SUMMARY
Osmotic stress imposed by soil salinity and drought stress significantly affects plant growth and development, but osmotic stress sensing and tolerance mechanisms are not well understood. Forward genetic screens using a root-bending assay have previously identified salt overly sensitive (sos) mutants of Arabidopsis that fall into five loci, SOS1 to SOS5. These loci are required for the regulation of ion homeostasis or cell expansion under salt stress, but do not play a major role in plant tolerance to the osmotic stress component of soil salinity or drought. Here we report an additional sos mutant, sos6-1, which defines a locus essential for osmotic stress tolerance. sos6-1 plants are hypersensitive to salt stress and osmotic stress imposed by mannitol or polyethylene glycol in culture media or by water deficit in the soil. SOS6 encodes a cellulose synthase-like protein, AtCSLD5. Only modest differences in cell wall chemical composition could be detected, but we found that sos6-1 mutant plants accumulate high levels of reactive oxygen species (ROS) under osmotic stress and are hypersensitive to the oxidative stress reagent methyl viologen. The results suggest that SOS6/AtCSLD5 is not required for normal plant growth and development but has a critical role in osmotic stress tolerance and this function likely involves its regulation of ROS under stress.
Keywords: Arabidopsis, cell wall, cellulose synthase-like protein, osmotic stress tolerance, reactive oxygen species, SOS6
INTRODUCTION
Salinity is a major abiotic stress that severely limits the productivity of crop plants world wide. An estimated 20% of cultivated land and approximately 40% of irrigated land are affected by salinity (Rhoades and Loveday, 1990). Understanding the molecular basis of salt stress signaling and tolerance mechanisms is critical for developing rational breeding and genetic engineering strategies for improving salt tolerance of crops.
Salinity causes ionic stress for plants. Plants have evolved mechanisms to maintain ion homeostasis in the cell cytoplasm by coordinating the activities of various ion transporters. Genetic, molecular and biochemical analysis has led to the identification of the Salt Overly Sensitive (SOS) pathway that regulates ion homeostasis in plants (Zhu, 2003). The calcium-binding protein, SOS3, is suggested to sense changes in cytosolic free calcium elicited by salt stress (Liu and Zhu, 1998; Ishitani et al., 2000). SOS3 interacts with and activates the protein kinase SOS2 (Halfter et al., 2000; Liu et al., 2000; Guo et al., 2001). The SOS3–SOS2 protein kinase complex regulates the expression and activity of a plasma membrane-localized Na+/H+ antiporter SOS1, which exports Na+ to the apoplast (Shi et al., 2000; Qiu et al., 2002; Quintero et al., 2002). Transgenic plants that overexpress SOS1 showed improved tolerance to salt stress (Shi et al., 2003a). Recent studies suggest that SOS2 also positively regulates the activities of tonoplast Na+/H+ anti-porters (Qiu et al., 2004), which sequester Na+ ions in the vacuole and of a vacuolar H+/Ca2+exchanger CAX1 (Cheng et al., 2004). Forward genetic screens in tomato also led to the isolation of several salt-hypersensitive mutants, although the mutated genes have yet to be cloned (Borsani et al., 2001a).
Salinity, as well as drought, also imposes osmotic stress on plants, resulting in cellular dehydration and growth inhibition. Salt and drought stress induces abscisic acid (ABA), which has a major role in various aspects of stress tolerance (Zhu, 2002). Besides ABA, little is known about the determinants of osmotic stress tolerance and forward genetic screens in Arabidopsis have thus far not yielded informative mutants. A large body of evidence shows that various protein kinases are activated by osmotic stress, although in many cases the function of the kinases in osmotic stress response pathways has yet to be defined (Zhu, 2002).
The cell wall is a unique feature which distinguishes plants from animals. The major polysaccharides in the primary cell wall consist of cellulose, hemicelluloses and pectins. The cellulose microfibrils are linked via hemicellulosic tethers to form the cellulose-hemicellulose network, which is embedded in the pectin matrix. Cellulose is a polysaccharide composed of 1,4-linked β-D-glucose residues accounting for 15% to 30% of the dry mass of primary cell wall and provides a rigid cellular environment. The most common hemicellulose in the primary cell wall is xyloglucan. In contrast, pectins are a mixture of heterogenous, branched and highly hydrated polysaccharides rich in D-galacturonic acid. A group of genes encoding cellulose synthases (CesA), which belong to the glycosyltransferase family, are responsible for the biosynthesis of cellulose in plants (Somerville, 2006). Report by Chen et al. (2005) has shown that mutation in one of the 10 Arabidopsis CesAs, AtCesA8, results in enhanced drought and osmotic stress tolerance. Plants also contain a large number of genes encoding cellulose synthase-like (CSL) proteins. The Arabidopsis genome encodes six subfamilies of CSL proteins (AtCSLA, AtCSLB, AtCSLC, AtCSLD, AtCSLE and AtCSLG) (Richmond and Somerville, 2000), most of which have not been assigned a physiological function. Two additional subfamilies (OsCSLF and OsCSLH) have been identified in rice (Oryza sativa). CSL proteins might function in the biosynthesis of non-cellulosic polysaccharides such as xylans, xyloglucans, galactans and mannans (Richmond and Somerville, 2001). This conjecture is supported by recent findings. For example, the rice CSL proteins OsCSLFs were shown to function in (1,3;1,4)-β-D-glucan biosynthesis (Burton et al., 2006). Liepman et al. (2005) reported that three members of the AtCSLA subfamily, AtCSLA2, AtCSLA7 and AtCSLA9, could function as β-mannan synthases when expressed in Drosophila Schneider 2 (S2) cells. Genetic characterization of an Arabidopsis mutant carrying a transposon insertion in the gene encoding AtCSLA7 implicated a physiological role for AtCSLA7 in pollen tube growth and embryogenesis (Goubet et al., 2003).
Among all the CSL subfamilies, the CSLDs are the most similar to the CESAs, with sequence identity in the range of 6–42%. Thus far, no biochemical function has been assigned to any CSLD protein, although AtCSLD3 was found to have a role in root hair growth by forward genetic analysis (Favery et al., 2001).
Here we report the isolation and characterization of a new sos mutant, sos6-1. sos6-1 plants are hypersensitive to NaCl and KCl but not LiCl. Furthermore, the sos6-1 plants are hypersensitive to general osmotic stress imposed by mannitol or polyethylene glycol (PEG). Map-based cloning revealed that SOS6 encodes a cellulose synthase-like protein (AtCSLD5). sos6-1 mutant plants accumulate higher levels of reactive oxygen species (ROS) under osmotic stress than the wild type and are hypersensitive to the oxidative stress reagent methyl viologen (MV). The results suggest that SOS6 and SOS6-dependent cell wall components may control osmotic stress tolerance in part by regulating stress-induced ROS levels in plant cells.
