Abstract
CD1 is an MHC class I-like antigen-presenting molecule consisting of a heavy chain and β2-microglobulin light chain. The in vitro refolding of synthetic MHC class I molecules has always required the presence of ligand. We report here the use of a folding method using an immobilized chaperone fragment, a protein disulphide isomerase, and a peptidyl-prolyl cis-trans isomerase (oxidative refolding chromatography) for the fast and efficient assembly of ligand-free and ligand-associated CD1a and CD1b, starting with material synthesized in Escherichia coli. The results suggest that “empty” MHC class I-like molecules can assemble and remain stable at physiological temperatures in the absence of ligand. The use of oxidative refolding chromatography thus is extended to encompass complex multisubunit proteins and specifically to members of the extensive, functionally diverse and important immunoglobulin supergene family of proteins, including those for which a ligand has yet to be identified.
Keywords: minichaperone, protein folding, GroEL, DsbA, PPI
Human CD1a, -b, -c, -d, and -e molecules are a family of MHC class I-like transmembrane glycoproteins that map to chromosome 1 and have restricted polymorphism (1–4). Although CD1d may exist as a β2-microglobulin- (β2m) independent form (5), CD1 proteins are generally heterodimers of a CD1 heavy chain in noncovalent association with a β2m light chain. CD1-mRNA molecules are spliced alternatively to generate cell surface, intracellular, and secretory isoforms (6).
Unlike MHC class I molecules that bind peptides (7), CD1 molecules bind and present a diverse range of nonclassical hydrophobic lipid and glycolipid ligands that include lipoglycan, lipoarabinomannan, mycolic acid, phosphatidylinositolmannan, glucose monomycolate, α-glucosylceramide, hexosyl-1-phosphoisoprenoids, and mannosyl-β1-phosphodolichols (8–13). They also present hydrophobic peptides (14, 15). This specialization of function is reflected in the three-dimensional structure of mouse CD1d (the only published CD1 structure). The antigen-binding pocket is narrow, deep, hydrophobic, and lined by residues with minimal polymorphism (16). The ligand–CD1 trimeric complexes are recognized by specific T cell antigen receptors (TCR) located on the surface of TCRγδ+ CD4−CD8−, TCRαβ+ CD4−CD8−, CD4+CD8+, and NK CD4+ CD4−CD8− T cells of the immune system (8–12). Although apparently ligand-free CD1 and MHC class I molecules have been synthesized in Drosophila melanogaster cells that lack the proteins necessary for intracellular peptide loading (17), crystallographic studies of mouse CD1d made in this system demonstrate electron density in the ligand-binding groove, suggesting that they may not be ligand-free (16).
Foldases and molecular chaperones are integral components of the in vivo cellular folding machinery (18). For MHC class I (19) and CD1 (20), calnexin and calreticulin provide chaperoning activity. The mechanism by which CD1 molecules are loaded with lipid antigen has not been elucidated fully, but because it is independent of the transporter associated with antigen process-ing (21) and occurs in endosomes or on the cell surface rather than in the endoplasmic reticulum, it is likely that CD1 employs a different pathway from that used by MHC class I (13, 22–24).
The cell-free assembly of MHC class I heterodimers requires the presence of exogenous ligand to form a ternary complex with the MHC class I heavy chain and β2m. Refolding in the presence of irrelevant ligands or the absence of ligands does not yield stable complexes (25–28). We found, similarly, that denatured CD1 heavy chains refold inefficiently in a cell-free environment containing β2m but lacking ligand. Oxidative refolding chromatography using an aqueous suspension of an equimolar mixture of agarose-gel bead immobilized prokaryotic miniGroEL (a minichaperone containing the apical domain of GroEL), DsbA (a protein disulphide isomerase), and a peptidyl-prolyl cis-trans isomerase (PPI) proved efficient in restoring the native conformation of scorpion toxin, which is a single polypeptide consisting of 63 amino acids (29). In this paper, we show that this artificial folding system allows refolding of CD1a and -b heterodimers.
Materials and Methods
Preparation of the Refolding Matrix.
GroEL minichaperone consisting of a 191–345 peptide fragment from Escherichia coli, PPI, and E. coli DsbA, were expressed, purified, and immobilized (29).
Expression of CD1a and -b.
Human CD1a and -b heavy chains were amplified by reverse transcription–PCR from a human dendritic-cell cDNA library by using 5′ oligonucleotides priming after the leader sequence and 3′ oligonucleotides priming between the α3 and TM domains. The primer sequences were: CD1A[B], (5′-TTCTCGAGCATATGAATGCAGACGGGCTC) and CD1A[F] (5′-AAGGATCCGTGATGCTCCCAGTAGAGGAC) for CD1a, and CD1-B[B] (5′-TTTCTAGACATATGAGTGAACATGCCTT) and CD1B[F] (5′-AAGGATCCGGGGGTTTCTCCAGTAG) for CD1b. The back primers incorporated XbaI and NdeI restriction sites, whereas the forward primers contained a BamHI restriction site. Following sequence verification, the cDNAs were digested and subcloned into expression-vector pET-23d (Novagen) containing a BirA enzymatic biotinylation site (30). An identical construct was prepared containing a full-length human β2m cDNA amplified from the same library with primers: B2M[B] (5′-GTGGATCGAGACATGTAAGGATTCTTT and B2M[F] (5′-TTTCATATGATCCAGCGTACTCCAAAG).
