Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1; EC 2.4.2.30) is an abundant nuclear enzyme, activated by DNA strand breaks to attach up to 200 ADP-ribose groups to nuclear proteins. As retroviral infection requires integrase-catalyzed DNA strand breaks, we examined infection of pseudotyped HIV type I in fibroblasts from mice with a targeted deletion of PARP-1. Viral infection is almost totally abolished in PARP-1 knockout fibroblasts. This protection from infection reflects prevention of viral integration into the host genome. These findings suggest a potential for PARP inhibitors in therapy of HIV type I infection.
Productive infection by the HIV type I (HIV-1) requires efficient integration of the viral genome into the host DNA (1–3). After HIV-1 enters susceptible host cells, the viral enzyme reverse transcriptase synthesizes a double-stranded DNA copy from the genomic HIV-1 RNA. The resulting HIV-1 DNA exists as part of a large preintegration complex. After nuclear entry of the preintegration complex, the HIV-1 DNA integrates into a host chromosome. The virion-associated viral enzyme integrase (IN) associates with the ends of the linear double-stranded viral DNA and catalyzes integration processes. At each end of the linear viral DNA molecule, IN removes two terminal 3′ nucleotides, exposing recessed 3′ hydroxyl groups. IN then catalyzes a nucleophilic attack of the recessed 3′ hydroxyl groups on phosphodiester bonds in each target cellular DNA strand. Each strand of the viral DNA becomes joined to cellular DNA, leaving a four- to six-base gap and a two-base mismatch at the end. Gap repair provides four to six base duplications of the target DNA at each host–virus DNA junction, completing formation of an integrated provirus. DNA-binding proteins have been proposed to accomplish the gap repair. On integration, the proviral DNA is maintained as part of the host genome, allowing productive infection.
Poly(ADP-ribose) polymerase-1 (PARP-1; EC 2.4.2.30) is a predominantly nuclear enzyme that occurs in several isoforms derived from distinct genes (for reviews, see refs. 4 and 5). PARP-1 has been implicated in the DNA repair process and thus might be a candidate to participate in HIV viral integration, especially because PARP-1 activation is initiated by DNA strand breaks (6, 7). PARP-1 catalyzes the attachment of branched chains of up to 200 ADP-ribose units to nuclear proteins, including PARP-1 itself. PARP-1 contains a DNA-binding domain with two zinc fingers that attach to DNA strand breaks. PARP-1 uses β-nicotinamide adenine dinucleotide (NAD+) as its substrate. Because PARP-1 is an abundant enzyme (2% of nuclear protein) and has a high turnover number, its activation can deplete cellular NAD+ levels (8–10). Cellular perturbations that lead to necrosis cause such “overactivation,” leading to NAD+ depletion. In efforts to resynthesize NAD+, ATP is depleted, and cells die from energy deficit (11, 12). The development of mice with targeted deletion of PARP-1, the major form of PARP in tissues, has clarified biological functions of PARP-1 (13–15). PARP-1 knockout (PARP-1−/−) mice are dramatically protected from tissue damage associated with vascular stroke (16, 17), myocardial ischemia (18), and pancreatic damage elicited by diabetes-inducing toxins (15, 19, 20). A role for PARP-1 in DNA repair processes is indicated by abnormalities in sister chromatid exchange in PARP-1−/− tissues (21, 22).
A role for PARP-1 in viral infection is suggested by findings that benzamide derivatives and benzopyrone analogues that inhibit PARP diminish retroviral infection in some (23–25) but not other studies (26). Antisense and dominant-negative constructs also diminish retroviral infection of cells (23). Benzamide derivatives are weak inhibitors of PARP, acting in the millimolar range, and can exert toxic effects of their own (27–29). Overexpression of antisense and dominant-negative constructs may produce nonspecific effects (for reviews, see refs. 30 and 31). To clarify the role of PARP-1 in retroviral integration, we have examined HIV-1 integration in cells from PARP-1−/− mice. We demonstrate a profound reduction of infection in PARP-1−/− fibroblasts and show that this stems from an inhibition of viral integration.
Materials and Methods
Cells.
Mouse embryonic fibroblasts (MEFs) derived from both PARP-1 wild-type (PARP-1+/+) and PARP-1−/− mice were kindly provided by Z. Q. Wang (Institute of Molecular Pathology, Vienna). MEFs were cultured at 37°C (5% CO2) in DMEM supplemented with 10% FBS/2 mM l-glutamine/penicillin (100 units/ml)/and streptomycin (100 μg/ml). MEFs were seeded in six-well plates at approximately 50% confluence before the infection. Cell numbers were similar for PARP-1−/− and PARP-1+/+ MEFs within a given experiment.
Virus Production and Infection.
