Abstract
Secretins are a family of large bacterial outer membrane protein complexes mediating the transport of complex structures, such as type IV pili, DNA and filamentous phage, or various proteins, such as extracellular enzymes and pathogenicity determinants. PilQ of the thermophilic bacterium Thermus thermophilus HB27 is a member of the secretin family required for natural transformation. Here we report the isolation, structural, and functional analyses of a unique PilQ from T. thermophilus. Native PAGE, gel filtration chromatography, and electrophoretic mobility shift analyses indicated that PilQ forms a macromolecular homopolymeric complex that binds dsDNA. Electron microscopy showed that the PilQ complex is 15 nm wide and 34 nm long and consists of an extraordinary stable “cone” and “cup” structure and five ring structures with a large central channel. Moreover, the electron microscopic images together with secondary structure analyses combined with structural data of type II protein secretion system and type III protein secretion system secretins suggest that the individual rings are formed by conserved domains of alternating α-helices and β-sheets. The unprecedented length of the PilQ complex correlated well with the distance between the inner and outer membrane of T. thermophilus. Indeed, PilQ was found immunologically in both membranes, indicating that the PilQ complex spans the entire cell periphery of T. thermophilus. This is consistent with the hypothesis that PilQ accommodates a PilA4 comprising pseudopilus mediating DNA transport across the outer membrane and periplasmic space in a single-step process.
Keywords: DNA Transformation, Membrane Proteins, Protein DNA-Interaction, Protein Purification, Protein Structure, Thermus thermophilus, Electron Microscopy, Membrane Protein Complex, Secretin
Introduction
Natural genetic transformation permits the exchange of DNA among organisms of different domains, and therefore, is suggested to be a driving force for bacterial adaptation and genome evolution (1–3). DNA uptake involves the assembly, action, and regulation of macromolecular DNA transport machineries that span the entire cell wall as well as the cytoplasmic membrane of bacteria. DNA translocators are similar to and sometimes even share components with type IV pili (T4P),2 filamentous phage secretion, type II protein secretion systems (T2SS), and type III protein secretion systems (T3SS) (4). Moreover, in many Gram-negative bacteria, such as Neisseria, Pseudomonas, and Thermus, natural transformation is coupled to pilus expression (5–9), although the molecular basis for this finding is not clear.
Despite the broad analyses of DNA translocator proteins, information on the structure and function of these complex transport machineries is scarce (3, 4, 10–12). Extensive studies performed with different Gram-negative mesophilic bacteria suggest at least three distinct substructures: a pilus-like rod that extends from the cytoplasmic membrane through the periplasm to the outer membrane; a ring-like structure, made by multiple copies of the secretin protein, guiding the rod through the outer membrane; and an inner membrane transporter. A cytoplasmic motor domain that powers assembly of the rod is associated with the transporter.
In analogy to the dynamic T4P, it is widely assumed that DNA translocators are highly dynamic structures (3, 10, 13–15). DNA is suggested to bind to the pilus-like rod, and then the translocator might retract, thus transporting DNA through a pore in the outer membrane composed of secretins, although it has to be noted that the mechanism of DNA binding and transport is still an open question. The preceding hypothesis is corroborated by the observation that T4P are highly dynamic machineries, which have been shown to bind DNA (16–19).
To get insights into the structure and function of natural transformation machineries, we chose the thermophilic bacterium Thermus thermophilus HB27 as model bacterium for molecular analyses of DNA uptake systems. A whole genome approach implying insertional mutagenesis followed by characterization of the mutants led to the identification of 16 competence proteins (9, 20, 21), which can be assigned to three different groups based on similarities to known proteins. 1) The first group consists of DNA translocator-specific proteins, such as ComEA and ComEC, which are suggested to be involved in DNA binding and transport across the inner membrane. 2) The second group consists of T4P-related proteins, such as four pilin-like proteins, PilA1–4, which might form a pseudopilus DNA translocator. This DNA translocator is suggested to be guided through the outer membrane (OM) by the secretin-like competence protein PilQ. These suggestions are supported by the finding that a pilQ mutant is devoid of PilA4 in the OM (22). Furthermore, PilF, a traffic NTPase that has been purified and found to exhibit NTPase activities, is suggested to power the DNA translocator (23). 3) In addition to these T4P-related proteins, PilM, a member of the actin family, and four novel competence proteins, ComZ, PilN, PilO, and PilW, were found to be essential for natural transformation in T. thermophilus. The inner membrane (IM) localization together with the results from mutant studies suggest that PilM, PilN, PilO, and PilW are components of the DNA translocator platform in the IM implicated in the biogenesis of the transporter complex and/or that they aid transport of DNA across the IM channel (22, 24).
In this study, we have isolated a homooligomeric secretin from T. thermophilus HB27 encoded by pilQ. Its structure, as determined by electron microscopy, revealed a unique shape and size that is discussed in view of the unusual architecture of the T. thermophilus cell periphery and of its thermophilic lifestyle. In addition, its function as a potential DNA-binding protein was addressed with the purified protein.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
For the purification of PilQ, T. thermophilus HB27 was grown at 68 °C on TM+ complex medium containing 4 g of yeast extract, 8 g of Tryptone peptone, and 3 g of NaCl/liter, pH 7.5 (25), and harvested in the late exponential growth phase.
Gradient SDS-PAGE, Blue Native Gradient PAGE, and Immunoblotting
Washed cells were resuspended in Sol buffer (20 mm Tris/HCl, pH 7.5, 400 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol) and disrupted by sonification (30 s; duty cycle, 50%; output control, 5) followed by centrifugation at 13,000 rpm for 10 min to remove unbroken cells. The resulting whole cell lysate was analyzed on denaturing 3–12% polyacrylamide gradient SDS gels or on Blue native 3–12% polyacrylamide gradient gels (26, 27). The presence of PilQ was detected using rabbit polyclonal antibodies raised against recombinant T. thermophilus HB27 PilQ (22). Conditions for sample preparation, SDS-PAGE, electroblotting, and antigen detection (dilution of the antisera: 1:6666) have been described elsewhere (22).
