Abstract
Interferon (IFN) treatment induces tyrosine phosphorylation and nuclear translocation of Stat1 (signal transducer and activator of transcription) to activate or repress transcription. We report here that a member of the protein inhibitor of activated STAT family, PIASy, is a transcriptional corepressor of Stat1. IFN treatment triggers the in vivo interaction of Stat1 with PIASy, which represses Stat1-mediated gene activation without blocking the DNA binding activity of Stat1. An LXXLL coregulator signature motif located near the NH2 terminus of PIASy, although not involved in the PIASy–Stat1 interaction, is required for the transrepression activity of PIASy. Our studies identify PIASy as a transcriptional corepressor of Stat1 and suggest that different PIAS proteins may repress STAT-mediated gene activation through distinct mechanisms.
Interferon signals to the nucleus by activating the latent cytoplasmic transcription factor Stat1 to induce IFN-responsive genes (1, 2). In addition to function as a transcriptional activator, the transcriptional repression activity of Stat1 has also been documented (3, 4). Furthermore, the involvement of Stat1 in the constitutive transcription of several genes in the absence of ligand stimulation has recently been reported. It is believed that these distinct activities of Stat1 in gene regulation are achieved, in part, by the association of Stat1 with different transcriptional modulators (5, 6). p300 and CREB-binding protein transcriptional coactivators interact with Stat1 to enhance Stat1-mediated gene activation (7, 8). The constitutive expression of the low molecular mass polypeptide 2 (LMP2) gene is mediated by a complex of unphosphorylated Stat1 and interferon regulatory factor 1 (IRF1) (9, 10). However, a transcriptional corepressor of a STAT protein has not been uncovered.
Two members of the protein inhibitor of activated STAT (PIAS) protein family (5), PIAS1 and PIAS3, have been shown to act as inhibitors of Stat1- and Stat3-mediated gene activation, respectively. PIAS1 and PIAS3 can block the DNA binding activity of STAT in vitro (11, 12). Two related proteins named PIASx (consisting of two splicing variants PIASxα and PIASxβ) and PIASy have been identified. The role of PIASx or PIASy in STAT signaling is not understood.
We report here the characterization of PIASy in STAT signaling. PIASy is localized in the nucleus. Upon IFN stimulation, PIASy becomes associated with Stat1 in vivo. PIASy represses Stat1-mediated gene activation without inhibiting the DNA binding activity of Stat1. An LXXLL coregulator signature motif present at the NH2 terminus of PIASy, although not involved in PIASy–Stat1 interaction, is required for the repressive activity of PIASy on Stat1-mediated gene activation. Our results suggest that PIASy is a transcriptional corepressor of Stat1.
Materials and Methods
Plasmids and Antibodies.
Flag-PIASy and Flag-PIASy-AA were constructed by insertion of the human wild-type PIASy cDNA or a mutant PIASy cDNA containing substitutions of the CTC and CTG codons for Leu-23 and Leu-24 to GCC (Ala) and GCG (Ala) into the BamHI and SalI sites of pCMV-Flag vector, respectively. pcDNA3-PIASy and pcDNA3-PIASy-AA were constructed by insertion of the human wild-type or L23L24→AA mutant PIASy cDNA into the EcoRI and NotI sites of pcDNA3 vector (Invitrogen).
Anti-PIASy antiserum was raised against a recombinant fusion protein of glutathione S-transferase (GST) with the 121 COOH-terminal amino acid residues of murine PIASy, the sequence of which was obtained by sequencing EST clone 444894. The resulting antibody can recognize both human and murine PIASy proteins. Anti-pStat1 is from New England Biolabs.
Immunofluorescence Analysis.
Cells were plated on cover slips coated with fibronectin (10 μg/ml) in a 24-well plate and incubated at 37°C for 16 h. Cells were then washed once with 1× PBS, fixed with 3.7% formaldehyde for 10 min. After two washes with 1× PBST (1× PBS, 0.1% Tween 20), cells were treated with 0.2% Triton-X-100 for 5 min. Cells were then washed twice with 1× PBST and incubated in the blocking solution (1× PBST, 10% goat serum) at room temperature for 30 min. The first antibody was added into the blocking solution, and cells were incubated at room temperature for 1 h. Cells were then washed three times with 1× PBST at 10 min each time and incubated with the secondary antibody (anti-rabbit IgG-FITC or anti-mouse IgG-Cy3, Jackson ImmunoResearch) at a 1:500 dilution in the blocking solution at room temperature for 1 h. After three washes with 1× PBST, cover slips were mounted onto slides for observation under the fluorescence microscope.
