Abstract
The Plasmodium vivax Merozoite Surface Protein-3α (PvMSP-3α) is considered as a potential vaccine candidates. However, the detailed investigations of the type of immune responses induced in naturally exposed populations are necessary. Therefore, we aim to characterize the naturally induced antibody to PvMSP-3α in 282 individuals with different levels of exposure to malaria infections residents in Brazilian Amazon. PvMSP3 specific antibodies (IgA, IgG and IgG subclass) to five recombinant proteins and the epitope mapping by Spot-synthesis technique to full-protein sequence of amino acids (15aa sequence with overlapping sequence of 9aa) were performed. Our results indicates that PvMSP3 is highly immunogenic in naturally exposed populations, where 78% of studied individuals present IgG immune response against the full-length recombinant protein (PVMSP3-FL) and IgG subclass profile was similar to all five recombinant proteins studied with a high predominance of IgG1 and IgG3. We also observe that IgG and subclass levels against PvMSP3 are associated with malaria exposure. The PvMSP3 epitope mapping by spot-synthesis shows a natural recognition of at least 15 antigenic determinants, located mainly in the two blocks of repeats, confirming the high immunogenicity of this region. In conclusion, PvMSP-3α is immunogenic in naturally exposed individuals to malaria infections and that antibodies to PvMSP3 are induced to several B cell epitopes. The presence of PvMSP3 cytophilic antibodies (IgG1 and IgG3), suggest that this mechanisms could also occur in P. vivax.
Keywords: Plasmodium vivax, Merozoite Surface Protein, B cell epitope, vaccine, malaria, immunity
1. Introduction
Plasmodium vivax is a leading cause of human malaria and, together with Plasmodium falciparum, accounts for the majority of malaria cases worldwide. Although P.falciparum is dominant in most of Sub-Saharan Africa, P. vivax causes approximately 50% of all malaria cases in endemic regions outside of Africa, with 2.5 billion inhabitants of the Middle East, Asia, Eastern Africa, Central and South America, and Oceania exposed to P. vivax, resulting in an estimated 71–391 million cases of vivax malaria each year [1–3]. Critically, P. vivax causes significant economic and social damage [4] and evidence of severe illness and death due to P. vivax is being reported with increasing frequency [4–9]. While considerably greater investments have been made over the last 30 years to research and control P.falciparum, there have been recent attempts to call attention to the need for increased resources for P. vivax vaccine and drug research and development [10]. Technological advances enabling the sequencing and analysis of the P. vivax genome [11–12] and the call for worldwide malaria eradication [13], have together placed new emphasis on the importance of addressing P. vivax as a major public health problem.
Multiple antigens from the asexual P. vivax parasites have been identified and immunologically characterized and a number of merozoite surface or apical organellar localized proteins have been receiving the most attention. These include P. vivax Merozoite Surface Protein-1 (PvMSP-1) [14], the PvMSP-3 family[15], PvMSP-9 [16], Reticulocyte Binding Protein-1 (PvRBP-1) [17], Apical Membrane Antigen-1 (PvAMA-1) [18] and Duffy Binding Protein (PvDBP) [19]. Among the merozite proteins, those with known essential functions that can be disrupted by antibodies, represent the most promising candidates for vaccine development. PvMSP-3α is a merozoite surface protein expressed during schizogony and it appears to become intimately associated with the surface of the merozoite [15, 20]. Moreover, PvMSP-3α is a member of a multi-gene family [20], which includes 11 members [12]). The initially discovered family members, PvMSP-3α, PvMSP-3β and PvMSP-3γ share 35–38% identity and 48 53% similarity in pair-wise comparisons [15, 20–22]. Structurally, these proteins lack a transmembrane domain or a GPI-lipid modification to anchor them in the outer membrane of the merozoite. The bulk of these proteins is an alanine-rich central domain containing a series of heptad repeats predicted to form a coiled-coil tertiary peptide structure, which may secure them on the merozoite surface through interaction with other surface proteins [15, 21]. Due to the remarkable diversity, particularly noted in the central domain [22], the PvMSP-3α gene sequence has become a highly regarded polymorphic marker for population based studies [23-25]; the acidic C-terminal domain and a smaller hydrophilic N-terminus are relatively conserved, while the central domain containing two annotated blocks of coiled-coil heptad repeats (Block I and Block II) is highly polymorphic and in some isolates of P. vivax is partially deleted [22].
PvMSP-3α has homologs in the simian malaria P. knowlesi [26–28], and in P.falciparum. The initially discovered P.falciparum MSP-3 contains a small series of alanine-based heptad repeats [29–30]. PfMSP-3 has been of considerable interest as a vaccine candidate, mainly because anti-PfMSP-3 antibodies significantly decrease parasitemia through an antibody-dependent cellular inhibition mechanism [29] and partially protected New World monkeys against lethal P.falciparum infectionin a pre-clinical vaccine trial [31]. PfMSP-3 long synthetic peptides have also been shown to be safe and immunogenic in a phase I clinical vaccine trial [32–33]. The predicted structural importance of PvMSP-3α and other PvMSP-3 family members at the surface of merozoites, the high relative conservation of the C-terminal regions, and the relationship of PvMSP-3 to a similar merozoite protein which has been highly regarded as a vaccine candidate in P.falciparum are reasons to investigate these P. vivax antigens as natural immunogens and possible vaccine candidates. The present study evaluates the naturally acquired immune response to PvMSP-3α in individuals exposed to malaria infections in Rondonia State, in the Amazon region of Brazil, and provides important information regarding PvMSP-3α immune responses generated in natural infections in support ofthis antigen as a P. vivax vaccine candidate.
2. Material and Methods
2.1 Study area and volunteers
A cross-sectional cohort study was conducted involving 282 individuals from communities in the malaria endemic region of Rondonia State, in the western Amazon region of Brazil, where in the last five years P. vivax malaria accounted for more than 70% of all malaria cases. The majority of the studied population consists of rain forest natives or transmigrants from several non-endemic areas of Brazil that have lived in the region for 10 years or more. Samples and survey data were collected during the dry months of June-August, coinciding with the period of increased malaria transmission in Rondonia State. In addition, we also have included as control subjects 24 naive individuals living in non endemic regions of Brazil and the USA (Rio de Janeiro and Atlanta, respectively). Written informed consent was obtained from all adult donors or from parents of donors in the case of minors. The study was reviewed and approved by the Fundação Oswaldo Cruz Ethical Committee and the National Ethical Committee of Brazil.
2.2 Epidemiological survey
In order to evaluate epidemiological factors that may influence the immune response against PvMSP-3α, all donors were interviewed upon informed consent. The survey included questions related to demographics, time of residence in the endemic area, personal and family histories of malaria, use of malaria prophylaxis, presence of malaria symptoms, and personal knowledge of malaria. Survey data was entered into a database created with Epi Info 2002 (Centers for Disease Control and Prevention, Atlanta, GA).
2.3 Malaria diagnosis and blood sampling
Venous peripheral blood was drawn into heparinized tubes and plasma collected after centrifugation (350 x g, 10 minutes). Plasma samples were stored at −20 ºC and transported to our laboratory. Thin and thick blood smears of all donors were examined for malaria parasites. Parasitological evaluations were done by examination of 200 fields at 1000X magnification under oil-immersion; all slides were examined by a research expert in malaria diagnosis. Donors positive for P. vivax and/or P.falciparum at the time of blood collection were subsequently treated using the chemotherapeutic regimen recommended by the Brazilian Ministry of Health.
2.4 Cloning, expression and purification of recombinant PvMSP3 α antigens
Five subfragments of the pvmsp3α gene, including sequences encoding the near full length protein (nucleotides 73-2520), the N-terminal (Nt) region (nucleotides 73-309), Block I (nucleotides 316-1242), Block II (nucleotides 1246-2058), and the C-terminal (Ct) region (nucleotides 2059-2523), were amplified from Plasmodium vivax (Belem strain) genomic DNA with the Expand High FidelityPLUS PCR System (Roche). The five PCR products were double digested with Nco I and Xho I, and ligated into the same restriction sites from the pET24d(+) kanamycin resistant expression vector containing a C-terminal 6x His Tag comprised of the amino acids ELHHHHH for purification purposes. The recombinant expression vectors were confirmed by Big Dye terminator v3.1 sequencing. All five recombinant proteins were successfully expressed at high levels in a soluble state using BL-21 (DE3) or BL21-AITM cells after 3 hour induction at 37º with 1mM IPTG. The soluble recombinant proteins were initially purified using HisTrapTM HP columns under native conditions, and further purified by gel-filtration using either a Sephacryl S-200 HR chromatography column or HiLoad Superdex 75 pg column. The final proteins were evaluated on SDS-PAGE gels and via western imunoblotting using standard conditions.
