Abstract
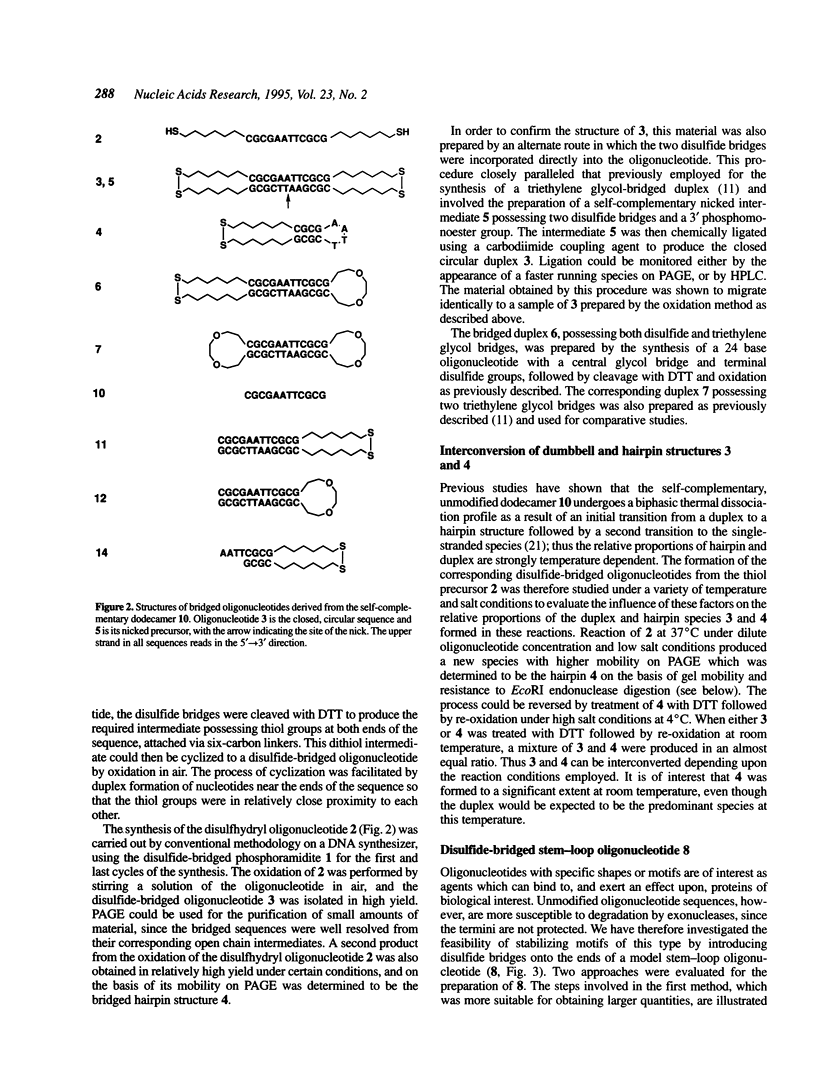
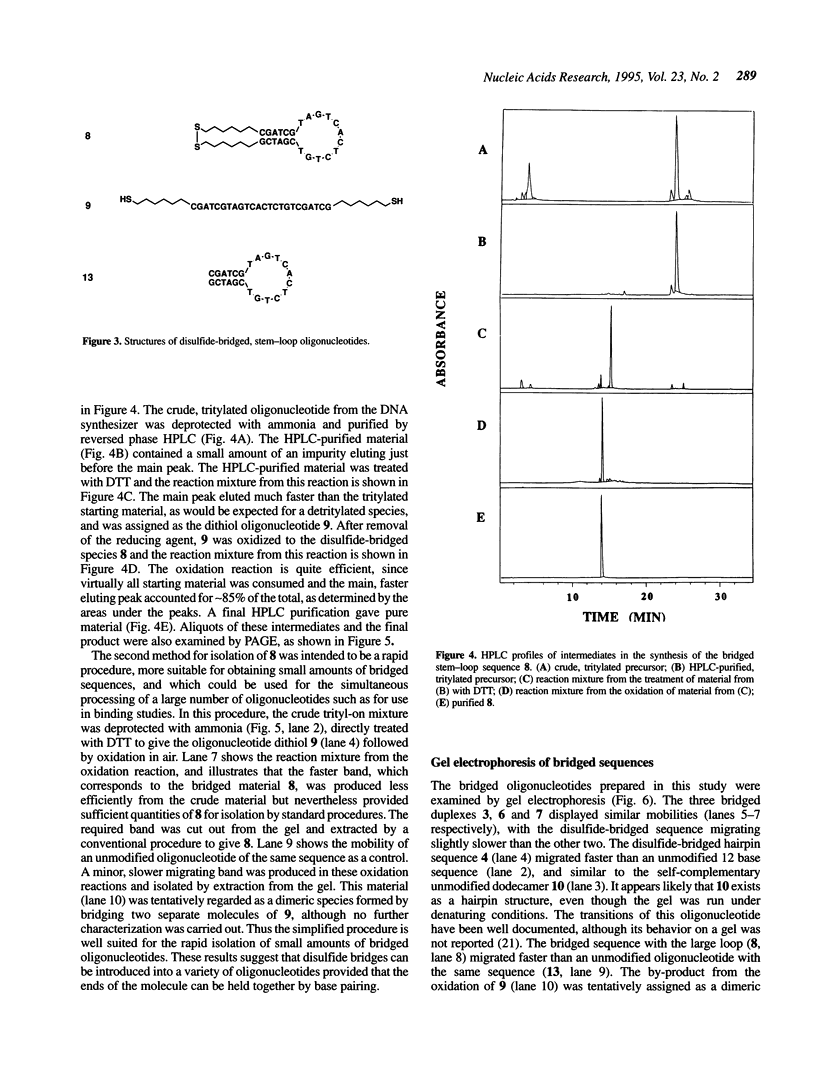
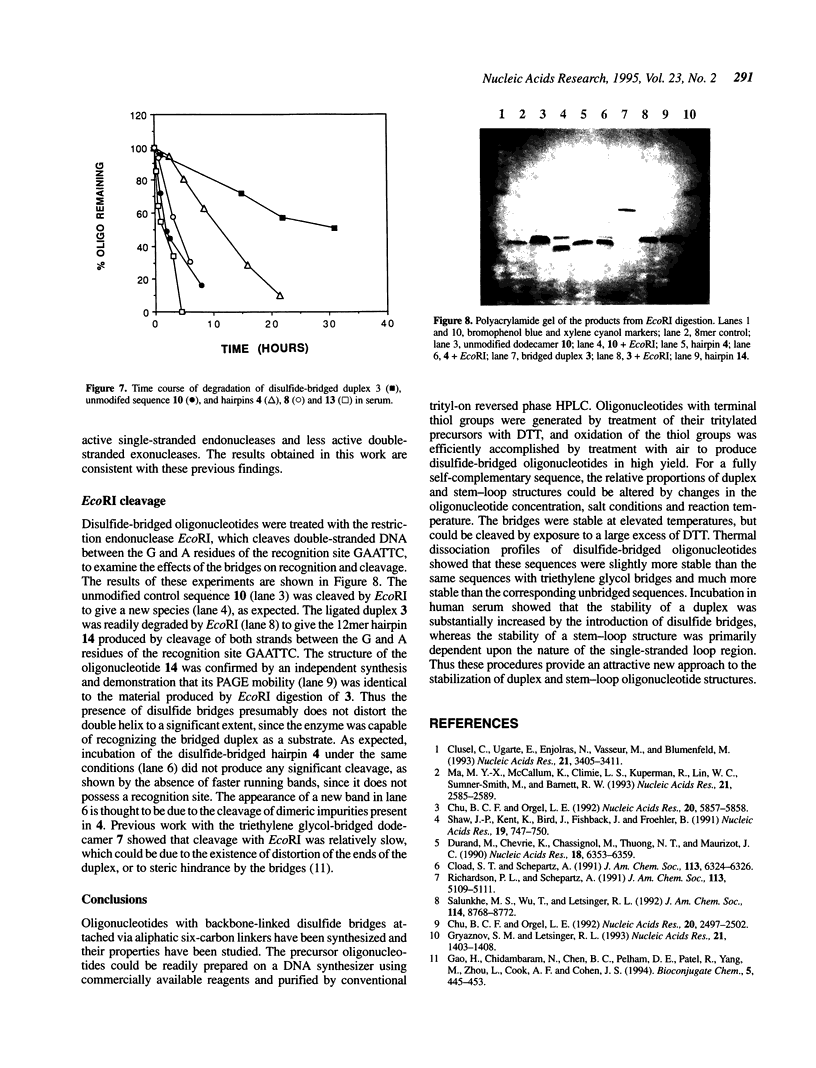
A convenient, practical route to the synthesis of disulfide-bridged oligonucleotides has been developed. Aliphatic linkers with terminal thiol groups have been attached to the phosphodiester backbones of partially or fully complementary oligonucleotide sequences and oxidized to yield covalently closed oligonucleotides with disulfide bridges. This procedure has been used to prepare a duplex with disulfide bridges at both ends and stem-loop sequences with single disulfide bridges. Oxidation of a self-complementary duplex possessing terminal thiol groups produced both hairpin and duplex structures with disulfide bridges, the relative proportions of each being dependent upon the reaction conditions. These bridged hairpin and duplex structures were shown to be interconvertible by reduction and re-oxidation. The melting profiles of disulfide-bridged oligonucleotides were compared with the same sequences without bridges and with sequences possessing triethylene glycol bridges, and in all cases the introduction of disulfide bridges resulted in a considerable increase in thermal stability. EcoRI endonuclease was capable of cleaving a disulfide-bridged duplex possessing a recognition site for this enzyme, thus supporting a lack of distortion of the recognition site. The disulfide bridges could be cleaved using a large excess of DTT to regenerate the corresponding sulfhydryl compounds. A study of the serum stabilities of disulfide-bridged oligonucleotides showed that the bridged duplexes were much more stable than their unmodified counterparts, whereas the rate of degradation of the stem-loop