Abstract
The biosynthetic pathway for selenocysteine (Sec), the 21st amino acid in the genetic code whose codeword is UGA, was recently determined in eukaryotes and archaea. Sec tRNA, designated tRNA[Ser]Sec, is initially aminoacylated with serine by seryl-tRNA synthetase and the resulting seryl moiety is converted to phosphoserine by O-phosphoseryl-tRNA kinase to form O-phosphoseryl-tRNA[Ser]Sec. Sec synthase (SecS) then uses O-phosphoseryl-tRNA[Ser]Sec and the active donor of selenium, selenophosphate, to form Sec-tRNA[Ser]Sec. Selenophosphate is synthesized from selenide and ATP by selenophosphate synthetase 2 (SPS2). Sec was the last protein amino acid in eukaryotes whose biosynthesis had not been established and the only known amino acid in eukaryotes whose biosynthesis occurs on its tRNA. Interestingly, sulfide can replace selenide to form thiophosphate in the SPS2-catalyzed reaction that can then react with O-phosphoseryl-tRNA[Ser]Sec in the presence of SecS to form cysteine-(Cys-)tRNA[Ser]Sec. This novel pathway of Cys biosynthesis results in Cys being decoded by UGA and replacing Sec in normally selenium-containing proteins (selenoproteins). The selenoprotein, thioredoxin reductase 1 (TR1), was isolated from cells in culture and from mouse liver for analysis of Cys/Sec replacement by MS. The level of Cys/Sec replacement in TR1 was proportional to the level of selenium in the diet of the mice. Elucidation of the biosynthesis of Sec and Sec/Cys replacement provides novel ways of regulating selenoprotein functions and ultimately better understanding of the biological roles of dietary selenium.
Introduction
Selenocysteine (Sec) is the 21st amino acid in the genetic code and this selenium containing amino acid is cotranslationally incorporated into selenium-containing proteins, designated selenoproteins, in response to the codon, UGA (1–3). Although UGA is normally a termination codon that dictates the cessation of protein synthesis, it is also used as a Sec codon by numerous organisms in each of the 3 domains of life: eubacteria, archaea, and eukaryotes. Of the >500 genomes sequenced in eubacteria, only ∼20% encode the machinery for inserting Sec into protein, and in archaea, ∼10% have this machinery (4, 5). In eukaryotes, the Sec insertion machinery has been found in a number of lower organisms such as green algae, kinetoplastida, and slime molds and it is widespread in animals but absent in fungi and higher plants (4, 5).
The mechanisms responsible for designating UGA as a Sec codon instead of termination involve a stem-loop structure in the 3′-untranslated region of selenoprotein mRNA in eukaryotes known as the Sec insertion sequence (SECIS)6 element (6) and a SECIS binding protein, designated SBP2 (7). SECIS-binding protein 2 (SBP2) binds to the SECIS element and forms a complex with the specific elongation factor for Sec tRNA[Ser]Sec, EFSec, for incorporation of Sec into protein in response to the UGA Sec codon. Because the synthesis of some selenoproteins is terminated by a UGA stop signal, the Sec insertion machinery has the capability of distinguishing between the UGA for amino acid incorporation and the UGA for termination. The distance between the Sec codon and the SECIS element plays an essential role in this distinction (8, and refs. therein).
Even though the biosynthesis of Sec was established in eubacteria in the early 1990s (9–11), only in the last several years was the complete biosynthetic pathway of this selenium-containing amino acid determined in eukaryotes and archaea (12). Very recently, it was also shown that cysteine (Cys) can be synthesized de novo by replacing sulfide with selenide in the Sec biosynthetic pathway, forming Cys on tRNA[Ser]Sec (13). The biosynthesis of Sec and the novel pathway of Cys biosynthesis are the subjects of this review.
Biosynthesis of Sec
The biosynthesis of Sec in Escherichia coli proceeds as follows. The enzyme that synthesizes Sec, Sec synthase (designated SelA in eubacteria and SecS in eukaryotes and archaea), is a pyridoxal phosphate dependent protein (9–11). This enzyme interacts with seryl-tRNA[Ser]Sec and removes the hydroxyl group from the seryl moiety to yield aminoacrylyl- (dehydroalanyl-) tRNA[Ser]Sec as an intermediate. Dehydroalanyl-tRNA[Sec]Sec then accepts the active selenium donor, which was identified in eubacteria as monoselenophosphate (9, 11), to form selenocysteyl-tRNA[Ser]Sec. Selenophosphate is synthesized from selenide and ATP by E. coli selenophosphate synthetase (SelD) (14).
