Abstract
Translation initiation in bacteria involves a stochastic binding mechanism in which the 30S ribosomal subunit first binds either to mRNA or to initiator tRNA, fMet-tRNAfMet. Leaderless λ cI mRNA did not form a binary complex with 30S ribosomes, which argues against the view that ribosomal recruitment signals other than a 5′-terminal start codon are essential for translation initiation of these mRNAs. We show that, in Escherichia coli, translation initiation factor 2 (IF2) selectively stimulates translation of λ cI mRNA in vivo and in vitro. These experiments suggest that the start codon of leaderless mRNAs is recognized by a 30S–fMet-tRNAfMet–IF2 complex, an intermediate equivalent to that obligatorily formed during translation initiation in eukaryotes. We further show that leaderless λ cI mRNA is faithfully translated in vitro in both archaebacterial and eukaryotic translation systems. This suggests that translation of leaderless mRNAs reflects a fundamental capability of the translational apparatus of all three domains of life and lends support to the hypothesis that the translation initiation pathway is universally conserved.
Keywords: evolution/IF2/initiator tRNA/leaderless mRNA/translation initiation
Introduction
Formation of the bacterial translation initiation complex involves interactions between the 30S subunit, fMet-tRNAfMet and mRNA. Three initiation factors, IF1, IF2 and IF3, affect the kinetics of formation of the ternary complex by promoting selection of the correct start codon of the mRNA (for reviews see Gold, 1988; Gualerzi and Pon, 1990; Gualerzi et al., 2000).
IF2 selects fMet-tRNAfMet by virtue of the formylated α-amino group (Hartz et al., 1989; RajBhandary and Chow, 1995; Schmitt et al., 1996), and stimulates its positioning in the ribosomal P-site by accelerating the codon–anticodon interaction (Pon et al., 1985; Gualerzi and Wintermeyer, 1986; La Teana et al., 1996).
IF1 occupies the ribosomal A-site (Moazed et al., 1995); according to the molecular mimicry hypothesis (Brock et al., 1998) it may, with IF2, form a complex that mimics the aminoacyl-tRNA–EF-Tu–GTP complex. Alone or within this complex, IF1 could occlude access of the latter ternary complex to the ribosome until the initiation steps are completed.
IF3 is known to act as a fidelity factor in selection of the initiator tRNA by preventing binding of tRNAs other than initiator tRNA to the ribosomal P-site and discriminating against unusual start codons, such as AUU (Brombach and Pon, 1987; Butler et al., 1987; Hartz et al., 1989; La Teana et al., 1993; Sacerdot et al., 1996; Sussman et al., 1996). These IF3 activities are probably the result of a conformational change imposed on the 30S subunit (Pon and Gualerzi, 1974; de Cock et al., 1999).
Most of our knowledge about the mechanism of translation initiation stems from work carried out on canonical mRNAs comprising a 5′ untranslated region (UTR), which generally contains a Shine–Dalgarno (SD) sequence. Nevertheless, leaderless mRNAs starting directly with a 5′-terminal AUG occur frequently in bacteria and other organisms (Janssen, 1993); it is not known whether the sequence of events characterizing translation initiation of ‘leadered’ mRNAs also applies to leaderless mRNAs. Several lines of evidence indicate that the initiation codon is the only constant, necessary and sufficient element of the translation initiation region (TIR) of both ‘leadered’ and leaderless mRNAs (Wu and Janssen, 1996, 1997). In fact, the 5′-UTR, including the SD sequence, is not essential for translation of canonical mRNAs (Calogero et al., 1988; Melançon et al., 1990), whereas the 5′-terminal start codon AUG is the only essential recognition element allowing translation of leaderless mRNAs (Wu and Janssen, 1996; Winzeler and Shapiro, 1997; van Etten and Janssen, 1998). We have recently shown that IF3 discriminates against 5′-terminal start codons and that high levels of this factor decrease the translational efficiency of a leaderless tetR–lacZ construct in vivo (Tedin et al., 1999). We interpreted these results as showing that, in the absence of the SD–anti-SD interaction, the ternary complex between leaderless mRNA, fMet-tRNAfMet and the 30S subunit cannot withstand the destabilization caused by IF3 (Moll et al., 1998; Tedin et al., 1999).
In the present study, we have examined the effect of IF2 on the recruitment of ribosomes to leaderless mRNAs. Our results demonstrate that ternary complex formation at 5′-terminal AUGs is stimulated by IF2 and that, in the presence of elevated concentrations of IF2, a leaderless mRNA can efficiently compete for translation with an mRNA containing a canonical ribosome binding site (rbs). Our results also suggest that the selection of the start codon of leaderless mRNAs is accomplished by ribosomes carrying the initiator tRNA bound in their P-site in the presence of IF2, through a mechanism resembling the obligatory translational initiation pathway in eukaryotes (Kozak, 1991; Merrick, 1992).
Results
30S subunit binding to leaderless λ cI mRNA depends on fMet-tRNAf Met
A large body of evidence demonstrates that an mRNA and the 30S ribosomal subunit can form binary complexes whose stability depends, to a large extent, on the presence or absence of a canonical SD sequence and on the extent of its complementarity with the anti-SD sequence of 16S rRNA (for reviews see Gualerzi and Pon, 1996; Gold, 1988; Gualerzi et al., 2000). Therefore, it seemed reasonable to test whether a leaderless mRNA has sufficient intrinsic affinity for the 30S ribosomal subunit to form a stable binary complex with it.
Nitrocellulose filter-binding assays were performed to compare the 30S binding capacity for leaderless cI34 mRNA with that for leadered cI34SD mRNA. As shown in Table I, one of these two model mRNAs starts with the 5′-terminal AUG and contains the first 34 nucleotides of λ cI mRNA, whereas the second contains the same coding sequence preceded by the upstream translation initiation signal(s) of phage T7 gene 10. When the mRNA was added to a 10-fold molar excess of 30S subunits (conditions in which the amount of each mRNA bound reflects the relative affinity constants of the corresponding complexes), ∼46% of cI34SD mRNA and 0.37% of cI34 mRNA formed a binary complex with 30S ribosomes (Table I). Thus, under these experimental conditions and in the absence of fMet-tRNAfMet, the affinity of the 30S subunit is over two orders of magnitude higher for the ‘leadered’ mRNA than for the leaderless mRNA. Upon addition of fMet-tRNAfMet, the amount of bound mRNA increased 5.3-fold for cI34 mRNA and 1.6-fold for cI34SD mRNA (Table I). These experiments showed that the immediate coding region downstream of the start codon of λ cI mRNA can hardly provide sufficient interactions with the 30S subunit to ensure the formation of a thermodynamically stable binary complex. In addition, in contrast to the cI34SD mRNA, the recognition of the leaderless cI34 mRNA by the ribosome depends apparently to a much larger extent on the presence of fMet-tRNAfMet.
Table I. Binding affinities of 30S ribosomes for c134 and c134SD mRNAs.
Both mRNAs are deplicted; the start codon(s) and the SD sequence are indicated by bars. The total c.p.m. values corrected for the number of Cs in each mRNA were set to 100% (=mRNA input). Ternary complexes were allowed to form in the presence of a 2-fold excess of fMet-tRNAfMet over ribosomes (see Materials and methods).
