Abstract
The catabolite repressor/activator (Cra) protein is a global sensor and regulator of carbon fluxes through the central metabolic pathways of Gram-negative bacteria. To examine the nature of the effector (or effectors) that signal such fluxes to the protein of Pseudomonas putida, the Cra factor of this soil microorganism has been purified and characterized and its three-dimensional structure determined. Analytical ultracentrifugation, gel filtration, and mobility shift assays showed that the effector-free Cra is a dimer that binds an operator DNA sequence in the promoter region of the fruBKA cluster. Furthermore, fructose 1-phosphate (F1P) was found to most efficiently dissociate the Cra-DNA complex. Thermodynamic parameters of the F1P-Cra-DNA interaction calculated by isothermal titration calorimetry revealed that the factor associates tightly to the DNA sequence 5′-TTAAACGTTTCA-3′ (KD = 26.3 ± 3.1 nm) and that F1P binds the protein with an apparent stoichiometry of 1.06 ± 0.06 molecules per Cra monomer and a KD of 209 ± 20 nm. Other possible effectors, like fructose 1,6-bisphosphate, did not display a significant affinity for the regulator under the assay conditions. Moreover, the structure of Cra and its co-crystal with F1P at a 2-Å resolution revealed that F1P fits optimally the geometry of the effector pocket. Our results thus single out F1P as the preferred metabolic effector of the Cra protein of P. putida.
Keywords: Bacterial Genetics, Bacterial Metabolism, Bacterial Transcription, DNA-Protein Interaction, X-Ray Crystallography, Glycolysis, Pseudomonas putida, Regulatory Networks
Introduction
The central metabolism of bacteria is controlled through the interplay of global and specific regulators, the outcome of which depends on the nature of the carbon and energy sources and the particular culture conditions (1). The catabolite repressor/activator (Cra) protein (also known as FruR) is a pleiotropic regulatory protein that plays a key role in the control of carbon flow in Escherichia coli and Salmonella typhimurium (2). This factor was first identified as a repressor that inhibited expression of the fructose operon of these two bacteria when the sugar was not available, thereby the earlier name FruR (3). Later work, however, revealed that the same protein represses genes for many other enzymes of the central metabolism (pfkA, pykA, pykF, acnB, edd, eda, mtlADR, and gapB; Refs. 2 and 4–6) and activates others (ppsA, fbp, pckA, acnA, icd, aceA, and aceB; Refs. 2 and 7–10), suggesting a dual character of Cra as both a transcriptional repressor and an activator.
From a structural point of view, the Cra protein has been classified as a member of the GalR-LacI superfamily of DNA-binding transcriptional regulators (11, 12). In E. coli, the Cra monomer is organized in two functional domains. The N-terminal helix-turn-helix module accounts for the binding of the protein to the cognate DNA operator sequence, whereas the C-terminal portion mediates the interactions between subunits and triggers changes in the protein upon effector binding (12).
Earlier studies of the Cra proteins of E. coli and S. typhimurium suggested that the regulator is a tetramer that recognizes an imperfect palindromic DNA sequence to which it binds asymmetrically (2). It is generally believed that promoters containing a Cra operator upstream of the RNA polymerase binding site might become activated by the factor, thereby stimulating transcription of downstream genes. In contrast, Cra binding sites overlapping those bound by RNA polymerase originate a typical negative regulation scenario (2). Whether positive or negative, the effects of Cra on transcription can be counteracted in vitro by micromolar concentrations of fructose 1-phosphate (F1P)3 or millimolar concentrations of fructose-1,6-bisphosphate (FBP, Refs. 2, 3, 5, 13). In fact, the nature of the metabolic effector(s) alleged to make Cra release target DNA sites is intriguing. This is because F1P and FBP are originated by different metabolic routes and produced at levels that depend on the consumed carbon source. Furthermore, electronic densities of these two chemicals are not alike, making it puzzling that the same effector recognition pocket can bind both of them. Conflicting publications on this matter bear witness of such a problem in the relevant literature. Gel retardation experiments with a labeled DNA fragment bearing a Cra operator (3, 5) could authenticate F1P as one bona fide effector, but not FBP. Some studies suggest that the in vitro response of Cra to FBP is due to a possible contamination of FBP with F1P (3). Yet, other authors have produced evidence that FBP is an authentic effector of Cra (5, 14). The consensus at this point is that F1P is the major agonist of the enterobacterial regulator, but FBP can do the same at much higher concentrations. But what is the situation in other types of bacteria?
This work addresses the nature of the metabolic signal(s) that control(s) binding of Cra to its genomic binding sites in the soil microorganism Pseudomonas putida. This bit of information is important for setting reliable connections between the central fluxes for catabolism/anabolism of carbohydrates (which are often reported by FBP levels, Ref. 3) and the metabolic and non-metabolic transcriptome (14) in this bacterium. The Cra regulator of P. putida is an ortholog of the virtually identical gene/protein found in E. coli. But in contrast to the protein of E. coli, crystals of the purified CraP. putida diffracted at a very high resolution (see below), thereby revealing an unprecedented structural detail of the effector-protein interaction that could not be attained with the enterobacterial variant (11). As shown below, analysis of such a structure along with other biochemical and biophysical tests indicates that F1P is the preferred metabolic effector of CraP. putida. As a sidelight, we could produce evidence that this Cra variant represses expression of the fructose-related phosphoenol pyruvate phosphotransferase carbohydrate system of P. putida in a fashion (predictably) dependent on F1P binding.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Protein Purification
The DNA sequence encoding the Cra protein of P. putida KT2440 (PP0792) was amplified from the chromosome of this bacterium with primers Cra-F (5′-GGGAATTCCATATGAAACTCAGCGATATCGCCCGT-3′) and Cra-R (5′-CCGGCTCGA GTCAGGCCACTGAGAT-3′). The resulting PCR product was digested with NdeI/XhoI and cloned into the bacterial expression vector pET28a (Novagen), which adds six histidines to the N terminus of the encoded protein. Note that we included a stop codon (TCA) in the Cra-R primer immediately after the XhoI sequence to avoid addition of more His residues in the C terminus by the vector. The resulting plasmid (pET28aCra) was transformed into the specialized host strain E. coli BL21(DE3). For expression of the His6-tagged protein, the transformed strain was grown at 37 °C in LB to A600 = 0.5, and 0.5 mm of isopropyl-β-d-thiogalactoside was added, after which the cultures were grown for an additional 3 h. Cells were then harvested by centrifugation and the pellets frozen at −80 °C until further use. For purification, cells were thawed, resuspended in 50 mm sodium phosphate buffer (pH 7.0) and 200 mm NaCl (1 ml of culture, A600 = 0.5 in 50 μl), and mechanically lysed using a French press (Thermo Electron Corp.). Cell debris was removed by centrifugation (20,000 × g for 30 min at 4 °C) and the native His6-Cra protein isolated to apparent homogeneity from the supernatant of the cell lysate using TALON metal affinity resin (Clontech) according to the manufacturer's instructions (see supplemental Fig. S1 for details). The purified protein was kept in the elution buffer at 4 °C until its utilization as indicated in each case. On the other hand, selenium-methionine (Se/Met)-derivatized P. putida Cra was purified as before from cells grown in M9 minimal medium (15) in the presence of 0.2% glucose, selenium-methionine, and other amendments known to inhibit methionine biosynthesis (for protocol details see Ref. 16). All buffers for (Se/Met)-Cra purification were supplemented with 5 mm dithiothreitol, but the isolation procedures were otherwise identical to those employed for the non-derivatized protein.