The biosynthesis of Sec in eukaryotes and archaea (Fig. 1) proceeds by a similar pathway as in eubacteria except that an immediate, O-phosphoseryl-tRNA[Ser]Sec, is involved and it arises by action of the kinase, O-phosphoseryl-tRNA[Ser]Sec kinase (PSTK), on seryl-tRNA[Ser]Sec (15). SecS then acts on phosphoseryl-tRNA[Ser]Sec, converting it to the proposed intermediate, dehydroalanyl-tRNA[Ser]Sec (12), accepts the active selenium donor, also identified as monoselenophosphate in eukaryotes, and generates selenocysteyl-tRNA[Ser]Sec from the dehydroalanyl-tRNA[Ser]Sec intermediate (12). In our studies, we relied on bioinformatics for identification of genes involved in Sec biosynthesis. One distinct benefit aiding us in identifying these genes was that several factors were previously found in higher vertebrates that were associated with selenium metabolism in some manner. For example, in 1970, a kinase that phosphorylated a minor rooster liver seryl-tRNA forming O-phosphoseryl-tRNA (16) and a minor mammalian and chicken liver seryl-tRNA recognizing specifically the stop codon, UGA (17), were reported. The minor seryl-tRNAUGA was subsequently shown to form phosphoseryl-tRNA (18) and identified as Sec tRNA[Ser]Sec (19), and the kinase originally found by Maaenpa and Bernfield (15) was shown to be PSTK. In addition, a 48-kDa protein that bound Sec tRNA[Ser]Sec in human liver and designated the soluble liver antigen (SLA) was found to be targeted by antibodies in patients with an autoimmune chronic hepatitis (20). SLA was found to occur in a separate family within a large superfamily of diverse pyridoxal phosphate dependent transferases (21), had been proposed to be the SecS in mammals (21–24), and was subsequently identified as SecS (12).
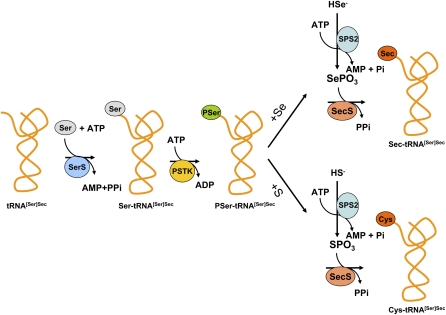
Figure 1.
Biosynthesis of Sec and de novo synthesis of Cys. The complete synthesis of Sec on its tRNA in eukaryotes and archaea wherein selenite and ATP are substrates for SPS2 yielding selenophosphate that interacts with SecS, and the intermediate, likely dehydroalanine, to generate Sec-tRNA[Ser]Sec is shown on the right in the upper pathway. In the lower pathway, the de novo synthesis of Cys is shown in mammals wherein sulfide and ATP are substrates for SPS2 yielding thiophosphate that interacts with SecS, and the intermediate, likely dehydroalanine, to yield Cys-tRNA[Ser]Sec. See text for further details.
To identify the enzyme responsible for making the active selenium donor in mammals, we examined the protein products of 2 previously identified genes, sps1 and sps2 (25–28), in their ability to generate selenophosphate (12, 29). Both genes were previously found in mammals as having homology to SelD and their products were proposed to be SPS (25–28). The product of one of these genes, SPS2, could serve as an autoregulator of selenoprotein synthesis because it was a selenoprotein (25, 27). Several other studies involving the roles of SPS1 and SPS2 in Sec biosynthesis should also be noted. For example, the Sec residue in SPS2 was changed to Cys and the resulting mutant was found to have enzyme activity (25, 27, 29) but could complement SelD deficient E. coli cells following their transfection with the Cys mutant form (29). In addition, transformation of SelD deficient E. coli cells with sps1 or sps2 suggested that SPS1 was involved in Sec recycling via a selenium salvage pathway, whereas SPS2 played a role in the synthesis of selenophosphate (30).
Further elucidation of the roles of SPS1 and SPS2 in the biosynthesis of Sec and characterization of seryl-tRNA[Ser]Sec synthetase, tRNA[Ser]Sec, seryl-tRNA[Ser]Sec, O-phosphoseryl-tRNA[Ser]Sec, PSTK, SLA, and selenophosphate in the pathway of Sec biosynthesis are discussed below.