2.5 Antibody Assays
Plasma samples from study participants were screened for the presence of naturally acquired antibodies against the five recombinant proteins PvMSP3-FL, PvMSP3-BLI, PvMSP3-BLII, PvMSP3-CT and PvMSP3-NT by ELISA. Briefly, maxisorp 96-well plates (Nunc, Rochester, NY) were coated with 2 μg per ml of each recombinant protein After overnight incubation at 4°C the plates were washed with PBS containing 0.05% Tween 20 (PBS-Tween) and blocked with PBS-Tween containing 5% non fat dry milk (PBS-Tween-M) for 2 h at 37°C. Individual plasma samples diluted 1:100 PBS-Tween-M were added in duplicate wells and the plates incubated at room temperature for 1 h. After four washes with PBS-Tween, bound antibodies were detected with peroxidase-conjugated goat anti-human IgG (Sigma, St Louis) followed by o-Phenylenediamine and hydrogen peroxide. The absorbance was read at 492 nm using an ELISA reader (Spectramax 250, Molecular Devices, Sunnyvale, CA). The results for total IgG were expressed as Reactivity Indexes (RI), which were calculated by dividing the mean optical density of tested samples by the mean optical density plus 3 standard deviations of 24 non-exposed control individuals living in non-endemic areas of malaria. Subjects were scored as positive for serum IgG to antigen if the RI was higher than 1.
For determination of IgA and IgG subclasses the following peroxidase conjugated monoclonal mouse anti-human antibodies were used: clone HP-6001 for IgG1, HP-6002 for IgG2, HP-6050 for IgG3, HP-6023 for IgG4 and B1524 for IgA (Sigma St. Louis, MO). This set of antibodies has been used previously to characterize IgG subclasses within a similar cohort of individuals living in Rondonia [34]. Subclass-specific prevalence for each antigen was determined from OD values using 3 standard deviations above the appropriate mean OD of 24 non-exposed controls as the cut-off for positive reactivity. To adjust the affinity differences between the IgG subclasses, standard curves were prepared using human IgG kappa myeloma proteins (Sigma, St. Louis, MO) from each of the four IgG subclasses and human IgA purified from human colostrum (Sigma, St. Louis, MO). In addition, standard curves enable conversion of OD values to concentration (μg/ml) for the comparison of different subclasses. Purified human antibodies from each of the four subclasses and IgA (Sigma St Louis, MO) were coated overnight at 4°C in PBS at 100 μl per well onto 96-well plates in 1:2 serial dilutions from 24 μg/ml to 2−11 μg/ml. After washing four times, the plates were incubated with the appropriate anti-human IgG subclass-specific mAb, washed four times, incubated with peroxidase-labeled goat anti-mouse antibody, washed a final four times, developed with o-phenylenediamine and hydrogen peroxide, and measured as described above for subclass-specific ELISA. Subclass-specific OD values were converted to concentration values (μg/ml) using sigmoidal curve-fit equations derived from subclass-specific standard curves.
2.6 Spot-synthesis
B-cell linear epitope mapping was carried out by the synthesis of an overlapping 15-mer peptide library covering the entire amino acid sequence of P. vivax PvMSP-3α. The peptides were simultaneously synthesized by the SPOT-method [35] on cellulose membranes with an Ala-Ala linker, for preparation of immobilized peptides. The assembly of the peptides was carried out utilizing 9-fluorenylmethoxycarbonyl (Fmoc) chemistry as previously described [35–36]. The prepared membrane consists of overlapping 15 amino acid peptide residues with an offset of four. For epitope analysis, a pool of 10 serum samples with high reactivity indices to all 5 recombinant proteins in ELISA test at 1:100 dilutions was used. Bound antibodies were detected with alkaline phosphatase (AP)-conjugated secondary antibody (anti-human IgG) followed by a color reaction with CDP-Star Chemiluminescent Substrate (Sigma; St Louis).
2.7 Statistical analysis
Statistical analyses were done using Epi Info 2002 (CDC, Atlanta, GA), Prism 4.0 and Instat (GraphPad Software, San Diego, CA) applying the statistical test necessary. Differences in medians for the study population data were tested by the non-parametric Mann-Whitney test where appropriate. Student’s t test was used to compare the means of normally distributed data, or normalized transformations were performed on raw data before testing by one-way analysis of variance where appropriate. Differences in proportions were evaluated by the chi-square (χ2) test. Relationships between years of residence in the endemic area and the number of past malaria infections or months since last known malaria episode were assessed with Spearman’s rank correlation. Multivariate logistic regression was used to assess the relationship between antigen-specific total IgG responses and the independent variables of gender, age, years of residence in the endemic area, number of past malaria episodes, and months since last known malaria infection.
3. Results
3.1 Characteristics of studied population
Among our sample set, the studied population did not differ significantly in gender ratio (χ2=1.4201; p=0.2222). Our epidemiological survey, summarized in table 1, shows that all individuals studied are exposed to malaria infections throughout the year. The time of residence in the malaria endemic area, number of past malaria episodes and past months since the last infection vary greatly in studied individuals. However, a significant proportion of studied individuals (52.9%) reported a prior experience with both P. vivax and P.falciparum malaria when compared with individuals that had in the past, a single species infection (32.1%) or individuals that could not recall infections in the past and mentioned that they never had malaria even though they were born in the endemic area (p<0.0001). Among donors with a previous malaria infection(s), years of residence in the endemic area correlated positively with the past months since last malaria episode (Spearman r = 0.2344, p<0.0001, n=249). Therefore, we used years of residence in the endemic area and number of past malaria episodes reported by donors as indices of malaria exposure, and past months since last malaria episode and infections during the year of blood collection as a crude approximation of clinical protection. At the time of blood collection 34 (12%) individuals were infected, 25 with P. vivax and 9 with P.falciparum (thus,74% P. vivax and 26% P.falciparum), consistent with the current local case distribution data for these two species reported by the Ministry of Health.
Table 1.
Summary of the epidemiological characteristics of studied individuals enrolled in this work
| Epidemiological characteristics | |
|---|---|
| Gender a | |
| Male (n) | 158 |
| Female (n) | 124 |
| TOTAL (n) | 282 |
| Age (Mean ± SD) | 36.1 + 16.9 |
| Time of residence in malaria endemic area (Mean ± SD) | 28.5 + 17.0 |
| Number of past malaria infections (Mean ± SD) | 7.0 + 9.1 |
| Number of malaria infections in the last 6 months (Mean ± SD) | 0.5 + 1.1 |
| Past months since the last malaria infection (Mean ± SD) | 41.0 + 50.1 |
| Hospitalization in malaria past infections b (n/%) | 56 / 19.8% |
| Use of prophylactic measures e | 131 / 46.5% |
| Previous malaria species contracted c | |
| Negative (n) | 33 / 11.8% |
| P. vivax (n) | 56 / 20% |
| P.falciparum (n) | 34 / 12.1% |
| Both species (n) | 148 / 52.9% d |
Differences in gender proportions were not statistically significant. Χ2=1,4201; p=0,2222
We could not determine if clinical criteria were compatible with severe malaria,
11 (3,2%) individuals did not remember the previous malaria parasite species contracted
Bold typeface indicates that the prevalence was significantly higher when compared with all other observations (P<0.0001)
Were considered as prophylactic measure; i.e., the use of residual insecticide, bednets or insect repellent
3.2 Expression and purification of recombinant PvMSP3α antigens
Five recombinant proteins representing segments of PvMSP3α (Figure 1A) were expressed in the pET24d (+) vector and purified (see materials and methods), and run in a standard SDS-PAGE gel under reducing and non-reducing conditions (Figure 1B). Each recombinant protein was soluble, and showed a higher molecular weight than projected from the calculated molecular weight, as observed before for this family of proteins [15, 20]. The near full length protein (FL), N-terminus protein (Nt), Block I (BLI), Block II (BLII), and C-terminus (Ct) proteins (Figure 1A) ran at > 95%, 116%, 28%, 46% and 79% of their expected molecular weights under reduced conditions (Figure 1B). Additionally, the FL and Block I proteins formed higher molecular weight structures through predicted disulfide bond formation under the non-reducing conditions (Figure 1B). The results of western immunoblotting with anti-His antibody and anti-PvMSP3α antibodies indicated that each of these purified recombinant protein bands correspond to the PvMSP3α subfragments (not shown). With the exception of the near full length recombinant PvMSP3α, which always appears as two protein bands on a reduced SDS-PAGE gel, each of the other four recombinant proteins appear as a single protein band as shown in the reduced SDS-PAGE gel. Based on the primary amino acid sequences, the recombinant proteins representing the NT, Block I and the near FL have cysteine residues and may therefore form higher molecular weight structures through disulfide bond. This seems to be the case for each of these proteins, as shown in the non-reduced SDS-PAGE gel (Figure 1B).