Identification and Synthesis of the Cra Operator
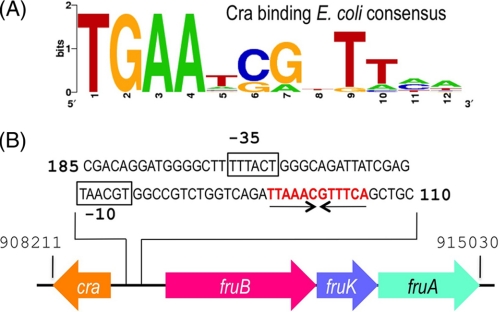
The reference E. coli consensus sequence for Cra binding sites was retrieved from the PRODORIC database (17). The resulting DNA motif (Fig. 1A) was then searched in the genome of P. putida KT2440 (18) by means of the Virtual Footprint software (version 3.0, Ref. 19), which revealed the presence of at least 89 possible Cra binding sites. One of these sites (5′TTAAACGTTTCA3′) happened to occur within the intergenic and divergent promoter region of the fruR/fruBKA gene cluster (P. putida chromosomal coordinates 908211–915030, Fig. 1B) which, because of its proximity to the cra (fruR) gene of this bacterium was likely to be functional. This plausible operator was synthesized as part of a 22-bp double-stranded DNA fragment (see below) and employed in the mobility shift and isothermal microcalorimetry (ITC) assays described under “Results.”
FIGURE 1.
E. coli Cra binding consensus sequence and regulatory region of the fructose operon of P. putida. A, the E. coli consensus sequence for the Cra binding site was retrieved from the PRODORIC database (17). This sequence forms an incomplete palindrome in which the left half-site is considerably better conserved than the right half-site. B, the regulatory region of the fruBKA operon in P. putida was analyzed and one single operator identified as shown. The sequence TTAAACGTTTCA (in red) corresponds to the palindromic Cra-binding motif, whereas the black boxes show the putative −35/-10 promoter region upstream of the fruBKA operon.
NonRadioactive EMSA
The 22-bp DNA fragment used for EMSA with the Cra protein was prepared by heating an equimolar (50 nmol) mixture of complementary oligonucleotides containing the presumed Cra binding site of the fruR/fruBKA promoter region (forward, 5′-TCAGATTAAACGTTTCAGCTGC-3′; and reverse, 5′-GCAGCTGAAACGTTTAATCTGA-3′) in 1 ml of TE buffer (10 mm Tris/HCl, pH 8.0; 1 mm EDTA) at 95 °C for 10 min and then keeping the annealed product chilled on ice until further use. The Cra-DNA binding reaction was assembled in a final volume of 20 μl containing 2 μl of TGED binding buffer 10× (final concentrations, 10 mm Tris-HCl (pH 8.0), 5% v/v glycerol, 0.1 mm EDTA, 1 mm dithiothreitol, 20 μg/ml poly (deoxyinosinic-deoxycytidylic), 200 μg/ml BSA, 0.75 μm Cra site-containing DNA fragment) and increasing amounts of the pure Cra. Mixtures were incubated for 15 min at 30 °C and electrophoresed in a nondenaturing 10% (w/v) polyacrylamide (bisacrylamide gel in Tris-glycine buffer 1× (pH 8.9)). Finally, the gels were stained with ethidium bromide and visualized using a Gel Doc XR (Bio-Rad). In the experiments for identification of potential effectors, Cra was incubated with the DNA fragment in the presence of 1 mm fructose (Fluka, 99% purity), 1 mm F1P (Sigma, 100% purity), FBP (Sigma, 98% purity), fructose 6-phosphate (F6P, Sigma, 98% purity), glucose 6-phosphate (G6P, Sigma, 100% purity), and then the samples were run in a gel and analyzed as described before.
ITC
ITC experiments were performed on a VP microcalorimeter (MicroCal, Northampton, MA) at 25 °C. Prior to experiments, Cra was thoroughly dialyzed in 25 mm Tris-HCl, 50 mm NaCl, and 1 mm DTT (pH 8.0). After dialysis, the protein solution was filtered through a 0.45-μm filter, and its concentration was determined by UV absorption spectroscopy using an extinction coefficient of 1.217 × 105 cm M−1 at 280 nm (20). Effectors were prepared by diluting pure powdered products in filtered dialysis buffer so that the ligand and protein solvent were the same. Each titration involved a single 1.6-μl injection and a series of 4.8-μl injections of a 250 μm preparation of each of the effectors into a 7.5 μm protein solution deposited in the 1.4-ml chamber of the apparatus. For DNA binding studies, the 22-bp DNA fragment used in EMSA experiments (see above) was dialyzed so that the DNA and protein solvent were the same. In this case, the assays consisted of the injection of 4.8-μl aliquots of 60 μm DNA into a protein solution of 10 μm in the same 1.4-ml chamber. The mean enthalpies measured from injection of the ligands into the buffer were subtracted from raw titration data prior to data fitting using the One Binding Site model of the MicroCal version of the ORIGIN software. From the curves thus fitted, the parameters ΔH (reaction enthalpy), KA (binding constant, KA = 1/KD), and n (reaction stoichiometry) were determined. From the values of KA and ΔH, the change in free energy (ΔG) and in entropy (ΔS) were calculated with the equation: ΔG = -RT lnKA = ΔH - TΔS, where R is the universal molar gas constant and T is the absolute temperature.