Sec tRNA[Ser]Sec and its aminoacylation
As noted above, Sec tRNA[Ser]Sec was discovered in 1970 and characterized as a minor seryl-tRNA that decoded specifically the stop codon, UGA, in bovine and chicken liver (17). Because there was considerable interest at that time in assessing whether nonsense suppressor tRNA occurred in mammalian cells, this minor “seryl-tRNA” was thought to be a nonsense suppressor tRNA. Subsequent studies demonstrated numerous unique features of this tRNA, including that it: 1) formed O-phosphoseryl-tRNA (17); 2) has a primary sequence unlike that of other tRNA in that it is 90 nucleotides long (and the longest eukaryotic sequenced) with relatively few base modifications (reviewed in 24); 3) suppresses the UGA termination codon in rabbit β-globin mRNA (31); 5) is transcribed unlike any other known tRNA in that transcription begins at the first nucleotide within the coding sequence (32); 6) is transcribed through the regulation of several novel transcription regulatory elements encoded both 5′-proximally and 5′-distally of its gene, Trsp (reviewed in 33); 7) has a unique cloverleaf model in that there are 9 bp in the acceptor stem and 4 in the TψC stem compared with a 7/5 cloverleaf model in other tRNA (24, and refs. therein); and 8) exists in 2 isoforms that differ from each other by a single methyl group, Um34 (34). In addition, the first tRNA transgenic mice involving tRNA were generated with wild-type or mutant Trsp transgenes (35). Overexpression of the wild-type Sec tRNA[Ser]Sec population did not appear to result in any detectable alteration in selenoprotein expression in the cells, tissues, and organs examined, suggesting that the Sec tRNA population was not limiting in selenoprotein expression. Furthermore, rescue of the knockout of wild-type Trsp, which is embryonic lethal (36, 37), with a mutant Sec tRNA[Ser]Sec transgene lacking Um34 demonstrated that the noncontaining Um34 isoform synthesized housekeeping (essential) selenoproteins, whereas the Um34 isoform was responsible for synthesizing stress related selenoproteins that are nonessential to survival (38).
In addition to the many unique features of Sec tRNA discussed above, transgenic mouse models encoding a mutant Sec tRNA transgene were developed as well as numerous conditional Trsp knockout mouse models targeting specific tissues and organs that demonstrated roles for selenoproteins in development and in a variety of health issues (reviewed in 39). What was initially thought to be a nonsense suppressor seryl-tRNA (17) was subsequently found to be Sec tRNA[Ser]Sec that expanded the genetic code by including Sec as the 21st amino acid (19, 40) and the only known protein amino acid in eukaryotes that is biosynthesized on its tRNA. Sec tRNA[Ser]Sec is aminoacylated with serine in the presence of seryl-tRNA synthetase (SerS), ATP, and Mg2+ to begin the biosynthesis of Sec:
Identifying the gene for PTSK and characterizing the gene product
Our rationale in identifying the PSTK gene (pstk) involved an initial proposal that Sec biosynthesis proceeded by identical pathways in organisms making selenoproteins within eukaryotes and archaea and that O-phosphoseryl-tRNA[Ser]Sec is an intermediate in the pathway (15). The strategy involved a computational and genomic comparison approach by initially searching for kinase-like genes that were present in Methanococcus jnnaschii and Methanopyrus kandleri, archaea that encode the Sec insertion machinery, and comparing all such genes to the other archaea whose genomes had been sequenced but did not make selenoproteins (15, and refs. therein). Two kinase-like genes were detected in the 2 archaea synthesizing selenoproteins that were not found in the other 12 archaea not synthesizing selenoproteins. The sequenced eukaryotic genomes that did (nematodes, Drosophila, and mice) and did not (yeasts) encode the Sec insertion machinery were scanned for homologous sequences of the 2 candidate genes found in archaea. A single candidate kinase (gi: 20095115) was detected and this putative pstk from mice was cloned into an expression vector, expressed, and the product isolated and characterized (15). The coding sequence specifically used seryl-tRNA[Ser]Sec as a substrate to form phosphoseryl-tRNA[Ser]Sec, unequivocally demonstrating that the putative kinase indeed coded for PSTK. The phosphorylation of seryl-tRNA[Ser]Sec proceeded as follows:
The function of PSTK was not resolved by the above studies, because the biosynthetic pathway in E. coli (i.e., in eubacteria encoding the Sec insertion machinery) did not involve O-phosphoseryl-tRNA[Ser]Sec as an intermediate (9–11) and this form of tRNA[Ser]Sec had been proposed as a storage form (41). The hypothesis that O-phosphoseryl-tRNA[Ser]Sec was as an intermediate in Sec biosynthesis (15) was shown to be correct when the biosynthesis in eukaryotes and archaea was resolved (see below).