HIV-1 infection of mouse cells was examined by using a replication-defective HIV-1 vector in which the env gene was disrupted. Pseudotyping with vesicular stomatitis virus protein was done essentially as described by Reiser and colleagues (32, 33). The HIV-1 vector was constructed from the reference NL4–3 proviral clone by removing the env sequence between the KpnI and NheI sites and inserting in frame a modified form of enhanced green fluorescent protein (EGFP) with a carboxyl-terminal KDEL sequence followed by a stop codon. Detailed characterization of this vector will be presented elsewhere (Y.Z. and R. F. Siliciano, unpublished data). In brief, Env plasmid DNA and HIV-EGFPΔE vector plasmid DNA were cotransfected into subconfluent human embryonic kidney 293T cells in T150 flasks by Lipofectamine 2000 (Life Technologies, Rockville, MD). The medium was replaced 12–14 h later. Virus stocks were harvested 60–65 h posttransfection, filtered through a 0.22-μm pore-size filter, and concentrated by ultracentrifugation at 60,000 × g for 1.7 h to ≈5 × 107 infectious units/ml. Concentrated virus was aliquoted and subsequently frozen at −80°C. Jurkat cells were infected with serially diluted virus for 48 h. Infected cells were analyzed by fluorescence-activated cell sorter (FACS) to determine the titer of virus. MEFs were split into six-well plates the day before infection to give approximately 50% confluence at the time of infection. Infections were performed with virus stock at a multiplicity of infection ≈1 in a total of 0.5 ml of DMEM. After 6 h at 37°C, 1.5 ml of DMEM was added, and the plates were incubated at 37°C for an additional 42 h. To analyze 72- and 120-h infections, MEFs were respectively split at 48 and 96 h.
Flow Cytometry.
MEFs for FACS were detached from the plate by using trypsin-EDTA (0.05% and 0.53 mM), washed twice with 1× PBS, and resuspended in 4% paraformaldehyde for 30 min on ice. After washing with 1× PBS, MEFs were subjected to FACS analysis, and MEFs expressing EGFP were quantitated by cellquest software (BD Immunocytometry Systems, San Jose, CA).
Assay for Integrated HIV-1.
HIV-1 integration assay was performed as described, with some modifications (34). Genomic DNA was isolated from MEFs infected with or without HIV-EGFPΔE for 48 h. All samples were diluted to the same final concentration. The region of the junction between repeat elements in the cellular DNA and the 5′ end of the integrated proviral DNA was amplified by using a B2, a mouse repeating genomic sequence (GenBank accession no. AF115851) PCR strategy with the second nested PCR. In the second nested PCR, the 25-cycle B2 PCR products were used as a template to amplify the integrated HIV-1 incrementally from 21 to 32 PCR cycles. PCR products were analyzed by gel electrophoresis (GAPDH) and Southern hybridization (HIV-1). The nucleotide sequences of the oligonucleotide primers used for HIV-1 DNA detection were derived from the nucleotide sequence of HIV-1JR-CSF (35). The nucleotide sequences of B2 were 5′-TTCACAACTCTCGGTGGATGGTGG-3′. Primers for the first PCR were B2 and M661. The second nested PCR primers were M667 and AA55, targeted toward the long terminal repeat (LTR) sequence, amplifying a 140-bp product. The sequences of oligonucleotides used for probe were 5′-GTGCTTCAAGTAGTGTGTGCCC-3′. The sequences of oligonucleotides used to amplify a region of GAPDH were 5′-AGTCTGATATCGCGGCCGCATGGTGAAGGTCGGTGTG-3′ and 5′-TGCCGTTGAATTTGCCGTGAG-3′.
Assay for HIV-1 Entry.
HIV-1 entry assay was performed as described, with some modifications (36). Genomic DNA was isolated from MEFs infected with or without HIV-EGFPΔE for 2 h. A virus stock was treated with DNase for 1 h to remove the possible contamination of viral-associated DNA before infection. All samples were diluted to the same final concentration. The amplified product resulting from the M667/M661 oligonucleotide pair was a 200-bp fragment. A pair of oligonucleotide primers complementary to the mouse GAPDH gene was used in some PCR analyses to normalize the total amount of cellular DNA present. The amplified product resulting in the 200-bp fragment was analyzed by electrophoresis on 2% agarose gels.
Results
Lack of HIV-1 Expression in PARP-1−/− MEFs Infected with HIV-EGFPΔE.