IF2 stimulates ternary complex formation on mRNAs with a canonical rbs and leaderless mRNA to a different degree
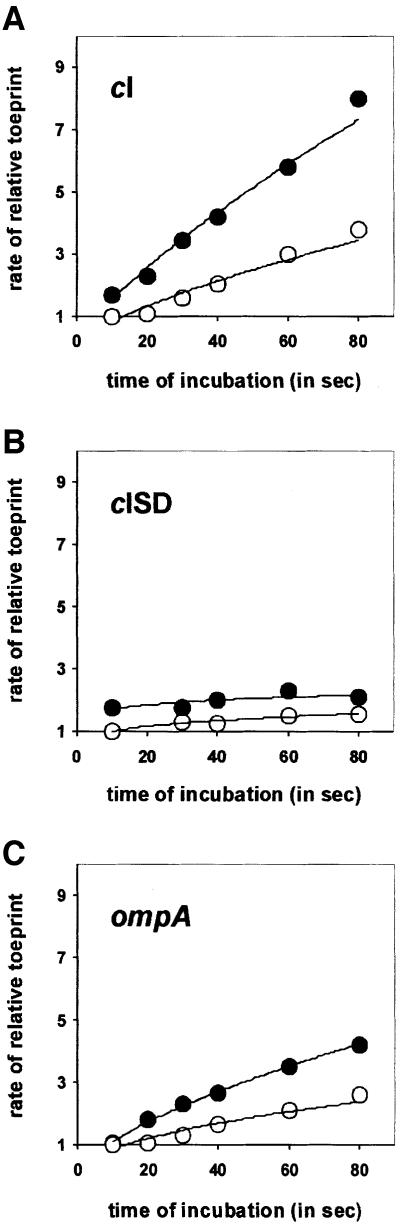
The results shown in Table I and those from our previous work (Resch et al., 1996) argued against the presence in the leaderless λ cI mRNA of particular cis elements, such as the downstream box sequence, which has been suggested to function in a manner analogous to that of the SD sequence (Sprengart and Porter, 1997). Translation initiation of canonical mRNAs entails a stochastic binding mechanism in which the ribosome first binds either to mRNA via SD–anti-SD interaction (Hartz et al., 1991) or to fMet-tRNAfMet (Jay and Kaempfer, 1974; Wu et al., 1996). Therefore, a reasonable assumption to explain the mechanism of translational start site selection on leaderless mRNAs was that the 5′-terminal start codon is recognized by a 30S–fMet-tRNAfMet complex. IF2 is known to stimulate both coded and non-coded binding of fMet-tRNAfMet to the 30S ribosomal P-site (La Teana et al., 1996). We therefore anticipated that the presence of IF2 would further increase ternary complex formation on ‘leadered’ as well as on leaderless mRNAs. However, since a 30S–initiator tRNA intermediary complex seemed to be instrumental for recognition of the start codon of a leaderless mRNA (see Table I), we expected the IF2 effect to be more pronounced for leaderless mRNAs. Ternary complex formation in the presence of IF2 was studied by kinetic toeprinting assays on leaderless λ cI mRNA, on cISD mRNA containing the 5′-terminal extension comprising the translational initiation region derived from phage T7 gene 10 (see Table I), and on Escherichia coli ompA mRNA containing a canonical rbs. Briefly, since a 30S subunit–initiator tRNA complex bound at the rbs can stop the progress of a downstream primed reverse transcriptase, toeprinting experiments are suited to test the efficiency of ternary complex formation at a given start codon (Hartz et al., 1988). In the experiment shown in Figure 1, fMet-tRNAfMet was incubated for 20 s with a 2.5-fold stoichiometric excess of 30S subunits, and, when present, with IF2. Then, either cI, cISD or ompA mRNA was added, the incubation was continued for various lengths of time (see Figure 1) and the samples were subjected to toeprinting analysis. Figure 1 shows the increase in the relative toeprints [toeprint signal/(readthrough signal + toeprint signal)] obtained for either mRNA in the presence or absence of IF2 plotted against incubation time. The relative toeprint signal obtained at 10 s was set to 1. The presence of IF2 resulted in an increase in ternary complex formation with both ‘leadered’ and leaderless mRNAs. However, the increase in ternary complex formation was more pronounced for leaderless mRNA. The presence of IF2 stimulated ternary complex formation on the leaderless mRNA ∼2.1-fold, whereas only 1.5- and 1.7-fold stimulation was observed for the cISD and ompA mRNA, respectively (Figure 1).
Fig. 1. Rate of relative toeprints obtained with leaderless cI mRNA (A), cISD mRNA (B) and ompA mRNA (C) in the presence (filled symbols) and absence (open symbols) of IF2. The kinetic toeprinting reactions contained 2 pmol of 30S subunits, 5 pmol of IF2 (when present), 0.8 pmol of fMet-tRNAfMet and 1 mM GTP, and were pre-incubated for 20 s before the mRNA(s) was added for the times (given in seconds) indicated. The toeprint reaction was then started with MMLV as described in Materials and methods.
The competition of leaderless λ cI mRNA with ompA mRNA for ribosomes in an in vitro translation system depends on IF2
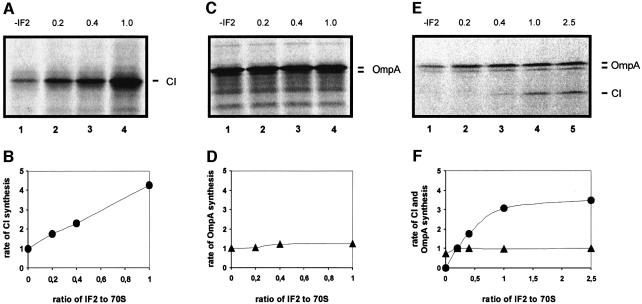
We repeatedly found using in vitro translation assays that leaderless cI mRNA cannot efficiently compete with SD-containing ‘leadered’ mRNAs for a limiting amount of ribosomes (data not shown). In the following in vitro experiments we used ompA mRNA containing a canonical rbs rather than cISD mRNA as a control because the ompA product, outer membrane protein A (OmpA), has an electrophoretic mobility different from that of CI protein. To quantify the effect of IF2, a defined E.coli in vitro translation system was utilized in which either cI mRNA (Figure 2A and B) or ompA mRNA (Figure 2C and D) was translated in the presence of increasing molar IF2:ribosome ratios. The increase in this ratio resulted in a concomitant increase in cI mRNA translation. Relative to the translation rate in the absence of exogenously added IF2, the increase was ∼4.5-fold at an IF2:ribosome molar ratio of 1:1 (Figure 2B). Under the same conditions, only 1.4-fold stimulation of ompA mRNA translation was observed (Figure 2C and D). In addition, we measured the rate of CI and OmpA synthesis as a function of increasing IF2:ribosome molar ratios using an in vitro translation system programmed with both cI and ompA mRNAs. When all three IFs were present at an IF:ribosome molar ratio of 0.2, translation of cI mRNA was hardly detected (Figure 2E) but, in contrast to that of ompA mRNA, translation of cI mRNA increased steadily with an increase in the IF2:ribosome ratio and reached a plateau when IF2 was in a 2.5-fold molar excess over ribosomes (Figure 2E and F). The maximum level of cI mRNA translation corresponded to an ∼5-fold stimulation over the translation rate obtained at an IF:ribosome ratio of 0.2 (Figure 2F). These translation competition assays demonstrated that the translational efficiency of the leaderless mRNA is selectively increased by IF2.