Recombinant CD1a and -b and β2m chains were expressed in E. coli (DE3) BL21 LysS. BL21 cells were electroporated and colonies inoculated into 2× TY growth medium (1.6% tryptone/1% yeast extract/0.5% NaCl, pH 7.4) containing 100 μg⋅ml−1 ampicillin and incubated at 27°C. Expression was induced with 1.0 mM isopropyl-β-D-thiogalactopyranoside and growth was continued for 3 h at 37°C. Cell pellets were lysed in a French pressure cell and centrifuged at 10,000 × g for 10 min. Inclusion bodies were washed several times in 10 mM Tris/0.1 mM EDTA (pH 8.0) (40 ml) containing PMSF (50 μg⋅ml−1), washed once in 1.0 M urea, flash frozen, and stored at −70°C. Protein was quantified by using Bio-Rad kits.
Refolding Procedure.
Batchwise refolding was performed with an equimolar suspension of miniGroEL agarose, DsbA agarose, and PPI agarose (29). Inclusion bodies were solubilized in freshly prepared 6 M GuHCl [containing 100 mM potassium phosphate buffer (pH 8.0)] and reduced with 0.1 M DTT.
The extent of unfolding and reduction was assessed by circular dichroism (CD) spectroscopy, measurement of turbidity, and quantification of free-SH groups by using 5,5′-dithiobis (2-nitrobenzoic acid). The refolding matrix was equilibrated with refolding buffer [100 mM potassium phosphate/0.3 M L-arginine HCl/8 mM oxidized glutathione/1.0 mM EDTA/0.1 M PMSF (pH 8.0)]. Freshly denatured/reduced CD1 heavy (-a or -b chains) and β2m light chains were mixed together in varying molar ratios (1:1 to 1:10) immediately before refolding. A molar ratio of 1:3 (heavy/light) was optimal. The mixture of heavy and light chains was added slowly, mixed, and diluted (1:100) into an aqueous suspension of the ternary refolding matrix. This mixture was rotated at 4°C for a range of incubation times (6 min to 12 h) and centrifuged at <1,000 × g for 5 min. The soluble fraction was concentrated by using dialysis membranes covered with D-trehalose (Sigma) and Ultrafree-15 centrifugal filter devices (Millipore). In conditions with ligand (+L), the glycolipid sulfatide (ceramide galactoside 3-sulfate, a newly established ligand of CD1a; A.S. and G.D., unpublished data) for CD1a and monosialoganglioside for CD1b were solubilized in PBS and sonicated (31). Ligand was added to the refolding buffer–ternary matrix suspension immediately before refolding in a 10-fold (final) molar excess giving a final molar ratio of 1:3:10 (heavy chain/β2m/synthetic peptide). In −L conditions, no ligand was added.
Analysis of Refolded Protein.
Gel filtration reverse-phase HPLC was performed on a Superdex-75 (Amersham Pharmacia) column equilibrated with 50 mM potassium phosphate buffer/150 mM KCl/2% (vol/vol) glycerol (29). Light-scattering analysis was done at 350 nm with a Hitachi 4000 spectrofluorometer (29). Immunogenicity was tested in an inhibition immunoassay (6) by using the high-CD1a-expressing mutant NH17 (32) or the high-CD1b-expressing mutant ER1 (3) as targets. Tissue-culture supernatant containing soluble CD1a was used as a positive control (6). Monoclonal antibodies (mAbs) NA1/34, 19H39, B17, and 10D12 were used as inhibitors. In the case of CD1b, mAb NU-T2 was used in the inhibition step. Rabbit anti-mouse HRP conjugate was used for detection. C/D spectra were obtained by using a Jasco (Easton, MD) Model J-720 spectrometer. Calibration was performed by using (1S)-(+)-10-camphorsulfonic acid (Aldrich) with a molar-extinction coefficient of 34.5 M−1⋅cm−1 at 285 nm and dichroism of 2.36 M−1⋅cm−1 at 290.5 nm. Samples from various refolding conditions (1.0 ml each) were analyzed by using refolding buffer as a blank. Spectra were recorded with protein concentrations of 0.15–0.30 mg⋅ml−1 in 25 mM potassium phosphate buffer (pH 8.0) and 0.1-cm quartz cuvettes at room temperature.
NH2-Terminal Sequencing and Stoichiometry.
HPLC-purified CD1a and -b (+L and −L) was run on an SDS/PAGE gel and electroblotted with a Bio-Rad Trans-Blot system. After Ponceau-S staining, bands corresponding to CD1a and -b heavy and β2m light chains were excised. NH2-terminal sequence analysis was done on a Procise-494 protein sequencer (Applied Biosystems). Stoichiometry of the chains was determined by amino acid composition analysis of HPLC-purified bands. Norleucine was added to a solution of the bands and the mixture was concentrated in a centrifugal evaporator. Hydrolyzed material (gas phase HCl at 115°C for 22 h) was analyzed on an Amersham-Pharmacia Alpha Plus Series II.
Ultracentrifugation.