Figure 1.
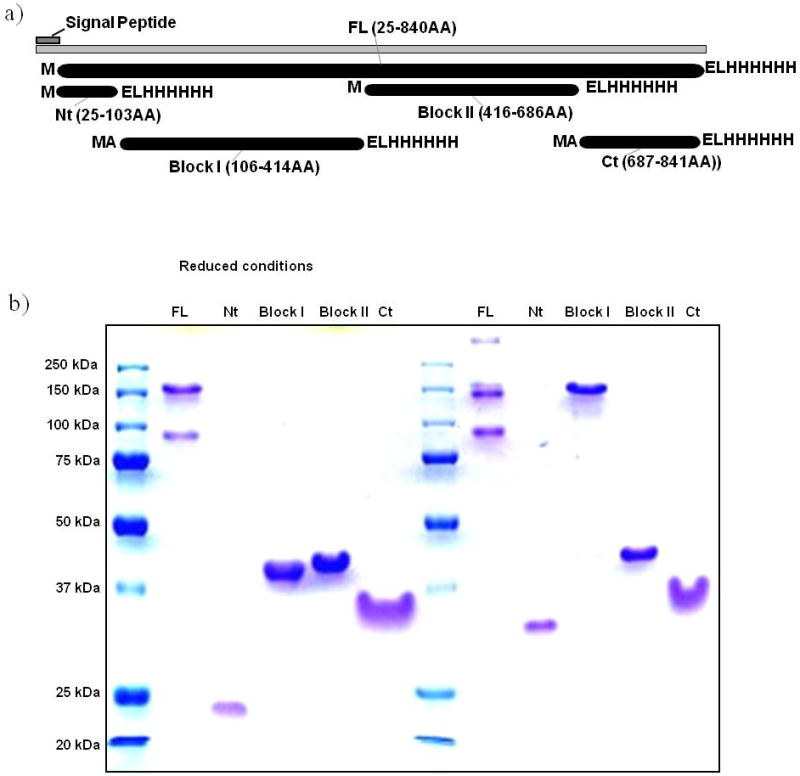
Expression and purification of recombinant PvMSP3α proteins in E. coli. 1A, Schematic depicting the near-full length, N-terminal (Nt), Block I, Block II and C-terminal (Ct) PvMSP3α segments designed for recombinant protein expression. Each recombinant protein was developed with a C-terminal His tag (with the ELHHHH amino acid sequence) for protein purification purposes. The Block I and Ct recombinant proteins have one additional alanine (A) after the start methionine (M). 1B, Purified recombinant proteins were separated by SDS-PAGE under reducing and non-reducing conditions, as indicated. The protein molecular weight standards are Precision Plus Protein All Blue Standards (Bio-Rad).
3.2 Frequency of IgG antibodies to recombinant proteins derived from PvMSP-3α
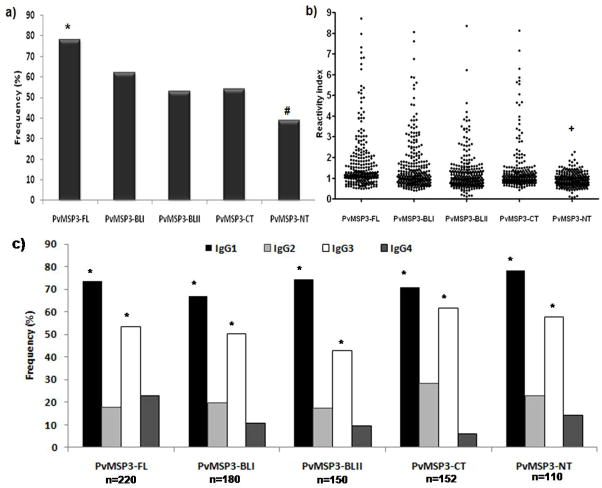
The prevalence of naturally acquired antibodies specific to the five recombinant proteins derived from PvMSP-3 was determined with plasma of all 282 studied individuals (Figure 2a). The results show that IgG antibody reactivity to PvMSP3-FL (full-length protein) was present in 78% of all 282 donors. Moreover, when we evaluated the prevalence of IgG antibodies to different regions of PvMSP-3, we observed that the two blocks of repeats (PvMSP3-BLI, 64% and PvMSP3-BLII; 53%) and the C-terminus (PvMSP3-CT, 54%) were significantly more recognized (p<0.01) than the N-terminal region (39%). As expected, all individuals positive for the PvMSP3-FL recombinant also presented an IgG response to at least one of the 4 recombinant proteins. In this context, among the PvMSP3-FL responders (n=220) 26.8% also presented an IgG response to all four recombinant proteins, 25.4% to 3 of the recombinant proteins (mainly PvMSP3-BLI, PvMSP3-BLII and PvMSP3-CT), 37.7% to 2 of the recombinant proteins, and 10% of individuals to 1 of the recombinant proteins. Only 22% did not present antibodies to any of the recombinant proteins.
Figure 2.
Frequency of IgG responders (a) reactivity indexes and (b) frequency of IgG subclass responders (c) against the five recombinant proteins tested in the studied population. Chi squared test for proportions analyses were performed to determine if a statistical difference existed for each antigen (*). The frequency of IgG responders to PvMSP3-FL was significantly higher when compared with all others recombinants (p<0.05), while the frequencies to PvMSP3-NT were the lowest when compared with all others (#) (p<0.01). The frequency of IgG1 and IgG3 subclasses statistically predominated (*) over IgG2 and IgG4.
3.3 Magnitude of IgG response against the five PvMSP-3α recombinant proteins
To compare the relative magnitude of antibody responses, we determined IgG reactivity indexes (RI) based on negative control cut-off values. As shown in Figure 2b, the RI of individuals against the recombinant proteins varied from 0.21 to 8.91. The average RI of responders against PvMSP3-FL (2.10±1.61), PvMSP3-BLI (2.06±1.45), PvMSP3-BLII (1,81±1.05) and PvMSP3-CT (1.87±1.37) did not differ significantly (p>0.05). However the average of RI against PvMSP3-NT (1.27±0.27) was significantly lower when compared with all of the other recombinants tested (PvMSP3-FL: t=5.365; df=328; p<0.0001; PvMSP3-BLI: t=5.651; df=288; p<0.0001; PvMSP3-BLII: t=5.265; df=258; p<0.0001; PvMSP3-CT: t=4.528; df=260, p<0.0001). It is important to mention that all of the serum samples from the 24 healthy individuals without previous history of exposure to malaria infections were negative for all five recombinant PvMSP-3 proteins.