Fig. 2. Selective stimulation of CI synthesis by IF2. (A–D) In vitro translation of equimolar amounts of cI and ompA mRNA, respectively, with an E.coli S100 extract as described in Materials and methods. The different ratios of IF2 to added 70S ribosomes are indicated above the panels. The ratio of IF1 or IF3 to 70S was 0.2:1 in all reactions. The samples were loaded on a 12% SDS–polyacrylamide gel and the rate of CI and OmpA synthesis at different molar ratios of IF2 to 70S ribosomes was calculated by ImageQuant analysis (B and D). In the absence of exogenously added IF2 (A and C, lane 1), the CI and OmpA synthesis rate was set to 1. (E and F) In vitro translation competition assay with cI and ompA mRNA at different molar IF2:70S ribosome ratios. The conditions were as in (A–D), except that the in vitro translation system was programmed with equimolar amounts of the two mRNAs. The CI and OmpA synthesis rate is set to 1 at a 0.2:1 molar ratio of IF1, IF2 or IF3 to ribosomes (E, lane 2).
Influence of IF2 on the competition for 30S subunits between a 5 ′-terminal AUG and an internal AUG preceded by a canonical SD sequence
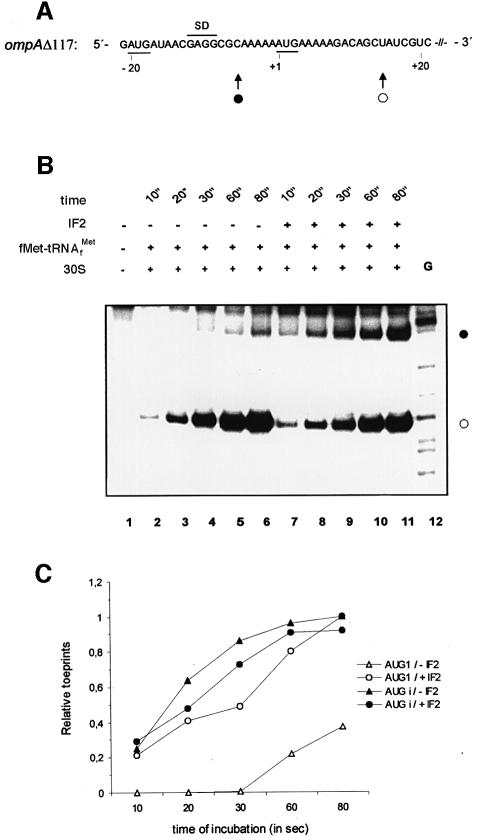
To study further the influence of IF2 on the choice of start codon by ribosomes (i.e. 5′-terminal versus internal), a kinetic toeprinting experiment was performed on ompAΔ117 mRNA, which contains a 5′-terminal AUG codon and a downstream AUG preceded by the canonical SD sequence of ompA mRNA (Figure 3A). The ribosomal choice of either AUG triplet as a start codon is mutually exclusive on this mRNA (Moll et al., 1998). The 30S subunits were pre-incubated for 20 s in a 2.5-fold stoichiometric excess over fMet-tRNAfMet and, when present, of IF2. Then, ompAΔ117 mRNA was added and incubation was continued for various lengths of time (see Figure 3B). Finally, the samples were subjected to toeprinting analysis. The electrophoretic pattern of the products obtained by reverse transcription (Figure 3B) and their quantification (Figure 3C) showed that, in the absence of IF2, the ternary complex readily formed at the internal AUG codon (AUGi) and increased steadily with incubation time. In contrast, after 30 s incubation, hardly any toeprint signal was visible for the 5′-terminal AUG (AUG1), and even after 80 s the intensity of the signal for AUG1 was only 20% of that obtained for AUGi. However, a different picture was observed in the presence of IF2. Under these conditions, the 5′-terminal AUG1 was able to compete efficiently with AUGi, its toeprint signal being ∼40% of that for AUGi at 20 s and reaching almost the same intensity after 80 s. These data demonstrated that IF2 shifted the 30S subunits to form a ternary complex at AUG1 at the expense of AUGi. Taken together with the results shown in Figure 2E and F, these data support the view that, in the presence of IF2, a 5′-terminal AUG can efficiently compete for 30S subunits with a canonical initiation site.
Fig. 3. Selection of the 5′-terminal and the internal AUG on ompAΔ117 mRNA in the presence of IF2. (A) Depiction of the ompAΔ117 mRNA. The start codon(s) and the authentic SD sequence of ompA mRNA are indicated by bars. For ompAΔ117, two toeprint signals are obtained, which result from formation of a ternary complex over AUG1 (nucleotide –20 to –18) and AUGi (nucleotide 1–3), which represents the authentic start codon of ompA mRNA. The positions of the toeprint signals are indicated by filled (AUG1) and open circles (AUGi). (B) Kinetic toeprints on ompAΔ117 mRNA in the presence and absence of IF2. The final concentration of 30S subunits, IF2, fMet-tRNAfMet and ompAΔ117 mRNA was 0.5, 1, 0.2 and 0.005 pmol/µl, respectively. Lane 1, primer extension in the absence of 30S subunits and fMet-tRNAfMet. Lanes 2–6, kinetic toeprint analysis in the absence of IF2. Lanes 7–11, kinetic toeprint analysis in the presence of IF2. 30S subunits, fMet-tRNAfMet, IF2 and mRNA were incubated for different times (given in seconds) as indicated at the top. Lane 12, G sequencing reaction. The toeprinting signals obtained for AUG1 and AUGi are indicated by a filled and open circle, respectively. (C) Relative toeprints on ompAΔ117 mRNA for AUG1 and AUGi. The relative toeprints at either start codon obtained in the absence of IF2 were normalized to that of AUG1 and AUGi obtained in the presence of IF2 after 80 s of incubation.
Increased intracellular IF2 concentrations stimulate translation of a leaderless cI–lacZ mRNA
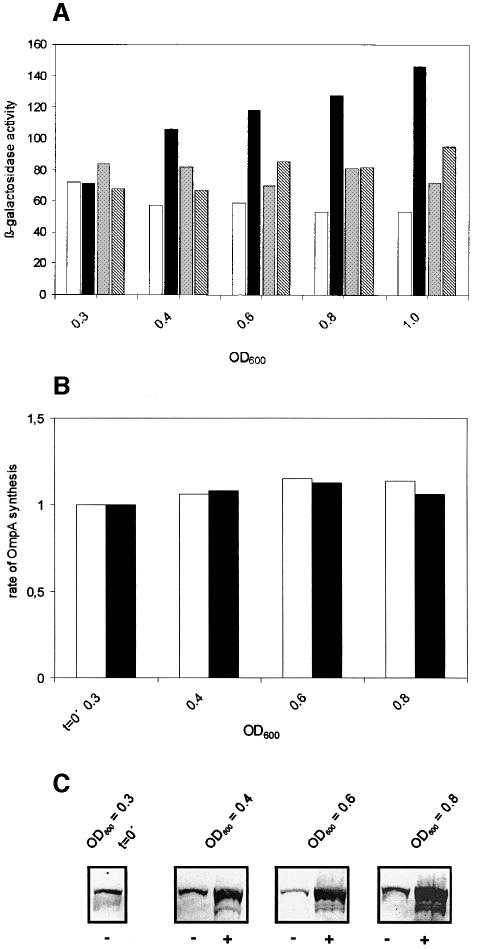
We next asked whether an artificially increased intracellular concentration of IF2 would result in a selective advantage for the translation of a leaderless mRNA in vivo. Escherichia coli strain UB89-1 harboring pKTplaccI, a plasmid encoding a leaderless cI–lacZ gene, was co-transformed with pinfB, which carries the infB (IF2) gene under the control of the λpL promoter. As shown in Figure 4A and C, upon induction of infB overexpression, the rate of translation of the cI–lacZ gene increased ∼2.5-fold above the basal level. In the control strain transformed with the parental plasmid pLc2833, the cI–lacZ gene was expressed with approximately the same efficiency at 28 and 42°C. In contrast, the expression of E.coli ompA mRNA (Figure 4B) and other control mRNAs (not shown) that contained a canonical rbs did not significantly increase with higher concentrations of IF2.