RESULTS
Identification of the sos6-1 mutant
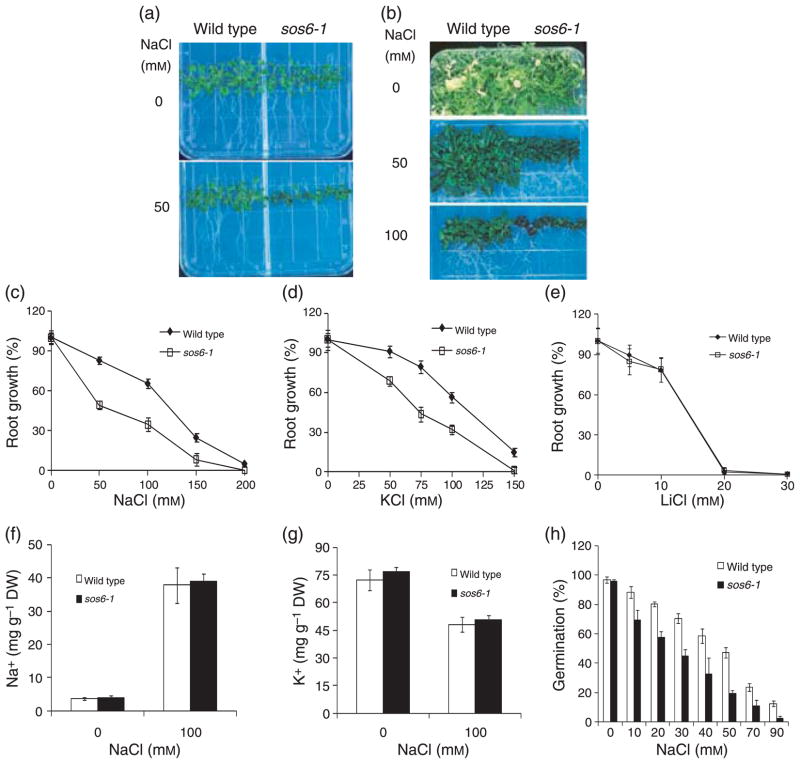
An Arabidopsis salt hypersensitive mutant, designated as sos6-1 (salt overly sensitive 6), was isolated from an ethyl methanesulfonate (EMS)-mutagenized M2 population using the previously reported root-bending assay (Wu et al., 1996) on medium containing 100 mM NaCl. The sos6-1 seedlings grow relatively normally in control medium containing MS salts and 3% sucrose (Figure 1). On medium supplemented with 50 mM NaCl, growth of both roots and shoots of sos6-1 seedlings was substantially more inhibited than that of the wild type (Figure 1a). sos6-1 mutant plants displayed an even greater decrease in shoot growth and development after prolonged exposure (e.g. 1 month) to media containing various levels of NaCl (Figure 1b).
Figure 1. Response of sos6-1 plants to different salts and ion content measurement.
(a) Four-day-old wild type and sos6-1 seedlings grown on germination media were transferred to media containing 0 or 50 mM NaCl and allowed to grow an additional 8 days. (b) Four-day-old wild type and sos6-1 seedlings grown on germination media were transferred to media containing 0, 50 or 100 mM NaCl and allowed to grow additional 30 days. (c) Response of sos6-1 plants to various levels of NaCl. The root growth was represented as proportion of growth compared with growth without NaCl (100%). (d) Response of sos6-1 plants to different levels of KCl. (e) Response of sos6-1 plants to various concentrations of LiCl. Error bars in (c–e) indicate standard deviation (n = 18). (f) Na+ content in sos6-1 plants as described (Zhu et al., 2007a). (g) K+ content in sos6-1 plants as described (Zhu et al., 2007a). (h) Sensitivity of sos6-1 seed germination to NaCl as described (Zhu et al., 2002). Error bars in (f–h) indicate standard deviation (n = 6). All experiments presented here and in the subsequent figures were performed at least three times and similar results were obtained.
sos6-1 mutant plants were backcrossed with wild type plants. All the F1 plants displayed a wild type phenotype and about three-quarters of the F2 progeny from self-pollinated F1 showed a wild type phenotype (data not shown). The result suggests that sos6-1 is a recessive mutation in a single nuclear gene.
sos6-1 plants are hypersensitive to NaCl and KCl but not LiCl
We tested whether the salt hypersensitivity of sos6-1 plants is specific to certain salts by measuring the response of root growth. Root growth of sos6-1 plants is hypersensitive to NaCl and KCl (Figure 1c,d). We also examined the effect of sos6-1 mutation on sensitivity to a more toxic analog of Na+, Li+, which can be used at a low concentration that does not cause significant osmotic stress. However, root growth of both the wild type and sos6-1 plants was similarly inhibited by LiCl (Figure 1e). This result indicates that sos6-1 is likely not hypersensitive to ionic stress. Consistent with this notion, ion absorbance spectrometry analysis (Zhu et al., 2002) revealed that the sos6-1 mutant plants accumulated essentially the same levels of Na+ or K+ as the wild type with or without NaCl treatment (Figure 1f,g). These results indicate that the salt hypersensitivity of sos6-1 plants was not due to disrupted Na+ homeostasis or impaired K+ acquisition.
We also tested the effect of sos6-1 mutation on seed germination under salt stress. Germination of sos6-1 seeds was also more susceptible to NaCl inhibition than was the wild type (Figure 1h) suggesting that salt hypersensitivity of sos6-1 plants does not depend on developmental stage.
sos6-1 plants are hypersensitive to mannitol and polyethylene glycol
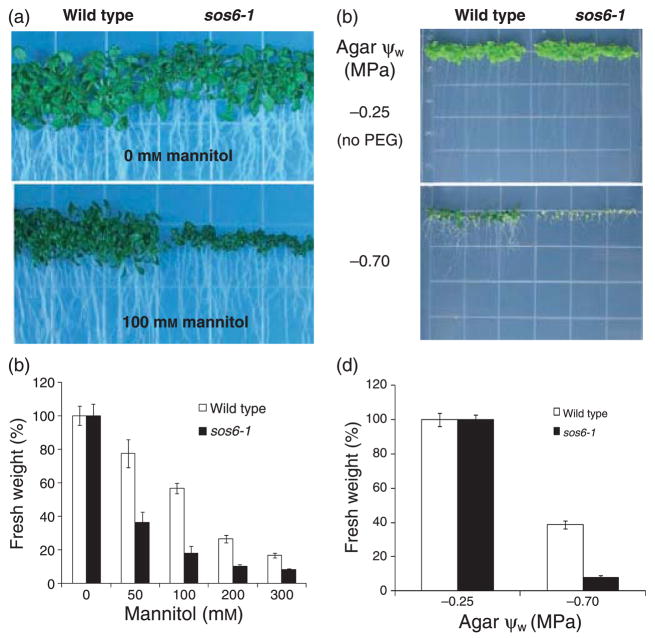
The increased sensitivity of sos6-1 plants to NaCl and KCl but not LiCl raises the possibility that sos6-1 mutation mainly affects tolerance to general osmotic stress. Indeed, we tested and found that sos6-1 plants are hypersensitive to osmotic stress imposed by mannitol. The increased inhibition by mannitol is especially obvious in the mutant shoot (Figure 2a,b). We then tested the response of sos6-1 to another osmotic stress agent, polyethylene glycol (PEG). When germinated in medium with low water potential imposed by PEG, sos6-1 seedlings were severely impaired in development and had significantly less fresh weight as compared with the wild type (Figure 2c,d). Thus, we conclude that the sos6-1 mutation confers hypersensitivity to general osmotic stress in Arabidopsis.
Figure 2. sos6-1 plants are hypersensitive to general osmotic stress.