Luciferase Assay.
293T cells were transfected by calcium precipitation and analyzed for luciferase activities as described (11). The relative luciferase units were corrected for relative expression of β-galactosidase. U2OS and HeLa cells were transfected with Lipofectamine reagent (Life Technologies, Grand Island, NY) and assayed for luciferase activities with a duel-luciferase reporter assay system (Promega), where a plasmid-encoding Renilla luciferase (pRLTK) was included to correct for differences in transfection efficiency, as instructed by the manufacturers.
GST Pull-Down Assays.
GST pull-down assays were performed as described (13). Briefly, PIASy and PIASy-AA proteins were in vitro translated and 35S labeled with a TNT (T7) quick-coupled transcription/translation system (Promega) by using pcDNA3-PIASy and pcDNA3-PIASy-AA as templates. Tyrosine-phosphorylated Stat1 (pStat1) was purified from RR1 strain expressing Stat1 and TrpE-v-Abl (J.t.H. and K.S., unpublished work). Non-tyrosine-phosphorylated Stat1 was purified from the same strain in the absence of TrpE-v-Abl. Approximately 1.6 μg of GST or GST-pStat1 proteins immobilized to GST beads were incubated with 20 μl of 35S-labeled proteins in 250 μl of the binding buffer (1× PBS, pH 7.5/0.02% Nonidet P-40/1 mM DTT/1 mM PMSF/1 μg/ml aprotonin/1 μg/ml leupeptin) at 4°C overnight with rotation. The beads were then washed four times with the binding buffer, and proteins bound to the beads were eluted by heating at 95°C for 5 min in 1× sample buffer, subjected to SDS/PAGE, and visualized by autoradiography.
Results
PIASy Is Localized in the Nucleus.
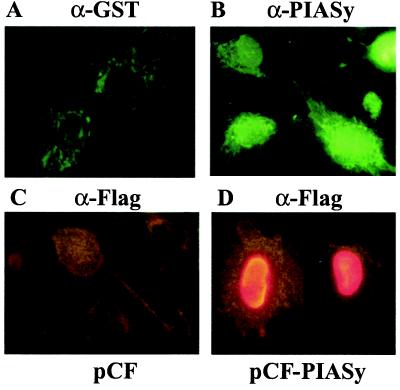
To study the function of PIASy, we prepared a specific antibody (anti-PIASy) against a recombinant fusion protein of GST to the 121 COOH-terminal amino acid residues of PIASy. This COOH-terminal region of PIASy does not display significant sequence homology with other PIAS proteins. Immunofluorescence analysis with anti-PIASy antibody indicated that endogenous PIASy is present in the nucleus in human 2fTGH fibroblasts (Fig. 1A). Furthermore, 2fTGH cells transiently transfected with a FLAG-tagged PIASy expression vector displayed exclusive nuclear staining when analyzed by anti-FLAG monoclonal antibody (Fig. 1B). Thus, PIASy is expressed in the nucleus.
Figure 1.
PIASy is localized in the nucleus. Human 2fTGH cells were stained with anti-GST (A) or anti-PIASy (B). 2fTGH cells were transfected with an empty vector (C) or pCF-PIASy (D) and stained with anti-Flag.
PIASy Is Associated with Stat1 in Vivo in Response to IFN Stimulation.