Characterization of SecS
We also used a computational and genomic comparison strategy to identify a SecS gene (SecS), because no sequences with homology to SelA could be detected in eukaryotes that synthesize selenoproteins. The genomes of archaea and eukaryotes that had been sequenced were examined for the presence of selenoproteins and genes that co-occur with the identified selenoprotein genes (12). Genes corresponding to the Sec insertion machinery were identified in addition to another gene that might be SecS in mammals. Homologous sequences of this gene were found in all archaea and eukaryotes encoding selenoproteins and not in the examined organisms in these kingdoms that did not make selenoproteins. The sequence of this putative SecS gene corresponded to the sequence of a protein in patients with autoimmune chronic hepatitis that coprecipitated with Sec tRNA[Ser]Sec in cell extracts from such individuals, designated SLA (20; and see above). SLA was found to bind other proteins involved in Sec metabolism, further suggesting a role of this protein in Sec biosynthesis (42).
The putative mouse SecS (mSecS) was then cloned and characterized for binding to a number of potential tRNA[Ser]Sec substrates, including tRNA[Ser]Sec, seryl-tRNA[Ser]Sec, and O-phosphoseryl-tRNA[Ser]Sec by using tRNASer and seryl-tRNASer as controls (12). O-Phosphoseryl-tRNA[Ser]Sec bound tightly to the expressed product. tRNA[Ser]Sec also bound, but less well, and seryl-tRNA[Ser]Sec bound poorly, whereas tRNASer and seryl-tRNASer did not bind at all, suggesting that O-phosphoseryl-tRNA[Ser]Sec is a substrate for mSecS. In addition, mSecS rapidly removed the phosphate group from O-phosphoserine, suggesting that the intermediate product was not seryl-tRNA[Ser]Sec because seryl-tRNA[Ser]Sec bound poorly to this protein. These results provided strong evidence that mSecS was indeed the missing SecS in eukaryotic Sec biosynthesis (12).
Characterization of mammalian SPS
Mouse sps2 (msps2), wherein the Sec codeword was changed to a Cys codeword, and mouse sps1 (msps1) were cloned into an expression vector, along with Caenorhabditis elegans sps2 (csps2), because this gene normally contains Cys in place of Sec in msps2. E. coli selD was cloned as a control to elucidate whether 1 or both of the mammalian SPS proteins were responsible for making the active selenium donor (12). The resulting proteins from each clone were expressed, purified, and their ability to generate monoselenophosphate from ATP and selenite examined. mSPS2(Cys), cSPS2, and SelD were all capable of synthesizing selenophosphate, but mSPS1 was not, providing strong evidence that eukaryotic SPS2 is responsible for making the active selenium donor and SPS1 must have another role (12, 43).
Sec biosynthesis
In vitro studies.
One important issue that needed to be resolved was whether selenophosphate could itself serve as the active selenium donor, because previous studies had not shown that this compound per se could be used to directly donate active selenium to reactions synthesizing Sec (10, 11, 14). We chemically synthesized monoselenophosphate and incubated it with O-phosphoseryl-tRNA[Ser]Sec and mSecS, which produced Sec attached to its tRNA (12). This study unequivocally demonstrated that selenophosphate is the active selenium donor in Sec biosynthesis and that SLA is the missing SecS and O-phosphoseryl-tRNA[Ser]Sec is an intermediate (12).
We also reconstituted the entire Sec biosynthetic pathway in vivo that included cloning and expressing mouse seryl-tRNA synthetase (mSerS) as well as each of the other components and substrates involved in the biosynthesis of the 21st amino acid (12). The only component not fully characterized was the intermediate attached to SecS that accepted activated selenium to form Sec attached to its tRNA. Dehydroalanine had been identified as the intermediate in E. coli wherein SelA acted on seryl-tRNA[Ser]Sec to yield dehydroalanyl-tRNA[Ser]Sec (9, 11); however, the available evidence suggested that the intermediate formed by the action of SecS on O-phosphoseryl-tRNA[Ser]Sec was indeed the same as in E. coli (12). The Sec biosynthetic pathway, based on these in vitro studies and on additional in vivo studies, is presented in Figure 1.
In vivo studies.