Fig. 4. In vivo expression of cI–lacZ and ompA in the presence of elevated concentrations of IF2. (A) Increased intracellular concentrations of IF2 stimulate cI–lacZ expression. β-galactosidase measurements were performed at various times after heat induction of the infB gene. White and black bars represent the β-galactosidase synthesis obtained with strain UB89-1 (pKTplaccI/pinfB) at 28 and 43°C, respectively. The β-galactosidase activity obtained with strain UB89-1 (pKTplaccI/plc2833) at 28 and 43°C is indicated by gray and hatched bars, respectively. The cultures were grown at 28°C and shifted to 43°C at an OD600 of 0.3. The β-galactosidase values are given in Miller units. (B) Determination of OmpA synthesis at basal and increased concentrations of IF2. Aliquots of strain UB89-1 (pKTplaccI/pinfB) were withdrawn at 28 or 43°C at the indicated OD values; 0.1 OD600 units of total cellular protein were loaded on to a 12% SDS–polyacrylamide gel for a given OD600 value. OmpA synthesis was determined by quantitative immunoblotting as described in Materials and methods. Black and white bars correspond to OmpA synthesis at 43 and 28°C, respectively. (C) Plasmid pinfB-directed synthesis of IF2. –, no induction of the plasmid-borne infB gene; +, heat induction of the infB gene at an OD600 of 0.3. As described in (B), aliquots of strain UB89-1 (pKTplaccI/pinfB) were withdrawn at OD600 values of 0.3, 0.4, 0.6 and 0.8. For a given OD, 0.1 OD600 unit of total cellular protein was subjected to SDS–PAGE and subsequent quantitative immunoblotting as described in Material and methods. Only the relevant section of the immunoblot showing the IF2-specific band(s) is depicted.
In vitro translation of λ cI and E.coli ompA mRNA in systems derived from Sulfolobus solfataricus and rabbit reticulocytes
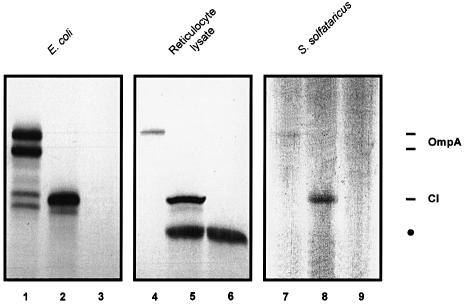
The results presented above strongly support the view that, in E.coli, a leaderless mRNA is recognized by a 30S–fMet-tRNAfMet complex formed in the presence of IF2, through a mechanism resembling the obligatory translation initiation pathway typical of eukaryotes. It is interesting to note that leaderless mRNAs are quite common in archaebacteria (Bult et al., 1996; Sensen et al., 1996) and occur in the lower eukaryote Giardia lamblia (Adam, 1991). To investigate whether the capability to translate leaderless mRNAs is a fundamental property of all translation systems, we asked whether the λ cI mRNA can be translated in heterologous in vitro translation systems derived from the archaebacterium S.solfataricus and from rabbit reticulocytes. The translation systems were programmed with equimolar amounts of cI mRNA and ompA mRNA. In both systems the cI mRNA was translated much more efficienctly than ompA mRNA (Figure 5). After normalization for the number of methionines present in the CI and OmpA proteins, the translational efficiency of cI mRNA was estimated to be 6- and 6.5-fold higher than that of ompA mRNA in the archaebacterial and eukaryotic system, respectively. To ensure that translation of cI mRNA started at the 5′-terminal AUG in either translation system, the AUG start codon of cI mRNA was changed to CUG and the corresponding mRNA was translated in vitro in either system. As shown in Figure 5 (lanes 3, 6 and 9), in the absence of a 5′-terminal AUG codon the band corresponding to the full-length CI protein disappeared. The additional product with a molecular weight lower than that of CI synthesized by the reticulocyte lysate programmed with cI mRNA (Figure 5, lanes 5 and 6) originates from an in-frame AUG at codon 43 (S.Grill, unpublished results), which is preceded by a G at position –3 and followed by a G at position +4; these are the major determinants of start codon selection in higher eukaryotes (Kozak, 1997).
Fig. 5. Translation of equimolar amounts of cI and ompA mRNA in the E.coli S30, rabbit reticulocyte and S.solfataricus in vitro translation systems. Samples from the in vitro translation reactions were analyzed on a 12% SDS–polyacrylamide gel. Translation of ompA mRNA with the E.coli S30 extract, reticulocyte system and S.solfataricus system is shown in lanes 1, 4 and 7, respectively. Translation of cI mRNA with the E.coli S30 extract, reticulocyte system and S.solfataricus system is shown in lanes 2, 5 and 8, respectively. Translation of cIAUG1→CUG mRNA with the E.coli S30 extract, reticulocyte system and S.solfataricus system is shown in lanes 3, 6 and 9, respectively. The positions of the bands corresponding to CI and OmpA are marked. The band marked with a filled circle in lane 5 results from an internal translation event in cI mRNA (see text).
Discussion
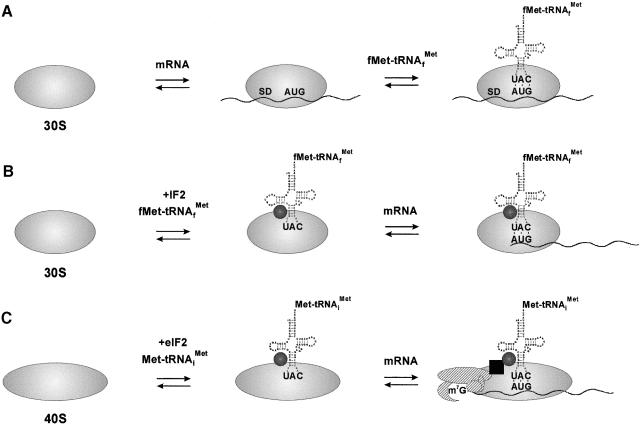
The data presented in this study strongly support a model in which translation of leaderless mRNA in bacteria occurs through a mechanism in which the initiation site of the mRNA is recognized by a 30S–initiator tRNA complex assisted by IF2 (Figure 6B). This mechanism is analogous to the obligatory initiation pathway in eukaryotes (Figure 6C). When compared with an mRNA containing a canonical 5′-UTR, a negligible amount of 30S–mRNA complex was formed with cI34 mRNA in the absence of initiator tRNA (see Table I). We conclude, therefore, that the 5′-terminal AUG is the only element of this leaderless mRNA that is recognized by the ribosome. Consistent with this observation is the lack of physical evidence that cis elements contribute to ribosomal recognition of leaderless mRNA (Resch et al., 1995, 1996; Tedin et al., 1997; Moll et al., 1998; Bläsi et al., 1999; O’Connor et al., 1999; La Teana et al., 2000). Further support for this notion comes from the studies of van Etten and Janssen (1998), who demonstrated that the translational efficiency of mRNAs deprived of their canonical 5′-UTR (which included the SD sequence) depended on the presence of an AUG start codon.