(a) Growth of wild type and sos6-1 plants in the presence of 0 or 100 mM mannitol for 21 days. The seeds were first germinated on media containing 0 mM mannitol and then transferred to media containing 0 or 100 mM mannitol. (b) Quantification of growth (at the end of day 21; indicated by fresh weight) of wild type and sos6-1 in response to different levels of mannitol. (c) Seed germination and seedling development of wild-type and sos6-1 plants in media with low ψW imposed by PEG for 21 days. Seeds were directly germinated in agar plates with low ψW. (d) Quantification of growth of wild type and sos6-1 as shown in (c). Error bars in (b) and (d) indicate standard deviation (n = 60–80).
sos6-1 plants accumulate more soluble sugars and proline under stress conditions
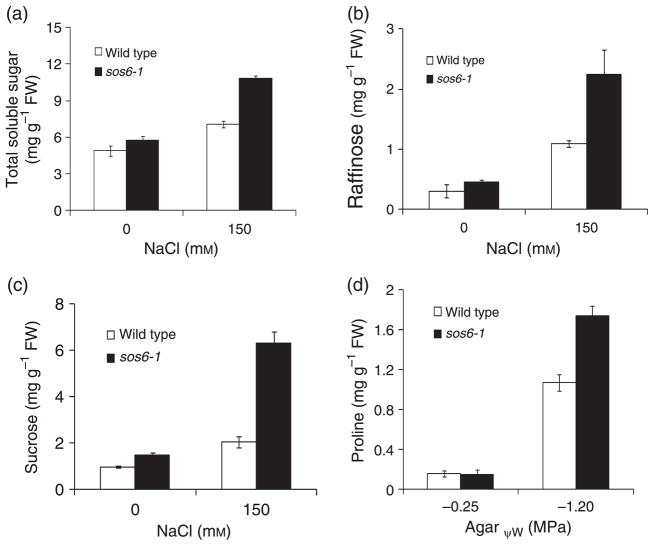
To investigate the physiological basis for the osmotic stress hypersensitivity in sos6-1 mutant plants, we determined the levels of compatible osmolytes such as soluble sugars and proline in sos6-1 and wild type plants. Under normal growth conditions, sos6-1 plants accumulated a similar level of total soluble sugars as the wild type (Figure 3a). When treated with 150 mM NaCl, total soluble sugar content was much higher in sos6-1 plants than in the wild type (Figure 3a). High pressure liquid chromatography (HPLC) analysis of soluble sugar content further revealed that sos6-1 plants accumulated a similar amount of glucose and fructose (data not shown) but much higher levels of raffinose and sucrose as compared with wild type plants (Figure 3b,c). Under the low water potential condition imposed by PEG (−1.2 MPa), the level of proline was higher in the sos6-1 mutant than in the wild type (Figure 3d). Clearly, the osmotic stress hypersensitivity of the mutant cannot be attributed to a deficiency in compatible osmolytes.
Figure 3. Content of soluble sugars and proline in sos6-1.
(a) Effect of sos6-1 mutation on soluble sugar levels. (b) Raffinose content of wild type and sos6-1 plants. (c) Sucrose content of wild type and sos6-1 plants. Error bars in (a–c) represent standard deviation (n = 16). (d) Effect of sos6-1 mutation on proline levels under low water-potential condition. Proline content was quantified as described (Verslues and Bray, 2004). Error bars represent standard deviation (n = 8).
sos6-1 plants are hypersensitive to salt and drought stress in soil
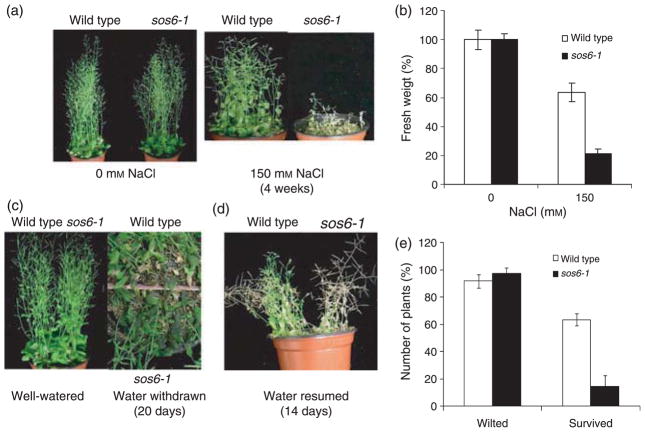
Without stress, the growth and development of sos6-1 plants were not distinguishable from those of the wild type (Figure 4a,c). When irrigated with 150 mM NaCl for 4 weeks, sos6-1 plants were more damaged, their growth was more severely inhibited and their survival was much more decreased than that of wild type plants, which continued to grow and develop to maturity (Figure 4a,b). At the seedling stage, sos6-1 plants showed no difference compared with wild type to growth inhibition by ABA in the media (Figure S1c,d). Both wild type and sos6-1 plants became wilty when water was withheld from soil for 20 days (Figure 4c,e). However, sos6-1 plants accumulated more anthocyanin under this dehydration condition (Figure 4c). When watering was resumed, most of the wilted sos6-1 plants failed to recover, whereas more than 50% of the wilted wild type plants survived (Figure 4d,e).
Figure 4. Soil-grown sos6-1 plants are hypersensitive to NaCl treatment and dehydration conditions.
(a) Two-week-old wild type and sos6-1 plants were treated with 0 or 150 mM NaCl and allowed to grow for additional 28 days. Plants were watered with solution containing 0 or 150 mM NaCl every 3 days. (b) Quantification of growth of wild type and sos6-1 at the end of salt treatment. Error bars represent standard deviation (n = 80–96). (c) Two-week-old wild type and sos6-1 plants grown in soil in the same pots were either well-watered or deprived of water for an additional 20 days. (d) Wild type and sos6-1 plants grown for 14 days when water was resumed. (e) Quantification of plants that were wilted when water was withdrawn (Wilted) and wilted plants that survived again when water was resumed (Survived). Error bars represent standard deviation (n = 74–90).
sos6-1 plants overaccumulate ROS and are hypersensitive to oxidative stress
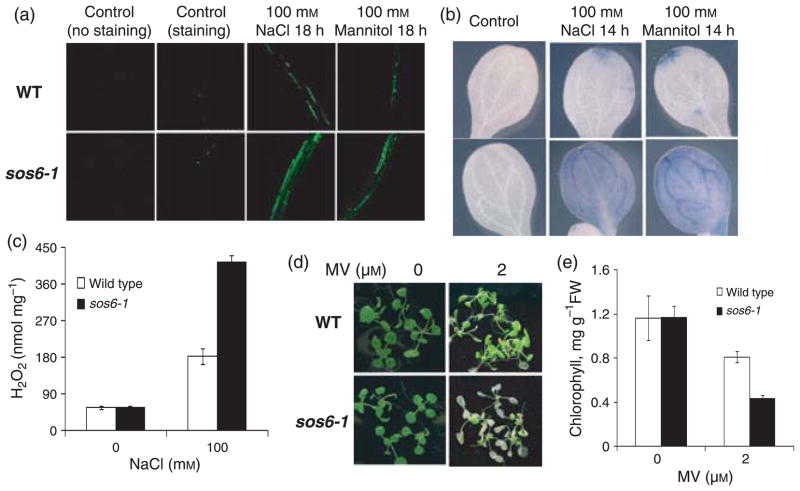
Salt and drought stress treatments can lead to the accumulation of ROS and cause oxidative stress (Price and Hendry, 1991; Borsani et al., 2001b). We examined the effect of sos6-1 mutation on ROS levels and the response to oxidative stress. Without stress, sos6-1 and wild type plants showed similar basal levels of ROS (Figure 5a,b). When treated with NaCl or mannitol, sos6-1 plants accumulated much higher amounts of total ROS (Figure 5a). Leaf staining with a superoxide-specific dye indicated that superoxide accumulated to higher levels in sos6-1 mutant plants compared with the wild type (Figure 5b). We quantified the levels of hydrogen peroxide (H2O2) using the Amplex red reagent (10-acetyl-3,7-dihydrophenoxazine). As shown in Figure 5c, under salt stress, sos6-1 plants accumulated a substantially higher amount of H2O2 than the wild type. Furthermore, we found that sos6-1 plants were hypersensitive to methyl viologen (mv), a chemical that generates superoxide radicals in chloroplasts, as indicated by substantial loss of chlorophyll in the mutant (Figure 5d,e). Taken together, these results suggest that SOS6 is an important regulator of ROS accumulation in plants under osmotic stress.
Figure 5. Detection of ROS and oxidative stress tolerance.