Protein samples were dialyzed against 50 mM Tris (pH 8.0)/60 mM GuHCl 2% glycerol/1.0 mM EDTA. Sedimentation equilibrium experiments were conducted in a Beckman Optima XLI analytical ultracentrifuge (An60Ti rotor, 4°C) and spun at 10,000 or 9,000 rpm. Runs were overspeeded at 27,000 rpm for 6 h (33, 34). Absorbance at 275 nm was measured until equilibrium was reached. The specific densities of β2m and CD1a were calculated as 0.7208 and 0.7212 ml⋅g−1, respectively (density = 1.009 g⋅ml−1) and corrected to 5°C by using SEDNTERP (34). Mass-average apparent molecular weights were calculated as described (35). Data were analyzed by direct fitting using ULTRASPIN software (http://www.mrc-cpe.cam.ac.uk/ultraspin).
Results
Folding of CD1a and -b With and Without Ligand.
Overproduction of CD1a and -b heavy chains and β2m light chains at high levels (30–40 mg⋅liter−1) as insoluble aggregates in the cytoplasm of E. coli was detected on SDS/PAGE gels. Refolding was carried out in the presence (+L) and absence (−L) of synthetic ligand (sulfatide in the case of CD1a and monosialoganglioside for CD1b). If mixtures of denatured/reduced synthetic chains of CD1a or -b (+L or −L) and β2m were refolded in the presence of minichaperone or foldases only (DsbA agarose or PPI agarose), the β2m remained in solution, whereas the CD1a or -b heavy chains formed a dense precipitate. In the presence of all three refolding components (+L and −L), the heavy chains were solubilized and there was no visible precipitate (Table 1).
Table 1.
Batchwise renaturation of synthetic denatured/reduced CD1a chains
| Conditions* | Soluble proteins, %
|
||||
|---|---|---|---|---|---|
| Total† | Aggregated (peak 1)‡ | Dimer of CD1a (peak 2a) | Monomer (peak 2b) | β2m (peak 3) | |
| Refolding buffer only§ | <10 | >80 | 0 | 0 | <20 |
| PPI-agarose¶ | 15–20 | 30–40 | 0 | 0 | <60 |
| DsbA-agarose‖ | 15–20 | <40 | 0 | 0 | >60 |
| Minichaperone−** | 15–20 | >40 | 0 | 0 | <60 |
| Refolding matrix‡‡ | 87 | 2.3 | 4–8 | 14.8 | 68 |
Yields were calculated as percentage of total protein recovered in the total soluble fraction.
Components of the refolding matrix were coupled independently to NHS-activated Sepharose.
Refers to soluble protein recovered after refolding.
Peak numbers (see Fig. 1). Large aggregates after concentration of peak 1 were removed by centrifugation.
0.12 mg of denatured/reduced CD1a were added to 4.4 mL of refolding buffer.
0.12 mg denatured/reduced CD1a was added to 2 mL of PPI-agarose/refolding buffer and made up to a final volume of 4.4 mL.
0.12 mg of denatured/reduced CD1a was added to 3 mL of DsbA-agarose/refolding buffer and made to a final volume of 4.4 mL.
0.12 mg of denatured/reduced CD1a was added to 2 mL of minichaperone–agarose/refolding buffer and made up to a final volume of 4.4 mL.
1.2 mg of denatured/reduced CD1a was added to 1.3 mL of minichaperone–agarose + 2.6 mL DsbA–agarose + 1.1 mL PPI–agarose/refolding buffer and made up to a final volume of 50 mL.
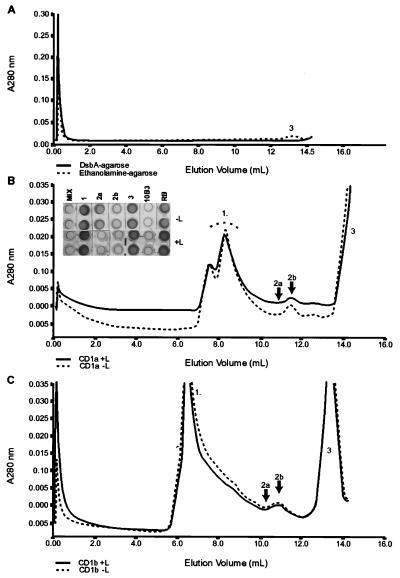
The ternary matrix (an equimolar mixture of miniGroEL agarose/DsbA agarose/PPI agarose) solubilized the greater majority of the sample (87%). Approximately 6% appeared as an HPLC shoulder (peak 2a in Fig. 1C; Table 1) with an elution volume corresponding to ≈90 kDa, consistent with dimeric CD1a. An additional shoulder corresponded to possible higher-order oligomers; 15% appeared as a symmetrical peak (2b) with an elution volume corresponding to the expected 44-kDa molecular mass of monomeric CD1a and 60% as a symmetrical chromatographic peak (peak 3) at an elution volume corresponding to 12 kDa (β2m) (Table 1; Fig. 1B). At this protein concentration, the peaks were absent if refolding was performed with either minichaperone alone (data not shown), DsbA alone (Fig. 1A), or ethanolamine agarose (Fig. 1A). Oxidative refolding of a mixture of denatured/reduced CD1b heavy chain and β2m (+L and −L) gave peaks corresponding to those seen with CD1a (Fig. 1C).