To examine the role of PARP-1 in HIV-1 infection, we used continuous lines of fibroblasts derived from PARP-1+/+ and PARP-1−/− mice. HIV-1 selectively infects human cells expressing CD4+ and appropriate coreceptors and will not infect mouse cells under normal circumstances. Therefore, we used a pseudotyped replication-defective HIV vector in which HIV-1 particles were pseudotyped with a vesicular stomatitis virus envelope glycoprotein. The pseudotyped virus has a broad host range, permitting the entry of HIV-1 into most mammalian as well as nonmammalian cells (37). To monitor HIV-1 infection, the envelope protein of HIV-1 was replaced with EGFP (HIV-EGFPΔE), disabling replication and permitting FACS analysis to study the requirements of HIV-1 integration and infection. EGFP was expressed only after the vector retrotranscribes and integrates its genome into the host chromosome.
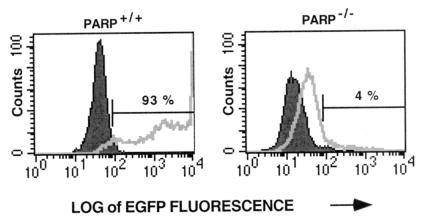
We exposed fibroblasts to pseudotyped HIV-EGFPΔE at a multiplicity of infection of ≈1. After 48 h of exposure, about 93% of PARP-1+/+ fibroblasts were infected (Fig. 1). By contrast, PARP-1−/− fibroblasts were almost completely protected from infection, with only about 4% of fibroblasts with HIV-1 gene expression.
Figure 1.
Analysis of the percentage of MEFs infected with HIV-EGFPΔE. Both PARP-1+/+ and knockout PARP-1−/− MEFs were infected with pseudotyped HIV-EGFPΔE vector stocks at a multiplicity of infection of ≈1. MEFs expressing EGFP were collected for FACS analysis after 48 h infection. About 93% of PARP-1+/+ fibroblasts were infected. By contrast, PARP-1−/− fibroblasts were almost completely protected from infection, with only about 4% of fibroblasts expressing HIV-1 genes. Cells with increased EGFP fluorescence intensity were indicative of HIV-1 infection. This analysis of infection was a representative of multiple experimental determinations.
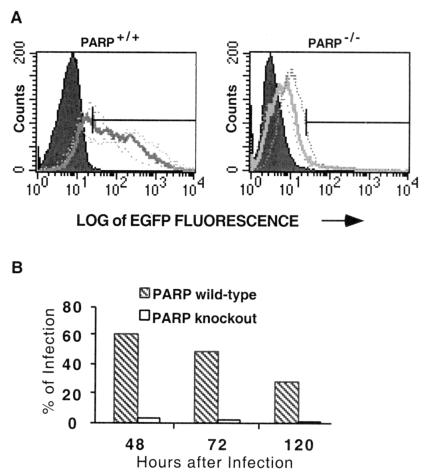
To determine whether the absence of PARP-1 abolishes HIV-1 infection or merely causes a delay, we examined multiple time points (Fig. 2A). We confirmed the differences in infection between PARP-1+/+ and PARP-1−/− fibroblasts at 48 h with 60% of PARP-1+/+ and 3% of PARP-1−/− cells, respectively, infected. Cells were split 48 and 72 h after infection to analyze 96- and 120-h time points, respectively. After passage, expression of EGFP remained high in PARP-1+/+ cells and low in PARP-1−/− cells. At 72 and 120 h, infection in PARP-1+/+ fibroblasts declined to 49 and 29%, respectively. The decline in infection over time presumably reflected dilution of the cells that were infected with replication-defective HIV-1 vectors. The virtual absence of infection in PARP-1−/− cells was also observed at 72 and 120 h (Fig. 2B). Accordingly, infection was severely reduced and not merely delayed in the absence of PARP-1.
Figure 2.
Extended analysis of the percentage of MEFs infected with HIV-EGFPΔE. (A) Both PARP-1+/+ and PARP-1−/− MEFs were infected with pseudotyped HIV-EGFPΔE vector stocks at a multiplicity of infection of ≈1. MEFs expressing EGFP were collected for FACS analysis after 48 (… … ), 72 (▄▄▄), and 120 (.. .) h infection. MEFs were split 48 h after infection to analyze 72-h infection and again split 96 h after infection to analyze 120-h infection. (B) The relative percentage of infection between PARP-1+/+ and PARP-1−/− MEFs after 48, 72, and 120 h was also shown in the bar graph. At 48 h, about 60% of PARP-1+/+ cells were infected, whereas only 3% of PARP-1−/− cells were infected. After passage, the expression of EGFP remained high in PARP-1+/+ cells and low in PARP-1−/− cells. At 72- and 120-h infection, infection in PARP-1+/+ fibroblasts declined to 49 and 29%, respectively. However, the virtually total loss of infection observed in PARP-1−/− cells was still evident at 72 and 120 h.
Lack of HIV-1 Integration in PARP-1−/− MEFs Infected with HIV-EGFPΔE.