3.4 IgG subclass distribution of anti-PvMSP-3α antibodies
We assessed the overall subclass distribution of the IgG antibody responses to each antigen using two different comparative analyses. Firstly, we determined subclass-specific prevalence of total IgG positive responders for each antigen using OD cutoffs determined from ODs of non-exposed controls. Secondly, antigen-specific IgG1, IgG2, IgG3, and IgG4 concentrations for total IgG positive responders were determined from OD values using subclass-specific standard curves (Table 2). Therefore, the results of both analyses were comparable with a noteworthy predominance of IgG1 and IgG3 cytophilic antibodies. As shown in figure 2c and table 2, the frequency and concentration respectively of IgG1 and IgG3 antibodies significantly predominated over the IgG2 and IgG4 antibodies against all five tested PvMSP-3 recombinant proteins (p<0.01).
Table 2.
Concentration and 95% confidence intervals of IgG subclass specific response positive individuals and frequency of IgA responders against PvMSP-3α recombinant proteins.
| PvMSP3-FL | PvMSP3-BLI | PvMSP3-BLII | PvMSP3-Ct | PvMSP3-Nt | |
|---|---|---|---|---|---|
| μg/ml (CI 95%) | μg/ml (CI 95%) | μg/ml (CI 95%) | μg/ml (CI 95%) | μg/ml (CI 95%) | |
| IgG subclassa | |||||
| IgG1 | 0.647 – 0.740b | 0.525 – 0.721 | 0.619 – 0.947 | 0.574 – 0.703 | 0.419 – 0.519 |
| IgG2 | 0.128 – 0.250 | 0.168 – 0.191 | 0.201 – 0.299 | 0.241 – 0.299 | 0.107 – 0.193 |
| IgG3 | 0.529 – 0.877 | 0.360 – 0.584 | 0.422 – 0.507 | 0.499 – 0.617 | 0.411 – 0.501 |
| IgG4 | 0.056 – 0.091 | 0.041 – 0.055 | 0.039 – 0.061 | 0.061 – 0.074 | 0.041 – 0.07 |
| IgAc | 0.125 – 0.293 | 0.083 – 0.174 | 0.120 – 0.246 | 0.121 – 0.276 | 0.122 – 0.275 |
| Frequency of IgA responders (n) | 28.4% (80) | 21.3% (60) | 24.5% (69) | 22.7% (64) | 23.0% (65) |
IgG1 and IgG3 predominate over IgG2 and IgG4 in PvMSP3-FL (P<0.001), PvMSP3-BLI (p<0.01), PvMSP3-CT (p<0.001) and PvMSP3-NT (p<0.0001), however in PvMSP-3-BLII the IgG1 predominate over all other subclasses (p<0.05).
Bold typeface indicates the predominant IgG subclass by Mann-Whitney test.
IgA antibody concentrations against all recombinant proteins were significantly lower (p<0.0001) when compared with cytophilic antibodies (IgG1 and IgG3), however they were similar to the non-cytophilic antibodies IgG2 and IgG4 (p>0.05)
3.5 Frequency and concentration of IgA antibodies against recombinant PvMSP- 3α antigens
The frequency of positive individuals with IgA antibodies against the five recombinant proteins were similar, presenting no statistically significant differences (P>0.05 in Chi-squared test). As observed in table 2, the frequencies ranged from 21.3% (PvMSP3-BLI) and 29.7% (PvMSP3-FL), however the comparison of IgA concentration with the IgG subclass shows a different profile, with the concentration of cytophilic antibodies (IgG1 and IgG3) significantly higher than IgA (P<0.001), on the other hand the concentration of non-cytophilic antibodies (IgG2 and IgG4) was similar to IgA concentration.
3.6 PvMSP-3α specific antibody responses and malaria exposure
All humoral immune response data from our 282 studied individuals (frequency, RI and antibody concentration) were matched with years of residence in the endemic area. Antibody RIs increased with years of residence in the endemic area for all recombinant proteins. However, the analysis by Spearman’s rank correlation showed that this association was significant only with IgG reactivity indexes against PvMSP3-FL (r=0.1115; p=0.0462) and PvMSP3-NT (r=0.1147; p=0.0480). Moreover, the concentration of IgG subclass antibodies also correlated with years of residence in endemic area for PvMSP3-FL (IgG1, r=0.2432; p=0.0021; IgG3, r=0.1231; p=0.0325), PvMSP3-BLI (IgG3, r=0.1672; p=0.0045) and PvMSP3-CT (IgG1, r=0.1990; p=0.0013). Interestingly, we also observed an inverse correlation between IgG4 concentration against the PvMSP3-CT recombinant and years of residence in the endemic area (r= −0.1213; p=0.0398).
3.7 PvMSP-3α specific antibody responses and previous malaria infections
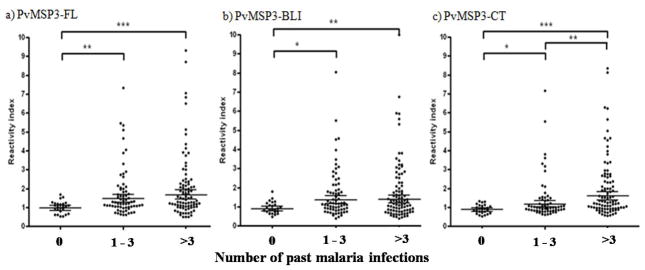
Antibody RIs to PvMSP3-FL (r=0.1680; p=0.0048) and PvMSP3-CT (r=0.1790; p=0.0026) were associated with the number of past malaria episodes. Moreover when we stratified the group of individuals according to the number of previous malaria infections (0, 1–3 or >3) we observed that the total IgG RIs to PvMSP3-FL, PvMSP3-BLI and PvMSP3-CT increased with the number of past malaria episodes and were significantly (p<0.05) higher in individuals who presented 3 or more previous malaria infections (Figure 4). The Spearman’s rank correlation analysis also showed that the IgG3 antibody concentration against PvMSP3-FL was correlated with the number of past malaria infections (r=0.1401; p=0.0415) and interestingly, the IgG2 antibody concentration against PvMSP3-CT was inversely correlated (r=-0.1490; p=0.0313).
Figure 4.
Stratification according to ratio between the concentration of cytophilic (IgG1 + IgG3) and non-cytophilic antibodies (IgG2 + IgG4) against PvMSP3-FL by past months since the last malaria infection.
* p<0.05 and *** p<0.001 by Mann-Whitney rank sum test.
3.8 Humoral response levels and protective immunity
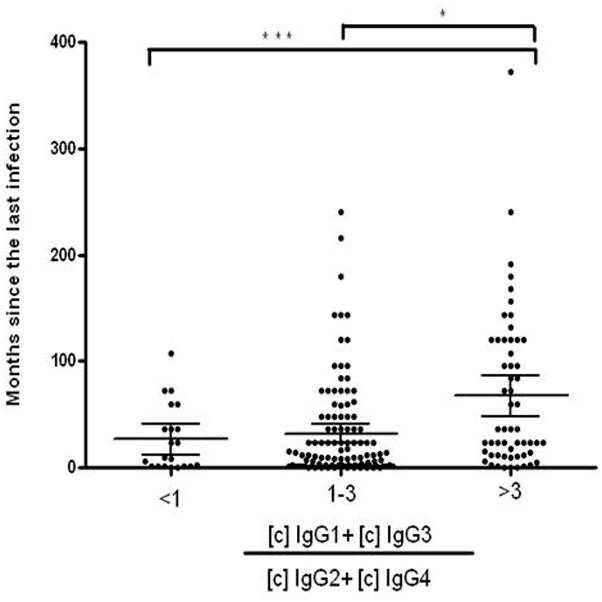
Time (in months) since the last malaria episode and the number of malaria infections in the last six months were used as a crude means to estimate the donor’s level of protection from clinical malaria. To assess if IgG and subclass levels correlated with this estimate, we calculated Spearman’s correlation coefficients between the IgG RI and IgG subclass concentrations and these two parameters. No association was detected with the total IgG RI. However, a positive correlation was observed between the IgG3 concentration for PvMSP3-FL (r=0.1199; p=0.0495) and IgG1 for PvMSP3-CT (r=0.1910; p=0.01333) and the past months since the last known malaria episode. Interestingly, as observed in Figure 5, individuals with a ratio between the concentration of cytophilic and non-cytophilic antibodies of at least 3 had a greater time since the last malaria infection when compared with individuals with a ratio of 1-3 or less than 1.