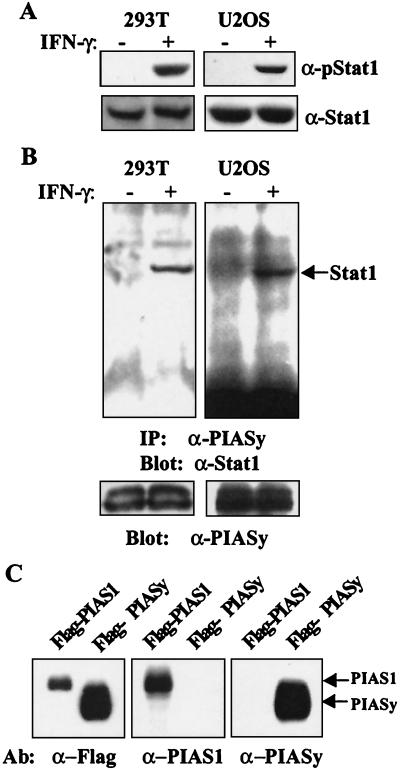
To demonstrate that PIASy, like PIAS1 and PIAS3, could interact with a STAT protein, coimmunoprecipitation analysis was performed. Protein extracts prepared from 293T cells untreated or treated with IFN-γ were analyzed by Western blot. As predicted, IFN-γ treatment induced the tyrosine phosphorylation of Stat1 as measured by an antibody that specifically recognizes tyrosine-phosphorylated Stat1 (Fig. 2A). Immunoprecipitation of the same extracts with anti-PIASy antibody, followed by immunoblot analysis, revealed that Stat1 was present in PIASy immunoprecipitates from cells treated with IFN-γ but not untreated cells (Fig. 2B). Similar results were obtained in human osteosarcoma U2OS cells (Fig. 2B). However, we did not detect the association of PIASy with other STATs (data not shown). These results demonstrate that IFN-γ treatment induces the association of endogenous PIASy and Stat1.
Figure 2.
The in vivo PIASy–Stat1 interaction is dependent on IFN stimulation. (A) Western blot analysis of protein extracts from 293T or U2OS cells with anti-pStat1 (New England Biolabs) or anti-Stat1. (B) Extracts from A were subjected to immunoprecipitation with anti-PIASy followed by Western blot analysis with anti-Stat1 (Upper). The same filter was reprobed with anti-PIASy (Lower). (C) Western blot analysis of protein extracts from 293T cells transfected with Flag-PIAS1 or Flag-PIASy. The filter was probed with anti-Flag, anti-PIAS1, or anti-PIASy as indicated.
To further validate the PIASy–Stat1 association, protein extracts from 293T cells transiently overexpressing Flag-PIAS1 or Flag-PIASy were analyzed by immunoblot by using anti-Flag, anti-PIAS1, or anti-PIASy antibodies (Fig. 2C). PIAS1, which had been shown previously to interact with Stat1, was used as a control. Anti-Flag antiserum detected both Flag-PIAS1 and Flag-PIASy. Anti-PIAS1 and anti-PIASy recognized only PIAS1 and PIASy, respectively. These results confirmed the specificity of these anti-PIAS antibodies and further validated the unique PIASy–Stat1 association.
PIASy Represses Stat1-Mediated Gene Activation.
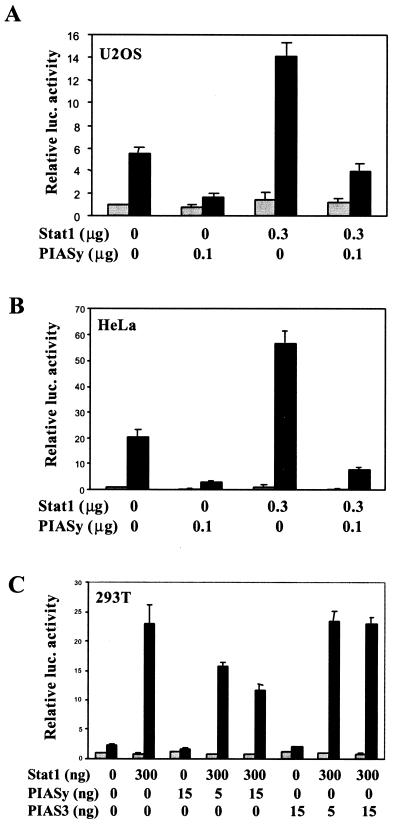
To determine the functional significance of the PIASy–Stat1 interaction, the effect of PIASy on Stat1-mediated gene activation was examined by using reporter assays. U2OS cells were transiently transfected with a luciferase construct containing three copies of the STAT binding sequence from the Ly6E gene (3xLy6E) (14), together with or without Stat1 or PIASy. IFN-γ treatment induced a 6-fold and 14-fold stimulation of luciferase expression by the endogenous and overexpressed Stat1, respectively (Fig. 3A). The coexpression of PIASy efficiently repressed the IFN-γ-induced Stat1-dependent gene activation. A similar inhibitory effect of PIASy on Stat1-mediated gene activation was also observed in other cell lines including HeLa cells (Fig. 3B) and 293T cells (Fig. 3C). Consistent with previously reported data (12), PIAS3 did not inhibit Stat1-mediated gene activation (Fig. 3C). These results imply that PIASy is a repressor of Stat1.