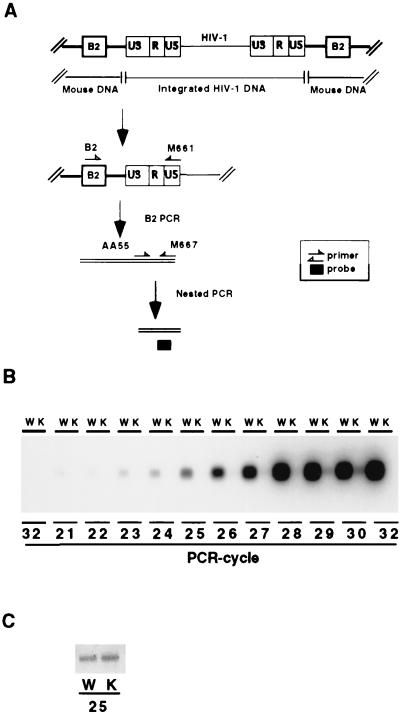
How might PARP-1 influence viral infection? The three principal processes involved in determining infection are viral entry into cells, viral integration into the host genome, and transcription of HIV-1 genome products. To differentiate among these processes, we developed an assay to monitor viral integration (Fig. 3A). We used a mouse form of a repeating genomic sequence, designated B2, which was analogous to the human Alu sequence (38, 39). We conducted B2 PCR, by using B2 and HIV primers, which selectively and unambiguously amplified the integrated form of HIV-1. Subsequently, we conducted a second nested PCR by using M667 and AA55 primers to permit amplification of a portion of the LTR region of HIV-1. We also used an incremental PCR, ranging from 21 to 32 cycles, to ensure detection of integrated HIV-1 quantitatively without overamplification. We observed amplified HIV-1 DNA in a PCR-cycle-dependent manner in PARP-1+/+ fibroblasts (Fig. 3B). Virtually no HIV-1 integration occurred in PARP-1−/− fibroblasts. At 29 and 30 cycles, some signal was evident for PARP-1−/− cells, which might represent amplification of unintegrated form by the second PCR. As a control, we evaluated genomic GAPDH DNA. At 25 PCR cycles, the product of PCR was identical in PARP-1+/+ and PARP-1−/− cells (Fig. 3C), indicating that the total amount of cellular DNA used for PCR amplification was the same.
Figure 3.
Selective assay for integrated HIV-1 DNA in MEFs infected with HIV-EGFPΔE. (A) B2 PCR assay for integrated HIV-1 DNA in mouse-derived cells. DNA was isolated from infected cells and amplified by PCR with the B2 and M661 primers. By using the first B2 PCR product as a template, the second nested PCR amplification was performed to detect the portion of the LTR region of HIV-1 DNA. Amplified products resulting from the PCR were analyzed by Southern analysis with internal probes. (B) Quantitative analysis of integration of HIV-1 in infected MEFs with HIV-EGFPΔE. Both PARP-1+/+ (W) and PARP-1−/− (K) MEFs were infected for 48 h, after which time DNA was isolated. After 25 cycles of B2 PCR to detect integrated HIV-1 DNA, the second nested PCR was performed to focus selectively on the LTR region of HIV-1 DNA. To detect integrated HIV-1 sequence quantitatively without overamplification, the second nested PCR was amplified incrementally from 21 to 32 cycles (deleting cycle 31). The first two lanes with 32 PCR cycles constituted the water control in PARP-1+/+ and PARP-1−/− MEFs. In PARP-1+/+ fibroblasts, HIV-1 DNA was amplified in a PCR-cycle-dependent manner. There was virtually complete absence of HIV-1 integration in PARP-1−/− fibroblasts. At 29 and 30 cycles, some signal was evident for PARP-1−/− cells. The odd-numbered lanes represent PARP-1+/+ MEFs, and the even-numbered lanes are PARP-1−/− MEFs. (C) The same sample was also amplified for 25 PCR cycles to detect GAPDH, normalizing for total cellular DNA. At 25 PCR cycles, the product of PCR was identical in PARP-1+/+ and PARP-1−/− cells, indicating that the total amount of cellular DNA used for PCR amplification was the same.
Normal HIV-1 Entry in PARP-1−/− MEFs Infected with HIV-EGFPΔE.