Figure 5.
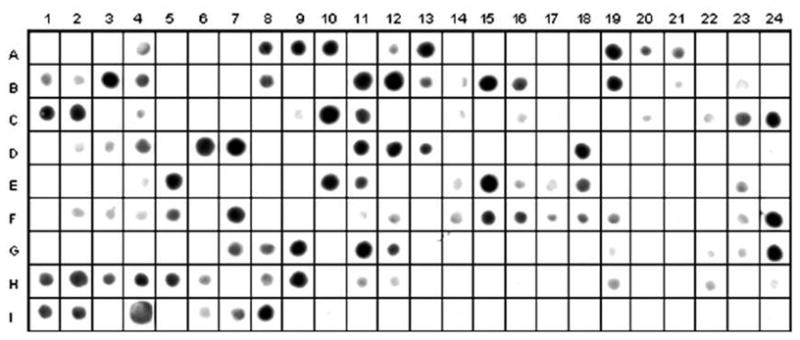
Reactivity of IgG antibodies from a pool of 10 serum samples from individuals with high reactivity indices to all 5 recombinant proteins against peptides covering the full-length sequence of PvMSP-3 (overlapping 15 amino acid peptide residues with an offset of 4).
3.9. Epitope mapping
To identify the epitopes present in the full-length protein, pooled serum from IgG responders to the full length protein were tested against 193 overlapping peptides corresponding to the complete sequence of PvMSP-3α synthesized in a solid phase by the spot method. According to spot-image intensities (Figure 6), 25 antigenic determinants were identified in the complete sequence of PvMSP-3α. The four regions represented in the recombinant proteins present antigenic determinants ranging in size from 10 to 25 amino-acids. The region corresponding to PvMSP3-NT presents three epitopes and PvMSP3-CT presents four. However, the central domain of PvMSP-3, which comprises the two blocks of tandem repeats, presents 11 and 7 antigenic determinants in PvMSP3-BLI and PvMSP3-BLII, respectively.
4. Discussion
The surface of the P. vivax merozoite is covered with a layer of proteins organized into a structurally complex coat, and the antigenic constituents of this surface coat are among the most frequently discussed vaccine targets [37–39], basically because antibody-mediated host immunity can limit the success of merozoite invasion into erythrocyte host cells [29, 40–41]. However, the merozoite presents a wide array of proteins that could be included in a vaccine. Therefore to include a merozoite protein as a vaccine candidate it is necessary to evaluate several features that include conservation of structure and function and immune response. In this context, PvMSP-3α has emerged as a potential candidate mainly because (I) it is expressed during schizogony and appears to become intimately associated with the merozoite surface; (II) it contains conserved regions; (III) it is related to P. knowlesi and P.falciparum MSP-3 proteins [15], which are classically known as vaccine candidates [31, 42–43]; and (IV) it induces antibodies that block merozoite invasion in vitro and induced partial protection in a preliminary trial to examine efficacy in a New World primate model system (J. W. Barnwell and M. R. Galinski, unpublished results). Consequently, the investigation of naturally acquired anti-PvMSP-3-α presented in this work becomes relevant with regards to the continued consideration of PvMSP-3α as a vaccine candidate.
In a cross-sectional study carried out in Porto Velho, Rondonia State, Brazil, we assessed naturally acquired humoral immune responses against five recombinant protein constructs representing the complete amino acid sequences of PvMSP-3α. The fact that the majority of studied individuals had reported P. vivax and P.falciparum infections in the past, suggests that both malaria parasites circulated in this region in recent decades. It is classically known that the natural exposure to malaria infections is closely related with acquisition of antibodies to Plasmodium spp. antigens [44–47]. The broad range of age, number of past malaria episodes, time of expositure and months since individual’s last reported malaria episode identified in our studied population would indicate that the inhabitants exhibit different degrees of immune reactivity against P. vivax and consequently to the PvMSP-3-α proteins tested here.
Our first set of data on antibody responses show that PvMSP-3α is a target of the immune response in individuals naturally exposed to P. vivax malaria transmission. We show a high frequency of IgG responders against the full length recombinant protein and recombinant proteins representing four different regions of the molecule, indicating that the pET E. coli expression systems successfully generated epitopes that share similar antigenic determinants as the native proteins. The high polymorphism observed in several regions of the world in the PvMSP-3α diverse central domain does not seem to occurs in the studied endemic area, since the frequency of individuals that recognized the recombinant proteins representing these domains (PvMSP3-BLI and PvMSP3-BLII) is higher that the frequency of individuals that recognized the recombinant proteins representing the conserved regions (PvMSP3-NT and PvMSP3- CT) [21, 48-49]. In addition, the frequencies of IgG responders against PvMSP-3α presented similar overall frequencies of naturally acquired IgG response against classical P. vivax candidate antigens like PvMSP-1 [50-51], PvAMA-1[52], PvMSP-9 [34] and PvRBP-1 [47], indicating that PvMSP-3 is highly immunogenic in naturally exposed individuals. However, recently, was demonstrated that the breadth and magnitude of IgG antibody responses to P.falciparum merozoite antigens (MSP-1, MSP-3 and AMA-1) are better associated with protection from clinical malaria than only the frequencies of responders [53]. In agreement with this finding, we observed that the RIs were high against the full-length protein, the two blocks of heptad repeats and the C-terminal region, while the lowest levels were observed in PvMSP3-NT. The observation that higher indexes of antibodies against PvMSP3-FL and PvMSP3-CT were present in individuals with more time of residence in malaria endemic areas and the IgG antibody levels against PvMSP3-FL, PvMSP3-BLI and PvMSP3-CT increased accordingly with the number of past malaria episodes indicate that repeated P. vivax infections and/or exposure could significantly increase, not only the frequency of responders, but also the magnitude of IgG antibodies against PvMSP-3α. This pattern of a cumulative immune response observed here against PvMSP-3α was also observed for several other P. vivax and P.falciparum candidate antigens [34, 44, 47, 54–58].
The naturally acquired protective immunity against P. vivax tested with vaccine candidate antigens is poorly explored. The association of clinical protection with specific antigens of P. vivax has been reported in only two prospective cohort longitudinal studies [59–60]. In the first, clinical protection was associated with IgG3 antibodies against the N-terminus of the merozoite surface protein 1 (PvMSP-1) in residents of the Brazilian Amazon region of Portuchuelo [60]. In the second, clinical protection was reported in children from Papua New Guinea [59] where the frequency of naturally-acquired binding inhibitory antibodies against the Duffy-binding protein region II (PvDBPII) were associated with protection against P. vivax infection. Studies on IgG responses against P.falciparum antigens have consistently showed that cytophilic subclasses IgG1 and IgG3 play an important role in a protective antibody response in humans [61]. The proposed mechanism involves parasitic inhibition mediated by engagement of Plasmodium-specific cytophilic Ig antibodies to Fc receptors on the surface of monocytes [62]. Interestingly, the first observation of this mechanism was reported by Oeuvray et al using antibodies against a PvMSP-3 homologue, the originally characterized P.falciparum MSP-3 [61]. Nevertheless, only a small number of studies have investigated cytophilic antibody responses against P. vivax [47, 52, 63]. Therefore, in our study, we have demonstrated that IgG positive individuals against all PvMSP-3α recombinant proteins studied here present a high frequency and concentrations of anti-PvMSP-3 cytophilic antibodies IgG1 and IgG3.
Although we demonstrate a general predominance of anti-PvMSP-3 cytophilic IgG antibodies, the correlation to clinical immunity is rather tenuous, based on currently available data. The cross-sectional design of our study limited the investigation to retrospective malaria histories, and the best approximation of an individual’s protection was the estimated amount of time that had passed since their last malaria episode. We were only able to observe positive correlations between IgG1 and IgG3 reactivities against PvMSP3-FL and PvMSP3-CT with the past months since malaria infection. These results suggest a possible role of PvMSP-3 cythophilic antibodies in protective immunity against P. vivax infection. However, prospective studies on humoral immune responses or biologic studies addressing the ability of these antibodies to inhibit merozoite invasion or the development of blood-stage parasites will provide more direct evidence with regards to their protective efficacy.