Figure 1.
Refolding of CD1a under different conditions. (A) Elution profile from a Superdex-75 gel-filtration column of soluble supernatants when refolding was performed in the presence of either DsbA agarose (full line) or Ethanolamine agarose (broken line). The estimated molecular mass for peak 3 is 12 kDa. (B) Elution profile from an analytical Superdex-75 gel-filtration column of refolded CD1a supernatant by using oxidative refolding chromatography in the presence (full line) or absence (broken line) of exogenous ligand. The refolded CD1a (indicated by arrows as peaks 2a and 2b) corresponds to estimated molecular masses of ≈90 and 44 kDa, respectively. Peak 1 is a high molecular-mass aggregate. The estimated molecular mass for peak 3 is 12 kDa. (Inset) Antigenic activity of the unfractionated refolding mixture, MIX; peak 1 corresponding to aggregate, 1; peak 2a corresponding to dimeric CD1a, 2a; peak 2b corresponding to monomeric CD1a, 2b; and peak 3 corresponding to β2m, 3. The sCD1a-positive control and refolding-buffer-only negative control are labeled 10B3 and RB, respectively. +L refers to samples refolded in the presence of ligand and −L to those refolded without ligand. Each sample was tested in duplicate. Protein concentrations of the samples and positive control were approximately 1.0 mg⋅ml−1. (C) Elution profile from an analytical Superdex-75 gel-filtration column of CD1b supernatant refolded with (+L) and without (−L) specific ligand. Peaks (including 2a and 2b) correspond to molecular masses approximately similar to those above.
It was possible to refold CD1a in the absence of ternary matrix but only in the presence of ligand and if the molarity of the denatured substrates was reduced by three orders of magnitude. The incubation times required for refolding (as indicated by the gain of reactivity to mAb NA1/34) were in the order of days, as opposed to minutes, when catalyzed by the ternary complex. The yield of refolded material was not, however, increased in the presence of refolding matrix (as compared with ligand only), and it is possible that the ternary matrix uses a similar folding pathway to that induced by the ligand. The ternary matrix enabled the refolding process to be performed with higher molarities of starting products, in smaller reaction volumes, and with much shorter incubation times.
Antigenic and Biochemical Characterization of Refolded CD1a.
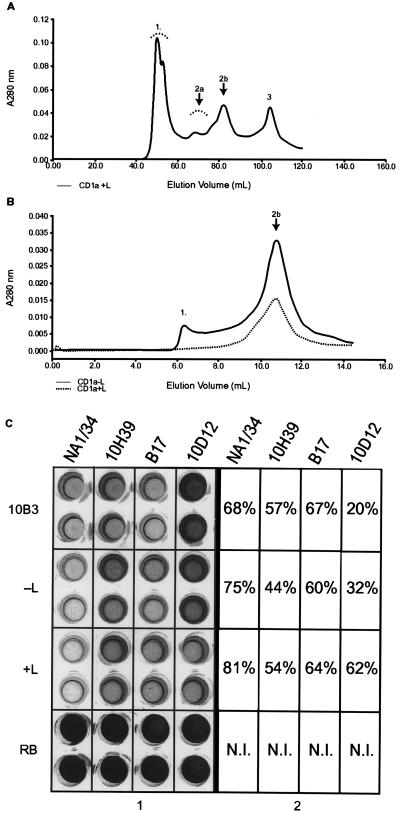
Soluble samples from refolding protocols (+L and −L) fractionated by gel-filtration chromatography were collected and tested for activity in an inhibition immunoassay by using the anti-CD1a conformation-sensitive mAb NA1/34 (Fig. 1B). Only the unfractionated refolding mixture, fractions 2b (monomeric CD1a ≈44 kDa) and 2a (dimeric CD1a ≈88 kDa) had activity. This activity was at least as high as that of the positive control (soluble human CD1a secreted by 10B3; Fig. 1B). The antigenic profile of the refolded CD1a was characterized further by using anti-CD1a conformation-sensitive mAbs (19H39.3, B17, and 10D12.2), each of which reacted with both monomeric (Fig. 2C) and dimeric (data not shown) CD1a. The activity of refolded CD1b was tested by using mAb NU-T2 and ER1 as a target. Only the unfractionated refolding mixture and HPLC-purified CD1b monomer and oligomer had activity (data not shown).
Figure 2.
Optimization of the refolding procedure. (A) Scale-up of the CD1a refolding experiments and elution profile in a preparative Superdex-75 gel-filtration column. Estimated molecular masses of peaks correspond to those seen in the analytical gel-filtration column. (B) Rerun of peak 2b (obtained from scale-up, see Fig. 2A) in an analytical Superdex-75 column. (C) Antigenic characterization of peak 2b (obtained from fractionation of the CD1a refolding mixture supernatant). Samples were tested with four different conformation-sensitive CD1a mAbs: NA1/34, 10H39, B17, and 10D12. Numbers in the boxes refer to percentage of inhibition as compared with the refolding buffer-only (RB) negative control. The positive control is labeled 10B3. −L refers to samples refolded without specific ligand and +L to those refolded with ligand. The concentration of CD1a in the 10B3 positive control was approximately 1 mg⋅ml−1. N.I. indicates that no inhibition was seen in the negative control reference standard.