Figure 3.
PIASy represses Stat1-mediated gene activation. (A) Luciferase assays. Human U2OS cells were transfected with a Stat1 luciferase reporter construct (3xLy6E) with or without Stat1 or PIASy as indicated. Cells were treated with (black bar) or without (gray bar) IFN-γ (5 ng/ml) for 6 h before harvesting. Dual-luciferase assays were performed. (B) Same as A except HeLa cells were used. (C) Same as A except 293T cells were transfected by calcium precipitation method, and PIAS3 was included as a control as indicated. luc., luciferase.
PIASy Does Not Inhibit the DNA Binding Activity of Stat1.
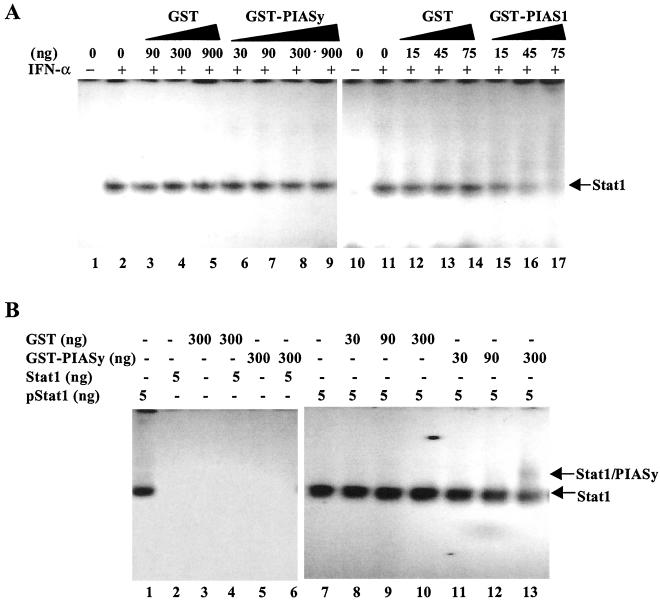
PIAS1 inhibits Stat1-mediated gene activation by blocking the DNA binding activity of Stat1 (11). To determine whether PIASy can also inhibit the DNA binding activity of Stat1, nuclear extracts from human Daudi B cells treated with IFN-α were analyzed by electrophoretic mobility-shift assays in the presence of GST or GST-PIASy proteins at various concentrations. PIASy had no effect on the DNA binding activity of Stat1 (Fig. 4A, lanes 1–9). In contrast, PIAS1 was found to be able to block the DNA binding activity of Stat1 in similar assays (Fig. 4A, lanes 10–17). We noted that a PIASy-Stat1 supershift was not detected in these assays, possibly because the PIASy–Stat1 complex was not stable under these conditions.
Figure 4.
PIASy does not block the DNA binding activity of Stat1. (A) Electrophoretic mobility-shift assay analysis. Nuclear extracts prepared from Daudi cells untreated or treated with IFN-α were mixed with a 32P-labeled Stat1 binding oligonucleotide (11) in the absence (lanes 1, 2, 10, and 11) or presence of various amounts of GST (lanes 3–5 and 12–14), GST-PIASy (lanes 6–9), or GST-PIAS1 (lanes 15–17) as indicated. (B) Same as A except purified tyrosine-phosphorylated Stat1 (lanes 1 and 7–13) or purified unphosphorylated Stat1 (lanes 2–6) was mixed with increasing concentrations of GST or GST-PIASy (30, 90, and 300 ng).