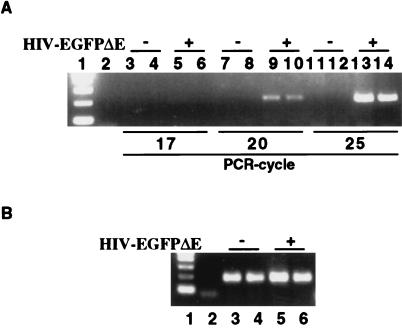
As differential entry of virus in PARP-1+/+ and PARP-1−/−, fibroblasts could lead to apparent differences in integration, we evaluated viral entry by monitoring HIV-1 DNA at 2 h after HIV-1 infection. To detect HIV-1 DNA, MEFs were infected with HIV-EGFPΔE for 2 h, permitting the viral genomic RNA to be reverse transcribed and yielding a double-stranded DNA copy of the viral RNA genome. In quantitative analysis of PCR from 17 to 25 cycles, equal amounts of HIV-1 DNA were detected in HIV-EGFP-infected PARP-1+/+ and PARP-1−/− MEFs. No HIV-1 DNA was detected in noninfected PARP-1+/+ and PARP-1−/− MEFs and in the water-control PCR (Fig. 4A). Also, at 20 PCR cycles, GAPDH DNA was equally amplified (Fig. 4B), indicating equal amounts of cellular DNA in PCR samples. Thus, we observed no difference in viral entry between PARP-1+/+ and PARP-1−/− fibroblasts. Accordingly, we concluded that PARP-1 was selectively required for HIV-1 viral integration into the host genome.
Figure 4.
PCR analysis of HIV-1 DNA in MEFs infected with HIV-EGFPΔE. HIV-1 and cellular GAPDH from infected MEFs were detected by PCR. (A) MEFs were infected with HIV-EGFPΔE for 2 h, after which time DNA was isolated. To detect HIV-1 sequence quantitatively without overamplification, the HIV-1 sequence was amplified for 17, 20, and 25 PCR cycles. Lane 1 represented the 1 kb DNA ladder marker. Lane 2 represented the water control with no DNA at 22 PCR cycles. Lanes 3, 4, 7, 8, 11, and 12 represented MEFs infected with HIV-EGFPΔE for 0 min, where cells were infected with HIV-EGFPΔE but immediately washed three times with 1 × PBS. Lanes 5, 6, 9, 10, 13, and 14 represented cells infected with HIV-EGFPΔE for 2 h. Lanes 3 ≈ 6 represented 17 PCR cycles, 7 ≈ 10, 20 PCR cycles, and 11 ≈ 14, 25 PCR cycles. The odd-numbered lanes represented PARP-1+/+ MEFs, whereas even-numbered lanes represented PARP-1−/− MEFs, except lanes 1 and 2. In quantitative analysis of PCR from 17 to 25 cycles, equal amounts of HIV-1 DNA were detected only in HIV-EGFP-infected PARP-1+/+ and PARP-1−/− MEFs. There was no HIV-1 DNA in the control PCR. (B) The same sample was also amplified for 20 PCR cycles to detect GAPDH, normalizing for total cellular DNA. Lane 1 represented 1-kb DNA ladder and lane 2, water control, lanes 3 and 5, PARP-1+/+ MEFs, and lanes 4 and 6, PARP-1−/− MEFs. Lanes 3 and 4 represented MEFs infected with HIV-EGFPΔE for 0 min and lanes 5 and 6, for 2 h. At 20 PCR cycles, GAPDH DNA was equally amplified, indicating equal amounts of cellular DNA in PCR samples. Amplified PCR products were analyzed by an ethidium-bromide-stained 2% agarose gel.
Discussion
The principal finding of this study is that targeted deletion of PARP-1 in mouse fibroblasts abolishes infection by pseudotyped HIV-1 vector and does this by preventing viral integration. IN catalyzes a nucleophilic attack of the recessed 3′ hydroxyl groups to phosphodiester bonds on each target cellular DNA strand. Each strand of the viral DNA becomes joined to cellular DNA, leaving a four- to six-base gap and a two-base mismatch at the end. Cellular DNA-binding proteins have been proposed to accomplish the gap repair. DNA-dependent protein kinase has been implicated as a host factor that completes retroviral integration (40). Reduced infection with massive cell death is observed in infected cell lines defective in the catalytic subunit of DNA-dependent protein kinase. At low viral titers, these cell lines are susceptible to infection, suggesting that DNA-dependent protein kinase is not critical for viral integration but somehow protects against cellular toxicity induced by high viral titers (26). Our findings establish that PARP-1 is required for the integration pathway.