Cellular Fractionation and Total Membrane Enrichment
Total membranes were separated from the soluble fraction by ultracentrifugation. Inner and outer membranes were prepared by N-laurylsarcosine extraction and by sucrose density gradient centrifugation as described previously (22, 28). The purity of outer and inner membranes was verified by Western blot analyses of membrane fractions with antibodies directed against the S-Layer protein (outer membrane) and with antibodies directed against the inner membrane protein PilN of T. thermophilus HB27.
Purification of PilQ
To purify PilQ, Thermus cells were grown to the late exponential growth phase and washed with Sol buffer (20 mm Tris/HCl, pH 7.5, 400 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol). 60 g of cells were resuspended in 200 ml of Sol buffer (including 0.5 mm PMSF) and disrupted by three passages through a French press followed by sonification (30 s; duty cycle, 50%; output control, 5). The membranes were collected by ultracentrifugation at 48,000 rpm for 1 h and washed once with Sol buffer containing 1% n-dodecyl-β-d-maltoside. Washed membranes were resuspended in Sol buffer again, and Triton X-100 was added (3 g/g of protein). Solubilized proteins were separated from membranes by ultracentrifugation (48,000 rpm, 4 °C, 1 h). 4% SDS was added to the solubilizate, and the sample was then loaded on a continuous 15–50% (w/v) sucrose gradient (in Sol buffer and 0.1% Triton X-100) and ultracentrifuged at 48,000 rpm and 4 °C for 24 h. PilQ-containing fractions were pooled and diluted with Sol buffer (including 0.1% Triton X-100) without NaCl in a 1:4 ratio to obtain a NaCl concentration of about 100 mm. Samples were loaded on a Q-Sepharose, and bound proteins were eluted by a gradient from 100 mm NaCl to 1 m NaCl (in Sol buffer containing 0.1% Triton X-100). Fractions that contained PilQ were pooled and concentrated using a Vivaspin 20 column. The concentrated samples were loaded on a Superose 6 column and eluted with 1.5 column volumes of Sol buffer containing 0.1% Triton X-100. For detection, the fractions were applied to 3–12% polyacrylamide gradient SDS gels and immunoblotted against the PilQ antiserum.
An alternative purification strategy was developed by taking advantage of the SDS resistance of PilQ. Therefore, membranes were treated with 10% SDS and incubated for 15 min at 100 °C. Membranes were separated via ultracentrifugation, and 0.1% Triton X-100 was added to the solubilized proteins, which were incubated at 4 °C overnight to remove the precipitating SDS. The supernatant was concentrated by a (NH4)2SO4 precipitation (20%) and solved in Sol buffer containing 0.1% Triton X-100. Finally, the highly stable PilQ complex was separated by a continuous 15–40% (w/v) sucrose gradient centrifugation, and PilQ-containing fractions were pooled.
Subunit Composition of the PilQ Complex
To analyze the subunit composition, the purified PilQ complex was separated by Blue native-PAGE and electroeluted (27) and treated with hot phenol as described by Hancock and Nikaido (29). The phenol-treated PilQ complex was subjected to SDS-PAGE and Western blot analyses.
Electron Microscopy and Image Analysis
PilQ complexes were negatively stained with 1% (w/v) uranyl acetate. Electron micrographs were collected using a Philips CM120 at 120 kV under low dose conditions, at a magnification of ×44,000 on Kodak SO-163 electron image film. Negatives checked by optical diffraction for correct defocus and a lack of drift and astigmatism were digitized on a PhotoScan scanner (Z/I Imaging, Aalen, Germany) at a pixel size of 7 μm. Subsequently, adjacent pixels were averaged to yield a pixel size on the specimen of 4.77 Å. Particles were selected using the boxer module from EMAN (30). The images were processed using Imagic V (31). The images were band-pass-filtered and subjected to reference-free alignment and multivariate statistical analysis and classification. Images assigned to the same class were averaged.
Electrophoretic Gel Mobility Shift Assay (EMSA)
Probes were made by PCR from genomic DNA of T. thermophilus using primers ComEA-1 (5′-TTTGGAATTCAAGTGCGGGTGGAGGCCCTGGGAAAGGTC-3′) and ComEA-2 (5′-GAATAAGCTTAGGTCCAAGAAGGCCTCATGGCCGGAGGTAG-3′) and from genomic DNA of Halobacillus halophilus using primers RT-reg forward (5′-GATCAGATGACACTTGATG-3′) and RT-reg reverse (5′-AGCTTCCGTAATGGCTTC-3′). The resulting 250- and 230-bp DNA fragments, respectively, were purified and backfilled using Klenow large fragment with α-[32P]CTP and purified using the PCR purification kit from Roche Applied Science.
For electromobility shift assays, the indicated quantities (5–200 nm) of the purified PilQ complex and 1 nm unlabeled dsDNA where resuspended in 50 mm MOPS, pH 7.5, 1 mm EDTA, 5 mm MgCl2, 1 mm DTT, and 5% glycerol. The dsDNA fragments (1 nm, 30,000 cpm) were incubated with or without various amounts of the PilQ complex for 30 min at 37 and 68 °C, respectively, and separated on nondenaturating 3% polyacrylamide gels (in 1-fold Tris acetate/EDTA buffer). Gels were dried and subjected to phosphorimaging analyses using a Typhoon 9100 (GE Healthcare). For competition experiments, PilQ was incubated for 15 min with a 20-fold excess of unlabeled probe.