Although earlier, conflicting reports suggested that SPS1 was either the mammalian enzyme responsible for making the active selenium donor (25–28) or was involved in recycling selenium in Sec biosynthesis (30), our in vitro studies demonstrated that SPS2 synthesizes selenophosphate and SPS1 has another role (12). To resolve the discrepancies between the earlier studies (25, 28) and ours (12), and to elucidate the roles of SPS1 and SPS2 intracellularly, these proteins were individually knocked down using RNAi technology and the consequences of their resulting loss examined (43). Selenoprotein biosynthesis was abolished in NIH 3T3 cells by loss of SPS2 expression, but loss of SPS1 had no effect. Complementation experiments were also carried out wherein selenoprotein synthesis was restored in SPS2 deficient NIH 3T3 cells with either mSPS2(Cys) or SelD but not with mSPS1. These studies unequivocally established that SPS2, which synthesizes selenophosphate in vivo, is essential to selenoprotein synthesis and that SPS1 most likely has another role in cells other than Sec or selenoprotein synthesis (12, 43). It should also be noted that sps1 is an essential gene in Drosophila development (44) and has been shown to have a role in preventing megamitochondrial formation (45). Interestingly, SPS1 has been retained in insects such as the red flour beetle and silkworm that have lost all selenoprotein synthetic machinery, providing further evidence that sps1 must have a role independent of Sec and selenoprotein biosynthesis (46).
The complete Sec biosynthetic pathway
The complete biosynthesis of Sec on its tRNA is shown in Figure 1. Initially, tRNA[Ser]Sec is aminoacylated with serine by SerS (17) and the seryl moiety is then converted to O-phosphoseryl-tRNA[Ser]Sec by PSTK (15). SecS acts on O-phosphoseryl-tRNA[Ser]Sec to hydrolyze the phosphate group forming the acceptor molecule, likely dehydroalanyl-tRNA[Ser]Sec, which in turn accepts the active selenium donor, selenophosphate, synthesized by SPS2 (12, 14, 43). SecS then converts the active acceptor to Sec (12), completing the biosynthesis of Sec, the 21st amino acid in the genetic code, the last known protein amino acid whose biosynthesis had not been established in eukaryotes, and the only known amino acid in eukaryotes whose biosynthesis occurs on its tRNA.
De novo synthesis of Cys and Cys/Sec replacement in protein synthesis
Cys is not synthesized by mammals and therefore is 1 of several of their essential amino acids. It is 1 of the 20 commonly used amino acids in protein synthesis and its genetic codewords are UGU/UGC. Although Cys and Sec use a different genetic language and have different biosynthetic mechanisms, they are structurally similar and the replacement of sulfur with selenium in methionine (Met) synthesis has been reported wherein selenomethionine can be inserted into protein in place of methionine (46,47). Cys in place of Sec in normally selenium containing proteins has also been reported in selenium deficient rodents (48), but the specific mechanism of how this occurred was not addressed. We examined whether SPS2 would engage sulfide in place of selenide in generating thiophosphate that would be used as an active sulfur donor in making Cys attached to tRNA[Ser]Sec (reaction catalyzed by SecS), whereby replacing Sec with Cys in normally selenium containing proteins. The replacement of Cys for Sec on tRNA[Ser]Sec and the incorporation of Cys into protein by this pathway is discussed below.
In vitro studies.
The incorporation of Cys in place of Sec was established in cell free reactions by adding all the appropriate enzymes and other components. For example, incubation of thiophosphate with O-phosphoseryl-tRNA[Ser]Sec and mSecS resulted in Cys-tRNA[Ser]Sec (13). Similarly, incubation of sodium sulfide, ATP, mSPS2(Cys), O-phosphoseryl-tRNA[Ser]Sec, and mSecS also yielded Cys-tRNA[Ser]Sec. Replacing mSecS with SelD in either of these reactions did not result in Cys-tRNA[Ser]Sec, demonstrating that the insertion of Cys in place of Sec on tRNA[Ser]Sec likely does not occur in E. coli.
In vivo studies.
Thiophosphate was added to NIH 3T3 cells in culture and the treated cells were examined for the insertion of Cys in place of Sec in thioredoxin reductase (TR)1 and TR3 (13). Initially, NIH 3T3 cells were labeled with 75Se either with or without thiophosphate and then a sample of protein extract was electrophoresed to separate the various labeled selenoproteins on polyacrylamide gels. The level of 75Se labeling in TR1 from thiophosphate-treated cells appeared to be slightly less than from control cells. On the other hand, analysis of TR1 by Western blotting of the same extracts showed a dramatic enrichment in TR1 levels treated with thiophosphate, suggesting that indeed TR1 must either be truncated and/or another amino acid was inserted in place of Sec. It should also be noted that the enrichment of TR1 shown by Western blotting in cells exposed to thiophosphate was dependent on the SECIS element in the 3′-untranslated region of TR1 mRNA (13). This experiment demonstrated that the insertion of Cys into selenoprotein, which was confirmed by MS analysis of purified TR1 (see below), was, like that of Sec incorporation, contingent on the presence of the SECIS element.