(a) Detection of total ROS accumulation in seedlings with or without stress treatment. Fluorescence indicates the presence of ROS. (b) Nitroblue tetrazolium staining for superoxide radical. Blue color indicates the presence of superoxide. (c) Quantitative measurement of H2O2 levels in sos6-1 and wild type plants. Error bars indicate standard deviation (n = 12). (d) Tolerance of sos6-1 and wild type plants to methyl viologen as indicated by chlorophyll content. (e) Chlorophyll content in plants as shown in (d) determined as described (Lichtenthaler and Wellburn, 1983). Error bars represent standard deviation (n = 10).
SOS6 encodes a cellulose synthase-like protein
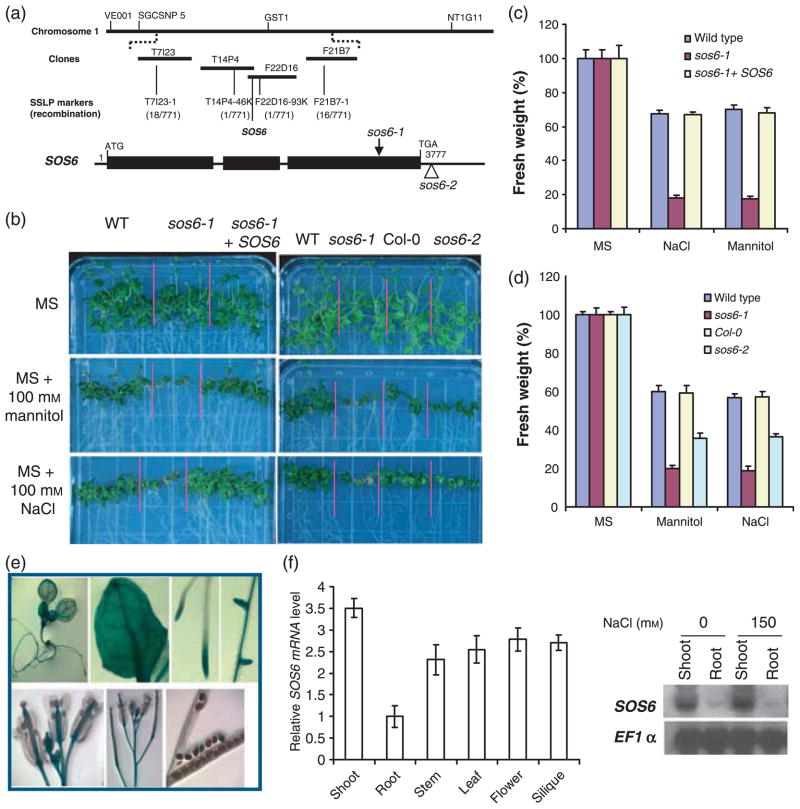
sos6-1 plants were crossed with Landsberg erecta wild type plants for genetic mapping of the SOS6 gene. SOS6 was mapped by using simple sequence length polymorphism (SSLP) markers. The gene was localized between BAC clones T7I23 and F21B7 on chromosome 1. The genomic region between T14P4–46K and F22D16–93K markers was sequenced and a mutation was identified in the gene At1g02730. This gene encodes a cellulose synthase-like protein (CSL) and is classified as AtCSLD5 (Figure S2). There is a duplication of 158 nucleotides (2626–2783 numbered from translation start codon ATG) in the mutant (Figure 6a). This mutation would insert three new amino acid (aa) residues followed by a premature stop codon, resulting in a truncated 933 amino acid polypeptide, as compared with a 1181 aa polypeptide in the wild type. The truncated residues are part of a putative cellulose synthase activity domain (aa 302–1181 in the wild type).
Figure 6. Molecular cloning of SOS6.
(a) SOS6 was identified through map-based cloning. SOS6 is located on bacterial artificial chromosome clone F22D16. The numbers of recombinant and total chromosomes tested (in parentheses) and developed sslp markers are indicated. The structure of SOS6 and the positions of sos6-1 and sos6-2 mutations are also indicated. Positions are relative to the translation initiation codon. Closed boxes indicate the open reading frame and lines between boxes indicate introns. (b) Gene complementation of sos6-1 and tolerance of sos6–2 to NaCl and mannitol. (c) Quantification of seedling development as shown in (b). (d) Quantification of growth of wild type, sos6-1, Col-0 and sos6-2 as shown in (b). Error bars in (c) and (d) represent standard deviation (n = 90–110). (e) Tissue-specific expression of SOS6::GUS. (f) Validation of SOS6::GUS expression patterns by real-time PCR analysis (left panel) and SOS6 is not regulated by salt stress as revealed by northern blot analysis (right panel). EF1α gene was used as loading controls. Error bars indicate standard deviation (n = 8).
To confirm that At1g02730 is SOS6, a genomic fragment containing the wild type At1g02730 gene was cloned and introduced into the sos6-1 mutant. Transformants in the T2 population showed wild type phenotypes, which indicates that At1g02730 is indeed SOS6 (Figure 6b,c). Plants of a second allele of At1g02730, designated as sos6-2, were also hypersensitive to NaCl or mannitol treatment compared with Col-0 plants (background of sos6-2) (Figure 6b,d). These results demonstrate that SOS6 is AtCSLD5.
The SOS6 promoter region was fused to a GUS reporter gene. SOS6::GUS expression was observed in all the tissues examined, with stronger expression in root tips, leaves, inflorescent stems and male and female organs of the flower (Figure 6e). Real-time RT-PCR analysis of SOS6 expression in different tissues (Figure 6f) suggested a similar pattern as SOS6::GUS expression. Real-time RT-PCR and northern blot analysis also indicated that SOS6 is constitutively expressed and not induced by salt stress and its transcript is more abundant in shoots than roots (Figure 6f).
The salt tolerance function is specific for CSLD5 in the CSLD subfamily
Among the five loci encoding additional members in the CSLD subfamily (CLSD1-4 and CSLD6), CSLD1-3 genes are induced by NaCl stress while CSLD4 and CSLD6 are essentially not responsive to salt stress (Figure S3). The sos6-1 mutation seems to have no effect on expression of these genes with or without NaCl stress (Figure S3) except that CSLD3 is more abundant in the untreated sos6-1 plants than wild type. We determined whether other members in the CSLD subfamily are involved in salt stress responses. For this purpose, homozygous T-DNA insertion lines with reduced expression levels in respective CSLD genes were identified and their ability to grow in the presence of NaCl or mannitol was measured (Figures S4–S8). None of other csld loss-of-function mutants displayed enhanced or decreased tolerance to NaCl or mannitol stress conditions. These results implicate that SOS6 in the CSLD subfamily is specifically required for osmotic stress tolerance.
Effect of the sos6-1/csld5 mutation on cell wall composition
We determined the glycosyl composition of the whole cell wall from shoots of sos6-1 and wild type plants, as an indication of their wall structures. Despite extensive analysis, we found only modest differences between the wild type and sos6-1 (Table 1). Under normal growth conditions, no statistically significant differences were detected between the cell wall from wild type and sos6-1 plants. Under 200 mM NaCl treatment, the mutant showed reduced levels of Ara and Rha and an increased level of Fuc relative to the wild type. Although these differences were statistically significant, together they represented only about 2 mol% of the glycosyl content of the cell wall. Larger change was detected when plants grown under 200 mM NaCl treatment were contrasted with plants grown under normal conditions. The mol% of GalU in the cell wall was significantly lower when sos6-1 plants were grown under 200 mM NaCl treatment than under normal conditions, while the analogous change in wild type plants was not statistically significant. To test for this change on an absolute, rather than mol%, basis, we performed a colorimetric assay of the total uronic acid (GalU + GlcU) content of the cell wall. Although seemingly in the same direction, differences between total uronic acids in the normal and NaCl conditions were not statistically significant (Table 1).
Table 1.