Analysis of the Cra Oligomerization State
Analytical ultracentrifugation experiments were performed in a Beckman Optima XL-A analytical ultracentrifuge with a UV-visible absorbance detection system. Sedimentation velocity measurements were carried out on an An50Ti eight-hole rotor in phosphate buffer with 150 mm imidazole and 200 mm NaCl (pH 7.0) (elution buffer in protein purification) at 45,000 rpm, with 400 μl of protein solution (0.5, 1, and 1.3 mg/ml) in each cell, using double-sector Epon-charcoal centerpieces. Differential sedimentation coefficient distributions c(s) were calculated by least squares boundary modeling of sedimentation velocity data using the SEDFIT program (21, 22). From this analysis, the experimental sedimentation coefficients (s) were corrected for solvent composition and temperature to obtain the corresponding standard s values (s20,w, standard solvent conditions, pure water at 20 °C) using the SEDNTERP program (23). Partial specific volume of 0.7312 ml g−1 at 20 °C was calculated from the amino acid composition (24). Sedimentation equilibrium experiments were performed at 20 °C by centrifugation of 80-μl samples of Cra at 11,000, 13,000, and 19,000 rpm in the Optima XL-A ultracentrifuge. Conservation of mass in the cell was verified in all the experiments. After the equilibrium scans, a high-speed centrifugation run (45,000 rpm) was done to estimate the corresponding baseline offsets. Weight-average buoyant molecular weights were determined by fitting a single-species model to the experimental data using the Hetero-Analysis program (25). The absolute molecular weight of the Cra protein was determined using the partial specific volume (0.7312 ml g−1) and the solvent density (1.0207 mg/ml). In addition, the oligomerization form of P. putida Cra was confirmed by size exclusion chromatography. To this end, a protein solution of 1.0 mg/ml was run through a Superdex 200 column (Amersham Biosciences) in 25 mm Tris-HCl and 50 mm NaCl (pH 8.0). The molecular weight markers were run under the same conditions. Proteins in the eluted fractions were monitored by SDS-PAGE.
Protein Crystallization
The affinity-purified His-tagged Cra protein (see above) was concentrated to 15 mg/ml for its crystallization. During concentration, the sample buffer was diluted ∼50 times against 25 mm Tris (pH 8.0), 50 mm NaCl, and 1 mm DTT. This procedure allowed a high concentration of the protein and avoided amorphous precipitation while simultaneously clearing the samples of the potentially disturbing imidazole left after eluting the protein from the metallo-affinity column. Crystals of the native Cra protein were then generated from a 15-mg/ml protein sample by the sitting-drop vapor-diffusion method. Drops were prepared by mixing 150 nl of protein with 150 nl of crystallization solution and equilibrated against 100 μl of the crystallization solution in Greiner plates. Crystals appeared after 2 days in drops prepared with a solution containing 15% (w/v) of polyethylene glycol 8000, 0.2 m sodium acetate, and 0.1 m MES buffer (pH 6.5). Selenium-methionine (Se/Met)-derivatized Cra protein was crystallized using identical conditions. The resulting crystals were dialyzed against the same solution supplemented with 20% glycerol before cryofreezing in liquid nitrogen (see below). For the ligand binding experiments, native Cra protein crystals were soaked for 4–7 h in the cryofreezing solution containing 1 mm of each tested candidate effectors (F1P, FBP, F6P, G6P, and fructose).
Data Collection and Structure Resolution
Datasets from the native, effector-less Cra protein, its (Se/Met)-derivatized variant, and the ligand-containing complexes F1P and FBP were collected at the European Synchrotron Radiation Facility (Grenoble, France) at beamlines ID14–1, ID23–1, and ID14–2 (crystals soaked with F6P, G6P, and fructose did not show any diffraction). The crystals, which belonged to the space group P21, contained two Cra molecules in the asymmetric unit and a solvent content around 43%. Diffraction data were processed with X-ray detector software (XDS) (26) and scaled with SCALA (27). The structure of the protein without effector was solved first by the Single-wavelength anomalous dispersion (SAD) method using the Autorickshaw server (28) and diffraction data extending up to a 2.5-Å resolution. Structure resolution and phasing in the server was carried out by the SHELX programs (29). The initial model built by RESOLVE (30) gave R-factor/R-free values of 22 and 28%, respectively. The model was manually adjusted using Coot (31) and refined using PHENIX 1.6-289 (32) with data up to a 2.0-Å resolution to a final R-factor and R-free of 18.4 and 21.8, respectively (supplemental Table S1). Crystal structures of the Cra protein in complex with F1P and FBP were determined by the molecular replacement method using the effector-less structure and the PHASER program (33), followed by refinement with PHENIX 1.6-289 (32, supplementary Table S1). The structure of the Cra protein prepared by soaking the crystal with FBP gave an electron density map in the effector binding site that was indistinguishable from the Cra-F1P complex and therefore has not been included separately in supplemental Table S1 (not shown). All residues are in the most favored region (91.7%) or the additional allowed regions (7.9%) of the Ramachandran plot, except the effector-binding Asp-148 that lies in a disallowed region in all structures. Coordinates of the Cra structure and its complex with F1P have been deposited in the Protein Data Bank with the following codes: PP-Cra, 3O74 and PP-Cra/F1P, 3O75.