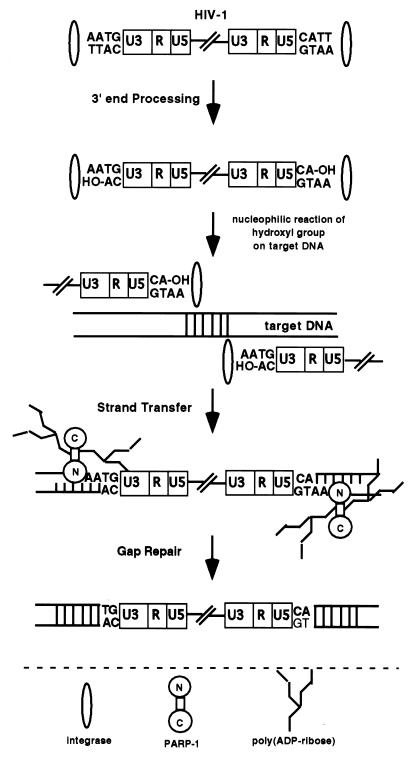
How does PARP-1 regulate HIV-1 integration? An abundant nuclear enzyme PARP-1 contains two zinc fingers that attach to the end of DNA strand breaks. On the basis of the known actions of PARP-1, it might bind the four- to six-base gap produced by the IN leading to PARP-1 activation. Activated PARP-1 synthesizes poly(ADP-ribose), which modifies histones, resulting in chromatin relaxation or decondensation. PARP-1 itself is also modified by poly(ADP-ribose), leading to its dissociation from the gap. These processes may mediate repair of the gap by facilitating access of the repair enzymes to the gap sites, allowing completion of the integration process. This resolution yields four to six base duplications of the target DNA at each end of the host–virus DNA junctions, completing the formation of an integrated provirus (Fig. 5).
Figure 5.
Possible role of PARP-1 in mediating HIV-1 integration. IN catalyzes 3′ end processing by removing two bases, leaving a hydroxyl group. IN then catalyzes a nucleophilic reaction of the hydroxyl group on the target cellular DNA. This strand-transfer reaction leaves a four- to six-base gap. PARP-1 recognizes and binds this gap through its two zinc fingers, leading to activation of PARP-1. Activated PARP-1 synthesizes poly(ADP-ribose). Modification of histones by poly(ADP-ribose) leads to chromatin decondensation. Autopoly(ADP-ribosyl)ation of PARP-1 leads to its dissociation from the gap. These processes may then repair the gap by facilitating access of the repair enzymes to the gap site. Without PARP-1, this gap is not resolved, leading to nonproductive infection. It is also possible that PARP-1 is involved in the initial integration reaction, as suggested by the B2 PCR results. N and C stand for the amino- and carboxyl-terminal domains of PARP-1, respectively.
Our observations that PARP-1 is required for HIV-1 infection are supported by the ability of PARP antisense and dominant-negative constructs to block retroviral infection (23). In studies of PARP inhibitors, some benzamide derivatives and benzopyrone analogues block retroviral replication (23–25), but one benzamide derivative fails to inhibit lentivirus vector-mediated transduction or HIV-1 replication (26). In our preliminary experiments, the effects of PARP inhibitors on HIV replication are inconclusive. These inconclusive effects of PARP inhibitors may indicate that viral integration requires only minimal PARP activity, so that prevention of viral infection demands virtually complete inhibition of enzyme activity.
PARP-1 may also affect HIV-1 transcription. However, we could not directly evaluate this possibility, as PARP-1 deletion abolishes integration, precluding an analysis of transcriptional activity. A possible interaction of PARP-1 and HIV-1 transcription might involve NF-κB, which enhances transcriptional activity by binding to the LTR of HIV-1 (41, 42). Transcription of NF-κB is markedly reduced in PARP-1−/− fibroblasts (43, 44). PARP-1 could increase HIV-1 transcription by facilitating actions of NF-κB (45, 46). Additional evidence suggesting a role of PARP-1 in transcription comes from studies demonstrating a coactivator role for PARP-1 in regulating transcription of the HTLV-1 TAX protein (47). Another link of PARP-1 to HIV-1 transcription involves Tat, an HIV-1-encoded transcriptional activator that is essential for viral replication (48, 49). PARP-1 can ADP-ribosylate the Tat protein in vitro (45).
Might the requirement of PARP-1 for HIV-1 integration have therapeutic consequences? Drugs that inhibit HIV-1 IN prevent HIV-1 infection in cells (50). The dramatic reduction of HIV-1 infection in PARP-1−/− MEFs suggests that PARP-1 may also be a therapeutic target. Recently, potent and selective inhibitors of PARP-1 have been developed (for review, see ref. 51). These agents inhibit PARP activity in intact mammals and are therapeutic in animal models of stroke (16, 17), myocardial infarction (18), diabetes (15, 19, 20), sepsis (44, 52), and inflammation (for review, see ref. 53). Such PARP inhibitors may prove useful in the treatment of HIV-1 infection.