Glycosyl composition (mol%) of the whole cell wall fraction from shoots of wild type and sos6-1 plants treated with 0 or 0.2 M NaCl
| Residuea | Glycosyl composition (mol%) (treatments/genotypes) |
|||
|---|---|---|---|---|
| 0 M NaCl |
0.2 M NaCl |
|||
| Wild type | sos6-1 | Wild type | sos6-1 | |
| Ara | 3.51d | 3.47d | 5.33e | 4.08d |
| Rha | 3.77d | 3.48d | 4.31e | 3.64d |
| Fuc | 0.79d,e | 0.79d,e | 0.69d | 0.87e |
| Gal | 5.43d | 5.59d | 7.00d | 7.26d |
| Xyl | 5.38d | 5.69d | 8.10e | 10.12e |
| Xyl-SAb | 1.46d | 0.95d | 1.16d | 2.00d |
| Man | 1.25d | 1.26d | 1.29d | 1.26d |
| Man-SAb | 1.20d | 1.07d | 1.26d | 1.00d |
| GlcU | 3.97d | 3.73d | 4.11d | 5.53d |
| GalU | 35.44d | 35.44d | 31.04d,e | 23.93e |
| Glc | 5.09d | 5.06d | 2.91e | 3.19e |
| Glc-SAb | 32.73d | 33.48d | 32.81d | 37.10d |
| Uronicsc (weight%) | 25.53d | 25.78d | 24.22d | 23.07d |
Each entry is the mean of n = 5 collections of plants.
From gas chromatographic analysis of trimethylsilyl ethers of methyl glycosides.
Additional amounts of glycosyl residues detected when the samples were subjected to H2SO4 (SA) swelling and hydrolysis, for cleavage of cellulose and tightly associated polysaccharides, prior to methanolysis.
From colorimetric analysis of total uronic acids.
Within each row, entries not followed by the same letter are statistically different by ANOVA (P < 0.05).
To examine the effect of sos6-1 mutation on cell wall in detail, we sequentially extracted different fractions of the cell wall based on their solubility in imidazole or NaOH and subsequently determined the glycosyl composition of each wall fraction. Among the three different cell wall fractions, main alterations between sos6-1 and wild type were found in the imidazole-soluble fraction (Table 2, principally pectic polysaccharides). Without stress, the levels of Rha, Fuc and Gal are significantly less in the sos6-1 mutant plants than wild type whereas Glc is more abundant in the mutant (Table 2). Under 200 mM NaCl treatment, the sos6-1 mutant displayed increased amount of Rha, reduced level of Xyl compared with wild type (Table 2). Although the treatment of salt stress in sos6-1 also causes increased level of Ara and reduced amounts of GalU and Glc (Table 2), there is no significant difference between wild type and sos6-1 for these three sugar residues. There are a few notable changes in the NaOH-soluble cell wall fraction (hemicelluloses and residual pectic polysaccharides) between sos6-1 and wild type plants (Table 3). We reasoned that this might be due to the incomplete extraction of pectic polysaccharides in the previous step (imidazole extraction) during the preparation of the cell wall fractions. Essentially there is no significant difference between sos6-1 and wild type in the NaOH-insoluble/cellulose fraction (Table 4).
Table 2.
Glycosyl composition of the imidazole-soluble cell wall fraction from shoots of wild type and sos6-1 plants treated with 0 or 0.2 M NaCl
| Residuea | Glycosyl composition (mol%) (treatments/genotypes) |
|||
|---|---|---|---|---|
| 0 M NaCl |
0.2 M NaCl |
|||
| Wild type | sos6-1 | Wild type | sos6-1 | |
| Ara | 7.98b | 9.16b | 10.85b,c | 13.96c |
| Rha | 9.60b | 7.60c | 9.28b,c | 11.66d |
| Fuc | 3.40b | 1.63c | 3.04b,c | 3.58b |
| Gal | 17.98b | 11.24c | 17.47b | 18.83b |
| Xyl | 10.12b | 8.93b | 10.98b | 8.24c |
| Man | 1.91b | 1.46b | 2.36b | 2.47b |
| GlcU | 10.12b | 8.94b | 10.98b | 8.26b |
| GlaU | 28.97b,c | 24.97b | 25.96b,c | 18.34c |
| Glc | 16.48b | 31.72c | 16.07b | 16.24b |
Each entry is the mean from n = 5 collections of plants.
From gas chromatographic analysis of trimethylsilyl ethers of methyl glycosides.
Within each row, entries not followed by the same letter are statistically different by ANOVA (P < 0.05).
Table 3.
Glycosyl composition of the NaOH-soluble cell wall fraction from shoots of wild type and sos6-1 plants treated with 0 or 0.2 M NaCl
| Residuea | Glycosyl composition (mol%) (treatments/genotypes) |
|||
|---|---|---|---|---|
| 0 M NaCl |
0.2 M NaCl |
|||
| Wild type | sos6-1 | Wild type | sos6-1 | |
| Ara | 8.53b | 10.60b | 8.86b | 9.44b |
| Rha | 8.43b | 9.29b,c | 10.17c | 14.82d |
| Fuc | 2.16b | 2.72b | 2.27b | 3.06b |
| Gal | 13.51b | 13.35b | 17.36c | 13.81b |
| Xyl | 20.48b | 23.21c | 22.08b,c | 30.84d |
| Man | 5.47b | 4.01b | 4.12b | 4.64b |
| GlcU | 4.05b | 3.53b | 3.77b | 3.97b |
| GlaU | 26.85b | 24.20b | 18.99c | 5.06d |
| Glc | 10.52b | 9.09b | 12.39b,c | 14.36c |
Each entry is the mean from n = 5 collections of plants.
From gas chromatographic analysis of trimethylsilyl ethers of methyl glycosides.
Within each row, entries not followed by the same letter are statistically different by ANOVA (P < 0.05).
Table 4.
Glycosyl composition of the NaOH-insoluble cell wall fraction from shoots of wild type and sos6-1 plants treated with 0 or 0.2 M NaCl
| Residuea | Glycosyl composition (mol%) (treatments/genotypes) |
|||
|---|---|---|---|---|
| 0 M NaCl |
0.2 M NaCl |
|||
| Wild type | sos6-1 | Wild type | sos6-1 | |
| Ara | 0.79b | 1.54c | 0.51b | 0.78b |
| Rha | 1.03b | 0.95b | 1.07b | 0.88b |
| Fuc | 0.42b | 0.43b | 0.92c | 1.21d |
| Gal | 1.65b | 2.62b | 2.35b | 2.42b |
| Xyl | 2.32b | 2.81b | 2.39b | 3.20b |
| Man | 0.67b | 0.65b | 0.72b | 0.52b |
| GlcU | 1.13b | 1.07b | 1.20b | 1.50b |
| GlaU | 1.88b | 1.02b | 0.63b | 0.80b |
| Glc | 90.10b | 88.92b | 90.21b | 88.68b |
Each entry is the mean from n = 5 collections of plants.
From the NaOH-insoluble cell-wall fraction, which was first hydrolyzed by H2SO4 and then subjected to methanolysis and trimethylsilylation followed by gas chromatographic analysis.
Within each row, entries not followed by the same letter are statistically different by ANOVA (P < 0.05).
Effect of sos6-1 mutation on gene expression
Since the sos6-1 plants carry a homozygous firefly luciferase report gene driven by the stress-inducible RD29A promoter (RD29A::LUC), we examined the expression of RD29A::LUC under different stress conditions (Figure S1a,b). These data indicate that sos6-1 mutation does not affect RD29A:: LUC expression.