RESULTS AND DISCUSSION
Identification and Gross Characterization of the Cra Protein of P. putida
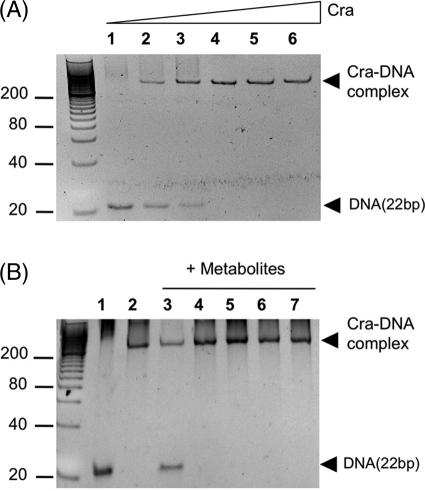
The genome of the soil bacterium P. putida bears an ORF for a protein (PP0792) of 331 amino acid residues (coordinates 909206–908211) and 38.4-kDa molecular mass that shares 74% similarity and 48% identity with the E. coli Cra protein (supplemental Fig. S2). This identity goes up to 70% in the predicted DNA binding domain of the proteins, suggesting that the target genomic sequences ought to be similar (if not identical) as well. All these criteria qualifies PP0792 as a bona fide ortholog of the reference enterobacterial counterpart. Yet, unlike E. coli and S. typhimurium, the craP. putida variant maps adjacent and divergently oriented to the fruBKA operon (Fig. 1B and supplemental Fig. S3). This gene cluster encodes proteins for phosphorylation and transport of fructose (34) through the phosphoenol pyruvate phosphotransferase carbohydrate system of P. putida (Ref. 34; PTSFru). Because of this proximity, PP0792 was formerly annotated as FruR, not as Cra (18). To ensure the functional assignment, the craP. putida sequence was amplified, a metallo-affinity His6-tag was added, and the protein was purified to apparent homogeneity as explained under “Experimental Procedures” (see supplemental Fig. S1). On the other hand, analysis of the intergenic region (Fig. 1B) revealed the presence of a motif (5′-TTAAACGTTTCA-3′) with a high score to the consensus binding site for the Cra regulator of E. coli and Salmonella (Fig. 1A, Refs. 3, 35, 36). To examine whether such a sequence was in fact the operator of CraP. putida, a 22-bp double-stranded DNA fragment containing the candidate target was synthesized and employed in the mobility shift assay shown in Fig. 2. The results indicated the formation of one specific complex between the DNA and the CraP. putida, the abundance of which increased with Cra concentration (Fig. 2A). The molar ratio where the DNA band disappeared entirely from the gel grossly corresponded to a molar DNA:Cra ratio of 1:2. These results verified the interaction of the protein with its predicted binding site within the fruR/fruBKA promoter region and laid the basis for determination of key parameters as explained below.
FIGURE 2.
Nonradioactive EMSA of Cra and its binding site in the intergenic fruR/fruBKA region of P. putida. A, EMSA experiment with increasing concentrations of pure Cra protein. Molar ratios of DNA:Cra were 1:0 (lane 1), 1:0.5 (lane 2), 1:1 (lane 3), 1:2 (lane 4), 1:4 (lane 5), and 1:8 (lane 6). B, Cra effector identification. DNA (0.75 μm) was incubated with pure protein (1.5 μm) in the presence of 1 mm of different metabolites: F1P (lane 3), FBP (lane 4), F6P (lane 5), G6P (lane 6), and fructose (lane 7). Lanes 1 and 2 correspond to DNA in the absence and presence of Cra, respectively, without any added metabolite. Note that only F1P prevents the binding of the protein to DNA.
The Cra Regulator of P. putida Is a Dimeric Protein
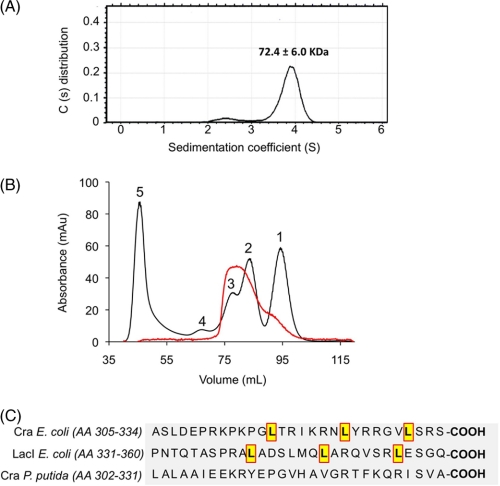
The oligomerization state of purified CraP. putida was determined by means of analytical ultracentrifugation experiments. The sedimentation profile (Fig. 3A) showed the occurrence of one main species to 3.9S with s20,w of 4.8 ± 0.1 (f/f0 = 1.35), which accounted for 88–94% of the observed molecules and which was compatible with a dimeric form of the protein. The sedimentation equilibrium results (data not shown) predicted a mass value of 72.4 ± 6.0 kDa, which was slightly lower than the theoretical mass for the dimeric form of the protein. But taken together, the velocity and equilibrium sedimentation results indicated that Cra existed mainly as a dimer in solution at the concentrations used. The protein oligomerization state was confirmed by size exclusion chromatography (Fig. 3B), in which one main peak of ∼60–80 kDa (i.e. a dimer) was followed by much less abundant species, tailing between the 21–50 kDa markers (i.e. a monomer). These results thus consistently documented that the Cra protein of P. putida occurs predominantly in a dimeric form, a feature that differs from the same regulator of E. coli, known to be a tetramer (37–39). However, this divergence is understandable because the leucine mini-zipper that causes tetramerization of proteins of the LacI family through their carboxyl-terminal regions is conserved in E. coli Cra (8) but not in the P. putida counterpart (Fig. 3C). On the other hand, the Cra binding site in the fruBKA promoter region of E. coli contains two nearby operators, O1 (TGAAAC|GTTTCA) and O2 (TGAATC|GTTTCA), which could accommodate the cognate repressor tetramer (3). In contrast, the same region of the P. putida genome has only one Cra site (Fig. 1B). A plausible consequence of this is that the affinity of the Cra dimer for the single site in the fruBKA promoter region of P. putida ought to be high to compensate the more extended range of interactions of the tetramer/two-operator scenario of the same regulatory device in E. coli. This possibility was addressed next.
FIGURE 3.
Oligomerization of the Cra protein of P. putida. A, distribution of sedimentation coefficient population (c(s)) against sedimentation coefficient (s), obtained by sedimentation velocity experiments. The assays were performed with 0.5, 1, and 1.3 mg/ml of pure Cra protein. The sedimentation profile was dominated by a species of 3.9S, accounting for 88–94% of the observed molecules, followed by a small peak at 2.4S (2.3–8.9%). The predominant species with sedimentation coefficient of 3.9S was corrected to s20,w using the viscosity value of the solution (measured in a viscometer Anton Paar AMVn). The corrected sedimentation coefficient was 4.8S, which is compatible with a globular dimer of the protein. Equilibrium sedimentation experiments predicted that the dominant peak has a molecular mass of 72.4 ± 6.0 kDa. B, gel filtration. The oligomerization form of P. putida Cra was confirmed by size exclusion chromatography as shown. The main peak of 60–80 kDa is consistent with a dimer, whereas the minor tail of 21–50 kDa surely corresponds to residual monomeric species. The chromatogram shows the superposition of two different runs: Cra alone (red line) and standards used as molecular mass marker (black line). The proteins in the elution fractions were monitored by SDS-PAGE. Molecular weight markers were run under the same conditions. Chymotrypsin (21 kDa) (1), albumin (50 kDa) (2), BSA (70 kDa) (3), Ab (170 kDa) (4), and Dextran (5). C, alignment of the C-terminal amino acid sequence of regulators Cra (E. coli), LacI (E. coli), and Cra (P. putida). Note that the E. coli tetrameric regulators Cra and LacI contain a leucine mini-zipper, which seems to be required to tetramerization. This mini-zipper is absent in the P. putida Cra protein.