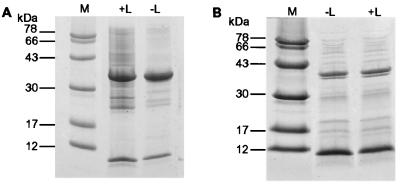
Refolded CD1a and -b supernatants were analyzed by SDS/PAGE (Fig. 3), which showed bands of the expected molecular masses. CD1a supernatant was loaded onto an NA1/34 immuno-affinity column. The high-molecular-mass soluble aggregate and β2m did not bind. Bound CD1a was eluted with 0.1 M glycine/HCl (pH 3.0) into 1.0 M Tris⋅HCl (pH 7.4) and found to react with all four mAbs in the inhibition immunoassay (data not shown). The material was fractionated by SDS/PAGE and the bands were electroblotted. The identity of the recovered products (bands of molecular mass ≈34 and 12 kDa corresponding to the CD1a and -b heavy chain and β2m, respectively) was confirmed by NH2-terminal amino acid sequence analysis (data not shown). Amino acid analysis of the bands from CD1a and -b was consistent with a 1:1 (heavy/light chain) stoichiometry (data not shown).
Figure 3.
SDS/PAGE analysis of CD1a and -b refolding mixtures. (A) 15% SDS/PAGE analysis of CD1a refolding mixtures carried out with (+L) and without (−L) synthetic ligand, demonstrating bands of the expected molecular massas for the CD1a heavy and β2m light chain. Molecular mass marker standards are as indicated. (B) As above, but for CD1b.
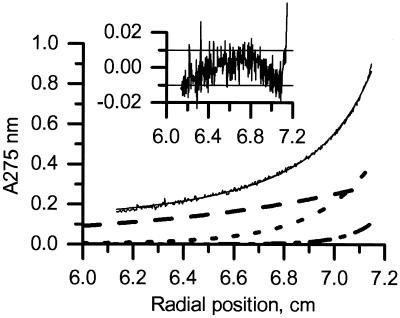
Sedimentation equilibrium runs of the fraction corresponding to peak 3 from the preparative gel filtration (Fig. 2A) demonstrated that, at protein concentrations of 20 and 70 mM, the refolded β2m light chain was monomeric and did not homooligomerize. Analytical ultracentrifugation of fractions corresponding to peak 2a and 2b from the scale-up was performed at concentrations of 20 and 70 μM (Fig. 4). Plots of protein concentration (A275 nm) against radial position (Fig. 4) were consistent with a mixture of β2m light-chain monomers and CD1a complexes that were mostly in a monomeric configuration but in equilibrium with a small quantity of oligomers. After 7 days of ultracentrifugation, the majority of the protein in all samples tested (+L and −L) remained soluble and active when retested in the inhibition assay (data not shown), indicating that the conformation of the material remained intact. The CD spectra of these samples were indistinguishable from those obtained before the run (data not shown), confirming that the secondary structural elements were intact.
Figure 4.
Equilibrium sedimentation of refolded CD1a. Experiments were performed at 10,000 rpm, 4°C, by using 50 mM Tris/60 mM GuHCl/2% glycerol/1 mM EDTA. Data are for the CD1a −L fraction. Data for the +L complex were very similar. Data were fitted to a model-free equation that assumes that the CD1a complex can oligomerize, and that there is an excess of β2m. Heavy chain alone was not seen, indicating that the CD1a/β2m association is very tight, or/and that nonassociated heavy chain is insoluble. Logarithms of oligomerization constants (for dimer formation) were estimated to be 4.63 ± 0.1 for empty (−L) and 5.07 ± 0.1 for ligand-loaded (+L) complexes (Kd = 23 and 8.5 μM, respectively). Black line, experimental data and theoretical fit; dashed line, β2m alone; dotted line, CD1a; dashed-dotted line, CD1a dimer. The baseline contribution (0.064) is not shown. (Inset) Residues (Aexp-Afit) of the fit (A275 units). The horizontal lines at ± 0.01 represent the typical limits of stability in the instrument baseline.
Optimizing Oxidative Refolding Chromatography for CD1.
To test whether the folding benefits from “priming” with refolded β2m, denatured/reduced CD1a heavy chain was added to a solution containing refolded β2m and ternary resin. A dense precipitate formed immediately, and we conclude that it is essential that the CD1 heavy chain and β2m light chain are mixed together in the denatured/reduced state before refolding. The duration of the denaturation step was critically important also. Yields of active refolded protein fell dramatically if denatured inclusion bodies were left at 4°C for more than 24 h (data not shown). It is essential, consequently, to work with freshly denatured material. A range of storage buffers were tested and the refolded molecules found to be stable in an aqueous environment containing 0.06 M GuHCl/1.0% glycerol. The efficiency of refolding was pH-dependent and favored above neutral values. Under more acidic conditions, the heterodimer dissociated, precipitated, and lost activity (data not shown). A pH of 8.0 was optimal, and all subsequent experiments were performed at this value. Time-course experiments indicated that refolding efficiency was limited by the time taken to implement the procedure (approximately 6 min) and not by a kinetic barrier. It should be noted, however, that our experiments were configured such that the molar ratios of immobilized chaperones and foldases were in significant (approximately 100-fold) excess relative to their substrates.