RESULTS
PilQ Forms an Extremely Stable Complex
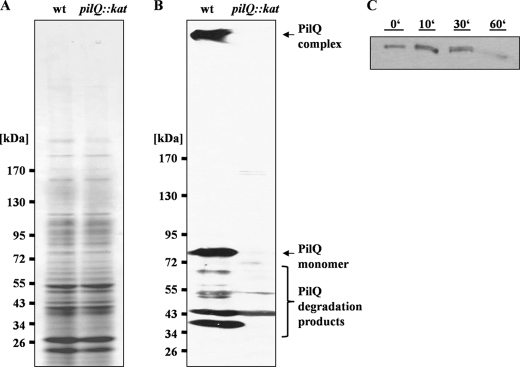
Cell-free extracts of T. thermophilus were separated on an SDS-PAGE gel and subjected to Western blot analysis with a PilQ antibody (Fig. 1, A and B). An 83-kDa protein reacted with the antibody (Fig. 1B). The apparent molecular mass fits well to the deduced mass of PilQ. In addition, three less abundant proteins of 72, 55, and 43 kDa reacted with the antibody, but they were also detected in a pilQ mutant, indicating that they do not represent degradation products of PilQ. Most interestingly, we reproducibly observed a large protein on top of the gel migrating very slowly. This protein reacted with the PilQ antibody and was not present in the pilQ mutant (Fig. 1B) and thus represents a PilQ-containing,-SDS resistant protein complex. This is consistent with secretin complexes of T2SS and phage secretion, such as PulD and pIV, which also exhibit SDS resistance (32, 33). SDS resistance has also been described for different secretins of T4P and T2SS/T3SS systems (34–39). However, secretins exhibit different resistances against heat treatment, such that XcpQ, BfpB, TcpC, or OutD secretin complexes do not withstand boiling in standard Laemmli sample buffer (40–42). In contrast, the T. thermophilus PilQ complex was even detected in the membranes after boiling the membranes for 30 min in 10% SDS, suggesting an extraordinary high stability of the secretin complex (Fig. 1C).
FIGURE 1.
Detection of a PilQ complex in T. thermophilus HB27. A and B, 3–12% polyacrylamide gradient SDS-PAGE of crude extracts of HB27 wild type (wt) and pilQ mutant (pilQ::kat) (A) followed by immunoblotting with antiserum against PilQ (B). C, membranes of HB27 were treated with 10% SDS and incubated at 100 °C. Samples were taken at the indicated time points and subjected to Western blot analyses with PilQ antibodies.
The PilQ Complex Is Associated with Outer Membranes
To determine the subcellular localization of the PilQ complex, total membranes were separated from the soluble fraction by ultracentrifugation, and IM and OM were further separated as described previously (22). Successful separation of IM and OM was verified using antibodies against the S-layer protein (OM) and PilN (IM). Immunoblotting of the total membranes led to the detection of a PilQ complex retaining on top of the separating gel (Fig. 2, lane M). In addition, substantial amounts of the PilQ monomer and small amounts of PilQ degradation products were detected, and minor amounts of the PilQ monomer and the immunoreactive degradation products were also detected in the soluble fraction. Immunoblotting revealed that PilQ was insoluble in N-laurylsarcosine, which is indicative for PilQ being in the OM; minor amounts of PilQ were found to be sarcosyl-soluble (IM). This may reflect solubility of PilQ in sarcosyl to some extent. On the other hand, when outer and inner membranes were separated from each other by sucrose gradient centrifugation, the PilQ complex was also found to some minor extent in the inner membrane fraction (data not shown).
FIGURE 2.
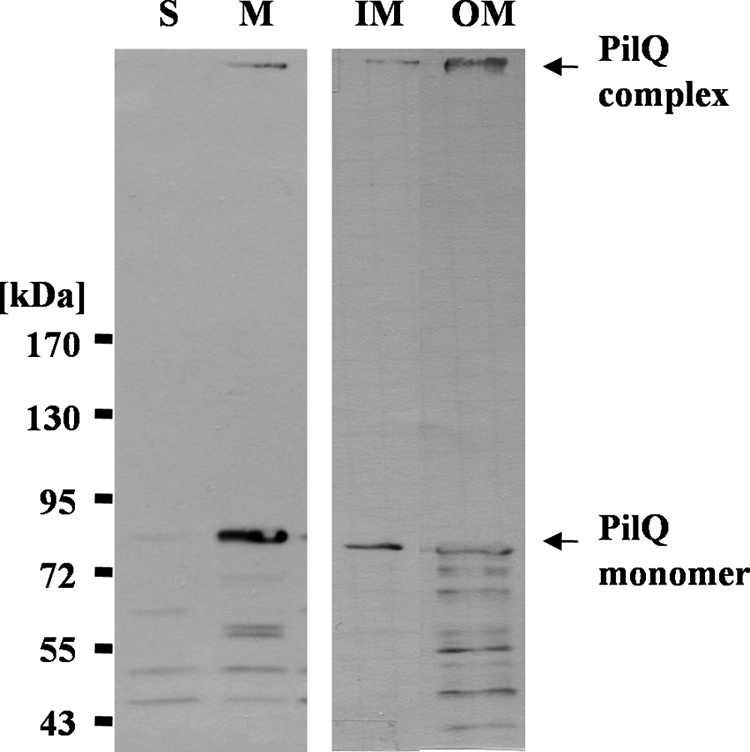
Subcellular localization of PilQ. T. thermophilus crude extracts were separated in soluble (S) and membrane (M) fractions. Total membranes were separated in IM and OM fractions. The fractions were subjected to SDS-PAGE in a 3–12% polyacrylamide gradient gel followed by immunoblotting with PilQ antibodies.
Purification of the PilQ Complex
The PilQ complex was purified from Thermus membranes by solubilization with the nonionic detergent Triton X-100, and the addition of 4% SDS to the membrane solubilizate prior to sucrose density gradient centrifugation was followed by anion exchange chromatography (Q-Sepharose) and gel filtration (Superose 6). The pooled PilQ fractions after anion exchange chromatography revealed that several proteins copurified with the PilQ complex (Fig. 3, lane 5). After gel filtration, an apparently homogeneous PilQ preparation was obtained (Fig. 3, lane 6). Minor contaminants in the preparation varied from preparation to preparation and contained pyruvate dehydrogenase subunit E1 (102 kDa), chaperonin GroEL (57.8 kDa), a nonconserved 49-kDa protein (TTC1295), and a potential dihydrolipoamide succinyltransferase (44.5 kDa).