Cra Strongly Binds Its Single Operator in the fruR/fruBKA Region of P. putida
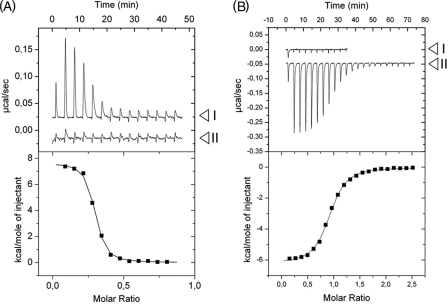
The interaction parameters of the CraP. putida protein with the operator DNA found within the fruBKA promoter segment (Fig. 1B) were examined by means of ITC. The corresponding results are shown in Fig. 4A. Note that in the upper panel peaks go upwards, which indicates that the binding of Cra to DNA is an endothermic process (ΔH = 7.66 ± 0.09 kcal/mol). Because enthalpy changes are unfavorable, binding ought to be entirely entropy-driven (TΔS = 17.68 ± 0.15 kcal/mol, ΔG = −10.02 ± 0.10 kcal/mol). This situation is not infrequent, as the binding of proteins to DNA is often endothermic but also fostered by the positive entropic effect of the displacement of DNA-bound water molecules (40). Subsequent calculations allowed us to determine a protein-DNA dissociation constant (KD) of 26.3 ± 3.1 nm that reflects a very strong interaction and that likely enables CraP. putida to exert a considerable repression on expression of the fruBKA genes.
FIGURE 4.
ITC assays with Cra, effectors, and DNA. The upper panels plot raw data from representative ITC experiments, whereas the lower panels show the fitted curves of the same results but integrated and corrected for dilution. A, titration of Cra protein with DNA in the absence (I) and presence (II) of F1P. Positive peaks represent an endothermal event. Note that no interaction between Cra and the DNA is detected in the presence of F1P. The thermodynamic values calculated for protein-DNA interaction were KD = 26.3 ± 3.1 nm, ΔH = 7.66 ± 0.09 kcal/mol, TΔS = 17.68 ± 0.15 kcal/mol, and ΔG = −10.02 ± 0.10 kcal/mol. B, titration of Cra protein with (I) FBP and (II) F1P. The experiment covered concentrations of the effectors in the chamber ranging from 300 nm to 20 μm. Note that only the titration with F1P produces a heat change in the experiment at the tested concentrations. Negative peaks are indicative of an exothermic event. The thermodynamic values calculated by curve fitting were KD = 209 ± 20 nm, ΔH = −5.94 ± 0.39 kcal/mol, TΔS = 3.16 ± 0.40 kcal/mol, and ΔG = −9.10 ± 0.06 kcal/mol. The sigmoidal shape of the binding curve allows determination of the protein-effector stoichiometry, which corresponds to the molar ratio (x axis in lower panel) at the point of inflection of the curve. In this case, such a molar ratio was 1:1, indicating that one molecule (calculated, 1.06 ± 0.06 molecules) of F1P interacts with one molecule of Cra monomer.
Surveying Metabolic Intermediates as CraP. putida Effector Candidates
To shed light on the nature of Cra effector(s), DNA samples were incubated with pure Cra protein in the presence of different metabolites and submitted to EMSA experiments as before, except for the addition of candidate molecules to the binding mixtures prior to loading the gel. The chemical species that were listed as possible Cra inducers able to release its strong binding to DNA included fructose, F1P, FBP, F6P, and G6P. The EMSA experiments shown in Fig. 2B (lanes 3–7) indicated that at 1.0 mm of the effector in the binding mixture, only F1P was able to prevent binding of the repressor to the fruBKA regulatory region (Fig. 1B), confirming this metabolite as a Cra binder.
The experiment shown in Fig. 1B, however, did not altogether rule out that the other small molecules could interact with the target protein at much higher concentrations not suitable for the conditions of the EMSA test. Furthermore, the gel of Fig. 2B revealed the persistence of a fraction of the Cra-DNA complex that did form even in the presence of F1P (Fig. 2B, lane 3). To elucidate these ambiguities, we again resorted to the ITC technology described before. First, we ran isothermal titration experiments with purified Cra and the various candidate effectors as the interaction partners. The results shown in Fig. 4B indicated that binding of F1P to Cra was both enthalpy-driven (ΔH = −5.94 ± 0.39 kcal/mol) and entropy-driven (TΔS = 3.16 ± 0.4 kcal/mol), producing a total free energy change (ΔG) of −9.10 ± 0.06 kcal/mol. Favorable (negative) enthalpy changes can be attributed to bond formation between F1P and amino acids in the cognate binding site, whereas favorable (positive) entropy changes are generally caused by the ligand-induced release into the medium of otherwise bound water, thereby increasing the disorder (entropy) of the system. Further calculations revealed not only an apparent stoichiometry of 1.06 ± 0.06 molecules of F1P per Cra monomer, but also demonstrated a high protein-effector affinity (KD = 209 ± 20 nm). It is noteworthy that this affinity value is significantly higher than the corresponding figures for other transcriptional regulators of the GalR-LacI family. For instance, the KD of isopropyl-β-d-thiogalactoside binding to LacI is 2.8 μm (41), that of 2-ketogluconate to PtxS, 15 μm (42), and so on. In contrast, ITC experiments indicated that Cra had little (if any) affinity for the other metabolites examined. Specifically, the test run with FBP as a Cra effector-to-be (assay I in Fig. 4B) did not produce any indication of interaction, at least at the concentrations employed in the assay. The same was true for other candidate effectors (e.g. G6P, glucose, and fructose; supplemental Fig. S4), indicating that the repressor is highly selective for F1P. Finally, to clarify whether some residual Cra-DNA complex could still be formed in the presence of F1P, a mixture of Cra and F1P was incubated with the same target DNA employed above for the repressor-operator binding experiments. The resulting ITC curves (Fig. 4A, assay II) confirmed that the Cra-F1P complex was altogether unable to bind DNA. This set of data clearly points at F1P as the preferred metabolic effector of Cra, whereas it raised doubts regarding whether the role FBP as an inducer of this repressor reported in the literature for the enterobacterial protein (5, 14) held true for the counterpart of P. putida.