When the residual 60 mM GuHCl was removed from the refolding mixture after dilution, the antigenic activity of monomeric and oligomeric CD1a (−L) was lost. Low molarity GuHCl seems to help maintain the conformational integrity of the “empty” molecules. All results obtained at the analytical level were reproducible at the preparative level (Fig. 2B, data not shown for CD1b). To characterize the stability of the monomeric CD1a complex further, peak 2b was concentrated and loaded onto an analytical HPLC column (Fig. 2C). Although the majority of the species present corresponded to the 44-kDa molecular mass of the monomeric complex, the peaks were not entirely symmetrical, confirming the ultracentrifugation results and indicating that in addition to free β2m, the refolded complex is in dynamic equilibrium with minor species that most likely represent a range of oligomeric CD1a complexes and relatively low-molecular-mass aggregates. The position of the equilibrium depended on protein concentration and could be shifted in a direction favoring production of monomeric CD1a by limiting starting concentrations. Refolding yields under all conditions were reduced significantly if the CD1 and β2m were synthesized with a nonnative N terminus (data not shown).
Circular Dichroism Spectroscopy of Refolded CD1a.
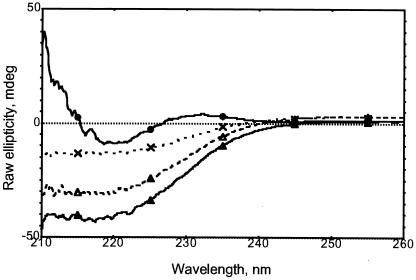
CD spectra of the refolded CD1a molecules were measured under native and denaturing conditions. Whereas the spectra of the denatured products were consistent with those expected for an unfolded state (data not shown), those from the refolded CD1a monomer (+L and −L) indicated the presence of secondary structural elements corresponding to α-helices and β-sheets (Fig. 5). Spectra obtained from the −L and + L conditions were indistinguishable. The presence of glycerol and GuHCl in the refolding buffer (needed to maintain the activity of the empty complex) prevented quantitative analysis of the β-sheets.
Figure 5.
C/D spectra (260–210 nm) of refolded/purified CD1a. Spectra were recorded in 25 mM potassium phosphate buffer (pH 8.0) containing 60 mM GuHCl and 2% glycerol. Black circles, β2m alone (0.15 mg⋅ml−1); crosses, CD1a (−L) (0.15 mg⋅ml−1); white triangles, CD1a (+L) (0.30 mg⋅ml−1); black triangles, CD1a dimer (−L) (0.35 mg⋅ml−1).
Discussion
The results demonstrate that the efficient in vitro refolding of a recombinant and fully denatured antigen-presenting molecule is possible in the absence of ligand and suggest that the ligand-binding pathway is not the only route by which stable molecules can be assembled in vivo. When denatured/reduced MHC class I and β2m light chains synthesized in E. coli are refolded in the absence of synthetic ligand, stable complexes are not produced (25, 26). Furthermore, evidence that the MHC and CD1 molecules expressed in Drosophila melanogaster and other insect expression systems are empty remains elusive. In our case, although we do not suggest that empty implies that the binding site is free of solvent molecules, the procedure used supports the view that the folded material did not harbor ligand or ligand-like components. Indeed, as the heavy and light chains were reduced fully and denatured, it is unlikely that components of the bacterial lysate remained bound to the starting material. The consistent presence of oligomeric forms observed in the chromatographic patterns and deduced from equilibrium centrifugation results raises the question of their functional significance in solution, inside the cell, or on the cell membrane.
The misfolding and aggregation of proteins expressed in the cytoplasm of E. coli to form inclusion bodies is a major problem in biotechnology and biomedical research (36). The Ig supergene family constitutes a significant percentage of the proteins encoded within the human genome (37). The in vitro synthesis of these functionally diverse proteins is limited by the fact that their expression in prokaryotic systems like E. coli often results in the production of insoluble inclusion bodies that cannot be refolded efficiently or at all. The alternative strategy, which involves expression in eukaryotic hosts, produces significantly lower yields of protein and is considerably more time consuming and expensive. As the CD1 family uses the canonical Ig fold, it may serve as a paradigm for the efficient refolding of other members of this superfamily. Indeed if, as seems likely, all Ig-like β-sandwich proteins share a common folding pathway (38), it is reasonable to suggest that it will be possible to refold other Ig domain-containing proteins by using the method we have described. These proteins include molecules of potential therapeutic, diagnostic, industrial, and biotechnological importance such as CD8, CD4, MHC proteins, and immunoglobulins. The method may, in addition, facilitate the in vitro production of Ig-fold-containing oligomeric complexes, both natural and artificial.
The folding is fast and most likely limited only by the time taken to perform the oxidative refolding chromatography procedure. The ternary resin can be denatured and renatured repeatedly with little or no loss of the immobilized proteins. The method therefore is suitable equally when initial refolding yields are low, because under appropriate conditions, the insoluble starting material could be denatured iteratively and renatured over a short timescale. The method, furthermore, is amenable to scale-up and automation, and if implemented in a bioreactor-type device, could allow large-scale production of recombinant multisubunit proteins.