To further confirm that PIASy does not affect the DNA binding activity of Stat1, we performed electrophoretic mobility-shift assay analysis with purified tyrosine-phosphorylated Stat1 protein with increasing concentrations of GST or GST-PIASy (Fig. 4B). Although GST had no effect on the DNA binding activity of Stat1 (Fig. 4B, lanes 8–10), the addition of GST–PIASy at a higher concentration supershifted Stat1 to a more slowly migrating complex (Fig. 4B, lane 13). As negative controls, no complexes were detected when unphosphorylated Stat1 was used (Fig. 4B, lanes 2–6). Thus, a weak Stat1–PIASy supershift complex could be detected in these assays when purified proteins were used. We conclude that PIASy does not inhibit the DNA binding activity of Stat1.
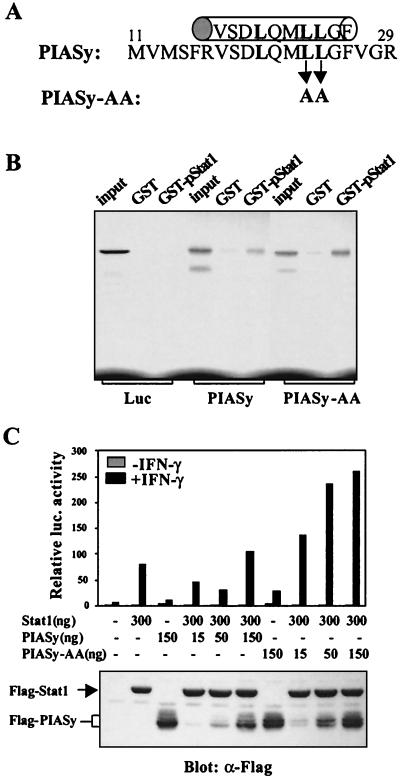
The N-terminal LXXLL Motif of PIASy Is Required for the Inhibitory Activity of PIASy.
In addition to the finding that PIASy is unable to block the DNA binding activity of Stat1, we also failed to detect an effect of PIASy on the tyrosine or serine phosphorylation status of Stat1 (data not shown). We next tested the hypothesis that PIASy may function as a nuclear corepressor of Stat1. A number of nuclear receptor (NR) coactivators contain the α-helical LXXLL signature motif (15, 16). Recently, sequences highly related to the LXXLL signature motif have also been found in NR corepressors (17–19). The LXXLL coactivator signature motif has been previously demonstrated as a protein interaction module that can mediate coactivator–NR interactions or coactivator–coactivator interactions (20–24). PIASy contains a putative α-helical LXXLL signature motif at its NH2 terminus (Fig. 5A). To test whether this putative LXXLL motif plays a role in PIASy-mediated transcriptional repression, a mutant PIASy (PIASy-AA) with point mutations in the LXXLL motif of PIASy (LXXLL to LXXAA) was generated. To test whether mutations of the LXXLL motif affect PIASy–Stat1 interaction, GST pull-down assays were performed with 35S-labeled PIASy and PIASy-AA proteins (Fig. 5B). Consistent with the in vivo coimmunoprecipitation assays, PIASy was bound to tyrosine-phosphorylated Stat1 (GST–pStat1) but not GST. Most importantly, PIASy-AA mutant protein was also capable of binding to Stat1 (Fig. 5B). We conclude that the LXXLL motif of PIASy is not required for PIASy–Stat1 interaction.
Figure 5.
The LXXLL motif of PIASy is required for the transrepression activity of PIASy. (A) The region containing the α-helical LXXLL motif of PIASy is indicated. (B) In vitro GST-pull-down assays. GST or GST-tyrosine-phosphorylated Stat1 (GST-pStat1) immobilized on glutathione beads were mixed with in vitro translated, 35S-labeled luciferase, PIASy or PIASy-AA as indicated. Proteins bound were analyzed on an 8% SDS/PAGE followed by autoradiography. (C) Luciferase assays. 293T cells were transfected with a Stat1 luciferase reporter construct together with or without Flag-Stat1 or PIASy or PIASy-AA as indicated (Upper). Protein extracts from the same samples were analyzed by Western blot with anti-Flag antibody (Lower). luc., luciferase.