FIGURE 3.
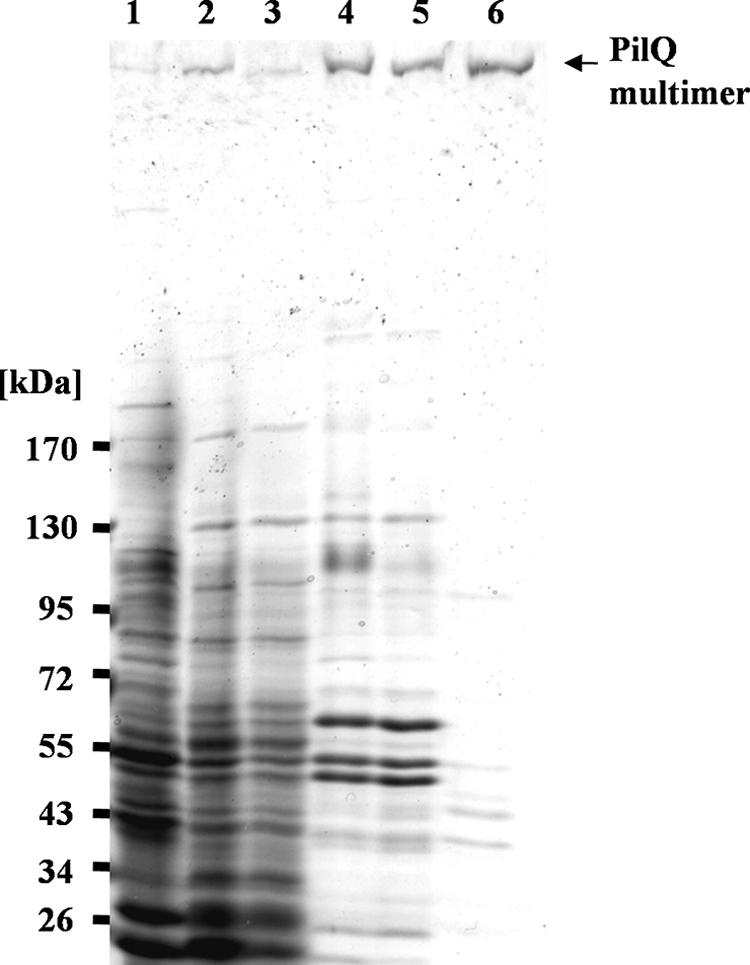
Purification of the PilQ complex. Protein fractions of the different steps of the purification procedure were analyzed on a 3–12% polyacrylamide gradient SDS-PAGE gel. Lane 1, crude extract; lane 2, membrane fraction; lane 3, solubilized proteins; lane 4, pooled PilQ-containing fractions of the density gradient; lane 5, pooled PilQ-containing fractions of the anion exchange chromatography (Q-Sepharose); lane 6, pooled PilQ-containing fractions of the gel filtration (Superose 6).
The PilQ Complex Is an Apparent Homomultimer
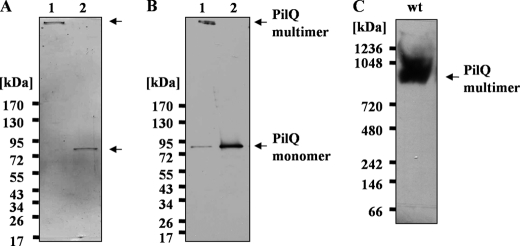
To elucidate its subunit composition, the purified PilQ complex was subjected to Blue native-PAGE, electroeluted, and treated with hot phenol (29). As shown in Fig. 4, A and B (lane 2), this treatment leads to a dissociation of the PilQ complex and formation of PilQ monomers. Only PilQ monomers were detected after phenol treatment, which provides clear evidence that the PilQ complex is a homomultimer. The apparent molecular mass of the PilQ complex, as determined by Blue native-PAGE, corresponds to ∼960 kDa, which is consistent with a homopolymeric complex comprising 12 monomers (Fig. 4C).
FIGURE 4.
Subunit composition of the PilQ complex. A and B, the purified PilQ complex was treated with hot phenol and subjected to 3–12% polyacrylamide gradient SDS-PAGE (A) and immunoblotting (B). Lane 1, untreated PilQ complex; lane 2, PilQ complex treated with hot phenol. C, the purified untreated PilQ complex was subjected to Blue native-PAGE followed by immunoblotting.
Electron Microscopy and Image Analysis
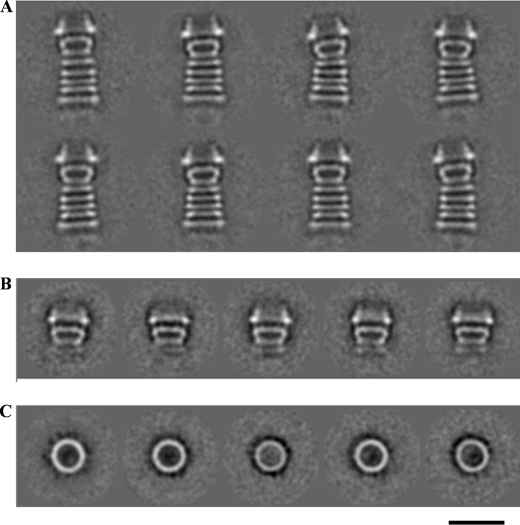
The purified PilQ complex was negatively stained and studied by electron microscopy. The sample shows long structures consisting of a number of stripes (Fig. 5A). About 2000 images were selected from 33 micrographs and aligned and classified, and images in the same class were averaged. All class averages showed the same view. The complex has a width of ∼15 nm and a length of 34 nm and consists of five distinct rings, each ∼3 nm thick, followed by a trapezoid density with a dense base (the “cup”) and a conical domain on the top.