Evidence suggests that calnexin and calreticulin assist the in vivo folding of MHC class I and CD1 molecules (19, 20). The mechanism by which the GroEL minichaperone assists CD1 folding is most likely different from that of calnexin and calreticulin, both of which recognize glycosylated polypeptides, as compared with GroEL, which interacts with substrate in a sugar-independent manner. Glycosylation may help stabilize the native molecule, but our results suggest that it is not essential for the correct folding of CD1. The fact that it is possible to assemble human CD1 molecules in vitro in the absence of ligand suggests that CD1 and possibly other antigen-presenting molecules may have access to ligand-independent folding pathways. A fraction of the heterodimers expressed in vivo may assemble empty, perhaps associating with ligand derived from exogenous sources or having a function dependent on their ligand-binding capacity. They might, for example, have a role in development, perhaps functioning as T cell selection elements in the cortical thymus. Further investigation will be necessary to establish whether empty CD1 molecules represent a folding intermediate whose α-helices are in a partially folded “molten globule” state (27, 28), or whether the CD1 binding site enables ligand-free complexes to fold completely and attain greater stability than their empty MHC class I counterparts. Sequence differences between CD1 isoforms are likely to produce differences in the relative stability of their respective ligand-free forms.
Our results may have therapeutic implications for conditions that affect protein folding and result in the formation of pathogenic intracellular aggregates such as Huntington's disease, Alzheimer's disease, and Parkinson's disease (39). Indeed, we recently demonstrated that bacterial GroEL fragments reduce aggregate formation and cell death in a mammalian cell model of Huntington's disease (40). We have shown also that synthetic human CD1d can be refolded with α-galactosylceramide ligand by using the oxidative refolding chromatography procedure outlined above. We have used this material to generate tetrameric CD1d molecules that stain T cell clones (41).
Acknowledgments
M.M.A. is a Biotechnology and Biological Sciences Research Council David Phillips Research Fellow, a Research Fellow at King's College, Cambridge, U.K., and formerly an European Molecular Biology Organization fellow. A.W. was a Wellcome Trust Clinical Training Fellow and a Charles and Katharine Darwin Research Fellow at Darwin College, Cambridge, U.K. A.W. and C.M. acknowledge the support of a grant from the National Foundation for Cancer Research. L.B.R. is a Consejo Nacional de Ciencia y Technología (Mexico) MSc Fellow and a Fundación Universidad Natl Autónoma de Mexico Fellow. D.B.V. is a recipient of a Human Frontier in Science Fellowship. G.D.L. is supported by the Swiss National Foundation, Human Science Frontier Program, and Swiss Multiple Sclerosis Foundation.
Abbreviations
- β2m
β2-microglobulin
- PPI
peptidyl-prolyl cis-trans isomerase
- CD
circular dichroism
Footnotes
See commentary on page 2950.
References
- 1.Calabi, F. and Milstein, C. (2001) Semin. Immunol., in press. [DOI] [PubMed]
- 2.Porcelli S A, Modlin R L. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 3.Calabi F, Yung Yu C, Bilsland C A G, Milstein C. In: CD1: From Structure to Function. Immunogenetics of the Major Histocompatibility Complex. Srivastava R, Ram B, Tyle P, editors. NY: VCH; 1991. [Google Scholar]
- 4.Burdin N, Kronenberg M. Curr Opin Immunol. 1999;3:326–331. doi: 10.1016/s0952-7915(99)80052-1. [DOI] [PubMed] [Google Scholar]
- 5.Balk S P, Burke S, Polischuk J E, Frantz M E, Yang L, Porcelli S, Colgan S P, Blumberg R S. Science. 1994;265:259–262. doi: 10.1126/science.7517575. [DOI] [PubMed] [Google Scholar]
- 6.Woolfson A, Milstein C. Proc Natl Acad Sci USA. 1994;91:6683–6687. doi: 10.1073/pnas.91.14.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.York I A, Rock K L. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 8.Sieling P A, Chatterjee D, Porcelli S A, Prigozy T I, Mazzaccaro R J, Soriano T, Bloom B R, Brenner M B, Kronenberg M, Brennan P J, Modlin R L. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 9.Joyce S, Woods A S, Yewdell J W, Bennink J R, Dharshan De Silva A, Boesteanu A, Balk S P, Cotter R J, Brutkiewicz R R. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 10.Moody D B, Ulrichs T, Muhlecker W, Young D C, Gurcha S S, Grant E, Rosat J P, Brenner M B, Costello C E, Besra G S, Porcelli S A. Nature (London) 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi T, Nieda M, Koezuka Y, Nicol A, Porcelli S A, Ishikawa Y, Tadokoro K, Hirai H, Juji T. J Immunol. 2000;164:4458–4464. doi: 10.4049/jimmunol.164.9.4458. [DOI] [PubMed] [Google Scholar]