Fig. 6. Translational initiation pathways in prokaryotes and eukaryotes. (A) Recruitment of the prokaryotic ribosome by an mRNA containing a canonical rbs through the SD–anti-SD interaction in the absence of P-site-bound initiator tRNA. (B) Recognition of leaderless mRNA by a prokaryotic 30S ribosome–initiator tRNA complex. IF2 is depicted by a closed circle. (C) Recognition of the cap complex by an eukaryotic 40S ribosome–initiator tRNA complex (reviewed by Gallie, 1998). The cap-binding complex eIF4F, consisting of eIF4E (crescent), eIF4G (oval), eIF4A (circle) and eIF4B (small oval); eIF3 (black square) and eIF2 (closed circle) are also depicted.
The formation of two binary complexes (30S–mRNA and 30S–fMet-tRNAfMet), which are intermediates in the random pathway leading first to the formation of a pre-ternary complex and subsequently to a 30S initiation complex, is supported by kinetic analyses (for reviews see Gualerzi and Pon, 1990; RajBhandary and Chow, 1995; Gualerzi et al., 2000). The unstable pre-ternary complex intermediate has been demonstrated in the presence of an excess of ribosomal protein S15 (Philippe et al., 1993). Binary complexes of 30S with mRNAs have been detected for mRNAs having SD sequences in their 5′-UTR (Hartz et al., 1991) as well as for mRNAs containing a 5′-UTR devoid of complementarity to the 16S rRNA (Calogero et al., 1988). The evidence for the formation of the 30S–fMet-tRNAfMet complex is also compelling (Jay and Kaempfer, 1974; Wu et al., 1996). The intrinsic randomness of the order of mRNA–fMet-tRNAfMet binding to the 30S ribosomal subunit during 30S initiation complex formation does not necessarily mean that both pathways are equally probable. In fact, depending on the nature of the mRNA, on the cellular concentration of the two ligands and, possibly, on the variation of environmental parameters, translation of individual mRNAs may preferentially proceed through the initial formation of an mRNA–30S complex followed by the binding of fMet-tRNAfMet or vice versa. In addition, the extent to which translation of different model and natural mRNAs depends on IF2 seems to vary (La Teana et al., 1993; Pediconi et al., 1995).
We have shown that increased concentrations of IF2 resulted in an increase in the translational efficiency of a leaderless mRNA in vitro (Figure 2) and in vivo (Figure 4) relative to that of ompA mRNA containing an internal AUG triplet preceded by a canonical SD sequence. The experimentally generated transient increase in IF2 concentration resembles that which is known to occur in vivo during the cold adaptation that follows cold shock in E.coli (Jones et al., 1987). It is interesting to note that, under cold-shock conditions, the ratio of leaderless tetR–lacZ mRNA to lacZ mRNA translation increased (K.Stiefel and U.Bläsi, unpublished data). The IF2-dependent stimulation of ternary complex formation on cI mRNA (see Figure 1) was less pronounced than the IF2 effect observed in the cI mRNA in vitro translation reaction (Figure 2A). The two assays differ in that, in the toeprint assay, the ternary complex has to withstand the elongating reverse transcriptase, whereas in the in vitro translation reaction, the stability of the ternary complex is modulated by the presence of the other initiation factors. It is conceivable that IF2, besides stimulating the formation of the 30S–fMet-tRNAfMet binary complex, also increases the stability of the 30S initiation complex in the translation reaction. This effect may not be as obvious in the toeprinting assays because it can be diminished by the elongating reverse transcriptase.
In the in vitro experiments presented in this study (Table I; Figures 1 and 3), we used 30S ribosomes to assess ternary complex formation on leaderless mRNAs. However, several lines of evidence suggest that translation of leaderless mRNAs may also be accomplished by 70S ribosomes (Balakin et al., 1992; Resch et al., 1996; Moll, 2000). Filter binding studies revealed that 70S monosomes, like 30S ribosomes (see Table I), do not form a binary complex with leaderless cI mRNA (Moll, 2000). Accordingly, binding of 70S ribosomes to different leaderless mRNAs strictly requires the presence of initiator tRNA (Moll, 2000). Thus, translation initiation of leaderless mRNAs requires a preformed ribosome–initiator tRNA complex, regardless of whether it is carried out by 30S or 70S ribosomes.
Translation in the kingdom Bacteria is accomplished by a different set of factors from that in Eukarya. However, homology searches performed by Kyrpides and Woese (1998a,b) have revealed that translation factors from all kingdoms share significant analogy, leading these authors to conclude that the rudiments of the present translational components (i.e. initiation factors) and mechanisms may have been present at the universal ancestor stage. Among these factors is the bacterial type IF2, which has been found in the kingdoms Archaea and Eukarya. Leaderless mRNAs are found in all three kingdoms. The results shown in Figure 5, and the faithful translation of leaderless rro mRNA derived from the lactococcal phage r1t in the same translation systems as used in this study (P.Londei, unpublished data), support the notion that, regardless of their origin, mRNAs with a 5′-terminal start codon can be translated by all three systems.
The translational behavior of the leaderless cI mRNA contrasted with that of the leadered ompA mRNA, which was poorly translated in both the archaeal and eukaryotic cell-free systems. The inefficient translation of ompA mRNA in the reticulocyte lysate is readily explained by the absence of a eukaryotic consensus sequence (Kozak, 1987, 1989) around the start codon and by the presence of extensive secondary structures in its 5′-UTR (Rosenbaum et al., 1993). According to the manufacturer, the reticulocyte lysate does not add a 7-methyl cap to the in vitro transcribed mRNA, which would be required for binding of the cap complex eIF4F comprising the helicase activity eIF4A.
Despite the near identity of the anti-SD sequence of S.solfataricus and E.coli 16S rRNA (Olsen et al., 1985), ompA translation was barely detectable in the S.solfataricus system (Figure 5, lane 7). However, since the S.solfataricus system operates at 73°C, it seems possible that the four-base complementarity of the ompA SD with the S.solfataricus anti-SD sequence is not sufficient for effective translation initiation. On the other hand, ompA translation depends on ribosomal protein S1 (Tedin et al., 1997), a homolog of which has not been found among the 30S ribosomal proteins of S.solfataricus (Londei et al., 1983).
Ribosomal protein S1 stimulates translation initiation in Gram-negative bacteria and in the low G+C branch of Gram-positive bacteria (Boni et al., 1982, 1991; Roberts and Rabinowitz, 1989). However, translation of leaderless mRNAs is independent of S1 (Tedin et al., 1997), which may suggest that these mRNAs were present before the the SD–anti-SD interaction and the S1 interaction developed. It is also worth noting that a leaderless mRNA derived from G.lamblia is translated in the absence of a 5′ cap structure and a consensus sequence surrounding the initiation AUG (Hughes and Andrews, 1997). It is tempting to speculate that, at the universal ancestor stage, mRNA recognition may have been achieved by a ribosome–initiator tRNA complex independently of ribosome recruitment signals. Hence, today’s leaderless mRNAs might be viewed as remnants of ancestral mRNAs.
Materials and methods
Bacterial strains and construction of plasmids
Escherichia coli strain UB89-1 (Bläsi et al., 1990) was grown in Luria–Bertani (LB) medium (Miller, 1972) supplemented with kanamycin (30 µg/ml), ampicillin (120 µg/ml) or tetracycline (20 µg/ml) where appropriate to maintain selection of plasmids. Growth of the liquid cultures was monitored photometrically by measuring the optical density at 600 nm.