Cra Crystallization and Structure Resolution
Although the results above confirmed the nature of F1P as a Cra effector, they did not discount that the effector-binding site of the repressor could accommodate other, thus far unidentified, metabolites. To shed light on this possibility, we set out to determine the actual three-dimensional structure of such a pocket in both the effector-less and the effector-bound Cra structure by x-ray crystallography. The protein was crystallized as described under “Experimental Procedures.” SDS-PAGE of extensively washed crystals showed a single, sharp band of ∼30 kDa for the crystallized sample, smaller than the theoretical size of 38 kDa. This suggested a precise excision of an ∼8-kDa fragment from the protein body during the crystallization process (see below). MALDI-time-of-flight analysis of the truncated protein produced a predominant peak of 30.4 kDa, confirming the previous observation. Proteolytic removal of nonstructured or unstable polypeptides often help the crystallization of the rest of the protein core (43). This appeared to be our case because the occurrence of such a cleavage during the concentration and crystallization of Cra was ultimately beneficial. Crystals were obtained that belonged to the P21 space group, contained two Cra molecules in the asymmetric unit, and did diffract to a resolution of 2 Å (see supplemental Table S1).
The structure of the Cra protein was determined for both the effector-free and the F1P-bound forms, in which last-case crystals were soaked in a solution containing a 1.0 mm concentration of the metabolite (see “Experimental Procedures”). The corresponding three-dimensional architectures are shown in Fig. 5A. Note that the dimers of the effector-less Cra and its F1P-bound counterpart were very similar, with a root mean square deviation of 0.48 Å for 270 residues. Note also that, as expected, the crystal structure lacks the leading N-terminal 59 amino acid residues. As mentioned above, this deletion is irrelevant (if not beneficial) for our purpose because the missing segment includes the DNA-binding domain and a short linker (supplemental Fig. S2) but leaves intact the effector-binding site of the protein.
FIGURE 5.
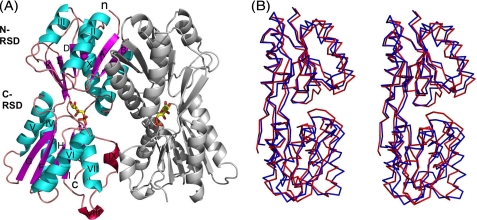
Crystal structure of Cra in complex with F1P. A, ribbon representation of the crystal structure of the regulatory subdomain of Cra in complex with its ligand F1P. β-strands are colored in magenta, helixes in cyan, 310 helixes in red, and loops in orange. F1P is shown with sticks, with carbons indicated in yellow, phosphorus in orange, and oxygens in red. The N and C termini are marked with lowercase letters and the secondary structure elements with uppercase letters. The N-regulatory subdomain consists of strand A (amino acids 63–68), helix I (74–89), strand B (93–98), helix II (103–115), strand C (120–123), helix III (133–139), strand D (144–147), strand E (157–164), helix X (293–310), and strand K (316–320). The C-regulatory subdomain is composed of helix IV (163–175), strand F (181–187), helix V (192–204), strand G (211–217), helix VI (222–236), strand H (242–245), helix VII (239–248), helix VIII (263–265), strand I (269–273), helix IX (277–280), strand J (286–290), and strand L (323–325). B, lateral stereo view (Cα representation) of the superimposed regulatory domains of Cra from P. putida (red) and E. coli (blue, PDB code 2IKS).
As shown in Fig. 5A, both Cra monomers are related by a pseudo 2-fold axis and share an interface surface of 1700 Å2 per monomer. The N-terminal regulatory subdomain (N-RSD) comprising residues from Thr-63 to Ser-161 and from His-293 to Val-320, specifically α helices I and II and β strand B (Fig. 5A), contributes most of the interactions to form the dimer. Residues from this region are implicated in 12 salt bridges and 80% of the hydrogen bonds between domains. In contrast, the C-terminal regulatory subdomain (C-RSD, residues Asp-163 to Ala-290 and Tre-323 to Lys-325) contributes only with six hydrogen bonds to the interaction through helices VII and IX (Fig. 5A). Structural comparisons using the DALI server (44) gave a top solution with a Z score of 33 and a sequence identity of 44% with the fructose repressor (Cra/FruR) of E. coli deposited in the RCSB data base (PDB code 2IKS) but not yet published. Structural comparison is shown in Fig. 5B. The folds observed in both proteins were close to the domain organization of other members of the family such as LacI (45) and PurR (46). These were defined by a central parallel β-sheet formed by six β strands (A–E and K) surrounded by four α helices (I–III and X) for the N-RSD, and a parallel β sheet composed of four β strands (G–I), an antiparallel β sheet with two β strands (J and L) with four α helices (IV–VII), and two 310 helices (VIII and IX) for the C-RSD.
Geometry of the Cra Inducer-binding Pocket
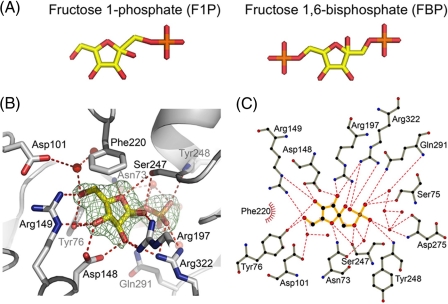
The F1P molecule could be coherently fitted in the electronic density maps obtained after refinement of the structures determined for the crystal soaked in F1P (Fig. 6). Yet, an attempt was made to generate an additional complex between FBP (instead of F1P) and Cra by soaking effector-less protein crystals in a 1 mm solution of this metabolite as above. The structures obtained from crystals exposed to F1P and FBP were virtually identical, with a root mean square deviation for their superposition in the range of 0.45 Å. But once the structure of the putative FBP-Cra complex was inspected, it was found that the molecule occupying the effector site was no other than F1P. These results suggested that it was the residual F1P present in the nominally 98% pure FBP preparation (see “Experimental Procedures”) the protein recognized as sole interaction partner. That such selective binding to F1P happened even in the presence of a large excess of FBP confirms the very high affinity for the first, consistently detected with the other procedures described above.
FIGURE 6.
Detail of the interaction between Cra and F1P. A, stick representation of F1P and FBP with the same color code as in Fig. 5A. B, F1P-Cra interactions in the protein binding pocket. The electron density map (Fo-Fc) determined in the absence of the F1P molecule is represented as a green mesh at a 3.5-σ contour level. C atoms are represented in gray, oxygens in red, and nitrogens in blue. Hydrogen bonds are indicated as dashed red lines and water molecules are shown as red spheres. C, Ligplot scheme of F1P interaction with Cra (54); the color code is as in B. C atoms are marked in black, and hydrophobic interactions are indicated by red arcs with spokes radiating toward the interacting atom.