We then performed Affymetrix near-full genome Gene-Chip analysis to determine the effect of sos6-1 mutation on gene expression. Statistical analysis of the microarray data revealed that after 150 mM NaCl treatment, there were only 23 genes that showed reduced levels of expression in roots of sos6-1 by at least two fold compared with the wild type (Table S1), whereas the transcript of only one gene was significantly increased in roots of sos6-1 (Table S2). Under the NaCl stress, we did not identify any gene with a significant reduction in transcript level in the shoot of sos6-1, whereas four genes showed increased transcript levels in the shoot of sos6-1 compared with wild type (Table S3). Real-time PCR analysis of transcript levels of six randomly selected genes from the Tables S1–S3 confirmed the microarray results (Figure S9). Consistent with the increased growth inhibition by salt stress in the mutant, several of the genes with reduced expression in salt stress-treated sos6-1 compared with the wild type (Table S1) have putative functions in cell expansion, e.g. the delta-TIP (Barkla et al., 1999), arabinogalactan-protein (Shi et al., 2003b), xyloglucan endotransglycosylase (Campbell and Braam, 1999) and expansin (Cosgrove, 1999). Relevant to the defect in ROS management and increased ROS sensitivity, oxidative stress related genes such as the putative glutathione transferase and peroxidase genes (Table S1) also showed reduced expression in salt stress-treated sos6-1 mutant plants compared with the wild type plants. Interestingly, a number of cytochrome P450 genes showed altered expression in the mutant (Tables S1 and S3). Cytochrome P450s are responsible for the biosynthesis and catabolism of a wide variety of compounds in plants including phytohormones such as ABA (Kushiro et al., 2004). It is possible that the altered level of one or more of these cytochrome P450s may contribute to the osmotic stress hypersensitivity of the sos6-1 mutant.
DISCUSSION
Forward genetic screening in Arabidopsis has been a powerful tool that helped identify several key loci important for the ion homeostasis aspect of plant salt tolerance (Zhu, 2003). However, mutants affected in the osmotic stress response pathway(s) have been difficult to isolate. The sos6-1 mutant isolated in this study showed a strong hypersensitive phenotype under osmotic stress imposed by salts, mannitol or PEG. Thus, SOS6 is a major locus for hyperosmotic stress tolerance in Arabidopsis. A few other mutants have been reported previously to be affected in their osmotic stress responses (Zhu et al., 2002; Koiwa et al., 2003), but their effects on osmotic stress tolerance are much smaller than that of sos6-1. Positional cloning led to the identification of the SOS6 gene as AtCSLD5 and this is confirmed by complementation of the sos6-1 mutant with the wild type AtCSLD5 gene (Figure 6). The results implicate AtCSLD5 and/or AtCSLD5-mediated cell wall components as critical for osmotic stress tolerance.
We analyzed the glycosyl composition of the whole cell wall and different cell wall fractions and found only moderate effects of sos6-1 mutation (Tables 1–4). No significant differences were detected between the wild type and sos6-1 under normal conditions using the whole cell wall fractions while four residues showed differences (decreases in Rha, Fuc, Gal; increase in Glc) in abundance in sos6-1 relative to wild type in the imidazole-soluble cell wall fraction (Table 2). Under NaCl stress, three residues – Ara, Rha and Fuc – differed slightly in abundance between the genotypes in the whole cell wall extracts (Table 1) and Rha and Xyl residues displayed altered levels between the genotypes in the imidazole-soluble cell wall fraction (Table 2). The two types of sugar analyses employed on the whole cell wall or different cell wall fractions complemented each other. Contrasting between normal and NaCl stress conditions within a genotype revealed that sos6-1 had significantly less GalU under NaCl stress (Tables 1 and 2). Likely relevant to this observation is the report of Zablackis et al. (1995) who found that 0.1–0.5 M potassium phosphate (pH 7) extracted remarkably more pectic polysaccharides from Arabidopsis leaves than from leaves of other plants. The lower level of GalU (the major residue in pectins) in the cell wall fraction and the lower levels of Ara and Rha (also found in pectins) in sos6-1 than in the wild type under NaCl stress together suggest a role for SOS6/AtCSLD5 in the incorporation of pectic polysaccharides into the cell wall. Overall, however, SOS6 does not appear to have a major role in controlling cell wall structure, consistent with the relatively normal growth and development of the sos6-1 mutant plants under unstressed conditions. A recent study of the T-DNA knockout lines of AtCSLD5 revealed that a null allele of AtCSLD5 shows a growth reduction under the growth conditions used (Bernal et al., 2007). Immunocytochemical analysis between wild type and the null allele of AtCSLD5 further indicates a role in xylan and homogalacturonan synthesis (Bernal et al., 2007). Our sugar analysis data also indicate that sos6-1 mutation has similar effects (Tables 1 and 2). Earlier evidence obtained by gene expression profiling and immunocytochemical analysis using isoxaben-habituated Arabidopsis suspension culture cells revealed that AtCLSD5 was highly induced and may contribute to the biosynthesis of the pectic component of the habituated cells (Manfield et al., 2004). Again, our data presented in this study are consistent with the previous observations.
In contrast to the apparent lack of function or minor function in unstressed conditions, SOS6 is required for plant growth under salt and drought stresses. What might be the mechanism of SOS6/AtCSLD5 function in osmotic stress tolerance? As the first line of defense of the cell, the cell wall and the plasma membrane attached to it are expected to be involved in the sensing and early signaling of osmotic stress (Zhu et al., 1997). CesA and possibly CSL proteins as well are transmembrane proteins that link with both wall polymers and microtubules and thus are part of the wall-membrane-cytoskeleton continuum, which is hypothesized to be important for turgor sensing (Zhu et al., 1993, 1997). It is conceivable that SOS6/AtCSLD5 may even have a specific role in osmotic stress sensing.
Alternatively, SOS6 may function in the synthesis of certain component(s) of the wall and this function may be related to ROS generation or scavenging. SOS6/AtCSLD5 itself or through the wall polymer it synthesizes may interact with and regulate proteins that generate ROS at the cell surface, such as plasma membrane NADPH oxidases, cell wall polyamine oxidases and peroxidases (Keller et al., 1998; Mittler, 2002; Sagi and Fluhr, 2006; Yoda et al., 2006). Upon osmotic stress, ROS accumulates to higher levels in the sos6-1 mutant (Figure 5), which indicates that SOS6 and/or related cell-wall components are indeed required for maintenance of ROS levels. Furthermore, the expression of a putative peroxidase (At2g39040) was less in sos6-1 relative to the wild type after stress treatment, as revealed by microarray analysis and validated by real-time PCR analysis (Figure S9 and Table S1). On the other hand, a germin-like protein (At5g39100) showed an elevated transcript level in sos6-1 upon salt stress (Figure S9 and Table S1). Germin and germin-like proteins have been shown to have superoxide dismutase activity (Yamahara et al., 1999; Carter and Thornburg, 2000; Christensen et al., 2004). Notwithstanding the unknown mechanism of function, our results open up new questions for salt and drought stress research and suggest an important role of SOS6/AtCSLD5 and related cell-wall components in regulating stress-induced ROS levels and osmotic stress tolerance.
EXPERIMENTAL PROCEDURES
Plant materials
A firefly luciferase reporter gene driven by the stress-responsive RD29A promoter (Ishitani et al., 1997) was introduced into Arabidopsis plants in the Columbia glabrous1 (gl1) background. Seeds from one homozygous line (referred to as the wild type) were mutagenized with ethyl methanesulfonate (EMS) and M2 seeds were used to screen for high throughput mutants with altered RD29A::LUC gene expression, for example STABILIZE 1 (Lee et al., 2006) and this same batch of seeds was also used to screen for hypersensitive mutants in the presence of 100 mM NaCl in this study with use of a root-bending assay (Wu et al., 1996).
Seeds of sos6-2 (SALK_002118), csld1 (SALK_097300), csld2 (SALK_119808), csld4 (SALK_059059) and csld6 (SALK_095234) were obtained from Arabidopsis Biological Resource Center (ABRC). Verification of insertions was performed using primers listed in Table S4. Homozygous csld3–1 (seed stock number CS899) was obtained from ABRC as described (Wang et al., 2001).