We next tested whether the PIASy LXXLL signature motif is required for the transrepression function of PIASy. 293T cells were transiently transfected with expression vectors encoding the wild-type PIASy or the PIASy-AA mutant together with Stat1 and 3xLy6E reporter constructs (Fig. 5C). Consistent with the earlier results (Fig. 3), the presence of low levels of wild-type PIASy inhibited Stat1-dependent transcription in response to IFN-γ stimulation (Fig. 5C). Interestingly, the inhibitory effect of wild-type PIASy was relieved when it was expressed at a high dose. This “squelching” phenomenon has also been observed with other coregulators and is believed to be the result of the uncoupling of transcription factors from other coregulator components (25). Of note, this squelching effect was not observed with PIAS1, which represses Stat1-mediated gene activation by blocking the DNA binding activity of Stat1 (11, 26). In contrast to the wild-type PIASy protein, coexpression of the PIASy-AA mutant at a low level failed to inhibit Stat1-mediated transcription. Most importantly, the presence of higher levels of PIASy-AA expression strongly enhanced Stat1-dependent gene activation, a dominant negative effect possibly resulting from the ability of the PIASy-AA mutant protein, which is able to bind to Stat1 but is defective in gene repression, to compete with the endogenous PIASy or other inhibitory molecules for binding to Stat1. A similar effect of PIASy-AA was also observed in HeLa and U2OS cells when PIASy-AA was overexpressed (unpublished observation). The proper expression of PIASy and Stat1 proteins was confirmed by Western blot analysis (Fig. 5C, Lower). We conclude that the PIASy LXXLL motif, although not involved in interacting with Stat1, is required for the transcriptional repression activity of PIASy.
Discussion
Transcriptional repression has increasingly been recognized to play an important role in regulating eukaryotic gene expression. Transcriptional repression can be achieved by protein factors that can interfere with the DNA binding activity of a transcription factor or by corepressors that influence transcription by interacting with the basal transcriptional machinery or remodeling chromatin (25). In this article, we report our studies on the role of PIASy in STAT signaling. Several lines of evidence suggest that PIASy is a transcriptional corepressor of Stat1. First, PIASy interacts with Stat1 both in vitro and in vivo. The in vivo PIASy–Stat1 interaction is dependent on cytokine stimulation. Second, PIASy can inhibit Stat1-mediated gene activation without blocking the DNA binding activity of Stat1. In fact, when PIASy was expressed at a high level, the inhibitory effect of PIASy on Stat1-mediated gene activation was relieved. This “squelching” phenomenon, which has been observed with many other coregulators, is consistent with our conclusion that PIASy does not inhibit the DNA binding activity of Stat1. PIASy also does not affect the tyrosine or serine phosphorylation of Stat1 (data not shown). Third, an LXXLL coregulator signature motif of PIASy is required for the transrepression activity of PIASy. The LXXLL signature motif was originally identified in several nuclear receptor coactivators. Recently, sequences highly related to the LXXLL signature motif have also been identified in NR corepressors. The α-helical LXXLL signature motif is a protein interaction module that has been demonstrated to mediate coregulator–NR interactions or coregulator–coregulator interactions. The LXXLL motif of PIASy is not involved in PIASy–Stat1 interaction but is absolutely required for the repression activity of PIASy. Consistently, the PIASy LXXLL mutant protein (PIASy-AA) significantly enhanced Stat1-mediated gene activation when overexpressed (Fig. 5C), possibly resulting from the competition of PIASy-AA with the endogenous PIASy or other inhibitory molecules to bind to Stat1. Based on these results, we propose that PIASy may act as an adaptor protein to recruit other corepressors to inhibit Stat1-mediated gene activation. The PIASy LXXLL motif is likely involved in the assembly of the PIASy-containing corepressor complex. In support of this model, our recent in vivo pull-down studies have clearly uncovered the specific association of several proteins with wild-type PIASy but not PIASy-AA (unpublished observation). Further biochemical characterization of the PIASy-containing protein complex is required to understand the detailed molecular mechanism of PIASy-mediated inhibition on Stat1. Nevertheless, our studies provide the first example that an LXXLL motif is involved in the modulation of STAT-mediated gene activation and suggest that this protein interaction motif may be more widely used in gene regulation than previously described.