The plasmid pinfB, provided by Dr C.Pon, University of Camerino, harbors the infB gene under transcriptional control of the λpL promoter. The parental plasmid pLc2833 has been described previously (Remaut et al., 1981). Plasmid pKTplaccI was constructed by inserting a PCR fragment containing the lac promoter (from nucleotide –60 to +1) from plasmid pUHE21-2 (Lanzer and Bujard, 1988) and the first 189 nucleotides of the λ cI gene into the XbaI and SmaI sites of plasmid pKT35 (Resch et al., 1995). In plasmid pKTplaccI, the lac promoter is abutted to the start codon of the cI gene via an NcoI site, which ensured that transcription of cI mRNA starts at the adenosine of the start codon. Plasmid pAXL7 harbors the full-length cI gene under transcriptional control of a T7 promoter (Tedin et al., 1997). Plasmid pKS0235 (Ried et al., 1994) served as a source for wild-type ompA mRNA and was provided by Dr R.Koebnik, MPI Tübingen, Tübingen, Germany.
Filter-binding assay
Filter-binding assays were performed using a Schleicher and Schuell SRC 072/0 Minifold II slot blot apparatus. The cI34SD and cI34 mRNAs were both obtained by PCR technology and T7 RNA polymerase-directed mRNA synthesis. Both mRNAs were labeled with [α-32P]CTP. In a reaction volume of 50 µl, 0.5 pmol of 32P-labeled mRNA was incubated with 30S ribosomes at a molar ratio of 1:10 (see Table I). The fMet-tRNAfMet was added in a 2-fold excess over 30S ribosomes. The reactions were incubated for 10 min at 37°C. The samples were added to the filter under vacuum and washed with 25 volumes of VD+ reaction buffer (Tedin et al., 1997). The filters were exposed to a Molecular Dynamics PhosphorImager screen to quantify the retained mRNA. The total c.p.m. values were corrected for the number of Cs in each mRNA.
β-galactosidase assays
The β-galactosidase activities were determined as described by Miller (1972). Triplicate aliquots of each culture were taken at different optical densities during logarithmic growth.
Quantitative immunoblotting
Samples were withdrawn from the cultures at different OD600 values as described in the legend to Figure 4C. Care was taken that, for a given OD600, the same amount of cellular protein was loaded on 12% SDS–polyacrylamide gels. The proteins were then transferred electrophoretically to a nitrocellulose sheet (pore size 0.2 µm) using standard procedures. OmpA was detected with anti-OmpA antibodies, provided by Dr U.Henning, MPI Tübingen, followed by visualization of the OmpA immunocomplex with the ECL kit of Amersham Life Science according to the manufacturer’s instructions. The intensity of the resulting bands was determined by ImageQuant analysis. The cellular level of OmpA before the temperature shift (OD600 = 0.3) was set to 1. The cellular level of OmpA after the shift and in the non-induced culture was normalized to that of OmpA at an OD600 of 0.3. The cellular level of IF2 at 28 and 42°C was examined by quantitative immunoblotting with anti-IF2 antibodies. The secondary antibody was labeled with alkaline phosphatase. IF2-specific bands were visualized with 5-bromo-4-chloro-3-indolylphosphate/ nitroblue tetrazolium.
In vitro translation
For full-length synthesis of cI and ompA mRNA, plasmids pAXL7 and pKS0235 were digested with PvuII and HindIII, respectively. The linearized plasmids served as templates for in vitro transcription reactions with T7 RNA polymerase.
In vitro translation of cI and ompA mRNAs with E.coli S100 extracts was performed essentially as described by Tedin et al. (1997). Mix I contained 16.6 mM MgOAc, 80 mM NH4Cl, 30 mM Tris–HCl pH 7.7, 3.3 mM dithiothreitol, 1.6 mg/ml E.coli tRNA, 0.2 mM citrovorum, 16.6 mM KCl, 0.33 mM amino acids (without lysine), 66.6 mM [14C]lysine, 3.3 mM ATP, 0.66 mM GTP, 16.6 mM phosphoenolpyruvate and 0.04 mg/ml pyruvate kinase. Mix II contained S100 extracts, 3.3 pmol/µl IF-free E.coli 70S ribosomes, 0.66 pmol/µl IF1, 0.66 pmol/µl IF3, 2 mM Tris–HCl pH 7.7, 60 mM NH4Cl and 10 mM MgOAc. Purified E.coli IF2 was added to Mix II at different IF:70S ratios (no IF2, or 0.2:1, 0.4:1, 1:1, 2.5:1 ratios of IF2:70S). The translation mixture contained, in a final volume of 25 µl, 18 µl of Mix I, 6 µl of Mix II (20 pmol of 70S, 4 pmol of IF1 and 4 pmol of IF3) and 10 pmol of mRNA. The translation reactions were started by adding mRNA and incubated at 37°C for 30 min.
The in vitro translation reactions with S.solfataricus extracts were performed according to Condo et al. (1999). The samples contained, in a final volume of 50 µl, 10 mM KCl, 20 mM Tris–HCl pH 7.0, 18 mM MgOAc, 7 mM β-mercaptoethanol, 3 mM ATP, 1 mM GTP, 5 µg of bulk S.solfataricus tRNA, 20 µM amino acids (without methionine), 2 µl of [35S]methionine (1200 Ci/mmol), 20 µl of S.solfataricus S30 extract (pre-incubated for 10 min at 73°C) and 10 pmol of mRNA. The samples were incubated at 73°C for 40 min.
The rabbit reticulocyte lysate was obtained from Promega. The reactions, which were set up according to the manufacturer’s instructions, contained 50 mM KOAc, 1 mM MgOAc and 5 pmol of mRNA in a final volume of 25 µl. The mixtures were incubated at 30°C for 60 min.
The labeled proteins were separated on 12% SDS–polyacrylamide gels. The gels were dried under vacuum and exposed to a Molecular Dynamics PhosphorImager screen for quantification.
Primer extension inhibition analysis (toeprinting)
IF-depleted 30S subunits were prepared as previously described (Spedding, 1990). They were judged as pure when 23S rRNA was absent from the preparations. Charged initiator tRNA was provided by Drs W.Wintermeyer and M.V.Rodnina, University of Witten/Herdecke.
The 189 nucleotide long cI mRNA (see Figure 1) was prepared exactly as described in Resch et al. (1996). The cISD (see Figure 1) mRNA, which comprises the same RNA sequence as cI mRNA except for the 5′-terminal extension (see Table I), was obtained by means of PCR technology using a plasmid containing the full-length cI gene abutted with its 5′-terminal start codon to the translation initiation region of phage T7 gene 10, followed by transcription with T7 RNA polymerase as described previously (Resch et al., 1996). Primer O8 (Moll et al., 1998) was used for the toeprinting reactions with both mRNAs.
Plasmid pUH100 (Lundberg et al., 1990) was used as a source of wild-type ompA mRNA (see Figure 1). The plasmid was digested with HincII and T7 RNA polymerase-generated run-off transcripts were used in the toeprint reactions together with the AvaII primer (Moll et al., 1998).
The ompAΔ117 mRNA (see Figure 3) was prepared from a PCR template (Tedin et al., 1997), and the AvaII primer (Moll et al., 1998) was used for toeprinting.