Acknowledgments
We thank Drs. Robert F. Siliciano and Janet D. Siliciano for valuable assistance and advice. We also thank Theodore C. Pierson, Xuefei Shen, and Christopher B. Buck for helpful suggestions. This work was supported by U. S. Public Health Service Grants DA00266 (S.H.S.) and Research Scientist Award DA00074 (S.H.S.) Under an agreement between the Johns Hopkins University and Guilford, S.H.S. is entitled to a share of sales royalties related to PARP received by the University from Guilford. The University owns stock in Guilford, with S.H.S. having an interest in the University Share under University policy. S.H.S. serves on the Board of Directors and the Scientific Advisory Board of Guilford, is a consultant to the company, and owns additional equity in Guilford. This arrangement is being managed by the University in accordance with its conflict-of-interest policies.
Abbreviations
- PARP-1
poly(ADP-ribose) polymerase-1
- PARP-1+/+
PARP-1 wild type
- PARP-1−/−
genetic deficiency of PARP-1
- NAD+
β-nicotinamide adenine dinucleotide
- HIV-1
HIV type I
- IN
integrase
- LTR
long terminal repeat
- MEFs
mouse embryonic fibroblasts
- FACS
fluorescence-activated cell sorter
- EGFP
enhanced green fluorescent protein
- HIV-EGFPΔE
envelope protein of HIV-1 replaced with EGFP
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Farnet C M, Bushman F D. AIDS. 1996;10:S3–S11. [PubMed] [Google Scholar]
- 2.Goodarzi G, Im G J, Brackmann K, Grandgenett D. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulkosky J, Skalka A M. Pharmacol Ther. 1994;61:185–203. doi: 10.1016/0163-7258(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 4.D'Amours D, Desnoyers S, D'Silva I, Poirier G G. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 5.Ha H C, Snyder S H. Neurobiol Dis. 2000;7:225–239. doi: 10.1006/nbdi.2000.0324. [DOI] [PubMed] [Google Scholar]
- 6.Lautier D, Lagueux J, Thibodeau J, Menard L, Poirier G G. Mol Cell Biochem. 1993;122:171–193. doi: 10.1007/BF01076101. [DOI] [PubMed] [Google Scholar]
- 7.de Murcia G, Schreiber V, Molinete M, Saulier B, Poch O, Masson M, Niedergang C, Menissier de Murcia J. Mol Cell Biochem. 1994;138:15–24. doi: 10.1007/BF00928438. [DOI] [PubMed] [Google Scholar]
- 8.Oleinick N L, Evans H H. Radiat Res. 1985;101:29–46. [PubMed] [Google Scholar]
- 9.Berger N A. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 10.Carson D A, Seto S, Wasson D B, Carrera C J. Exp Cell Res. 1986;164:273–281. doi: 10.1016/0014-4827(86)90028-5. [DOI] [PubMed] [Google Scholar]
- 11.Ha H C, Snyder S H. Proc Natl Acad Sci USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herceg Z, Wang Z Q. Mol Cell Biol. 1999;19:5124–5133. doi: 10.1128/mcb.19.7.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 14.de Murcia J M, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver F J, Masson M, Dierich A, LeMeur M, et al. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masutani M, Suzuki H, Kamada N, Watanabe M, Ueda O, Nozaki T, Jishage K, Watanabe T, Sugimoto T, Nakagama H, et al. Proc Natl Acad Sci USA. 1999;96:2301–2304. doi: 10.1073/pnas.96.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endres M, Wang Z Q, Namura S, Waeber C, Moskowitz M A. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Eliasson M J, Sampei K, Mandir A S, Hurn P D, Traystman R J, Bao J, Pieper A, Wang Z Q, Dawson T M, Snyder S H, Dawson V L. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 18.Zingarelli B, Salzman A L, Szabo C. Circ Res. 1998;83:85–94. doi: 10.1161/01.res.83.1.85. [DOI] [PubMed] [Google Scholar]
- 19.Burkart V, Wang Z Q, Radons J, Heller B, Herceg Z, Stingl L, Wagner E F, Kolb H. Nat Med. 1999;5:314–319. doi: 10.1038/6535. [DOI] [PubMed] [Google Scholar]
- 20.Pieper A A, Brat D J, Krug D K, Watkins C C, Gupta A, Blackshaw S, Verma A, Wang Z Q, Snyder S H. Proc Natl Acad Sci USA. 1999;96:3059–3064. doi: 10.1073/pnas.96.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison C, Smith G C, Stingl L, Jackson S P, Wagner E F, Wang Z Q. Nat Genet. 1997;17:479–482. doi: 10.1038/ng1297-479. [DOI] [PubMed] [Google Scholar]