The present work describes for the first time the fine B cell epitope mapping of a full-length protein of P. vivax using spot-synthesis. Information at the amino acid level about the epitopes of proteins recognized by antibodies is important for their use as biological tools, therapeutic molecules, and for understanding molecular recognition events in general [64]. In this context, T and B cell epitope prediction programs are largely used in malaria research [65–66]. However, the use of chemically prepared arrays of short peptides has emerged as a powerful tool to identify and characterize epitopes recognized by antibodies [35–36]. Our results from the spot synthesis assays suggest that PvMSP-3α can present as many as 25 antigenic determinants to the immune system. However, several events are important in determining whether an antibody against a specific peptide will bind to the native protein from which the peptide sequence is derived; in this process the length of the immunizing peptide is critical. To raise antibodies to a peptide, a minimum length of six amino acids is required, and peptides of >10 amino acids are generally required for the induction of antibodies that may bind to the native protein [67]. In this context, the synthesis of 15 amino acid peptides, with 9 overlapping, has allowed the identification of PvMSP-3α B-cell epitopes encompassed in sequences ranging from 10 to 25 amino acids in length. The majority (18/25) of the sequences containing the B-cell epitopes are localized in the two blocks of heptad repeats in the diverse central domain of PvMSP-3α. On the other hand, 7 epitopes were assigned to sequences in the N and C terminal flanking regions, which are relatively conserved [22]. This finding could be important, in a future vaccine composition based on these conserved regions, rather than the highly polymorphic central domains, as so far confirmed for PvMSP-3α and PvMSP-3β [21–25] and which may be the rule for other members of this gene family[12].
In conclusion, PvMSP-3α is highly immunogenic in individuals living in malaria endemic areas of the Brazilian Amazon. This protein presents several linear B-cell epitopes in its sequence, and the immune response generated is mainly mediated by cytophilic antibodies, which are associated with time of exposure to infections and apparent protective immunity. However, studies on the functional activity of these antibodies and further characterization of the B cell epitopes described here are required to further assess the potential of PvMSP-3α as a vaccine candidate. Moreover, the complexity of the immune response against the surface of the P. vivax merozoite must be re-considered, particularly if many or all of the members of the 11 member pvmsp3 gene family [12] prove to be similarly diverse, expressed and immunogenic.
Figure 3.
Stratification according to numbers of past malaria infections reported, 0 (n=33), 1–3 (n=102) and >3 (n=138), in the epidemiological interview of donors enrolled in our study and the reactivity indexes against PvMSP3-FL (a) and PvMSP3-BLI (b) and PvMSP3-CT (c).
* p<0.05, ** p<0.01 and *** p<0.001 by Mann-Whitney rank sum test.
Table 3.
Epitopes identified by IgG reactivity detected in the spot-synthesis assay with serum from positive responders against the PvMSP-3α recombinant proteins.
| PvMSP-3α Region | Epitopes | AA position | Position in Spot- synthesis assay |
|---|---|---|---|
| PvMSP3-Nt | EAPNSSRHHLRNGF | E32-F46 | A8-A10 |
| KNDSLPHEEPNNLE | K49-E62 | A12-A13 | |
| DQVTKKEKKTIKKA | D77-A90 | A19-A21 | |
| PvMSP3-BLI | AEQIQAELQKVKTA | A105-A118 | B1-B4 |
| SATAAETAKNNAVS | S125-S138 | B8 | |
| KGLDAAKTAIKKAKAAAEEAKKEAAI | K141-I166 | B11-B16 | |
| KAEKDAEAAQ | K168-Q178 | B19 | |
| AKTAATNAEKKKTK | A193-K206 | C1-C2 | |
| EAQKACEKAKKAHA | K229-A242 | C10-C11 | |
| KSTEDLSVAKDK | K282-K294 | C23-C24 | |
| EIAAEVAKAKVAK | E313-K326 | D2-D4 | |
| KKAEEAKKIVDKI | K333-I345 | D6-D7 | |
| AAEFATEVKKAT | A357-T370 | D11-D13 | |
| GAKKAAGEAKKASI | G401-I409 | E5 | |
| PvMSP3-BLII | EVAKAEVLNAEVK | E421-K434 | E10-E11 |
| NDATEAKKQAEKAKAAAEEAKTHGEK | N441-K466 | E14-E18 | |
| KAYAVEAHLAKTKN | K505-N518 | F2-F5 | |
| DAANIAHQKWLKAT | D541-T554 | F14-F19 | |
| KAQKEATAAKLKA | K573-A585 | F23-F24 | |
| AEDAAEEAKEAAKK | A645-K658 | G7-G12 | |
| DKTIAAAKKAKKARE | D673-E686 | G24-H2 | |
| PvMSP3-Ct | KAAYGLLKTKNQYVL | K687-L702 | H3-H7 |
| LDISPESADNITSK | L705-K718 | H8-H9 | |
| QTGGNRERRNTSDTVDDT | Q773-T790 | I1-I4 | |
| GDEFDTYDDIKKVT | G796-T810 | I6-I8 |
Acknowledgments
This work was supported by Brazilian National Research Council–CNPq/PAPES, Fiocruz, National Institute of Health, the Yerkes National Primate Research Center Base Grant # RR00165 awarded by the National Center for Research Resources of the National Institutes of Health, and NIH Grant #RO1 AI0555994 (MRG). Josué da Costa Lima Junior was the recipient of a FAPERJ Fellowship. We are grateful to all individuals that participate in this study for their cooperation and generous donation of blood, which made this study possible. We thank Eileen Farnon and Jennie Larson for the assistance during the sample collection and Paloma Napoleão Pêgo for the Spot-synthesis technique. We thank the Secretary of Health of Rondonia State and the Laboratorio Central–LACEN of Rondonia for providing fieldwork support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guerra CA, Snow RW, Hay SI. Mapping the global extent of malaria in 2005. Trends Parasitol. 2006 Aug;22(8):353–8. doi: 10.1016/j.pt.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001 Jan–Feb;64(1–2 Suppl):97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 3.Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007 Dec;77(6 Suppl):79–87. [PMC free article] [PubMed] [Google Scholar]
- 4.Baird JK. Neglect of Plasmodium vivax malaria. Trends Parasitol. 2007 Nov;23(11):533–9. doi: 10.1016/j.pt.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Genton B, D'Acremont V, Rare L, Baea K, Reeder JC, Alpers MP, et al. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med. 2008 Jun 17;5(6):e127. doi: 10.1371/journal.pmed.0050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picot S, Bienvenu AL. [Plasmodium vivax infection: not so benign] Med Sci (Paris) 2009 Jun–Jul;25(6–7):622–6. doi: 10.1051/medsci/2009256-7622. [DOI] [PubMed] [Google Scholar]
- 7.Price RN, Douglas NM, Anstey NM. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis. 2009 Oct;22(5):430–5. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 8.Sharma A, Khanduri U. How benign is benign tertian malaria? J Vector Borne Dis. 2009 Jun;46(2):141–4. [PubMed] [Google Scholar]