FIGURE 5.
Electron microscopic analysis of PilQ. A, class averages from a data set of 2000 PilQ particles after multivariate statistical analysis and classification. B and C, class averages of SDS-treated PilQ from a data set of 4000 particles. B, side views; C, top views. The scale bar represents 20 nm for all panels.
When membranes were treated with 10% SDS at 100 °C and PilQ was subsequently purified by ammonium sulfate precipitation and sucrose density gradient centrifugation, the complex was smaller, with a height of ∼20 nm, consisting of the cone and cup domains as well as a faint image of the first of the five rings, but lacking the lower four rings (Fig. 5B). Presumably, SDS treatment denatures the protein domains forming the lower rings, but not the domains that retain the cone, the cup, and the first ring structure.
The SDS-treated PilQ exhibited ring-shaped top views (Fig. 5C), which were not seen in negatively stained samples of the untreated complex with its 34-nm-high complexes. These top views are similar in appearance to other secretin complexes. The rings have a diameter of 15 nm and a thickness of ∼3 nm with a ∼6-nm hole in the center. Weak densities can be seen extending from the edge of the ring, indicating a subunit number of 12–14, which is consistent with the apparent molecular mass of the PilQ complex (Fig. 4C), but the exact number could not be determined unambiguously.
The PilQ Complex Binds DNA
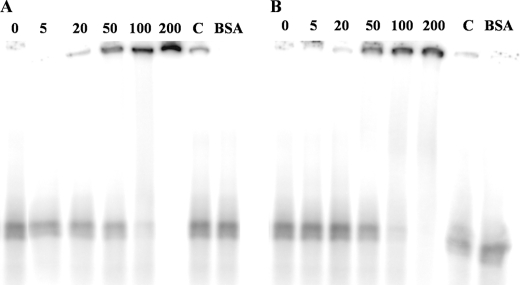
In a previous work, we showed that T. thermophilus HB27 is able to bind and transport DNA from members of all three domains of life (24). Mutant studies revealed that PilQ is implicated in DNA binding. To investigate the ability of the PilQ complex to bind DNA directly, bandshift assays were performed using radioactively labeled DNA from T. thermophilus and H. halophilus. As shown in Fig. 6, A and B, Thermus PilQ is shifting dsDNA of H. halophilus in a concentration-dependent manner at 37 and 68 °C. Competition experiments indicate a specific interaction of the PilQ complex with dsDNA. Bandshift assays with Thermus DNA led to the same results (data not shown). Thus, the PilQ complex binds homologous and heterologous DNA.
FIGURE 6.
DNA binding of the PilQ complex measured by bandshift analyses. Reaction mixtures contained 1 nm (30,000 cpm) of dsDNA. DNA was incubated with 5, 20, 50, 100, and 200 nm purified PilQ complex at 37 °C (A) and 68 °C (B), respectively. Competitor DNA was added to 200 nm PilQ complex in 20-fold excess (lane C). 200 nm BSA was used as negative control.
DISCUSSION
The secretin family is known to form oligomeric ring-like structures in the outer membranes of Gram-negative bacteria, which are specialized in translocating their particular substrates (36, 37, 43, 44). Here we show that Thermus PilQ forms oligomeric complexes in the membrane fraction and binds DNA. Through these PilQ complexes, pilus structures and the DNA translocator are thought to be extruded because deletion of the pilQ gene leads to a loss of transformation, piliation, and absence of PilA4 in the OM (9, 22). The PilQ complex was found to be associated with the OM.
Because the PilQ complex is part of the DNA translocator in Thermus, which is composed of several inner and outer membrane proteins and protein complexes, an association with the inner membrane is imaginable and might be important for translocating DNA across both membranes. Association with the inner membrane has been found for several secretins of secretion and pilus systems and is explained by trafficking to the outer membrane or interactions at the outer face of the inner membrane (41, 45–48). Indeed, we also found PilQ complex present to some minor extent in the sarcosyl fraction as well as in the inner membrane fraction after separation of IM and OM by density gradient centrifugation.
Furthermore, we show that PilQ of T. thermophilus has the most complex structure of a secretin known to date; it measures ∼34 nm in height, and the height of this complex corresponds to the diameter of the extraordinary thick cell wall layers of T. thermophilus, comprising peptidoglycan covered by a thick layer of strongly bound secondary cell wall polymers (49–51) with a diameter of ∼40 nm (24). These molecular dimensions are consistent with the hypothesis that the PilQ complex spans the OM and the periplasmic space, mediating DNA binding on the cell surface on one side and interacting with IM components of the DNA translocator on the other side. Potential interacting IM components of the DNA translocator could be PilM, PilN, PilO, and/or PilW, which have been detected in the inner membrane of T. thermophilus (22). Moreover, the diameter of the channel in the PilQ ring structure (∼6 nm), together with our recent finding that in a PilQ mutant no pilin-like PilA4 competence protein is detected in the outer membrane, is consistent with the hypothesis that the stacked PilQ ring structures accommodate a pseudopilus-like DNA translocator, composed of pilin-like proteins, through the outer membrane and the periplasmic space (22).
Secretins are known to be involved in a wide variety of systems, for example in protein secretion, pilus assembly, and filamentous phage assembly and secretion, and for several of them, electron microscopic structures have been published (36, 39, 52–60). The structures of several T4P and T2SS/T3SS secretins have been determined by electron microscopy and partially by crystallization, and although some differences in symmetry and the nature of the constriction or plug were found, they share a similar basic overall structure (61, 62). Members of the secretin family form large, ring-shaped, oligomeric complexes with a central cavity, which can mediate protein export (63). The C termini of members of the secretins are homologous (61). These domains, designated the “secretin homology region,” were found to be important for species-specific functioning, oligomerization of the ring-shaped structure, and channel formation, whereas the N-terminal domains are only less conserved in secretins of similar transport systems (53, 63–67).