- 23.Gaken J A, Tavassoli M, Gan S U, Vallian S, Giddings I, Darling D C, Galea-Lauri J, Thomas M G, Abedi H, Schreiber V, et al. J Virol. 1996;70:3992–4000. doi: 10.1128/jvi.70.6.3992-4000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole G A, Bauer G, Kirsten E, Mendeleyev J, Bauer P I, Buki K G, Hakam A, Kun E. Biochem Biophys Res Commun. 1991;180:504–514. doi: 10.1016/s0006-291x(05)81093-9. [DOI] [PubMed] [Google Scholar]
- 25.Kameoka M, Tanaka Y, Ota K, Itaya A, Yoshihara K. Biochem Biophys Res Commun. 1999;262:285–289. doi: 10.1006/bbrc.1999.1146. [DOI] [PubMed] [Google Scholar]
- 26.Baekelandt V, Claeys A, Cherepanov P, De Clercq E, De Strooper B, Nuttin B, Debyser Z. J Virol. 2000;74:11278–85. doi: 10.1128/jvi.74.23.11278-11285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milam K M, Cleaver J E. Science. 1984;223:589–591. doi: 10.1126/science.6420886. [DOI] [PubMed] [Google Scholar]
- 28.Milam K M, Thomas G H, Cleaver J E. Exp Cell Res. 1986;165:260–268. doi: 10.1016/0014-4827(86)90550-1. [DOI] [PubMed] [Google Scholar]
- 29.Morris S M, Heflich R H. Mutat Res. 1984;126:63–71. doi: 10.1016/0027-5107(84)90170-2. [DOI] [PubMed] [Google Scholar]
- 30.Bennett C F. Biochem Pharmacol. 1998;55:9–19. doi: 10.1016/s0006-2952(97)00214-1. [DOI] [PubMed] [Google Scholar]
- 31.Summerton J, Weller D. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 32.Reiser J, Harmison G, Kluepfel-Stahl S, Brady R O, Karlsson S, Schubert M. Proc Natl Acad Sci USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mochizuki H, Schwartz J P, Tanaka K, Brady R O, Reiser J. J Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Pescador R, Power M D, Barr P J, Steimer K S, Stempien M M, Brown-Shimer S L, Gee W W, Renard A, Randolph A, Levy J A, et al. Science. 1985;227:484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- 36.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 37.Bartz S R, Vodicka M A. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 38.Bollag R J, Crawford K B, Stadt H, Kumiski D, Zdanowicz M, Baptista C, Herlea V, Kirby M L. Exp Cell Res. 1999;248:75–78. doi: 10.1006/excr.1999.4401. [DOI] [PubMed] [Google Scholar]
- 39.Kass D H, Kim J, Rao A, Deininger P L. Genetica. 1997;99:1–13. doi: 10.1007/BF02259494. [DOI] [PubMed] [Google Scholar]
- 40.Daniel R, Katz R A, Skalka A M. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 41.Nabel G, Baltimore D. Nature (London) 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 42.Nabel G J, Rice S A, Knipe D M, Baltimore D. Science. 1988;239:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- 43.Hassa P O, Hottiger M O. Biol Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 44.Oliver F J, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet J C, de Murcia G. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kameoka M, Tanaka Y, Ota K, Itaya A, Yamamoto K, Yoshihara K. Biochem Biophys Res Commun. 1999;261:90–94. doi: 10.1006/bbrc.1999.0964. [DOI] [PubMed] [Google Scholar]
- 46.Kameoka M, Ota K, Tetsuka T, Tanaka Y, Itaya A, Okamoto T, Yoshihara K. Biochem J. 2000;346:641–649. [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson M G, Scoggin K E, Simbulan-Rosenthal C M, Steadman J A. J Virol. 2000;74:2169–2177. doi: 10.1128/jvi.74.5.2169-2177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dayton A I, Sodroski J G, Rosen C A, Goh W C, Haseltine W A. Cell. 1986;44:941–947. doi: 10.1016/0092-8674(86)90017-6. [DOI] [PubMed] [Google Scholar]
- 49.Fisher A G, Feinberg M B, Josephs S F, Harper M E, Marselle L M, Reyes G, Gonda M A, Aldovini A, Debouk C, Gallo R C, et al. Nature (London) 1986;320:367–371. doi: 10.1038/320367a0. [DOI] [PubMed] [Google Scholar]
- 50.Hazuda D J, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler J A, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller M D. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 51.Szabo C, Dawson V L. Trends Pharmacol Sci. 1998;19:287–298. doi: 10.1016/s0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 52.Kuhnle S, Nicotera P, Wendel A, Leist M. Biochem Biophys Res Commun. 1999;263:433–438. doi: 10.1006/bbrc.1999.1393. [DOI] [PubMed] [Google Scholar]
- 53.Szabo C. Eur J Pharmacol. 1998;350:1–19. doi: 10.1016/s0014-2999(98)00249-0. [DOI] [PubMed] [Google Scholar]