Genetic analysis and genetic mapping of sos6-1 locus
sos6-1 plants were backcrossed with the wild type. F1 and F2 seedlings were scored for salt sensitivity by the root-bending assay (Wu et al., 1996). For mapping of the sos6-1 locus, homozygous sos6-1 plants were crossed to wild type plants of Landsberg erecta. From the segregating F2 population, 771 homozygous sos6-1 mutants were selected and DNA was extracted from each of these plants for mapping with SSLP markers between Columbia and Landsberg erecta.
Complementation of sos6-1 mutant
The genomic DNA fragment of At1g02730 was amplified by PCR with F22D16 BAC clone used as a template with the primers listed in Table S4. The PCR product was cloned into pCAMBIA1200 between KpnI and XbaI sites and sequenced. The resulting binary vector was transferred to sos6-1 through Agrobacterium (strain GV3101)-mediated transformation. The transgenic plants (T1) were selected on hygromycin (25 mg/L) and transferred to soil to maturity. T2 seedlings were examined for sensitivity to 100 mM NaCl and 100 mM mannitol, respectively.
SOS6 promoter::GUS construct
The DNA fragment containing the SOS6 promoter was amplified by PCR with BAC F22D16 clone DNA used as a template with the primers listed in Table S4. The PCR product was cloned into the binary vector pCAMBIA1381Z between EcoRI and PstI sites and sequenced. The resulting binary vector was transferred to wild-type (Columbia) plants through an Agrobacterium (strain GV3101)-mediated transformation by a floral dip method. Seedlings or tissues from the T1 and T2 populations were first emerged in 5-bromo-4-chloro-3-indoyl glucuronide (X-Gluc) solution (2 mM X-Gluc, 100 mM sodium phosphate buffer (pH 7.5), 0.5% Triton X-100, 2 mm K3[Fe(CN)6], 2 mM K4[Fe(CN)6], 0.02% NaN3) and vacuum infiltrated for 10 min and then incubated at 37°C for at least 12 h in dark, followed by incubation in 70% ethanol to remove chlorophyll as described (Jefferson et al., 1987).
RNA gel analysis, microarray analysis and real-time PCR analysis
Wild type and sos6-1 seedlings were grown on separate halves of the same 0.6% agar MS medium for 14 days and then left untreated or treated with NaCl. Total RNA was extracted from whole seedlings and RNA gel analysis was conducted as described (Zhu et al., 2007a).
Total RNA (20 μg) extracted with use of the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) from 28-day-old wild type and sos6-1 seedlings grown on 1.2% agar MS medium after salt treatment (150 mM NaCl for 24 h) was used to make biotin-labeled cRNA targets. Affymetrix GeneChip arrays were performed as described (Chinnusamy et al., 2003). Statistical analysis of the microarray data were performed as described (Li and Wong, 2001; Tusher et al., 2001; Wu et al., 2004; Johnson et al., 2007). Data were adjusted for batch effect (due to the fact that experiments were done at two different locations: University of Arizona and University of California-Riverside) and compared by the same statistical method. Fold change indicates relative expression level in sos6-1 (value <0 indicates downregulation in sos6-1; value >0 indicates upregulation in sos6-1). Q-value (%) stands for chance of false positive. We used a threshold q-value of 0.05 to select the genes whose expression levels are statistically significantly changed in sos6-1.
Total RNA isolation, first strand cDNA synthesis and real-time PCR analysis were carried out as previously described (Zhu et al., 2007b). The primers used for real-time PCR reactions were listed in Table S4.
Soluble sugar measurement
Ground fresh tissues (0.5 g) from 1-month-old wild-type and sos6-1 plants grown in soil treated or untreated with 150 mM NaCl for 10 days were extracted with 10 ml 80% ethanol and incubated in a 80°C water bath for 30 min. The extract was centrifuged for 5 min at 17 300 g (Beckman, J2-MC) and the supernatant was dried in a speed vacuum concentrator (Savant, Farmingdale, NY, USA). The dry samples were then extracted with 5.8 ml of methanol/water/chloroform (1:0.9:1) and the extract was then dried in the Savant speed vacuum concentrator. The extract was suspended in 1 ml deionized water and deionized by passage through 1 ml anion and cation resin columns (AG1-X8/formate and AG50W-X8/H+, BIORAD). Sugars were eluted with 10 ml water, dried and redissolved in 200 μl water and filtered (0.45 μm pore size). Then 20 μl of the soluble sugar extract was analyzed with HPLC by use of a Sugar-pak (Waters, Milford, MA, USA) column as described by Liu et al. (1999). Total soluble sugar was also assayed with the remaining soluble sugar extract as described (Dubois et al., 1956).
Determination of ROS levels
One-week-old seedlings grown on 1.2% agar MS medium were used to determine ROS levels as described (Zhu et al., 2007a).
Glycosyl composition analysis of cell wall
One-month-old plants grown in soil under long day photoperiod (16 h light, 8 h dark) were treated with 0 or 200 mM NaCl for 10 day. Preparation of the cell wall was performed as described (Fry, 2000). Briefly, 2 g of fresh shoots were harvested and ground into fine powder in liquid N2. Proteins and starch were removed by extracting twice with phenol-acetic acid-water (approximately first 1:2:0.5 and then 1:2:1) and twice with 90% DMSO, respectively. Residual lipids were extracted twice with 2:1 (v/v) chloroform-methanol and twice with acetone. The cell wall fraction was then dried in air and again in a vacuum desiccator over P2O5. The dry cell wall fraction was then analyzed by methanolysis and trimethylsilylation combined with gas liquid chromatography (GC) for glycosyl composition of non-cellulosic polysaccharides as described (Komalavilas et al., 1990). For analysis including cellulosic polysaccharides, an aliquot of the cell-wall fraction was first hydrolyzed by H2SO4 (Fry, 2000) and then subjected to methanolysis and trimethylsilylation followed by GC. Different cell wall fractions were also sequentially extracted from total cell wall prepared with procedures described above. Briefly, two grams of dry cell wall were extracted twice in 500 mM imidazole-HCl, pH 7.0 (40 ml each time) and stirred overnight. The pooled supernatant after centrifugation was filtered through GF/A glass fiber filter (Whatman, 3.7 cm), dialyzed against distilled water and freeze dried. The imidazole-insoluble residue was further extracted twice in 30 ml each time of 6 M NaOH (with addition of 1% (w/v) NaBH4) and stirred overnight. The pooled supernatant after centrifugation was filtered through GF/A glass fiber filter (3.7 cm), titrated to pH 6 by addition of acetic acid, dialyzed against distilled water and freeze-dried. The NaOH-insoluble residue was repeatedly washed by centrifugation with distilled water until the pH was neutral and then freeze-dried. The dry cell wall fractions (imidazole soluble and NaOH soluble) were analyzed by methanolysis and trimethylsilylation combined with gas liquid chromatography (GC) for glycosyl composition as described (Komalavilas et al., 1990). The NaOH-insoluble cell-wall fraction was first hydrolyzed by H2SO4 (Fry, 2000) and then subjected to methanolysis and trimethylsilylation followed by GC analysis.
One-way Analysis of Variance (ANOVA) of the cell wall glycosyl composition data was performed using InStat (version 2.0, GraphPad Software, San Diego, CA, USA) and the same software was used to perform the Tukey–Kramer multiple comparison post-test when ANOVA indicated the existence of statistically significantly differences.