Top views of secretins showed ring-like structures ranging from 12 to 20 nm in diameter, with large central channels ranging from 5 to 10 nm in diameter (43). Side views revealed the presence of several stacked ring-shaped domains with a height of about 10–20 nm, where two of the rings form a cup-like structure, which is stacked on another ring-like structure. Such a structure is also part of the Thermus PilQ complex and builds the SDS-resistant core (Fig. 5B). However, in contrast to the cup-like structures of PulD and pIV (54, 59), the PilQ complex has five additional, well resolved rings below the cup-like part of the structure, as shown in our electron micrographs, and therefore, it differs from all T2SS/T3SS and T4P secretin complexes known to date (Fig. 5A). The PilQ complex exhibits a remarkable SDS stability; for example, treatment of the membrane solubilizate with 4% SDS did not destroy the multiple ring structures of the PilQ complex. The cup-like part of the PilQ complex was found to exhibit an even higher SDS stability because it is still assembled after treatment of the membranes with 10% SDS at 100 °C (Fig. 5B). Presumably, the lower rings get denatured by boiling in SDS and are no longer structured and thus not visible in averaged EM images.
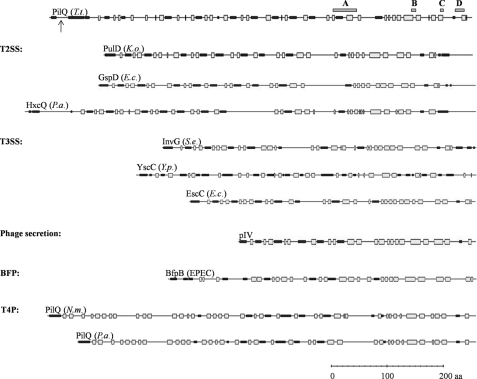
It has been proposed for T3SS and T2SS secretins such as PulD, GspD, and pIV that the upper ring and the bottom of the cup-like structure, which is termed the periplasmic gate, are embedded in the outer membrane (24, 54, 56, 59–61). These parts are formed by the C-terminal, protease-resistant secretin homology region, which contains potentially amphipathic transmembrane β-strands. To compare structural similarities of the secretins, the secondary structures were predicted (Fig. 7). The similarities of the C terminus of PilQ to membrane-embedded secretin homology domains of other secretins, as well as the large number of predicted β-strands in the C-terminal region and the extraordinary SDS resistance of a cup-like PilQ subcomplex, lead to the assumption that the C-terminal conserved secretin homology domain spanning the conserved domains A to D is also embedded in the outer membrane, whereas the additional five rings are formed by the nonconserved N-terminal domains of PilQ. Although the N termini of secretins do not exhibit significant amino acid similarities, they share conserved modular architectures (Fig. 7). The N termini of T2SS secretins (PulD, GspD, and HxcQ), the secretins of T3SS, such as InvG, YscC, and EscC, and those of phage secretion, such as pIV and secretins of BfpB, share alternating α-helical and β-sheet domains (Fig. 7). However, the N termini of Neisseria and Pseudomonas T4P secretins (PilQ) differ with respect to their secondary structure (Fig. 7). Interestingly, the secondary structure differences correspond to the finding that the meningococcal PilQ complex, which is essential for T4P biogenesis and natural transformation, differs in several important aspects from T2SS/T3SS and phage secretin complexes (52, 54, 68). The Neisseria PilQ exhibits a rounded, conical profile, whereas secretins of T2SS, such as the dodecameric PulD complex of Klebsiella oxytoca, which is remarkably similar in electron microscopic structure to the pIV complex of phage secretion, are composed of two rings that sandwich a closed disc (54, 58, 59, 68).
FIGURE 7.
Secondary structure prediction of secretins. The PredictProtein server was used to predict potentially β-sheet (gray boxes) and α-helical (black boxes) regions within the secretins shown. Conserved C-terminal domains are indicated as dark gray boxes A, B, C, and D (61) for PilQ of T. thermophilus; the processing site is indicated by an arrow. T. t., T. thermophilus; K. o., K. oxytoca; E. c., E. coli; P. a., Pseudomonas aeruginosa; S. e., Salmonella enterica; Y. p., Yersinia pestis; EPEC, enteropathogenic E. coli; BFP, bundle-forming pilus system; N. m. Neisseria meningitidis.
It has been suggested that the conserved architecture of secretins might facilitate structural rearrangements in the fully assembled secretin complexes through variations in the connecting linkers (56). Recently, a conserved structural motif mediating the formation of periplasmic rings in secretin complexes of T3SS has been identified by the crystal structure of the periplasmic domain (amino acids 21–174) of the outer membrane secretin EscC of the T3SS of the enteropathogenic Escherichia coli (56). The conserved structural motif exhibits a wedge-shaped structure and consists of a two-domain fold (spanning 153 amino acids); each domain fold comprises about 70–80 amino acids forming two α-helices folded against a β-sheet (56). The striking similarity in fold and architecture between EscC and inner membrane ring components of the T3SS led to the suggestion that the wedge-shaped fold might provide a common ring-building motif of the T3SS basal body. The secondary structure of the nonconserved N terminus of the mature PilQ protein (amino acids 21–505) exhibits an EscC-analogous modular organization with alternating α-helical and β-sheet domains (Fig. 7). In comparison with the EscC secretin, the mature T. thermophilus secretin PilQ exhibits a significantly extended N terminus spanning ∼500 amino acids. Due to the conserved modular architecture of the N terminus together with the length of the nonconserved N terminus of PilQ, it is tempting to speculate that the conserved architecture of the N terminus facilitates the assembly of the five-ring system underneath the cup-like structure of the fully assembled PilQ complex and that these rings are localized in the periplasmic space. This assumption is further affirmed by the published crystal structure of the N-terminal part of GspD (T2SS), which also revealed a modular architecture, composed of consecutively linked domains with mixed α-helical and β-sheet topology (60, 69).