Toeprinting assays were performed essentially as described by Hartz et al. (1988). For the kinetic toeprinting reactions, 2 pmol of 30S subunits, 5 pmol of IF2 (when present), 0.8 pmol of fMet-tRNAfMet and 1 mM GTP were pre-incubated for 20 s in a 10 µl reaction before the mRNAs were added at a final concentration of 0.005 pmol/µl. The incubation times varied (see legends to Figures 1 and 3B). The Moloney murine leukemia virus reverse transcription reaction was then performed at 37°C for 3 min. The relative toeprints shown in Figures 1 and 3C were calculated as described by Hartz et al. (1991).
Acknowledgments
Acknowledgements
We thank Drs U.Henning, R.Koebnik, C.L.Pon, M.Rodnina and W.Wintermeyer for the gift of materials and suggestions. M.Huber is acknowledged for carrying out the filter-binding studies. A part of this work was performed by S.G. in the laboratory of C.O.G., who is supported by MURST-CNR 95/95, CNR (Progetto Strategico ST74) and the EC Biotechnology Programme. The work in U.B.’s laboratory was supported by grant P-12065 MOB from the Austrian Science Foundation.
References
- Adam R. (1991) The biology of Giardia spp. Microbiol. Rev., 55, 706–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin A.G., Skripkin,E.A., Shatsky,I.N. and Bogdanov,A.A. (1992) Unusual ribosome binding properties of mRNA encoding bacteriophage λ repressor. Nucleic Acids Res., 20, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsi U., Kalousek,S. and Lubitz,W. (1990) A bifunctional vector system for controlled expression and subsequent release of the cloned gene product by ΦX174 lysis protein-E. Appl. Microbiol. Biotechnol., 33, 564–568. [DOI] [PubMed] [Google Scholar]
- Bläsi U., O’Connor,M., Squires,C.L. and Dahlberg,A.E. (1999) Misled by sequence complementarity: does the DB–anti-DB interaction withstand scientific scrutiny? Mol. Microbiol., 33, 439–441. [DOI] [PubMed] [Google Scholar]
- Boni I.V., Zlatkin,I.V. and Budowsky,E.I. (1982) Ribosomal protein S1 associates with the Escherichia coli ribosomal 30S subunit by means of protein–protein interactions. Eur. J. Biochem., 121, 371–376. [DOI] [PubMed] [Google Scholar]
- Boni I.V., Isaeva,D.M., Musychenko,M.L. and Tzareva,N.V. (1991) Ribosome–messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res., 19, 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock S., Szkaradkiewicz,K. and Sprinzl,M. (1998) Initiation factors of protein biosynthesis in Bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol., 29, 409–417. [DOI] [PubMed] [Google Scholar]
- Brombach M. and Pon,C.L. (1987) The unusual translation initiation codon AUU limits the expression of the infC (initiation factor IF3) gene of Escherichia coli. Mol. Gen. Genet., 208, 94–100. [DOI] [PubMed] [Google Scholar]
- Bult C.J. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- Butler J.S., Springer,M. and Grunberg-Manago,M. (1987) AUU-to-AUG mutation in the initiator codon of the translation initiaton factor IF3 abolishes translation autocontrol of its own gene (infC) in vivo. Proc. Natl Acad. Sci. USA, 84, 4022–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero R.A., Pon,C.L., Canonaco,M.A. and Gualerzi,C.O. (1988) Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA, 85, 6427–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condo I., Ciammaruconi,A., Benelli,D., Ruggero,D. and Londei,P. (1999) Cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus.Mol. Microbiol., 34, 377–384. [DOI] [PubMed] [Google Scholar]
- De Cock E., Springer,M. and Dardel,F. (1999) The interdomain linker of Escherichia coli initiation factor IF3: a possible trigger of translation initiation specificity. Mol. Microbiol., 32, 193–202. [DOI] [PubMed] [Google Scholar]
- Gallie D.R. (1998) A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene, 216, 1–11. [DOI] [PubMed] [Google Scholar]
- Gold L. (1988) Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem., 57, 199–233. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1996) mRNA–ribosome interaction during initiation of protein synthesis. In Zimmermann,R. and Dahlberg,A.E. (eds), Ribosomal RNA: Structure, Evolution, Processing and Function in Protein Synthesis. CRC Press, Boca Raton, FL, pp. 259–276. [Google Scholar]
- Gualerzi C.O. and Wintermeyer,W. (1986) Prokaryotic initiation factor 2 acts at the level of the 30S ribosomal subunit. A fluorescence stopped-flow study. FEBS Lett., 202, 1–6. [Google Scholar]
- Gualerzi C.O., Brandi,L., Caserta,E., La Teana,A., Spurio,R., Tomsic,J. and Pon,C.L. (2000) Translation initiation in bacteria. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 477–494. [Google Scholar]
- Hartz D., McPheeters,D.S., Traut,R. and Gold,L. (1988) Extension inhibition analysis of translation initiation complexes. Methods Enzymol., 164, 419–425. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters,D.S. and Gold,L. (1989) Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev., 3, 1899–1912. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters,D.S., Green,L. and Gold,L. (1991) Detection of Escherichia coli ribosome binding at translation initiation sites in the absence of tRNA. J. Mol. Biol., 218, 99–105. [DOI] [PubMed] [Google Scholar]
- Hughes M.J.G. and Andrews,D.W. (1997) A single nucleotide is a sufficient 5′-untranslated region for translation in an eukaryotic in vitro system. FEBS Lett., 414, 19–22. [DOI] [PubMed] [Google Scholar]
- Janssen G.R. (1993) Eubacterial, archaebacterial, and eukaryotic genes that encode leaderless mRNA. In Baltz,R.H., Hegeman,G.D. and Skatrud,P.L. (eds), Industrial Microorganisms: Basic and Applied Molecular Genetics. ASM Press, Washington, DC, pp. 59–67. [Google Scholar]
- Jay G. and Kaempfer,R. (1974) Sequence of events in initiation of translation: a role for initiator transfer RNA in the recognition of mRNA. Proc. Natl Acad. Sci. USA, 71, 3199–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.G., van Bogelen,R.A. and Neidhardt,F.C. (1987) Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol., 169, 2092–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1989) Context effects and (inefficient) initiation at non-AUG codons in eukaryotic cell-free translation systems. Mol. Cell. Biol., 9, 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem., 266, 19867–19870. [PubMed] [Google Scholar]
- Kozak M. (1997) Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J., 16, 2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides N.C. and Woese,C.R. (1998a) Universally conserved translation factors. Proc. Natl Acad. Sci. USA, 95, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides N.C. and Woese,C.R. (1998b) Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B α–β–δ subunit families. Proc. Natl Acad. Sci. USA, 95, 3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzer M. and Bujard,H. (1988) Promoters determine largely the efficiency of repressor action. Proc. Natl Acad. Sci. USA, 85, 8973–8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A., Pon,C.L. and Gualerzi,C.O. (1993) Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc. Natl Acad. Sci. USA, 90, 4161–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A., Pon,C.L. and Gualerzi,C.O. (1996) Late events in translation initiation. Adjustment of fMet-tRNAfMet in the ribosomal P-site. J. Mol. Biol., 256, 667–675. [DOI] [PubMed] [Google Scholar]
