. Biol. 210, 429–438 [DOI] [PubMed] [Google Scholar]
- 36. Pfeifer G. P. (1997) Photochem. Photobiol. 65, 270–283 [DOI] [PubMed] [Google Scholar]
- 37. Suquet C., Smerdon M. J. (1993) J. Biol. Chem. 268, 23755–23757 [PubMed] [Google Scholar]
- 38. Park H., Zhang K., Ren Y., Nadji S., Sinha N., Taylor J. S., Kang C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15965–15970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kosmoski J. V., Smerdon M. J. (1999) Biochemistry 38, 9485–9494 [DOI] [PubMed] [Google Scholar]
- 40. Kosmoski J. V., Ackerman E. J., Smerdon M. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10113–10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Svedruzić Z. M., Wang C., Kosmoski J. V., Smerdon M. J. (2005) J. Biol. Chem. 280, 40051–40057; Correction (2010) J. Biol. Chem.285, 39574 [DOI] [PubMed] [Google Scholar]
- 42. Thoma F. (2005) DNA Repair 4, 855–869 [DOI] [PubMed] [Google Scholar]
- 43. Wang Z. G., Wu X. H., Friedberg E. C. (1991) J. Biol. Chem. 266, 22472–22478 [PubMed] [Google Scholar]
- 44. Hara R., Sancar A. (2003) Mol. Cell. Biol. 23, 4121–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu X., Smerdon M. J. (2000) J. Biol. Chem. 275, 23729–23735 [DOI] [PubMed] [Google Scholar]
- 46. Ober M., Lippard S. J. (2007) J. Am. Chem. Soc. 129, 6278–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Q., Wrange O. (1993) Genes Dev. 7, 2471–2482 [DOI] [PubMed] [Google Scholar]
- 48. Li Q., Wrange O. (1995) Mol. Cell. Biol. 15, 4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Balasubramanian B., Pogozelski W. K., Tullius T. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9738–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J. (2002) J. Mol. Biol. 319, 1097–1113 [DOI] [PubMed] [Google Scholar]
- 51. Sohail A., Hayes C. S., Divvela P., Setlow P., Bhagwat A. S. (2002) Biochemistry 41, 11325–11330 [DOI] [PubMed] [Google Scholar]
- 52. Carpenter M., Divvela P., Pingoud V., Bujnicki J., Bhagwat A. S. (2006) Nucleic Acids Res. 34, 3762–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cannistraro V. J., Taylor J. S. (2010) Nucleic. Acids. Res. 38, 6943–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jing Y., Kao J. F., Taylor J. S. (1998) Nucleic. Acids. Res. 26, 3845–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mu D., Tursun M., Duckett D. R., Drummond J. T., Modrich P., Sancar A. (1997) Mol. Cell. Biol. 17, 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sugasawa K., Okamoto T., Shimizu Y., Masutani C., Iwai S., Hanaoka F. (2001) Genes Dev. 15, 507–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kreutzer D. A., Essigmann J. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3578–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.