A major question was whether Cys occurred naturally in place of Sec in selenoproteins of animals. To resolve this issue, TR1 and TR3 were purified from livers of mice that had been fed selenium deficient (0 ppm selenium), selenium adequate (0.1 ppm selenium), or selenium enriched (2.0 ppm selenium) diets (13). MS analysis of TR1 and TR3 showed that the ratio of Sec:Cys was ∼1:1 in mice maintained on a selenium deficient diet and this ratio changed to ∼9:1 in mice receiving a selenium adequate diet, whereas Cys was not detected in these selenoproteins on a selenium enhanced diet. In NIH 3T3 cells, the ratio of Sec:Cys was 9:1 in control cells and 1:24 in cells treated with thiophosphate (17). Thus, Cys is inserted into normally selenium containing proteins in vivo that occurs by sulfide competing with selenide in generating the active donor catalyzed by SPS2. The active donor, thiophosphate, interacts with the acceptor molecule generated from O-phosphoseryl-tRNA[Ser]Sec by SecS, wherein SecS in turn converts to Cys-tRNA[Ser]Sec. In mammals, de novo synthesis of Cys on tRNA[Ser]Sec is shown in Figure 1.
Conclusions
A computational, comparative genomics approach was used to identify a single kinase that had close homologs only in those archaea and eukaryotes that encode the Sec protein insertion machinery. This protein was confirmed to be PSTK by biochemical and genetic studies (15). Furthermore, a similar comparative genomics approach was used to predict SLA as the likely SecS that was indeed confirmed to serve this function (12). With few exceptions, orthologs of both SLA and PSTK were found in eukaryotes and archaea that utilize the Sec protein insertion machinery, but not in organisms that do not, suggesting that this pathway is used by all eukaryotes and archaea that synthesize selenoproteins (12, 15). Establishing the mechanism by which selenium makes its way into proteins demonstrates how this essential element becomes a component of Sec that is incorporated into selenoproteins as the 21st amino acid in the genetic code in eukaryotes and archaea. Many health benefits of selenium have been reported that include roles in decreasing incidence of cancer and heart disease, development and immune function, and delaying the aging process and the onset of AIDS in HIV positive patients. Selenoproteins are likely the components responsible for most of these health benefits.
Cys, an essential amino acid in mammals, was also found to be synthesized de novo by replacing Sec in the synthesis of selenium containing proteins. The precise means of how this synthesis occurs was determined in vitro and the presence of Cys in place of Sec was shown to occur in vivo in both cells in culture and in mice. In addition to establishing the pathway of Sec biosynthesis, the replacement of Sec with Cys provides unique possibilities for regulating the expression of selenoproteins and their functions as well as elucidating the biological roles of dietary selenium.
Acknowledgments
All authors analyzed data, wrote the paper, and had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Supported by the Intramural Research Program of the NIH, National Cancer Institute, and Center for Cancer Research (to D.L.H.) and by NIH grants awarded to V.N.G.
Author disclosures: A. Turanov, X-M. Xu, B. A. Carlson, M-H. Yoo, V. N. Gladyshev, and D. L. Hatfield, no conflicts of interest.
Present address: GeneCopoeia, Inc., Rockville, MD 20850.
Abbreviations used: SECIS, Sec insertion sequence; Um34, methyl group located on the 2′-hydroxylribose at wobble position of selenocysteine tRNA.