In conclusion, our work has unraveled a novel, very complex structure of a secretin complex, the first DNA translocator secretin complex detected in a thermophilic bacterium. Given the conservation in the C-terminal primary and secondary structure and the modular architecture of the nonconserved N terminus matching the domains of other secretins, it is likely that the 500 N-terminal amino acids are forming a complex structure comprising five staged ring systems spanning the periplasmic space and guiding the DNA translocator pseudopilus through the OM and periplasmic space. The strikingly complex structure of the PilQ complex sets the frame for the future structural, biochemical, and functional studies of secretins in DNA translocator systems.
Acknowledgments
We are grateful to José Berenguer (Universidad Autonomes de Madrid, Spain) for providing antibodies against T. thermophilus HB27 S-layer protein, which were used to analyze the purity of membrane fractionation. We also thank Tobias Beckhaus (Karas group, Goethe University, Frankfurt, Germany) for MADLI-TOF analyses of copurified proteins. Volker Müller (Goethe University, Frankfurt, Germany) is gratefully acknowledged for stimulating discussions.
Footnotes
- T4P
- type IV pili
- T2SS
- type II protein secretion systems
- T3SS
- type III protein secretion systems
- IM
- inner membrane
- OM
- outer membrane.
REFERENCES
- 1. Johnsborg O., Eldholm V., Håvarstein L. S. (2007) Res. Microbiol. 158, 767–778 [DOI] [PubMed] [Google Scholar]
- 2. Averhoff B. (2009) FEMS Microbiol. Rev. 33, 611–626 [DOI] [PubMed] [Google Scholar]
- 3. Averhoff B. (2004) J. Bioenerg. Biomembr. 36, 25–33 [DOI] [PubMed] [Google Scholar]
- 4. Hobbs M., Mattick J. S. (1993) Mol. Microbiol. 10, 233–243 [DOI] [PubMed] [Google Scholar]
- 5. Tønjum T., Freitag N. E., Namork E., Koomey M. (1995) Mol. Microbiol. 16, 451–464 [DOI] [PubMed] [Google Scholar]
- 6. Seifert H. S., Ajioka R. S., Paruchuri D., Heffron F., So M. (1990) J. Bacteriol. 172, 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koomey M. (1998) APMIS Suppl. 84, 56–61 [DOI] [PubMed] [Google Scholar]
- 8. Graupner S., Frey V., Hashemi R., Lorenz M. G., Brandes G., Wackernagel W. (2000) J. Bacteriol. 182, 2184–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friedrich A., Prust C., Hartsch T., Henne A., Averhoff B. (2002) Appl. Environ. Microbiol. 68, 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen I., Christie P. J., Dubnau D. (2005) Science 310, 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen I., Dubnau D. (2004) Nat. Rev. Microbiol. 2, 241–249 [DOI] [PubMed] [Google Scholar]
- 12. Tønjum T., Koomey M. (1997) Gene 192, 155–163 [DOI] [PubMed] [Google Scholar]
- 13. Fussenegger M., Rudel T., Barten R., Ryll R., Meyer T. F. (1997) Gene 192, 125–134 [DOI] [PubMed] [Google Scholar]
- 14. Collins R. F., Frye S. A., Balasingham S., Ford R. C., Tønjum T., Derrick J. P. (2005) J. Biol. Chem. 280, 18923–18930 [DOI] [PubMed] [Google Scholar]
- 15. Chen I., Provvedi R., Dubnau D. (2006) J. Biol. Chem. 281, 21720–21727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maier B., Koomey M., Sheetz M. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10961–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maier B., Potter L., So M., Long C. D., Seifert H. S., Sheetz M. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16012–16017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Schaik E. J., Giltner C. L., Audette G. F., Keizer D. W., Bautista D. L., Slupsky C. M., Sykes B. D., Irvin R. T. (2005) J. Bacteriol. 187, 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Assalkhou R., Balasingham S., Collins R. F., Frye S. A., Davidsen T., Benam A. V., Bjørås M., Derrick J. P., Tønjum T. (2007) Microbiology 153, 1593–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Friedrich A., Hartsch T., Averhoff B. (2001) Appl. Environ. Microbiol. 67, 3140–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedrich A., Rumszauer J., Henne A., Averhoff B. (2003) Appl. Environ. Microbiol. 69, 3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rumszauer J., Schwarzenlander C., Averhoff B. (2006) FEBS J. 273, 3261–3272 [DOI] [PubMed] [Google Scholar]
- 23. Rose I., Biukovic G., Aderhold P., Müller V., Grüber G., Averhoff B. (January 6, 2011) Extremophiles [DOI] [PubMed] [Google Scholar]
- 24. Schwarzenlander C., Haase W., Averhoff B. (2009) Environ. Microbiol. 11, 801–808 [DOI] [PubMed] [Google Scholar]
- 25. Koyama Y., Hoshino T., Tomizuka N., Furukawa K. (1986) J. Bacteriol. 166, 338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 27. Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 28. Maier E., Polleichtner G., Boeck B., Schinzel R., Benz R. (2001) J. Bacteriol. 183, 800–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hancock R. E., Nikaido H. (1978) J. Bacteriol. 136, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 31. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 32. Hardie K. R., Lory S., Pugsley A. P. (1996) EMBO J. 15, 978–988 [PMC free article] [PubMed] [Google Scholar]