The cavity where F1P molecules bind Cra is formed by portions of the N-RSD and C-RSD subdomains (Fig. 5A), at the intermodule cleft. Incubation of Cra native crystals with 1 mm F1P for 1 h was enough to obtain an equimolecular 1:1 association of the ligand to each domain present in the asymmetric unit of the crystals. The F1P molecule fitted to perfection the electron density map of the cognate protein cavity determined in the absence of the effector (Fig. 6B). The F1P molecule has a solvent-accessible area of 372 Å2, which is completely buried by contacts with the interacting protein residues and solvent molecules. Interactions occur with polar and aromatic residues in that cavity through direct hydrogen bonds with residues Tyr-76, Ser-75, Asn-73, and Asp-148 of the N-RSD, as well as Arg-149, Arg-197, Ser-247, and Tyr-248 of the C-RSD (Fig. 6, B and C). Moreover, two residues (Gln-291 and Gln-322) of the hinge regions connecting both domains contribute with two extra direct hydrogen bonds to the interactions. In addition, water-mediated hydrogen bonds connect F1P with Asn-73 and Asp-101 of the N-RSD and Asp-275 and Gln-291 of the C-RSD. Hydrophobic interactions also play a role in the strong affinity and specificity of Cra for F1P. These involve the furanose ring of the effector, which sits between the side chains of Tyr-76 (part of Ser-75 and Asp-148) and Phe-220. These amino acids provide not less than five interactions that trap the sugar ring between two hydrophobic surfaces. The dense constellation of interactions between F1P and Cra shown in Fig. 6 creates a high degree of specificity that accounts for the extraordinary preference of the P. putida regulator for this (and not for other) possible effectors.
CONCLUSION
The biochemical and structural results presented above constitute strong evidence that F1P is the most favored effector of the Cra protein of P. putida. The extensive contacts of F1P with Cra lead to a high affinity binding interaction (KD ∼ 200 nm). In contrast, we found no proof of binding to either FBP or other metabolites at the concentrations employed in the assays, a body of (negative) data, which somehow diverges from the results reported for the enterobacterial protein (5, 14). Furthermore, the extensive similarity between the proteins of P. putida and E. coli suggest that they could also share the same structural transmission pathway to regulate its function, which is likely to be identical to that of known members of the family (46, 47).
That F1P is the preferred effector of CraP. putida opens interesting questions because current metabolic models for this microorganism entertain F1P appearance only as a result of fructose phosphorylation by the PTSFru system (48–50). Furthermore, in contrast to E. coli, P. putida lacks a real glycolytic route, which makes virtually impossible any significant accumulation of FBP when cells grow in glucose (51). Although the regulatory scope of Cra in P. putida remains to be explored, it could well happen that (similarly to E. coli) the factor controls a large number of genes (4, 52), in which case there must be alternative ways of producing F1P as a descriptor of metabolic activity (14) in this bacterium. One way or the other, it is likely that the first regulatory duty of Cra was that of a repressor of fructose metabolism which was later co-opted as a global regulator (2).
A sidelight of the results presented in this work is the evidence that Cra regulates the PTSFru system in P. putida by binding strongly as a dimer to a single palindromic sequence within the cra/fruB intergenic region (which is different to those of E. coli and S. typhimurium), and that only F1P can lift such down-regulation. Because there is cross-talk between PTSFru and the nitrogen-related branch of the PTS system in this bacterium (53), it is possible that Cra closes the loop that connects N consumption and metabolic C fluxes. These hypotheses are being actively examined in our laboratory.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility (ESRF) for provision of synchrotron radiation facilities through the Block Allocation Group (BAG)-Madrid project.
This work was supported by grants of the CONSOLIDER program of the Spanish Ministry of Science and Innovation, by the BACSINE and MICROME Contracts of the European Union, and by funds of the Autonomous Community of Madrid. This work was also supported by a doctoral scholarship of the Universidad de Costa Rica and the CSIC (to M. C.). C. S. was supported by the Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
The atomic coordinates and structure factors (codes 3O74, 3O75, and 2IKS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- F1P
- fructose 1-phosphate
- FBP
- fructose 1,6-phosphate
- PTSFru
- phosphoenol pyruvate phosphotransferase carbohydrate system of P. putida
- ITC
- isothermal microcalorimetry
- F6P
- fructose 6-phosphate
- G6P
- glucose 6-phosphate
- MES
- 4-morpholineethanesulfonic acid
- ΔH
- reaction enthalpy
- ΔG
- change in free energy
- ΔS
- change in entropy
- T
- absolute temperature
- KA
- binding constant.