- 9.Poespoprodjo JR, Fobia W, Kenangalem E, Lampah DA, Hasanuddin A, Warikar N, et al. Vivax malaria: a major cause of morbidity in early infancy. Clin Infect Dis. 2009 Jun 15;48(12):1704–12. doi: 10.1086/599041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galinski MR, Barnwell JW. Plasmodium vivax: who cares? Malar J. 2008;7( Suppl 1):S9. doi: 10.1186/1475-2875-7-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlton J. The Plasmodium vivax genome sequencing project. Trends Parasitol. 2003 May;19(5):227–31. doi: 10.1016/s1471-4922(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 12.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008 Oct 9;455(7214):757–63. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stratton L, O'Neill MS, Kruk ME, Bell ML. The persistent problem of malaria: addressing the fundamental causes of a global killer. Soc Sci Med. 2008 Sep;67(5):854–62. doi: 10.1016/j.socscimed.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 14.del Portillo HA, Longacre S, Khouri E, David PH. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci U S A. 1991 May 1;88(9):4030–4. doi: 10.1073/pnas.88.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galinski MR, Corredor-Medina C, Povoa M, Crosby J, Ingravallo P, Barnwell JW. Plasmodium vivax merozoite surface protein-3 contains coiled-coil motifs in an alanine-rich central domain. Mol Biochem Parasitol. 1999 Jun 25;101(1–2):131–47. doi: 10.1016/s0166-6851(99)00063-8. [DOI] [PubMed] [Google Scholar]
- 16.Barnwell JW, Galinski MR, DeSimone SG, Perler F, Ingravallo P. Plasmodium vivax, P. cynomolgi, and P. knowlesi: identification of homologue proteins associated with the surface of merozoites. Exp Parasitol. 1999 Mar;91(3):238–49. doi: 10.1006/expr.1998.4372. [DOI] [PubMed] [Google Scholar]
- 17.Galinski MR, Medina CC, Ingravallo P, Barnwell JW. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992 Jun 26;69(7):1213–26. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Q, Saul A. Sequence analysis of the apical membrane antigen I (AMA-1) of Plasmodium vivax. Mol Biochem Parasitol. 1994 May;65(1):183–7. doi: 10.1016/0166-6851(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 19.Adams JH, Fang X, Kaslow DC, Miller LH. Identification of a cryptic intron in the Plasmodium vivax Duffy binding protein gene. Mol Biochem Parasitol. 1992 Nov;56(1):181–3. doi: 10.1016/0166-6851(92)90166-h. [DOI] [PubMed] [Google Scholar]
- 20.Galinski MR, Ingravallo P, Corredor-Medina C, Al-Khedery B, Povoa M, Barnwell JW. Plasmodium vivax merozoite surface proteins-3beta and-3gamma share structural similarities with P. vivax merozoite surface protein-3alpha and define a new gene family. Mol Biochem Parasitol. 2001 Jun;115(1):41–53. doi: 10.1016/s0166-6851(01)00267-5. [DOI] [PubMed] [Google Scholar]
- 21.Rayner JC, Huber CS, Feldman D, Ingravallo P, Galinski MR, Barnwell JW. Plasmodium vivax merozoite surface protein PvMSP-3 beta is radically polymorphic through mutation and large insertions and deletions. Infect Genet Evol. 2004 Dec;4(4):309–19. doi: 10.1016/j.meegid.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Rayner JC, Corredor V, Feldman D, Ingravallo P, Iderabdullah F, Galinski MR, et al. Extensive polymorphism in the Plasmodium vivax merozoite surface coat protein MSP-3alpha is limited to specific domains. Parasitology. 2002 Nov;125(Pt 5):393–405. doi: 10.1017/s0031182002002317. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z, Miao J, Huang Y, Li X, Putaporntip C, Jongwutiwes S, et al. Genetic structures of geographically distinct Plasmodium vivax populations assessed by PCR/RFLP analysis of the merozoite surface protein 3beta gene. Acta Trop. 2006 Dec;100(3):205–12. doi: 10.1016/j.actatropica.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mascorro CN, Zhao K, Khuntirat B, Sattabongkot J, Yan G, Escalante AA, et al. Molecular evolution and intragenic recombination of the merozoite surface protein MSP-3alpha from the malaria parasite Plasmodium vivax in Thailand. Parasitology. 2005 Jul;131(Pt 1):25–35. doi: 10.1017/s0031182005007547. [DOI] [PubMed] [Google Scholar]
- 25.Han ET, Song TE, Park JH, Shin EH, Guk SM, Kim TY, et al. Allelic dimorphism in the merozoite surface protein-3alpha in Korean isolates of Plasmodium vivax. Am J Trop Med Hyg. 2004 Dec;71(6):745–9. [PubMed] [Google Scholar]
- 26.David PH, Hudson DE, Hadley TJ, Klotz FW, Miller LH. Immunization of monkeys with a 140 kilodalton merozoite surface protein of Plasmodium knowlesi malaria: appearance of alternate forms of this protein. J Immunol. 1985 Jun;134(6):4146–52. [PubMed] [Google Scholar]
- 27.Hudson DE, Wellems TE, Miller LH. Molecular basis for mutation in a surface protein expressed by malaria parasites. J Mol Biol. 1988 Oct 5;203(3):707–14. doi: 10.1016/0022-2836(88)90204-5. [DOI] [PubMed] [Google Scholar]
- 28.Klotz FW, Hudson DE, Coon HG, Miller LH. Vaccination-induced variation in the 140 kD merozoite surface antigen of Plasmodium knowlesi malaria. J Exp Med. 1987 Feb 1;165(2):359–67. doi: 10.1084/jem.165.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oeuvray C, Bouharoun-Tayoun H, Gras-Masse H, Bottius E, Kaidoh T, Aikawa M, et al. Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood. 1994 Sep 1;84(5):1594–602. [PubMed] [Google Scholar]
- 30.McColl DJ, Silva A, Foley M, Kun JF, Favaloro JM, Thompson JK, et al. Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol Biochem Parasitol. 1994 Nov;68(1):53–67. doi: 10.1016/0166-6851(94)00149-9. [DOI] [PubMed] [Google Scholar]
- 31.Hisaeda H, Saul A, Reece JJ, Kennedy MC, Long CA, Miller LH, et al. Merozoite surface protein 3 and protection against malaria in Aotus nancymai monkeys. J Infect Dis. 2002 Mar 1;185(5):657–64. doi: 10.1086/339187. [DOI] [PubMed] [Google Scholar]
- 32.Audran R, Cachat M, Lurati F, Soe S, Leroy O, Corradin G, et al. Phase I malaria vaccine trial with a long synthetic peptide derived from the merozoite surface protein 3 antigen. Infect Immun. 2005 Dec;73(12):8017–26. doi: 10.1128/IAI.73.12.8017-8026.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Druilhe P, Spertini F, Soesoe D, Corradin G, Mejia P, Singh S, et al. A malaria vaccine that elicits in humans antibodies able to kill Plasmodium falciparum. PLoS Med. 2005 Nov;2(11):e344. doi: 10.1371/journal.pmed.0020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lima-Junior JC, Tran TM, Meyer EV, Singh B, De-Simone SG, Santos F, et al. Naturally acquired humoral and cellular immune responses to Plasmodium vivax merozoite surface protein 9 in Northwestern Amazon individuals. Vaccine. 2008 Dec 2;26(51):6645–54. doi: 10.1016/j.vaccine.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank R, Overwin H. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol Biol. 1996;66:149–69. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- 36.Frank R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports--principles and applications. J Immunol Methods. 2002 Sep 1;267(1):13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 37.Holder AA. Malaria vaccines. Proc Natl Acad Sci U S A. 1999 Feb 16;96(4):1167–9. doi: 10.1073/pnas.96.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holder AA. Developments with anti-malarial vaccines. Ann N Y Acad Sci. 1993 Dec 21;700:7–21. doi: 10.1111/j.1749-6632.1993.tb26301.x. [DOI] [PubMed] [Google Scholar]
- 39.Galinski MR, Barnwell JW. Plasmodium vivax: Merozoites, invasion of reticulocytes and considerations for malaria vaccine development. Parasitol Today. 1996 Jan;12(1):20–9. doi: 10.1016/0169-4758(96)80641-7. [DOI] [PubMed] [Google Scholar]
- 40.McGregor IA. Studies in the Acquisition of Immunity of Plasmodium falciparum Infections in Africa. Trans R Soc Trop Med Hyg. 1964 Jan;58:80–92. doi: 10.1016/0035-9203(64)90073-2. [DOI] [PubMed] [Google Scholar]
- 41.Bouharoun-Tayoun H, Oeuvray C, Lunel F, Druilhe P. Mechanisms underlying the monocyte-mediated antibody-dependent killing of Plasmodium falciparum asexual blood stages. J Exp Med. 1995 Aug 1;182(2):409–18. doi: 10.1084/jem.182.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh S, Soe S, Mejia JP, Roussilhon C, Theisen M, Corradin G, et al. Identification of a conserved region of Plasmodium falciparum MSP3 targeted by biologically active antibodies to improve vaccine design. J Infect Dis. 2004 Sep 1;190(5):1010–8. doi: 10.1086/423208. [DOI] [PubMed] [Google Scholar]