structures was more dependent upon the size of the loop than the presence or absence of the disulfide bridge. In summary, we have described a novel methodology, employing commercially available reagents, for the stabilization of oligonucleotide duplexes or stem-loop structures by disulfide bridge formation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chu B. C., Orgel L. E. Crosslinking transcription factors to their recognition sequences with PtII complexes. Nucleic Acids Res. 1992 May 25;20(10):2497–2502. doi: 10.1093/nar/20.10.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B. C., Orgel L. E. The stability of different forms of double-stranded decoy DNA in serum and nuclear extracts. Nucleic Acids Res. 1992 Nov 11;20(21):5857–5858. doi: 10.1093/nar/20.21.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clusel C., Ugarte E., Enjolras N., Vasseur M., Blumenfeld M. Ex vivo regulation of specific gene expression by nanomolar concentration of double-stranded dumbbell oligonucleotides. Nucleic Acids Res. 1993 Jul 25;21(15):3405–3411. doi: 10.1093/nar/21.15.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B. A., Rider P. Chemical synthesis of oligonucleotides containing a free sulphydryl group and subsequent attachment of thiol specific probes. Nucleic Acids Res. 1985 Jun 25;13(12):4485–4502. doi: 10.1093/nar/13.12.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M., Chevrie K., Chassignol M., Thuong N. T., Maurizot J. C. Circular dichroism studies of an oligodeoxyribonucleotide containing a hairpin loop made of a hexaethylene glycol chain: conformation and stability. Nucleic Acids Res. 1990 Nov 11;18(21):6353–6359. doi: 10.1093/nar/18.21.