REFERENCES
- 1. Downs D. M. (2006) Annu. Rev. Microbiol. 60, 533–559 [DOI] [PubMed] [Google Scholar]
- 2. Saier M. H., Jr., Ramseier T. M. (1996) J. Bacteriol. 178, 3411–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramseier T. M., Nègre D., Cortay J. C., Scarabel M., Cozzone A. J., Saier M. H., Jr. (1993) J. Mol. Biol. 234, 28–44 [DOI] [PubMed] [Google Scholar]
- 4. Ow D. S., Lee R. M., Nissom P. M., Philp R., Oh S. K., Yap M. G. (2007) J. Biotechnol. 131, 261–269 [DOI] [PubMed] [Google Scholar]
- 5. Bledig S. A., Ramseier T. M., Saier M. H., Jr. (1996) J. Bacteriol. 178, 280–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sarkar D., Siddiquee K. A., Araúzo-Bravo M. J., Oba T., Shimizu K. (2008) Arch. Microbiol. 190, 559–571 [DOI] [PubMed] [Google Scholar]
- 7. Nègre D., Oudot C., Prost J. F., Murakami K., Ishihama A., Cozzone A. J., Cortay J. C. (1998) J. Mol. Biol. 276, 355–365 [DOI] [PubMed] [Google Scholar]
- 8. Cortay J. C., Nègre D., Scarabel M., Ramseier T. M., Vartak N. B., Reizer J., Saier M. H., Jr., Cozzone A. J. (1994) J. Biol. Chem. 269, 14885–14891 [PubMed] [Google Scholar]
- 9. Prost J. F., Nègre D., Oudot C., Murakami K., Ishihama A., Cozzone A. J., Cortay J. C. (1999) J. Bacteriol. 181, 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramseier T. M., Chien S. Y., Saier M. H., Jr. (1996) Curr. Microbiol. 33, 270–274 [DOI] [PubMed] [Google Scholar]
- 11. Penin F., Geourjon C., Montserret R., Böckmann A., Lesage A., Yang Y. S., Bonod-Bidaud C., Cortay J. C., Nègre D., Cozzone A. J., Deléage G. (1997) J. Mol. Biol. 270, 496–510 [DOI] [PubMed] [Google Scholar]
- 12. Scarabel M., Penin F., Bonod-Bidaud C., Nègre D., Cozzone A. J., Cortay J. C. (1995) Gene 153, 9–15 [DOI] [PubMed] [Google Scholar]
- 13. Ramseier T. M., Bledig S., Michotey V., Feghali R., Saier M. H., Jr. (1995) Mol. Microbiol. 16, 1157–1169 [DOI] [PubMed] [Google Scholar]
- 14. Kotte O., Zaugg J. B., Heinemann M. (2010) Mol. Syst. Biol. 6, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller J. H. (1972) Experiments in Molecular Genetics, pp. 431–432, Cold Spring Harbor, New York [Google Scholar]
- 16. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 17. Münch R., Hiller K., Barg H., Heldt D., Linz S., Wingender E., Jahn D. (2003) Nucleic Acids Res. 31, 266–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nelson K. E., Weinel C., Paulsen I. T., Dodson R. J., Hilbert H., Martins dos Santos V. A., Fouts D. E., Gill S. R., Pop M., Holmes M., Brinkac L., Beanan M., DeBoy R. T., Daugherty S., Kolonay J., Madupu R., Nelson W., White O., Peterson J., Khouri H., Hance I., Chris Lee P., Holtzapple E., Scanlan D., Tran K., Moazzez A., Utterback T., Rizzo M., Lee K., Kosack D., Moestl D., Wedler H., Lauber J., Stjepandic D., Hoheisel J., Straetz M., Heim S., Kiewitz C., Eisen J. A., Timmis K. N., Düsterhöft A., Tümmler B., Fraser C. M. (2002) Environ. Microbiol. 4, 799–808 [DOI] [PubMed] [Google Scholar]
- 19. Münch R., Hiller K., Grote A., Scheer M., Klein J., Schobert M., Jahn D. (2005) Bioinformatics 21, 4187–4189 [DOI] [PubMed] [Google Scholar]
- 20. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M. ed) pp. 571–607, Humana Press [Google Scholar]
- 21. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schuck P. (2004) Biophys. Chem. 108, 187–200 [DOI] [PubMed] [Google Scholar]
- 23. Lebowitz J., Lewis M. S., Schuck P. (2002) Protein. Sci. 11, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laue T. M., Shah B., Ridgeway T. M., Pelletier S. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Horton J. C., Rowe A. J. eds) pp. 90–125, Royal Society of Chemistry, Cambridge, England [Google Scholar]
- 25. Cole J. L. (2004) Methods Enzymol. 384, 212–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 27. (1994) Acta Crystallogr D Biol Crystallogr 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 28. Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A. (2005) Acta Crystallogr. D. Biol. Crystallogr. 61, 449–457 [DOI] [PubMed] [Google Scholar]
- 29. Sheldrick G. M. (2002) Z. Kristallogr. 217, 644–650 [Google Scholar]
- 30. Terwilliger T. C. (2000) Acta Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Velázquez F., Pflüger K., Cases I., De Eugenio L. I., de Lorenzo V. (2007) J. Bacteriol. 189, 4529–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nègre D., Bonod-Bidaud C., Geourjon C., Deléage G., Cozzone A. J., Cortay J. C. (1996) Mol. Microbiol. 21, 257–266 [DOI] [PubMed] [Google Scholar]
- 36. Shimada T., Fujita N., Maeda M., Ishihama A. (2005) Genes Cells 10, 907–918 [DOI] [PubMed] [Google Scholar]
- 37. Chakerian A. E., Tesmer V. M., Manly S. P., Brackett J. K., Lynch M. J., Hoh J. T., Matthews K. S. (1991) J. Biol. Chem. 266, 1371–1374 [PubMed] [Google Scholar]
- 38. Chen J., Matthews K. S. (1992) J. Biol. Chem. 267, 13843–13850 [PubMed] [Google Scholar]
- 39. Alberti S., Oehler S., von Wilcken-Bergmann B., Müller-Hill B. (1993) EMBO J. 12, 3227–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krell T., Busch A., Guazzaroni M., Lacal J., Gallegos M., Terán W. (2007) in Pseudomonas: A Model System in Biology (Ramos J. L., Filloux A. eds) pp. 255–277, Springer, Utrecht, The Netherlands [Google Scholar]
- 41. Wilson C. J., Das P., Clementi C., Matthews K. S., Wittung-Stafshede P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14563–14568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Daddaoua A., Krell T., Alfonso C., Morel B., Ramos J. L. (2010) J. Bacteriol. 192, 4357–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wernimont A., Edwards A. (2009) PLoS ONE 4, e5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lewis M., Chang G., Horton N. C., Kercher M. A., Pace H. C., Schumacher M. A., Brennan R. G., Lu P. (1996) Science 271, 1247–1254 [DOI] [PubMed] [Google Scholar]
- 46. Schumacher M. A., Choi K. Y., Zalkin H., Brennan R. G. (1994) Science 266, 763–770 [DOI] [PubMed] [Google Scholar]
- 47. Schumacher M. A., Choi K. Y., Lu F., Zalkin H., Brennan R. G. (1995) Cell 83, 147–155 [DOI] [PubMed] [Google Scholar]
- 48. Sawyer M. H., Baumann P., Baumann L., Berman S. M., Cánovas J. L., Berman R. H. (1977) Arch. Microbiol. 112, 49–55 [DOI] [PubMed] [Google Scholar]
- 49. Puchalka J., Oberhardt M. A., Godinho M., Bielecka A., Regenhardt D., Timmis K. N., Papin J. A., Martins dos Santos V. A. (2008) PLoS Comput. Biol. 4, e1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nogales J., Palsson B. Ø., Thiele I. (2008) BMC Syst. Biol. 2, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Velázquez F., di Bartolo I., de Lorenzo V. (2004) J. Bacteriol. 186, 8267–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shimada. T., Yamamoto K., Ishihama A. (2010) J. Bacteriol. 193, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pflüger K., de Lorenzo V. (2008) J. Bacteriol 190, 3374–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.