Literature Cited
- 1.Birringer M, Pilawa S, Flohe L. Trends in selenium biochemistry. Nat Prod Rep. 2002;19:693–718 [DOI] [PubMed] [Google Scholar]
- 2.Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40 [DOI] [PubMed] [Google Scholar]
- 3.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Gladyshev VN. General trends in trace element utilization revealed by comparative genomic analyses of Co, Cu, Mo, Ni, and Se. J Biol Chem. 2010;285:3393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Gladyshev VN. Comparative genomics of trace elements: emerging dynamic view of trace element utilization and function. Chem Rev. 2009;109:4828–61 [DOI] [PubMed] [Google Scholar]
- 6.Berry MJ, Banu L, Larsen PR. Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature. 1991;349:438–40 [DOI] [PubMed] [Google Scholar]
- 7.Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin GW III, Harney JW, Berry MJ. Selenocysteine incorporation in eukaryotes: insights into mechanism and efficiency from sequence, structure, and spacing proximity studies of the type 1 deiodinase SECIS element. RNA. 1996;2:171–82 [PMC free article] [PubMed] [Google Scholar]
- 9.Forchhammer K, Leinfelder W, Boesmiller K, Veprek B, Bock A. Selenocysteine synthase from Escherichia coli. Nucleotide sequence of the gene (selA) and purification of the protein. J Biol Chem. 1991;266:6318–23 [PubMed] [Google Scholar]
- 10.Forchhammer K, Boesmiller K, Bock A. The function of selenocysteine synthase and SELB in the synthesis and incorporation of selenocysteine. Biochimie. 1991;73:1481–6 [DOI] [PubMed] [Google Scholar]
- 11.Forchhammer K, Bock A. Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence. J Biol Chem. 1991;266:6324–8 [PubMed] [Google Scholar]
- 12.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu XM, Turanov AA, Carlson BA, Yoo MH, Everley RA, Nandakumar R, Sorokina I, Gygi SP, Gladyshev VN, et al. Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc Natl Acad Sci USA. 2010;107:21430–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass RS, Singh WP, Jung W, Veres Z, Scholz TD, Stadtman TC. Monoselenophosphate: synthesis, characterization, and identity with the prokaryotic biological selenium donor, compound SePX. Biochemistry. 1993;32:12555–9 [DOI] [PubMed] [Google Scholar]
- 15.Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl tRNA[Ser]Sec kinase. Proc Natl Acad Sci USA. 2004;101:12848–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maenpaa PH, Bernfield MR. A specific hepatic transfer RNA for phosphoserine. Proc Natl Acad Sci USA. 1970;67:688–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatfield D, Portugal FH. Seryl-tRNA in mammalian tissues: chromatographic differences in brain and liver and a specific response to the codon, UGA. Proc Natl Acad Sci USA. 1970;67:1200–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatfield D, Diamond AM, Dudock B. Opal suppressor serine tRNAs from bovine liver form phosphoseryl-tRNA. Proc Natl Acad Sci USA. 1982;79:6215–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee BJ, Worland PJ, Davis JN, Stadtman TC, Hatfield DL. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem. 1989;264:9724–7 [PubMed] [Google Scholar]
- 20.Gelpi C, Sontheimer EJ, Rodriguez-Sanchez JL. Autoantibodies against a serine tRNA-protein complex implicated in cotranslational selenocysteine insertion. Proc Natl Acad Sci USA. 1992;89:9739–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kernebeck T, Lohse AW, Grotzinger J. A bioinformatical approach suggests the function of the autoimmune hepatitis target antigen soluble liver antigen/liver pancreas. Hepatology. 2001;34:230–3 [DOI] [PubMed] [Google Scholar]
- 22.Allmang C, Krol A. Selenoprotein synthesis: UGA does not end the story. Biochimie. 2006;88:1561–71 [DOI] [PubMed] [Google Scholar]
- 23.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, et al. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26:2337–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog Nucleic Acid Res Mol Biol. 2006;81:97–142 [DOI] [PubMed] [Google Scholar]
- 25.Guimarães MJ, Peterson D, Vicari A, Cocks BG, Copeland NG, Gilbert DJ, Jenkins NA, Ferrick DA, Kastelein RA, et al. Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in selenocysteine metabolism? Proc Natl Acad Sci USA. 1996;93:15086–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim IY, Stadtman TC. Selenophosphate synthetase: detection in extracts of rat tissues by immunoblot assay and partial purification of the enzyme from the archaean Methanococcus vannielii. Proc Natl Acad Sci USA. 1995;92:7710–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim IY, Guimaraes MJ, Zlotnik A, Bazan JF, Stadtman TC. Fetal mouse selenophosphate synthetase 2 (SPS2): characterization of the cysteine mutant form overproduced in a baculovirus-insect cell system. Proc Natl Acad Sci USA. 1997;94:418–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Low SC, Harney JW, Berry MJ. Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J Biol Chem. 1995;270:21659–64 [DOI] [PubMed] [Google Scholar]