- 33. Spagnuolo J., Opalka N., Wen W. X., Gagic D., Chabaud E., Bellini P., Bennett M. D., Norris G. E., Darst S. A., Russel M., Rakonjac J. (2010) Mol. Microbiol. 76, 133–150 [DOI] [PubMed] [Google Scholar]
- 34. Helm R. A., Barnhart M. M., Seifert H. S. (2007) J. Bacteriol. 189, 3198–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drake S. L., Sandstedt S. A., Koomey M. (1997) Mol. Microbiol. 23, 657–668 [DOI] [PubMed] [Google Scholar]
- 36. Bitter W., Koster M., Latijnhouwers M., de Cock H., Tommassen J. (1998) Mol. Microbiol. 27, 209–219 [DOI] [PubMed] [Google Scholar]
- 37. Koster M., Bitter W., de Cock H., Allaoui A., Cornelis G. R., Tommassen J. (1997) Mol. Microbiol. 26, 789–797 [DOI] [PubMed] [Google Scholar]
- 38. Guilvout I., Hardie K. R., Sauvonnet N., Pugsley A. P. (1999) J. Bacteriol. 181, 7212–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collins R. F., Davidsen L., Derrick J. P., Ford R. C., Tønjum T. (2001) J. Bacteriol. 183, 3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Daniel A., Singh A., Crowther L. J., Fernandes P. J., Schreiber W., Donnenberg M. S. (2006) Microbiology 152, 2405–2420 [DOI] [PubMed] [Google Scholar]
- 41. Bose N., Taylor R. K. (2005) J. Bacteriol. 187, 2225–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shevchik V. E., Robert-Baudouy J., Condemine G. (1997) EMBO J. 16, 3007–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thanassi D. G. (2002) J. Mol. Microbiol. Biotechnol. 4, 11–20 [PubMed] [Google Scholar]
- 44. Koster M., Bitter W., Tommassen J. (2000) Int. J. Med. Microbiol. 290, 325–331 [DOI] [PubMed] [Google Scholar]
- 45. Clock S. A., Planet P. J., Perez B. A., Figurski D. H. (2008) J. Bacteriol. 190, 980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. d'Enfert C., Reyss I., Wandersman C., Pugsley A. P. (1989) J. Biol. Chem. 264, 17462–17468 [PubMed] [Google Scholar]
- 47. Ramer S. W., Bieber D., Schoolnik G. K. (1996) J. Bacteriol. 178, 6555–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Russel M., Kaźmierczak B. (1993) J. Bacteriol. 175, 3998–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Caston J. R., Carrascosa J., de Pedro M. A., Berenguer J. (1988) FEMS Microbiol. Lett. 51, 225–230 [Google Scholar]
- 50. Faraldo M. M., de Pedro M. A., Berenguer J. (1992) J. Bacteriol. 174, 7458–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fernández-Herrero L. A., Olabarría G., Castón J. R., Lasa I., Berenguer J. (1995) J. Bacteriol. 177, 5460–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nouwen N., Ranson N., Saibil H., Wolpensinger B., Engel A., Ghazi A., Pugsley A. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8173–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nouwen N., Stahlberg H., Pugsley A. P., Engel A. (2000) EMBO J. 19, 2229–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Opalka N., Beckmann R., Boisset N., Simon M. N., Russel M., Darst S. A. (2003) J. Mol. Biol. 325, 461–470 [DOI] [PubMed] [Google Scholar]
- 55. Burghout P., van Boxtel R., Van Gelder P., Ringler P., Müller S. A., Tommassen J., Koster M. (2004) J. Bacteriol. 186, 4645–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Spreter T., Yip C. K., Sanowar S., André I., Kimbrough T. G., Vuckovic M., Pfuetzner R. A., Deng W., Yu A. C., Finlay B. B., Baker D., Miller S. I., Strynadka N. C. (2009) Nat. Struct. Mol. Biol. 16, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schmidt S. A., Bieber D., Ramer S. W., Hwang J., Wu C. Y., Schoolnik G. (2001) J. Bacteriol. 183, 4848–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Collins R. F., Ford R. C., Kitmitto A., Olsen R. O., Tønjum T., Derrick J. P. (2003) J. Bacteriol. 185, 2611–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chami M., Guilvout I., Gregorini M., Rémigy H. W., Müller S. A., Valerio M., Engel A., Pugsley A. P., Bayan N. (2005) J. Biol. Chem. 280, 37732–37741 [DOI] [PubMed] [Google Scholar]
- 60. Reichow S. L., Korotkov K. V., Hol W. G., Gonen T. (2010) Nat. Struct. Mol. Biol. 17, 1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Genin S., Boucher C. A. (1994) Mol. Gen. Genet. 243, 112–118 [DOI] [PubMed] [Google Scholar]
- 62. Bayan N., Guilvout I., Pugsley A. P. (2006) Mol. Microbiol. 60, 1–4 [DOI] [PubMed] [Google Scholar]
- 63. Bitter W., van Boxtel R., Groeneweg M., Carballo P. S., Zähringer U., Tommassen J., Koster M. (2007) J. Bacteriol. 189, 2967–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brok R., Van Gelder P., Winterhalter M., Ziese U., Koster A. J., de Cock H., Koster M., Tommassen J., Bitter W. (1999) J. Mol. Biol. 294, 1169–1179 [DOI] [PubMed] [Google Scholar]
- 65. Bouley J., Condemine G., Shevchik V. E. (2001) J. Mol. Biol. 308, 205–219 [DOI] [PubMed] [Google Scholar]
- 66. Daefler S., Guilvout I., Hardie K. R., Pugsley A. P., Russel M. (1997) Mol. Microbiol. 24, 465–475 [DOI] [PubMed] [Google Scholar]
- 67. Korotkov K. V., Krumm B., Bagdasarian M., Hol W. G. (2006) J. Mol. Biol. 363, 311–321 [DOI] [PubMed] [Google Scholar]
- 68. Collins R. F., Frye S. A., Kitmitto A., Ford R. C., Tønjum T., Derrick J. P. (2004) J. Biol. Chem. 279, 39750–39756 [DOI] [PubMed] [Google Scholar]
- 69. Korotkov K. V., Pardon E., Steyaert J., Hol W. G. (2009) Structure 17, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]