- 12.Rosat J P, Grant E P, Beckman E M, Dascher C C, Sieling P A, Frederique D, Modlin R L, Porcelli S A, Furlong S T, Brenner M B. J Immunol. 1999;162:366–371. [PubMed] [Google Scholar]
- 13.Shamshiev A, Donda A, Prigozy T I, Mori L, Chigorno V, Benedict C A, Kappos L, Sonnino S, Kronenberg M, De Libero G. Immunity. 2000;13:255–264. doi: 10.1016/s1074-7613(00)00025-x. [DOI] [PubMed] [Google Scholar]
- 14.Ulanova M, Tarkowski A, Hahn-Zoric M, Hanson L A. Scand J Immunol. 1999;50:387–393. doi: 10.1046/j.1365-3083.1999.00607.x. [DOI] [PubMed] [Google Scholar]
- 15.Castano A R, Tangri S, Miller J E W, Holcombe H R, Jackson M R, Huse W D, Kronenberg M, Peterson P A. Science. 1995;269:223–226. doi: 10.1126/science.7542403. [DOI] [PubMed] [Google Scholar]
- 16.Zeng Z-H, Castano A R, Segelke B W, Stura E A, Peterson P A, Wilson I A. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 17.Jackson M R, Song E S, Yang Y, Peterson P A. Proc Natl Acad Sci USA. 1992;89:12117–12121. doi: 10.1073/pnas.89.24.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 19.Williams D B, Watts T H. Curr Opin Immunol. 1995;7:77–84. doi: 10.1016/0952-7915(95)80032-8. [DOI] [PubMed] [Google Scholar]
- 20.Huttinger R, Staffler G, Majdic O, Stockinger H. Int Immunol. 1999;11:1615–1623. doi: 10.1093/intimm/11.10.1615. [DOI] [PubMed] [Google Scholar]
- 21.Brutkiewicz R R, Bennink J R, Yewdell J W, Bendelac A. J Exp Med. 1995;182:1913–1919. doi: 10.1084/jem.182.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugita M, Grant E P, van Donselaar E, Hsu V W, Rogers R A, Peters P J, Brenner M B. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 23.Jackman R M, Moody D B, Porcelli S A. Crit Rev Immunol. 1999;19:49–63. [PubMed] [Google Scholar]
- 24.Tangri S, Brossay L, Burdin N, Lee D J, Corr M, Kronenberg M. Proc Natl Acad Sci USA. 1998;95:14314–14319. doi: 10.1073/pnas.95.24.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garboczi D N, Hung D T, Wiley D C. Proc Natl Acad Sci USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid S W, Smith K J, Jakobsen B K, O'Callaghan C A, Reyburn H, Harlos K, Stuart D I, McMichael A J, Bell J I, Yvonne Jones E. FEBS Lett. 1996;383:119–123. doi: 10.1016/0014-5793(96)00226-8. [DOI] [PubMed] [Google Scholar]
- 27.Bouvier M, Wiley D C. Nat Struct Biol. 1998;5:377–383. doi: 10.1038/nsb0598-377. [DOI] [PubMed] [Google Scholar]
- 28.Hansen T. Nat Struct Biol. 1998;5:340–341. doi: 10.1038/nsb0598-340. [DOI] [PubMed] [Google Scholar]
- 29.Altamirano M M, Garcia C, Possani L D, Fersht A R. Nat Biotechnol. 1999;17:187–191. doi: 10.1038/6192. [DOI] [PubMed] [Google Scholar]
- 30.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. J Exp Med. 2000;191:1895–1904. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shamshiev A, Donda A, Carena I, Mori L, Ludwig Kappos L, De Libero G. Eur J Immunol. 1999;29:1667–1675. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Burrone O R, Calabi F, Kefford R F, Milstein C. EMBO J. 1983;2:1591–1595. doi: 10.1002/j.1460-2075.1983.tb01629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Holde K E, Baldwin R E. Annu Rev Biochem. 1958;65:441–473. [Google Scholar]
- 34.Laue T M, Shah B D, Rigewat T M, Pelletier S L. In: Analytical Utracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A O, Hatan J C, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 35.Poget S F, Legge D B, Proctor M R, Butler P J G, Williams R L. J Mol Biol. 1999;290:867–879. doi: 10.1006/jmbi.1999.2910. [DOI] [PubMed] [Google Scholar]
- 36.Speed M A, Wang D I, King J. Nat Biotechnol. 1996;14:1283–1287. doi: 10.1038/nbt1096-1283. [DOI] [PubMed] [Google Scholar]
- 37.Halaby D M, Mornon J P E. J. Mol. Evol. 1997. 389–400. [DOI] [PubMed] [Google Scholar]
- 38.Clarke J, Cota E, Fowler S B, Hamill S J. Struct Fold Des. 1999;7:1145–1153. doi: 10.1016/s0969-2126(99)80181-6. [DOI] [PubMed] [Google Scholar]
- 39.Dobson C M. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 40.Carmichael J, Chatellier J, Woolfson A, Milstein C, Fersht A R, Rubinsztein D C. Proc Natl Acad Sci USA. 2000;97:9701–9705. doi: 10.1073/pnas.170280697. . (First Published August 1, 2000; 10.1073/pnas.170280697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karadimitris A, Gadola S, Altamirano M, Brown D, Woolfson A, Klenerman P, Chen J-L, Koezuka Y, Roberts I A G, Price D A, et al. Proc Natl Acad Sci USA. 2001;98:3294–3298. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]