Four mammalian members of the PIAS family have been identified including PIAS1, PIAS3, PIASx, and PIASy. Additional PIAS family members may also exist (R. Schreiber, personal communication). Recently, a Drosophila PIAS homologue named Zimp has also been described (27). Genetic studies indicate that the Drosophila PIAS protein is a negative regulator of dSTAT (J. Darnell, personal communication). We have previously reported that PIAS1 and PIAS3 are inhibitors of Stat1 and Stat3, respectively. In this article, we demonstrated that PIASy is a transcriptional corepressor of Stat1. Thus, it seems that members of the PIAS family can differentially modulate the activity of STATs by at least three ways. (i) A PIAS protein can specifically interact with a STAT molecule, e.g., PIAS1–Stat1 and PIAS3–Stat3 interactions. (ii) PIAS can inhibit STAT-mediated gene activation through different mechanisms, as in the case of the inhibition of Stat1 activity by PIAS1 and PIASy. (iii) Different PIAS proteins may be involved in the regulation of STAT signaling in different tissues because of their differential tissue distribution. For example, PIAS1 is enriched in thymus, where PIASy is barely detectable. Instead, PIASy is highly expressed in spleen (unpublished observations). We believe that these interesting properties of PIAS proteins contribute significantly to the complexity and flexibility in the negative regulation of STAT-mediated gene activation.
Acknowledgments
We thank M. Carey for comments and discussion, X. Rao for technical assistance, and T. Graeber for help in structure analysis. B. L. is a Cancer Research Institute fellow. This work was supported by National Institutes of Health Grants AI39612 and AI43438 and the Valvano Foundation (to K.S.).
Abbreviations
- GST
glutathione S-transferase
- NR
nuclear receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 3.Ramana C V, Grammatikakis N, Chernov M, Nguyen H, Goh K C, Williams B R, Stark G R. EMBO J. 2000;19:263–272. doi: 10.1093/emboj/19.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramana C V, Chatterjee-Kishore M, Nguyen H, Stark G R. Oncogene. 2000;19:2619–2627. doi: 10.1038/sj.onc.1203525. [DOI] [PubMed] [Google Scholar]
- 5.Shuai K. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee-Kishore M, van den Akker F, Stark G R. Trends Cell Biol. 2000;10:106–111. doi: 10.1016/s0962-8924(99)01709-2. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee-Kishore M, Wright K L, Ting J P, Stark G R. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee-Kishore M, van Den Akker F, Stark G R. J Biol Chem. 2000;275:20406–20411. doi: 10.1074/jbc.M001861200. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, Liao J, Rao X, Kushner S A, Chung C D, Chang D D, Shuai K. Proc Natl Acad Sci USA. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung C D, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 13.Tan J, Hall S H, Hamil K G, Grossman G, Petrusz P, Liao J, Shuai K, French F S. Mol Endocrinol. 2000;14:14–26. doi: 10.1210/mend.14.1.0408. [DOI] [PubMed] [Google Scholar]
- 14.Khan K D, Shuai K, Lindwall G, Maher S E, Darnell J E, Jr, Bothwell A L. Proc Natl Acad Sci USA. 1993;90:6806–6810. doi: 10.1073/pnas.90.14.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 16.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 17.Hu X, Lazar M A. Nature (London) 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 18.Perissi V, Staszewski L M, McInerney E M, Kurokawa R, Krones A, Rose D W, Lambert M H, Milburn M V, Glass C K, Rosenfeld M G. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy L, Kao H Y, Love J D, Li C, Banayo E, Gooch J T, Krishna V, Chatterjee K, Evans R M, Schwabe J W. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass C K, Rosenfeld M G. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 21.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, et al. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neaear A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Nature (London) 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 24.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Neaear A M, Erdjument-Bromage H, Tempst P, Freedman L P. Nature (London) 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 25.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 26.Liao J, Fu Y, Shuai K. Proc Natl Acad Sci USA. 2000;97:5267–5272. doi: 10.1073/pnas.97.10.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohr S E, Boswell R E. Gene. 1999;229:109–116. doi: 10.1016/s0378-1119(99)00033-5. [DOI] [PubMed] [Google Scholar]