- La Teana A., Brandi,A., O’Connor,M. and Pon,C.L. (2000) Translation during cold adaptation does not involve mRNA–rRNA base-pairing to the downstream-box. RNA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londei P., Teichner,A., Cammarano,P., de Rosa,M. and Gambacorta,A. (1983) Particle weights and protein composition of the ribosomal subunits of the extremely thermoacidophilic archaebacterium Caldariella acidophila. Biochem. J., 209, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg U., von Gabain,A. and Melefors,Ö. (1990) Cleavages in the 5′ region of the ompA and bla mRNA control stability: studies with an Escherichia coli mutant altering mRNA stability and a novel endoribonuclease. EMBO J., 9, 2731–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melançon P., Leclerc,D., Destroismaisons,N. and Brakier-Gingras,L. (1990) The anti-Shine–Dalgarno region in Escherichia coli 16S ribosomal RNA is not essential for the correct selection of translational starts. Biochemistry, 29, 3402–3407. [DOI] [PubMed] [Google Scholar]
- Merrick W.C. (1992) Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev., 56, 291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Moazed D., Samaha,R.R., Gualerzi,C.O. and Noller,H.F. (1995) Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol., 248, 207–210. [DOI] [PubMed] [Google Scholar]
- Moll I. (2000) Studies on translation initiation of leaderless mRNAs. PhD thesis, University of Vienna, Vienna, Austria. [Google Scholar]
- Moll I., Resch,A. and Bläsi,U. (1998) Discrimination of 5′-terminal start codons by translation initiation factor 3 is mediated by ribosomal protein S1. FEBS Lett., 436, 213–217. [DOI] [PubMed] [Google Scholar]
- O’Connor M., Asai,T., Squires,C.L. and Dahlberg,A.E. (1999) Enhancement of translation by the downstream box does not involve mRNA–rRNA base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc. Natl Acad. Sci. USA, 96, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G.J., Pace,N.R., Nuell,M., Daine,B.P., Gupta,R. and Woese,C.R. (1985) Sequence of the 16S rRNA gene from the thermoacidophilic archaebacterium Sulfolobus solfataricus and its evolutionary implications. J. Mol. Evol., 22, 301–307. [DOI] [PubMed] [Google Scholar]
- Pediconi D., Spurio,R., La Teana,A., Jemiolo,D., Gualerzi,C.O. and Pon,C.L. (1995) Translational regulation of infC operon in Bacillus stearothermophilus.Biochem. Cell Biol., 73, 1071–1078. [DOI] [PubMed] [Google Scholar]
- Philippe C., Eyermann,F., Bénard,L., Portier,C., Ehresmann,B. and Ehresmann,C. (1993) Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the RNA initiation loading site. Proc. Natl Acad. Sci. USA, 90, 4394–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon C.L. and Gualerzi,C.O. (1974) Effect of initiation factor 3 binding on the 30S ribosomal subunits of Escherichia coli.Proc. Natl Acad. Sci. USA, 71, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon C., Paci,M., Pawlik,R. and Gualerzi,C.O. (1985) Biochemical and biophysical characterization of the interaction between IF2 and guanosine nucleotides. J. Biol. Chem., 260, 8918–8924. [PubMed] [Google Scholar]
- RajBhandary U.L. and Chow,C.M. (1995) Initiator tRNAs and initiation of protein synthesis. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis, and Function. ASM Press, Washington, DC, pp. 511–528. [Google Scholar]
- Remaut E., Stanssens,P. and Fiers,W. (1981) Plasmid vectors for high-efficiency expression controlled by the pL promoter of coliphage λ. Gene, 15, 81–93. [DOI] [PubMed] [Google Scholar]
- Resch A., Tedin,K., Graschopf,A., Haggård-Ljungquist,E. and Bläsi,U. (1995) Ternary complex formation on leaderless phage mRNA. FEMS Microbiol. Rev., 17, 151–157. [DOI] [PubMed] [Google Scholar]
- Resch A., Tedin,K., Gründling,A., Mündlein,A. and Bläsi,U. (1996) Downstream/anti-downstream–box interactions are dispensable for translation initiation of leaderless mRNAs. EMBO J., 15, 4740–4748. [PMC free article] [PubMed] [Google Scholar]
- Ried G., Koebnik,R., Hindenach,I., Mutschler,B. and Henning,U. (1994) Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coli K12. Mol. Gen. Genet., 243, 127–135. [DOI] [PubMed] [Google Scholar]
- Roberts M.W. and Rabinowitz,J.C. (1989) The effect of Escherichia coli ribosomal protein S1 on the translational specificity of bacterial ribosomes. J. Biol. Chem., 264, 2228–2235. [PubMed] [Google Scholar]
- Rosenbaum V., Klahn,T., Lundberg,U., Holmgren,E., von Gabain,A. and Riesner,D. (1993) Co-existing structures of an mRNA stability determinant. The 5′-region of the Escherichia coli and Serratia marcescens ompA mRNA. J. Mol. Biol., 229, 656–670. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Chiaruttini,C., Engst,K., Graffe,M., Milet,M., Mathy,N., Dondon,J. and Springer,M. (1996) The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol., 21, 331–346. [DOI] [PubMed] [Google Scholar]
- Sensen C.W. et al. (1996) Organizational characteristics and information content of an archaeal genome: 156 kb of sequence from Sulfolobus solfataricus P2. Mol. Microbiol., 22, 175–191. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Guillon,J.M., Meinnel,T., Mechulam,Y., Dardel,F. and Blanquet,S. (1996) Molecular recognition governing the initiation of translation in Escherichia coli.Biochimie, 78, 543–554. [DOI] [PubMed] [Google Scholar]
- Spedding G. (1990) Isolation and analysis of ribosomes from prokaryotes, eukaryotes and organelles. In Spedding,G. (ed.), Ribosomes and Protein Synthesis: a Practical Approach. IRL Press at Oxford University Press, New York, NY, pp. 1–29. [Google Scholar]
- Sprengart M.L. and Porter,A.G. (1997) Functional importance of RNA interactions in selection of translation initiation codons. Mol. Microbiol., 24, 19–28. [DOI] [PubMed] [Google Scholar]
- Sussman J.K., Simons,E.L. and Simons,R.W. (1996) Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol., 21, 347–360. [DOI] [PubMed] [Google Scholar]
- Tedin K., Resch,A. and Bläsi,U. (1997) Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5′-leader sequence. Mol. Microbiol., 25, 189–199. [DOI] [PubMed] [Google Scholar]
- Tedin K., Moll,I., Grill,S., Resch,A., Gualerzi,C.O. and Bläsi,U. (1999) Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol., 31, 67–77. [DOI] [PubMed] [Google Scholar]
- Van Etten W.J. and Janssen,G.R. (1998) An AUG initiation codon, not codon–anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol. Microbiol., 27, 987–1001. [DOI] [PubMed] [Google Scholar]
- Winzeler E. and Shapiro,L. (1997) Translation of the leaderless Caulobacter dnaX mRNA. J. Bacteriol., 179, 3981–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J. and Janssen,G.R. (1996) Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5′-untranslated leader. Mol. Microbiol., 22, 339–355. [DOI] [PubMed] [Google Scholar]
- Wu C.J. and Janssen,G.R. (1997) Expression of a streptomycete leaderless mRNA encoding chloramphenicol acetyltransferase in Escherichia coli.J. Bacteriol., 179, 6824–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.Q., Iyengar,P. and RajBhandary,U.L. (1996) Ribosome–initiator tRNA complex as an intermediate in translation initiation in Escherichia coli revealed by use of mutant initiator tRNAs and specialized ribosomes. EMBO J., 15, 4734–4739. [PMC free article] [PubMed] [Google Scholar]