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Chidambaram N., Chen B. C., Pelham D. E., Patel R., Yang M., Zhou L., Cook A., Cohen J. S. Double-stranded cyclic oligonucleotides with non-nucleotide bridges. Bioconjug Chem. 1994 Sep-Oct;5(5):445–453. doi: 10.1021/bc00029a011. [DOI] [PubMed] [Google Scholar]
- Gryaznov S. M., Letsinger R. L. Template controlled coupling and recombination of oligonucleotide blocks containing thiophosphoryl groups. Nucleic Acids Res. 1993 Mar 25;21(6):1403–1408. doi: 10.1093/nar/21.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- Shaw J. P., Kent K., Bird J., Fishback J., Froehler B. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 1991 Feb 25;19(4):747–750. doi: 10.1093/nar/19.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N. D., Cook R. M. The preparation and application of functionalised synthetic oligonucleotides: III. Use of H-phosphonate derivatives of protected amino-hexanol and mercapto-propanol or -hexanol. Nucleic Acids Res. 1988 Mar 25;16(6):2659–2669. doi: 10.1093/nar/16.6.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- X Ma M. Y., McCallum K., Climie S. C., Kuperman R., Lin W. C., Sumner-Smith M., Barnett R. W. Design and synthesis of RNA miniduplexes via a synthetic linker approach. 2. Generation of covalently closed, double-stranded cyclic HIV-1 TAR RNA analogs with high Tat-binding affinity. Nucleic Acids Res. 1993 Jun 11;21(11):2585–2589. doi: 10.1093/nar/21.11.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckermann R., Corey D., Schultz P. Efficient methods for attachment of thiol specific probes to the 3'-ends of synthetic oligodeoxyribonucleotides. Nucleic Acids Res. 1987 Jul 10;15(13):5305–5321. doi: 10.1093/nar/15.13.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]