- 43.Kaushik NK, Ananthanarayanan M, Subrahmanyam D, Sehgal S. Protection of rhesus monkeys against Plasmodium knowlesi infection with merozoite vaccine. Indian J Med Res. 1986 May;83:471–9. [PubMed] [Google Scholar]
- 44.Baird JK. Age-dependent characteristics of protection v. susceptibility to Plasmodium falciparum. Ann Trop Med Parasitol. 1998 Jun;92(4):367–90. doi: 10.1080/00034989859366. [DOI] [PubMed] [Google Scholar]
- 45.Ceravolo IP, Bruna-Romero O, Braga EM, Fontes CJ, Brito CF, Souza JM, et al. Anti-Plasmodium vivax duffy binding protein antibodies measure exposure to malaria in the Brazilian Amazon. The American journal of tropical medicine and hygiene. 2005 Jun;72(6):675–81. [PubMed] [Google Scholar]
- 46.Pratt-Riccio LR, Lima-Junior JC, Carvalho LJ, Theisen M, Espindola-Mendes EC, Santos F, et al. Antibody response profiles induced by Plasmodium falciparum glutamate-rich protein in naturally exposed individuals from a Brazilian area endemic for malaria. The American journal of tropical medicine and hygiene. 2005 Dec;73(6):1096–103. [PubMed] [Google Scholar]
- 47.Tran TM, Oliveira-Ferreira J, Moreno A, Santos F, Yazdani SS, Chitnis CE, et al. Comparison of IgG reactivities to Plasmodium vivax merozoite invasion antigens in a Brazilian Amazon population. Am J Trop Med Hyg. 2005 Aug;73(2):244–55. [PubMed] [Google Scholar]
- 48.Zakeri S, Safi N, Afsharpad M, Butt W, Ghasemi F, Mehrizi AA, et al. Genetic structure of Plasmodium vivax isolates from two malaria endemic areas in Afghanistan. Acta Trop. 2010 Jan;113(1):12–9. doi: 10.1016/j.actatropica.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 49.Cristiano FA, Perez MA, Nicholls RS, Guerra AP. Polymorphism in the Plasmodium vivax msp 3: gene in field samples from Tierralta, Colombia. Mem Inst Oswaldo Cruz. 2008 Aug;103(5):493–6. doi: 10.1590/s0074-02762008000500015. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Becerra C, Sanz S, Brucet M, Stanisic DI, Alves FP, Camargo EP, et al. Naturally-acquired humoral immune responses against the N- and C-termini of the Plasmodium vivax MSP1 protein in endemic regions of Brazil and Papua New Guinea using a multiplex assay. Malar J. 2010;9:29. doi: 10.1186/1475-2875-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soares IS, Levitus G, Souza JM, Del Portillo HA, Rodrigues MM. Acquired immune responses to the N- and C-terminal regions of Plasmodium vivax merozoite surface protein 1 in individuals exposed to malaria. Infect Immun. 1997 May;65(5):1606–14. doi: 10.1128/iai.65.5.1606-1614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodrigues MH, Rodrigues KM, Oliveira TR, Comodo AN, Rodrigues MM, Kocken CH, et al. Antibody response of naturally infected individuals to recombinant Plasmodium vivax apical membrane antigen-1. Int J Parasitol. 2005 Feb;35(2):185–92. doi: 10.1016/j.ijpara.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Osier FH, Fegan G, Polley SD, Murungi L, Verra F, Tetteh KK, et al. Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect Immun. 2008 May;76(5):2240–8. doi: 10.1128/IAI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahlborg N, Ling IT, Howard W, Holder AA, Riley EM. Protective immune responses to the 42-kilodalton (kDa) region of Plasmodium yoelii merozoite surface protein 1 are induced by the C-terminal 19-kDa region but not by the adjacent 33-kDa region. Infect Immun. 2002 Feb;70(2):820–5. doi: 10.1128/IAI.70.2.820-825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mertens F, Levitus G, Camargo LM, Ferreira MU, Dutra AP, Del Portillo HA. Longitudinal study of naturally acquired humoral immune responses against the merozoite surface protein 1 of Plasmodium vivax in patients from Rondonia, Brazil. Am J Trop Med Hyg. 1993 Sep;49(3):383–92. doi: 10.4269/ajtmh.1993.49.383. [DOI] [PubMed] [Google Scholar]
- 56.Seth RK, Bhat AA, Rao DN, Biswas S. Acquired immune response to defined Plasmodium vivax antigens in individuals residing in northern India. Microbes Infect. 2010 Mar;12(3):199–206. doi: 10.1016/j.micinf.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 57.Hudson Keenihan SN, Ratiwayanto S, Soebianto S, Krisin, Marwoto H, Krishnegowda G, et al. Age-dependent impairment of IgG responses to glycosylphosphatidylinositol with equal exposure to Plasmodium falciparum among Javanese migrants to Papua, Indonesia. Am J Trop Med Hyg. 2003 Jul;69(1):36–41. [PubMed] [Google Scholar]
- 58.Xainli J, Baisor M, Kastens W, Bockarie M, Adams JH, King CL. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J Immunol. 2002 Sep 15;169(6):3200–7. doi: 10.4049/jimmunol.169.6.3200. [DOI] [PubMed] [Google Scholar]
- 59.King CL, Michon P, Shakri AR, Marcotty A, Stanisic D, Zimmerman PA, et al. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci U S A. 2008 Jun 17;105(24):8363–8. doi: 10.1073/pnas.0800371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nogueira PA, Alves FP, Fernandez-Becerra C, Pein O, Santos NR, Pereira da Silva LH, et al. A reduced risk of infection with Plasmodium vivax and clinical protection against malaria are associated with antibodies against the N terminus but not the C terminus of merozoite surface protein 1. Infect Immun. 2006 May;74(5):2726–33. doi: 10.1128/IAI.74.5.2726-2733.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oeuvray C, Bouharoun-Tayoun H, Grass-Masse H, Lepers JP, Ralamboranto L, Tartar A, et al. A novel merozoite surface antigen of Plasmodium falciparum (MSP-3) identified by cellular-antibody cooperative mechanism antigenicity and biological activity of antibodies. Mem Inst Oswaldo Cruz. 1994;89( Suppl 2):77–80. doi: 10.1590/s0074-02761994000600018. [DOI] [PubMed] [Google Scholar]
- 62.Druilhe P, Khusmith S. Epidemiological correlation between levels of antibodies promoting merozoite phagocytosis of Plasmodium falciparum and malaria-immune status. Infect Immun. 1987 Apr;55(4):888–91. doi: 10.1128/iai.55.4.888-891.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soares IS, Oliveira SG, Souza JM, Rodrigues MM. Antibody response to the N and C-terminal regions of the Plasmodium vivax Merozoite Surface Protein 1 in individuals living in an area of exclusive transmission of P. vivax malaria in the north of Brazil. Acta Trop. 1999 Jan 15;72(1):13–24. doi: 10.1016/s0001-706x(98)00078-3. [DOI] [PubMed] [Google Scholar]
- 64.Reineke U, Sabat R. Antibody epitope mapping using SPOT peptide arrays. Methods Mol Biol. 2009;524:145–67. doi: 10.1007/978-1-59745-450-6_11. [DOI] [PubMed] [Google Scholar]
- 65.Lima-Junior JC, Banic DM, Tran TM, Meyer VS, De-Simone SG, Santos F, et al. Promiscuous T-cell epitopes of Plasmodium merozoite surface protein 9 (PvMSP9) induces IFN-gamma and IL-4 responses in individuals naturally exposed to malaria in the Brazilian Amazon. Vaccine. 2010 Feb 26; doi: 10.1016/j.vaccine.2010.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics. 2008;9( Suppl 12):S22. doi: 10.1186/1471-2105-9-S12-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dyrberg T, Oldstone MB. Peptides as antigens. Importance of orientation. J Exp Med. 1986 Oct 1;164(4):1344–9. doi: 10.1084/jem.164.4.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]