- 29.Kim TS, Yu MH, Chung YW, Kim J, Choi EJ, Ahn K, Kim IY. Fetal mouse selenophosphate synthetase 2 (SPS2): biological activities of mutant forms in Escherichia coli. Mol Cells. 1999;9:422–8 [PubMed] [Google Scholar]
- 30.Tamura T, Yamamoto S, Takahata M, Sakaguchi H, Tanaka H, Stadtman TC, Inagaki K. Selenophosphate synthetase genes from lung adenocarcinoma cells: Sps1 for recycling L-selenocysteine and Sps2 for selenite assimilation. Proc Natl Acad Sci USA. 2004;101:16162–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diamond A, Dudock B, Hatfield D. Structure and properties of a bovine liver. UGA suppressor serine tRNA with a tryptophan anticodon. Cell. 1981;25:497–506 [DOI] [PubMed] [Google Scholar]
- 32.Lee BJ, de la Peña P, Tobian JA, Zasloff M, Hatfield D. Unique pathway of expression of an opal suppressor phosphoserine tRNA. Proc Natl Acad Sci USA. 1987;84:6384–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatfield D, Gladyshev V, Park JM, Park SI, Chittum HS, Huh JR, Carlson BA, Kim M, Moustafa ME, et al. Biosynthesis of selenocysteine and its incorporation into protein as the 21st amino acid. : Kelly JF, editor Comprehensive natural products chemistry. New York: Elsevier Sc. Ltd; 1999. p. 353–80 [Google Scholar]
- 34.Diamond AM, Choi IS, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, et al. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec). J Biol Chem. 1993;268:14215–23 [PubMed] [Google Scholar]
- 35.Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, et al. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol Cell Biol. 2001;21:3840–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bösl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc Natl Acad Sci USA. 1997;94:5531–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumaraswamy E, Carlson BA, Morgan F, Miyoshi K, Robinson GW, Su D, Wang S, Southon E, Tessarollo L, et al. Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol Cell Biol. 2003;23:1477–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlson BA, Novoselov SV, Kumaraswamy E, Lee BJ, Anver MR, Gladyshev VN, Hatfield DL. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J Biol Chem. 2004;279:8011–7 [DOI] [PubMed] [Google Scholar]
- 39.Carlson BA, Yoo MH, Xu XM, Tsuji P, Tobe R, Naranjo-Suarez S, Gladyshev V, Hatfield DL. Mouse models that target removal or over-expression of the selenocysteine tRNA[Ser]Sec gene to elucidate the role of selenoproteins in health and development. : Hatfield DL, Berry MJ, Gladyshev VN, Selenium: its molecular biology and role in human health. New York: Springer; In press 2011 [Google Scholar]
- 40.Leinfelder W, Stadtman TC, Böck A. Occurrence in vivo of selenocysteyl-tRNA(SERUCA) in Escherichia coli. Effect of sel mutations. J Biol Chem. 1989;264:9720–3 [PubMed] [Google Scholar]
- 41.Amberg R, Mizutani T, Wu XQ, Gross HJ. Selenocysteine synthesis in mammalia: An identity switch from tRNA(Ser) to tRNA(Sec). J Mol Biol. 1996;263:8–19 [DOI] [PubMed] [Google Scholar]
- 42.Xu XM, Mix H, Carlson BA, Grabowski PJ, Gladyshev VN, Berry MJ, Hatfield DL. Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen, in the selenoprotein synthesis machinery. J Biol Chem. 2005;280:41568–75 [DOI] [PubMed] [Google Scholar]
- 43.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404:115–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alsina B, Corominas M, Berry MJ, Baguñà J, Serras F. Disruption of selenoprotein biosynthesis affects cell proliferation in the imaginal discs and brain of Drosophila melanogaster. J Cell Sci. 1999;112:2875–84 [DOI] [PubMed] [Google Scholar]
- 45.Shim MS, Kim JY, Jung HK, Lee KH, Xu XM, Carlson BA, Kim KW, Kim IY, Hatfield DL, et al. Elevation of glutamine level by selenophosphate synthetase 1 knockdown induces megamitochondrial formation in Drosophila cells. J Biol Chem. 2009;284:32881–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobanov AV, Hatfield DL, Gladyshev VN. Selenoproteinless animals: selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci. 2008;17:176–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrauzer GN. Selenomethionine: a review of its nutritional significance, metabolism and toxicity. J Nutr. 2000;130:1653–6 [DOI] [PubMed] [Google Scholar]
- 48.Lu J, Zhong L, Lönn ME, Burk RF, Hill KE, Holmgren A. Penultimate selenocysteine residue replaced by cysteine in thioredoxin reductase from selenium-deficient rat liver. FASEB J. 2009;23:2394–402 [DOI] [PMC free article] [PubMed] [Google Scholar]