Total uronic acid analysis was performed as described (Blumenkrantz and Asboe-Hansen, 1973). Briefly, an aliquot of the dry cell-wall fraction (~300 μg) was suspended in 1 ml H2O in a glass tube and 6 ml of 0.0125 M sodium tetraborate in concentrated H2SO4 were added. The mixture in the glass tube was boiled in a water bath for 7 min and then cooled in an ice-H2O bath. One hundred μl of 0.15% (w/v) meta-hydroxydiphenyl reagent in 0.5% NaOH were added and uronic acid was quantified by measuring the absorbance at 520 nm, with D-galacturonic acid used as a standard.
Supplementary Material
Acknowledgments
We thank Xianwu Zheng, Chun-Hai Dong, Becky Stevenson and Woody Smith for excellent technical assistance. This work was supported by National Institutes of Health Grant R01GM059138 to J.-K. Zhu and by National Science Foundation Grant IOS0919745 to J. Zhu and by Sogang University Research Grant (200810022) and Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology of the Korean Government (2009–0089383) to B.-H. Lee.
References
- Barkla BJ, Vera-Estrella R, Pantoja O. Towards the production of salt-tolerant crops. Adv Exp Med Biol. 1999;464:77–89. doi: 10.1007/978-1-4615-4729-7_7. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Jensen JK, Harholt J, et al. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 2007;52:791–802. doi: 10.1111/j.1365-313X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G. New method for quantitative determination of uronic acids. Anal Biochem. 1973;54:484–489. doi: 10.1016/0003-2697(73)90377-1. [DOI] [PubMed] [Google Scholar]
- Borsani O, Cuartero J, Fernandez JA, Valpuesta V, Botella MA. Identification of two loci in tomato reveals distinct mechanisms for salt tolerance. Plant Cell. 2001a;13:873–887. doi: 10.1105/tpc.13.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Valpuesta V, Botella MA. Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol. 2001b;126:1024–1030. doi: 10.1104/pp.126.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999;4:361–366. doi: 10.1016/s1360-1385(99)01468-5. [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. Tobacco nectarin I. Purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J Biol Chem. 2000;275:36726–36733. doi: 10.1074/jbc.M006461200. [DOI] [PubMed] [Google Scholar]
- Chen Z, Hong X, Zhang H, Wang Y, Li X, Zhu JK, Gong Z. Disruption of the cellulose synthase gene, AtCesA9/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 2005;43:273–283. doi: 10.1111/j.1365-313X.2005.02452.x. [DOI] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Zhu JK, Hirsschi KD. The protein kinase SOS2 activates the Arabidopsis H+/Ca2+antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem. 2004;279:2922–2926. doi: 10.1074/jbc.M309084200. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal A, Zhu JK. ICE1, a master regulator of cold induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AB, Thordal-Christensen H, Zimmermann G, Gjetting T, Lynkjær MF, Dudler R, Schweizer P. The germin-like protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol Plant Microbe Interact. 2004;17:109–117. doi: 10.1094/MPMI.2004.17.1.109. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:391–417. doi: 10.1146/annurev.arplant.50.1.391. [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001;15:79–89. doi: 10.1101/gad.188801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. The Growing Plant Cell Wall: Chemical and Metabolic Analysis. Caldwell, New Jersey: The Blackburn Press; 2000. Reprint Edition. [Google Scholar]
- Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P. AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 2003;131:547–557. doi: 10.1104/pp.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–1400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK. SOS3 function in plant salt tolerance requires N-myristoylation and calcium-binding. Plant Cell. 2000;12:1667–1677. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Rabinovic A, Li C. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+binding motifs. Plant Cell. 1998;10:255–266. doi: 10.1105/tpc.10.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003;15:2273–2284. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komalavilas P, Zhu JK, Nothnagel EA. Arabinogalactan-protein from the suspension culture medium and plasma membrane of rose cells. J Biol Chem. 1990;266:15956–15965. [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Kapoor A, Zhu J, Zhu JK. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell. 2006;18:1736–1749. doi: 10.1105/tpc.106.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophyll a and b of leaf extracts in different solvents. Biochem Soc Trans. 1983;603:591–592. [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA. 2005;102:2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Liu X, Robinson PW, Wadore MA, Witney GW, Arpaia ML. ‘Hass’ Avocado carbohydrate fluctuations. I. Growth and phenology. J Amer Soc Hort Sci. 1999;124:671–675. [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Orfila C, McCartney L, Harholt J, Bernal AJ, Scheller HV, Gilmartin PM, Mikkelsen JD, Paul Knox J, Willats WG. Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J. 2004;40:260–275. doi: 10.1111/j.1365-313X.2004.02208.x. [DOI] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Price AH, Hendry GAF. Iron-catalyzed oxygen radical formation and its possible contribution to drought damage in nine native grasses and three cereals. Plant Cell Environ. 1991;14:477–484. [Google Scholar]
- Qiu Q, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane N+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem. 2004;279:207–215. doi: 10.1074/jbc.M307982200. [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Nat1 Acad Sci USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades JD, Loveday J. American Society of Civil Engineers. Salinity in irrigated agriculture. In: Steward BA, Nielsen DR, editors. Irrigation of Agricultural Crops (Monograph 30) Madison: American Society of Agronomists; 1990. pp. 1089–1142. [Google Scholar]
- Richmond TA, Somerville C. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville C. Integrative approaches to determining Csl function. Plant Mol Biol. 2001;47:131–143. [PubMed] [Google Scholar]
- Sagi M, Fluhr R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006;141:336–340. doi: 10.1104/pp.106.078089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wu SJ, Zhu JK. Overexpression of a plasma membrane Na+/H+ antiporter improves salt tolerance in Arabidopsis. Nat Biotechnol. 2003a;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- Shi H, Kim YS, Guo Y, Stevenson B, Zhu JK. The Arabidopsis SOS5 locus encodes a cell surface adhesion protein and is required for normal cell expansion. Plant Cell. 2003b;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Bray EA. LWR1 and LWR2 are required for osmo-regulation and osmotic adjustment in Arabidopsis. Plant Physiol. 2004;136:2831–2842. doi: 10.1104/pp.104.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M. AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol. 2001;126:575–586. doi: 10.1104/pp.126.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Lei D, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez Murillo F, Spencer F. A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- Yamahara T, Shiono T, Suzuki T, Tanaka K, Takio S, Sato K, Yamazaki S, Satoh T. Isolation of a germin-like protein with manganese superoxide dismutase activity from cells of a moss, Barbula unguiculata. J Biol Chem. 1999;274:33274–33278. doi: 10.1074/jbc.274.47.33274. [DOI] [PubMed] [Google Scholar]
- Yoda H, Hiroi Y, Sano H. Polyamine oxidase is one of the key elements for oxidative burst to induce programmed cell death in tobacco cultured cells. Plant Physiol. 2006;142:193–206. doi: 10.1104/pp.106.080515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis E, Huang J, Müller B, Darvill AG, Albersheim P. Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 1995;107:1129–1138. doi: 10.1104/pp.107.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- Zhu JK, Shi J, Singh U, Wyatt SE, Bressan RA, Hasegawa PM, Carpita NC. Enrichment of vitronectin- and fibronectin-like proteins in NaCl-adapted plant cells and evidence for their involvement in plasma membrane-cell wall adhesion. Plant J. 1993;3:637–646. [PubMed] [Google Scholar]
- Zhu JK, Hasegawa PM, Bressan RA. Molecular aspects of osmotic stress in plants. CRC Crit Rev Plant Sci. 1997;16:253–277. [Google Scholar]
- Zhu J, Gong Z, Zhang C, Song CP, Damsz B, Inan G, Koiwa H, Zhu JK, Hasegawa PM, Bressan RA. OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid–mediated and non-abscisic acid–mediated responses to abiotic stress. Plant Cell. 2002;14:3009–3028. doi: 10.1105/tpc.006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fu X, Koo YD, et al. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol Cell Biol. 2007a;27:5214–5224. doi: 10.1128/MCB.01989-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007b;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.